Abstract
Akebia trifoliata subsp. australis is a well-known medicinal and potential woody oil plant in China. The limited genetic information available for A. trifoliata subsp. australis has hindered its exploitation. Here, a high-quality chromosome-level genome sequence of A. trifoliata subsp. australis is reported. The de novo genome assembly of 682.14 Mb was generated with a scaffold N50 of 43.11 Mb. The genome includes 25,598 protein-coding genes, and 71.18% (485.55 Mb) of the assembled sequences were identified as repetitive sequences. An ongoing massive burst of long terminal repeat (LTR) insertions, which occurred ~1.0 million years ago, has contributed a large proportion of LTRs in the genome of A. trifoliata subsp. australis. Phylogenetic analysis shows that A. trifoliata subsp. australis is closely related to Aquilegia coerulea and forms a clade with Papaver somniferum and Nelumbo nucifera, which supports the well-established hypothesis of a close relationship between basal eudicot species. The expansion of UDP-glucoronosyl and UDP-glucosyl transferase gene families and β-amyrin synthase-like genes and the exclusive contraction of terpene synthase gene families may be responsible for the abundant oleanane-type triterpenoids in A. trifoliata subsp. australis. Furthermore, the acyl-ACP desaturase gene family, including 12 stearoyl-acyl-carrier protein desaturase (SAD) genes, has expanded exclusively. A combined transcriptome and fatty-acid analysis of seeds at five developmental stages revealed that homologs of SADs, acyl-lipid desaturase omega fatty acid desaturases (FADs), and oleosins were highly expressed, consistent with the rapid increase in the content of fatty acids, especially unsaturated fatty acids. The genomic sequences of A. trifoliata subsp. australis will be a valuable resource for comparative genomic analyses and molecular breeding.
Subject terms: Genomics, Lipids
Introduction
Akebia trifoliata (Thumb.) Koidz. subsp. australis is a perennial woody plant that belongs to the genus Akebia (Lardizabalaceae) and is mainly distributed in East Asia1. A. trifoliata subsp. australis (abbreviated as A. trifoliata hereafter) as well as A. trifoliata subsp. trifoliata and A. quinata, which are two other members of the genus Akebia, are listed in the Chinese Pharmacopoeia2 and have been used as traditional herbal medicines for >2000 years. An example is “Akebiae Fructus”, which is made from the dry fruit of A. trifoliata, including seeds, peel, and flesh3,4. Experimental and clinical studies have demonstrated that A. trifoliata possesses anti-inflammatory, antimicrobial, antioxidative, and anticancer properties4,5. Lu et al.5 reported that the ethanol extract of A. trifoliata seeds has antimetastatic potency against hepatocellular carcinoma cells. The pharmacological properties of A. trifoliata are attributed to numerous bioactive compounds, including triterpenoid saponins, triterpenes, and flavonoids, in dried fruits, stems, leaves, and seeds of A. trifoliata6. The content of oleanane-type triterpenoids, especially oleanolic acid, is high in the dry fruit of A. trifoliata7. These natural bioactive compounds with minimal or no side effects have attracted attention from chemists and pharmacologists due to their complex structural features and multiple biological effects.
Studies have indicated that the upstream reactions of triterpenoid biosynthesis through the mevalonate (MVA) or methylerythritol phosphate (MEP) pathways are involved in making building blocks and intermediates such as isopentenylpyrophosphate and farnesyl diphosphate (FPP)8. The downstream reactions of triterpenoid biosynthesis start from the condensation of two molecules of FPP, and the triterpenoid backbone undergoes oxidation, substitution, and glycosylation to generate various triterpenoids, involving many key enzymes, including squalene synthase (SQS), squalene monoxidase, oxidosqualene cyclase (OSC), cytochrome P450 monooxygenase (CYP), and uridine diphosphate glycosyltransferase (UGT)9. Among these enzymes, β-amyrin synthase (β-AS), a kind of OSC, is a unique key enzyme that catalyzes oleanane-type triterpenoid biosynthesis, and its expression is positively correlated with oleanane-type saponin content10.
As a potential oilseed medicinal plant, A. trifoliata seeds contain up to 39% oil with 77% unsaturated fatty acids3. Owing to the high UFA content, the seed oil of A. trifoliata is used as a quality edible oil and dietary supplement in China. In contrast to saturated fatty acids (SFAs), the dietary potency of UFAs has health benefits by reducing the risk of cardiovascular disease, obesity, and cancer11 and promoting the absorption of lipophilic nutritional compounds12. Fatty acids (FAs) are synthesized in plastids from acetyl-CoA up to 18:1Δ9, the first desaturation catalyzed by stearoyl-acyl-carrier protein desaturase (SAD). After export to the cytosol, FAs can be elongated and desaturated to produce long-chain and polyunsaturated fatty acids (PUFAs) by various enzymes within the ER and eventually processed into the storage lipid triacylglycerol (TAG)13. TAG is stored in oil bodies that serve as a natural protective system against fatty-acid oxidation and maintain lipid stability14. In olive oil, monounsaturated oleic acid (C18:1) makes up 75% of all TAGs, followed by saturated palmitic acid (C16:0; ~13.5%), polyunsaturated linoleic acid (C18:2; ~5.5%), and α-linolenic acid (C18:3; ~0.75%)15. In sesame seed oil, both oleic acid and linoleic acid are more evenly present (~40%)16. The differential accumulation of oleic and linoleic acids in olive compared with sesame is attributed to the functional divergence of oil biosynthesis pathway genes, such as ω-6 fatty-acid desaturase 2 (FAD2) and SAD, following duplication17. Oleosin, the most abundant oil body-associated protein, is a major determinant of oil body size18 and has been suggested to contribute to the stability of oil bodies and their synthesis and metabolism19. The heterologous expression of castor bean oleosin in Arabidopsis led to a 20% increase in the ricinoleic acid content in TAGs20. Moreover, coexpression of oleosins with other TAG biosynthesis genes increased the oil content21.
In addition to the usages mentioned above, the cultivated fruit of A. trifoliata is consumed as a delicacy due to its delicious taste and abundance of nutrients. Akebia trifoliata belongs to the genus Akebia (Lardizabalaceae) and, like Aquilegia coerulea and Papaver somniferum, is a member of the basal eudicots, which possess more diverse floral morphologies than the core eudicots and monocots. The identification and investigation of floral genes in basal eudicots can be used as an evolutionary link between core eudicots and grasses22.
Despite the considerable importance of A. trifoliata, genomic information for the species is limited, which has hindered its study and utilization. Here, we report the sequencing and assembly of a high-quality chromosomal-level genome sequence of A. trifoliata subsp. australis, which is the first sequenced species in the genus Akebia. Furthermore, we identified key genes involved in triterpene synthesis and the accumulation of UFAs. The availability of this genomic information will be helpful for comparative genomic analysis and the molecular breeding and engineering of A. trifoliata.
Results and discussion
De novo genome sequencing, assembly, and quality assessment
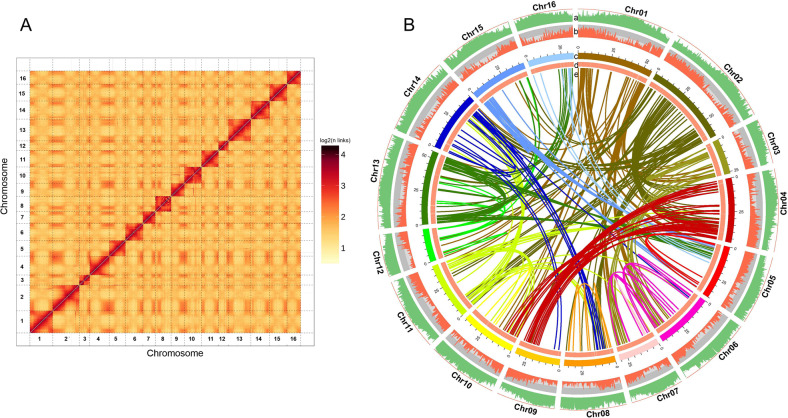
The genome of A. trifoliata was initially sequenced and assembled using the HiSeq X Ten sequencing platform from Illumina and PacBio single-molecule real-time (SMRT) sequencing technology, and the assembled contigs were anchored to pseudochromosomes using the Hi-C technique. K-mer analysis revealed that the estimated genome size of A. trifoliata was 669.76 Mb with a heterozygosity of 0.89% (Fig. S1). Furthermore, the genome size of A. trifoliata was estimated to be 654.34 Mb using flow cytometry (Fig. S2). In total, 193.71 Gb of high-quality sequences with a depth of 284-fold of the A. trifoliata genome were used to assemble the genome (Table S1). To obtain further chromosomal information about A. trifoliata, the sequences were then scaffolded and corrected using optical mapping data, and the resulting scaffolds were clustered into 16 pseudochromosomes (2n = 32), accounting for 98.05% (668.89/682.14 Mb) of the genome (Fig. 1, Fig. S3). The final chromosome-scale genome was 682.14 Mb in length with 689 contigs (contig N50 = 6.20 Mb) and 109 scaffolds (scaffolds N50 = 43.11 Mb) (Table 1). The final assembled sequence 682.14 Mb for A. trifoliata was close to the calculated estimated size (669.76 Mb) and to the size estimated by flow cytometry (654.34 Mb). The assembled size was marginally larger than the estimated size, probably because of the relatively high content of repetitive sequences (71.18%).
