Abstract
Biomedical institutions rely on data evaluation and are turning into data factories. Big-data storage centers, supercomputing systems, and increased algorithmic efficiency allow us to analyze the ever-increasing amount of data generated every day in biomedical research centers. In network science, the principal intrinsic problem is how to integrate the data and information from different experiments on genes or proteins. Data curation is an essential process in annotating new functional data to known genes or proteins, undertaken by a biobank curator, which is then reflected in the calculated networks. We provide an example of how protein–protein networks today have space-time limits. The next step is the integration of data and information from different biobanks. Omics data and networks are essential parts of this step but also have flawed protocols and errors. Consider data from patients with cancer: from biopsy procedures to experimental tests, to archiving methods and computational algorithms, these are continuously handled so require critical and continuous “updates” to obtain reproducible, reliable, and correct results. We show, as a second example, how all this distorts studies in cellular hepatocellular carcinoma. It is not unlikely that these flawed data have been polluting biobanks for some time before stringent conditions for the veracity of data were implemented in Big data. Therefore, all this could contribute to errors in future medical decisions.
Supplementary Information
The online version contains supplementary material available at 10.1007/s40291-020-00505-3.
Key Points
| Large-scale multimodal data on patient health is an important factor in the development of personalized medicine. |
| Biomedical Big data is inherently versatile. However, generation, storage, and analysis processes can incorporate inaccuracies that occur when the different structure–function relationships of the intrinsically disordered proteins and the post-translational modifications of proteins, used without a spatiotemporal interpretation, generate inaccurate interpretations in hub genes’ protein–protein interaction networks. |
| We encourage the biomedical community to search for a solution that can rectify data retrospectively, as pitfalls and flaws must be avoided when generating clinical decisions in personalized medicine.. |
Introduction
In this review, we illustrate the consequences that occur when data and information from two unrelated scientific fields converge into common biomedical repositories if not properly “annotated”. This may lead to alterations in the chain of processes that define decision making and models in Big-data systems for medicine.
Although the term “Big data” is a commercial term, we use it to show the ability to analyze and relate an enormous amount of often heterogeneous data to discover new correlations/patterns between different events with the possibility of accurate predictions [1, 2]. Network science, with the support of artificial intelligence, promises to make a significant contribution to medicine, but we must realize that it is a very complex technical-scientific enterprise that must integrate heterogeneous processes, information, and data. As such, it must be considered still in its infancy.
In particular, we explain how the structural properties of intrinsically disordered proteins (IDPs) and the intrinsic phenotypic heterogeneity of genes in tumor tissues may introduce and propagate errors when involved in metabolic networks as omics data without consideration of spatiotemporal events. The discovery of IDPs [3] changed the study of living systems. It is clear today that these multifunctional flexible fluctuating proteins manage key roles in many biological functions. Recent estimates [4] suggest that databases contain about 40 million compact and globular proteins, of which about 70,000 are IDPs and the remaining are “mixed proteins” that have fixed-in-time and disordered structures in the same molecule. These numbers show the importance and weight of “disorder” in functional activities. Their “broad functions spectrum” depends on the many molecular forms generated by post-translational covalent modifications [5]. Therefore, we should consider their particular functional and structural properties entirely in the specific cancer tissue and metabolic context in which they are active. Any attribution outside the correct context introduces uncertainties. Even the intrinsic phenotypic heterogeneity generated in tumor tissues by disease progression [6, 7] may introduce critical inaccuracies in biomedical data, as we show through the disconcerting discovery of hundreds of different hub genes from networks reported in recent literature for human hepatocellular carcinoma (HCC). We also present some tips for solving these issues.
Biomedical researchers are faced with high-dimensional data from a number of sources, including microarrays and next-generation sequencing [8, 9]. The analysis and accurate interpretation of biological information is crucial because, in such data, the number of variables (genes and their coded products) is many times greater than the number of tested samples. The challenge in genomic data is in applying appropriate computational methods to the ever-increasing size of high-throughput multiomics data [10]. Processing and integration of multiomics data requires in-depth technical and biological knowledge of how these data are generated [11]. Even the extraction of large-scale genomic experiments from the scientific literature through data mining relies on computational approaches and statistical control [12] to transform it into a reduced set of accurate information as, for example, when contextualizing molecular changes associated with a physiological mechanism [13], functional links, experimentally found for a disease, should merge in the human proteomics network because of common genetic relationships [14, 15]. We explain how compromised functional correlations also derive from chemical–physical modifications of the same proteins encoded by hepatocellular carcinoma (HCC) genes. This happens because no accurate logical-semantic classification is performed during archiving processes in the computational analysis systems. This involves imperfect interactions because of the incorrect attribution of the space-time context in which they are operating. Through fast and massive transfers from biobank to Big data biomedical systems, it is very likely that we will find these metabolic models in medicine-related decision-making processes.
Big Data in Biomedicine
Semantic Logic
Before dealing with the biomedical aspects, a general, albeit minimal, picture of what Big data is and its logic is necessary. Big data is characterized by extensive data collections, in terms of volume, speed, and complexity, and thus requires specific technologies and analytical methods for information extraction [16]. Therefore, a Big-data system has at least two fundamental characteristics: a continuous increase in data volume and an ability to manage a high-speed information flow. Data are gained from widely heterogeneous sources, both from structured meta-data (data from information systems organized as archives) and non-structured mega-data (images, annotations, etc.) [17, 18]. These Big-data systems continuously growing, so adequate tools and methodologies to extract and process the information are needed. The reliability of the data must be controlled during their generation, acquisition, extraction, archiving, integration, analysis, and modeling to avoid erroneous interpretations [19]. The value of the data is in its quality rather than its quantity. When the algorithms analyze large masses of data, both structured and unstructured, to search for all existing relations between them, they need to know the precise meaning of each node, term, and entity, otherwise it becomes impossible to establish precise models (see Box 1 in the Electronic Supplementary Material for more detail and explanations). Thus, ontologies, semantics, and interoperability become essential logical issues and must be applied consistently [20]. For example, when two (or more) different functions could be applied to the same node (e.g., a protein) in a network, they must first be assessed ontologically and semantically to define which is appropriate in that context, certainly not all of them. The application of Big data has extended into many fields of human knowledge, but what we want to briefly explain here is the fundamental role of these systems in modern biomedical disciplines [2, 21].
Type and Purpose of Biomedical Big Data
Multiomics, medical imaging, device data, and electronic health records (EHRs) represent the central data types in biomedicine. EHRs are an appropriate example of medical Big data because they represent a powerful tool for improving the quality of healthcare [22]. In fact, clinical data are the fundamental basis of medical information to integrate with specific biobanks to implement precision medicine [23]. These medical records are an electronic compilation of longitudinal data (data collected through a series of repeated observations of the same patient over some extended time) related to the healthcare of individuals. For example, by coupling EHRs with genomic biobanks, EHRs could provide clinical phenotypes for genomic studies [24]. However, the integration of multiomics data with EHRs is currently still in a nascent stage. Using biomedical Big data in precision medicine [25] will require significant scientific and technical developments, including infrastructure, engineering, and project and financial management. Substantial challenges remain in the provision of an accurate interpretation of EHR data to allow for its repurposing in clinical and genomic research, where algorithms will be used to accurately find cases of disease through specific controls. Such algorithms integrate a good deal of heterogeneous information, such as laboratory and test result data, medication records, billing codes, and clinical notes, to accomplish necessary recall and precision [26]. Clinical records contain unstructured (narrative) clinical documentation so requires natural language processing through ontological dictionaries and semantics.
All this should work to support clinicians in making a diagnosis. It is crucial to understand that they face a complex pyramid of data and analytics. At the bottom of the pyramid is the input of raw clinical data and information about patients; the next level is data organization with specific semantic codes; next is the coupling with biobanks for critical evaluation of hospitalized patients; at the top is a personalized diagnosis aided by the clinical decisional support. Every level of the pyramid has a clear relationship with the next, where the goal is to have certain and accurate knowledge in the preceding levels as the foundation for forecasting outcomes at the higher levels. Thus, it is important that data imported from biobanks are error free and that information transfer between the levels occurs without error. Data collection is faster than the ability to process and analyze information. In fact, the gap between the rather slow and sometimes imprecise semantic and functional interpretation of biomedical information and its rapid acquisition is increasing. If the semantic analysis is inaccurate or slow, errors are generated that the system is no longer able to recognize [27]. Therefore, correct integration of molecular information, such as multiomics data or phenotypic information relating to individual patients from EHRs, is becoming crucial [28]. Recent advances in single-cell genome and RNA sequencing [29] are having a substantial impact in medicine because of their greater precision in defining the space-time context, thus are the biobanks destined to be integrated in standard medical practice. Therefore, we can imagine “biomedical Big data” as a complex system of heterogeneous information where groups of functional correlations and patterns exist among specific events in this mass of data. Extracting these groups and their hidden relationships is one of the major purposes of using Big data because this is how new medical models can support clinical decisions. The best way to represent these relationships is through graphs (network medicine) where the entirety of their correlations acts as a complex system [30, 31]. We define a system as complex when it comprises many interactions among the constituent components (including interactions between the system and external environment), even in their evolution over time. One of the main features is that the individual knowledge of each component is insufficient to establish the overall evolution of the system [32]. Furthermore, a complex system is characterized by a considerable number of variables, where only their interactions determine the global behavior by showing unpredictable non-linear behaviors unrelated to the single elements or the sum of their properties [29, 32]. Without further explanation outside the context of this review, we want to highlight that, in medicine, interactive networks can also be used for a new representation of patient care.
Perspective for Biomedical Big Data
A new stream of research is beginning to investigate the digital health networks originating from digital medical records (i.e., EHRs) to test whether the whole structural organization generated by patient care as nodes shares properties similar to those of biological networks that show a power law characteristic of scale-free networks. The networks are scale free if the fraction of nodes with degree k follows a power law distribution K−α, where α is greater than 1. This law requires that, in the representation of the distribution of the degrees of connectivity of each node present in the calculated network, the nodes with low connectivity are in the majority and the nodes with high connectivity, and therefore with the role of aggregators, are very few [33]. First attempts have shown that, despite differences in theme, content, and data type, the net of digital medical records follows properties of power laws [34]. For example, EHR’s connect patient, doctor, diagnosis, drugs, medical processes, instrumental and biochemical-clinical analyses, and administrative processes, as functional nodes. Through specific coding of these nodes (e.g., the Logical Observation Identifiers Names and Codes [LOINC] [35] used for laboratory digitized records), we can implement the correct semantic and functional relationships among all the players through a network representation with scale-free properties. Even if it does not appear as such to most people, the individual patient’s medical record can be considered as a node (or, better, a module) of the social hospital network [34]. Through these networks, patients’ biomedical data, the capabilities of individual doctors, and the costs of certain medical protocols can be extracted and analyzed; they are also transferred to the archives of the biomedical Big data system. In a network, we can grasp the features of the individual nodes, but they are of no use when we analyze the nonlinear global biomedical behaviors that emerge from relational aggregations between nodes [36].
