Abstract
Background
Colorectal cancer (CRC), characterized by high mortality and incidence rate, is one of the most common types of rectum tumors in the gastrointestinal tract worldwide. An increasing number of investigations indicated that long noncoding RNAs (lncRNAs) have been implicated in the growth of a wide range of cancers. Although it has obtained general acceptance that lncRNA LINC00511 plays a significant role in numerous cancers, the regulatory mechanism of LINC00511 in CRC still needs to be explored.
Materials and Methods
Bioinformatics analysis and a wide range of experiments of sphere formation assay, cell proliferation assay, RT-qPCR, colony formation assay, Transwell assay and Western blot assays investigated the function and mechanism of LINC00511 in CRC tissues and cells.
Results
Our results manifested that the expression level of LINC00511 was obviously upregulated in CRC tissues and cells and it accelerated CRC development through facilitating cell proliferation, metastasis and stemness. Molecular mechanism exploration uncovered that LINC00511 acted as a ceRNA competing with NFIA to bind with miR-29c-3p. Through rescue experiments, we discovered that NFIA upregulation partly counteracted the inhibitive effect induced by LINC00511 silencing on CRC progression.
Conclusion
These results revealed that LINC00511 participated in the progression of CRC by targeting the LINC00511/miR-29c-3p/NFIA axis, indicating that LINC00511 may be a potential therapeutic target for CRC treatment.
Keywords: LINC00511, miR-29c-3p, NFIA, colorectal cancer
Introduction
Colorectal cancer (CRC) has long been one of the most frequent cancers and the second reason for cancer-associated death worldwide.1,2 The mortality rate and incidence remain evidently high among the middle-aged and the elderly in developed countries on account of therapy resistance and distant metastasis.3 Additionally, in young patients, the incidence of CRC is continuously increasing.4 It has been verified that the occurrence of CRC mainly results from human lifestyle, dietary environmental factors.5 Meanwhile, cancer stem cells (CSCs) possess the ability of self-renew, differentiation and actuates the occurrence and development of tumors, becoming an important reason for the failure of cancer treatment.6,7 Although there have been many researches on a variety of biomarkers in CRC, the underlying molecular regulatory mechanisms of these biomarkers are also not very explicit. Therefore, it is of great significance to seek out the biological targets of CRC to advance the diagnosis, therapy, and survival rate for CRC patients.
Long noncoding RNA (lncRNA) is a kind of noncoding RNA with more than 200 nucleotides in length.8 More and more evidences show that lncRNA is the main regulatory factor of cancers.9 An increasing number of researchers have devoted themselves to disclose the function of lncRNAs in cancers. For example, lncRNA LINC00337 inhibits TIMP2 expression by sponging DNMT1 to accelerate the development of NSCLC.10 Upregulation of lncRNA SNHG3 expression promotes the invasive and metabolic capacity of lung cancer cells.11 Evidence shows that LINC00511 could act as a carcinogenic gene in several human cancers. For instance, overexpression of LINC00511 facilitates cell proliferation, migration and invasion in lung cancer.12 Through interacting with Snail, LINC00511 accelerates cell proliferation and invasion in triple-negative breast cancer.13 A recent study has found that in thyroid carcinoma, LINC00511 sponges JAK2 to exert oncogenic function.14 Nevertheless, the biological function and molecular mechanism of LINC00511 in CRC remain obscure.
In this study, we intended to probe whether LINC00511 played oncogenic role in the progression of CRC. Our study demonstrated that LINC00511 regulated the progression of CRC through mediating LINC00511/miR-29c-3p/NFIA axis, which may become a promising therapeutic target for the treatment of patients with CRC.
Materials and Methods
Clinical Samples
Twelve pairs of CRC tissues and noncancerous samples were obtained from patients with CRC at China-Japan Union Hospital of Jilin University (Jilin, China). All collected tissues were instantly stored at −80°C for the following studies. None of the patients accepted treatment before the operation. All written informed consents of this study were obtained from the patients with CRC. The approval of this research was obtained from the Ethics Committee of China-Japan Union Hospital of Jilin University (Jilin, China).
Cell Lines
Four CRC cell lines (HT-29, HCT8, HCE8693, SW620) and a normal human colorectal cell line NCM460 were provided by Chinese Academy of Sciences Cell Bank (Shanghai, China). All these cells were incubated in Roswell Park Memorial Institute (RPMI) 1640 medium including 10% fetal bovine serum (FBS). The cells were cultivated at 37°C with 5% CO2.
Transfection
Cells were seeded on 24‐well plates, 1 × 105 per well, and cultured at 37°C with 5% CO2 in a humid incubator. In order to knockdown the expression of LINC00511, sh-LINC00511#1 and sh-LINC00511#2 vectors were, respectively, designed and synthesized and then transfected to HT-29 and HCT8 cells, with sh-NC vectors as negative control (NC). MiR-29c-3p mimics were designed and synthesized to overexpress miR-29c-3p, and NC mimics served as negative control. Similarly, pcDNA3.1 vector was used to overexpress NFIA. Plasmids transfections were carried out by Lipofectamine 2000 (Invitrogen, USA). GenePharma (Shanghai, China) provided the synthetical plasmids including sh-NC, sh-LINC00511#1/2, NC mimics, miR-29c-3p mimics, pcDNA3.1 and pcDNA3.1/NFIA.
Sphere Formation Assay
In 6-well ultra-low attachment plates (Corning Incorporated, Corning, NY, USA), CRC cells were digested, counted and seeded at a density of 5000 cells/well in serum-free RPMI 1640 medium (Invitrogen, Shanghai, China) including 20 ng/mL human FGF (Gibco, USA), 20 ng/mL human EGF (Gibco), 1% N2 supplement (Gibco) and 1% B27 (Gibco). Cells were cultivated in an atmosphere at 37°C, involving 5% CO2 to form spheres for 2 weeks. After 2 weeks, a light microscope was used for observation and evaluation of the spheres.
Cell Viability Assay
Cell Counting Kit‐8 (CCK‐8; Dojindo Molecular Technologies, Japan) was used to measure the viability of cells. 96-well plates were applied to incubate the cells. After adding CCK-8 solution at the time points of 0, 24, 48, and 72 h, cells were then incubated for 2 h at 37°C. An automatic microplate reader was used for detection of the OD values at the wavelength of 450 nm.