Fig. 1. Chromosomal features of Akebia trifoliata subsp. australis.
Hi-C map of the A. trifoliata subsp. australis genome showing genome-wide all-by-all interactions (A) and characterization of the A. trifoliata subsp. australis genome (B). a repeat density; b gene density; c chromosomes; d GC content; and e relationship between synteny blocks
Table 1.
Statistics of A. trifoliata subsp. australis genome sequencing, assembly, and annotation
| Genomic feature | A. trifoliata subsp. australis |
|---|---|
| Assembled genome size (Mb) | 682.14 |
| PacBio reads (Gb) | 65.96 (96.7 Xa) |
| Illumina reads (Gb) | 24.75 (36.28 X) |
| Hi-C (Gb) | 103.00 (151.03 X) |
| Total reads (Gb) | 193.71 (284.02 X) |
| GC content (%) | 35.02 |
| Percentage of anchoring | 98.05% |
| Number of contigs | 689 |
| Contigs N50 (kb) | 6,198 |
| Contigs N90 (kb) | 1,563 |
| Number of scaffolds | 109 |
| Scaffold N50 (kb) | 43,106 |
| Scaffold N90 (kb) | 30,967 |
| Longest sequence length (kb) | 64,151 |
| Total repetitive sequences (Mb) | 485.55 (71.18%) |
| Total protein-coding genes | 25,598 |
| Annotated protein-coding genes | 25,008 |
| Average length per gene (kb) | 6.47 |
| Total exon length (kb) | 41.34 |
| Average exon length (kb) | 1.62 |
| Total intron length (kb) | 124.39 |
| Average intron length (kb) | 4.86 |
aSequence coverage
In the sequencing and assembly quality assessment, 98.56% of Illumina short-insert reads could be aligned back to the final assembly. Moreover, BUSCO analysis showed that 1518 (94%) of the 1614 orthologs from the Embryophyta database were completely captured in our assembly, and CEGMA analysis showed that the assembled genome completely recalled 424 (93%) of the 458 core eukaryotic genes (CEGs) and 186 (75%) of the 248 highly conserved CEGs (Fig. S4).
Genome annotation
A total of 25,598 protein-coding genes were predicted in the A. trifoliata genome through a combination of ab initio prediction, homology search, and RNA-Seq prediction. Of all the predicted protein-coding genes, 24,814 (97%) were annotated based on homology search and RNA-Seq reads, with only 3.1% deriving solely from ab initio gene prediction, suggesting that the results of the protein-coding gene prediction were high quality (Fig. S5a). The A. trifoliata genome had an average gene length of 6,47 kb and average exon and intron lengths of 1,62 kb and 4,86 kb, respectively (Table 1). By similarity search, 25,008 protein-coding genes (98%) had functional annotations in public databases from several species, including N. nucifera (57.51%), V. vinifera (13.91%), Theobroma cacao (3.05%), and others (25.52%) (Fig. S5b, c and d, Tables S2 and S3). With regard to nonprotein-coding genes, we identified 431 tRNAs, 97 miRNAs, 222 rRNAs, 94 snRNAs, and 332 snoRNAs in our assembly (Table S4).
Repetitive sequences generally account for a substantial part of plant genomes and have a close relationship with genome size variation and functional adaptation23. A total of 485.55 Mb (71%) of the A. trifoliata genome was identified as repetitive sequences, which is the same as the proportion in P. somniferum (71%)24 and higher than that in M. cordata (63%)25, both of which are medicinal plants and basal eudicots. Approximately 87% of A. trifoliata repetitive sequences were classified as transposable elements (TEs) (Table S5). TEs occupy a significant fraction of many eukaryotic genomes and play an important role in the increase in genome size among angiosperms26. In our assembly, retrotransposon (Class I) and DNA transposon (Class II) TEs accounted for 72% and 7% of the genome, respectively. Among all TEs, long terminal repeats (LTRs) were the most abundant category of TEs, with 32% gypsy and 10% copia. Interestingly, the proportion of Penelope-like element/large retrotransposon derivative (LARD) elements in A. trifoliata was higher than that in most sequenced plant species, accounting for 21% of the genome. LARDs are considered to be nonautonomous elements and are the remnants of the deletion of autonomous LTR retrotransposons27. Recent lineage-specific radiation of LARDs (13% of the whole genome) in the pomegranate genome is responsible for fruit development, such as coloration, by affecting the expression of putative UDP-glucose:flavonoid glucosyltransferase (UFGT) and MYB genes27. The abundant LARD elements in the A. trifoliata genome might also have a close relationship with many important characteristics, including secondary metabolite accumulation, as in pomegranate.
Gene family analysis and phylogenetic tree construction
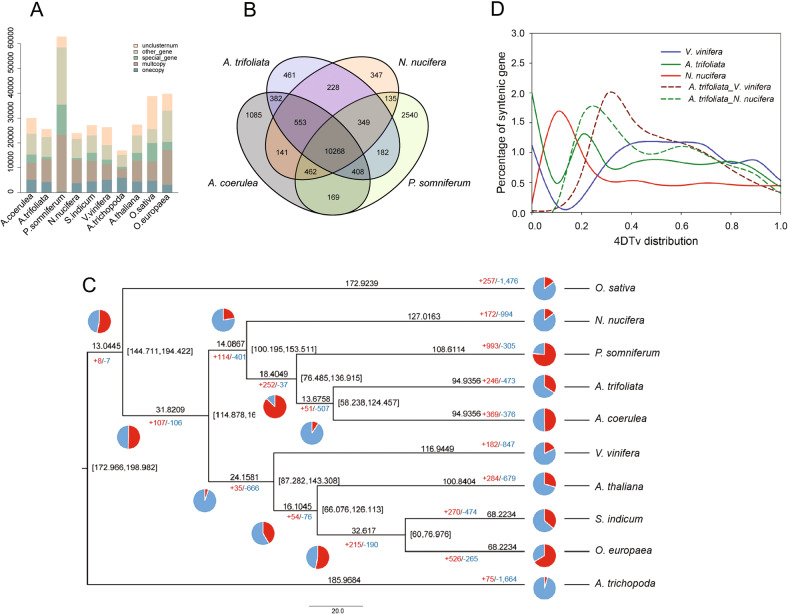
We examined the evolutionary relationships among A. trifoliata, two eurosid species (Arabidopsis thaliana, Vitis vinifera), three basal eudicot species (Aquilegia coerulea, Papaver somniferum, and Nelumbo nucifera), two core eudicot oilseed species (Sesamum indicum, Olea europaea), a monocot species (Oryza sativa), and Amborella trichopoda. A. trichopoda, one of the basal angiosperm species, represents a sister group to other flowering plants28. By using gene family cluster analysis, we identified 12,831 gene families in A. trifoliata, of which 399 were unique gene families containing 1,028 genes (Fig. 2A, Table S6). Of those species, P. somniferum possessed the most genes (62,879) and unique gene families (2,394) owing to a relatively recent whole-genome duplication (WGD) event24. Then, we further compared the gene families among the four basal eudicot species. As shown in Fig. 2B, 10,268 gene families were shared by A. coerulea, P. somniferum, N. nucifera, and A. trifoliata. Compared with two other basal eudicot species, P. somniferum (2,662) and A. coerulea (1,085), A. trifoliata had fewer unique gene families (461). There were more shared gene family clusters between A. trifoliata and A. coerulea (11,611) than with any two of the other three species. Hence, we inferred a relatively close taxonomic relationship between the two species.
Fig. 2. Comparative genomics analysis of A. trifoliata subsp. australis and other representative plant species.
A Classification and statistics of common and lineage-specific genes in A. trifoliata and other representative plant species. B Venn diagram of gene families in A. trifoliata and three other basal eudicot species. C Phylogenetic analysis, gene family expansion/contraction analyses and divergence time estimations. The number of gene family contraction and expansion events are indicated by blue (the blue part of the pie chart) and red (the red part of the pie chart) numbers, respectively. Divergence times (million years ago, Mya) are indicated by the numbers beside the branch nodes. D Distribution of 4DTv among A. trifoliata, N. nucifera, and V. vinifera in intra- and intergenomic comparisons
In total, 197 conserved single-copy orthologs were identified from the 10 species and were used to construct the phylogenetic tree with O. sativa and A. trichopoda serving as outgroups (Fig. 2C). Phylogenetic analysis revealed that A. trifoliata is closely related to A. coerulea and forms a clade with P. somniferum and N. nucifera, which supports the well-established hypothesis of a close relationship between basal eudicot species25. Based on the phylogenetic tree, we determined that 246 and 473 gene families were expanded and contracted, respectively, in A. trifoliata (Fig. 2C, Table S7). Gene ontology analysis also revealed that “supramolecular complex” and “cell-killing” appeared exclusively in expanded gene families, while “extracellular region part”, “cell proliferation”, and “biological adhesion” arose exclusively in contracted gene families (Fig. S6, Table S8).
