Abstract
Circular RNAs (circRNAs) are a class of endogenous single-stranded covalently closed RNAs, primarily produced from pre-mRNAs via non-canonical back-splicing. circRNAs are highly conserved, stable, and expressed in tissue- and development-specific pattern. circRNAs play essential roles in physiological process as well as cancer biology. By the advances of deep sequencing and bioinformatics, the number of circRNAs have increased explosively. circRNAs function as miRNA/protein sponge, protein scaffold, protein recruitment, enhancer of protein function, as well as templates for translation involved in the regulation of transcription/splicing, translation, protein degradation, and pri-miRNA processing in human cancers and contributed to the pathogenesis of cancer. Numerous circRNAs may function in diverse manners. In this review, we survey the current understanding of circRNA functions in human cancer including miRNA sponge, circRNA-protein interaction, and circRNA-encoded protein, and summarize available databases for circRNA annotation and functional prediction.
Keywords: circRNA, miRNA, transcription, translation, peptide
Introduction
Circular RNAs (circRNAs) are a class of endogenous single-stranded covalently closed RNAs, primarily produced from pre-mRNAs via non-canonical back-splicing (1). circRNAs were first discovered in the 1970s, and were ignored as byproducts of RNA splicing (2, 3). In 2013, circRNAs were found to act as microRNA (miRNA) sponge (4, 5). circRNAs are highly conserved, and exhibit tissue-specific and development stage-dependent expression patterns (6). Both the expression and functions of circRNAs may be independent of their host genes (7). However, experimental functional evidence is still missing for most of the discovered circRNAs. Recently, by the advances in deep sequencing technologies and bio-informatic tools, the number of circRNAs have increased explosively.
Several types of circRNAs have been identified, including exonic circRNA (EcRNA), intronic circRNA (CiRNA), and exon-intron circRNA (EIcRNAs). Recently, tRNA intron derived circRNA (tricRNA) via pre-tRNA splicing has been reported (8). circRNAs can bind to primary miRNAs (pri-miRNA) or mature miRNA, coding region and 3’-untranslated region (3’-UTR) of mRNA directly, or recruit proteins to these regions. The multi-functions of circRNAs have been widely investigated, including miRNA or protein sponges, protein scaffold, protein recruitment, enhancer of protein function, as well as templates for translation (1). A variety of databases have been reported, including circRNA annotation, internal ribosome entry site (IRES) assessment, and open reading frame (ORF) prediction (9).
In this review, we survey the current understanding of circRNA biogenesis, miRNA sponge, circRNA-protein interaction, circRNA-encoded protein, and available databases in human cancer.
Biogenesis of circRNAs
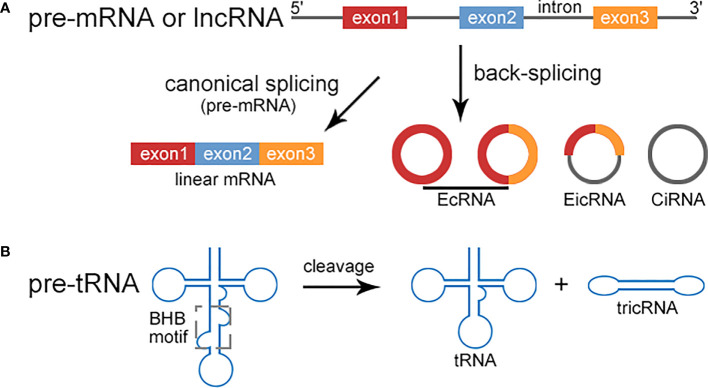
circRNAs are usually generated from non-canonical back-splicing of pre-mRNA, ranging from hundreds to thousands of nucleotides in length. circRNAs are resistant to exonucleases (RNase R), therefore highly stable. circRNAs can be classified into three types: EcRNAs, CiRNAs, and EIcRNAs. Besides of pre-mRNAs, recent studies have revealed that circRNAs may be derived from long noncoding RNAs (lncRNAs) (Figure 1A). LINC00632 was found to be the host gene for CDR1as (10). circLNC-PINT was originated from the lncRNA LNC-PINT (11). In addition, tricRNAs are a novel type of circRNAs generated during pre-tRNA splicing (Figure 1B). A number of proteins are involved in the regulation of circRNA biogenesis, including muscleblind protein (MBL), ADAR family of RNA binding proteins (RBPs), quaking (QKI) (12).
Figure 1.
Biogenesis of circRNA. (A) Pre-mRNA can be transcribed into linear mRNA via canonical splicing or produce circRNAs including EcRNA, EIcRNA, and CiRNA through back-splicing. EcRNAs and CiRNAs only contain exons and introns respectively, while EIcRNAs consist of both. circRNAs can also be originated from lncRNAs via back-splicing, such as CDR1as and circLINC-PINT. (B) The intron excision of pre-tRNA occurs when the tRNA splicing endonuclease complex (TSEN) binds to the bulge-helix-bulge (BHB) motif to release tRNA and tricRNA.
Mechanisms of Actions of circRNAs in Human Cancer
miRNA sponge is the most widely investigated function of circRNAs. Besides, circRNAs may function as protein sponges, protein scaffold, protein recruitment, enhancer of protein function, as well as templates for translation. Moreover, circRNAs may contribute to cancer progression in more than one way (13, 14).
circRNAs Act as miRNA Sponges in Human Cancer
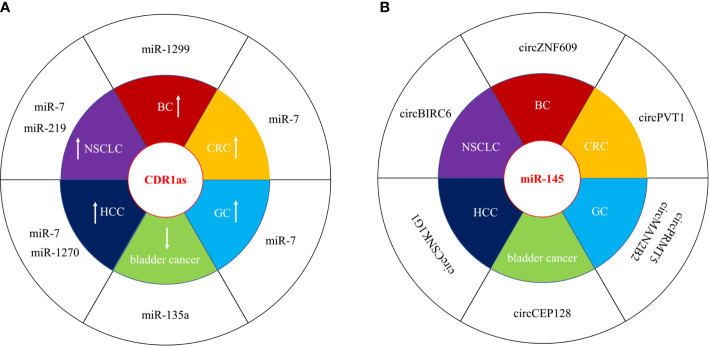
circRNAs can function as competing endogenous RNA (ceRNAs) or miRNA sponges to inhibit miRNA by interacting with the miRNA response elements (MREs), leading to the up-regulation of miRNA target gene. Numerous circRNAs participate to the progression of human cancer via acting as miRNA sponges in oncogenic or suppressive way. We have listed dys-regulated circRNAs with miRNA sponge potential in human cancer published in recent 5 years in Table 1, including bladder cancer, breast cancer (BC), colorectal cancer (CRC), gastric cancer (GC), hepatocellular carcinoma (HCC), and non-small-cell lung cancer (NSCLC). circRNAs show tissue specific expression and function in different human cancers. CDR1as (ciRS-7) was increased in BC, CRC, GC, HCC, and NSCLC, and acted as oncogenic, however, it was decreased and inhibited the progression of bladder cancer by sponging different target miRNAs (Figure 2A) (27, 29, 40, 56, 69, 87). One miRNA could be sponged by diverse circRNAs in different types of cancer. MiR-145 acted as a tumor suppressor in several cancers, and could be inhibited by a number of circRNAs, including circBIRC6, circCEP128, circMAN2B2, circPRMT5, circPVT1, circCSNK1G1, and circZNF609 (Figure 2B) (15, 52, 60, 65, 91, 107). The function of miRNA sponge is the most extensively studied role of circRNAs, although multi-functions of circRNAs have been identified.
Table 1.
circRNAs act as miRNA sponges in human cancer.
| Cancer | circRNA | Change | Target miRNA | Effect | Ref |
|---|---|---|---|---|---|
| Bladder cancer | circCEP128 | Up | miR-145 | oncogenic | (15, 16) |
| circFNTA | Up | miR-370-3p, miR-451a | oncogenic | (17, 18) | |
| circHIPK3 | Down | miR-558 | suppressive | (19) | |
| circITCH | Down | miR-17, miR-224 | suppressive | (20) | |
| circMTO1 | Down | miR-221 | suppressive | (21) | |
| circPTPRA | Down | miR-636 | suppressive | (22) | |
| circVANGL1 | Up | miR-605-3p, miR-1184 | oncogenic | (23, 24) | |
| circZFR | Up | miR-377 | oncogenic | (25) | |
| circZKSCAN1 | Down | miR-1178-3p | suppressive | (26) | |
| CDR1as | Down | miR-135a | suppressive | (27) | |
| circFNDC3B | Down | miR-1178-3p | suppressive | (28) | |
| BC | CDR1as | Up | miR-1299 | oncogenic | (29) |
| circABCB10 | Up | miR-1271 | oncogenic | (30) | |
| circAGFG1 | Up | miR-195 | oncogenic | (31) | |
| circANKS1B | Up | miR-148a, miR-152-3p | oncogenic | (32) | |
| circCDYL | Up | miR-1275 | oncogenic | (33) | |
| circDENND4C | Up | miR-200b, miR-200c | oncogenic | (34) | |
| circEPSTI1 | Up | miR-4753, miR-6809 | oncogenic | (35) | |
| circGFRA1 | Up | miR-34a | oncogenic | (36) | |
| circITCH | Down | miR-214, miR-17 | suppressive | (37) | |
| circUBAP2 | Up | miR-661 | oncogenic | (38) | |
| circZNF609 | Up | miR-145 | oncogenic | (39) | |
| CRC | CDR1as | Up | miR-7 | oncogenic | (40) |
| circANKS1B | Up | miR-149 | oncogenic | (41) | |
| circCDYL | Down | miR-150 | suppressive | (42) | |
| circCSPP1 | Up | miR-361 | oncogenic | (43) | |
| circDENND4C | Up | miR-760 | oncogenic | (44) | |
| circHIPK3 | Up | miR-7, miR-1207-5p | oncogenic | (45, 46) | |
| circITGA7 | Down | miR-370-3p, miR-3187-3p | suppressive | (47, 48) | |
| circMTO1 | Down | miR-19b | suppressive | (49) | |
| circPIP5K1A | Up | miR-1273a | oncogenic | (50) | |
| circPRMT5 | Up | miR-377 | oncogenic | (51) | |
| circPVT1 | Up | miR-145 | oncogenic | (52) | |
| circVAPA | Up | miR-101, miR-125a | oncogenic | (53, 54) | |
| circZNF609 | Up | miR-150 | oncogenic | (55) | |
| GC | CDR1as | Up | miR-7 | oncogenic | (56) |
| circHIAT1 | Down | miR-21 | suppressive | (57) | |
| circHIPK3 | Up | miR-107 | oncogenic | (58) | |
| circLARP4 | Down | miR-424 | suppressive | (59) | |
| circMAN2B2 | Up | miR-145 | oncogenic | (60) | |
| circMTO1 | Down | miR-3200-5p | suppressive | (61) | |
| circPIP5K1A | Up | miR-376c, miR-671 | oncogenic | (62, 63) | |
| circPRKCI | Up | miR-545 | oncogenic | (64) | |
| circPRMT5 | Up | miR-145 | oncogenic | (65) | |
| circPVT1 | Up | miR-125 | oncogenic | (66) | |
| circZFR | Down | miR-130a, miR-107 | suppressive | (67) | |
| circZNF609 | Up | miR-145 | oncogenic | (68) | |
| HCC | CDR1as | Up | miR-7, miR-1270 | oncogenic | (69, 70) |
| circABCB10 | Up | miR-670-3p | oncogenic | (71) | |
| circBIRC6 | Up | miR-3918 | oncogenic | (72) | |
| circCDYL | Up | miR-892a, miR-328-3p | oncogenic | (73) | |
| circHIAT1 | Down | miR-3171 | suppressive | (74) | |
| circHIPK3 | Up | miR-124 | oncogenic | (75) | |
| circLARP4 | Down | miR-761 | suppressive | (76) | |
| circMAN2B2 | Up | miR-217 | oncogenic | (77) | |
| circMTO1 | Down | miR-9 | suppressive | (78) | |
| circMYLK | Up | miR-362-3p | oncogenic | (79) | |
| circPRKCI | Up | miR-545 | oncogenic | (80) | |
| circPRMT5 | Up | miR-188 | oncogenic | (81) | |
| circPVT1 | Up | miR-203, miR-3666 | oncogenic | (82, 83) | |
| circVAPA | Up | miR-377 | oncogenic | (84) | |
| circZFR | Up | miR-511 | oncogenic | (85) | |
| circZNF652 | Up | miR-203, miR-502 | oncogenic | (86) | |
| NSCLC | CDR1as | Up | miR-7, miR-219 | oncogenic | (87–89) |
| circAGFG1 | Up | miR-203 | oncogenic | (90) | |
| circBIRC6 | Up | miR-145 | oncogenic | (91) | |
| circFOXM1 | Up | miR-614, miR-1304-5p | oncogenic | (92, 93) | |
| circGFRA1 | Up | miR-183-3p | oncogenic | (94) | |
| circHIPK3 | Up | miR-149 | oncogenic | (95) | |
| circNT5E | Up | miR-134 | oncogenic | (96) | |
| circPIP5K1A | Up | miR-136, miR-142-5p | oncogenic | (97, 98) | |
| circPRMT5 | Up | miR-377, miR-382, miR-498 | oncogenic | (99) | |
| circPTPRA | Down | miR-96 | suppressive | (100) | |
| circPVT1 | Up | miR-125b, miR-497 | oncogenic | (101, 102) | |
| circSMARCA5 | Down | miR-19b | suppressive | (103) | |
| circVANGL1 | Up | miR-195 | oncogenic | (104) | |
| circZFR | Up | miR-101 | oncogenic | (105) | |
| circZKSCAN1 | Up | miR-330-5p | oncogenic | (106) |
CRC, colorectal cancer; HCC, hepatocellular carcinoma; NSCLC, non-small-cell lung cancer.