Western Blot Analysis
Cell lysis was obtained using Radioimmunoprecipitation assay (RIPA) buffer. Proteins were quantified utilizing the BCA™ protein analysis kit (Pierce, Appleton, USA). Proteins were separated by SDS-PAGE and then electrophoretically devolved onto the PVDF membranes. The membrane was enclosed in skim milk and cultivated with primary antibodies overnight at 4°C, and then incubated with secondary antibodies for 1 hour. Protein levels were detected by the chemiluminescent imager (Shanghai, China). GAPDH acted as an internal control. The primary antibodies were displayed as follows: Sox-2 (ab97959); Oct-4 (ab19857); CD44 (ab51037); Nanog (ab80892); GAPDH (ab8245).
Colony Formation Assay
Transfected cells (1 × 103 cells/well) were planted on 6-well plates. The medium needed to be changed every 3 days. Cultivation of the cells was conducted for 14 days. Then, the cells were rinsed by PBS, immobilized with methanol and dyed by crystal violet. Finally, the number of colonies greater than 50 cells per colony could be counted manually.
RNA Extraction and Quantitative Real-Time PCR
Total RNA extraction was performed with the employment of TRIzol reagent (Takara, Otsu, Japan) following the manufacturer’s protocols. RNAs were reverse transcribed into complementary DNA (cDNA) using a SYBR Green Master Mix kit (Takara, Otsu, Japan). The RT-qPCR was conducted by SYBR-Green Real-Time PCR Kit (Takara, Otsu, Japan) in a CFX96™ Real-Time System (Bio-Rad). PCR was conducted under the following conditions: 40 cycles of 94°C for 15 s, 60°C for 10 s, and 72°C for 20 s. NCBI online tool Primer-BLAST (www.ncbi.nlm.nih.gov/tools/primer-blast) was adopted to design the primers which were listed below: LINC00511, forward: 5ʹ-CTAACAAGAGGGTAAGTGTCAG-3ʹ, reverse: 5ʹ-AAGTCGACAACCCCATCGTTAC-3ʹ; miR-29c-3p forward: 5ʹ-GCTGGTTTCATATGGTGG-3ʹ, reverse: 5′-GAACATGTCTGCGTATCTC −3ʹ; NFIA, forward: 5ʹ-ACCAGCTCAAAAAACCTGTGGA-3ʹ, reverse: 5ʹ-TGTTGTGAAACGAAACACCCC-3ʹ; Glyceraldehyde 3-phosphate dehydrogenase (GAPDH), forward: 5ʹ-CATCAAGAAGGTGGTGAAGCAG-3ʹ, reverse: 5ʹ-CGTCAAAGGTGGAGGAGTGG-3ʹ; U6, forward: 5ʹ-GCGCGTCGTGAAGCGTTC-3ʹ, reverse: 5ʹ-GTGCAGGGTCCGAGG-3ʹ. GAPDH was used as the internal reference for lncRNA and mRNA, while U6 was used as an internal reference for miRNA. Calculation of relative genes expressions was performed with the 2−ΔΔCt method.
Transwell Assay
Transwell assays were conducted to detect cell invasion and migration abilities. Transfected cells were added to the upper chambers while the lower chambers were added with 10% FBS. Forty-eight hours later, we respectively immobilized and dyed the invaded or migrated cells with the employment of methanol and crystal violet (Amresco Co, USA). Afterwards, the stained cells were counted from 5 random fields. The operation procedures of cell migration and invasion assay were the same except that there was no Matrigel coated in the upper chambers in migration assay.
Subcellular Fractionation Location
A Nuclear/Cytosol Fractionation Kit (Biovision, USA) was utilized to separate the RNAs from nuclear or cytoplasm fraction. Subcellular fractionation location assay was conducted to detect the levels of LINC00511, GAPDH, and U6 in cytoplasm or nuclear components. GAPDH and U6 were separately functioned as nuclear or cytoplasm control.
RNA Pull-Down Assay
Pull-down assay was adopted to measure the potential binding capacity between LINC00511 and miR-29c-3p. MiR-29c-3p was biotinylated to be miR-29c-3p probe-no biotin and miR-29c-3p probe-biotin by GenePharma Company (Shanghai, China). Cells were lysed by RIPA lysis buffer and incubated with miR-29c-3p probe-no biotin or miR-29c-3p probe-biotin in HT-29 or HCT8 cells. Afterwards cell lysate was cultivated with Dynabeads M-280 Streptavidin (Invitrogen, USA). Purified RNA complex was subjected to RT-qPCR for miR-29c-3p expression detection.
Luciferase Reporter Assay
The 3′-UTR sequences of NFIA including the predicted miR-29c-3p binding sites and full-length sequences of NFIA-Mut were subcloned into the pmirGLO vectors (Promega, Madison, USA) to produce the wild-type NFIA reporter (NFIA-WT) and the mutant NFIA reporter (NFIA-Mut). NC mimics or miR-29c-3p mimics were co-transfected with pmirGLO-NFIA-WT or pmirGLO-NFIA-Mut into HT-29 or HCT8 cells. After 48 h of incubation, the luciferase activities were tested using a dual-luciferase reporter assay system (Promega, USA).
Statistical Analysis
Student’s t-test and One‐way/two-way analysis of variance were employed to compare differences of each group. Data are shown as the mean ± standard deviation (SD). Statistical analysis was analyzed by SPSS 20.0 software (SPSS, USA). Differences were considered as statistically significant while the p-value was less than 0.05. All experiments were repeated three times.
