Abstract
Colorectal cancer (CRC) is a common clinical cancer that remains incurable in most cases. miRNAs are reported to play a part in the development of various tumors. In the present study, we found that miR-324-5p was downregulated in CRC cells, while ELAV (embryonic lethal, abnormal vision, Drosophila)-like protein 1 (ELAVL1) showed a higher expression. miR-324-5p transfection significantly inhibited the proliferation as well as invasion in both SW620 and SW480 cells. miR-324-5p mimic transfection markedly decreased the expression of ELAVL1. Luciferase reporter gene assay confirmed that ELAVL1 is a direct target of miR-324-5p. Furthermore, cancer invasion factors uPA, uPAR, and MMP-9 were found to drop significantly in miR-324-5p-transfected groups. To conclude, our findings indicate that miR-324-5p may play a suppressive role in colorectal cell viability and invasion, at least in part, through directly targeting ELAVL1. Therefore, miR-234-5p might function as a promising candidate for CRC treatment and deserves deeper research.
Key words: Colorectal cancer (CRC); miR-324-5p; Proliferation; Invasion; (Embryonic lethal, abnormal vision, Drosophila)-like protein 1 (ELAVL1)
INTRODUCTION
Colorectal cancer (CRC) is one of the most common malignant digestive tract tumors1 worldwide. This cancer risk usually rises after the age of 50. Development of CRC involves multiple factors, including old age, genetic disorders, and lifestyle factors like diet and obesity. The current treatment of CRC primarily consists of surgery, chemotherapy, and radiation. Although great progress has been made to increase the survival of patients2–4, in most cases the disease remains incurable. So studies searching for novel targets or therapies are still in urgent need.
Over the recent decades, a type of non-protein-coding RNA molecule known as microRNA (miRNA) has been involved in cancer development via functioning as tumor suppressors or oncogenes5. miRNAs can participate in a variety of cellular biological processes including cell survival, apoptosis, proliferation, differentiation, and cell cycle progression6,7. Cao et al.8 found that miR-324-5p could suppress hepatocellular carcinoma cell invasion via inhibiting extracellular matrix (ECM) degradation. In addition, miR-324-5p was recently reported to have essential biological functions in breast cancer and CRC9.
ELAVL1, also known as human antigen R (HuR), belongs to a family of proteins analogous to the Drosophila embryonic-lethal abnormal vision (ELAV)10. This is an RNA-binding protein (RBP) containing an AU-rich element (ARE) and a U-rich element (URE). ELAVL1 increases the stability of mRNAs, thereby promoting gene expression11. A previous study revealed that ELAVL1 overexpression was observed in 76% of colorectal adenomas and 94% of colorectal adenocarcinomas12. Furthermore, a therapeutic strategy to target ELAVL1 in CRC was shown to be highly promising13. However, no reports were observed to research the connection between miR-324-5p and ELAVL1 and their applicative potential in suppressing CRC.
In this study, we predicted that miR-324-5p can directly target ELAVL1 and investigated whether miR-324-5p can influence CRC cells via targeting ELAVL1. The final aim was to elucidate the therapeutic potential of miR-324-5p/ELAVL1 in CRC and bring hope to the suffering patients.
MATERIALS AND METHODS
Cell Culture and Transfection
Two colon cancer cell lines, SW620 and SW480 (ATCC, Manassas, VA, USA)14, were studied along with the normal human colon mucosal epithelial cell line NCM-460, which was used as a control. All cell lines were incubated with RPMI-1640 (Gibco, Gaithersburg, MD, USA) containing 10% fetal bovine serum (FBS; Thermo Fisher Scientific, Carlsbad, CA, USA), 100 U/ml penicillin, and 100 μg/ml streptomycin (Sigma-Aldrich, St. Louis, MO, USA) at 37°C in a 5% CO2 atmosphere. For transfection, SW620 and SW480 cells were seeded in six-well plates at a cell density of 70%–90% and transfected with Lipofectamine™ 3000 reagent (Thermo Fisher) and miR-324-5p mimic (5′-UGUGGU-UACGGGAUCCCCUACGC-3′)15 or miR-324-5p inhibitor (5′-ACACCAAUGCCCUAGGGGAUGCG-3′)16 according to the manufacturer’s instructions. Nonhomologous miRNA mimic control was used as the negative control (NC).