Figure 2.
circRNAs act as miRNA sponges in human cancer. (A) CDR1as was both oncogenic and suppressive in different types of cancer. “↑” represented upregulated and oncogenic, “↓” represented downregulated and suppressive. (B) miR-145 wad sponged by several circRNAs in different types of cancer.
circRNA-Protein Interaction in Human Cancer
circRNA-protein interaction is considered as the second-most important function of circRNAs (108). RBPs are the most well-known circRNA interacting molecules, and a number of RBPs are involved in circRNAs-mediated oncogenic or suppressive functions in human cancer. circRNAs may act as protein sponge, protein recruitment, or protein scaffolding to regulate transcription/translation directly or indirectly. Moreover, circRNA-protein interaction also contributes to the post-transcriptional regulation of target protein, including ubiquitination and phosphorylation mediated degradation. These functions are described in Table 2.
Table 2.
circRNA-protein interaction in human cancer.
| circRNA | Type | Cancer | Change | RBP | Location | Effect | Ref |
|---|---|---|---|---|---|---|---|
| circERBB2 (hsa_circ_0007766) | EcRNA | Gallbladder cancer | Up | – | nucleus | transcription regulation | (109) |
| circHuR (hsa_circ_0049027) | EcRNA | Gastric cancer | Down | CNBP | nucleus | transcription regulation | (110) |
| circDONSON (hsa_circ_0004339) | EcRNA | Gastric cancer | Up | – | nucleus | transcription regulation | (111) |
| circDNMT1 (hsa_circ_0049224) | EcRNA | Breast cancer | Up | – | nucleus | transcription regulation | (112) |
| circXIAP (hsa_circ_0005276) | EcRNA | Prostate cancer | Up | FUS | cytoplasm/nucleus | transcription regulation | (113) |
| circCUX1 (hsa_circ_0132813) | EcRNA | Neuroblastoma | Up | EWSR1 | nucleus | transcription regulation | (114) |
| circCTNNB1 (hsa_circ_0123778) | CiRNA | Gastric cancer | Up | DDX3 | nucleus | transcription regulation | (115) |
| circSMARCA5 (hsa_circ_0001445) | EIcRNA | Glioblastoma | Down | SRSF1 | – | transcription regulation | (116) |
| circAGFG1 | – | Cervical cancer | Up | EZH2 | nucleus | transcription regulation | (117) |
| circRHOT1 | – | HCC | Up | – | nucleus | transcription regulation | (118) |
| circSMURF2 | – | PTC | Up | – | nucleus | transcription regulation | (119) |
| circCTIC1 | – | Colon cancer | Up | – | nucleus | transcription regulation | (120) |
| circSOX4 (hsa_circ_0131457) | EcRNA | NSCLC | – | – | – | Wnt/β-catenin pathway | (121) |
| circZKSCAN1 | EcRNA | HCC | Down | FMRP | cytoplasm | Wnt/β-catenin pathway | (122) |
| circE2F3 (hsa_circ_0075804) | EcRNA | Retinoblastoma | Up | HNRNPK | cytoplasm | translation regulation | (123) |
| circBACH1 (hsa_circ_0061395) | EcRNA | HCC | Up | HuR | cytoplasm/nucleus | translation regulation | (124) |
| circFOXO3 (hsa_circ_0006404) | EcRNA | Breast cancer | Down | – | – | ubiquitination | (125) |
| circADD3 (hsa_circ_0020007) | EcRNA | HCC | Down | EZH2 | – | ubiquitination | (126) |
| circNOL10 | EcRNA | Lung cancer | Down | – | nucleus | ubiquitination | (127) |
| circGSK3β (hsa_circ_0007986) | EcRNA | ESCC | Up | – | cytoplasm | phosphorylation | (128) |
| circPTK2 (hsa_circ_0005273) | EcRNA | CRC | Up | – | – | EMT | (129) |
| circSKA3 | – | Breast cancer | Up | – | cytoplasm | invasion | (130) |
| circAGO2 (hsa_circ_0135889) | CiRNA | Gastric cancer | Up | HuR | cytoplasm/nucleus | Ago2/miRNA | (131) |
PTC, papillary thyroid carcinoma; ESCC, esophageal squamous cell carcinoma. “–” means none or unavailable.
circRNA-Protein Interaction Regulates Transcription
EcRNAs are usually located in cytoplasm, while CiRNAs are mainly located in nucleus, and regulate Pol II transcription (132). circCTNNB1, a CiRNA derived from introns of β-catenin (CTNNB1), bound DDX3 to facilitate DDX3-mediated transactivation of transcription factor Yin Yang 1 (YY1) (115). EIcRNAs can regulate the transcription of host gene. circCUX1 interacted with EWSR1 and facilitated EWSR1-mediated MYC-associated zinc finger protein (MAZ) transactivation, resulting in transcriptional activation of its host gene CUX1 in neuroblastoma (114).
Accumulating evidence shows that ecircRNAs may also be enriched in nucleus and regulate transcription or splicing. circERBB2, containing exons 3 to 7 of ERBB2, bound and promoted nucleolus localization of PA2G4, a modulator of ribosomal DNA (rDNA) transcription, to form a circERBB2-PA2G4-TIFIA complex and increase Pol I activity and rDNA transcription in gallbladder cancer (109). circDONSON interacted with SNF2L subunit of the nucleosome-remodeling factor (NURF) complex and recruited the complex to the promoter region of SOX4 and initiates SOX4 transcription in gastric cancer (111). circHuR, consisting of exons 3 to 5 of HuR, interacted with the RBP CCHC-type zinc finger nucleic acid binding protein (CNBP) and restrained its binding to HuR promoter, resulting in down-regulation of HuR and repression of tumor progression (110).
circRNA-Protein Interaction Regulates Translation
EcRNAs may be involved in the regulation of translation via interacting with RBPs in cytoplasm. Has_circ_0075804 enhanced E2F transcription factor 3 (E2F3) mRNA stability through binding with HNRNPK, and promoted the proliferation of retinoblastoma in E2F3 dependent manner (123). circBACH1 bound HuR and induced its translocation from the nucleus to the cytoplasm, where HuR disrupted the IRES in the 5’-UTR of p27 and inhibited p27 translation, leading to the promotion of HCC growth (124).
circRNA-Protein Interaction Regulates Protein Degradation
Post-transcriptional regulation of proteins, including ubiquitination and phosphorylation, are linked to protein degradation. circFOXO3 recruited both MDM2 and p53 as protein scaffolding, enhancing MDM2 mediated poly-ubiquitination and degradation of p53. The increased interaction of MDM2 and p53 released Foxo3, another target protein of MDM2, and induced apoptosis by up-regulation of the Foxo3 downstream target in breast cancer (125). circADD3 interacts with EZH2 and CDK1 to promote CDK1-mediated EZH2 ubiquitination, increasing the expression of metastasis suppressors due to EZH2 induced H3K27me3 (126).
circRNA can also prevent protein degradation by inhibiting ubiquitination or phosphorylation. circNOL10 interacted with sex comb on midleg-like 1 (SCML1) to prevent its ubiquitination, and promoted the SCML1 mediated transcriptional regulation of the HN family to inhibits lung cancer development (127). circGSK3β interacted with GSK3β to release β-catenin from GSK3β mediated phosphorylation and degradation, promoting metastasis of ESCC (128).
circRNAs Act as Template for Translation in Human Cancer
circRNAs are considered as non-coding RNAs (ncRNAs), due to the absence of 5’ and 3’ ends. Emerging evidence shows circRNAs can encode regulatory peptides. Chen et al. firstly observed Initiation of protein synthesis in artificial constructs of circRNAs in vitro (133). Endogenous circRNAs can also function as translation templates (134, 135). The IRES, initiation codon AUG, and ORF have been found in numerous circRNAs, indicating their protein-coding potentials. Mechanically, IRES- and N6-methyladenosines (m6A)-mediated translation initiation contribute to circRNA translation (136). We have listed circRNAs acting as template for translation in human cancer in Table 3.
Table 3.
circRNAs encoded in human cancer.
| circRNA | Type | Cancer | Change | Length | ORF | Peptide | Effect | Ref |
|---|---|---|---|---|---|---|---|---|
| circFNDC3B (hsa_circ_0006156) | EcRNA | CRC | Down | 526bp | 651bp | 218aa | suppressive | (137) |
| circFBXW7 | EcRNA | Glioma | Down | 620bp | 552bp | 185aa | suppressive | (138) |
| circAKT3 (hsa_circ_0017250) | EcRNA | Glioblastoma | Down | 524bp | 519bp | 174aa | suppressive | (139) |
| circSHPRH (hsa_circ_0001649) | EcRNA | Glioma | Down | 440bp | 435bp | 146aa | suppressive | (140) |
| circLNC-PINT | EcRNA | Glioblastoma | Down | 1084bp | 258bp | 87aa | suppressive | (11) |
| circβ-catenin (hsa_circ_0004194) | EcRNA | Liver cancer | Up | 1129bp | 1080bp | 370aa | oncogenic | (141) |
| circPPP1R12A (hsa_circ_0000423) | EcRNA | Colon cancer | Up | 1138bp | 216bp | 73aa | oncogenic | (142) |
| circLgr4 | – | CRC | Up | – | 54bp | 19aa | oncogenic | (143) |
| circGprc5a | – | Bladder cancer | Up | – | 30bp | 11aa | oncogenic | (144) |
“–” means none or unavailable.
circFBXW7, down-regulated in both glioma and breast cancer, was able to encode a 185 amino-acid (aa) peptide named FBXW7-185aa. FBXW7-185aa competitively interacted with USP28, release c-Myc from USP28 induced de-ubiquitination, leading to c-Myc ubiquitination and degradation (138, 145). CircSHPRH, another suppressive circRNA decreased in glioma, carried a unique sequence “UGAUGA” which contained both the initiation codon “UGAUGA” and termination codon is “UGAUGA” with overlapping. circSHPRH encoded a 146aa peptide named SHPRH-146aa, which protected SHPRH from ubiquitination mediated degradation, leading to suppression of cell proliferation and tumorigenicity (140). circAKT3 was the second circRNA contained overlapped start-stop codons, which carried a unique sequence “UAAUGA” including both the initiation codon “UAAUGA” and termination codon is “UAAUGA” with overlapping. circAKT3 encoded a 174aa peptide named AKT3-174aa, which negatively modulated the PI3K/AKT signal pathway in glioblastoma by competitively interacting with phosphorylated PDK1 (139). circFNDC3B, a 526bp ecircRNA derived from exons 5 and 6 of FNDC3B, harbored a predicted ORF of 651bp and took more than one whole circle to translate a novel 218aa peptide named circFNDC3B-218aa. circFNDC3B-218aa alleviated the suppressive effect of Snail on FBP1 to inhibit EMT in colon cancer (137). These studies suggest that the ORFs in circRNAs are quite different from linear mRNAs, and may be even longer than the circRNA itself due to the circle structure.
