Abstract
Colorectal cancer (CRC) remains one of the deadliest diseases in the whole world. Cancer recurrence and chemotherapeutic drug resistance limit the overall survival rate of patients with CRC. This study aimed to discover the latent miRNAs and genes associated with oxaliplatin resistance in CRC cells. The study found that miR-1254 is upregulated in oxaliplatin-resistant CRC cell line HCT116-R compared with its parental cell line HCT116 by transcriptome sequencing and small RNA sequencing. Meanwhile, MEGF6 (multiple EGF-like domains 6) was downregulated in HCT116-R cells. Transient transfection of miR-1254 mimics significantly reduced cell apoptosis, increased HCT116 tolerance to oxaliplatin, and enhanced MEGF6 expression. Furthermore, transfection of miR-1254 inhibitor increased apoptosis, decreased HCT116-R tolerance to oxaliplatin, and reduced MEGF6 expression. In addition, transient transfection of SiMEGF6 enhanced HCT116 cell resistance to oxaliplatin and reduced cell apoptosis. In summary, MEGF6 is a latent functional target of miR-1254 in regulating oxaliplatin resistance and apoptosis in human CRC cells, suggesting a potential therapeutic target for CRC.
Keywords: Colorectal cancer, chemoresistance, miR-1254, MEGF6, oxaliplatin
Introduction
Cancer imposes a tremendous burden on society, especially in developing countries. Colorectal cancer (CRC) is considered as the commonest and serious malignancies worldwide, and it currently ranks as the second most common cancer among women (at 614,000 cases per year) and the third most common cancer among men (746,000 cases per year) [1]. The incidence rate of CRC is increasing due to the increasing risk of population growth, smoking, overweight, obesity and lack of exercise. The prognosis of CRC has gradually improved owing to multidisciplinary and comprehensive treatment under its surgical treatment [2]. Sadly, the survival rate of CRC patients is really low yet; about 25% of patients have disseminated stage IV disease, and 10%-15% of patients with the initial regional disease have transferred within 5 years after the discovery of cancer [3,4]. Current research shows that the first-line drugs for the cytotoxic treatment of metastatic colorectal cancer (mCRC) comprise fluoropyrimidine combine with oxaliplatin or together with irinotecan [5]. However, many patients have no positive response to combination therapy, which is mostly due to chemotherapy resistance and recurrence [6]. The recurrence was mainly due to cells resistance to oxaliplatin. Therefore, understanding the mechanisms of oxaliplatin resistance will lay a foundation for the recognition of predicted biomarkers such as microRNA (miRNA) in cancer cells.
MiRNA generally refers to microRNA, which have become the main regulatory factors in the occurrence and development of human cancer [7], including CRC [8]. There is considerable evidence that abnormally expressed miRNAs regulate the oncogenic mRNAs (targeting tumor suppressor genes), leading to cancer cell dysfunction [9], such as cell proliferation, apoptosis, differentiation, and migration [10]. In addition, miR-1254 dysregulation has been reported in different types of cancer, and it has been reported that miR-1254 accelerates breast cancer cell growth and suppresses apoptosis [4]; furthermore, it may might inhibit oral squamous cell carcinoma progression [11], and suppresses non-small cell lung cancer (NSCLC) development in vivo [12,13]. Moreover, miR-1254 overexpression can suppress gastric cancer cell proliferation, migration, and invasion [14]. An increasing number of studies have linked miRNAs to anticancer drug resistance [15]. Meanwhile, some miRNAs have been contacted to 5-fluorouracil (5-FU) resistance [16]. However, the functional relationship between miR-1254 and oxaliplatin resistance remains unclear.
To expound the relationship between miRNA and oxaliplatin resistance, the study compared the differentially expressed miRNAs and their target genes in two types of CRC cells using transcriptional sequencing and microRNA sequencing. Previous studies have concluded that miR-1254 regulates some target genes in some types of tumors. For example, miR-1254 accelerates breast cancer cell proliferation and suppresses their apoptosis through the regulation of p53 expression via RASSF9 [17]; furthermore, mutual regulation of miR-1254 in oral squamous cell carcinoma restrain epithelial-mesenchymal transition-mediated metastasis and tumor-initiating function by mitogen-activated protein kinase (MAPK) signaling [18]. Besides, studies have indicated that miR-1254 suppresses heme oxygenase-1 (HO-1) expression to attenuate NSCLC growth [12]. However, we found MEGF6 (multiple EGF-like domains 6) was regulated by miR-1254 in CRC cells. As one of the MEGF family, homo sapiens MEGF6 is touches in many known diseases [19]. Moreover, MEGF6 controls Purkinje cells adhesion and degeneration in patients with hereditary ataxia [19,20]. The results of these previous studies indicate that miRNAs and their target genes are potential biomarkers of cancer development. Therefore, it is necessary to explore how imbalanced miRNAs regulate drug resistance and other biological characteristics of CRC.
The study explored the mechanism of action of miR-1254 in CRC cells. We proved that miR-1254 expression is upregulated in HCT116-R cells. Furthermore, miR-1254 inhibition potently promotes CRC cell apoptosis and decreases resistance to oxaliplatin. In particular, we demonstrated that MEGF6 was regulated by miR-1254 and that knockdown of MEGF6 increases CRC cell resistance to oxaliplatin and decreases apoptosis. These results indicate that miR-1254 represents a promising treatment target for CRC therapy.
Materials and methods
CRC cell culture
Human CRC cell line HCT116 was derived from the Cell Bank of the Chinese Academy of Science (Shanghai, China). The cell was tested and authenticated by the Cell Bank of Genechem. Oxaliplatin-resistant cell line HCT116-R was established from their parental cells by gradually increasing concentration of oxaliplatin. HCT116 and HCT116-R were growing in Roswell Park Memorial Institute (RPMI) 1640 medium (Gibco, Carlsbad, CA, USA) aided with 10% fetal bovine serum (FBS) (Gibco), 100 μg/mL penicillin, and 100 μg/mL streptomycin (Gibco). The cell was grown in a constant temperature incubator filled with 5% CO2 at 37°C. HCT116-R cells were cultured in 10% FBS RPMI1640 medium with oxaliplatin concentration of 10 μg/mL.