RNA Isolation and RT-qPCR
Cells were transferred into six-well plates, and the total RNA was isolated with Invitrogen TRIzol reagent (Thermo Fisher). Total RNA was then reverse transcribed into cDNA using PrimeScript™ RT Master Mix (TaKaRa, Dalian, P.R. China). RT-qPCR was performed at 95°C for an initial 10 min followed by 40 cycles of denaturation for 30 s at 95°C, annealing at 55°C for 30 s, and extension at 72°C for 30s with the SYBR Premix Ex Taq (Takara). The mRNA relative expression levels were then calculated with the 2−ΔΔCt method after normalization by glyceraldehyde 3-phosphate dehydrogenase (GAPDH). U6 was used as a control for miRNA expression. Sequences (Thermo Fisher) of the PCR primers are as follows: U6, 5′-GCTTCGGCAGCACATATACT-3′ (forward) and 5′-AACGCTTCACGAATTTGCGT-3′ (reverse); GAPDH, 5′-GGAGCGAGATCCCTCCAAAAT-3′ (forward) and 5′-GGCTGTTGTCATACTTCTCATGG-3′ (reverse); miR-324-5p, 5′-GAGGCCAAGCCCTGGTATG-3′ (forward) and 5′-CGGGCCGATTGATCTCAGC-3′ (reverse); ELAVL1, 5′-AGCTACGAATCTCCGACCAC-3′ (forward) and 5′-CGTTATCCCATGTGTCGAAGAA-3′ (reverse).
MTT Assay
Forty-eight hours after transfection, SW620 and SW480 cells were cultured at a density of 5 × 103 cells/well in 96-well plates17. Then cells were incubated with 20 μl of MTT solution (0.5 mg/ml; Sigma-Aldrich) for a further 4 h at 37°C. The formazan crystals were solubilized in 100 μl of dimethyl sulfoxide (DMSO; Sigma-Aldrich). Absorbance of each well was measured by a microplate reader (Molecular Devices LLC, Sunnyvale, CA, USA) at a wavelength of 490 nm.
Western Blot
Briefly, total protein was extracted with RIPA buffer containing 1% protease inhibitor PMSF, and a BCA Protein Assay Kit (Beyotime, Shanghai, P.R. China) was applied to determine the protein concentration. Ten percent sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gel was used to separate the proteins, and the proteins were then transferred to polyvinylidene fluoride (PVDF) membranes (Millipore, Billerica, MA, USA). The membranes were blocked with 5% bovine serum albumin (BSA), and then the membranes were incubated with primary antibodies ELAVL1 (1:1,000; ab200342; Abcam, Cambridge, MA, USA), matrix metalloproteinase 9 (MMP-9; 1:1,000; ab73734; Abcam), urokinase-type plasminogen activator (uPA; 1:1,000; ab169754; Abcam), uPA receptor (uPAR; 1:500; ab103791; Abcam), and GAPDH (1:5,000; ab8245; Abcam), respectively, at 4°C overnight. After washing, corresponding horse radish peroxidase (HRP)-conjugated secondary antibodies (1:5,000; Abcam) were used, and then chemiluminescence (Beyotime) was detected.
Wound Healing Assay
Cells were seeded into six-well dishes at a density of 1 × 105 cells per well and incubated in a humidified incubator at 37°C with 5% CO2. A wound was generated using a sterilized 1-ml pipette in the cell monolayer. The debris was washed with PBS and incubated with serum-free medium. Photos were taken at 0 and 24 h after the scraping, respectively. Experiments were carried out in triplicate at least three times.