Relationships Between a Node and Its Network
From this general overview, we further examine the molecular level of the genes and proteins, where omics data are born. In the network, the node, as a single entity, does not have great meaning, because it expresses its value only through the entire network. This implies that the properties of a network depend on the constitutive functional relationships that each node implements with the other nodes in that biological system [37]. Hub nodes are particularly important. They are highly connected nodes, present in real-world and biological networks, which include many representations of networks such as protein–protein interaction networks (PPINs), gene regulation networks, and residue interaction networks, among others. Hub nodes occupy important functional network positions; for example, in PPINs, they are often key targets for drugs [38]. Their role is to coordinate relationships between nodes by determining the robustness of the network. Therefore, the few hubs in a network [31] represent the weak points for the resilience of the whole system. If a hub is targeted by a drug, functional relationships fail and the network collapses. A net is a heavily intertwined and mutually dependent dynamical system in which its functional modules are organized around individual hubs. When direct connections between hubs change, the net changes its topology and the modules’ specificity. In fact, topologically, the modules’ specificity can be changed by changing interactions between hubs, but the net is destroyed if some hubs are eliminated [39].
Computational Consequences of Post-Translational Modifications
Incorrect attribution of functional capacities to a physical node that it does not possess leads to emergence of a very different set of characteristics. Translating into molecular terms, the attribution of different metabolic functions to the same node, for example those associated with post-translational modifications (PTM) of a protein, leads to functional information that often has a different metabolic meaning, with possible distortions in the structural topology of the network. In fact, a PTM form is a covalently modified molecular form of a protein, thus, a new biomolecule performing different functions in different spatiotemporal contexts in respect to the native protein [40]. Therefore, the function of a PTM protein should be associated only with the specific context, i.e. tissue, in which the modified protein implements its constitutive functional relationships as a node of that specific network. This allows it to relate to the other node proteins that exist in that metabolic context, allowing the generation of a graph with the topology more suitable for that specific temporal event [41]. Therefore, it is necessary to briefly explain what special properties biological evolution has given to hub proteins to make them possess a high degree of connectivity. Given the nature of this review, this is a point we cannot disregard. Proteome-wide screening approaches have provided information about interacting proteins by discovering that the intrinsic disorder is a common feature of hub nodes. In particular, hub nodes with nuclear co-localization encode for IDPs [42, 43]. Intrinsic disorder favors interactions with many molecular partners and therefore their inclusion in functionally complex metabolic contexts.
A Concise Description of the Evolutionary Mechanisms Used for PTMs in Intrinsically Disordered Proteins
Evolution uses two main molecular mechanisms by which a protein containing intrinsic disorder can drive the disorder towards new biological functions: PTMs and alternative splicing (AS). Both mechanisms change the covalent molecular organization of the protein, albeit differently.
While AS is a mechanism that derives from gene expression, PTMs perform a chemical modification of proteins. We know about 300 distinct PTMs of eukaryote proteins as a consequence of the action of as many enzymes, and the human proteome contains up to a million modified polypeptides, where intrinsically disordered proteins (IDPs) and mixed proteins are preferential targets of multiple PTMs [5]. The quality and quantity of sequence modification changes the physico-chemical properties of the protein, even inducing changes in the charge distribution. Thus, IDPs represent excellent functional hubs able to switch their modified molecular forms to new functional states. Unfortunately, the characterization of these “functional states” and how they change, when the context evolves, are not yet the focus of many studies because homogeneous samples of the modified IDPs from the specific tissue are required. Although various experimental approaches exist [5], they are not being pursued because of the complexity of purifying the large quantity of sample needed.
Structure–Function Relationships in the Operative Context of Modified IDPs
PTMs change the chemical-physical properties of the molecule. Consequently, if we do not associate each molecular form to its time- and context-dependent functional activity, our metabolic models will be incorrect. In fact, the different molecular forms will act differently, depending on the tissue and cell type, and vary according to the metabolic context. Therefore, when we collapse new functions, derived from the PTMs, to the native IDP sequence through annotations, we generate a flawed functional meaning of the network. When this information flows to a biobank, we generate complications in the logical-semantic interpretation of the data because of the lack of the correct logical parameters that allow the attribution to each entity, word, molecule, and event, of the correct molecular and functional meaning. If the analytics of the biomedical Big-data system does not intercept the correct spatiotemporal functional activities, it will consider some biological functions that do not exist in a specific metabolic context.
Sequence chemical modification encodes for a form of the IDP that performs a precise informational function [44–46], so acting as an encoder and transmitter of the biological information. In fact, we can say that the modified polypeptide carries a piece of peculiar biological information merely when it gives actual form to this peculiar function. Therefore, the view that the conformational ensemble distributions could be random contrasts with the great diversity of the many biological and pathological roles exerted by IDPs. From the point of view of information theory, this means the informational entropy of the modified polypeptide must be reduced, otherwise the biological information cannot be sent through the transmission channel [44–46], and the receiver (the interaction partner) cannot get the biological information to make it work. From a structural point of view, this means that the molecular partner can only implement new biological information through the one-to-one recognition of a specific PTM polypeptide. Without a correct logical-semantic classification of the properties of the transmitting node, the recognition of the molecular partner becomes a completely random event.
The Case of Human SELK and Its Disordered C-Domain: An Example of Multifunctional Spatiotemporal Events Exerted by PTMs
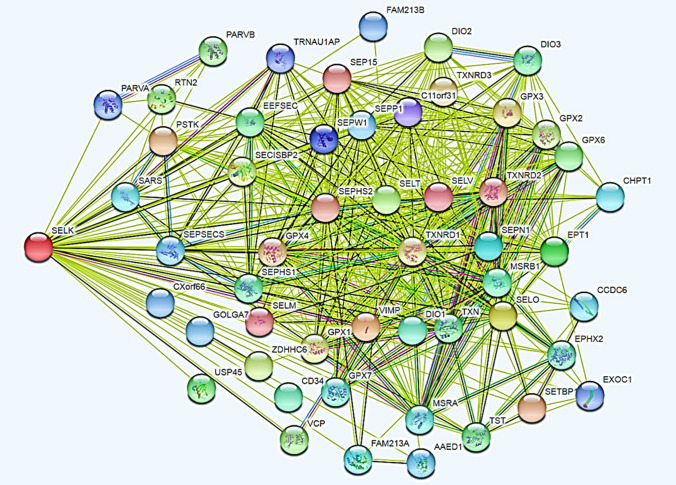
Here, we examine recent investigations on the structural and functional characterization of selenium-containing proteins [47, 48]. These proteins show disordered domains in the terminal regions. In particular, a segment of 51 residues found in the C-terminal region of human SELK is a disordered domain [49]. This segment shows six phosphorylation sites, of which three were found in experiments. The combinatorial calculation with n sites occupied by K similar objects can estimate the action of the phosphorylating enzymes (kinases) on the six SELK sites. Thus, we can calculate how many total molecular forms we might get. We can calculate six single forms, 15 pairs, 20 triples, 15 quadruples, 6 quintuples, and one sextuple for 63 likely molecular forms. We used STRING (Search Tool for the Retrieval of Interacting Genes/Proteins) to check how many molecular forms are known for SELK.
STRING is an annotated database and web resource of protein interactions, based on PPINs. STRING aims to collect and integrate this information, consolidating the known and expected PPI data for a large number of organisms. Associations in STRING include physical and functional interactions [50, 51]. In STRING, a score is provided for each PPI, indicating the estimated probability that an interaction is biologically significant, specific, and reproducible given the supporting evidence. STRING imports data from two channels: experiments (experimentally proven data) and databases (from text mining and biobanks). The score is a cutoff to limit the number of interactions. The highest score (0.9) is more likely to select true positives. Setting the cutoff lower (e.g., at 0.4) will increase the coverage but also the fraction of false positives.
Using STRING, through the first-order enrichment (direct interaction) partners (PPI enrichment p value: < 1.0e−16), we found 52 different functional known partners (Fig. 1). This value is in good agreement with the 63 potential molecular forms. At the same time, we are representing a non-existent metabolic context in which functional relationships that should take place in specific cellular districts for different biological situations add up, so it is a misleading metabolic image since the conclusions we can draw from it are wrong. This metabolic representation is created by algorithms that do not recognize the logic and semantics that characterize specific biological events and rests on the absence of experimental studies aimed at characterizing the structural and functional knowledge necessary to position those same biological events in time and space.
Fig. 1.
The total amount of first-order functional interactions (52 interactors) found for human SELK through STRING. The graph shows the maximum enrichment for SELK (53 nodes; 540 edges; average node degree: 20.4; average local clustering coefficient: 0.833; p < 1.0e−16).
In Table 1, we illustrate the structural effects of the variable amount of PTMs (phosphorylation) on conformational states of the C-terminal segment alone. We can see that the native polypeptide at neutral pH populates a conformational state represented by ensembles of coils and chimeras of globules. When SELK accumulates phosphorylation to its six sites, the protein changes its conformational class with a sequential migration to ensembles populated by coils, hairpins, or chimeras of coils. This means that the PTM forms of SELK, by changing their chemical-physical and structural parameters, change the polypeptide structure. Therefore, when we annotate extra features to a native protein, this is reflected in the network because these annotations collect all the functions found for a specific node with no context discrimination. This means that, in the network datasets, we consider the functional interactions independent from their biological context. STRING tries to minimize these effects by listing some known biological roles exerted by a node to select the most adequate role for the biological context under study. But it is a cat that bites its tail, because the interaction network from gene expression experiments is calculated without knowing the functional interactions involving a specific node. This favors interactions with incorrect functional partners. Today, this is poorly considered, but, in the meantime, the functional entropy increases among Big data, where, instead of stabilizing the information, we introduce misleading metabolic hypotheses with a high probability of propagation [52].
Table 1.