Cytotoxicity assay
HCT116 and HCT116-R cells were cultured in a 96-well plate (Thermo Fisher Scientific, Wilmington, DE, USA), respectively. Each cell line has three replications. After incubation for 24 h, they were cultured in fresh medium containing 0, 12.5, 25, 50, 100, 200, 400, 600, 800 and 1000 μg/mL concentrations of oxaliplatin for 24 h. Cell survival rate was detected using the Cell Counting Kit-8 (CCK-8; Dojindo, Kumamoto, Japan). Next, 10 μL CCK-8 was supplemented to each well with 90 μL RPMI-1640, followed by incubation for 2 h. Cell survival rate was tested by detecting the absorbance of each well at 450 nm [21]. The inhibition ratio was the ratio of apoptotic cells in each group contrasted with the control group.
Transfection of miR-1254 and siRNA oligo
SiRNA and its control were obtained from GenePharma Biotech (Shanghai, China) and used to knockdown mRNA. The SiMEGF6 sequences were MEGF6-Si-490 (sense: 5’-GGAGAACCGUCUACUACAUTT-3’, antisense: 5’-AUGUAGUAGACGGUUCUCCTT-3’); MEGF6-Si-1763 (sense: 5’-GGAUGACUCCUUUGGCCAUTT-3’, antisense: 5’-AUGGCCAAAGGAGUCAUCCTT-3’); and MEGF6-Si-4599 (sense: 5’-GCCACCUGUAACCUGGAUUTT-3’, antisense: 5’-AAUCCAGGUUACAGGUGGCTT-3’). For SiMEGF6 negative control (NC), (sense 5’-UUCUCCGAACGUGUCACGUTT-3’, antisense 5’-ACGUGACACGUUCGGAGAATT-3’). The NC for miR-1254 mimics (sense 5’-UUCUCCGAACGUGUCACGUTT-3’, antisense 5’-ACGUGACACGUUCGGAGAATT-3’) and miR-1254 mimics (sense 5’-AGCCUGGAAGCUGCAGCCUGCAGU-3’, antisense 5’-UGCAGGCUCCAGCUUCCAGGCUUU-3’) were designed and synthesized by GenePharma Biotechand used for miRNA upregulation. NC for miR-1254 inhibitor (5’-CAGUACUUUUGUGUAGUACAA-3’) and miR-1254 inhibitor (5’-ACUGCAGGCUCCAGCUUCCAGGCU-3’) were designed and synthesized by GenePharma Biotech and used for miRNA downregulation. These oligonucleotides were introduced into CRC cells cultured in six-well plates using liposome 2000 reagent (Invitrogen, Carlsbad, CA, USA). After 48 h of incubation, the cell was used for polymerase chain reaction (PCR) analysis or subsequent experiments.
RNA isolation and qRT-PCR
Extraction of total RNA from CRC cells was performed according to the protocols provided by the manufacturer (TRIzol reagent, Invitrogen). To examine gene expression, the samples were amplified with PCR by One Step SYBR PrimeScript™ TMRT-PCR Kit II (Takara Bio, Shiga, Japan). For miRNA detection, the samples amplified using the ABI ViiA™ 7 according to the instructions by Mir-X miRNA qRT-PCR SYBR Kit (Clontech Laboratories Inc. Takara Bio, Japan). The cDNA was synthesized according to the manufacturer’s instructions for the reverse transcriptase kit (Takara, China). Then, the mRNA expression level was normalized using glyceraldehyde 3-phosphate dehydrogenase, whereas U6 as a control for miR-1254 expression levels.
Cell apoptosis analysis
The cell was cultured in 12-well plates (5×105 cells/well), then transfected with miR-1254 mimics and its NC, miR-1254 inhibitor and its NC, or SiMEGF6 and its NC to induce apoptosis. After 24 h of incubation at 37°C, different concentrations of oxaliplatin were used to treat the cells for 48 h. Apoptosis (expressed as a percentage) was tested using an Annexin V-FITC/propidium iodide staining assay after washing with phosphate-buffered saline via flow cytometry. The experiments were performed at least three times.
Western blot analysis
The cell was growing to about 80% in 6-well plates, then transfected with miR-1254 mimics and its NC or miR-1254 inhibitor and its NC, and SiMEGF6 and its NC. After these specific treatments and washed with PBS three times in a 6-well plate (Thermo Fisher Scientific, Wilmington, DE, USA), cells were lysed in 150 μL RIPA buffer (Thermo Fisher Scientific, Wilmington, DE, USA). Protein concentrations were tested by Pierce™ Rapid Gold BCA Protein Assay Kit (Thermo Fisher Scientific, Wilmington, DE, USA). Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was used to isolated proteins; the specific method is described in the earlier study [24].
Small RNA and transcriptome sequencing analysis
Three replication samples were collected from HCT116 and HCT116-R for small RNA or transcriptome sequencing [22]. Small RNA libraries were constructed and sequenced on the Illumina HiSeq platform. Sequencing analysis identified 82 known miRNAs and 107 potentially novel miRNAs. |log2FC|≥1.5 and false discovery rate (FDR)≥0.005 were used as screening criteria to screen differentially expressed miRNAs. The transcriptome sequencing library was generated using NEB Next Ultra™ RNA Library Prep Kit for Illumina (NEB), and 47,956 genes were identified, including 1679 known genes [23]. The original sequence data of the sample was uploaded to the Genome Sequence Archive (GSA) of BIG Data Center in Beijing institute of Genomics (http://bigd.big.ac.cn/). The accession number is CRA001292.