Transwell Assay
Cells were serum starved for 24 h and then plated at the density of 1 × 105 cells per well in the serum-free medium. RPMI-1640 medium containing 10% FBS was added into the bottom chamber (8-μm pore size; BD Pharmingen, San Diego, CA, USA) as a chemoattractant. After incubation for 24 h18, nonmigrating cells were removed from the upper chamber with a cotton swab, and the cells that migrated through the membrane were fixed with 4% formaldehyde (Sigma-Aldrich) and stained with crystal violet staining solutions (Sigma-Aldrich)19.
Dual-Luciferase Reporter Gene Assay
miR-324-5p targets were predicted with the TargetScan database (https://www.targetscan.org). Luciferase reporter vectors for the wild type and mutant type of ELAVL1 3′-untranslated regions (3′-UTRs) were then constructed. Either miR-324-5p mimics or control mimics [negative control (NC)] were cotransfected with the wild-type or mutant-type luciferase reporter vector into SW620 and SW480 cells by Lipofectamine 2000 (Invitrogen, Grand Island, NY, USA). The pmirGLO Dual-Luciferase miRNA Target Expression vector (Promega, Madison, WI, USA) was transfected and served as the control. Forty-eight hours after cell transfection, the luciferase activity was assessed with the Dual-Luciferase Reporter Assay System (Promega)20.
Statistical Analysis
Data are expressed as mean ± standard error of the mean (SEM) of triplicate samples. Data significance was measured through Student’s t-test and one-way analysis of variance. Statistical analysis was performed by IBM SPSS Statistics v20.0 (SPSS Inc., Chicago, IL, USA). A value of p < 0.05 was considered as a statistically significant difference.
RESULTS
miR-324-5p Is Downregulated While ELAVL1 Is Overexpressed in CRC Cells
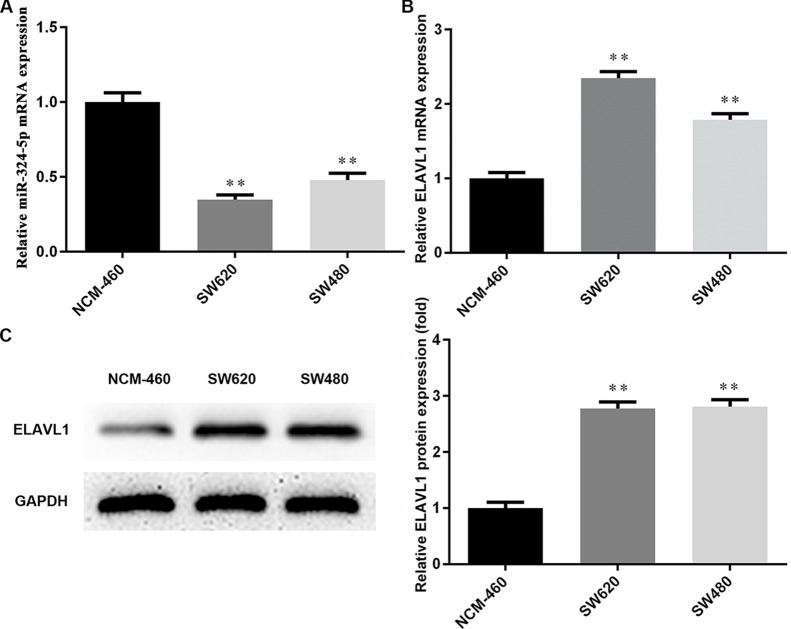
A couple of isogenic human colon cancer cell lines derived from a lymph node metastasis (SW620) or the primary tumor (SW480) was used in this study. First, the expression of miR-324-5p and ELAVL1 in CRC cells was measured via qRT-PCR and Western blotting. As shown in Figure 1A, compared with the normal human colon mucosal epithelial cell line NCM-460, miR-324-5p was at a significantly lower level in SW620 and SW480 cells. Conversely, ELAVL1 exhibited an obvious increase in both mRNA and protein levels in CRC cells (Fig. 1B and C). Our results provide novel evidence of miR-234-5p downregulation and ELAVL1 upregulation in CRC cells, indicating that miR-324-5p and ELAVL1 may play a part in CRC development.