Results
LINC00511 Expression is Prominently Upregulated in CRC Tissues and Cells
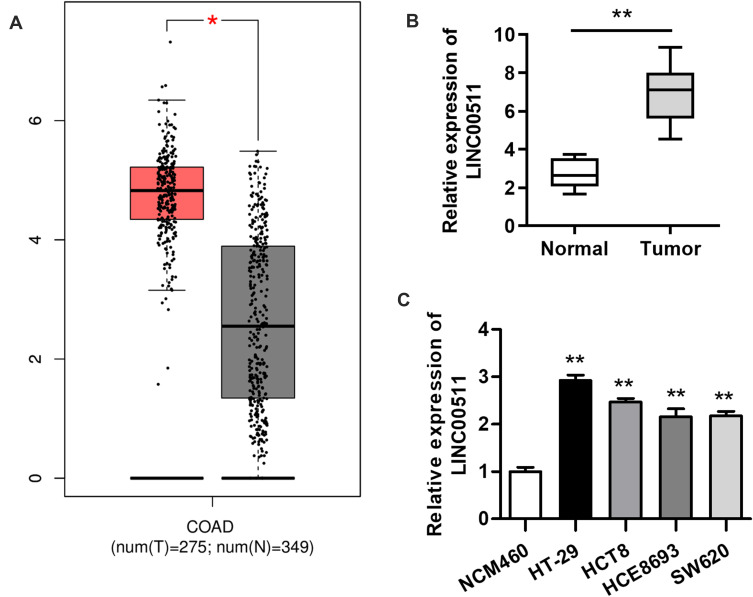
Although LINC00511 upregulation has been discovered in numerous cancers, its expression status in CRC remains to be explored. GEPIA database displayed that the expression of LINC00511 in colorectal tumor tissues was much higher than in non-tumor tissues, and then this result was further determined by RT-qPCR assay (Figure 1A and B). Consistently, the expression of LINC00511 was evidently upregulated in CRC cell lines (HT-29, HCT8, HCE8693, SW620) in comparison with the normal human colorectal cell line NCM460 (Figure 1C). Overall, the above results delineated that LINC00511 was highly expressed in CRC tissues and cells.
Figure 1.
LINC00511 level is observably elevated in CRC tissues and cells. (A) LINC00511 expression in CRC tissues and adjacent normal tissues was found at the GEPIA website. (B) RT-qPCR analysis was conducted to determine LINC00511 expression in 12 pairs of CRC tissues and normal colorectal tissues. (C) The expression of LINC00511 in normal colorectum cell line NCM460 and in CRC cell lines (HT-29, HCT8, HCE8693 and SW620) was measured by RT-qPCR assay. *P<0.05, **P<0.01.
LINC00511 Contributes to the Malignant Phenotypes of CRC Cells
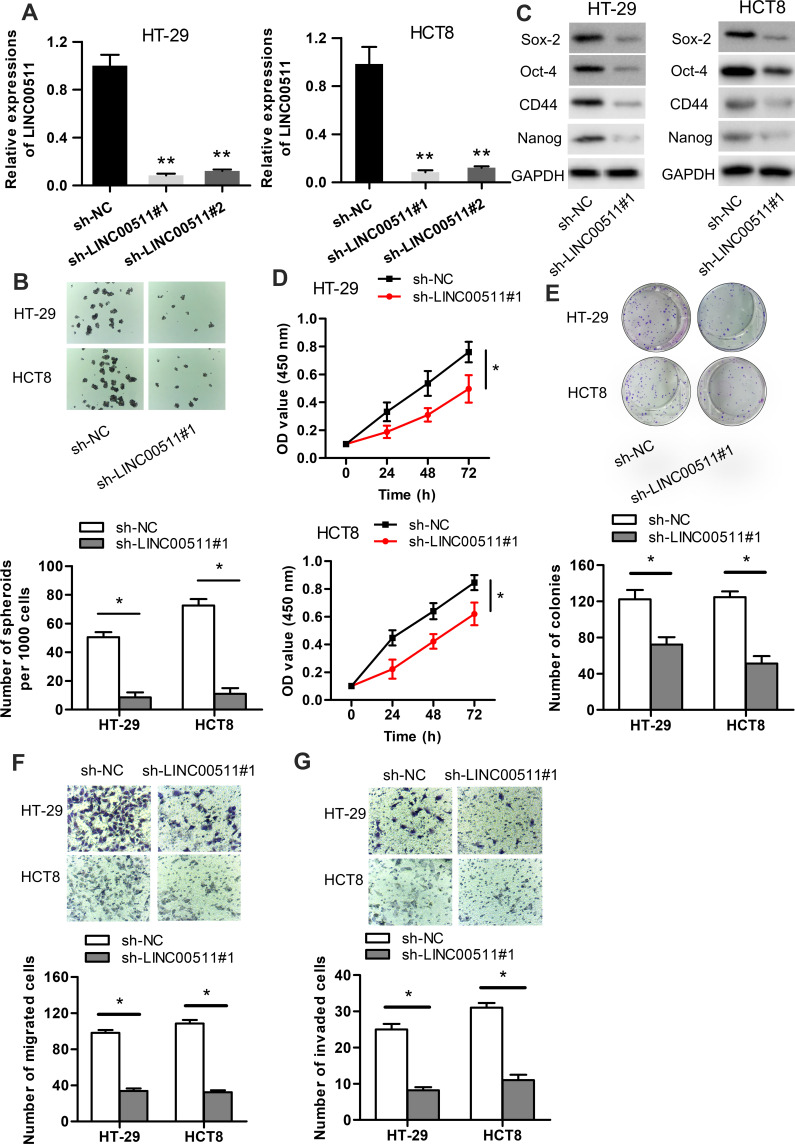
In order to investigate the possible biological function of LINC00511 in CRC development, LINC00511 was knocked down through the utilization of sh-LINC00511#1 or sh-LINC00511#2 with sh-NC as a scramble control. Then, the interference efficiency of sh-LINC00511#1/2 in HT-29 and HCT8 cells was confirmed by RT-qPCR assay. As presented in Figure 2A, the introduction of sh-LINC00511#1/2 caused a significant reduction of LINC00511 expression in HT-29 and HCT8 cells compared with scramble control, suggesting that sh-LINC00511#1/2 were suitable for the following functional experimentations. Then, the influence of LINC00511 down-regulation on cell stemness, viability, proliferation, migration and invasion were further tested in HT-29 and HCT8 cells. Sphere formation assay exhibited that LINC00511 knockdown induced a notable reduction on the number of spheroids (Figure 2B). Western blot assay indicated that LINC00511 deficiency reduced the protein expression of Sox-2, Oct-4, CD44 and Nanog which were associated with stemness in HT-29 and HCT8 cells (Figure 2C). CCK-8 assay demonstrated that LINC00511 knockdown significantly inhibited cell proliferation (Figure 2D). In addition, colony formation assay illustrated that LINC00511 suppression dramatically decreased the number of colonies formed by HT-29 and HCT8 cells (Figure 2E). Moreover, transwell assays revealed that silence of LINC00511 conspicuously hampered the migration and invasion abilities of CRC cells (Figure 2F–G). Taken together, LINC00511 facilitates the malignant phenotypes of CRC through promoting cell stemness, viability, proliferation, migration, and invasion.