Effect of phosphorylation on the structural organization of SELK
| ID | PTM | K | FCR | NPCR | Hydropathy | Isoelectric point | Plot region |
|---|---|---|---|---|---|---|---|
| Seq1 | Native polypeptide | 0.265 | 0.275 | 0.118 | 2.914 | 10.86 | 2 |
| Seq2 | + 1 phosphate | 0.239 | 0.314 | 0.078 | 2.914 | 10.24 | 2 |
| Seq3 | +2 phosphate | 0.265 | 0.353 | 0.039 | 2.808 | 9.68 | 3 |
| Seq4 | + 3 phosphate | 0.262 | 0.392 | 0.000 | 2.686 | 8.49 | 3 |
| Seq5 | + 4 phosphate | 0.219 | 0.431 | -0.039 | 2.573 | 7.36 | 3 |
| Seq6 | + 5 phosphate | 0.196 | 0.471 | -0.078 | 2.451 | 6.91 | 3 |
| Seq7 | + 6 phosphate | 0.177 | 0.510 | -0.118 | 2.365 | 6.52 | 3 |
K, FCR, and NCPR values have been calculated according to Pappulab (http://pappulab.wustl.edu/CIDER/analysis/) [109], hydropathy values according to Kyte and Doolittle [110], and the isoelectric point on the platform Bachem (https://www.bacandhem.com/de/service-support/peptide-calculator/). K (charge patterning parameter) is a parameter to describe the extent of charged amino acid mixing in a sequence; for a sequence of fixed composition, K goes from 0 to 1. FCR is the fraction of charged residues. As the fraction of charged residues increases, the relative impact of how those charges are spread across a sequence becomes more significant. Hydropathy is the 0–9 scaled Kyte–Doolittle hydropathy score for the sequence (9 most hydrophobic, 0 least hydrophobic) [110]. Phase plot region is the location where the sequence falls on the Das–Pappu phase plot. Region 2 is the collapsed or expanded structure, where their behavior may depend on other factors (salt concentration, ligand binding, interactions, etc.), and region 3 includes strong polyampholytes: coils, hairpins and chimeras – here the types of structures that form may depend on the K value
ID sequence identification, NCPR net charge per residue, PTM post-translational modification
In the following, we illustrate how the different information existing in different databases for the same protein can affect the topology of a graph. As an example, we compare BioGRID (Biological General Repository for Interaction Datasets) and STRING. BioGRID is an annotated database that shows the PPIs, genetic interactions, chemical interactions, and PTM with a significant weight towards the physical interactions. In BioGRID, these interactions have been determined experimentally with classical methods (affinity chromatography and mass spectroscopy, two-yeast-hybrid, immunoprecipitations, and others) in solutions comprising ground cells [53, 54].
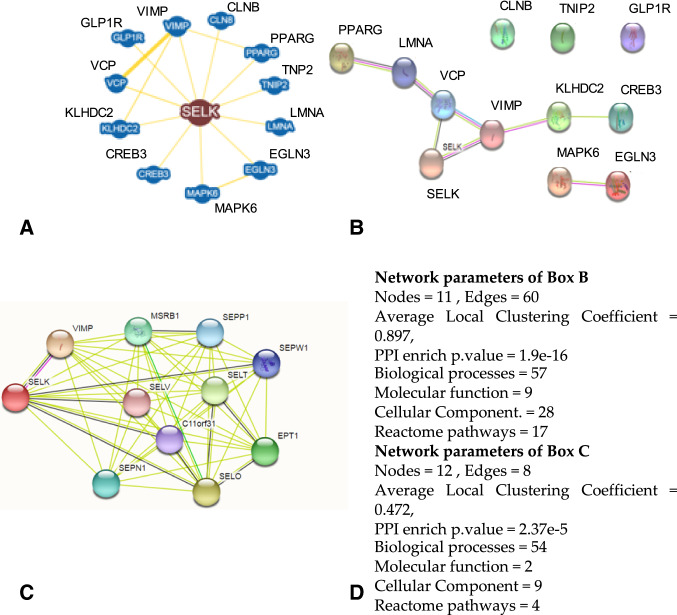
In Fig. 2a, we show the graph of the main first-order interactions of SELK calculated by BioGRID. The graph shows 12 nodes with first-order interactions, reported as physical (80% significant and 20% low). If we launch the same search for SELK on STRING, we have 11 nodes (Fig. 2b) but with different proteins. Only VIMP was present in both networks, and STRING gives the SELK-VIMP interaction a score of 0.966. The remaining interactions, all experimental, get high scores of 0.946–0.805. Yet, STRING’s interactions are functional, whereas those from BioGRID are physical and obtained from solutions of fragmented cells. Neither approach reflects the real metabolic context in which the molecular partners should have been present or the correct molecular partner. The physical interactions used by BioGRID show the ability of two proteins to interact, as determined by biophysical techniques in a non-physiological context.
Fig. 2.
The graphs obtained for the SELK protein using BioGRID and STRING. a Shows the BioGRID result for SELK only. BioGRID, unlike STRING, does not allow node enrichments but reports all the nodes that physically interact with SELK. b Shows the STRING result for SELK enriched with several interactors, similar to that from BioGRID. c Shows the STRING result for multiple searches with the set of proteins (including SELK) as reported by BioGRID in (a). d Shows the network parameters and functions reported by STRING for (b) and (c)
If instead we analyze through STRING the entire set of 12 proteins found with BioGRID, we have a new graph (Fig. 2c) that shows a different organization from the previous ones with some proteins not functionally connected into the graph. The two analysis systems give different results, although the STRING network parameters for the Fig. 2 graphs are still significant. The two graphs in Fig. 2b and c also show different biological behaviors. In Fig. 2d, we report the network parameters in the number of biological processes, molecular functions, cellular components, and reactome pathways, as calculated by STRING, for networks in Fig. 2b and c. Parameters reported in the figure show how differences in the organization of the metabolic networks derive from the context in which the properties of nodes have been determined as well as from the network topology.
The two computational platforms should reflect the functions of cells in the multicellular human system. Interactions assemble proteins into modules that drive the spatial and functional organization of tissues, defining PPINs whose topologies should encode each protein’s cellular environment, but these organizations vary with cell state. Thus, at present, network analyses do not show the actual functioning of cells, but only a static view of their functioning. They reflect these limits in the metabolic networks and knowledge we can draw from them. The PTM forms, the isoforms of many proteins [55, 56], the short duration of many interactions, the interactions, and the metabolic context, or even the variability of protein expression, represent the barriers that do not allow us yet to have complete knowledge of the whole repertoire of protein interactions and the metabolic contexts in which they occur. In this framework, errors are likely and must be taken into account in precision medicine. In fact, we can find them in biobanks where they will interact with medical data with unpredictable consequences.
The Case of the Hepatocellular Carcinoma Hub Genes: An Example of Cancer Phenotype Heterogeneity, Where Knowledge of the Spatiotemporal Action of the Genes is Lacking
Although the clinical management of HCC has evolved considerably over the last decade, and the use of some drugs has shown some survival benefit, surgical resection or liver transplantation are still the recommended therapy for accurately selected patients. Immunotherapy approaches are awaited for these patients, but this cancer is peculiar because the coexistence of viral infections develops a complex immunobiology, generating many hepatic mechanisms of immune regulation and tolerance. However, reliable markers to discriminate mechanisms behind tumor progression remain to be identified. This prevents a formal clinical approach for the continuation of treatment during tumor progression. Researchers are still trying to obtain clear information, but even if a picture of its molecular pathogenesis is defined, the underlying molecular mechanisms will remain unclear. Next-generation sequencing has improved our understanding of the heterogeneous cell subpopulations within tumor tissue; however, we require not only the identification of a gene or protein and its associated signaling but also its correct spatiotemporal location in the pathophysiological events that characterize the progression of this cancer. We need this knowledge to develop targeted therapies.
In this context, many research teams are actively trying to identify this potential pharmacological target to design a molecule capable of treating this type of cancer. They look for genes that code for proteins that play key roles in tumor progression and with high connectivity, i.e., hub nodes. Our literature search for a reliable marker of progression or a target protein resulted in a confusing array of targets. We noticed that several teams reported the discovery of many hub genes for HCC. Therefore, we performed a systematic literature review in MEDLINE, with keywords referring to articles containing data on “hub genes in hepatocellular carcinoma”.
We searched for all articles published between 2014 and 2018 that identified genes/proteins with a key role as target or hub genes in this cancer. We selected articles reporting hub gene expression data [56–87]. In total, 324 hub genes were isolated from from patients with HCC (Table 2).
Table 2.