Roles of circRNAs in Human Cancer
circRNAs are responsible for various cellular processes including proliferation, differentiation, and apoptosis. A number of literatures have explained the role of circRNAs in cancer progression, including proliferation and apoptosis, angiogenesis, EMT and metastasis, tumor microenvironment, and drug resistance. These circRNAs act as tumor promoters or suppressors. circRNAs are highly stable in serum and exempted from endonuclease degradation, showing remarkable potential of cancer diagnostic and prognostic biomarkers.
circRNAs and Cell Proliferation and Apoptosis in Human Cancer
CircRNA expression profiling by RNA sequencing or microarray is indispensable to explore novel oncogenic and tumor suppressive circRNAs and to elucidate the underlying mechanisms. By using circRNA microarray, Sun et al. identified that circLRIG3 (hsa_circ_0027345), derived from the back splicing of pre-LRIG3 mRNA exons 2–11, was increased in HCC. They further tested the expression of circLRIG3 in 130 pairs of HCC and para-cancerous normal tissues. circLRIG3 was remarkably upregulated in HCC patients and HCC cell lines. High expression of circLRIG3 was associated with poor prognosis in HCC patients. Overexpression of circLRIG3 increased HepG2 cell viability by promoting proliferation and reducing apoptosis, and circLRIG3 depletion led to opposite effects. The gene-set enrichment analysis (GSEA) results revealed the enrichment of STAT3 pathway after circLRIG3 overexpression. circLRIG3 facilitated EZH2-induced STAT3 methylation and phosphorylation by forming a ternary complex with EZH2 and STAT3, leading to the activation of STAT3 signaling to promote proliferation and suppress apoptosis of HCC cells (146).
circRNAs and Angiogenesis in Human Cancer
Tumor angiogenesis is a process of new blood vessels formation derived from already existing ones and it is a critical process for tumor growth and metastasis, as the newly formed blood vessels provide oxygen, nutrition, and growth factors. Angiogenesis is a consequence of hypoxic and ischemic signals under physiological conditions, whereas, it is uncontrolled and upregulated under pathological conditions. Pathological angiogenesis is characterized by increased proliferation of the endothelial cells and atypical morphology of tumor vasculature (147). Chen et al. found that circERBIN (has_circ_0001492), formed by circularization of exons 2–4 of the ERBIN gene, was significantly increased in CRC cells. circERBIN was higher in stage III/IV tissues than in stage I/II CRC samples. Tumors formed by CRC cells stably expressing circERBIN grew larger and more quickly. The number of microvessels were greatly increased in circERBIN overexpression tumors as analyzed by staining CD31, a sensitive vascular marker to evaluate tumor angiogenesis. They further confirmed that circERBIN promoted tumor angiogenesis by increasing a cap-independent protein translation of HIF-1α through miR-125a5p/miR-138-5p/4EBP-1 signaling (148).
circRNAs and EMT in Human Cancer
EMT is an essential prerequisite for cancer metastasis, which facilitates cancer cells to transfer from epithelial phenotype to mesenchymal traits and to enhance the mobility of cancer cells to invade and metastasize. Liu et al. screened differently expressed circRNAs in oral squamous cell carcinoma (OSCC) with circRNA microarray, and found that circIGHG (hsa_circ_0000579) was upregulated by 109 folds in OSCC with the largest number of potential miRNA targets. circIGHG was increased in both OSCC patients and cell lines. High expression of circIGHG was correlated with poor prognosis in OSCC patients. miR-142-5p/IGF2BP3 pathway was further confirmed as the downstream target of circIGHG in OSCC cell lines. circIGHG promoted the expression of several EMT markers to influence OSCC metastasis, including ZEB-2 and SNAI. circIGHG was also found to promote EMT in vivo through xenograft tumors. Finally, circIGHG promoted OSCC metastasis via inducing IGF2BP3-mediated EMT by sponging miR-142-5p (149).
circRNAs and Tumor Microenvironment
The tumor microenvironment (TME) consists of stromal cells, extracellular matrix components, endothelial cells, vasculature, immune cells, and various signaling entities. Different cell types within the TME form organ-like structures and play specific roles during tumor metastasis, including adipocytes, endothelial cells, fibroblasts, immune cells, and neuroendocrine cells. The crosstalk between TME components and tumor cells and drives tumor metastasis. Endothelial cells are a crucial component of the TME, which participate in angiogenesis and development of tumors. Yan et al. screened differently expressed circRNAs using RNA sequencing in human umbilical vein endothelial cells (HUVECs) under an HCC microenvironment. Two of the most downregulated circRNAs were validated by qPCR, circ_4911 and circ_4302. Overexpression of the two circRNAs both arrested cells at the GO/G1 stage and suppressed the proliferation and migration of HUVECs under an HCC microenvironment (150).
circRNAs and Drug Resistance in Human Cancer
Chemotherapy and molecular targeted drug therapy are the main treatment options for cancers, as most cancers have progressed to the middle or late stages when diagnosed (151). The most common therapeutic drugs include 5-fluorouracil, cisplatin, epidermal growth factor receptor-tyrosine kinase inhibitors (EGFR-TKIs), oxaliplatin, and sorafenib. Drug resistance occurred after long-term therapy and leads to poor prognosis (152). circRNA expression profiles analysis have identified differentially expressed circRNAs between drug resistant and drug sensitive cancer patients and cell models (153), and functional studies have further explored the underling mechanisms of circRNAs involved in drug resistance in human cancer (154). By using RNA sequencing and qPCR, Huang et al. found that circAKT3 (hsa_circ_0000199) was increased in cisplatin-resistant GC cell lines. Consistent with the cell lines results, circAKT3 was significantly higher in the cisplatin-resistant GC tissues than in the sensitive tissues. Mechanically, circAKT3 increased the expression of PIK3R1and activated the PI3K/AKT signaling pathway by sponging miR-198, to promote DNA damage repair and to suppress apoptosis of GC cells (155). On the other hand, some circRNAs may act to sensitize cancer cells to drugs. circMCTP2 was found to be decreased in both cisplatin-resistant GC tissues and cells compared to cisplatin-sensitive GC tissues and cells. Moreover, a high level of circMCTP2 was correlated with a better prognosis of GC patients. circMCTP2 inhibited proliferation and autophagy while promoting apoptosis of cisplatin-resistant GC cells via sponging the miR-99a-5p/MTMR3 pathway (156).
Circulatory circRNAs in Human Cancer
Screening biomarkers in extracellular body fluids is essential in the era of precision medicine. Whole circulating transcriptome has been applied in search of cancer biomarkers. In the era of precision medicine, the main objective of medical oncology is to promote the diagnosis and treatment of cancer. Liquid biopsy” (LB), aiming to screen highly sensitive and specific biomarkers for diagnosis and prognosis by non-invasive or minimally invasive means, has attracted much attention. A recent review on circRNAs including 77 studies in several types of cancers suggested the potential role of serum circRNAs as diagnostic biomarkers for cancer. This meta-analysis found that plasma circRNAs showed higher diagnostic accuracy than tissue. Moreover, combined circRNAs panel had good diagnostic efficacy for GC (157). Exosomes are phospholipid bilayer nanovesicles with a diameter of 30–150 nm in length which contain a range of molecules, including proteins, miRNAs, and circRNAs. Exosomes secreted from cancer cells often reflect the molecular characteristics of cancer cells and affect their tumorigenic potential. The biomarkers detected within exosomes were highly correlate with the tissue analysis. Furthermore, circRNAs within exosomes are protected from being degraded during circulation due to the lipid bilayer membrane of exosomes. Exosomes are relatively more stable than serum at room temperature. These aspects may suggest exosomal circRNAs good diagnostic and prognostic biomarkers (158).
circRNA Databases
By the advances of bioinformatics, a number of databases have been provided including comprehensive databases with circRNA annotation, primer design, IRES and ORF prediction. There are also tissue specific and cancer specific circRNA databases (159, 160). CircBase, circRNADb, and CircInteractome are the most commonly used comprehensive databases (161–163). Recently, a variety of bioinformatic tools for predicting circRNA coding potential have been reported, including IRES prediction and ORF assessment (164, 165). We have listed these circRNA databases in Table 4.
Table 4.
circRNA databases.
| Database | Website | Function | Ref |
|---|---|---|---|
| circBase | http://www.circbase.org/ | circRNA database | (161) |
| circAtlas 2.0 | http://circatlas.biols.ac.cn/ | comprehensive database of circRNAs across species | (166) |
| TSCD | http://gb.whu.edu.cn/TSCD/ | tissue-specific circRNA database | (159) |
| CSCD | http://gb.whu.edu.cn/CSCD/ | cancer-specific circRNA database | (160) |
| deepBase v2.0 | http://biocenter.sysu.edu.cn/deepBase/ | circRNA database | (167) |
| circRNADisease | http://cgga.org.cn:9091/circRNADisease/ | circRNA and disease associations database | (168) |
| CircBank | http://www.circbank.cn/index.html | annotation and function prediction. | (169) |
| CircInteractome | https://circinteractome.nia.nih.gov/ | primer design and function prediction | (163) |
| circRNADb | http://reprod.njmu.edu.cn/cgi-bin/circrnadb/circRNADb.php | annotation and function prediction | (162) |
| Circ2Traits | http://gyanxet-beta.com/circdb/ | annotation and function prediction | (170) |
| circNet | http://syslab5.nchu.edu.tw/CircNet/ | circRNA-miRNA prediction | (171) |
| RegRNA2.0 | http://regrna2.mbc.nctu.edu.tw/index.html | platform for functional RNA motif prediction | (172) |
| RNAstructure | http://rna.urmc.rochester.edu/RNAstructureWeb/ | secondary structure and RNA-RNA binding prediction | (173) |
| starBase v2.0 | http://starbase.sysu.edu.cn/ | circRNA-RNA and circRNA-RBP interaction | (174) |
| catRAPID | http://s.tartaglialab.com/page/catrapid_group | circRNA-protein interaction prediction | (175) |
| RPISeq | http://pridb.gdcb.iastate.edu/RPISeq/ | circRNA-protein interaction prediction | (176) |
| IRESite | http://www.iresite.org | database of experimentally verified IRES structures | (177) |
| IRESfinder | https://github.com/xiaofengsong/IRESfinder | IRES prediction | (178) |
| IRESPred | http://bioinfo.net.in/IRESPred/ | IRES prediction | (164) |
| SORFS | http://www.sorfs.org/ | ORF database | (179) |
| ORFfinder | https://www.ncbi.nlm.nih.gov/orffinder/ | ORF prediction | – |
| PhyloCSF | http://compbio.mit.edu/PhyloCSF | ORF prediction | (180) |
| OrfPredictor | http://bioinformatics.ysu.edu/tools/OrfPredictor.html | ORF prediction | (165) |
| SMS : ORF Finder | http://www.bioinformatics.org/sms2/orf_find.html | ORF prediction | (181) |
| CPC | http://cpc.cbi.pku.edu.cn | coding-potential assessment | (182) |
| CPAT | http://lilab.research.bcm.edu/cpat/index.php | coding-potential assessment | (183) |
| CircPro | http://bis.zju.edu.cn/CircPro | coding-potential assessment | (184) |
| CircCode | https://github.com/PSSUN/CircCode | coding-potential assessment | (185) |
Conclusions and Perspectives
Advances of deep sequencing and algorithms result in the rapid increasement of circRNA number. Developments in molecular biology, bioinformatics and computational analysis, and proteomics have driven the function of circRNAs from miRNA sponge to circRNA-RNA/circRNA-protein interaction, and circRNA coding potential. Many traditional ideas need to be corrected. Traditionally, EcRNAs are thought to be expressed and function in cytoplasm, while EIcRNAs and CiRNAs are in nucleus. However, accumulating evidence shows that EcRNAs were also enriched in nucleus and regulate transcription or splicing in human cancer, such as circERBB2 (109), circHuR (110), circDONSON (111), circDNMT1 (112), and circXIAP (113). Moreover, CiRNA can also function in cytoplasm. CircAGO2 interacted with HuR to facilitate its shuttling from nucleus to cytoplasm, where HuR was activated and enriched on the 3’-UTR of target mRNA, preventing AGO2 binding and subsequent AGO2/miRNA-mediated gene silencing (131). Beside of miRNA, circRNAs can bind to pri-miRNA, promoter/coding region/3’-UTR of linear mRNA directly (186–189). These studies suggest the regulatory effects of circRNAs on transcription, translation, and pri-miRNA processing as important RNA-binding molecules.