Figure 2.
LINC00511 knockdown inhibits the malignant phenotypes of CRC. (A) The knockdown efficiency of LINC00511 in HT-29 and HCT8 cells was examined by RT-qPCR. (B) The number of spheroids was determined by performing sphere formation assay. (C) Western blot assay was adopted to determine the levels of proteins (Sox-2, Oct-4, CD44 and Nanog) associated with stemness. (D–E) CCK-8 and colony formation assays were separately used to detect cell viability and proliferation. (F–G) Cell migration and invasion abilities were ensured by transwell assays. *P<0.05, **P<0.01.
LINC00511 Serves as a Sponge for miR-29c-3p in CRC Cells
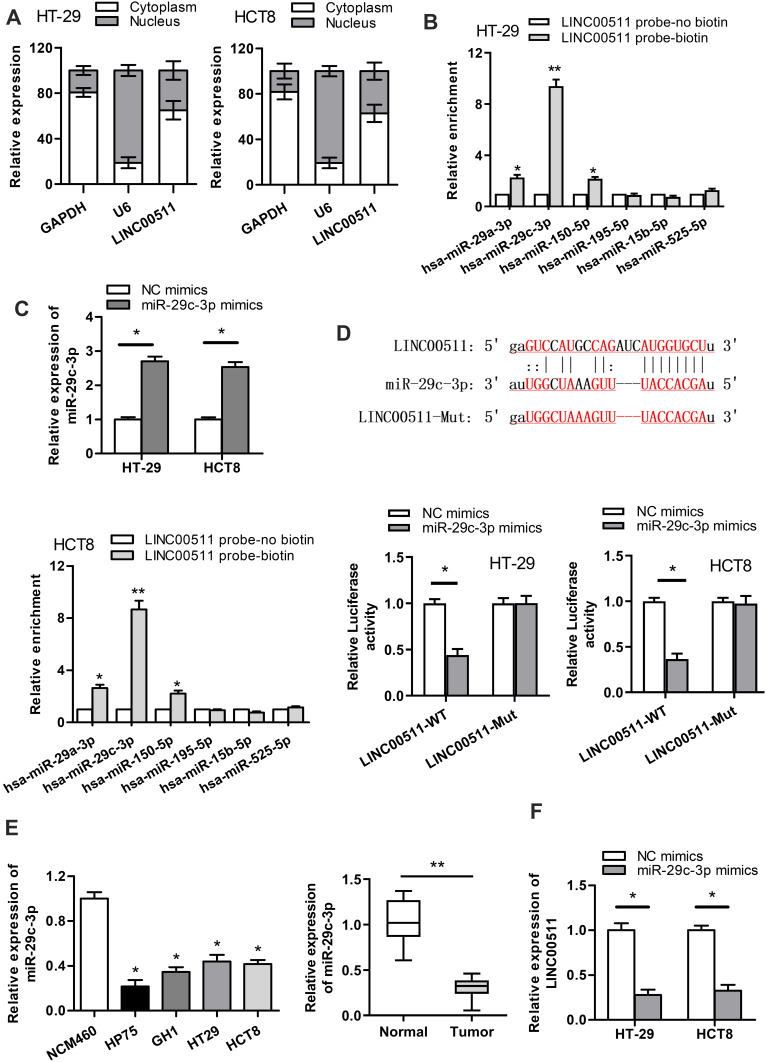
As LINC00511 was reported to function as a ceRNA in multiple cancers,15–18 we designed to figure out whether LINC00511 could combine with specific miRNAs in CRC. As shown in Figure 3A, LINC00511 level was upregulated in cell cytoplasm than that in nucleus based on subcellular fraction assay. By searching starBase website, six candidate miRNAs (miR29a-3p, miR-29c-3p, miR-150-5p, miR-15b-5p, miR-195-5p, miR-525-5p) were speculated to have binding sites with LINC00511 (Table 1). Moreover, we discovered that miR-29c-3p expression was the most enriched in the LINC00511 probe-biotin group in HT-29 and HCT8 cell lines relative to other candidate miRNAs by RNA pulldown analysis (Figure 3B). RT-qPCR assay showed that the expression of miR-29c-3p was significantly elevated by transfection of miR-29c-3p mimics vector (Figure 3C). Then, we validated the interaction between LINC00511 and miR-29c-3p. Luciferase reporter assay showed that the luciferase activity of LINC00511-WT reporters was considerably decreased in miR-29c-3p mimics transfected cells, while no significant change was found in LINC00511-Mut reporters (Figure 3D). As illustrated in Figure 3E, the expression of miR-29c-3p was significantly lower in CRC cell lines (HP75, GH1, HT-29, HCT8) than in human normal colorectal cell line NCM460. Besides, miR-29c-3p expression was also found to be downregulated in CRC tumor tissues relative to adjacent normal tissues. Moreover, miR-29c-3p could negatively regulate LINC00511 expression (Figure 3F). Overall, these results suggested that LINC00511 sponged miR-29c-3p in CRC.
Figure 3.
LINC00511 sponges miR-29c-3p in CRC. (A) The cellular localization of LINC00511 was detected by subcellular fraction assay. (B) RNA pulldown assay was carried out to detect the enrichment of predicted miRNAs pulled down by LINC00511 probe-biotin. (C) RT-qPCR assay was adopted to examine the overexpression efficiency of miR-29c-3p. (D) The combination correlation between miR-29c-3p and LINC00511 was validated by luciferase reporter assay. (E) The expression of miR-29c-3p in normal colorectum cell line NCM460 and CRC cell lines (HP75, GH1, HT-29, HCT8) was measured by RT‐qPCR. The expression of miR-29c-3p in CRC tumor tissues and adjacent normal tissues was detected by RT-qPCR. (F) The effect of miR-29c-3p overexpression on LINC00511 expression was examined by RT‐qPCR. *P<0.05, **P<0.01.