Divergence time estimation and whole-genome duplication analysis
Using MCMCtree, we estimated that the divergence between A. trifoliata and A. coerulea, which occurred ~94.94 Mya, whereas P. somniferum divergence occurred ~108.61 Mya (Fig. 2C). The previous estimates indicate that the divergence between A. coerulea and M. cordata occurred approximately 115.24 Mya25, suggesting a closer relationship between A. coerulea and A. trifoliata than between A. coerulea and M. cordata. The constructed tree provides the overall divergence time of important basal eudicot species. WGD has been found in almost all fundamental lineages of land plants and is considered a driver of diversity and adaptation29. The fourfold degenerate site transversion (4DTv) value revealed one peak at ~0.2 in A. trifoliata, which implies that it may have undergone WGD after divergence from V. vinifera and N. nucifera (Fig. 2D). An ancient paleohexaploidy (γ) event, occurring ~125 Mya, was detected in the common ancestor of all sequenced eudicot genomes30. According to the phylogenetic analysis and 4DTv values, we inferred that the γ event was absent in A. trifoliata, which is a basal eudicot species, as well as in the basal eudicot species lotus and M. cordata25,31. The WGD of basal eudicot species should be further analyzed.
LTR insertion time
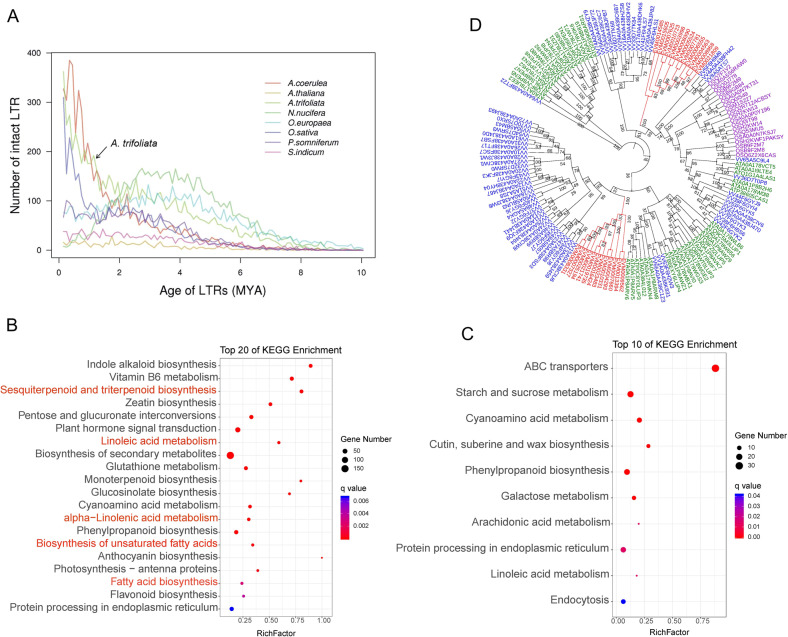
LTR retrotransposons play an important role in genome instability and evolution, which affect the expression and profiles of nearby genes and have significant consequences for phenotypic variation32. To investigate the insertion time of LTRs in the A. trifoliata genome, we estimated the intrasequence divergence of identified full-length LTR elements. We discovered that massive recent insertion events of LTRs occurred in A. trifoliata within the last one million years and in A. coerulea and O. sativa, which explains the accumulation of many recent LTRs (Fig. 3A). The LTR amplifications were inferred to have taken place during the Pleistocene epoch in which freezing occurred and there was limited atmospheric CO2. Changes in the climate and environment cause serious survival stresses that force evolutionary adaptation by reorganization of genomes, represented by activated TEs33. Such an ongoing amplification of LTRs may have contributed to an especially large proportion of LTRs in A. trifoliata. Furthermore, the estimation of insertion times of LTRs suggests that N. nucifera and O. europaea experienced amplification events 2–5 million years earlier than A. trifoliata.
Fig. 3. LTR insertion time estimation, enrichment analysis of expanded and contracted genes, and phylogenetic analysis of β-amyrin synthase genes.
A LTR insertion time analysis in A. trifoliata and other representative plant species. B, C KEGG enrichment analysis of expanded and contracted genes in A. trifoliata. Red indicates pathways related to triterpenoid biosynthesis and fatty-acid metabolism. D Phylogenetic analysis of β-amyrin synthase genes in A. trifoliata (red), rice, Arabidopsis and grape. The protein sequences of β-amyrin synthase in Arabidopsis, rice and grape were downloaded from the UniProt database. The phylogenetic tree was constructed using the ML method with 1000 bootstrap replicates
Determination of functional genes involved in terpenoid biosynthesis
KEGG enrichment analysis indicated that the sesquiterpenoid and triterpenoid biosynthesis pathways and monoterpenoid biosynthesis pathway were significantly enriched in expanded genes (Fig. 3B, C). Notably, the 24 β-amyrin synthase-like (Atrβ-AS) genes involved in the sesquiterpenoid and triterpenoid biosynthesis pathways catalyzing the conversion of oxidosqualene to β-amyrin, the proposed aglycone of oleanane-type saponins10, were exclusively identified in the expanded gene set (Tables S7 and 8). Suppression of β-AS expression by RNA interference led to a reduced content of β-amyrin and oleanane-type saponins10. The systematic identification, prediction, and evolutionary analysis of Atrβ-ASs in A. trifoliata have been of great significance to our understanding of the synthesis and regulation of triterpene saponins. Phylogenetic analysis revealed that 24 Atrβ-AS genes were classified into two clusters, which formed two monophyletic groups. One Atrβ-AS gene (EMV0021809) was grouped together with three Vitβ-AS genes (Fig. 3D). The expansion of Atrβ-AS genes might be an explanation for the high content of hederasaponin and oleanolic acid in many tissues of A. trifoliata.
In the A. trifoliata genome, 12 UDP-glucoronosyl and 3 UDP-glucosyltransferase gene families were expanded and contracted, respectively, while 7 and 14 cytochrome P450 gene families were expanded and contracted, respectively. The downstream reactions in the biosynthesis of saponins are believed to include a set of cytochrome P450-dependent hydroxylation/oxidations and several glycosyl transfer reactions catalyzed by glycosyltransferases34. The frequent expansion and contraction of the two gene families indicate a rapid evolution of these gene families, which might be the cause of the great variety of triterpenes in A. trifoliata. Unexpectedly, terpene synthase (TPS) gene families (PF01397, PF03936), which are responsible for the synthesis of various terpene molecules, were contracted in the A. trifoliata genome (Table S7). We performed genome-wide identification of the TPS gene family and identified 34 AtrTPS genes (Table S9), which is much fewer than that of V. vinifera with 95 TPS genes. Moreover, the TPS genes of A. trifoliata and the other three species were used to construct a phylogenetic tree (Fig. S7), and they grouped into four distinct clusters. All AtrTPS genes were distributed in different branches of Cluster I, while Clusters II, III and IV comprised TPS genes of rice, V. vinifera and Arabidopsis, respectively. These results suggest that there is functional diversification among the TPS genes in those species and the AtrTPS genes in A. trifoliata. Evolutionary plasticity is evident in the TPS family, represented by different product profiles, subcellular locations, activities, and substrates.
Molecular foundation for UFA accumulation
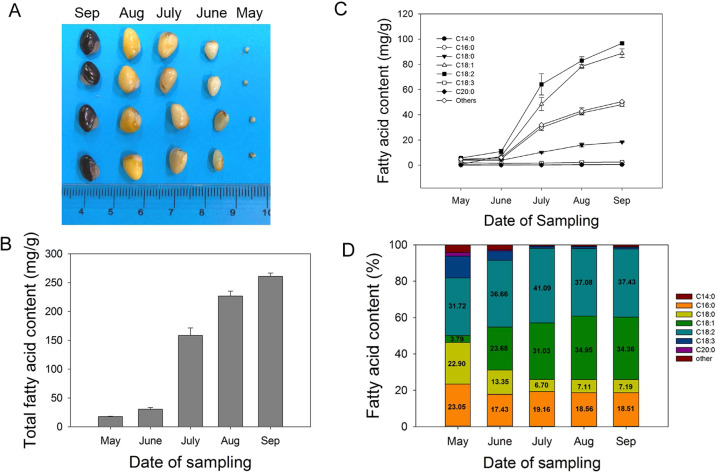
The oil of A. trifoliata seeds has been suggested to have health benefits because of its high UFA content3. To further study FAs metabolism in A. trifoliata seeds, we determined the FAs composition and gene expression profiles at different stages of seed development (Fig. 4A). The total FA content increased 15-fold from May (17.8 mg g−1) to September (260.9 mg g−1), and FA accumulated dramatically from June (30.4 mg g−1) to July (158.3 mg g−1) (Fig. 4B). The time courses for the levels of the individual FAs were similar to that of the total FA content (Fig. 4C). A sharp rise (six- to sevenfold) in the contents of oleic acid, linoleic acid, and palmitic acid was also observed from June to July. The linolenic acid levels remained stable at ~2 mg g−1 during seed development. In May, linoleic acid (32%), palmitic acid (23%), and stearic acid (23%) were the dominant FAs in seeds, whereas oleic acid (3.8%) was much less abundant. With increasing seed maturity, we observed a significant increase in the proportion of oleic acid (34%) and linoleic acid (37%) and a reduced proportion of palmitic acid (19%), stearic acid (7%), and linolenic acid (0.95%) (Fig. 4D). In summary, the FA contents increased with seed development, and UFAs, including linoleic acid and oleic acid, accumulated especially rapidly during the early stages of seed development.