There are still some issues needed to be clarified urgently. A standard nomenclature system and updated comprehensive databases for circRNAs are quite necessary due to the rapid growth in the number of circRNAs. Although circBase is the most frequently used database, its last update was in 2017. CircAtlas, recently published by Wu et al., includes 1,007,087 highly reliable circRNAs, and over 81.3% of these circRNAs have been assembled into full-length sequences (166). Currently, the nomenclature was inconsistent. circFOXM1 was found as circFOXM1, circ-FOXM1, and circ_0025033 in Pubmed. circFBXW7 has been reported to encode a novel 185aa peptide driven by IRES. Yang et al. found that the 185aa peptide was encoded by circFBXW7 in glioma, derived from exons 3 and 4 of FBXW7 with a length of 620 bp (138). Ye et al. later reported that hsa_circ_0001451, derived from FBXW7, also encoded this peptide, while hsa_circ_0001451 was 1,227 bp in length longer than 620 bp (145). A standard nomenclature system may help circRNA research.
In the future, the mystery of circRNAs will be further uncovered in the following aspects. First, the regulation of biogenesis and degradation of circRNAs, which may be involved in the tissue specific expression of circRNAs. RBPs, host gene, and methylation have been reported to regulate the biogenesis of circRNAs (12, 127), but the detailed molecular mechanisms remain unclear. How circRNAs are degraded also remains unclear. Fischer et al. revealed a structure-mediated circRNA decay by UPF1 and G3BP1 (190). Second, the driving mechanisms translatable circRNAs and functions of their translation products. IRES-driven or m6A-mediated initiation have been found to mediate circRNA translation (191). Third, circRNA-host gene interaction. The generation of circRNAs can be independent of host linear mRNA or compete with host mRNA splicing. circRNAs regulate the expression of host gene via sponging miRNAs which target host genes. Pre-mRNA methylation can also modify circRNA expression. How these circRNAs interact with their host genes in human cancer deserves further investigation. Last, circRNA as biomarker for diagnosis and treatment of cancers due to its stability.
In summary, circRNAs function as miRNA/protein sponge, protein scaffold, protein recruitment, enhancer of protein function, as well as templates for translation involved in the regulation of transcription/splicing, translation, protein degradation, and pri-miRNA processing in human cancers and contributed to the pathogenesis of cancer. By the advance of deep sequencing, bioinformatic algorithms, molecular biology, as well as proteomics, the mystery of circRNAs in human cancer will be uncovered gradually.
Author Contributions
LC and YL designed this review. XW and HL collected the related paper. XW wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by grants from the National Natural Science Foundation of China (81572071 and 81000331).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Abbreviations
circRNA, circular RNA; miRNA, microRNA; EcRNA, exonic circRNA; EIcRNA, exon-intron circRNA; CiRNA, intronic circRNA; tricRNA, tRNA intron derived circRNA; pri-miRNA, primary miRNA; 3’-UTR, 3’-untranslated region; IRES, internal ribosome entry site; ORF, open reading frame; RBP, RNA binding protein; ceRNA, competing endogenous RNA; MRE, miRNA response element; BC, breast cancer; CRC, colorectal cancer; GC, gastric cancer; HCC, hepatocellular carcinoma; NSCLC, non-small-cell lung cancer; m6A, N6-methyladenosine; CSC, cancer stem cell; LSCC, laryngeal squamous cell carcinoma; PDAC, pancreatic ductal adenocarcinoma; ESCC, esophageal squamous cell carcinoma; OSCC, oral squamous cell carcinoma; EOC, epithelial ovarian cancer.
References
- 1. Kristensen LS, Andersen MS, Stagsted LVW, Ebbesen KK, Hansen TB, Kjems J. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet (2019) 20(11):675–91. 10.1038/s41576-019-0158-7 [DOI] [PubMed] [Google Scholar]
- 2. Hsu MT, Coca-Prados M. Electron microscopic evidence for the circular form of RNA in the cytoplasm of eukaryotic cells. Nature (1979) 280(5720):339–40. 10.1038/280339a0 [DOI] [PubMed] [Google Scholar]
- 3. Capel B, Swain A, Nicolis S, Hacker A, Walter M, Koopman P, et al. Circular transcripts of the testis-determining gene Sry in adult mouse testis. Cell (1993) 73(5):1019–30. 10.1016/0092-8674(93)90279-y [DOI] [PubMed] [Google Scholar]
- 4. Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, et al. Natural RNA circles function as efficient microRNA sponges. Nature (2013) 495(7441):384–8. 10.1038/nature11993 [DOI] [PubMed] [Google Scholar]
- 5. Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature (2013) 495(7441):333–8. 10.1038/nature11928 [DOI] [PubMed] [Google Scholar]
- 6. Sun J, Li B, Shu C, Ma Q, Wang J. Functions and clinical significance of circular RNAs in glioma. Mol Cancer (2020) 19(1):34. 10.1186/s12943-019-1121-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Li Y, Zhang J, Huo C, Ding N, Li J, Xiao J, et al. Dynamic Organization of lncRNA and Circular RNA Regulators Collectively Controlled Cardiac Differentiation in Humans. EBioMedicine (2017) 24:137–46. 10.1016/j.ebiom.2017.09.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Schmidt CA, Giusto JD, Bao A, Hopper AK, Matera AG. Molecular determinants of metazoan tricRNA biogenesis. Nucleic Acids Res (2019) 47(12):6452–65. 10.1093/nar/gkz311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wu P, Mo Y, Peng M, Tang T, Zhong Y, Deng X, et al. Emerging role of tumor-related functional peptides encoded by lncRNA and circRNA. Mol Cancer (2020) 19(1):22. 10.1186/s12943-020-1147-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hanniford D, Ulloa-Morales A, Karz A, Berzoti-Coelho MG, Moubarak RS, Sanchez-Sendra B, et al. Epigenetic Silencing of CDR1as Drives IGF2BP3-Mediated Melanoma Invasion and Metastasis. Cancer Cell (2020) 37(1):55–70 e15. 10.1016/j.ccell.2019.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhang M, Zhao K, Xu X, Yang Y, Yan S, Wei P, et al. A peptide encoded by circular form of LINC-PINT suppresses oncogenic transcriptional elongation in glioblastoma. Nat Commun (2018) 9(1):4475. 10.1038/s41467-018-06862-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Barrett SP, Salzman J. Circular RNAs: analysis, expression and potential functions. Development (2016) 143(11):1838–47. 10.1242/dev.128074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zang Y, Li J, Wan B, Tai Y. circRNA circ-CCND1 promotes the proliferation of laryngeal squamous cell carcinoma through elevating CCND1 expression via interacting with HuR and miR-646. J Cell Mol Med (2020) 24(4):2423–33. 10.1111/jcmm.14925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wang S, Zhang Y, Cai Q, Ma M, Jin LY, Weng M, et al. Circular RNA FOXP1 promotes tumor progression and Warburg effect in gallbladder cancer by regulating PKLR expression. Mol Cancer (2019) 18(1):145. 10.1186/s12943-019-1078-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sun M, Zhao W, Chen Z, Li M, Li S, Wu B, et al. Circular RNA CEP128 promotes bladder cancer progression by regulating Mir-145-5p/Myd88 via MAPK signaling pathway. Int J Cancer (2019) 145(8):2170–81. 10.1002/ijc.32311 [DOI] [PubMed] [Google Scholar]
- 16. Wu Z, Huang W, Wang X, Wang T, Chen Y, Chen B, et al. Circular RNA CEP128 acts as a sponge of miR-145-5p in promoting the bladder cancer progression via regulating SOX11. Mol Med (2018) 24(1):40. 10.1186/s10020-018-0039-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chen J, Sun Y, Ou Z, Yeh S, Huang CP, You B, et al. Androgen receptor-regulated circFNTA activates KRAS signaling to promote bladder cancer invasion. EMBO Rep (2020) 21(4):e48467. 10.15252/embr.201948467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tian J, Fan J, Xu J, Ren T, Guo H, Zhou L. circ-FNTA accelerates proliferation and invasion of bladder cancer. Oncol Lett (2020) 19(1):1017–23. 10.3892/ol.2019.11150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Li Y, Zheng F, Xiao X, Xie F, Tao D, Huang C, et al. CircHIPK3 sponges miR-558 to suppress heparanase expression in bladder cancer cells. EMBO Rep (2017) 18(9):1646–59. 10.15252/embr.201643581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yang C, Yuan W, Yang X, Li P, Wang J, Han J, et al. Circular RNA circ-ITCH inhibits bladder cancer progression by sponging miR-17/miR-224 and regulating p21, PTEN expression. Mol Cancer (2018) 17(1):19. 10.1186/s12943-018-0771-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Li Y, Wan B, Liu L, Zhou L, Zeng Q. Circular RNA circMTO1 suppresses bladder cancer metastasis by sponging miR-221 and inhibiting epithelial-to-mesenchymal transition. Biochem Biophys Res Commun (2019) 508(4):991–6. 10.1016/j.bbrc.2018.12.046 [DOI] [PubMed] [Google Scholar]
- 22. He Q, Huang L, Yan D, Bi J, Yang M, Huang J, et al. CircPTPRA acts as a tumor suppressor in bladder cancer by sponging miR-636 and upregulating KLF9. Aging (2019) 11(23):11314–28. 10.18632/aging.102530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zeng Z, Zhou W, Duan L, Zhang J, Lu X, Jin L, et al. Circular RNA circ-VANGL1 as a competing endogenous RNA contributes to bladder cancer progression by regulating miR-605-3p/VANGL1 pathway. J Cell Physiol (2019) 234(4):3887–96. 10.1002/jcp.27162 [DOI] [PubMed] [Google Scholar]
- 24. Yang D, Qian H, Fang Z, Xu A, Zhao S, Liu B, et al. Silencing circular RNA VANGL1 inhibits progression of bladder cancer by regulating miR-1184/IGFBP2 axis. Cancer Med (2020) 9(2):700–10. 10.1002/cam4.2650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhang WY, Liu QH, Wang TJ, Zhao J, Cheng XH, Wang JS. CircZFR serves as a prognostic marker to promote bladder cancer progression by regulating miR-377/ZEB2 signaling. Biosci Rep (2019) 39(12):1–10. 10.1042/BSR20192779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bi J, Liu H, Dong W, Xie W, He Q, Cai Z, et al. Circular RNA circ-ZKSCAN1 inhibits bladder cancer progression through miR-1178-3p/p21 axis and acts as a prognostic factor of recurrence. Mol Cancer (2019) 18(1):133. 10.1186/s12943-019-1060-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Li P, Yang X, Yuan W, Yang C, Zhang X, Han J, et al. CircRNA-Cdr1as Exerts Anti-Oncogenic Functions in Bladder Cancer by Sponging MicroRNA-135a. Cell Physiol Biochem Int J Exp Cell Physiol Biochem Pharmacol (2018) 46(4):1606–16. 10.1159/000489208 [DOI] [PubMed] [Google Scholar]