Table 1.
The Potential miRNAs for LINC00511 Were Predicted by starBase Website
| miRNA ID | miRNA Name | Gene ID | Gene Name | Gene Type | Pancancer Num |
|---|---|---|---|---|---|
| MIMAT0000086 | hsa-miR-29a-3p | ENSG00000227036 | LINC00511 | lincRNA | 10 |
| MIMAT0000681 | hsa-miR-29c-3p | ENSG00000227036 | LINC00511 | lincRNA | 19 |
| MIMAT0000451 | hsa-miR-150-5p | ENSG00000227036 | LINC00511 | lincRNA | 12 |
| MIMAT0000417 | hsa-miR-15b-5p | ENSG00000227036 | LINC00511 | lincRNA | 10 |
| MIMAT0000461 | hsa-miR-195-5p | ENSG00000227036 | LINC00511 | lincRNA | 11 |
| MIMAT0002838 | hsa-miR-525-5p | ENSG00000227036 | LINC00511 | lincRNA | 10 |
NFIA is a Downstream Target Gene of miR‐29c-3p in CRC Cells
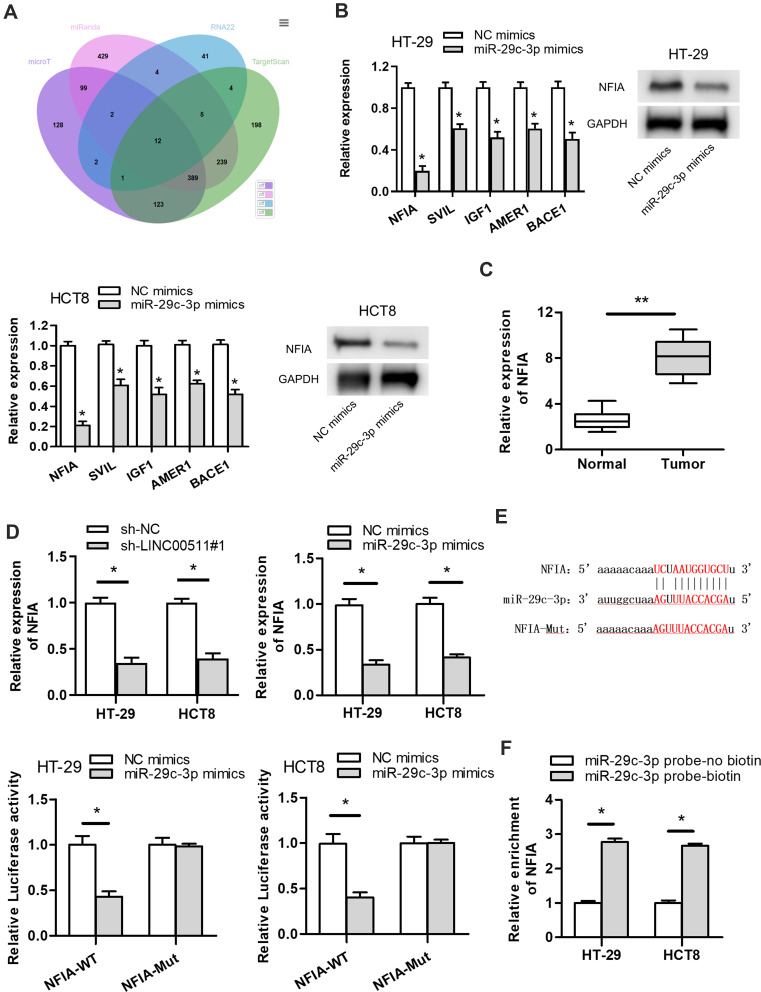
To find out the downstream target genes of miR‐29c-3p, we searched starBase website first. Then, the potential target genes (NFIA, DIP2C, SVIL, BACE1, IGF1, RNF138, KMT5C, BCL11A, C1QTNF6, ADAMTS9, KLHDC3, AMER1) of miR‐29c-3p were presented by Veen diagram (Figure 4A). RT-qPCR assay revealed that among all predicted mRNAs, the relative expressions of the five mRNAs (NFIA, SVIL, IGF1, AMER1 and BACE1) were dramatically decreased in miR-29c-3p mimics transfected cells. NFIA protein expression was obviously decreased by miR-29c-3p mimics (Figure 4B). Furthermore, we discovered the marked upregulated level of NFIA in 12 pairs of CRC tumor tissues compared with non-tumor tissues (Figure 4C). RT-qPCR assay declared the downregulated NFIA expression in sh-LINC00511#1 or miR-29c-3p mimics transfected cells (Figure 4D). Based on the above research and through searching starBase website, we hypothesized that NFIA shared binding sites with miR-29c-3p (Figure 4E). Luciferase reporter assay revealed that luciferase activity of NFIA-WT reporters was significantly decreased by transfection of miR‐29c-3p mimics, while no significant difference was found in NFIA-Mut reporters, indicating that NFIA could bind with miR-29c-3p (Figure 4E). RNA pulldown assay further revealed the abundant enrichment of NFIA precipitated by miR-29c-3p probe-biotin, compared with the miR-29c-3p probe-no biotin, suggesting that miR-29c-3p could directly bind with NFIA (Figure 4F). In conclusion, these data suggested that NFIA could bind to miR-29c-3p in CRC cells.
Figure 4.
NFIA is a downstream target gene of miR-29c-3p. (A) The predicated mRNAs for miR-29c-3p were displayed by Veen diagram. (B) The influence of miR-29c-3p overexpression on target genes expression levels were detected by RT-qPCR. The effect of miR-29c-3p overexpression on NFIA protein expression was detected by Western blot assay. (C) NFIA expression in 12 pairs of CRC tissues and normal colorectal tissues was detected by RT‐qPCR assay. (D) The impact of LINC00511 knockdown or miR-29c-3p upregulation on NFIA expression was measured by RT‐qPCR assay. (E) The predicated binding sites between miR-29c-3p and NFIA were displayed. Luciferase reporter assay was employed to verify whether miR-29c-3p could combine with NFIA. (F) RNA pulldown assay was conducted to further verify the combination correlation between miR-29c-3p and NFIA. *P<0.05, **P<0.01.