Fig. 4. Changes in the seed content and percentage of total fatty acids (FAs) at different developmental stages.
A Photos of seeds at different developmental stages. The scale bar is in cm. B Changes in the content of total FAs. C Changes in FA contents. D Changes in the percentage of FAs
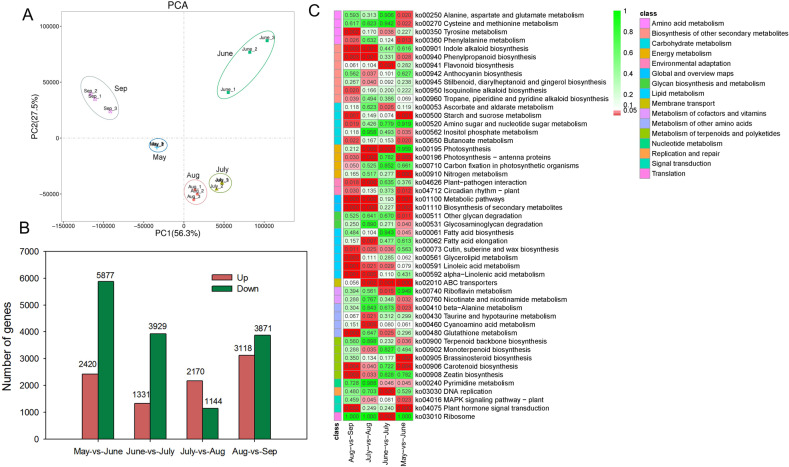
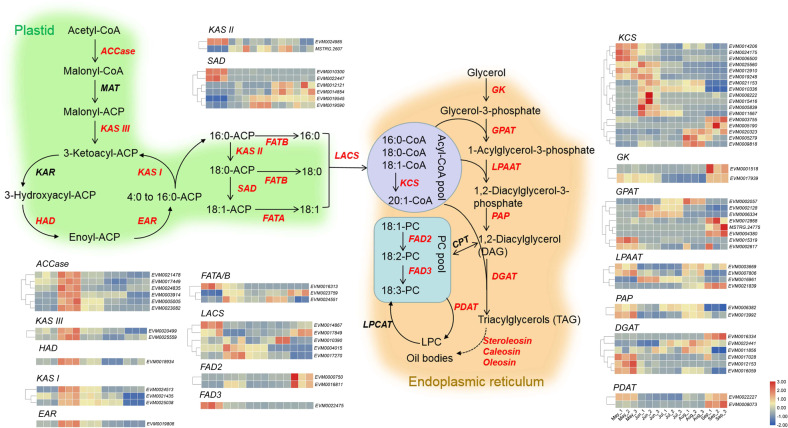
Transcriptomic analysis revealed that the gene expression profiles of seeds in July and August were closer to each other and obviously differed from those of the three other stages (Fig. 5A). A total of 2420, 1331, 2170, and 3118 upregulated differentially expressed genes (DEGs) and 5877, 3929, 1144, and 3871 downregulated DEGs were identified compared with those of the previous month (Fig. 5B). There were more DEGs between May vs June and August vs September, which agrees with the principal component analysis results, demonstrating more marked changes in gene expression profiles at the early and later stages of seed development. KEGG enrichment analysis indicated that many DEGs were enriched in lipid metabolism pathways (Fig. 5C, Table S10). An expression heat map of these DEGs linked to FA metabolism, integrating the changes in FA contents during the development of A. trifoliata seeds, can provide valuable information about the regulation of oil accumulation and UFA synthesis. The reconstruction of the FA and TAG biosynthetic pathways includes de novo formation of acyl chains in the plastid and TAG assembly in the endoplasmic reticulum (Fig. 6). Almost all DEGs involved in the biotin carboxyl carrier protein (BCCP) of acetyl-CoA carboxylase (ACCase) and fatty-acid synthase (FAS) complexes including β-ketoacyl-ACP synthase (KAS), β-hydroxacyl-ACP dehydratase (HAD), and enoyl-ACP reductase (EAR), showed maximal transcription in June, followed by July. The transcription peak of the core FA biosynthetic machinery coincided with the onset of oil accumulation in the seed at the early developmental stage. ACCase catalyzes the conversion of acetyl-CoA into malonyl-CoA and is the rate-limiting enzyme in fatty-acid biosynthesis35,36. It has been proposed that lipid biosynthesis can be increased by overexpressing ACCase37,38. KASI catalyzes the elongation of de novo fatty acids, and KASI mutation results in a significant reduction in FA contents in seeds39. EAR catalyzes a key regulatory step in FA biosynthesis and shows the highest expression during the early stages of seed development40. In the oil palm mesocarp, the key FA biosynthesis genes were highly expressed at 120 days after pollination when oil accumulation began in the mesocarp41. We inferred that the significant upregulation of ACCase and FAS in seeds in June has a close relationship with the rapid increase in the oil content in July. Additionally, three acyl-ACP thioesterases (FATA/B) and three long-chain acyl-CoA synthetases (LACS) were also highly expressed in June. FATA and FATB play an essential role in chain termination during fatty-acid synthesis. LACS mediates FA transport and conversion to acyl-CoA41. However, many DEGs involved in triacylglycerol (TAG) biosynthesis, such as glycerol kinase (GK), glycerol-3-phosphate acyltransferase (GPAT), 1-acylglycerol-3-phosphate acyltransferase (LPAAT), phosphatidate phosphatase (PAP), acyl-CoA:diacylglycerol acyltransferase (DGAT), and phospholipid:diacylglycerol acyltransferase (PDAT), were highly expressed throughout seed development.
Fig. 5. Transcriptomic analysis of seeds at different developmental stages.
A PCA of different expression profiles in seeds. B Statistics on the numbers of differentially expressed genes (DEGs) in seeds at different developmental stages. C KEGG enrichment pathways in seeds at different developmental stages. The bar indicates the p values
Fig. 6. The expression levels of enzymes involved in FA synthesis and TAG assembly in developing A. trifoliata seeds.
Gene expression levels in May, June, July, August, and September are indicated with colored bars (log2 FPKM). ACCase acetyl-CoA carboxylase, CPT diacylglycerol cholinephosphotransferase, DGAT acyl-CoA:diacylglycerol acyltransferase, EAR enoyl-acyl carrier protein (ACP) reductase, FAD2 oleate desaturase, FAD3 linoleate desaturase, FATA/B acyl-ACP thioesterase A/B, GK glycerol kinase, GPAT glycerol-3-phosphate acyltransferase, HAD hydroxyacyl-ACP dehydrase, KAR ketoacyl-ACP reductase, KASI ketoacyl-ACP synthase I, KAS II ketoacyl-ACP synthase II, KAS III ketoacyl-ACP synthase III, KCS 3-ketoacyl-CoA synthase, LACS long-chain acyl-CoA synthetase, LPAAT 1-acylglycerol-3-phosphate acyltransferase, PAP phosphatidate phosphatase, LPCAT 1-acylglycerol-3-phosphocholine acyltransferase, MAT malonyl-CoA:ACP malonyltransferase, PDAT phospholipid:diacylglycerol acyltransferase, SAD stearoyl-ACP desaturase. The colored boxes represent log2 FPKM values of DEGs in developing seeds, and sample names are shown on the lower right-hand side
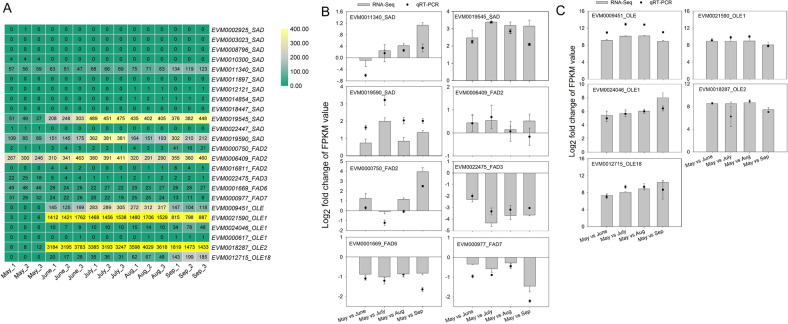
Comparative genomics and enrichment analyses indicated that linoleic acid metabolism, α-linolenic acid metabolism, UFA, and FA biosynthesis were enriched in an expanded gene set (Fig. 3B). The expansion of those genes involved in lipid metabolism may be responsible for the high UFA and oil contents in A. trifoliata seeds. Among those expanded gene families, an acyl-ACP desaturase gene family (PF03405.9), including 12 AtrSAD genes involved in UFA biosynthesis, was expanded in the A. trifoliata genome. Furthermore, two AtrSADs (EVM0019545 and EVM0019590) were highly expressed during seed development and showed significantly increased expression levels in seeds in July (Fig. 7), which may contribute to the high oleic acid content in A. trifoliata seeds. Three omega FAD genes, FAD2 (EVM0006409), FAD6 (EVM0001669), and FAD7 (EVM0000977), also remained at a high expression level during seed development (Fig. 7A, B), especially FAD2. SAD and FAD2 play key roles in the synthesis of UFAs42. The suppression of FAD2 expression by siRNA leads to a low linoleic acid content (5.5%) in olive oil17. It is noteworthy that homologs of oleosin (EVM0009451, EVM0021590, EVM0024046, EVM0018287, and EVM0012715), encoding oil body proteins that assist with packaging of TAG and determining oil body size, showed a sharp increase and a significantly higher expression in seeds from June to September compared with that in May (Fig. 7A, C). For example, the FPKM values of the two AtrOLEs (EVM0021590 and EVM0018287) in seeds were 441- and 366-fold higher in June than in May, respectively, which was confirmed by qRT-PCR (Fig. 7C). The results indicate that the significant upregulation of ACCase and FAS in FA biosynthesis, the high levels of SAD and FAD2 in FA desaturation, and the stabilization of oil bodies by oleosin allow A. trifoliata to accumulate high levels of oil. In Jatropha, increased expression of FA biosynthesis genes and oleosins synergistically results in the accumulation of high levels of oil in kernels (~63%)43.