Statistical analysis
All experiments were performed at least three times. The study use SPSS version 18.0 (SPSS Inc, Chicago, IL, USA) and GraphPad Prism (GraphPad Software, Inc., San Diego, CA, USA) software for statistical analyses. The students’ t-test (double tail) was used for statistical evaluation. A p-value < 0.05 was considered a significant difference.
Results
Oxaliplatin toxicity profile of HCT116 and HCT116-R cell lines
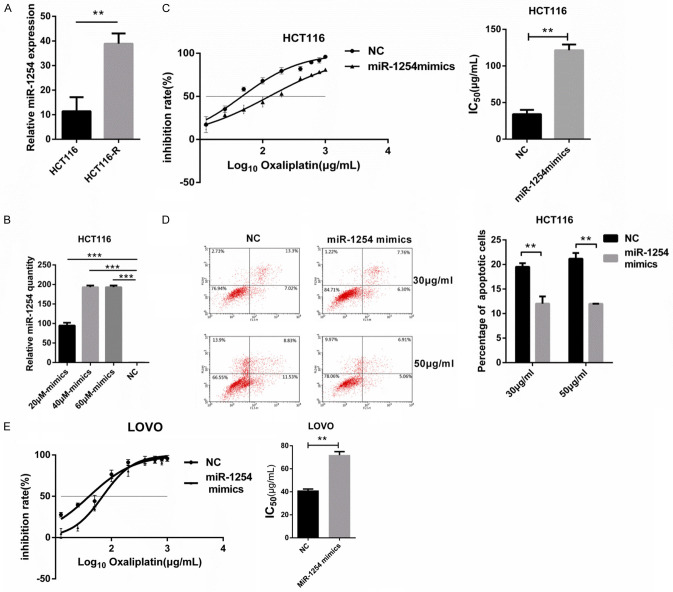
The HCT116-R cell model was established by the stepwise increasing method. The resistance index of HCT116-R was determined. CCK-8 kit was used to detect the number of living cells at different concentrations. After the CCK-8 solution reacted with cells for 2 hours, the OD value of CCK-8 was detected at 450 nm to obtain the cell survival rate. The oxaliplatin toxicity profiles of HCT116 and HCT116-R cell lines were assessed. The 50% inhibitory concentration values (IC50) of HCT116 and HCT116-R cells were 21.87 and 107.1 μg/mL, respectively, and the resistance index was 4.89 (P < 0.05) (Figure 1).
Figure 1.

HCT116 and HCT116-R cells were cultured in fresh medium containing 0, 12.5, 25, 50, 100, 200, 400, 600, 800 and 1000 μg/ml oxaliplatin for 24 hours. The IC50 of HCT116 and HCT116-R cells was detected by CCK-8 kit and the drug resistance index was calculated.
Screening of differentially expressed miRNAs and genes
To investigate the potential mechanism of oxaliplatin resistance, the small-RNA-Sequence analyses and RNA-Sequence of HCT116 and HCT116-R cells were analyzed. The results indicated that 189 miRNAs (Figure 2A, 2B) and 1671 genes (Figure 2C, 2D) [22]. Then randomly selected 5 miRNAs and 5 genes to check their differential expression by quantitative reverse transcription (qRT)-PCR. The consequence of this experiment showed that the 5 miRNAs and 5 genes displayed the similar expression trend in sequencing results and qRT-PCR analysis (Figure 2E, 2F). To further screen miRNA and target genes, the screening criteria was defined as log fold change >1 or < -1, (P < 0.05) and FDR (q) < 0.05. Some unknown new genes and small RNAs were not considered. The candidate miRNAs and potential target genes were exhibited opposite expression trends. Finally, we selected 7 miRNAs that were downregulated and 4 miRNAs that were upregulated in HCT116-R by querying the functions of miRNA and target genes (Tables 1, 2).
Figure 2.
Identification of differentially expressed miRNAs/genes in small RNA-Seq/RNA-Seq data. A. Cluster diagram of differentially expressed miRNAs. The columns represent different samples, and the rows represent different miRNAs. The cluster is based on log10 (TPM + 1) value. Red indicates high expression miRNA, and green indicates low expression miRNA. B. Volcano map of differentially expressed miRNA. Each point represents a miRNA, the abscissa is the logarithm of the difference multiple of miRNA expression in the two samples; ordinate is the negative logarithm of the error detection rate. C. Differential expression MA map, each dot represents a gene. The abscissa is the logarithm of the mean expression amount in the two samples; the ordinate is the logarithm of the multiple of gene expression difference between the two samples. B and C. The green dots represent down regulated differentially expressed genes/miRNAs, red dots represent up-regulated differentially expressed genes/miRNAs, and blue/black dots represent non differentially expressed genes/miRNAs. D. Cluster diagram of differentially expressed miRNAs. Different columns represent different samples, and different rows represent different genes. The color represents the level of gene expression in the sample, log10 (FPKM + 0.000001). E. Five miRNAs were randomly selected to validate their differential expression using quantitative reverse transcription (qRT)-PCR and compare with microRNA-seq results. F. Five genes were randomly selected to validate their differential expression using qRT-PCR and compare with mRNA-seq results. Log2FC/RQ is the logarithm of the fold change or relative quantity to base 2. Error bars represent the standard deviation (STDEV) of the qRT-PCR analysis (n = 3).
Table 1.