LINC00511 Promotes CRC Malignant Phenotypes via Regulating NFIA
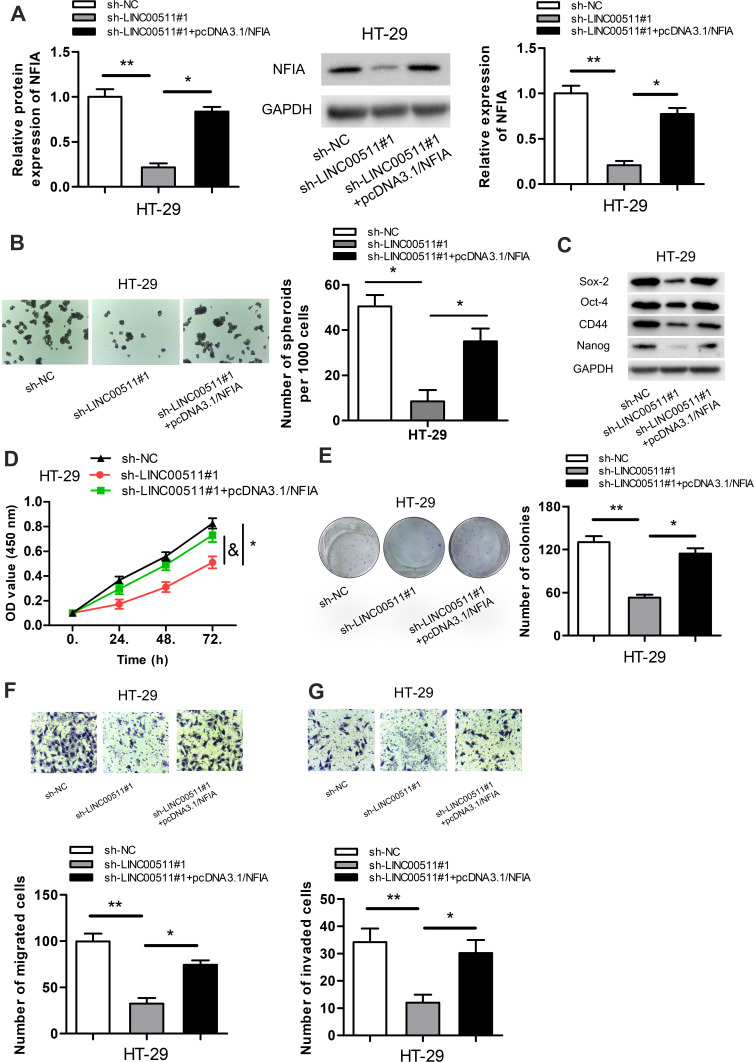
To certify whether LINC00511 made a cancerogenic impact on CRC progression through regulating NFIA expression, the follow-up explorations were conducted. As exhibited in Figure 5A, overexpression of NFIA reversed the inhibitive function of LINC00511 silencing on the expression of NFIA. As displayed in Figure 5B, NFIA overexpression offset the inhibitory effect of LINC00511 silencing on sphere formation. Furthermore, compared with sh-LINC00511#1 transfected cells, the protein expression of Sox-2, Oct-4, CD44 and Nanog were also partly recovered by transfection with sh-LINC00511#1+pcDNA3.1/NFIA (Figure 5C). LINC00511 knockdown repressed cell viability and proliferation of CRC cells, but this effect was rescued by NFIA overexpression (Figure 5D and E). Likewise, NFIA overexpression reversed the inhibitory effect on cell migration and invasion caused by LINC00511 knockdown (Figure 5F–G). The above results led to the conclusion that knockdown of LINC00511 inhibited cell stemness, viability, proliferation, migration and invasion in CRC cells by mediating LINC00511/miR-29c-3p/NFIA axis.
Figure 5.
LINC00511 promotes CRC malignant phenotypes via regulating NFIA expression. (A) NFIA expression in cells transfected with sh-LINC00511#1 or sh-LINC00511+pcDNA3.1/NFIA vectors was detected by RT‐qPCR and Western blot assays. (B) The number of spheroids was tested by sphere formation assay. (C) Relative expressions of proteins associated with stemness were examined by Western blot assay. (D and E) Cell viability and proliferation were separately confirmed by CCK-8 and colony formation assays. (F and G) Transwell assays were carried out to assess cell migration and invasion. *P<0.05, **P<0.01.
Discussion
Previous researches implied that lncRNAs have taken part in numerous cancers like glioblastoma, gastric cancer and ovarian cancer.19–21 Additionally, it was found that a certain amount of lncRNAs were associated with poor prognosis in CRC patients.22–24 Experimental investigations indicate that LINC00511 exerted oncogenic function in many cancers.12–15 Although the biological role of LINC00511 in CRC has been studied once, its biological function and regulatory mechanisms still deserve further exploration. In this research, highly expressed LINC00511 expression was discovered in CRC tissues and cell lines. Besides, LINC00511 silencing cut down cell viability, proliferation, migration, invasion and stemness in CRC. All evidences demonstrated that LINC00511 facilitated the malignant phenotypes of CRC.
MicroRNAs (miRNAs) are a class of short non-coding single-stranded RNA molecules with approximately 22 nucleotides, exerting an influence on regulating gene expressions.25 MiR-29c-3p has been reported to have anti-tumor effect in many cancers. For example, miR-29c-3p inhibits tumor progression in hepatocellular carcinoma by regulating DNMT3B and LATS1 methylation.26 MiR-29c-3p inhibits gastric cancer by regulating MCL-1.27 Recently, it has been reported that lncRNAs sponge miRNAs and then regulate a wide variety of diseases.28 For example, lncRNA GAPLINC facilitates the progression of glioblastoma via binding with miR-331-3p.29 LncRNA NNT-AS1 promotes the proliferation and invasion of cholangiocarcinoma cells via modulating miR-485 expression.30 Besides, lncRNA LINC00511 sponges miR-185 to exert an oncogenic role in breast cancer.31 However, the interaction between LINC00511 and its target miRNAs in CRC cells still needs to be explored. In our current study, we selected miR-29c-3p for LINC00511 from the potential target miRNAs. Despite the fact that miR-29c-3p was upregulated in gastric cancer,32 documents have also confirmed that miR-29c-3p was downregulated and served as a tumor suppressor gene in various cancers like esophageal carcinoma, hepatocellular carcinoma and ovarian cancer.26,33,34 In this research, we certified that miR-29c-3p was downregulated in CRC tissues and cells. LINC00511 and miR-29c-3p can interact with each other in CRC cells.