Fig. 7. The expression levels of SADs, FADs, and oleosins in seeds at different developmental stages.
A The log2 (FPKM) values. B, C The expression levels during seed development determined by RNA-Seq and qRT-PCR
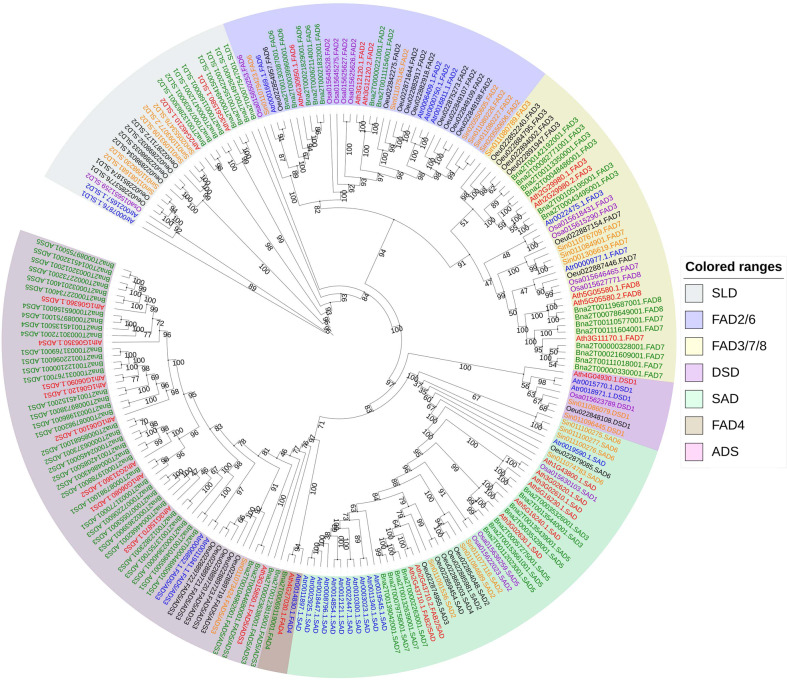
A total of 25 FADs (acyl-lipid desaturase and acyl-ACP desaturase) in A. trifoliata were identified (Table S11). To evaluate their evolutionary relationships and predict their gene functions, all the FAD genes from A. trifoliata, Arabidopsis, rice, Brassica napus, S. indicum, and O. europaea were aligned to construct an unrooted ML phylogenetic tree (Fig. 8). All AtrSADs, except for Atr0019590, formed a well-defined monophyletic group, suggesting that SAD extension occurred after the divergence of A. trifoliata and other species. Notably, all identified AtrFAD2 and AtrFAD6 genes, as well as one AtrSAD (Atr0019590) gene, clustered with the homologs of S. indicum and O. europaea, which suggests that these genes perform similar functions in the three species. The expansion and neofunctionalization of SADs in oleasters are likely also responsible for the higher oleic acid contents in A. trifoliata than in sesame17.
Fig. 8. Phylogenetic relationship of FAD genes from A. trifoliata subsp. australis, Arabidopsis thaliana, Oryza sativa, Sesamum indicum, Olea europaea, and Brassica rapa.
An ML phylogenetic tree of all detected FADs was constructed using RAxML with 1000 bootstrap replicates
In summary, genome sequencing of A. trifoliata has provided crucial information for the systematic study of the biosynthesis and metabolism of triterpenes in this medicinally and economically important nonmodel plant species. The A. trifoliata genome represents a useful resource for the genetic improvement of this plant species and for better understanding its genome evolution.
Materials and methods
Sampling and sequencing
Young A. trifoliata subsp. australis leaves were collected from Huaihua, Hunan Province, China (N27°33’17.95”, E109°59’54.70”). A modified cetyl trimethylammonium bromide method44 was used for DNA extraction. The DNA was used to construct a 20 kb insert-sized SMRTbell library for PacBio sequencing and 350 bp insert-size paired-end libraries for Illumina short-read sequencing. These libraries were prepared according to the manufacturer’s protocol (PacBio, CA, USA and Illumina, CA, USA). For RNA-seq, total RNA was extracted from the leaves, roots, and stems of A. trifoliata, and a mixture was made by combining an equal amount from each. After removing genomic DNA using DNase I (Takara), mRNAs were obtained using oligo (dT) beads and broken into short fragments, followed by cDNA synthesis. Paired-end sequencing was conducted on the HiSeq X Ten platform (Illumina, CA, USA). All PacBio and Illumina sequencing procedures were performed by the BioMarker Technologies Company (Beijing, China).
Genome size estimation
Flow cytometry was used to estimate the genome size of A. trifoliata according to the method described by Huang et al.45. First, young fresh leaves of A. trifoliata were chopped in nuclear isolation buffer. The supernatant was filtered through a 50-μm CellTrics filter. After being treated with RNase, cell nuclei were stained with propidium iodide in the dark. The fluorescence intensity of the sample was determined using a flow cytometer (BD FACSVerse). Maize B73 was used as an internal standard. The genome size of A. trifoliata was evaluated by k-mer frequency analysis using Illumina short reads. After removing contaminants, the optimal k-mer size was analyzed using KmerGenie46. Then, Jellyfish was used to analyze the k-mer counts, which were used to estimate the genome size and heterozygosity47.
Genome assembly and evaluation of genome quality
The PacBio Sequel long reads and Illumina short reads were combined to perform a de novo assembly of the A. trifoliata genome. Canu (v1.4) was first used to assemble the genome with the corrected-error-rate parameter48. Then, the corrected reads were independently assembled with WTDBG (v1.2.3)49. The well-assembled Canu and WTDBG results were merged by Quickmerge50. The merged genome was corrected with the Illumina data using Pilon51.
For the evaluation of assembly coverage, all paired-end short reads were mapped to our assembly using BWA52. RNA-Seq data from leaf, root, and stem tissues were assembled by Trinity53. HISAT2 was used to map all expressed sequence tags generated from RNA-Seq to the assembly with the default settings54 to evaluate gene completeness. The completeness of the assembly was assessed by the CEG mapping approach (v2.5) (CEGMA)55 and benchmarking universal single-copy ortholog (v4.0) (BUSCO) analysis56.
Chromosome-scale assembly with Hi-C data
Hi-C libraries were constructed from DNA extracted from fresh leaf tissue of A. trifoliata, similar to what was used for genome assembly, as previously described57. The purified and enriched DNA was used for sequencing using the Illumina HiSeq X ten platform. A total of 103 Gb of clean data (151-fold the estimated genome size) was obtained and aligned to the PacBio assembly contigs using BOWTIE258. The valid paired reads required for genome assembly were defined as uniquely mapped paired-end reads. Hi-C unique mapped paired-end reads were then applied to scaffold the assembled genome using the LACHESIS program59. The mapped read pairs were clustered into different chromosomal groups based on agglomerative hierarchical clustering. LACHESIS iterated all the possibilities of scaffold orientation and generated finely oriented scaffolds using a weighted directed acyclic graph.
Genome annotation
Repeat elements were identified by combining de novo- and homology-based approaches. RepeatModeler60, LTR_Finder61, and RepeatScout62 were used to construct a repeat library for de novo prediction. Based on the repeat sequence database, homology prediction was conducted using RepeatProteinMask and RepeatMasker63 against the Repbase TE library64 and the TE protein database. Noncoding RNA was annotated using tRNAscan-SE65 (for tRNAs) or INFERNAL66 (for miRNAs and snRNAs). The rRNAs were identified using BLASTN alignment and RNAammer67.