Primers of mRNAs used for quantitative real-time polymerase chain reaction (qRT-PCR)
| mRNA | 5’-Forward primer-3’ | 5’-Reverse primer-3’ |
|---|---|---|
| MEGF6 | AGATGTGGACGAATGCCGAACC | AGCGATGCCGAGTGATTGTGAG |
| FBLIM1 | GGCCTCACTCATTGCAGACT | TGTCTGTGGATGCCCCTTTC |
| CD163L1 | GGGAATGTCAACACCGGGAA | GGCTTCACCTTGCCTTCCTAC |
| CREBBP | CTCAGAGCCAGTTTCTGCCA | GGTGACTGTGTCACTGGAGG |
| CTCF | GGCTTCACCTTGCCTTCCTAC | TCCTCTTCCTCTCCCTCTGC |
Table 2.
Expression of 11 differentially expressed candidate miRNAs in colorectal cancer cells
| MiRNA | FDR | log2FC | Regulated |
|---|---|---|---|
| hsa-miR-483-3p | 4.6867E-06 | -2.20032561859457 | Down |
| hsa-miR-642a-5p | 1.8056E-04 | -1.68781419682841 | Down |
| hsa-miR-1247-5p | 4.0109E-04 | -2.03822301098972 | Down |
| hsa-miR-3158-3p | 4.5863E-04 | -1.37506200850894 | Down |
| hsa-miR-151a-5p | 4.8531E-04 | -1.22775674500255 | Down |
| hsa-miR-139-3p | 5.4357E-04 | -1.62603783557166 | Down |
| hsa-miR-139-5p | 9.5704E-04 | -1.2444273831688 | Down |
| hsa-miR-92a-1-5p | 3.2123E-05 | 2.01266561204892 | Up |
| hsa-miR-1254 | 2.2470E-04 | 1.70615911301397 | Up |
| hsa-miR-212-5p | 5.5935E-04 | 1.81023845875748 | Up |
| hsa-miR-409-3p | 6.3684E-08 | 2.55861025723854 | Up |
Note: False discovery rate (FDR) control is a statistical method used in multiple hypotheses testing to correct for multiple comparisons. Fold change (FC) is a measure describing how much a quantity changes going from an initial to a final value. The Log2 fold-change (log2FC) is an estimate of the log2 ratio of expression in a cluster to that in all other cells. A value of 1.0 indicates a 2-fold higher expression in the cluster of interest.
MiR-1254 overexpression enhanced oxaliplatin resistance
The differential expression of miR-1254 in various tumors has been documented in previous studies [24], miR-1254 expression was upregulated in HCT116-R contrasted with in HCT116 cells, (Figure 3A). Regarding the role of miR-1254 in CRC, it has been reported that miR-1254 suppresses colon adenocarcinoma cell migration. However, whether miR-1254 affects CRC oxaliplatin resistance has not been investigated to date. Particularly, among the upregulated miRNAs in HCT116-R cells, it was noticed that miR-1254 was significantly upregulated in HCT116-R cells compared with HCT116 cells. Therefore, the study regarded it as the target. To examine the function of miR-1254 in HCT116 cells, different concentrations of miR-1254 mimics and its NC were transfected into the colon adenocarcinoma cell line HCT116. Finally, 40 μΜ miR-1254 mimics were selected as the subsequent transfection conditions (Figure 3B). Cell survival assays were used to test the IC50 of oxaliplatin in HCT116 cells by Cell Counting Kit-8 (CCK-8). As expected, the IC50 of HCT116 cells increased significantly after being transfected with 40 μΜ miR-1254 mimics contrasted with that of NC (IC50 = 46.76 and 127.8 μg/mL, respectively; **P < 0.01) (Figure 3C). The results of this experiment indicated that miR-1254 overexpression enhanced HCT116 cell resistance to oxaliplatin. Besides, flow cytometry was used to detect the influence of miR-1254 on HCT116 cell apoptosis. After 48 h of oxaliplatin treatment (30 and 50 μg/mL), the apoptosis of HCT116 cells treated with miR-1254 mimics decreased. In other words, the percentage of apoptotic cells was reduced (Figure 3D).
Figure 3.
MiR-1254 overexpression enhanced the resistance of HCT116 to oxaliplatin. A. miR-1254 expression in HCT116 and HCT116-R cells was analyzed by qRT-PCR. According to the expression level of miR-1254 in HCT116- R cells, the expression of miR-1254 in HCT116-R cells was increased. B. Negative control (NC) and different concentrations (20 μM, 40 μM and 60 μM) of miR-1254 mimics were transfected into HCT116. The expression of miR-1254 was detected by qRT-PCR. C. After transient transfection of miR-1254 and negative control (NC), HCT116 cells were cultured in fresh medium containing 0, 12.5, 25, 50, 100, 200, 400, 600, 800 and 1000 μg/ml oxaliplatin for 24 hours. The IC50 was detected by CCK-8 and calculated from the inhibition curves. D. miR-1254 mimics and negative control (NC) were transfected into HCT116 respectively. Under different concentrations of oxaliplatin (30, 50 μg/ml), the apoptosis rate of transfected cells decreased significantly after overexpression of miR-1254 compared with NC group. E. miR-1254 mimics and negative control (NC) were transfected into HCT116 respectively. LOVO cells were cultured in fresh medium with different concentrations oxaliplatin for 24 hours. The IC50 was detected by CCK-8 and calculated from the inhibition curves. Each experiment was conducted at least three times. The error bar represents the standard deviation (n = 3). **P < 0.01 and ***P < 0.001.
Last, to examine whether miR-1254 affect other CRC cells in oxaliplatin resistance, the study used another CRC cell lines (LOVO). The LOVO cells were transfected with miR-1254 mimics and its NC. The IC50 of LOVO increased after miR-1254 mimics treatment showed by drug sensitivity assay (40.03 and 69.71 μg/mL; P < 0.05; Figure 3E). This result is consistent with HCT116 cell lines. To sum up, these results indicated that miR-1254 overexpression reduces HCT116 cell apoptosis and enhances CRC cells oxaliplatin resistance.