It is well known that NFIA plays a carcinogenic role in many human cancers.35–37 For example, high NFIA expression was reported to be firmly related to lymph node metastasis in patients with ESCC.35 NFIA leads to adverse prognosis and recurrence for GBM patients.36 Our study verified that miR-29c-3p combined with NFIA in CRC. Rescue assays illuminated that NFIA upregulation partially counteracted LINC00511 silencing-induced inhibition on CRC progression.
In conclusion, our study certified that silenced LINC00511 inhibited the malignant phenotypes of CRC via modulating the LINC00511/miR-29c-3p/NFIA axis, suggesting that this axis might provide a novel therapeutic target for CRC treatment.
Acknowledgment
We appreciate all participants in this work.
Data Sharing Statement
The datasets used during the current study are available from the corresponding author on reasonable request.
Disclosure
The authors report no conflicts of interest for this work.
References
- 1.Xu W, Zhou G, Wang H, et al. Circulating lncRNA SNHG11 as a novel biomarker for early diagnosis and prognosis of colorectal cancer. Int J Cancer. 2019. doi: 10.1002/ijc.32747 [DOI] [PubMed] [Google Scholar]
- 2.Rapado-Gonzalez O, Alvarez-Castro A, Lopez-Lopez R, Iglesias-Canle J, Suarez-Cunqueiro MM, Muinelo-Romay L. Circulating microRNAs as promising biomarkers in colorectal cancer. Cancers (Basel). 2019;11. doi: 10.3390/cancers11070898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen Y, Zhang B, Jin Y, Wu Q, Cao L. MiR-27b targets PI3K p110alpha to inhibit proliferation and migration in colorectal cancer stem cell. Am J Transl Res. 2019;11:5988–5997. [PMC free article] [PubMed] [Google Scholar]
- 4.Ulanja MB, Beutler BD, Rishi M, et al. Colorectal cancer presentation and survival in young individuals: a retrospective cohort study. Cancers (Basel). 2018;10. doi: 10.3390/cancers10120472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Murphy N, Moreno V, Hughes DJ, et al. Lifestyle and dietary environmental factors in colorectal cancer susceptibility. Mol Aspects Med. 2019;69:2–9. doi: 10.1016/j.mam.2019.06.005 [DOI] [PubMed] [Google Scholar]
- 6.Kleffel S, Schatton T. Tumor dormancy and cancer stem cells: two sides of the same coin? Adv Exp Med Biol. 2013;734:145–179. doi: 10.1007/978-1-4614-1445-2_8 [DOI] [PubMed] [Google Scholar]
- 7.Lin CH, Li HY, Liu YP, et al. High-CLDN4 ESCC cells harbor stem-like properties and indicate for poor concurrent chemoradiation therapy response in esophageal squamous cell carcinoma. Ther Adv Med Oncol. 2019;11:1758835919875324. doi: 10.1177/1758835919875324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhao Y, Wang N, Zhang X, Liu H, Yang S. LncRNA ZEB1-AS1 down-regulation suppresses the proliferation and invasion by inhibiting ZEB1 expression in oesophageal squamous cell carcinoma. J Cell Mol Med. 2019;23(12):8206–8218. doi: 10.1111/jcmm.14692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yan S, Wang P, Wang J, et al. Long non-coding RNA HIX003209 promotes inflammation by sponging miR-6089 via TLR4/NF-kappaB signaling pathway in rheumatoid arthritis. Front Immunol. 2019;10:2218. doi: 10.3389/fimmu.2019.02218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang X, Gong J, Lu J, et al. Long noncoding RNA LINC00337 accelerates the non-small-cell lung cancer progression through inhibiting TIMP2 by recruiting DNMT1. Am J Transl Res. 2019;11:6075–6083. [PMC free article] [PubMed] [Google Scholar]
- 11.Shi J, Li J, Yang S, et al. LncRNA SNHG3 is activated by E2F1 and promotes proliferation and migration of non-small-cell lung cancer cells through activating TGF-beta pathway and IL-6/JAK2/STAT3 pathway. J Cell Physiol. 2019. doi: 10.1002/jcp.29194 [DOI] [PubMed] [Google Scholar]
- 12.Jiang L, Xie X, Ding F, Mei J, Bi R. Silencing LINC00511 inhibits cell proliferation, migration and EMT via PTEN/AKT/FOXO1 signaling pathway in lung cancer. Biochem Cell Biol. 2019;1–8. doi: 10.1139/bcb-2018-0364 [DOI] [PubMed] [Google Scholar]
- 13.Liu R, Wang L, Gan T, Pan T, Huang J, Bai M. Long noncoding RNA LINC00511 promotes cell growth and invasion in triple-negative breast cancer by interacting with Snail. Cancer Manag Res. 2019;11:5691–5699. doi: 10.2147/cmar.S203455 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 14.Chen Y, Bao C, Zhang X, Lin X, Fu Y. Knockdown of LINC00511 promotes radiosensitivity of thyroid carcinoma cells via suppressing JAK2/STAT3 signaling pathway. Cancer Biol Ther. 2019;20:1249–1257. doi: 10.1080/15384047.2019.1617569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lu G, Li Y, Ma Y, et al. Long noncoding RNA LINC00511 contributes to breast cancer tumourigenesis and stemness by inducing the miR-185-3p/E2F1/Nanog axis. J Exp Clin Cancer Res. 2018;37:289. doi: 10.1186/s13046-018-0945-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shi H, Shen H, Xu J, Zhao S, Yao S, Jiang N. MiR-143-3p suppresses the progression of ovarian cancer. Am J Transl Res. 2018;10:866–874. [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao X, Liu Y, Li Z, et al. Linc00511 acts as a competing endogenous RNA to regulate VEGFA expression through sponging hsa-miR-29b-3p in pancreatic ductal adenocarcinoma. J Cell Mol Med. 2018;22:655–667. doi: 10.1111/jcmm.13351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ding J, Yang C, Yang S. LINC00511 interacts with miR-765 and modulates tongue squamous cell carcinoma progression by targeting LAMC2. J Oral Pathol Med. 2018;47:468–476. doi: 10.1111/jop.12677 [DOI] [PubMed] [Google Scholar]