Multiple gene prediction methods integrating homolog-, de novo-, and transcriptome-based gene prediction were used to annotate protein-coding genes. For homologous prediction, a gene set including proteins from four plant genomes (Arabidopsis thaliana, A. coerulea, Opium poppy, and M. cordata) was mapped to the assembly of the A. trifoliata genome by BLAST68, and then GeneWise software was used to provide an accurate gene structure prediction69. RNA-Seq reads were first aligned to our genome assembly using HISAT254, and then StringTie70 was used to assemble the alignments into gene models in a transcriptome-based prediction. De novo identification was performed using Augustus (v2.5.5)71, GENSCAN (v1.0)72, SNAP73, and GlimmerHMM (v3.0.1)74. All the resulting genes mentioned above were integrated using EVidenceModeler75.
The protein-coding genes in A. trifoliata were blasted with an E value cutoff of 1.0 × 10−5 against SwissProt, NR, and the Kyoto Encyclopedia of Genes and Genomes (KEGG)76 for functional annotation. InterProScan (v4.8)77 and HMMER (v3.1)78 were used to annotate protein domains against the InterPro (v32.0)79 and Pfam (v27.0)80 databases, respectively. GO81 terms were obtained and grouped into three categories based on the results from the InterPro and Pfam entries.
Dating of LTR retrotransposon elements
Intact LTR retrotransposons were identified by searching the genomes of A. trifoliata with LTR_Finder61 and LTR_STRUC82. According to the sequence divergence, the insertion times of the identified full-length LTR retrotransposons were estimated with Dismat (EMBOSS package)83. The average base substitution rate of 1.3E-08 per site per year was used to calculate the insertion times84.
Gene family, phylogenetic analysis, and divergence time estimation
We collected the protein sequences from A. trifoliata and nine other plant species, namely, A. thaliana, O. sativa, V. vinifera, A. coerulea, N. nucifera, P. somniferum, O. europaea, S. indicum, and A. trichopoda, for gene family clustering. We conducted all-versus-all protein sequence queries through BLASTP with an E value of 1.0 × 10−5. OrthoMCL85 was used to cluster paralogous and orthologous groups. The four basal eudicot species, A. trifoliata, A. coerulea, P. somniferum, and N. nucifera, were further analyzed to explore their species-specific and shared gene families. The expansion and contraction of the gene family were analyzed using CAFE software86.
Following alignment by MUSCLE alignment software87, all single-copy genes identified and shared by the ten abovementioned species were used for evolutionary analysis and phylogenetic tree (ML Tree) reconstruction using RAxML software88. The divergence time was estimated using the MCMCtree program within the PAML package89. The divergence times were calibrated with the TimeTree database90.
Whole-genome duplication analysis
Protein sequences were aligned against each other with BLASTP with an E value ≤ 1 × 10−5 to identify conserved paralogous and orthologous genes in A. trifoliata, N. nucifera, and Vitis vinifera. The 4DTv values were calculated using the HKY model23. Then, potential WGD events in the genome were evaluated based on the 4DTv value.
Transcriptome analysis of developing seeds and qRT-PCR verification
A. trifoliata seeds were collected at 20, 50, 80, 110, and 140 days after flowering in May, June, July, August, and September, respectively. Library construction and RNA-Seq were performed as mentioned above. Clean reads were mapped to the reference genomes by HISAT254. The expression level (fragments per kilobase of transcript per million fragments mapped, FPKM value) of unigenes was calculated by StringTie65. DEGs were identified by DESeq291 (adjusted P value, FDR < 0.05). To clarify the biological functions of the DEGs, KEGG enrichment analysis was performed (http://www.genome.jp/kegg). Pathways with P < 0.05 were considered significantly enriched. A quantitative real-time polymerase chain reaction (qRT-PCR) assay was performed as described in Zahn et al.22. The actin gene was used as a reference in all experiments. Primers used for qRT-PCR are listed in Table S11. The qRT-PCR results were derived from three repeated reactions for each gene and sample. Fold change was calculated using the formula 2−ΔΔCt.
Genome-wide identification and phylogenetic analysis of gene families
The genome and protein sequences of A. thaliana, O. sativa, Vitis vinifera, O. europaea, S. indicum, and Brassica napus were downloaded from the NCBI database. The hmmsearch program of HMMER software (version 3.2.1) (http://hmmer.org/download.html) was also applied to the identification of TPSs (PF01397, PF03936) and FADs (PF00487, PF03405) in Pfam 32.0 data (http://pfam.xfam.org/). To classify and investigate the phylogenetic relationships of the amyrin synthase-like, TPS, and FAD genes, the predicted genes were aligned using MUSCLE. Hence, two ML (maximum-likelihood) phylogenetic trees were constructed using RAxML software. The bootstrap test was performed with 1000 replicates to obtain high reliability of interior branches. The phylogenetic tree was imported to iTOL (https://itol.embl.de/) for visualization92.
Assay of fatty-acid composition
The seeds at different developmental stages used in transcriptomic analysis were used to determine the FA composition. FA extraction and analysis were conducted according to Liu et al.93. Fifty milligrams of seed powder (fresh weight) was treated using chloroform-methanol solution (V/V = 2:1). Ultrasonic technology was used for FA extraction. Fatty acyl methyl esters (FAMEs) were prepared by direct transesterification of FA with 1% sulfuric acid in methanol at 80 °C for 30 min. The FAMEs were extracted with 1 mL hexane and analyzed by gas chromatography-mass spectrometry (GC-MS) with methyl heptadecanoate as an internal standard. GC-MS analysis was performed on an Agilent 6890 N/5975B (Agilent, USA) equipped with an Agilent HP-INNOWAX column (30 m × 0.25 mm ID × 0.25 µm). The column temperature was raised from 150 °C to 230 °C at a rate of 10 °C min−1 and then increased to 250 °C and maintained for 10 min. Peaks were identified by comparing the retention times with those of the corresponding standards (Sigma), and their identities were also confirmed by comparing mass spectra to the National Institute of Standards and Technology mass spectral library. The concentration of each sample was normalized according to the internal control.
Supplementary information accompanies the manuscript on the Horticulture Research website http://www.nature.com/hortres
Supplementary information
Acknowledgements
We are very grateful to professor Jay J. Thelen (University of Missouri, USA) for constructive criticism and helpful suggestions. This work was funded by the Natural Science Foundation of Hunan Province (2019JJ50475), Key Scientific Research Projects of Hunan Education Department (18A448), Foundation of Hunan Double First-rate Discipline Construction Projects of Bioengineering and Key Laboratory of Research and Utilization of Ethnomedicinal Plant Resources of Hunan Province, and the National Science Foundation (81874334).
Author contributions
S.Q.S. and H.H. conceived and designed the experiments. H.H., J.L., Q.T., L.F.O., X.L.L., C.H.Z., H.L.H., and W.X.J. collected materials and performed the experiments. H.H. and I.M.M. analyzed the data. H.H. wrote the paper, which was modified by I.M.M. and S.Q.S. All authors read and approved the final manuscript.
Data availability
The A. trifoliata subsp. australis genome sequences, gene annoataion information, raw sequence data of genome sequencing and RNA-seq have been deposited under BioProject accession number PRJNA685604.
Conflict of interest
The authors declare that they have no conflict of interest.
Supplementary information
Supplementary Information accompanies this paper at (10.1038/s41438-020-00458-y).
References
- 1.Liu GY, Ma SC, Zheng J, Zhang J, Lin RC. Two new triterpenoid saponins from Akebia quinata (Thunb.) Decne. J. Integr. Plant Biol. 2007;49:196–201. doi: 10.1111/j.1744-7909.2007.00362.x. [DOI] [Google Scholar]
- 2.Chinese Pharmacopoeia Commission. The Pharmacopoeia of the People’s Republic of China. Beijing: China medicine science and technology press (2015).