MiR-1254 was significantly upregulated in HCT116-R cells and inhibition of miR-1254 restored oxaliplatin responsiveness
To further confirm the role of miR-1254 in oxaliplatin resistance, different concentrations of miR-1254 inhibitor and its NC were transfected into the colon adenocarcinoma cell line HCT116-R cells. Finally, 40 μΜ miR-1254 inhibitor was selected as the subsequent transfection conditions (Figure 4A). Later, drug sensitivity assays were performed, indicating that HCT116-R treated with miR-1254 inhibitor restored oxaliplatin responsiveness compared with treated with its NC (IC50 = 33.16 and 90.06 μg/mL, respectively; P < 0.001; Figure 4B). Moreover, flow cytometry results suggested that after 48 h of oxaliplatin treatment (30 and 50 μg/mL), the apoptosis of HCT116-R cells treated with miR-1254 inhibitor was increased contrasted with that of HCT116-R cells treated with its NC. In other words, the ratio of apoptotic cells increased (Figure 4C). In a few words, these results of these experiments suggested that the inhibition of miR-1254 may recover oxaliplatin responsiveness by increased apoptosis in HCT116-R cells.
Figure 4.
MiR-1254 down expression restored oxaliplatin responsiveness in HCT116-R cells. A. Negative control (NC) and different concentrations (20 μM, 40 μM and 60 μM) of miR-1254 inhibitor were transfected into HCT116-R by lip2000. MiR-1254 expression was detected by qRT-PCR. B. After transient transfection of miR-1254 inhibitor and negative control (NC), HCT116-R cells were cultured in fresh medium containing 0, 12.5, 25, 50, 100, 200, 400, 600, 800 and 1000 μg/ml oxaliplatin for 24 hours. The IC50 was detected by CCK-8 and calculated from the inhibition curves. C. miR-1254 inhibitor and negative control (NC) were transfected into HCT116-R respectively. After treated with different concentrations of oxaliplatin (30, 50 μg/ml), the apoptosis rate of transfected cells decreased significantly after inhibition of miR-1254 compared with NC group. Each experiment was conducted at least three times, n = 3. *P < 0.05 and **P < 0.01.
MEGF6 is regulated by miR-1254 in CRC cells
To identify the relationship between miR-1254 and MEGF6, MEGF6 expression was determined to be higher in HCT116 cells than in HCT116-R cells by qRT-PCR and western blot (Figure 5A, 5B). Then, qRT-PCR and western blot were used to explore the correlation between miR-1254 and MEGF6. MiR-1254 inhibitor or mimics and their NC were transfected into CRC cells. The results demonstrated that MEGF6 expression was reduced after miR-1254 mimics treatment compared with that after NC treatment in HCT116 cells (Figure 5C, 5D). In contrast, the mRNA and protein level of MEGF6 was increased in HCT116-R cells with miR-1254 inhibitor treatment (Figure 5E, 5F). Besides, several publicly available online databases such as MiRanda and RNAhybrid were used in the prediction and MEGF6 was considered as a potential downstream target of miR-1254. According to the results of the miRNA target gene website prediction (http://www.targetscan.org/vert_72/), miR-1254 combined with the 368-375 position of MEGF6 3’UTR (Figure 5G), indicating that miR-1254 may regulate MEGF6 expression through this site. These results suggest that MEGF6 was modulated by miR-1254.
Figure 5.

MEGF6 is a target of miR-1254 in HCT116 cell lines. A. The expression levels of MEGF6 in HCT116 and HCT116-R cells tested by qRT-PCR. B. The expression levels of MEGF6 in HCT116 and HCT116-R cells test by western blot. C. Relative quantity of MEGF6 was detection by qRT-PCR in HCT116 cells transfected with different concentrations (20 μM, 40 μM and 60 μM) of miR-1254 mimics and its negative control. D. Western blot was used to test the expression levels of MEGF6 in HCT116 transfected with 40 μM miR-1254 mimics and its negative control. E. Relative levels of MEGF6 were detection by qRT-PCR in HCT116-R cells transfected with different concentrations (40 μM, 80 μM and 120 μM) of miR-1254 inhibitor and its negative control. F. Western blot was used to test the expression levels of MEGF6 in HCT116-R transfected with 120 μM miR-1254 inhibitor and its negative control. G. Prediction binding sites of miR-1254 and MEGF6. Each experiment was performed in triplicate, n = 3 experiments. *P < 0.05, **P < 0.01, ***P <0.001.
Knockdown MEGF6 enhanced oxaliplatin resistance
To further explore the relationship between miR-1254 and MEGF6, next, we explore whether knockdown MEGF6 regulated oxaliplatin resistance as the same as overexpression miR-1254. We used specific SiRNAs oligo targeting human MEGF6 (SiR490, SiR1573, and SiR4955) and a control (Si-NC) to transfect HCT116 cells, in which MEGF6 expression was higher than that in HCT116-R. According to qRT-PCR analysis suggested that the finest knockdown efficiency of MEGF6 was about 67% in HCT116 cells (Figure 6A), western blot analysis suggested that the knockdown efficiency of MEGF6 was about 51% in HCT116 cells (Figure 6B). CCK-8 analysis was performed to determine the IC50 value of HCT116 cells, and the results demonstrated that IC50 of HCT116 cells treated with SIMEGF6 and Si-NC were 71.31 and 26.07 μg/mL, respectively (Figure 6C). In addition, after 48 h of oxaliplatin treatment (30 and 50 μg/mL), the apoptosis of SiMEGF6-treated HCT116 cells was decreased compared with its NC. In other words, the knockdown of MEGF6 in HCT116 cells declined the apoptosis percentage (Figure 6D). These results indicate that MEGF6 was associated with HCT116 cell resistance to oxaliplatin.
Figure 6.