- 28. Liu H, Bi J, Dong W, Yang M, Shi J, Jiang N, et al. Invasion-related circular RNA circFNDC3B inhibits bladder cancer progression through the miR-1178-3p/G3BP2/SRC/FAK axis. Mol Cancer (2018) 17(1):161. 10.1186/s12943-018-0908-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sang M, Meng L, Liu S, Ding P, Chang S, Ju Y, et al. Circular RNA ciRS-7 Maintains Metastatic Phenotypes as a ceRNA of miR-1299 to Target MMPs. Mol Cancer Res MCR (2018) 16(11):1665–75. 10.1158/1541-7786.MCR-18-0284 [DOI] [PubMed] [Google Scholar]
- 30. Liang HF, Zhang XZ, Liu BG, Jia GT, Li WL. Circular RNA circ-ABCB10 promotes breast cancer proliferation and progression through sponging miR-1271. Am J Cancer Res (2017) 7(7):1566–76. [PMC free article] [PubMed] [Google Scholar]
- 31. Yang R, Xing L, Zheng X, Sun Y, Wang X, Chen J. The circRNA circAGFG1 acts as a sponge of miR-195-5p to promote triple-negative breast cancer progression through regulating CCNE1 expression. Mol Cancer (2019) 18(1):4. 10.1186/s12943-018-0933-7 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 32. Zeng K, He B, Yang BB, Xu T, Chen X, Xu M, et al. The pro-metastasis effect of circANKS1B in breast cancer. Mol Cancer (2018) 17(1):160. 10.1186/s12943-018-0914-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Liang G, Ling Y, Mehrpour M, Saw PE, Liu Z, Tan W, et al. Autophagy-associated circRNA circCDYL augments autophagy and promotes breast cancer progression. Mol Cancer (2020) 19(1):65. 10.1186/s12943-020-01152-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ren S, Liu J, Feng Y, Li Z, He L, Li L, et al. Knockdown of circDENND4C inhibits glycolysis, migration and invasion by up-regulating miR-200b/c in breast cancer under hypoxia. J Exp Clin Cancer Res CR (2019) 38(1):388. 10.1186/s13046-019-1398-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chen B, Wei W, Huang X, Xie X, Kong Y, Dai D, et al. circEPSTI1 as a Prognostic Marker and Mediator of Triple-Negative Breast Cancer Progression. Theranostics (2018) 8(14):4003–15. 10.7150/thno.24106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. He R, Liu P, Xie X, Zhou Y, Liao Q, Xiong W, et al. circGFRA1 and GFRA1 act as ceRNAs in triple negative breast cancer by regulating miR-34a. J Exp Clin Cancer Res CR (2017) 36(1):145. 10.1186/s13046-017-0614-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wang ST, Liu LB, Li XM, Wang YF, Xie PJ, Li Q, et al. Circ-ITCH regulates triple-negative breast cancer progression through the Wnt/beta-catenin pathway. Neoplasma (2019) 66(2):232–9. 10.4149/neo_2018_180710N460 [DOI] [PubMed] [Google Scholar]
- 38. Wang S, Li Q, Wang Y, Li X, Wang R, Kang Y, et al. Upregulation of circ-UBAP2 predicts poor prognosis and promotes triple-negative breast cancer progression through the miR-661/MTA1 pathway. Biochem Biophys Res Commun (2018) 505(4):996–1002. 10.1016/j.bbrc.2018.10.026 [DOI] [PubMed] [Google Scholar]
- 39. Wang S, Xue X, Wang R, Li X, Li Q, Wang Y, et al. CircZNF609 promotes breast cancer cell growth, migration, and invasion by elevating p70S6K1 via sponging miR-145-5p. Cancer Manage Res (2018) 10:3881–90. 10.2147/CMAR.S174778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Tang W, Ji M, He G, Yang L, Niu Z, Jian M, et al. Silencing CDR1as inhibits colorectal cancer progression through regulating microRNA-7. OncoTargets Ther (2017) 10:2045–56. 10.2147/OTT.S131597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Li D, Yang R, Yang L, Wang D, Zhou X, Sun Y. circANKS1B regulates FOXM1 expression and promotes cell migration and invasion by functioning as a sponge of the miR-149 in colorectal cancer. OncoTargets Ther (2019) 12:4065–73. 10.2147/OTT.S201310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Cui W, Dai J, Ma J, Gu H. circCDYL/microRNA-105-5p participates in modulating growth and migration of colon cancer cells. Gen Physiol Biophys (2019) 38(6):485–95. 10.4149/gpb2019037 [DOI] [PubMed] [Google Scholar]
- 43. Yang W, Xie T. Hsa_circ_CSPP1/MiR-361-5p/ITGB1 Regulates Proliferation and Migration of Cervical Cancer (CC) by Modulating the PI3K-Akt Signaling Pathway. Reprod Sci (2020) 27(1):132–44. 10.1007/s43032-019-00008-5 [DOI] [PubMed] [Google Scholar]
- 44. Zhang ZJ, Zhang YH, Qin XJ, Wang YX, Fu J. Circular RNA circDENND4C facilitates proliferation, migration and glycolysis of colorectal cancer cells through miR-760/GLUT1 axis. Eur Rev Med Pharmacol Sci (2020) 24(5):2387–400. 10.26355/eurrev_202003_20506 [DOI] [PubMed] [Google Scholar]
- 45. Zeng K, Chen X, Xu M, Liu X, Hu X, Xu T, et al. CircHIPK3 promotes colorectal cancer growth and metastasis by sponging miR-7. Cell Death Dis (2018) 9(4):417. 10.1038/s41419-018-0454-8 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 46. Yan Y, Su M, Qin B. CircHIPK3 promotes colorectal cancer cells proliferation and metastasis via modulating of miR-1207-5p/FMNL2 signal. Biochem Biophys Res Commun (2020) 524(4):839–46. 10.1016/j.bbrc.2020.01.055 [DOI] [PubMed] [Google Scholar]
- 47. Li X, Wang J, Zhang C, Lin C, Zhang J, Zhang W, et al. Circular RNA circITGA7 inhibits colorectal cancer growth and metastasis by modulating the Ras pathway and upregulating transcription of its host gene ITGA7. J Pathol (2018) 246(2):166–79. 10.1002/path.5125 [DOI] [PubMed] [Google Scholar]
- 48. Yang G, Zhang T, Ye J, Yang J, Chen C, Cai S, et al. Circ-ITGA7 sponges miR-3187-3p to upregulate ASXL1, suppressing colorectal cancer proliferation. Cancer Manage Res (2019) 11:6499–509. 10.2147/CMAR.S203137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Fan M, Wang Y, Gao S. Circular RNA circMTO1 acts as an antitumor factor in rectal cancer cell lines by downregulation of miR-19b-3p. J Cell Biochem (2019) 1:1. 10.1002/jcb.29570 [DOI] [Google Scholar]
- 50. Zhang Q, Zhang C, Ma JX, Ren H, Sun Y, Xu JZ. Circular RNA PIP5K1A promotes colon cancer development through inhibiting miR-1273a. World J Gastroenterol (2019) 25(35):5300–9. 10.3748/wjg.v25.i35.5300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Yang B, Du K, Yang C, Xiang L, Xu Y, Cao C, et al. CircPRMT5 circular RNA promotes proliferation of colorectal cancer through sponging miR-377 to induce E2F3 expression. J Cell Mol Med (2020) 24(6):3431–7. 10.1111/jcmm.15019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wang Z, Su M, Xiang B, Zhao K, Qin B. Circular RNA PVT1 promotes metastasis via miR-145 sponging in CRC. Biochem Biophys Res Commun (2019) 512(4):716–22. 10.1016/j.bbrc.2019.03.121 [DOI] [PubMed] [Google Scholar]
- 53. Li XN, Wang ZJ, Ye CX, Zhao BC, Huang XX, Yang L. Circular RNA circVAPA is up-regulated and exerts oncogenic properties by sponging miR-101 in colorectal cancer. Biomed Pharmacother = Biomed Pharmacother (2019) 112:108611. 10.1016/j.biopha.2019.108611 [DOI] [PubMed] [Google Scholar]
- 54. Zhang X, Xu Y, Yamaguchi K, Hu J, Zhang L, Wang J, et al. Circular RNA circVAPA knockdown suppresses colorectal cancer cell growth process by regulating miR-125a/CREB5 axis. Cancer Cell Int (2020) 20:103. 10.1186/s12935-020-01178-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Wu L, Xia J, Yang J, Shi Y, Xia H, Xiang X, et al. Circ-ZNF609 promotes migration of colorectal cancer by inhibiting Gli1 expression via microRNA-150. J BUON Off J Balkan Union Oncol (2018) 23(5):1343–9. [PubMed] [Google Scholar]
- 56. Pan H, Li T, Jiang Y, Pan C, Ding Y, Huang Z, et al. Overexpression of Circular RNA ciRS-7 Abrogates the Tumor Suppressive Effect of miR-7 on Gastric Cancer via PTEN/PI3K/AKT Signaling Pathway. J Cell Biochem (2018) 119(1):440–6. 10.1002/jcb.26201 [DOI] [PubMed] [Google Scholar]
- 57. Quan J, Dong D, Lun Y, Sun B, Sun H, Wang Q, et al. Circular RNA circHIAT1 inhibits proliferation and epithelial-mesenchymal transition of gastric cancer cell lines through downregulation of miR-21. J Biochem Mol Toxicol (2020) 34(4):e22458. 10.1002/jbt.22458 [DOI] [PubMed] [Google Scholar]
- 58. Wei J, Xu H, Wei W, Wang Z, Zhang Q, De W, et al. circHIPK3 Promotes Cell Proliferation and Migration of Gastric Cancer by Sponging miR-107 and Regulating BDNF Expression. OncoTargets Ther (2020) 13:1613–24. 10.2147/OTT.S226300 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 59. Zhang J, Liu H, Hou L, Wang G, Zhang R, Huang Y, et al. Circular RNA_LARP4 inhibits cell proliferation and invasion of gastric cancer by sponging miR-424-5p and regulating LATS1 expression. Mol Cancer (2017) 16(1):151. 10.1186/s12943-017-0719-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Sun B, Sun H, Wang Q, Wang X, Quan J, Dong D, et al. Circular RNA circMAN2B2 promotes growth and migration of gastric cancer cells by down-regulation of miR-145. J Clin Lab Anal (2020) 34:e23215. 10.1002/jcla.23215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Hu K, Qin X, Shao Y, Zhou Y, Ye G, Xu S. Circular RNA MTO1 suppresses tumorigenesis of gastric carcinoma by sponging miR-3200-5p and targeting PEBP1. Mol Cell Probes (2020) 52:101562. 10.1016/j.mcp.2020.101562 [DOI] [PubMed] [Google Scholar]
- 62. Ma Y, Cong X, Zhang Y, Yin X, Zhu Z, Xue Y. CircPIP5K1A facilitates gastric cancer progression via miR-376c-3p/ZNF146 axis. Cancer Cell Int (2020) 20:81. 10.1186/s12935-020-1122-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Song H, Xu Y, Xu T, Fan R, Jiang T, Cao M, et al. CircPIP5K1A activates KRT80 and PI3K/AKT pathway to promote gastric cancer development through sponging miR-671-5p. Biomed Pharmacother = Biomed Pharmacother (2020) 126:109941. 10.1016/j.biopha.2020.109941 [DOI] [PubMed] [Google Scholar]
- 64. Wu L, Li Y, Xu XM, Zhu X. Circular RNA circ-PRKCI promotes cell proliferation and invasion by binding to microRNA-545 in gastric cancer. Eur Rev Med Pharmacol Sci (2019) 23(21):9418–26. 10.26355/eurrev_201911_19435 [DOI] [PubMed] [Google Scholar]
- 65. Du W, Li D, Guo X, Li P, Li X, Tong S, et al. Circ-PRMT5 promotes gastric cancer progression by sponging miR-145 and miR-1304 to upregulate MYC. Artif cells Nanomed Biotechnol (2019) 47(1):4120–30. 10.1080/21691401.2019.1671857 [DOI] [PubMed] [Google Scholar]
- 66. Chen J, Li Y, Zheng Q, Bao C, He J, Chen B, et al. Circular RNA profile identifies circPVT1 as a proliferative factor and prognostic marker in gastric cancer. Cancer Lett (2017) 388:208–19. 10.1016/j.canlet.2016.12.006 [DOI] [PubMed] [Google Scholar]
- 67. Liu T, Liu S, Xu Y, Shu R, Wang F, Chen C, et al. Circular RNA-ZFR Inhibited Cell Proliferation and Promoted Apoptosis in Gastric Cancer by Sponging miR-130a/miR-107 and Modulating PTEN. Cancer Res Treat Off J Korean Cancer Assoc (2018) 50(4):1396–417. 10.4143/crt.2017.537 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 68. Liu Z, Pan HM, Xin L, Zhang Y, Zhang WM, Cao P, et al. Circ-ZNF609 promotes carcinogenesis of gastric cancer cells by inhibiting miRNA-145-5p expression. Eur Rev Med Pharmacol Sci (2019) 23(21):9411–7. 10.26355/eurrev_201911_19433 [DOI] [PubMed] [Google Scholar]