- 3.Du YX, et al. Physicochemical and functional properties of the protein isolate and major fractions prepared from Akebia trifoliata var. australis seed. Food Chem. 2012;133:923–929. doi: 10.1016/j.foodchem.2012.02.005. [DOI] [Google Scholar]
- 4.Wang XY, et al. The profiling of bioactives in Akebia trifoliata pericarp and metabolites, bioavailability and in vivo anti-inflammatory activities in DSS-induced colitis mice. Food Funct. 2019;10:3977–3991. doi: 10.1039/C9FO00393B. [DOI] [PubMed] [Google Scholar]
- 5.Lu WL, et al. Akebia trifoliata (Thunb.) Koidz seed extract inhibits human hepatocellular carcinoma cell migration and invasion in vitro. J. Ethnopharmacol. 2019;234:204–215. doi: 10.1016/j.jep.2018.11.044. [DOI] [PubMed] [Google Scholar]
- 6.Jiang D, Shi SP, Cao JJ, Gao QP, Tu PF. Triterpene saponins from the fruits of Akebia quinata. Biochem. Syst. Ecol. 2008;36:138–141. doi: 10.1016/j.bse.2007.06.004. [DOI] [Google Scholar]
- 7.Wang J, et al. Antibacterial oleanane-type triterpenoids from pericarps of Akebia trifoliata. Food Chem. 2015;168:623–629. doi: 10.1016/j.foodchem.2014.07.105. [DOI] [PubMed] [Google Scholar]
- 8.Vranova E, Coman D, Gruissem W. Network analysis of the MVA and MEP pathways for isoprenoid synthesis. Annu. Rev. Plant Biol. 2013;64:665–700. doi: 10.1146/annurev-arplant-050312-120116. [DOI] [PubMed] [Google Scholar]
- 9.Kushiro, T. & Ebizuka, Y. “Triterpenes”. In Mander L., Liu H. W. B. (eds) Comprehensive Natural Products II: Chemistry and Biology. p 673–708 (Elsevier, Oxford, 2010).
- 10.Zhao C, et al. Functional analysis of β-amyrin synthase gene in ginsenoside biosynthesis by RNA interference. Plant Cell Rep. 2015;8:1307–1315. doi: 10.1007/s00299-015-1788-7. [DOI] [PubMed] [Google Scholar]
- 11.Milićević D, et al. The role of total fats, saturated/unsaturated fatty acids and cholesterol content in chicken meat as cardiovascular risk factors. Lipids Health Dis. 2014;13:42. doi: 10.1186/1476-511X-13-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Failla M, Chitchumronchokchai C, Ferruzzi MG, Goltz SR, Campbell WW. Unsaturated fatty acids promote bioaccessibility and basolateral secretion of carotenoids and α-tocopherol by Caco-2 cells. Food Funct. 2014;5:1101–1112. doi: 10.1039/C3FO60599J. [DOI] [PubMed] [Google Scholar]
- 13.Baud S, Lepiniec L. Physiological and developmental regulation of seed oil production. Prog. Lipid Res. 2010;49:235–249. doi: 10.1016/j.plipres.2010.01.001. [DOI] [PubMed] [Google Scholar]
- 14.Shimada TL, Shimada T, Takahashi H, Fukao Y, Hara-Nishimura I. A novel role for oleosins in freezing tolerance of oilseeds in Arabidopsisthaliana. Plant J. 2008;55:798–809. doi: 10.1111/j.1365-313X.2008.03553.x. [DOI] [PubMed] [Google Scholar]
- 15.Rueda, A., et al. Characterization of fatty acid profile of argan oil and other edible vegetable oils by gas chromatography and discriminant analysis. J. Chem. 2014, 843908 (2014).
- 16.Wang L, et al. Genome sequencing of the high oil crop sesame provides insight into oil biosynthesis. Genome Biol. 2014;15:R39. doi: 10.1186/gb-2014-15-2-r39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Unver T, et al. Genome of wild olive and evolution of oil biosynthesis. Proc. Natl Acad. Sci. USA. 2017;9:9413–9422. doi: 10.1073/pnas.1708621114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ting JTL, et al. Oleosin genes in maize kernels having diverse oil contents are constitutively expressed independent of oil contents. Planta. 1996;199:158–165. doi: 10.1007/BF00196892. [DOI] [PubMed] [Google Scholar]
- 19.Frandsen GI, Mundy J, Tzen JTC. Oil bodies and their associated proteins, oleosin and caleosin. Physiol. Plant. 2001;112:301–307. doi: 10.1034/j.1399-3054.2001.1120301.x. [DOI] [PubMed] [Google Scholar]
- 20.Lu C, Fulda M, Wallis JG, Browse J. A high-throughput screen for genes from castor that boost hydroxy fatty acid accumulation in seed oils of transgenic Arabidopsis. Plant J. 2006;45:847–856. doi: 10.1111/j.1365-313X.2005.02636.x. [DOI] [PubMed] [Google Scholar]
- 21.Bhatla SC, Kaushik V, Yadav MK. Use of oil bodies and oleosins in recombinant protein production and other biotechnological applications. Biotechnol. Adv. 2010;28:293–300. doi: 10.1016/j.biotechadv.2010.01.001. [DOI] [PubMed] [Google Scholar]
- 22.Zahn LM, et al. Comparative transcriptomics among floral organs of the basal eudicot Eschscholzia californica as reference for floral evolutionary developmental studies. Genome Biol. 2010;11:101. doi: 10.1186/gb-2010-11-10-r101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hasegawa M, Kishino H, Yano T. Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J. Mol. Evol. 1985;22:160–174. doi: 10.1007/BF02101694. [DOI] [PubMed] [Google Scholar]
- 24.Guo L, et al. The opium poppy genome and morphinan production. Science. 2018;362:343–347. doi: 10.1126/science.aat4096. [DOI] [PubMed] [Google Scholar]
- 25.Liu X, et al. The genome of medicinal plant Macleaya cordata provides new insights into benzylisoquinoline alkaloids metabolism. Mol. Plant. 2017;10:975–989. doi: 10.1016/j.molp.2017.05.007. [DOI] [PubMed] [Google Scholar]
- 26.Tenaillon MI, Hollister JD, Gaut BS. A triptych of the evolution of plant transposable elements. Trends Plant Sci. 2010;15:471–478. doi: 10.1016/j.tplants.2010.05.003. [DOI] [PubMed] [Google Scholar]
- 27.Yuan Z, et al. The pomegranate (Punica granatum L.) genome provides insights into fruit quality and ovule developmental biology. Plant Biotechnol. J. 2018;16:1363–1374. doi: 10.1111/pbi.12875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Amborella Genome Project. The Amborella genome and the evolution of flowering plants. Science. 2013;342:1241089. doi: 10.1126/science.1241089. [DOI] [PubMed] [Google Scholar]
- 29.Tank DC, et al. Nested radiations and the pulse of angiosperm diversification: increased diversification rates often follow whole genome duplications. N. Phytol. 2015;207:454–467. doi: 10.1111/nph.13491. [DOI] [PubMed] [Google Scholar]
- 30.Vanneste K, Baele G, Maere S, Van de Peer Y. Analysis of 41 plant genomes supports a wave of successful genome duplications in association with the Cretaceous-Paleogene boundary. Genome Res. 2014;24:1334–1347. doi: 10.1101/gr.168997.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ming R, et al. Genome of the long-living sacred lotus (Nelumbo nucifera Gaertn.) Genome Biol. 2013;14:41. doi: 10.1186/gb-2013-14-5-r41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Casacuberta E, González J. The impact of transposable elements inenvironmental adaptation. Mol. Ecol. 2013;22:1503–1517. doi: 10.1111/mec.12170. [DOI] [PubMed] [Google Scholar]
- 33.Grandbastien MA. Activation of plant retrotransposons under stress conditions. Trends Plant Sci. 1998;3:181–187. doi: 10.1016/S1360-1385(98)01232-1. [DOI] [Google Scholar]
- 34.Achnine L, et al. Genomics-based selection and functional characterization of triterpene glycoyltransferases from the model legume Medicago truncatula. Plant J. 2005;41:875–887. doi: 10.1111/j.1365-313X.2005.02344.x. [DOI] [PubMed] [Google Scholar]
- 35.Salie MJ, Zhang N, Lancikova V, Xu D, Thelen JJ. A family of negative regulators targets the committed step of de Novo fatty acid biosynthesis. Plant Cell. 2016;28:2312–2325. doi: 10.1105/tpc.16.00317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Salie MJ, Thelen JJ. Regulation and structure of the heteromeric acetyl-CoA carboxylase. Biochim. Biophys. Acta. 2016;186:1207–1213. doi: 10.1016/j.bbalip.2016.04.004. [DOI] [PubMed] [Google Scholar]
- 37.Lu J, Sheahan C, Fu P. Metabolic engineering of algae for fourth generation biofuels production. Energy Environ. Sci. 2011;4:2451–2466. doi: 10.1039/c0ee00593b. [DOI] [Google Scholar]
- 38.Chaturvedi, S. et al. Overexpression and repression of key rate-limiting enzymes (acetyl CoA carboxylase and HMG reductase to enhance fatty acid production from Rhodotorula mucilaginosa. J. Basic Microbiol. 1–11 (2020). [DOI] [PubMed]
- 39.Wu GZ, Xue HW. Arabidopsis β-ketoacyl-[acyl carrier protein] synthase I is crucial for fatty acid synthesis and plays a role in chloroplast division and embryo development. Plant Cell. 2010;22:3726–3744. doi: 10.1105/tpc.110.075564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.González-Thuillier I, Venegas-Calerón M, Garcés R, Wettstein-Knowles P, Martínez-Force E. Sunflower (Helianthus annuus) fatty acid synthase complex: enoyl-[acyl carrier protein]-reductase genes. Planta. 2015;241:43–56. doi: 10.1007/s00425-014-2162-7. [DOI] [PubMed] [Google Scholar]
- 41.Tranbarger TJ, et al. Regulatory mechanisms underlying oil palm fruit mesocarp maturation, ripening, and functional specialization in lipid and carotenoid metabolism. Plant Physiol. 2011;156:564–584. doi: 10.1104/pp.111.175141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hernández ML, Sicardo MD, Alfonso M, Martínez-Rivas JM. Transcriptional regulation of stearoyl-acyl carrier protein desaturase genes in response to abiotic stresses leads to changes in the unsaturated fatty acids composition of olive mesocarp. Front. Plant Sci. 2019;10:251. doi: 10.3389/fpls.2019.00251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ha J, et al. Genome sequence of Jatropha curcas L., a non-edible biodiesel plant, provides a resource to improve seed-related traits. Plant Biotech. J. 2019;17:517–530. doi: 10.1111/pbi.12995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tel-Zur N, Abbo S, Myslabodski D, Mizrahi Y. Modified CTAB procedure for DNA isolation from epiphytic cacti of the genera hylocereus and selenicereus (Cactaceae) Plant Mol. Biol. Rep. 1999;17:249–254. doi: 10.1023/A:1007656315275. [DOI] [Google Scholar]
- 45.Huang H, Tong Y, Zhang QJ, Gao LZ. Genome size variation among and within Camellia species by using flow cytometric analysis. PLoS ONE. 2014;8:e64981. doi: 10.1371/journal.pone.0064981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chikhi R, Medvedev P. Informed and automated k-mer size selection for genome assembly. Bioinformatics. 2013;30:31–37. doi: 10.1093/bioinformatics/btt310. [DOI] [PubMed] [Google Scholar]
- 47.Marçais G, Kingsford C. A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics. 2011;27:764–770. doi: 10.1093/bioinformatics/btr011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Koren S, et al. Canu: scalable and accurate long-read assembly via adaptive -mer weighting and repeat separation. Genome Res. 2017;27:722–736. doi: 10.1101/gr.215087.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.WTDBG package. Accessed 10 Jan 2018. available from https://github.com/ruanjue/wtdbg2.