Hub genes in hepatocellular carcinoma extracted from the literature
| Set of 208 non-redundant genes extracted from a total of 324 hub genes found in the literature. The acronyms of the genes are listed alphabetically | Redundancy of the 61 hub genes. In parentheses the number of times that each gene has been found for a total of 177 times | |||
|---|---|---|---|---|
| A2M | COL1A1 | HSF1 | POP1 | DC20(9) |
| ABAT | COL1A2 | HSP | POTEF | BUB1(6) |
| ACAA1 | COL4A1 | HSP90AA1 | POU3F4 | CCNB2(6) |
| ACACA | COMMD5 | HSP90AB1 | PRC1 | TOP2A(6) |
| ACADM | COPS5 | HSPA1A | PRKCA | CCNB1(5) |
| ACSM3 | CRYL1 | IGF1 | PRKDC | CDK1(5) |
| ACTB | CSNK2A1 | ILF3 | PRKG2 | MAD2L1(5) |
| AGXT | CXCL1 | INCENP | PTEN | MYC(5) |
| AHSG | CXCL12 | ITGA2 | PTGS2 | HSP90AB1(4) |
| AKT1 | CYCS | JUN | PYCRL | ESR1(4) |
| ALB | CYP2B6 | KDM6B | Q12834 | PRKDC(4) |
| ALDH2 | CYP3A4 | KGK | RACGAP1 | ARHGAP39(3) |
| ALDH6A1 | CYP4A11 | KIF11 | RAP2A | AURKA(3) |
| APOA1 | DAO | KIF20A | RFC4 | BIRC5(3) |
| APOC3 | DCAF13 | KIF23 | SCNN1A | BUB1B(3) |
| AR | DKK1 | KIF2C | SFN | C8ORF33(3) |
| ARPC4 | DLGAP5 | KIF4A | SIRT1 | CCNA2(3) |
| ASL | DNMT1 | KNG1 | SLA | CDKN3(3) |
| ASPM | DSCC1 | KRAS | SPARC | CSNK2A1(3) |
| ATN | DTL | KRT18 | SPC24 | DSCC1(3) |
| AURKA | ECHDC2 | MAD2L1 | SPP2 | INTS8(3) |
| AURKB | ECHS1 | MAPK1 | SRC | NUDCD1(3) |
| AXIN2 | EFNA4 | MAPK8 | STAT3 | PCNA(3) |
| AZGP1 | EGFR | MCM10 | STIP1 | POP1(3) |
| BCL2 | EGR1 | MCM2 | SUCLA2 | PUSL1(3) |
| BHMT | EHHADH | MCM3 | SUMO | RFC4(3) |
| BIRC5 | ENO1 | MCM4 | SUMO1 | STIP1(3) |
| BMP4 | ERBB2 | MCM6 | SUMO2 | TGD5(3) |
| BUB1 | ESPL1 | MDM2 | TFRC | UBD(3) |
| BUB1B | ESR1 | MELK | TGFB1 | ACAA1(2) |
| CALM3 | F2 | MME | TIGD5 | ACADM(2) |
| CASP8 | F8 | MMP2 | TK1 | ACSM3(2) |
| CCNA2 | FGF2 | MT2A | TOP1MT | AURKB(2) |
| CCNB1 | FN1 | MUT | TOP2A | BCL2(2) |
| CCNB2 | FOS | MYC | TP53 | CDKN1A(2) |
| CCND1 | FOXO1 | NCAPG | TPX2 | CEP55(2) |
| CD8A | GCDH | NCOR1 | TTK | CHEK1(2) |
| CDC20 | GINS1 | NDRG1 | TTR | CKAP5(2) |
| CDC37L1 | GKB | NEK2 | TXNRD1 | COL1A1(2) |
| CDC45 | GLI1 | NPM1 | UBD | DLGAP5(2) |
| CDCA8 | GMPS | NSMCE2 | UBR5 | ERBB2(2) |
| CDH1 | GNAO1 | NUSAP1 | UQCRC2 | FOS(2) |
| CDK1 | HBA1 | OS | VCAM1 | FOXO1(2) |
| CDKN1A | HBA2 | PBK | VEGFA | HMMR(2) |
| CDKN3 | HBB | PCNA | VIM | HSF1(2) |
| CENPA | HBD | PEG10 | VTN | KIF20A(2) |
| CENPE | HDAC1 | PHF20L1 | VWF | KIF2C(2) |
| CENPF | HLAB | PIK3CD | YWHAZ | KNG1(2) |
| CEP55 | HMGA1 | PIK3CG | ZIC2 | KRAS(2) |
| CHEK1 | HMMR | PIK3R1 | ZNF16 | MCM2(2) |
| CKAP5 | HRAS | PLCB1 | ZNF250 | MCM4(2) |
| CLU | HRG | PLK1 | ZWINT | MELK(2) |
| MMP2(2) | ||||
| MUT(2) | ||||
| PLK1(2) | ||||
| PRC1(2) | ||||
| RACGAP1(2) | ||||
| SPARC(2) | ||||
| VWF(2) | ||||
| YDJC(2) | ||||
| ZNF623(2) | ||||
| ZWINT | ||||
What was most surprising was the wealth of hub genes found in the literature for the same cancer. The previously mentioned 324 hub genes were in stark contrast to Barabasi’s model [88, 89], which featured only very few hub genes in a metabolic network. We also noted the arbitrary way in which a node was defined as a hub. The scale-free degree distribution characterizes the PPINs so it was unnecessary to identify any special degree scale. These studies mainly searched for target-based selected genes but with few comprehensive examinations for hub genes. Even more surprising, the entire set of these hubs contained 61 redundant genes, each of which is repeated several times, for 177 redundancies, reducing the total number of genes to 208. This raises concerns about the functional significance of the network properties found for the HCC genes [90], even though the studies were peer reviewed and published in reputable journals. So far, a total of about 300 known cancer genes (around 1% of the human genome) are involved in all cancers as driver genes, and only 12 were predicted as drivers for HCC [91]. This shows we have found many more key genes than the literature and reinforces the concern that some conclusions about the role and functional significance of these proteins may not be robust. Recently, 560 genes associated with HCC were extracted from 1074 published articles [92] and another 1509 genes from biobanks [93]. Both groups presented a comprehensive analysis to select driver [92] and carbon metabolism genes [93]. The different purposes of these studies did not lead to a search for hub genes; however, even though the genomic datasets were well populated, 208 hub genes is an unexpectedly large number.
To gather more information, we used the 208 hubs from HCC as seed to extract from human interactome a subnet or “module” on the assumption that genes involved in this subnet will share similar gene expression patterns or functional modules in HCC [94]. The analysis on experimentally evidenced interactions (non-redundant, no self-loops ) of human proteome from various databases retrieved using in-house R scripts (https://bitbucket.org/datasetsPPI/ppi_humanproteome/src/master/) [95, 96] and visualized using Cytoscape [97] showed that the human interactome average degree was 43.65 and the hub nodes average degree is 943.10. The functional module [98] should be a set of genes whose function is separable from those of other modules. The members of the module should share genetic or cellular interactions, being members of the same PPI ensemble or of the same metabolic or signaling pathway. In fact, PPI data reflect the association of proteins to achieve a common goal. As a result, members should share more interactions among themselves than with members of other modules [96].
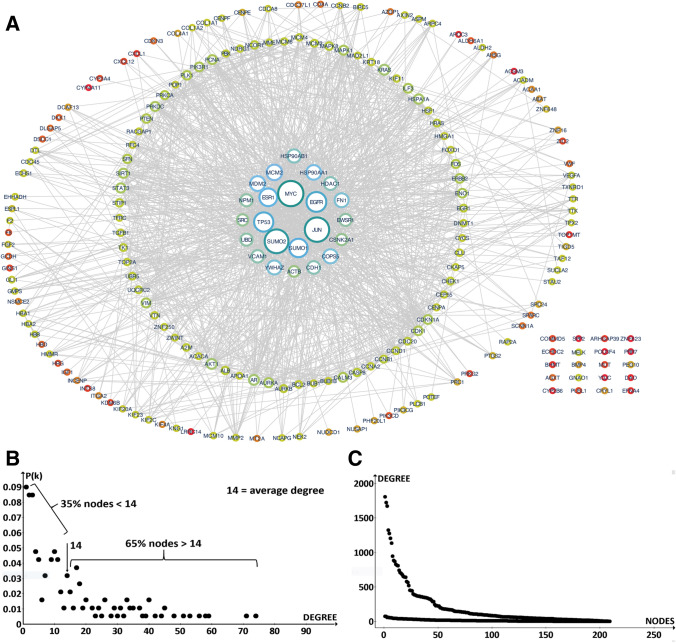
We extracted a subnetwork of 204 nodes out of the 208 HCC hubs, which showed experimentally validated interactions in human proteome (Fig. 3a). In the subnet, 23 major proteins showed hubness with a high degree of connections.
Fig. 3.
a The whole set of extracted proteins; b the 23 proteins with greater connectivity; c the distribution degree of the 209 nodes; d the comparison between the distribution of the 209 nodes with that in the human proteome
The average degree of the 204 nodes is 14, which is very low considering they are all hubs. It is also low if compared with non-hub nodes of the entire human interactome, and even lower given the average value of the proteome hub nodes is 943.1. In these 23 protein sets, only ten have a degree higher than 44. Their degrees range from 74 (JUN #1) to 34 (COPS5 #23), with the average of this subset being 43, approximately 21.1 times lower than their corresponding average degree in the human interactome. If we look at protein degree distribution (Fig. 3b), we see that the number of fractions with low degree (weakly interacting proteins) is very low, whereas the fractions with great interactivity are very high. But if we compare our net distribution with that of the entire human interactome we find that the gene expression profiles of the corresponding HCC proteins have a very flat distribution (Fig. 3c). In other words, many hubs found for HCC do not actually work as hubs. Altogether, the interaction parameters of the HCC module, when compared with the human interactome, suggest that the interactions are few with an apparently weak graph structure. An explanation is that many genes selected by researchers for HCC were not true hub nodes. In fact, this can be the effect of poor computational evaluations or experimental errors. For example, if the degree distribution of a PPIN is biased toward cancer proteins, because there are many well-characterized proteins in the experimental set, the average degree should increase, whereas if there are many poorly characterized proteins in the experimental set, with no interactions reported yet, then the topological pattern and the degree distribution can change, and the degree should decrease. This is because a well-studied protein has more molecular partners and thus a higher degree. However, the collapse of PTM annotations on a single node can also change the net topology.
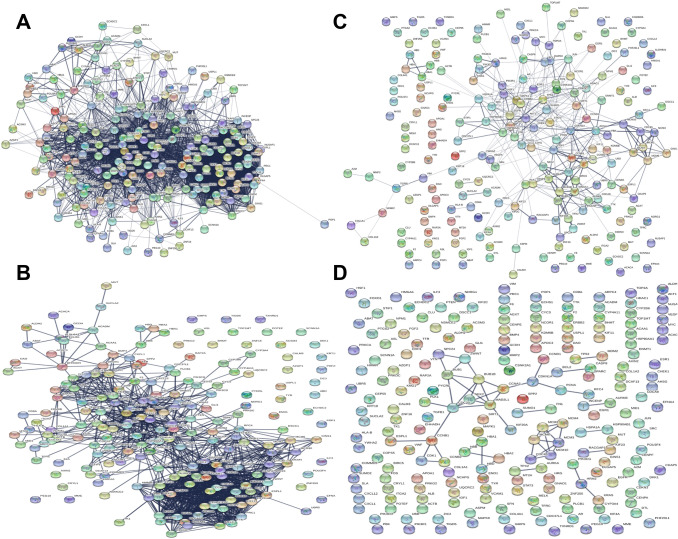
To show how these proteins interact, we analyzed through STRING and the experimentally evidenced interactome obtained from various databases visualized using Cytoscape, for the 208 non-redundant hub genes as a seed input node in Table 2. The network shows the gene acronym converted into those coded for the corresponding human proteins. The purpose of this evaluation is to ascertain how many proteins can form a compact network with significant functional relationships in pairs (or “of the first order,” i.e., without intermediates). In Fig. 4, we show the effect of confidence scores (0.4 for middle value and 0.9 for the highest) on the metabolic network involving the hub nodes. We show that a more stringent data analysis provides more meaningful results but with a significant loss of PPIs. In Fig. 4a, four proteins (2%) are shown as non-interacting, which means they have no functional relationships, but we have 204 nodes and 3502 interactions. Passing from score 0.4–0.9 (Fig. 4b) we lose another 31 nodes (− 15%) leaving 1385 PPIs (− 60.5%), but the decrease is even clearer when we analyze only data experimentally proven with the two previous scores (Fig. 4c and d). We lose 82 nodes (− 40.1%), leaving 359 PPIs (− 89.75%), and the network collapses with the highest score (52 PPIs in three small clusters; − 99.98%). This knockout perturbation of nodes shows that many interactions are loosely or not at all connected to HCC and that the evolutionary pressure does not favor the net robustness [99–101]. We show the corresponding parametric values calculated for the networks in Table 3.