- 69. Su Y, Lv X, Yin W, Zhou L, Hu Y, Zhou A, et al. CircRNA Cdr1as functions as a competitive endogenous RNA to promote hepatocellular carcinoma progression. Aging (2019) 11(19):8182–203. 10.18632/aging.102312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Yu L, Gong X, Sun L, Zhou Q, Lu B, Zhu L. The Circular RNA Cdr1as Act as an Oncogene in Hepatocellular Carcinoma through Targeting miR-7 Expression. PLoS One (2016) 11(7):e0158347. 10.1371/journal.pone.0158347 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 71. Fu Y, Cai L, Lei X, Wang D. Circular RNA ABCB10 promotes hepatocellular carcinoma progression by increasing HMG20A expression by sponging miR-670-3p. Cancer Cell Int (2019) 19:338. 10.1186/s12935-019-1055-z [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 72. Yang G, Wang X, Liu B, Lu Z, Xu Z, Xiu P, et al. circ-BIRC6, a circular RNA, promotes hepatocellular carcinoma progression by targeting the miR-3918/Bcl2 axis. Cell Cycle (2019) 18(9):976–89. 10.1080/15384101.2019.1601477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Wei Y, Chen X, Liang C, Ling Y, Yang X, Ye X, et al. A Noncoding Regulatory RNAs Network Driven by Circ-CDYL Acts Specifically in the Early Stages Hepatocellular Carcinoma. Hepatology (2020) 71(1):130–47. 10.1002/hep.30795 [DOI] [PubMed] [Google Scholar]
- 74. Wang Z, Zhao Y, Wang Y, Jin C. Circular RNA circHIAT1 inhibits cell growth in hepatocellular carcinoma by regulating miR-3171/PTEN axis. Biomed Pharmacother = Biomed Pharmacother (2019) 116:108932. 10.1016/j.biopha.2019.108932 [DOI] [PubMed] [Google Scholar]
- 75. Chen G, Shi Y, Liu M, Sun J. circHIPK3 regulates cell proliferation and migration by sponging miR-124 and regulating AQP3 expression in hepatocellular carcinoma. Cell Death Dis (2018) 9(2):175. 10.1038/s41419-017-0204-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Chen Z, Zuo X, Pu L, Zhang Y, Han G, Zhang L, et al. circLARP4 induces cellular senescence through regulating miR-761/RUNX3/p53/p21 signaling in hepatocellular carcinoma. Cancer Sci (2019) 110(2):568–81. 10.1111/cas.13901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Fu X, Zhang J, He X, Yan X, Wei J, Huang M, et al. Circular RNA MAN2B2 promotes cell proliferation of hepatocellular carcinoma cells via the miRNA-217/MAPK1 axis. J Cancer (2020) 11(11):3318–26. 10.7150/jca.36500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Han D, Li J, Wang H, Su X, Hou J, Gu Y, et al. Circular RNA circMTO1 acts as the sponge of microRNA-9 to suppress hepatocellular carcinoma progression. Hepatology (2017) 66(4):1151–64. 10.1002/hep.29270 [DOI] [PubMed] [Google Scholar]
- 79. Li Z, Hu Y, Zeng Q, Wang H, Yan J, Li H, et al. Circular RNA MYLK promotes hepatocellular carcinoma progression by increasing Rab23 expression by sponging miR-362-3p. Cancer Cell Int (2019) 19:211. 10.1186/s12935-019-0926-7 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 80. Qi SX, Sun H, Liu H, Yu J, Jiang ZY, Yan P. Role and mechanism of circ-PRKCI in hepatocellular carcinoma. World J Gastroenterol (2019) 25(16):1964–74. 10.3748/wjg.v25.i16.1964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Ding Z, Guo L, Deng Z, Li P. Circ-PRMT5 enhances the proliferation, migration and glycolysis of hepatoma cells by targeting miR-188-5p/HK2 axis. Ann Hepatol (2020) 19(3):269–79. 10.1016/j.aohep.2020.01.002 [DOI] [PubMed] [Google Scholar]
- 82. Zhu Y, Liu Y, Xiao B, Cai H, Liu M, Ma L, et al. The circular RNA PVT1/miR-203/HOXD3 pathway promotes the progression of human hepatocellular carcinoma. Biol Open (2019) 8(9):1–8. 10.1242/bio.043687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Li Y, Shi H, Yuan J, Qiao L, Dong L, Wang Y. Downregulation of circular RNA circPVT1 restricts cell growth of hepatocellular carcinoma through downregulation of Sirtuin 7 via microRNA-3666. Clin Exp Pharmacol Physiol (2020) 47:1291–300. 10.1111/1440-1681.13273 [DOI] [PubMed] [Google Scholar]
- 84. Liu C, Zhong X, Li J, Xu F. Circular RNA circVAPA Promotes Cell Proliferation in Hepatocellular Carcinoma. Hum Gene Ther Clin Dev (2019) 30(4):152–9. 10.1089/humc.2019.079 [DOI] [PubMed] [Google Scholar]
- 85. Yang X, Liu L, Zou H, Zheng YW, Wang KP. circZFR promotes cell proliferation and migration by regulating miR-511/AKT1 axis in hepatocellular carcinoma. Digest Liver Dis Off J Ital Soc Gastroenterol Ital Assoc Study Liver (2019) 51(10):1446–55. 10.1016/j.dld.2019.04.012 [DOI] [PubMed] [Google Scholar]
- 86. Guo J, Duan H, Li Y, Yang L, Yuan L. A novel circular RNA circ-ZNF652 promotes hepatocellular carcinoma metastasis through inducing snail-mediated epithelial-mesenchymal transition by sponging miR-203/miR-502-5p. Biochem Biophys Res Commun (2019) 513(4):812–9. 10.1016/j.bbrc.2019.03.214 [DOI] [PubMed] [Google Scholar]
- 87. Li Y, Zhang J, Pan S, Zhou J, Diao X, Liu S. CircRNA CDR1as knockdown inhibits progression of non-small-cell lung cancer by regulating miR-219a-5p/SOX5 axis. Thoracic Cancer (2020) 11(3):537–48. 10.1111/1759-7714.13274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Zhang X, Yang D, Wei Y. Overexpressed CDR1as functions as an oncogene to promote the tumor progression via miR-7 in non-small-cell lung cancer. OncoTargets Ther (2018) 11:3979–87. 10.2147/OTT.S158316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Su C, Han Y, Zhang H, Li Y, Yi L, Wang X, et al. CiRS-7 targeting miR-7 modulates the progression of non-small cell lung cancer in a manner dependent on NF-kappaB signalling. J Cell Mol Med (2018) 22(6):3097–107. 10.1111/jcmm.13587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Xue YB, Ding MQ, Xue L, Luo JH. CircAGFG1 sponges miR-203 to promote EMT and metastasis of non-small-cell lung cancer by upregulating ZNF281 expression. Thoracic Cancer (2019) 10(8):1692–701. 10.1111/1759-7714.13131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Yang H, Zhao M, Zhao L, Li P, Duan Y, Li G. CircRNA BIRC6 promotes non-small cell lung cancer cell progression by sponging microRNA-145. Cell Oncol (2020) 43:477–88. 10.1007/s13402-020-00503-x [DOI] [PubMed] [Google Scholar]
- 92. Yu C, Cheng Z, Cui S, Mao X, Li B, Fu Y, et al. circFOXM1 promotes proliferation of non-small cell lung carcinoma cells by acting as a ceRNA to upregulate FAM83D. J Exp Clin Cancer Res CR (2020) 39(1):55. 10.1186/s13046-020-01555-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Liu G, Shi H, Deng L, Zheng H, Kong W, Wen X, et al. Circular RNA circ-FOXM1 facilitates cell progression as ceRNA to target PPDPF and MACC1 by sponging miR-1304-5p in non-small cell lung cancer. Biochem Biophys Res Commun (2019) 513(1):207–12. 10.1016/j.bbrc.2019.03.213 [DOI] [PubMed] [Google Scholar]
- 94. Yao J, Xu G, Zhu L, Zheng H. circGFRA1 Enhances NSCLC Progression by Sponging miR-188-3p. OncoTargets Ther (2020) 13:549–58. 10.2147/OTT.S230795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Lu H, Han X, Ren J, Ren K, Li Z, Sun Z. Circular RNA HIPK3 induces cell proliferation and inhibits apoptosis in non-small cell lung cancer through sponging miR-149. Cancer Biol Ther (2020) 21(2):113–21. 10.1080/15384047.2019.1669995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Dong L, Zheng J, Gao Y, Zhou X, Song W, Huang J. The circular RNA NT5E promotes non-small cell lung cancer cell growth via sponging microRNA-134. Aging (2020) 12(4):3936–49. 10.18632/aging.102861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Geng Y, Bao Y, Zhang W, Deng L, Su D, Zheng H. Circular RNA hsa_circ_0014130 Inhibits Apoptosis in Non-Small Cell Lung Cancer by Sponging miR-136-5p and Upregulating BCL2. Mol Cancer Res MCR (2020) 18(5):748–56. 10.1158/1541-7786.MCR-19-0998 [DOI] [PubMed] [Google Scholar]
- 98. Wang M, Shi J, Jiang H, Xu K, Huang Z. Circ_0014130 Participates in the Proliferation and Apoptosis of Nonsmall Cell Lung Cancer Cells via the miR-142-5p/IGF-1 Axis. Cancer Biotherapy Radiopharmaceuticals (2020) 35(3):233–40. 10.1089/cbr.2019.2965 [DOI] [PubMed] [Google Scholar]
- 99. Wang Y, Li Y, He H, Wang F. Circular RNA circ-PRMT5 facilitates non-small cell lung cancer proliferation through upregulating EZH2 via sponging miR-377/382/498. Gene (2019) 720:144099. 10.1016/j.gene.2019.144099 [DOI] [PubMed] [Google Scholar]
- 100. Wei S, Zheng Y, Jiang Y, Li X, Geng J, Shen Y, et al. The circRNA circPTPRA suppresses epithelial-mesenchymal transitioning and metastasis of NSCLC cells by sponging miR-96-5p. EBioMedicine (2019) 44:182–93. 10.1016/j.ebiom.2019.05.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Li X, Zhang Z, Jiang H, Li Q, Wang R, Pan H, et al. Circular RNA circPVT1 Promotes Proliferation and Invasion Through Sponging miR-125b and Activating E2F2 Signaling in Non-Small Cell Lung Cancer. Cell Physiol Biochem Int J Exp Cell Physiol Biochem Pharmacol (2018) 51(5):2324–40. 10.1159/000495876 [DOI] [PubMed] [Google Scholar]
- 102. Qin S, Zhao Y, Lim G, Lin H, Zhang X, Zhang X. Circular RNA PVT1 acts as a competing endogenous RNA for miR-497 in promoting non-small cell lung cancer progression. Biomed Pharmacother = Biomed Pharmacother (2019) 111:244–50. 10.1016/j.biopha.2018.12.007 [DOI] [PubMed] [Google Scholar]
- 103. Wang Y, Li H, Lu H, Qin Y. Circular RNA SMARCA5 inhibits the proliferation, migration, and invasion of non-small cell lung cancer by miR-19b-3p/HOXA9 axis. OncoTargets Ther (2019) 12:7055–65. 10.2147/OTT.S216320 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 104. Wang L, Ma H, Kong W, Liu B, Zhang X. Up-regulated circular RNA VANGL1 contributes to progression of non-small cell lung cancer through inhibition of miR-195 and activation of Bcl-2. Biosci Rep (2019) 39(6):1–9. 10.1042/BSR20182433 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 105. Zhang H, Wang X, Hu B, Zhang F, Wei H, Li L. Circular RNA ZFR accelerates non-small cell lung cancer progression by acting as a miR-101-3p sponge to enhance CUL4B expression. Artif cells Nanomed Biotechnol (2019) 47(1):3410–6. 10.1080/21691401.2019.1652623 [DOI] [PubMed] [Google Scholar]