Knockdown MEGF6 enhanced oxaliplatin resistance. A. Relative MEGF6 levels were analyzed by qRT-PCR in HCT116-R cells after its different SiRNA (SiMEGF6-490, SiMEGF6-1753 and SiMEGF6-4599) treatments. B. Western blot was used to test the expression levels of MEGF6 in HCT116 transfected with SiMEGF6-490 or NC. C. CCK-8 was used to detect the drug sensitivity of the HCT116 cells transfected with SiMEGF6-490 or negative control (NC). The IC50 of HCT116-R cells was calculated from the inhibition curves. D. SiMEGF6-490 and its NC were transfected into HCT116 respectively. Under different concentrations of oxaliplatin (30, 50 μg/ml) treated, the apoptosis rate of transfected cells decreased significantly after knockdown MEGF6 compared with NC group. E. SiMEGF6-490 and its NC were transfected into LOVO respectively, then cultured in fresh medium with different concentrations oxaliplatin for 24 hours. The IC50 was detected by CCK-8 and calculated from the inhibition curves. F. High MEGF6 expression reduces the overall survival rate of CRC. Each experiment was performed in triplicate, n = 3 experiments. *P < 0.05, **P < 0.01, ***P <0.001.
Finally, to examine whether MEGF6 affect other CRC cells in oxaliplatin resistance, the LOVO cells were transfected with SiMEGF6 and its NC. The IC50 of LOVO increased after knockdown MEGF6 treatment showed by drug sensitivity assay (65.48 vs. 91.58 μg/mL; P < 0.05; Figure 6E). This result is consistent with HCT116 cell lines. Besides, high MEGF6 expression reduces the overall survival rate of CRC (Figure 6F), indicated that MEGF6 is associated with prognosis. To sum up, these results indicated that miR-1254 and MEGF6 regulates oxaliplatin resistance in CRC cells.
Discussion
The study results of all experiments showed that the miR-1254 expression level is remarkably lower in CRC cells than in oxaliplatin-resistant cells and that miR-1254 and MEGF6 regulates oxaliplatin resistance in human CRC cells. Firstly, we screened miRNAs and mRNAs differentially expressed between HCT116 and HCT116-R cell lines by small RNA sequencing and transcriptome sequencing. A total of 82 known miRNAs and 107 potentially novel miRNAs. These differentially expressed miRNAs targeted 47956 genes, including 1679 known genes. We used |log2FC|≥1.5 and FDR≥0.005 as the screening criteria and finally obtained 13 miRNAs and 41 mRNAs. The function of the differentially expressed miRNAs and mRNAs was annotated based on Gene Ontology and Kyoto Encyclopedia of Genes and Genomes analyses. Given these results, miR-1254 was identified to be involved in a potential resistance mechanism. Last, we performed qRT-PCR, western blot, and flow cytometry to validate miR-1254 and MEGF6 mRNA expression, their relationship with drug resistance and apoptosis was explored.
Oxaliplatin, which is first-line chemotherapy drugs used for CRC, and it represents a reasonably designed chemotherapeutic drug based on tumors molecular biology. Resistance has been related to altered drug metabolism, oncogenic signaling pathways bypass, apoptotic cascades suppression, an enhanced DNA repair capacity, and other numerous mechanisms. Furthermore, there are multiple molecular mechanisms responsible for oxaliplatin resistance; one of them is the abnormal miRNAs expression. Among the disparity expressed miRNAs between the HCT116 and HCT116-R cell lines, miR-1254 has been reported to be a prospects cancer predictive marker [14]. The mature sequence of hsa-miR-1254 is as follows: AGCCUGGAAGCUGGAGCCUGCAGU. Several studies have been conducted on the relationship between miR-1254 and drug resistance [18].
Previous studies have shown that miR-1254 inhibits cell proliferation and induces apoptosis in human breast cancer [17] and promotes cell proliferation in lung cancer [24]. Consequently, we further analyzed the relationship between miR-1254 expression and oxaliplatin resistance. We observed that miR-1254 expression was higher in HCT116-R than in HCT116. Besides, our results showed that miR-1254 knockdown increases sensitivity to oxaliplatin and induces apoptosis, whereas miR-1254 overexpression reduces sensitivity to oxaliplatin and inhibits apoptosis. However, we did not observe any influence of miR-1254 on cell proliferation in this study. Studies have also shown that miR-1254 is associated with CRC cell migration.
However, drug resistance was, is, and remains one of the key challenges in cancer treatment. Research has indicated that miR-1254 mutual with structured elements in cell cycle and apoptosis regulator (CCAR1) 5’UTR; in this relationship, CCAR1 5’UTR, as a natural miRancer of endogenous miR-1254, is related to tamoxifen resistance in breast cancer cells [25,27]. However, there are only a few studies that have focused on drug resistance associated with miR-1254, and there is little information about the differential expression of miRNAs and the role of drug resistance in CRC. In view of this, according to sequencing results we found that MEGF6 was regulated by miR-1254. Subsequently, we used CCK-8 analysis and found knockdown of miR-1254 decreased cell resistance to oxaliplatin. Conversely, miR-1254 overexpression or MEGF6 knockdown increased cell resistance to oxaliplatin. These results indicate that miR-1254 plays critical roles in many aspects of tumor biology, such as cancer cell proliferation, apoptosis, drug resistance, and metastasis, and it influences MEGF6 action.