Fig. 4.
The different PPI networks obtained from the hub genes of hepatocellular carcinoma. STRING has translated the names of the genes into those of the respective proteins. a The network obtained for the 208 hub genes found in the literature with a confidence score of 0.4. The proteins that have not shown relations with the rest of the network are visible in the upper-right part of the panel. b The same network but with a confidence score of 0.9 to obtain relationships with greater significance. The number of proteins that do not exchange interactions is increased (see also Table 3). c and d The PPI networks obtained using only the experimentally validated interactions as data sources. The networks have a confidence score of c 0.4 and d 0.9. Notice how the use of experimentally validated interactions reduces the significant relationships between proteins with the collapse of the network (d). High resolution figures of the networks are shown in supplementary material figures 1S to 4S
Table 3.
Network parameters in STRING
| Network parameters | Source: 2 channels Confidence Score = 0.4 |
Source: 2 channels Confidence Score = 0.9 | Source: 1 channel (Exp) Confidence score = 0.4 | Source: 1 channel (Exp) Confidence score = 0.9 |
|---|---|---|---|---|
| Interactions | 3502 | 1385 | 359 | 52 |
| Number of connected nodes | 204 | 173 | 91 | Collapsed network with three clusters of 28 nodes |
| Average node degree | 34.3 | 13.6 | 3.52 | 0.51 |
| Average local clustering coefficient | 0.652 | 0.542 | 0.283 | 0.163 |
An average node degree is a numerical value of how many interactions (at the score threshold) a protein has on the average in the network. The clustering coefficient is a measure of how connected the nodes in the network are. 2 channels denote data retrieved from experimentally proven data channel as well as databases and text mining channel
Exp stands for experimental channel only
We have to consider that the function pertains to the biomolecules between which a chemical reaction occurs in the cellular system, that is, a physical interaction aims at providing insights into molecular mechanisms of the function played by the molecules. To achieve complete and accurate information on the biomolecules involved in the studies, carefully planned methods and experiments should be adopted to confirm the molecular interaction (e.g., affinity chromatography, mass spectrometry, immune techniques). Functional molecules interacting through a direct physical interaction can be shown. In most cases, when we say that two molecules have a functional relationship, we do not mean that they physically interact. In fact, we should measure the function through products of the functional activity using wet biochemical tests. Therefore, without experiments, we do not know whether the function, analyzed in silico, occurs via direct interaction or intermediates, which leads to assumptions of an interaction being a functional interaction. However, in the last few years, the speed of new protein discovery, or prediction, has pushed towards high-throughput interaction-detection methods. Our examples show two possible conclusions: functional and physical. STRING cannot determine them with internal statistics or filtering. When the database channel drives functional data, this inherent uncertainty is involved. We can have actual knowledge of a functional relationship only, and only if, in the same experiment we measure the physical binding (binding curve and Kb, assessed by biophysical techniques) and functional quantitative parameters. Therefore, data and annotations, before being entered on a computational platform, should be classified according to their biological meaning through ontological and semantic processes. From a computational point of view, this lack introduces serious problems that change functional relationships and relative metabolic models.
Furthermore, the topology of molecular networks is organized according to common functional properties, where hub nodes should be part of the metabolic module that characterizes a disease [89, 90, 92, 94]. Here, we observe that genes, extracted from expression profiling data of the same disease from individual patients but different medical databases, show inconsistent functional relationships. These observations suggest that database contents are heterogeneous, perhaps because genes were not associated with the correct disease staging or cancerous cell phenotype. Thus, they have been stored in the databases with no adequate control of accuracy and veracity [102, 103]. Results show a great genomic heterogeneity of the original samples, where the spatiotemporal action of the genes is not known. In fact, we do not know the phenotypes of cancerous cells during disease progression to attribute the correct stage to genomic data. This also explains why the same protein has been associated with so many different metabolic partners, because it shows the functional relationships of different metabolic phenotypes [55]. In the same disease, a hub protein should always be associated with the same molecular partners with which it has metabolic continuity. Cancer reflects its genetic perturbations in the molecular network where interactions rest on the physical protein–protein associations of the effectors of the pathological phenotype. Indeed, we must consider that many cancerous phenotypes could also reflect interactions between molecular components not associated with the disease and, in addition, the high-throughput methods cover only a small part of the potential protein interactions in pairs of the more than 200 human cell types [104]. This means that we are trying to discover pathological mechanisms based on interactive maps that remain very incomplete. The proteins associated with HCC should not be randomly scattered in the HCC interactome but should interact with each other, forming one or more connected modules.
In conclusion, the logical-semantic organization of PTM data of the analysis platforms, together with the lack of knowledge of the spatiotemporal action of the HCC genes, and the incompleteness of the human interactome data, have generated flawed metabolic relationships between HCC proteins. This metabolic model might translate to biomedical Big data by introducing metabolic considerations that can change the robustness of precision medicine related to patients with HCC. Considering that many years have already passed, many of these relationships should already be an integral part of biomedical Big data.
Concluding Remarks
We have illustrated how some methodological limits in network medicine can generate flawed results. These results are stored in biobanks from which biomedical Big-data systems collect knowledge. At present, network biology remains focused on static models that do not cover the functional conditions in time and space, which is a limit to the understanding of human diseases and their treatment. The modern race for biomedical Big data does not take these limits into account because the need to obtain biomedical computer systems for precision medicine is strongly connected to marketing and competitiveness and so is constrained in a time-bound manner. Therefore, it is necessary that both the biomedical and the technological world understand these limits and the substantial vulnerabilities that these inaccuracies, which remain under-considered, can pollute the data core with unpredictable future errors.
Although the illustrated cases may appear to be specific, their frequency may be high because the evaluation of a single article in a specific field as a typical case apparently does not show the inconsistencies that the analysis of many similar articles does. The robustness of HCC networks regarding the perturbations of the system by using different levels of statistics based on the connectivity between disease–gene associations shows clear failure. This reflects an inadequate sampling or staging, but we cannot exclude that the many hub genes that show unusual weak interactions may instead reflect the presence of various methodological errors. Consequently, flawed scientific data may enter public international repositories through scientific articles and the databases of clinical institutions that collect genomic data and EHRs from their patients without adequate checks and controls (Fig. 5). The data extracted from public databases may produce scientific models that may be skewed by these intrinsic errors. In addition, articles that do not undergo peer review but are published by the thousands of pseudoscientific journals disseminate misleading models [105, 106]. These data also flow into the biomedical Big-data system. The result is that biomedical knowledge is afflicted by multifaceted pollution, which may slow its rigorous progression. Thus, data variety, veracity, and heterogeneity represent both an enormous challenge and a cause for concern for Big data [107, 108]. However, we can improve error detection by embracing a more comprehensive approach to monitoring the logic and semantics of data from so many different sources. Through logic and semantics, we interpret the different meaning of similar terms or actions by placing them in the correct context with the correct meaning.
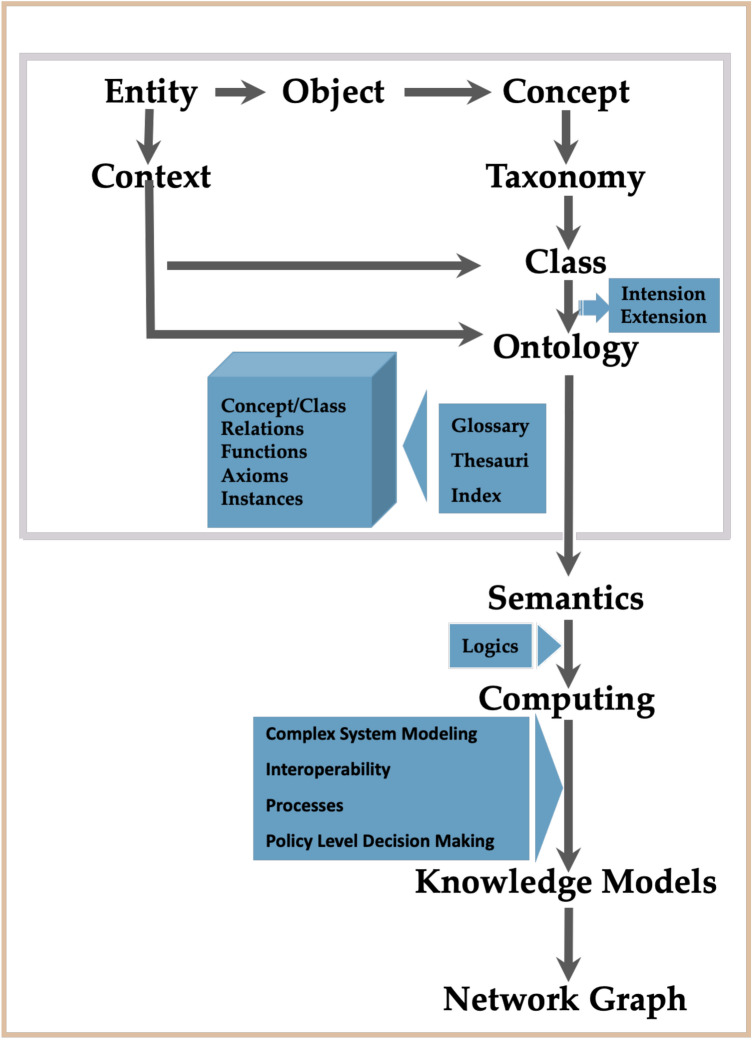
Fig. 5.