- 106. Wang Y, Xu R, Zhang D, Lu T, Yu W, Wo Y, et al. Circ-ZKSCAN1 regulates FAM83A expression and inactivates MAPK signaling by targeting miR-330-5p to promote non-small cell lung cancer progression. Trans Lung Cancer Res (2019) 8(6):862–75. 10.21037/tlcr.2019.11.04 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Ding B, Fan W, Lou W. hsa_circ_0001955 Enhances In Vitro Proliferation, Migration, and Invasion of HCC Cells through miR-145-5p/NRAS Axis. Mol Ther Nucleic Acids (2020) 22:445–55. 10.1016/j.omtn.2020.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Huang A, Zheng H, Wu Z, Chen M, Huang Y. Circular RNA-protein interactions: functions, mechanisms, and identification. Theranostics (2020) 10(8):3503–17. 10.7150/thno.42174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Huang X, He M, Huang S, Lin R, Zhan M, Yang D, et al. Circular RNA circERBB2 promotes gallbladder cancer progression by regulating PA2G4-dependent rDNA transcription. Mol Cancer (2019) 18(1):166. 10.1186/s12943-019-1098-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Yang F, Hu A, Li D, Wang J, Guo Y, Liu Y, et al. Circ-HuR suppresses HuR expression and gastric cancer progression by inhibiting CNBP transactivation. Mol Cancer (2019) 18(1):158. 10.1186/s12943-019-1094-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Ding L, Zhao Y, Dang S, Wang Y, Li X, Yu X, et al. Circular RNA circ-DONSON facilitates gastric cancer growth and invasion via NURF complex dependent activation of transcription factor SOX4. Mol Cancer (2019) 18(1):45. 10.1186/s12943-019-1006-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Du WW, Yang W, Li X, Awan FM, Yang Z, Fang L, et al. A circular RNA circ-DNMT1 enhances breast cancer progression by activating autophagy. Oncogene (2018) 37(44):5829–42. 10.1038/s41388-018-0369-y [DOI] [PubMed] [Google Scholar]
- 113. Feng Y, Yang Y, Zhao X, Fan Y, Zhou L, Rong J, et al. Circular RNA circ0005276 promotes the proliferation and migration of prostate cancer cells by interacting with FUS to transcriptionally activate XIAP. Cell Death Dis (2019) 10(11):792. 10.1038/s41419-019-2028-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Li H, Yang F, Hu A, Wang X, Fang E, Chen Y, et al. Therapeutic targeting of circ-CUX1/EWSR1/MAZ axis inhibits glycolysis and neuroblastoma progression. EMBO Mol Med (2019) 11(12):e10835. 10.15252/emmm.201910835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Yang F, Fang E, Mei H, Chen Y, Li H, Li D, et al. Cis-Acting circ-CTNNB1 Promotes beta-Catenin Signaling and Cancer Progression via DDX3-Mediated Transactivation of YY1. Cancer Res (2019) 79(3):557–71. 10.1158/0008-5472.CAN-18-1559 [DOI] [PubMed] [Google Scholar]
- 116. Barbagallo D, Caponnetto A, Brex D, Mirabella F, Barbagallo C, Lauretta G, et al. CircSMARCA5 Regulates VEGFA mRNA Splicing and Angiogenesis in Glioblastoma Multiforme Through the Binding of SRSF1. Cancers (2019) 11(2):194–205. 10.3390/cancers11020194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Wang XH, Li J. CircAGFG1 aggravates the progression of cervical cancer by downregulating p53. Eur Rev Med Pharmacol Sci (2020) 24(4):1704–11. 10.26355/eurrev_202002_20345 [DOI] [PubMed] [Google Scholar]
- 118. Wang L, Long H, Zheng Q, Bo X, Xiao X, Li B. Circular RNA circRHOT1 promotes hepatocellular carcinoma progression by initiation of NR2F6 expression. Mol Cancer (2019) 18(1):119. 10.1186/s12943-019-1046-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Bi W, Huang J, Nie C, Liu B, He G, Han J, et al. CircRNA circRNA_102171 promotes papillary thyroid cancer progression through modulating CTNNBIP1-dependent activation of beta-catenin pathway. J Exp Clin Cancer Res CR (2018) 37(1):275. 10.1186/s13046-018-0936-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Zhan W, Liao X, Wang Y, Li L, Li J, Chen Z, et al. circCTIC1 promotes the self-renewal of colon TICs through BPTF-dependent c-Myc expression. Carcinogenesis (2019) 40(4):560–8. 10.1093/carcin/bgy144 [DOI] [PubMed] [Google Scholar]
- 121. Wang L, Zheng C, Wu X, Zhang Y, Yan S, Ruan L, et al. Circ-SOX4 promotes non-small cell lung cancer progression by activating the Wnt/beta-catenin pathway. Mol Oncol (2020) 14:3253. 10.1002/1878-0261.12656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Zhu YJ, Zheng B, Luo GJ, Ma XK, Lu XY, Lin XM, et al. Circular RNAs negatively regulate cancer stem cells by physically binding FMRP against CCAR1 complex in hepatocellular carcinoma. Theranostics (2019) 9(12):3526–40. 10.7150/thno.32796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Zhao W, Wang S, Qin T, Wang W. Circular RNA (circ-0075804) promotes the proliferation of retinoblastoma via combining heterogeneous nuclear ribonucleoprotein K (HNRNPK) to improve the stability of E2F transcription factor 3 E2F3. J Cell Biochem (2020) 121:3516–25. 10.1002/jcb.29631 [DOI] [PubMed] [Google Scholar]
- 124. Liu B, Yang G, Wang X, Liu J, Lu Z, Wang Q, et al. CircBACH1 (hsa_circ_0061395) promotes hepatocellular carcinoma growth by regulating p27 repression via HuR. J Cell Physiol (2020) 235:6929–41. 10.1002/jcp.29589 [DOI] [PubMed] [Google Scholar]
- 125. Du WW, Fang L, Yang W, Wu N, Awan FM, Yang Z, et al. Induction of tumor apoptosis through a circular RNA enhancing Foxo3 activity. Cell Death Differentiation (2017) 24(2):357–70. 10.1038/cdd.2016.133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Sun S, Wang W, Luo X, Li Y, Liu B, Li X, et al. Circular RNA circ-ADD3 inhibits hepatocellular carcinoma metastasis through facilitating EZH2 degradation via CDK1-mediated ubiquitination. Am J Cancer Res (2019) 9(8):1695–707. [PMC free article] [PubMed] [Google Scholar]
- 127. Nan A, Chen L, Zhang N, Jia Y, Li X, Zhou H, et al. Circular RNA circNOL10 Inhibits Lung Cancer Development by Promoting SCLM1-Mediated Transcriptional Regulation of the Humanin Polypeptide Family. Adv Sci (2019) 6(2):1800654. 10.1002/advs.201800654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Hu X, Wu D, He X, Zhao H, He Z, Lin J, et al. circGSK3beta promotes metastasis in esophageal squamous cell carcinoma by augmenting beta-catenin signaling. Mol Cancer (2019) 18(1):160. 10.1186/s12943-019-1095-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Yang H, Li X, Meng Q, Sun H, Wu S, Hu W, et al. CircPTK2 (hsa_circ_0005273) as a novel therapeutic target for metastatic colorectal cancer. Mol Cancer (2020) 19(1):13. 10.1186/s12943-020-1139-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Du WW, Yang W, Li X, Fang L, Wu N, Li F, et al. The Circular RNA circSKA3 Binds Integrin beta1 to Induce Invadopodium Formation Enhancing Breast Cancer Invasion. Mol Ther J Am Soc Gene Ther (2020) 28:1287–98. 10.1016/j.ymthe.2020.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Chen Y, Yang F, Fang E, Xiao W, Mei H, Li H, et al. Circular RNA circAGO2 drives cancer progression through facilitating HuR-repressed functions of AGO2-miRNA complexes. Cell Death Differentiation (2019) 26(7):1346–64. 10.1038/s41418-018-0220-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Zhang Y, Zhang XO, Chen T, Xiang JF, Yin QF, Xing YH, et al. Circular intronic long noncoding RNAs. Mol Cell (2013) 51(6):792–806. 10.1016/j.molcel.2013.08.017 [DOI] [PubMed] [Google Scholar]
- 133. Chen CY, Sarnow P. Initiation of protein synthesis by the eukaryotic translational apparatus on circular RNAs. Science (1995) 268(5209):415–7. 10.1126/science.7536344 [DOI] [PubMed] [Google Scholar]
- 134. Pamudurti NR, Bartok O, Jens M, Ashwal-Fluss R, Stottmeister C, Ruhe L, et al. Translation of CircRNAs. Mol Cell (2017) 66(1):9–21 e7. 10.1016/j.molcel.2017.02.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Legnini I, Di Timoteo G, Rossi F, Morlando M, Briganti F, Sthandier O, et al. Circ-ZNF609 Is a Circular RNA that Can Be Translated and Functions in Myogenesis. Mol Cell (2017) 66(1):22–37 e9. 10.1016/j.molcel.2017.02.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Lei M, Zheng G, Ning Q, Zheng J, Dong D. Translation and functional roles of circular RNAs in human cancer. Mol Cancer (2020) 19(1):30. 10.1186/s12943-020-1135-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Pan Z, Cai J, Lin J, Zhou H, Peng J, Liang J, et al. A novel protein encoded by circFNDC3B inhibits tumor progression and EMT through regulating Snail in colon cancer. Mol Cancer (2020) 19(1):71. 10.1186/s12943-020-01179-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Yang Y, Gao X, Zhang M, Yan S, Sun C, Xiao F, et al. Novel Role of FBXW7 Circular RNA in Repressing Glioma Tumorigenesis. J Natl Cancer Institute (2018) 110(3):304–15. 10.1093/jnci/djx166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Xia X, Li X, Li F, Wu X, Zhang M, Zhou H, et al. A novel tumor suppressor protein encoded by circular AKT3 RNA inhibits glioblastoma tumorigenicity by competing with active phosphoinositide-dependent Kinase-1. Mol Cancer (2019) 18(1):131. 10.1186/s12943-019-1056-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Zhang M, Huang N, Yang X, Luo J, Yan S, Xiao F, et al. A novel protein encoded by the circular form of the SHPRH gene suppresses glioma tumorigenesis. Oncogene (2018) 37(13):1805–14. 10.1038/s41388-017-0019-9 [DOI] [PubMed] [Google Scholar]
- 141. Liang WC, Wong CW, Liang PP, Shi M, Cao Y, Rao ST, et al. Translation of the circular RNA circbeta-catenin promotes liver cancer cell growth through activation of the Wnt pathway. Genome Biol (2019) 20(1):84. 10.1186/s13059-019-1685-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Zheng X, Chen L, Zhou Y, Wang Q, Zheng Z, Xu B, et al. A novel protein encoded by a circular RNA circPPP1R12A promotes tumor pathogenesis and metastasis of colon cancer via Hippo-YAP signaling. Mol Cancer (2019) 18(1):47. 10.1186/s12943-019-1010-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Zhi X, Zhang J, Cheng Z, Bian L, Qin J. circLgr4 drives colorectal tumorigenesis and invasion through Lgr4-targeting peptide. Int J Cancer (2019) 1:1–30. 10.1002/ijc.32549 [DOI] [PubMed] [Google Scholar]
- 144. Gu C, Zhou N, Wang Z, Li G, Kou Y, Yu S, et al. circGprc5a Promoted Bladder Oncogenesis and Metastasis through Gprc5a-Targeting Peptide. Mol Ther Nucleic Acids (2018) 13:633–41. 10.1016/j.omtn.2018.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Ye F, Gao G, Zou Y, Zheng S, Zhang L, Ou X, et al. circFBXW7 Inhibits Malignant Progression by Sponging miR-197-3p and Encoding a 185-aa Protein in Triple-Negative Breast Cancer. Mol Ther Nucleic Acids (2019) 18:88–98. 10.1016/j.omtn.2019.07.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Sun S, Gao J, Zhou S, Li Y, Wang Y, Jin L, et al. A novel circular RNA circ-LRIG3 facilitates the malignant progression of hepatocellular carcinoma by modulating the EZH2/STAT3 signaling. J Exp Clin Cancer Res CR (2020) 39(1):252. 10.1186/s13046-020-01779-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Hoff PM, Machado KK. Role of angiogenesis in the pathogenesis of cancer. Cancer Treat Rev (2012) 38(7):825–33. 10.1016/j.ctrv.2012.04.006 [DOI] [PubMed] [Google Scholar]
- 148. Chen LY, Wang L, Ren YX, Pang Z, Liu Y, Sun XD, et al. The circular RNA circ-ERBIN promotes growth and metastasis of colorectal cancer by miR-125a-5p and miR-138-5p/4EBP-1 mediated cap-independent HIF-1alpha translation. Mol Cancer (2020) 19(1):164. 10.1186/s12943-020-01272-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Liu J, Jiang X, Zou A, Mai Z, Huang Z, Sun L, et al. circIGHG-induced epithelial-to-mesenchymal transition promotes oral squamous cell carcinoma progression via miR-142-5p/IGF2BP3 signaling. Cancer Res (2020) 17:1–25. 10.1158/0008-5472.CAN-20-0554 [DOI] [PubMed] [Google Scholar]