According to Hu et al. [20], the proteomic analysis of skin growth factor receptor-interacting shows that MEGF6 may affect cell growth and migration in CRC [26]. Another study has shown that MEGF6 is associated with epithelial-mesenchymal transformation [26,28]. These reports indicate that MEGF6 could influence the increase and migration of cells. However, there are only a few studies on the function of MEGF6 in drug resistance. This study found that MEGF6 was downregulated in HCT116-R cells compared with HCT116 cells, suggesting that MEGF6 is involved in CRC development and is even associated with drug resistance. Therefore, our investigation focused on the function of MEGF6 in CRC. The paper found that knockdown MEGF6 in HCT116 cells decreases the apoptosis rate, as determined via flow cytometry. Besides, we are the first to confirm, using CCK-8 analysis, that knockdown MEGF6 increases sensitivity to oxaliplatin. We observed that miR-1254 correlates with cell apoptosis and oxaliplatin-based chemoresistance in HCT116 cells. To demonstrate similar effects in other CRC cell lines, MEGF6 knockdown or miR-1254 overexpression was used in LOVO cells; as expected, both of them increased LOVO cell line resistance to oxaliplatin. Besides, miR-1254 combined with the 368-375 position of MEGF6 3’UTR, indicating that miR-1254 may regulate MEGF6 expression through this site. In fact, previous studies have indicated that increased miR-1254 expression can inhibit oral cancer invasion and proliferation [11]. However, to date, there have been only a few reports on the relationship between miR-1254 and drug resistance or apoptosis in CRC.
To sum up, we demonstrated for the first time that miR-1254 knockdown increased sensitivity to oxaliplatin and induced apoptosis in vitro, which indicated an that miR-1254 and MEGF6 were associated with oxaliplatin resistance in CRC cells. This study may supply a new insight into acquired resistance mechanisms to oxaliplatin and shows a latent target in CRC cells. The specific inhibition of miR-1254 expression may optimize oxaliplatin-based chemotherapy for CRC in the future. However, the molecular mechanism of miR-1254 in CRC and its significance in clinical diagnosis and treatment remain need to be elucidated.
Acknowledgements
We would like to acknowledge all individuals who contributed to the development of this research and provided input during the study. This work was supported by the scientific research plan projects of Shanxi Education Department, China [12Jk0836].
Disclosure of conflict of interest
None.
References
- 1.Tariq K, Ghias K. Colorectal cancer carcinogenesis: a review of mechanisms. Cancer Biol Med. 2016;13:120–135. doi: 10.28092/j.issn.2095-3941.2015.0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Steele RJC, Digby J, Chambers JA, O’Carroll RE. The impact of personalised risk information compared to a positive/negative result on informed choice and intention to undergo colonoscopy following colorectal Cancer screening in Scotland (PERICCS) - a randomised controlled trial: study protocol. BMC Public Health. 2019;19:411. doi: 10.1186/s12889-019-6734-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sun G, Li Y, Peng Y, Lu D, Zhang F, Cui X, Zhang Q, Li Z. Impact of the preoperative prognostic nutritional index on postoperative and survival outcomes in colorectal cancer patients who underwent primary tumor resection: a systematic review and meta-analysis. Int J Colorectal Dis. 2019;34:681–689. doi: 10.1007/s00384-019-03241-1. [DOI] [PubMed] [Google Scholar]
- 4.Rasmussen MH, Lyskjaer I, Jersie-Christensen RR, Tarpgaard LS, Primdal-Bengtson B, Nielsen MM, Pedersen JS, Hansen TP, Hansen F, Olsen JV, Pfeiffer P, Orntoft TF, Andersen CL. miR-625-3p regulates oxaliplatin resistance by targeting MAP2K6-p38 signalling in human colorectal adenocarcinoma cells. Nat Commun. 2016;7:12436. doi: 10.1038/ncomms12436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blower PE, Chung JH, Verducci JS, Lin S, Park JK, Dai Z, Liu CG, Schmittgen TD, Reinhold WC, Croce CM, Weinstein JN, Sadee W. MicroRNAs modulate the chemosensitivity of tumor cells. Mol Cancer Ther. 2008;7:1–9. doi: 10.1158/1535-7163.MCT-07-0573. [DOI] [PubMed] [Google Scholar]
- 6.Rasmussen MH, Jensen NF, Tarpgaard LS, Qvortrup C, Romer MU, Stenvang J, Hansen TP, Christensen LL, Lindebjerg J, Hansen F, Jensen BV, Hansen TF, Pfeiffer P, Brunner N, Orntoft TF, Andersen CL. High expression of microRNA-625-3p is associated with poor response to first-line oxaliplatin based treatment of metastatic colorectal cancer. Mol Oncol. 2013;7:637–646. doi: 10.1016/j.molonc.2013.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang S, Chen Z, Zhu S, Lu H, Peng D, Soutto M, Naz H, Peek R Jr, Xu H, Zaika A, Xu Z, El-Rifai W. PRDX2 protects against oxidative stress induced by H. pylori and promotes resistance to cisplatin in gastric cancer. Redox Biol. 2019;28:101319. doi: 10.1016/j.redox.2019.101319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Krivohlava R, Neuhferova E, Jakobsen KQ, Benson V. Knockdown of microRNA-135b in mammary carcinoma by targeted nanodiamonds: potentials and pitfalls of in vivo applications. Nanomaterials (Basel) 2019;9:866. doi: 10.3390/nano9060866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang X, Lan Z, He J, Lai Q, Yao X, Li Q, Liu Y, Lai H, Gu C, Yan Q, Fang Y, Zhang Y, Li A, Liu S. LncRNA SNHG6 promotes chemoresistance through ULK1-induced autophagy by sponging miR-26a-5p in colorectal cancer cells. Cancer Cell Int. 2019;19:234. doi: 10.1186/s12935-019-0951-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang M, Hu H, Wang Y, Huang Q, Huang R, Chen Y, Ma T, Qiao T, Zhang Q, Wu H, Chen Q, Han D, Wang G, Wang X. Long non-coding RNA TUG1 mediates 5-fluorouracil resistance by acting as a ceRNA of miR-197-3p in colorectal cancer. J Cancer. 