- 50.Chakraborty M, Baldwin-Brown JG, Long AD, Emerson J. Contiguous and accurate de novo assembly of metazoan genomes with modest long read coverage. Nucleic Acids Res. 2016;44:147–147. doi: 10.1093/nar/gkw419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Walker BJ, et al. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE. 2014;9:e112963. doi: 10.1371/journal.pone.0112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Grabherr MG, et al. Full-length transcriptome assembly from RNA-seq data without a reference genome. Nat. Biotechnol. 2011;29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kim D, Paggi JM, Park C, Bennett C, Salzberg SL. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019;37:907–915. doi: 10.1038/s41587-019-0201-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Parra G, Bradnam K, Korf I. CEGMA: a pipeline to accurately annotate coregenes in eukaryotic genomes. Bioinformatics. 2007;23:1061–1067. doi: 10.1093/bioinformatics/btm071. [DOI] [PubMed] [Google Scholar]
- 56.Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 2015;31:3210–3212. doi: 10.1093/bioinformatics/btv351. [DOI] [PubMed] [Google Scholar]
- 57.Lieberman-Aiden E, et al. Comprehensive mapping of long-rangeinteractions reveals folding principles of the human genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Burton JN, et al. Chromosome-scale scaffolding of de novogenome assemblies based on chromatin interactions. Nat. Biotechnol. 2013;31:1119–1125. doi: 10.1038/nbt.2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Smit, A. F. A. & Hubley, R. RepeatModeler Open-1.0. http://www.repeatmasker.org (2008–2015).
- 61.Xu Z, Wang H. LTR_FINDER: an efficient tool for the prediction of full-length LTR retrotransposons. Nucleic Acids Res. 2007;35:265–268. doi: 10.1093/nar/gkm286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Price AL, Jones NC, Pevzner PA. De novo identification of repeat families in large genomes. Bioinformatics. 2005;21:351–358. doi: 10.1093/bioinformatics/bti1018. [DOI] [PubMed] [Google Scholar]
- 63.Smit, A. F. A., Hubley, R. & Green, P. RepeatMasker Open-4.0. http://www.repeatmasker.org (1996–2015).
- 64.Jurka J, et al. Repbase update, a database of eukaryotic repetitive elements. Cytogenet Genome Res. 2005;110:462–467. doi: 10.1159/000084979. [DOI] [PubMed] [Google Scholar]
- 65.Lowe TM, Chan PP. tRNAscan-SE On-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 2016;44:54–57. doi: 10.1093/nar/gkw413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nawrocki EP, Eddy SR. Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics. 2013;29:2933–2935. doi: 10.1093/bioinformatics/btt509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lagesen K, et al. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 2007;35:3100–3108. doi: 10.1093/nar/gkm160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Altschul SF, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Birney E, Clamp M, Durbin R. GeneWise and genomewise. Genome Res. 2004;14:988–995. doi: 10.1101/gr.1865504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pertea M, et al. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotech. 2015;33:290–295. doi: 10.1038/nbt.3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Stanke M, Steinkamp R, Waack S, Morgenstern B. AUGUSTUS: a web server for gene finding in eukaryotes. Nucleic Acids Res. 2004;32:309–312. doi: 10.1093/nar/gkh379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Aggarwal G, Ramaswamy R. Ab initio gene identification: prokaryote genome annotation with GeneScan and GLIMMER. J. Biosci. 2002;27:7–14. doi: 10.1007/BF02703679. [DOI] [PubMed] [Google Scholar]
- 73.Bromberg Y, Rost B. SNAP: predict effect of non-synonymous polymorphisms on function. Nucleic Acids Res. 2007;35:3823. doi: 10.1093/nar/gkm238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Majoros WH, Pertea M, Salzberg SL. TigrScan and GlimmerHMM: twoopen source ab initio eukaryotic gene-finders. Bioinformatics. 2004;20:2878–2879. doi: 10.1093/bioinformatics/bth315. [DOI] [PubMed] [Google Scholar]
- 75.Haas BJ, et al. Automated eukaryotic gene structure annotation using EVidenceModeler and the program to assemble spliced alignments. Genome Biol. 2008;9:1. doi: 10.1186/gb-2008-9-1-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Quevillon E, et al. InterProScan: protein domains identifier. Nucleic Acids Res. 2005;33:116–120. doi: 10.1093/nar/gki442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Finn RD, Clements J, Eddy SR. HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 2011;39:29–37. doi: 10.1093/nar/gkr367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hunter S, et al. InterPro: the integrative protein signature database. Nucleic Acids Res. 2009;37:211–215. doi: 10.1093/nar/gkn785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Finn RD, et al. Pfam: the protein families database. Nucleic Acids Res. 2014;42:222–230. doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ashburner M, et al. Gene ontology: tool for the unification of biology. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.McCarthy EM, McDonald JF. LTR_STRUC: a novel search and identification program for LTR retrotransposons. Bioinformatics. 2003;19:362–367. doi: 10.1093/bioinformatics/btf878. [DOI] [PubMed] [Google Scholar]
- 83.SanMiguel P, Gaut BS, Tikhonov A, Nakajima Y, Bennetzen JL. The paleontology of intergene retrotransposons of maize. Nat. Genet. 1998;20:43–45. doi: 10.1038/1695. [DOI] [PubMed] [Google Scholar]
- 84.Ma J, Bennetzen JL. Rapid recent growth and divergence of rice nuclear genomes. Proc. Natl Acad. Sci. USA. 2004;101:12404–12410. doi: 10.1073/pnas.0403715101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Li L, Stoeckert CJ, Jr, Roos DS. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 2003;13:2178–2189. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Han MV, Thomas GW, Lugo-Martinez J, Hahn MW. Estimating gene gain and loss rates in the presence of error in genome assembly and annotation using CAFE 3. Mol. Biol. Evol. 2013;30:1987–1997. doi: 10.1093/molbev/mst100. [DOI] [PubMed] [Google Scholar]
- 87.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 90.Kumar S, Stecher G, Suleski M, Hedges SB. TimeTree: a resource for timelines, timetrees, and divergence times. Mol. Biol. Evol. 2017;34:1812–1819. doi: 10.1093/molbev/msx116. [DOI] [PubMed] [Google Scholar]
- 91.Anders, S. & Huber, W. Differential expression analysis for sequence count data. Genome Biol. 11, R106 (2010). [DOI] [PMC free article] [PubMed]
- 92.Letunic I, Bork P. Interactive tree of life (iTOL) v3: an online tool for the displayand annotation of phylogenetic and other trees. Nucleic Acids Res. 2016;44:242–245. doi: 10.1093/nar/gkw290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Liu J, Mao X, Zhou W, Guarnieri MT. Simultaneous production of triacylglycerol and high-value cartenoids by the astaxanthin-producing oleaginous green microalga Chlorella zofingiensis. Bioresour. Tech. 2016;214:319–327. doi: 10.1016/j.biortech.2016.04.112. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The A. trifoliata subsp. australis genome sequences, gene annoataion information, raw sequence data of genome sequencing and RNA-seq have been deposited under BioProject accession number PRJNA685604.