- 150. Yan K, Cheng W, Xu X, Cao G, Ji Z, Li Y. Circulating RNAs, circ_4911 and circ_4302, are novel regulators of endothelial cell function under a hepatocellular carcinoma microenvironment. Oncol Rep (2020) 44(4):1727–35. 10.3892/or.2020.7702 [DOI] [PubMed] [Google Scholar]
- 151. Ayati A, Moghimi S, Salarinejad S, Safavi M, Pouramiri B, Foroumadi A. A review on progression of epidermal growth factor receptor (EGFR) inhibitors as an efficient approach in cancer targeted therapy. Bioorganic Chem (2020) 99:103811. 10.1016/j.bioorg.2020.103811 [DOI] [PubMed] [Google Scholar]
- 152. Song H, Liu D, Dong S, Zeng L, Wu Z, Zhao P, et al. Epitranscriptomics and epiproteomics in cancer drug resistance: therapeutic implications. Signal Transduct Target Ther (2020) 5(1):193. 10.1038/s41392-020-00300-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Xiong W, Ai YQ, Li YF, Ye Q, Chen ZT, Qin JY, et al. Microarray Analysis of Circular RNA Expression Profile Associated with 5-Fluorouracil-Based Chemoradiation Resistance in Colorectal Cancer Cells. BioMed Res Int (2017) 2017:8421614. 10.1155/2017/8421614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Greene J, Baird AM, Casey O, Brady L, Blackshields G, Lim M, et al. Circular RNAs are differentially expressed in prostate cancer and are potentially associated with resistance to enzalutamide. Sci Rep (2019) 9(1):10739. 10.1038/s41598-019-47189-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Huang X, Li Z, Zhang Q, Wang W, Li B, Wang L, et al. Circular RNA AKT3 upregulates PIK3R1 to enhance cisplatin resistance in gastric cancer via miR-198 suppression. Mol Cancer (2019) 18(1):71. 10.1186/s12943-019-0969-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Sun G, Li Z, He Z, Wang W, Wang S, Zhang X, et al. Circular RNA MCTP2 inhibits cisplatin resistance in gastric cancer by miR-99a-5p-mediated induction of MTMR3 expression. J Exp Clin Cancer Res CR (2020) 39(1):246. 10.1186/s13046-020-01758-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Li J, Li H, Lv X, Yang Z, Gao M, Bi Y, et al. Diagnostic performance of circular RNAs in human cancers: A systematic review and meta-analysis. Mol Genet Genomic Med (2019) 7(7):e00749. 10.1002/mgg3.749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Baassiri A, Nassar F, Mukherji D, Shamseddine A, Nasr R, Temraz S. Exosomal Non Coding RNA in LIQUID Biopsies as a Promising Biomarker for Colorectal Cancer. Int J Mol Sci (2020) 21(4):1398–418. 10.3390/ijms21041398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Xia S, Feng J, Lei L, Hu J, Xia L, Wang J, et al. Comprehensive characterization of tissue-specific circular RNAs in the human and mouse genomes. Briefings Bioinf (2017) 18(6):984–92. 10.1093/bib/bbw081 [DOI] [PubMed] [Google Scholar]
- 160. Xia S, Feng J, Chen K, Ma Y, Gong J, Cai F, et al. CSCD: a database for cancer-specific circular RNAs. Nucleic Acids Res (2018) 46(D1):D925–9. 10.1093/nar/gkx863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Glazar P, Papavasileiou P, Rajewsky N. circBase: a database for circular RNAs. RNA (2014) 20(11):1666–70. 10.1261/rna.043687.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Chen X, Han P, Zhou T, Guo X, Song X, Li Y. circRNADb: A comprehensive database for human circular RNAs with protein-coding annotations. Sci Rep (2016) 6:34985. 10.1038/srep34985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Dudekula DB, Panda AC, Grammatikakis I, De S, Abdelmohsen K, Gorospe M. CircInteractome: A web tool for exploring circular RNAs and their interacting proteins and microRNAs. RNA Biol (2016) 13(1):34–42. 10.1080/15476286.2015.1128065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Kolekar P, Pataskar A, Kulkarni-Kale U, Pal J, Kulkarni A. IRESPred: Web Server for Prediction of Cellular and Viral Internal Ribosome Entry Site (IRES). Sci Rep (2016) 6:27436. 10.1038/srep27436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Min XJ, Butler G, Storms R, Tsang A. OrfPredictor: predicting protein-coding regions in EST-derived sequences. Nucleic Acids Res (2005) 33(Web Server issue):W677–80. 10.1093/nar/gki394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Wu W, Ji P, Zhao F. CircAtlas: an integrated resource of one million highly accurate circular RNAs from 1070 vertebrate transcriptomes. Genome Biol (2020) 21(1):101. 10.1186/s13059-020-02018-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Zheng LL, Li JH, Wu J, Sun WJ, Liu S, Wang ZL, et al. deepBase v2.0: identification, expression, evolution and function of small RNAs, LncRNAs and circular RNAs from deep-sequencing data. Nucleic Acids Res (2016) 44(D1):D196–202. 10.1093/nar/gkv1273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Zhao Z, Wang K, Wu F, Wang W, Zhang K, Hu H, et al. circRNA disease: a manually curated database of experimentally supported circRNA-disease associations. Cell Death Dis (2018) 9(5):475. 10.1038/s41419-018-0503-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Liu M, Wang Q, Shen J, Yang BB, Ding X. Circbank: a comprehensive database for circRNA with standard nomenclature. RNA Biol (2019) 16(7):899–905. 10.1080/15476286.2019.1600395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Ghosal S, Das S, Sen R, Basak P, Chakrabarti J. Circ2Traits: a comprehensive database for circular RNA potentially associated with disease and traits. Front Genet (2013) 4:283. 10.3389/fgene.2013.00283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Liu YC, Li JR, Sun CH, Andrews E, Chao RF, Lin FM, et al. CircNet: a database of circular RNAs derived from transcriptome sequencing data. Nucleic Acids Res (2016) 44(D1):D209–15. 10.1093/nar/gkv940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Chang TH, Huang HY, Hsu JB, Weng SL, Horng JT, Huang HD. An enhanced computational platform for investigating the roles of regulatory RNA and for identifying functional RNA motifs. BMC Bioinf (2013) 14 Suppl 2:S4. 10.1186/1471-2105-14-S2-S4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. DiChiacchio L, Mathews DH. Predicting RNA-RNA Interactions Using RNAstructure. Methods Mol Biol (2016) 1490:51–62. 10.1007/978-1-4939-6433-8_4 [DOI] [PubMed] [Google Scholar]
- 174. Li JH, Liu S, Zhou H, Qu LH, Yang JH. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res (2014) 42(Database issue):D92–7. 10.1093/nar/gkt1248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175. Agostini F, Zanzoni A, Klus P, Marchese D, Cirillo D, Tartaglia GG. catRAPID omics: a web server for large-scale prediction of protein-RNA interactions. Bioinformatics (2013) 29(22):2928–30. 10.1093/bioinformatics/btt495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176. Mann CM, Muppirala UK, Dobbs D. Computational Prediction of RNA-Protein Interactions. Methods Mol Biol (2017) 1543:169–85. 10.1007/978-1-4939-6716-2_8 [DOI] [PubMed] [Google Scholar]
- 177. Mokrejs M, Masek T, Vopalensky V, Hlubucek P, Delbos P, Pospisek M. IRESite–a tool for the examination of viral and cellular internal ribosome entry sites. Nucleic Acids Res (2010) 38(Database issue):D131–6. 10.1093/nar/gkp981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Zhao J, Wu J, Xu T, Yang Q, He J, Song X. IRESfinder: Identifying RNA internal ribosome entry site in eukaryotic cell using framed k-mer features. J Genet Genomics = Yi Chuan xue bao (2018) 45(7):403–6. 10.1016/j.jgg.2018.07.006 [DOI] [PubMed] [Google Scholar]
- 179. Olexiouk V, Crappe J, Verbruggen S, Verhegen K, Martens L, Menschaert G. sORFs.org: a repository of small ORFs identified by ribosome profiling. Nucleic Acids Res (2016) 44(D1):D324–9. 10.1093/nar/gkv1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180. Lin MF, Jungreis I, Kellis M. PhyloCSF: a comparative genomics method to distinguish protein coding and non-coding regions. Bioinformatics (2011) 27(13):i275–82. 10.1093/bioinformatics/btr209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181. Stothard P. The sequence manipulation suite: JavaScript programs for analyzing and formatting protein and DNA sequences. BioTechniques (2000) 28(6):1102, 4. 10.2144/00286ir0 [DOI] [PubMed] [Google Scholar]
- 182. Kong L, Zhang Y, Ye ZQ, Liu XQ, Zhao SQ, Wei L, et al. CPC: assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res (2007) 35(Web Server issue):W345–9. 10.1093/nar/gkm391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183. Wang L, Park HJ, Dasari S, Wang S, Kocher JP, Li W. CPAT: Coding-Potential Assessment Tool using an alignment-free logistic regression model. Nucleic Acids Res (2013) 41(6):e74. 10.1093/nar/gkt006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184. Meng X, Chen Q, Zhang P, Chen M. CircPro: an integrated tool for the identification of circRNAs with protein-coding potential. Bioinformatics (2017) 33(20):3314–6. 10.1093/bioinformatics/btx446 [DOI] [PubMed] [Google Scholar]
- 185. Sun P, Li G. CircCode: A Powerful Tool for Identifying circRNA Coding Ability. Front Genet (2019) 10:981. 10.3389/fgene.2019.00981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186. Han K, Wang FW, Cao CH, Ling H, Chen JW, Chen RX, et al. CircLONP2 enhances colorectal carcinoma invasion and metastasis through modulating the maturation and exosomal dissemination of microRNA-17. Mol Cancer (2020) 19(1):60. 10.1186/s12943-020-01184-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187. Jie M, Wu Y, Gao M, Li X, Liu C, Ouyang Q, et al. CircMRPS35 suppresses gastric cancer progression via recruiting KAT7 to govern histone modification. Mol Cancer (2020) 19(1):56. 10.1186/s12943-020-01160-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188. Chen L, Kong R, Wu C, Wang S, Liu Z, Liu S, et al. Circ-MALAT1 Functions as Both an mRNA Translation Brake and a microRNA Sponge to Promote Self-Renewal of Hepatocellular Cancer Stem Cells. Adv Sci (2020) 7(4):1900949. 10.1002/advs.201900949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189. Chen RX, Chen X, Xia LP, Zhang JX, Pan ZZ, Ma XD, et al. N(6)-methyladenosine modification of circNSUN2 facilitates cytoplasmic export and stabilizes HMGA2 to promote colorectal liver metastasis. Nat Commun (2019) 10(1):4695. 10.1038/s41467-019-12651-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190. Fischer JW, Busa VF, Shao Y, Leung AKL. Structure-Mediated RNA Decay by UPF1 and G3BP1. Mol Cell (2020) 78(1):70–84 e6. 10.1016/j.molcel.2020.01.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191. Kong S, Tao M, Shen X, Ju S. Translatable circRNAs and lncRNAs: Driving mechanisms and functions of their translation products. Cancer Lett (2020) 483:59–65. 10.1016/j.canlet.2020.04.006 [DOI] [PubMed] [Google Scholar]