2019;10:4603–4613. doi: 10.7150/jca.32065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Uddin MN, Li M, Wang X. Identification of transcriptional markers and microRNA-mRNA regulatory networks in colon cancer by integrative analysis of mrna and microrna expression profiles in colon tumor stroma. Cells. 2019;8:1054. doi: 10.3390/cells8091054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Taguchi YH. Identification of more feasible microRNA-mRNA interactions within multiple cancers using principal component analysis based unsupervised feature extraction. Int J Mol Sci. 2016;17:696. doi: 10.3390/ijms17050696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen JC, Hsieh YY, Lo HL, Li A, Chou CJ, Yang PM. In vitro and in silico mechanistic insights into miR-21-5p-mediated topoisomerase drug resistance in human colorectal cancer cells. Biomolecules. 2019;9:467. doi: 10.3390/biom9090467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen R, Zhang Y, Zhang X. MiR-1254 functions as a tumor suppressor in oral squamous cell carcinoma by targeting CD36. Technol Cancer Res Treat. 2019;18:1533033819859447. doi: 10.1177/1533033819859447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pu M, Li C, Qi X, Chen J, Wang Y, Gao L, Miao L, Ren J. MiR-1254 suppresses HO-1 expression through seed region-dependent silencing and non-seed interaction with TFAP2A transcript to attenuate NSCLC growth. PLoS Genet. 2017;13:e1006896. doi: 10.1371/journal.pgen.1006896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Venkatesan N, Deepa PR, Khetan V, Krishnakumar S. Computational and in vitro investigation of miRNA-gene regulations in retinoblastoma pathogenesis: miRNA mimics strategy. Bioinform Biol Insights. 2015;9:89–101. doi: 10.4137/BBI.S21742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jiang M, Shi L, Yang C, Ge Y, Lin L, Fan H, He Y, Zhang D, Miao Y, Yang L. miR-1254 inhibits cell proliferation, migration, and invasion by down-regulating Smurf1 in gastric cancer. Cell Death Dis. 2019;10:32. doi: 10.1038/s41419-018-1262-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kwon Y, Kim Y, Jung HS, Jeoung D. Role of HDAC3-miRNA-CAGE network in anti-cancer drug-resistance. Int J Mol Sci. 2018;20:51. doi: 10.3390/ijms20010051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim C, Hong Y, Lee H, Kang H, Lee EK. MicroRNA-195 desensitizes HCT116 human colon cancer cells to 5-fluorouracil. Cancer Lett. 2018;412:264–271. doi: 10.1016/j.canlet.2017.10.022. [DOI] [PubMed] [Google Scholar]
- 20.Li B, Chen P, Wang J, Wang L, Ren M, Zhang R, He J. MicroRNA-1254 exerts oncogenic effects by directly targeting RASSF9 in human breast cancer. Int J Oncol. 2018;53:2145–2156. doi: 10.3892/ijo.2018.4530. [DOI] [PubMed] [Google Scholar]
- 21.Lu M, Chen WH, Wang CY, Mao CQ, Wang J. Retracted: reciprocal regulation of miR-1254 and c-Myc in oral squamous cell carcinoma suppresses EMT-mediated metastasis and tumor-initiating properties through MAPK signaling. Biochem Biophys Res Commun. 2017;484:801–807. doi: 10.1016/j.bbrc.2017.01.170. [DOI] [PubMed] [Google Scholar]
- 22.Lim J, Hao T, Shaw C, Patel AJ, Szabo G, Rual JF, Fisk CJ, Li N, Smolyar A, Hill DE, Barabasi AL, Vidal M, Zoghbi HY. A protein-protein interaction network for human inherited ataxias and disorders of Purkinje cell degeneration. Cell. 2006;125:801–814. doi: 10.1016/j.cell.2006.03.032. [DOI] [PubMed] [Google Scholar]
- 23.Hu H, Wang M, Wang H, Liu Z, Guan X, Yang R, Huang R, Tang Q, Zou C, Wang G, Gao X, Wang X. MEGF6 promotes the epithelial-to-mesenchymal transition via the TGFbeta/SMAD signaling pathway in colorectal cancer metastasis. Cell Physiol Biochem. 2018;46:1895–1906. doi: 10.1159/000489374. [DOI] [PubMed] [Google Scholar]
- 24.Wen S, Wang X, Wang Y, Shen J, Pu J, Liang H, Chen C, Liu L, Dai P. Nucleoside diphosphate kinase 2 confers acquired 5-fluorouracil resistance in colorectal cancer cells. Artif Cells Nanomed Biotechnol. 2018;46:896–905. doi: 10.1080/21691401.2018.1439835. [DOI] [PubMed] [Google Scholar]
- 25.Liang H, Xu Y, Zhang Q, Yang Y, Mou Y, Gao Y, Chen R, Chen C, Dai P. MiR-483-3p regulates oxaliplatin resistance by targeting FAM171B in human colorectal cancer cells. Artif Cells Nanomed Biotechnol. 2019;47:725–736. doi: 10.1080/21691401.2019.1569530. [DOI] [PubMed] [Google Scholar]
- 26.Betel D, Wilson M, Gabow A, Marks DS, Sander C. The microRNA. org resource: targets and expression. Nucleic Acids Res. 2008;36:D149–153. doi: 10.1093/nar/gkm995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li G, Wu X, Qian W, Cai H, Sun X, Zhang W, Tan S, Wu Z, Qian P, Ding K, Lu X, Zhang X, Yan H, Song H, Guang S, Wu Q, Lobie PE, Shan G, Zhu T. CCAR1 5’ UTR as a natural miRancer of miR-1254 overrides tamoxifen resistance. Cell Res. 2016;26:655–673. doi: 10.1038/cr.2016.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Y, Song H, Wang W, Zhang Z. Generation and characterization of Megf6 null and Cre knock-in alleles. Genesis. 2019;57:e23262. doi: 10.1002/dvg.23262. [DOI] [PubMed] [Google Scholar]