- 19.Liao K, Lin Y, Gao W, et al. Blocking lncRNA MALAT1/miR-199a/ZHX1 axis inhibits glioblastoma proliferation and progression. Mol Ther Nucleic Acids. 2019;18:388–399. doi: 10.1016/j.omtn.2019.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen RY, Ju Q, Feng LM, Yuan Q, Zhang L. The carcinogenic complex lncRNA FOXP4-AS1/EZH2/LSD1 accelerates proliferation, migration and invasion of gastric cancer. Eur Rev Med Pharmacol Sci. 2019;23:8371–8376. doi: 10.26355/eurrev_201910_19148 [DOI] [PubMed] [Google Scholar]
- 21.Kong FR, Lv YH, Yao HM, Zhang HY, Zhou Y, Liu SE. LncRNA PCAT6 promotes occurrence and development of ovarian cancer by inhibiting PTEN. Eur Rev Med Pharmacol Sci. 2019;23:8230–8238. doi: 10.26355/eurrev_201910_19132 [DOI] [PubMed] [Google Scholar]
- 22.Zhang Y, Li KF. Karyopherin beta1 deletion suppresses tumor growth and metastasis in colorectal cancer (CRC) by reducing MET expression. Biomed Pharmacother. 2019;120:109127. doi: 10.1016/j.biopha.2019.109127 [DOI] [PubMed] [Google Scholar]
- 23.Shen X, Bai Y, Luo B, Zhou X. Upregulation of lncRNA BANCR associated with the lymph node metastasis and poor prognosis in colorectal cancer. Biol Res. 2017;50:32. doi: 10.1186/s40659-017-0136-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu C, Zhang F. LncRNA AC009022.1 enhances colorectal cancer cells proliferation, migration, and invasion by promoting ACTR3B expression via suppressing miR-497-5p. J Cell Biochem. 2019. doi: 10.1002/jcb.29428 [DOI] [PubMed] [Google Scholar]
- 25.D’Angelo B, Benedetti E, Cimini A, Giordano A. MicroRNAs: a puzzling tool in cancer diagnostics and therapy. Anticancer Res. 2016;36:5571–5575. doi: 10.21873/anticanres.11142 [DOI] [PubMed] [Google Scholar]
- 26.Wu H, Zhang W, Wu Z, et al. miR-29c-3p regulates DNMT3B and LATS1 methylation to inhibit tumor progression in hepatocellular carcinoma. Cell Death Dis. 2019;10:48. doi: 10.1038/s41419-018-1281-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Han Y, Wu N, Jiang M, et al. Long non-coding RNA MYOSLID functions as a competing endogenous RNA to regulate MCL-1 expression by sponging miR-29c-3p in gastric cancer. Cell Prolif. 2019;e12678. doi: 10.1111/cpr.12678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang X, Zhang J, Zheng K, et al. Long noncoding RNAs sustain high expression levels of exogenous octamer-binding protein 4 by sponging regulatory microRNAs during cellular reprogramming. J Biol Chem. 2019;294(47):17863–17874. doi: 10.1074/jbc.RA119.010284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen HH, Zong J, Wang SJ. LncRNA GAPLINC promotes the growth and metastasis of glioblastoma by sponging miR-331-3p. Eur Rev Med Pharmacol Sci. 2019;23:262–270. doi: 10.26355/eurrev_201901_16772 [DOI] [PubMed] [Google Scholar]
- 30.Huang L, Jiang X, Kang P, et al. Long non-coding RNA NNT-AS1 functions as an oncogenic gene through modulating miR-485/BCL9 in cholangiocarcinoma. Cancer Manag Res. 2019;11:7739–7749. doi: 10.2147/cmar.S207801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu L, Zhu Y, Liu AM, Feng Y, Chen Y. Long noncoding RNA LINC00511 involves in breast cancer recurrence and radioresistance by regulating STXBP4 expression via miR-185. Eur Rev Med Pharmacol Sci. 2019;23:7457–7468. doi: 10.26355/eurrev_201909_18855 [DOI] [PubMed] [Google Scholar]
- 32.Dong XZ, Song Y, Lu YP, Hu Y, Liu P, Zhang L. Sanguinarine inhibits the proliferation of BGC-823 gastric cancer cells via regulating miR-96-5p/miR-29c-3p and the MAPK/JNK signaling pathway. J Nat Med. 2019;73:777–788. doi: 10.1007/s11418-019-01330-7 [DOI] [PubMed] [Google Scholar]
- 33.Wang H, Fu L, Wei D, et al. MiR-29c-3p suppresses the migration, invasion and cell cycle in esophageal carcinoma via CCNA2/p53 axis. Front Bioeng Biotechnol. 2020;8:75. doi: 10.3389/fbioe.2020.00075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu H, Mao HL, Zhao XR, Li Y, Liu PS. MiR-29c-3p, a target miRNA of LINC01296, accelerates tumor malignancy: therapeutic potential of a LINC01296/miR-29c-3p axis in ovarian cancer. J Ovarian Res. 2020;13:31. doi: 10.1186/s13048-020-00631-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang B, Zhou ZH, Chen L, et al. Prognostic significance of NFIA and NFIB in esophageal squamous carcinoma and esophagogastric junction adenocarcinoma. Cancer Med. 2018;7:1756–1765. doi: 10.1002/cam4.1434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yu X, Wang M, Zuo J, et al. Nuclear factor I A promotes temozolomide resistance in glioblastoma via activation of nuclear factor kappaB pathway. Life Sci. 2019;236:116917. doi: 10.1016/j.lfs.2019.116917 [DOI] [PubMed] [Google Scholar]
- 37.Wu X, Chen H, Zhang G, et al. MiR-212-3p inhibits cell proliferation and promotes apoptosis by targeting nuclear factor IA in bladder cancer. J Biosci. 2019;44. doi: 10.1007/s12038-019-9903-5 [DOI] [PubMed] [Google Scholar]