Schematic representation of Ontology and Knowledge Modeling. The part enclosed in the red box shows the processes where human error may be more likely. The meaning of the terms is explained in the Electronic Supplementary Material (Box 1)
The widespread use of biomedical data is teaching us how to quantitate many aspects of human health. This, together with machine learning techniques, means we can build models of human health in ways we could never do before. We are using artificial intelligence and biomedical Big data to guide medicine with excellent decision systems or predictions, but they require data with high-quality controls. In Big data, veracity checks are particularly important in terms of the biases, noise, and abnormalities in data being analyzed [102, 103]. Flawed data transforms Big data into a giant with clay feet. A critical point is to integrate information through different knowledge domains to establish an interface or interaction between two biological events or objects. Thus, as the interactome (the synonym connectome is most used in brain research) is a map of all functional connections in an organism, and the interactomics is the mapping of these connections, biomedical Big-data systems try to discover new patterns through the complex interactome connected with patient care. These technologies are finding useful applications for humans in the medicine knowledge domain. Myriad biomedical information (biological, clinical, and patient care related) extrapolated from many and different archives through special algorithms converge in an immense system of archives that should provide us a diagnosis, a cure, or a prognosis for a single patient. To implement a suitable “connectome model” for patient care, it is necessary to understand the particular purposes and impact of a definite data category in establishing the ultimate decision. Although this remains unclear to many people, we are moving towards a level of more complex connectivity, where today's metabolic or genomic networks will be specialized modules of a larger and multifaced network of interactions in the service of medicine. As such, data should be as error free as possible. After the event, it is impossible to eliminate them.
A second critical point is that data science in medicine requires people with strong interdisciplinary skills for advanced analytics of fast data with the best understanding of the basic processes, both technological and scientific.
Furthermore, our analysis shows that wet and in silico researchers cannot work alone; such isolation favors imperfect analyses and conceptual errors and means that the methodological aspects of other disciplines are not considered, leading to partial and sometimes even erroneous results and analyses.
The enthusiastic rush to precision medicine involves all these problems without the understanding that the clinical models it will produce could be already flawed. A search of PubMed for “data pollution” or “information pollution in biomedicine”, found no articles (the only pollution found was atmospheric). Therefore, action is necessary before the increased volume of biomedical information makes it difficult, if not impossible, to separate out the waste. Our conclusion can only be that we must remove the weeds before harvesting.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
The authors are very thankful to Les Foster for proofreading the manuscript. We would also like to thank the referees and the editor for constructive comments.
Declarations
Funding
No sources of funding were used to conduct this study or prepare this manuscript. The authors would like to thank the University of Oslo for covering the open access publishing fees through an open access agreement with Springer Nature.
Conflicts of Interest
Ankush Sharma and Giovanni Colonna have no conflicts of interest that are directly relevant to the content of this article.
Ethics approval
Not applicable.
Consent
Not applicable.
Data availability
Not applicable.
References
- 1.Jastania R, Nageeti T, Al-Juhani H, Basahel A, Aljuraid R, Alanazi A, et al. Utilizing big data in healthcare, how to maximize ts value. Stud Health Technol Inform. IOS Press; 2019, pp. 356–9. http://www.ncbi.nlm.nih.gov/pubmed/31349341. Accessed 14 Mar 2020 [DOI] [PubMed]
- 2.Lee CH, Yoon HJ. Medical big data: promise and challenges. Kidney Res Clin Pract. 2017;36:3–11. doi: 10.23876/j.krcp.2017.36.1.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Uversky VN, Gillespie JR, Fink AL. Why are “natively unfolded” proteins unstructured under physiologic conditions? Proteins. 2000;41:415–427. doi: 10.1002/1097-0134(20001115)41:3<415::aid-prot130>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 4.Necci M, Piovesan D, Tosatto SCE. Large-scale analysis of intrinsic disorder flavors and associated functions in the protein sequence universe. Protein Sci. 2016;25:2164–2174. doi: 10.1002/pro.3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bah A, Forman-Kay JD. Modulation of intrinsically disordered protein function by post-translational modifications. J Biol Chem. 2016;291:6696–6705. doi: 10.1074/jbc.R115.695056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yin Z, Dong C, Jiang K, Xu Z, Li R, Guo K, et al. Heterogeneity of cancer-Associated fibroblasts and roles in the progression, prognosis, and therapy of hepatocellular carcinoma. J Hematol Oncol. BioMed Central Ltd.; 2019, p. 101. https://jhoonline.biomedcentral.com/articles/10.1186/s13045-019-0782-x. Accessed 4 Aug 2020 [DOI] [PMC free article] [PubMed]
- 7.Simpson KL, Stoney R, Frese KK, Simms N, Rowe W, Pearce SP, et al. A biobank of small cell lung cancer CDX models elucidates inter- and intratumoral phenotypic heterogeneity. Nat Cancer. 2020;1:437–451. doi: 10.1038/s43018-020-0046-2. [DOI] [PubMed] [Google Scholar]
- 8.Zhu X, Gerstein M, Snyder M. Getting connected: analysis and principles of biological networks. Genes Dev. 2007;21:1010–1024. doi: 10.1101/gad.1528707. [DOI] [PubMed] [Google Scholar]
- 9.Bommert A, Sun X, Bischl B, Rahnenführer J, Lang M. Benchmark for filter methods for feature selection in high-dimensional classification data. Comput Stat Data Anal. 2020;143:106839. [Google Scholar]
- 10.Venkataramana L, Jacob SG, Shanmuganathan S, Dattuluri VVP. Benchmarking gene selection techniques for prediction of distinct carcinoma from gene expression data: a computational study. Nat Inspired Comput Data Sci. 2019 doi: 10.1007/978-3-030-33820-6_10. [DOI] [Google Scholar]
- 11.Palsson B, Zengler K. The challenges of integrating multi-omic data sets. Nat Chem Biol. 2010;6:787–789. doi: 10.1038/nchembio.462. [DOI] [PubMed] [Google Scholar]
- 12.Navarro-Muñoz JC, Selem-Mojica N, Mullowney MW, Kautsar SA, Tryon JH, Parkinson EI, et al. A computational framework to explore large-scale biosynthetic diversity. Nat Chem Biol. 2020;16:60–68. doi: 10.1038/s41589-019-0400-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pearl J. Causality: models, reasoning, and inference. Econom Theory, vol. 19. Cambridge University Press (CUP); 2003. p. 675–85. https://assets.cambridge.org/97805218/95606/frontmatter/9780521895606_frontmatter.pdf.
- 14.Salas D, Stacey RG, Akinlaja M, Foster LJ. Next-generation interactomics: considerations for the use of co-elution to measure protein interaction networks. Mol Cell Proteomics. 2020;19:1–10. doi: 10.1074/mcp.R119.001803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Poverennaya EV, Kiseleva OI, Ivanov AS, Ponomarenko EA. Methods of computational interactomics for investigating interactions of human proteoforms. Biochemistry. 2020;85:68–79. doi: 10.1134/S000629792001006X. [DOI] [PubMed] [Google Scholar]
- 16.Kohavi R, Rothleder NJ, Simoudis E. Emerging trends in business analytics. Commun ACM. 2002;45:45–48. [Google Scholar]
- 17.Wang L. Heterogeneous data and big data analytics. Autom Control Inf Sci. 2017;3:8–15. [Google Scholar]
- 18.Billings SA. Nonlinear system identification: NARMAX methods in the time, frequency, and spatio-temporal domains. Hoboken: Wiley; 2013. [Google Scholar]
- 19.Lazer D, Kennedy R, King G, Vespignani A. The parable of google flu: traps in big data analysis. Science. 2014;343:1203–1205. doi: 10.1126/science.1248506. [DOI] [PubMed] [Google Scholar]
- 20.Obrst L. Ontologies for semantically interoperable systems. In: Proceedings of the twelfth international conference on Inf Knowl Manag—CIKM’03. New York, New York, USA: ACM Press; 2003.
- 21.Househ MS, Aldosari B, Alanazi A, Kushniruk AW, Borycki EM. Big data, big problems: a healthcare perspective. Stud Health Technol Inform. 2017;238:36–39. [PubMed] [Google Scholar]
- 22.Boland MR, Hripcsak G, Shen Y, Chung WK, Weng C. Defining a comprehensive verotype using electronic health records for personalized medicine. J Am Med Inform Assoc. 2013;20:e232–e238. doi: 10.1136/amiajnl-2013-001932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lewis C, McQuaid S, Hamilton PW, Salto-Tellez M, McArt D, James JA. Building a ‘Repository of Science’: the importance of integrating biobanks within molecular pathology programmes. Eur J Cancer. 2016;67:191–199. doi: 10.1016/j.ejca.2016.08.009. [DOI] [PubMed] [Google Scholar]
- 24.Wu PY, Cheng CW, Kaddi CD, Venugopalan J, Hoffman R, Wang MD. Omic and electronic health record big data analytics for precision medicine. IEEE Trans Biomed Eng. 2017;64:263–273. doi: 10.1109/TBME.2016.2573285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Elsebakhi E, Lee F, Schendel E, Haque A, Kathireason N, Pathare T, et al. Large-scale machine learning based on functional networks for biomedical big data with high performance computing platforms. J Comput Sci. 2015;11:69–81. [Google Scholar]
- 26.Chen H, Chen W, Liu C, Zhang L, Su J, Zhou X. Relational network for knowledge discovery through heterogeneous biomedical and clinical features. Sci Rep. 2016;6:1–13. doi: 10.1038/srep29915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rashid M, Singh H, Goyal V, Ahmad N, Mogla N. Efficient big data-based storage and processing model in internet of things for improving accuracy fault detection in industrial processes. 2020. https://www.igi-global.com/gateway/chapter/239163. Cited 20 Apr 2020.
- 28.Gale NK, Heath G, Cameron E, Rashid S, Redwood S. Using the framework method for the analysis of qualitative data in multi-disciplinary health research. BMC Med Res Methodol. 2013;13:117. doi: 10.1186/1471-2288-13-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Borisov N, Sorokin M, Garazha A, Buzdin A. Quantitation of molecular pathway activation using RNA sequencing data. Methods Mol Biol. Humana Press Inc.; 2020, pp. 189–206. http://www.ncbi.nlm.nih.gov/pubmed/31667772. Accessed 20 Apr 2020 [DOI] [PubMed]
- 30.Fondi M, Liò P. Genome-scale metabolic network reconstruction. Methods Mol Biol. 2015;5:233–256. doi: 10.1007/978-1-4939-1720-4_15. [DOI] [PubMed] [Google Scholar]
- 31.Goh K-I, Cusick ME, Valle D, Childs B, Vidal M, Barabási A-L. The human disease network. Proc Natl Acad Sci USA. 2007;104:8685–8690. doi: 10.1073/pnas.0701361104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rong B, Rui X, Tao L, Wang G. Theoretical modeling and numerical solution methods for flexible multibody system dynamics. Nonlinear Dyn. 2019;98:1519–1553. doi: 10.1007/s11071-019-05191-3. [DOI] [Google Scholar]
- 33.Jeong H, Mason SP, Barabasi A-L, Oltvai ZN. Lethality and centrality in protein networks. Nature. 2001;411:41–42. doi: 10.1038/35075138. [DOI] [PubMed] [Google Scholar]
- 34.van Mierlo T, Hyatt D, Ching AT. Mapping power law distributions in digital health social networks: methods, interpretations, and practical implications. J Med Internet Res. 2015;17:e160–e160. doi: 10.2196/jmir.4297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Parr SK, Shotwell MS, Jeffery AD, Lasko TA, Matheny ME. Automated mapping of laboratory tests to LOINC codes using noisy labels in a national electronic health record system database. J Am Med Inform Assoc. 2018;25:1292–1300. doi: 10.1093/jamia/ocy110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Giuliani A. Networks as a privileged way to develop mesoscopic level approaches in systems biology. Systems. 2014;2:237–242. doi: 10.3390/systems2020237. [DOI] [Google Scholar]
- 37.Bernhardt BC, Bonilha L, Gross DW. Network analysis for a network disorder: the emerging role of graph theory in the study of epilepsy. Epilepsy Behav. 2015;50:162–170. doi: 10.1016/j.yebeh.2015.06.005. [DOI] [PubMed] [Google Scholar]
- 38.Hase T, Tanaka H, Suzuki Y, Nakagawa S, Kitano H. Structure of protein interaction networks and their implications on drug design. PLoS Comput Biol. 2009;5:e1000550–e1000550. doi: 10.1371/journal.pcbi.1000550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Maslov S, Sneppen K. Specificity and stability in topology of protein networks. Science. 2002;296:910–913. doi: 10.1126/science.1065103. [DOI] [PubMed] [Google Scholar]
- 40.Civillico EF, Contreras D. Spatiotemporal properties of sensory responses in vivo are strongly dependent on network context. Front Syst Neurosci. 2012;6:25. doi: 10.3389/fnsys.2012.00025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yamada T, Bork P. Evolution of biomolecular networks—lessons from metabolic and protein interactions. Nat Rev Mol Cell Biol. 2009;10:791–803. doi: 10.1038/nrm2787. [DOI] [PubMed] [Google Scholar]
- 42.Zhu L, Brangwynne CP. Nuclear bodies: the emerging biophysics of nucleoplasmic phases. Curr Opin Cell Biol. 2015;34:23–30. doi: 10.1016/j.ceb.2015.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Meng F, Na I, Kurgan L, Uversky VN. Compartmentalization and functionality of nuclear disorder: intrinsic disorder and protein–protein interactions in intra-nuclear compartments. Int J Mol Sci. 2015;17:24. doi: 10.3390/ijms17010024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bergstrom CT, Rosvall M. The transmission sense of information. Biol Philos. 2009;26:159–176. doi: 10.1007/s10539-009-9180-z. [DOI] [Google Scholar]
- 45.Hanus P, Goebel B, Dingel J, Weindl J, Zech J, Dawy Z, et al. Information and communication theory in molecular biology. Electr Eng. 2007;90:161–173. doi: 10.1007/s00202-007-0062-6. [DOI] [Google Scholar]
- 46.Shea N. What’s transmitted? Inherited information. Biol Philos. 2010;26:183–189. doi: 10.1007/s10539-010-9232-4. [DOI] [Google Scholar]
- 47.Polo A, Colonna G, Guariniello S, Ciliberto G, Costantini S. Deducing the functional characteristics of the human selenoprotein SELK from the structural properties of its intrinsically disordered C-terminal domain. Mol Biosyst. 2016;12:758–772. doi: 10.1039/c5mb00679a. [DOI] [PubMed] [Google Scholar]
- 48.Potenza N, Castiello F, Panella M, Colonna G, Ciliberto G, Russo A, et al. Human MiR-544a modulates SELK expression in hepatocarcinoma cell lines. PLoS ONE. 2016;11:e0156908. doi: 10.1371/journal.pone.0156908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Polo A, Guariniello S, Colonna G, Ciliberto G, Costantini S. A study on the structural features of SELK, an over-expressed protein in hepatocellular carcinoma, by molecular dynamics simulations in a lipid–water system. Mol Biosyst. 2016;12:3209–3222. doi: 10.1039/c6mb00469e. [DOI] [PubMed] [Google Scholar]
- 50.Szklarczyk D, Morris JH, Cook H, Kuhn M, Wyder S, Simonovic M, et al. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45:D362–D368. doi: 10.1093/nar/gkw937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Snel B, Lehmann G, Bork P, Huynen MA. STRING: a web-server to retrieve and display the repeatedly occurring neighbourhood of a gene—PubMed. Nucleic Acids Res. 2000;28:3442–3442. doi: 10.1093/nar/28.18.3442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schnoes AM, Brown SD, Dodevski I, Babbitt PC. Annotation error in public databases: misannotation of molecular function in enzyme superfamilies. PLoS Comput Biol. 2009;5:e1000605. doi: 10.1371/journal.pcbi.1000605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stark C, Breitkreutz B-J, Reguly T, Boucher L, Breitkreutz A, Tyers M. BioGRID: a general repository for interaction datasets. Nucleic Acids Res. 2006;34:D535–D539. doi: 10.1093/nar/gkj109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Breitkreutz B-J, Stark C, Tyers M. The GRID: the general repository for interaction datasets. Genome Biol. 2003;4:R23–R23. doi: 10.1186/gb-2003-4-3-r23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Murray-Zmijewski F, Lane DP, Bourdon J-C. p53/p63/p73 isoforms: an orchestra of isoforms to harmonise cell differentiation and response to stress. Cell Death Differ. 2006;13:962–972. doi: 10.1038/sj.cdd.4401914. [DOI] [PubMed] [Google Scholar]
- 56.Marcel V, Dichtel-Danjoy M-L, Sagne C, Hafsi H, Ma D, Ortiz-Cuaran S, et al. Biological functions of p53 isoforms through evolution: lessons from animal and cellular models. Cell Death Differ. 2011;18:1815–1824. doi: 10.1038/cdd.2011.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cheng Y, Ping J, Chen J. Identification of potential gene network associated with HCV-related hepatocellular carcinoma using microarray analysis. Pathol Oncol Res. 2017;24:507–514. doi: 10.1007/s12253-017-0273-8. [DOI] [PubMed] [Google Scholar]
- 58.Costantini S, Di Bernardo G, Cammarota M, Castello G, Colonna G. Gene expression signature of human HepG2 cell line. Gene. 2013;518:335–345. doi: 10.1016/j.gene.2012.12.106. [DOI] [PubMed] [Google Scholar]
- 59.Costantini S, Capone F, Maio P, Guerriero E, Colonna G, Izzo F, et al. Cancer biomarker profiling in patients with chronic hepatitis C virus, liver cirrhosis and hepatocellular carcinoma. Oncol Rep. 2013;29:2163–2168. doi: 10.3892/or.2013.2378. [DOI] [PubMed] [Google Scholar]
- 60.Di Stasio M, Volpe MG, Colonna G, Nazzaro M, Polimeno M, Scala S, et al. A possible predictive marker of progression for hepatocellular carcinoma. Oncol Lett. 2011;2:1247–1251. doi: 10.3892/ol.2011.378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Guariniello S, Di Bernardo G, Colonna G, Cammarota M, Castello G, Costantini S. Evaluation of the selenotranscriptome expression in two hepatocellular carcinoma cell lines. Anal Cell Pathol (Amst). 2015;2015:419561. doi: 10.1155/2015/419561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Guerriero E, Accardo M, Capone F, Colonna G, Castello G, Costantini S. Assessment of the Selenoprotein M (SELM) over-expression on human hepatocellular carcinoma tissues by immunohistochemistry. Eur J Histochem. 2014;58:2433. doi: 10.4081/ejh.2014.2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hu WQ, Wang W, Fang DL, Yin XF. Identification of biological targets of therapeutic intervention for hepatocellular carcinoma by integrated bioinformatical analysis. Med Sci Monit. 2018;24:3450–3461. doi: 10.12659/MSM.909290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Huang D, Yuan W, Li H, Li S, Chen Z, Yang H. Identification of key pathways and biomarkers in sorafenib-resistant hepatocellular carcinoma using bioinformatics analysis. Exp Ther Med. 2018;16:1850–1858. doi: 10.3892/etm.2018.6427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lin P, Wen DY, Dang YW, He Y, Yang H, Chen G. Comprehensive and integrative analysis reveals the diagnostic, clinicopathological and prognostic significance of polo-like kinase 1 in hepatocellular carcinoma. Cell Physiol Biochem. 2018;47:925–947. doi: 10.1159/000490135. [DOI] [PubMed] [Google Scholar]
- 66.Chen P, Wang F, Feng J, Zhou R, Chang Y, Liu J, et al. Co-expression network analysis identified six hub genes in association with metastasis risk and prognosis in hepatocellular carcinoma. Oncotarget. 2017;8:48948–48958. doi: 10.18632/oncotarget.16896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yin L, Cai Z, Zhu B, Xu C. Identification of key pathways and genes in the dynamic progression of HCC based on WGCNA. Genes (Basel). MDPI AG; 2018;9. http://www.ncbi.nlm.nih.gov/pubmed/29443924. Accessed 16 Mar 2020 [DOI] [PMC free article] [PubMed]
- 68.Singh S, Colonna G, Di Bernardo G, Bergantino F, Cammarota M, Castello G, et al. The gene expression profiling of hepatocellular carcinoma by a network analysis approach shows a dominance of intrinsically disordered proteins (IDPs) between hub nodes. Mol Biosyst. 2015;11:2933–2945. doi: 10.1039/c5mb00434a. [DOI] [PubMed] [Google Scholar]
- 69.Ardakani MJE, Safaei A, Oskouie AA, Haghparast H, Haghazali M, Shalmani HM, et al. Evaluation of liver cirrhosis and hepatocellular carcinoma using protein–protein interaction networks. Gastroenterol Hepatol. 2016;9:S14–22. [PMC free article] [PubMed] [Google Scholar]
- 70.Zheng Y, Long J, Wu L, Zhang H, Li L, Zheng Y, et al. Identification of hub genes involved in the development of hepatocellular carcinoma by transcriptome sequencing. Oncotarget. 2017;8:60358–60367. doi: 10.18632/oncotarget.19483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhang R, Zhang LJ, Yang ML, Huang LS, Chen G, Feng ZB. Potential role of microRNA-223-3p in the tumorigenesis of hepatocellular carcinoma: a comprehensive study based on data mining and bioinformatics. Mol Med Rep. 2018;17:2211–2228. doi: 10.3892/mmr.2017.8167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhang C, Peng L, Zhang Y, Liu Z, Li W, Chen S, et al. The identification of key genes and pathways in hepatocellular carcinoma by bioinformatics analysis of high-throughput data. Med Oncol. 2017;34:101. doi: 10.1007/s12032-017-0963-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Costantini S, Sharma A, Colonna G. The value of the cytokinome profile. In: Nagal A (eds) Inflammatory diseases—A modern perspective. InTech; 2011. Available from: http://www.intechopen.com/books/inflammatory-diseases-a-modern-perspective/the-value-of-the-cytokinome-profile.
- 74.Zhou L, Du Y, Kong L, Zhang X, Chen Q. Identification of molecular target genes and key pathways in hepatocellular carcinoma by bioinformatics analysis. Onco Targets Ther. 2018;11:1861–1869. doi: 10.2147/OTT.S156737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Polo A, Singh S, Crispo A, Russo M, Giudice A, Montella M, et al. Evaluating the associations between human circadian rhythms and dysregulated genes in liver cancer cells. Oncol Lett. 2017;14:7353–7359. doi: 10.3892/ol.2017.7109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Xu W, Rao Q, An Y, Li M, Zhang Z. Identification of biomarkers for Barcelona Clinic Liver Cancer staging and overall survival of patients with hepatocellular carcinoma. PLoS ONE. 2018;13:e0202763. doi: 10.1371/journal.pone.0202763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Xing T, Yan T, Zhou Q. Identification of key candidate genes and pathways in hepatocellular carcinoma by integrated bioinformatical analysis. Exp Ther Med. 2018;15:4932–4942. doi: 10.3892/etm.2018.6075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Xuo H, Luo L, Yao YT, Wei LL, Deng SP, Huang XL. Integrated analysis of the RNA-Seq data of liver hepatocellular carcinoma. Neoplasma. 2018;65:97–103. doi: 10.4149/neo_2018_170212n98. [DOI] [PubMed] [Google Scholar]
- 79.Sang L, Wang X-M, Xu D-Y, Zhao W-J. Bioinformatics analysis of aberrantly methylated-differentially expressed genes and pathways in hepatocellular carcinoma. World J Gastroenterol. 2018;24:2605–2616. doi: 10.3748/wjg.v24.i24.2605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Liang L, Gao L, Zou XP, Huang ML, Chen G, Li JJ, et al. Diagnostic significance and potential function of miR-338-5p in hepatocellular carcinoma: a bioinformatics study with microarray and RNA sequencing data. Mol Med Rep. 2018;17:2297–2312. doi: 10.3892/mmr.2017.8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wen D-Y, Lin P, Pang Y-Y, Chen G, He Y, Dang Y-W, et al. Expression of the long intergenic non-protein coding RNA 665 (LINC00665) gene and the cell cycle in hepatocellular carcinoma using the cancer genome atlas, the gene expression omnibus, and quantitative real-time polymerase chain reaction. Med Sci Monit. 2018;24:2786–2808. doi: 10.12659/MSM.907389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lou W, Chen J, Ding B, Chen D, Zheng H, Jiang D, et al. Identification of invasion-metastasis-associated microRNAs in hepatocellular carcinoma based on bioinformatic analysis and experimental validation. J Transl Med. 2018;16:266. doi: 10.1186/s12967-018-1639-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Li L, Lei Q, Zhang S, Kong L, Qin B. Screening and identification of key biomarkers in hepatocellular carcinoma: evidence from bioinformatic analysis. Oncol Rep. 2017;38:2607–2618. doi: 10.3892/or.2017.5946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Martincorena I, Fowler JC, Wabik A, Lawson ARJ, Abascal F, Hall MWJ, et al. Somatic mutant clones colonize the human esophagus with age. Science. 2018;362:911–917. doi: 10.1126/science.aau3879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yang M-R, Zhang Y, Wu X-X, Chen W. Critical genes of hepatocellular carcinoma revealed by network and module analysis of RNA-seq data. Eur Rev Med Pharmacol Sci. 2016;20:4248–4256. [PubMed] [Google Scholar]
- 86.Yang Y, Lu Q, Shao X, Mo B, Nie X, Liu W, et al. Development of a three-gene prognostic signature for hepatitis B virus associated hepatocellular carcinoma based on integrated transcriptomic analysis. J Cancer. 2018;9:1989–2002. doi: 10.7150/jca.23762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yan H, Li Z, Shen Q, Wang Q, Tian J, Jiang Q, et al. Aberrant expression of cell cycle and material metabolism related genes contributes to hepatocellular carcinoma occurrence. Pathol Res Pract. 2017;213:316–321. doi: 10.1016/j.prp.2017.01.019. [DOI] [PubMed] [Google Scholar]
- 88.Barabási A-L, Albert R. Emergence of scaling in random networks. Science. 1999;286:509–512. doi: 10.1126/science.286.5439.509. [DOI] [PubMed] [Google Scholar]
- 89.Barabási A-L, Oltvai ZN. Network biology: understanding the cell’s functional organization. Nat Rev Genet. 2004;5:101–113. doi: 10.1038/nrg1272. [DOI] [PubMed] [Google Scholar]
- 90.Vallabhajosyula RR, Chakravarti D, Lutfeali S, Ray A, Raval A. Identifying hubs in protein interaction networks. PLoS ONE. 2009;4:e5344. doi: 10.1371/journal.pone.0005344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kemp R, Godwin AK, Prados M, Zwarthoff EC, Disaia P, Evason K, et al. Comprehensive characterization of cancer driver genes and mutations. Cell. 2018;174:1034–1035. doi: 10.1016/j.cell.2018.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Chen W, Jiang J, Wang PP, Gong L, Chen J, Du W, et al. Identifying hepatocellular carcinoma driver genes by integrative pathway crosstalk and protein interaction network. DNA Cell Biol. 2019;38:1112–1124. doi: 10.1089/dna.2019.4869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhang J, Baddoo M, Han C, Strong MJ, Cvitanovic J, Moroz K, et al. Gene network analysis reveals a novel 22-gene signature of carbon metabolism in hepatocellular carcinoma. Oncotarget. 2016;7:49232–49245. doi: 10.18632/oncotarget.10249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ghiassian SD, Menche J, Barabási A-L. A DIseAse MOdule Detection (DIAMOnD) algorithm derived from a systematic analysis of connectivity patterns of disease proteins in the human interactome. PLOS Comput Biol. 2015;11:e1004120. doi: 10.1371/journal.pcbi.1004120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Sharma A, Costantini S, Colonna G. The protein–protein interaction network of the human Sirtuin family. Biochim Biophys Acta. 2013;1834:1998–2009. doi: 10.1016/j.bbapap.2013.06.012. [DOI] [PubMed] [Google Scholar]
- 96.Sharma A, Cinti C, Capobianco E. Multitype network-guided target controllability in phenotypically characterized osteosarcoma: role of tumor microenvironment. Front Immunol. 2017;8:918. doi: 10.3389/fimmu.2017.00918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Spirin V, Mirny LA. Protein complexes and functional modules in molecular networks. Proc Natl Acad Sci USA. 2003;100:12123–12128. doi: 10.1073/pnas.2032324100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Güell O, Sagués F, Serrano MÁ. Assessing the significance and predicting the effects of knockout cascades in metabolic networks. Birkhäuser: Cham; 2014. pp. 39–44. [Google Scholar]
- 100.Albert R, Jeong H, Barabasi A. Error and attack tolerance of complex networks. Nature. 2000;406:378–382. doi: 10.1038/35019019. [DOI] [PubMed] [Google Scholar]
- 101.Szalay MS, Kovács IA, Korcsmáros T, Böde C, Csermely P. Stress-induced rearrangements of cellular networks: consequences for protection and drug design. FEBS Lett. 2007;581:3675–3680. doi: 10.1016/j.febslet.2007.03.083. [DOI] [PubMed] [Google Scholar]
- 102.Demchenko Y, Grosso P, De Laat C, Membrey P. Addressing big data issues in scientific data infrastructure. In: Proceedings of 2013 international conference on Collab Technol Syst CTS 2013; 2013, pp. 48–55.
- 103.Buhl HU, Röglinger M, Moser F, Heidemann J. Big data: a fashionable topic with(out) sustainable relevance for research and practice? Bus Inf Syst Eng. 2013;5:65–69. [Google Scholar]
- 104.Regev A, Teichmann SA, Lander ES, Amit I, Benoist C, Birney E, et al. The human cell atlas. Elife. 2017;6:e27041. doi: 10.7554/eLife.27041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Clark J, Smith R. Firm action needed on predatory journals. BMJ. 2015;350:h210. doi: 10.1136/bmj.h210. [DOI] [PubMed] [Google Scholar]
- 106.Sorokowski P, Kulczycki E, Sorokowska A, Pisanski K. Predatory journals recruit fake editor. Nature. 2017;543:481–483. doi: 10.1038/543481a. [DOI] [PubMed] [Google Scholar]
- 107.Lokers R, Knapen R, Janssen S, van Randen Y, Jansen J. Analysis of Big Data technologies for use in agro-environmental science. Environ Model Softw. 2016;84:494–504. [Google Scholar]
- 108.Papalkar RR, Nerkar PR, Dhote CA. Issues of concern in storage system of IoT based big data. In: IEEE International conference on Information, Commun Instrum Control ICICIC 2017. Institute of Electrical and Electronics Engineers Inc.; 2018. pp. 1–6.
- 109.Holehouse AS, Das RK, Ahad JN, Richardson MOG, Pappu RV. CIDER: resources to analyze sequence-ensemble relationships of intrinsically disordered proteins. Biophys J. 2017;112:16–21. doi: 10.1016/j.bpj.2016.11.3200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kyte J, Doolittle RF. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Not applicable.