Figure 1.
Expression of microRNA (miR)-324-5p and ELAV (embryonic lethal, abnormal vision, Drosophila)-like protein 1 (ELAVL1) in colorectal cells. (A) The expression levels of miR-324-5p in colorectal cancer (CRC) cells were detected using RT-qPCR. (B) The expression of ELAVL1 in SW620 and SW480 cells was detected by RT-qPCR. (C) The protein expression levels of ELAVL1 in SW620 and SW480 cells were detected by Western blotting. **p < 0.01 versus NCM-460 group.
Overexpression of miR-324-5p in CRC Cells Significantly Inhibits Cell Proliferation
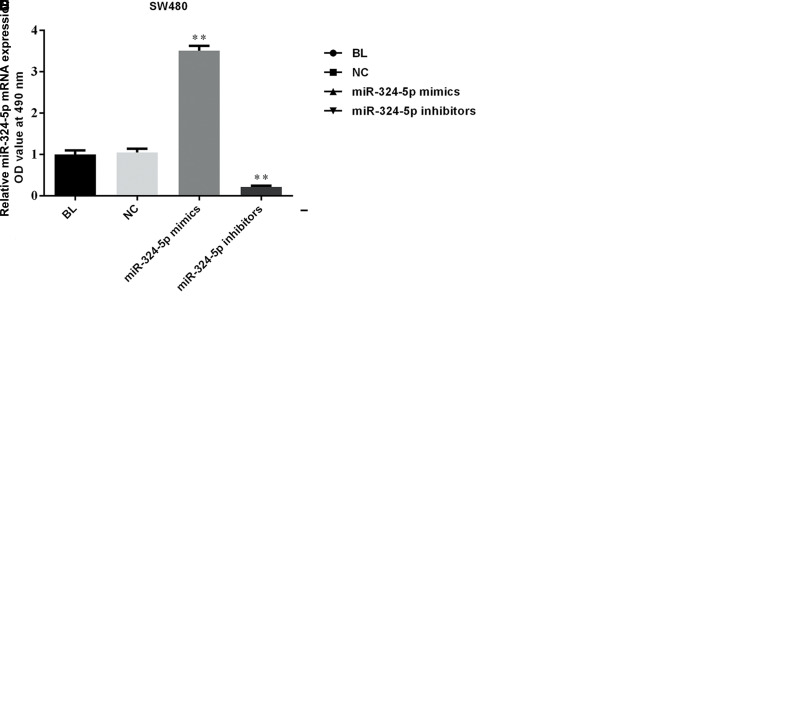
To explore whether miR-324-5p can affect the proliferation in CRC, we stably overexpressed miR-324-5p in SW620 and SW480 cells. Figure 2A and B shows that transfection of miR-324-5p mimics was a success in our experiments. As shown in Figure 2C and D, MTT assays revealed that miR-324-5p overexpression significantly reduced the proliferation, while miR-324-5p inhibitors obviously heightened the proliferation in both SW620 and SW480 cells. Results here indicated that miR-324-5p may help inhibit the viability of CRC cells.
Figure 2.
miR-324-5p suppresses proliferation of colorectal cells. RT-qPCR was used to verify the expression of miR-324-5p in SW620 (A) and SW480 (B) cells. MTT assay was used to determine the proliferation of SW620 (C) and SW480 (D) cells. OD values between miR-324-5p mimics and control groups were significantly different. **p < 0.01 versus negative control (NC) group, *p < 0.05 versus NC group.
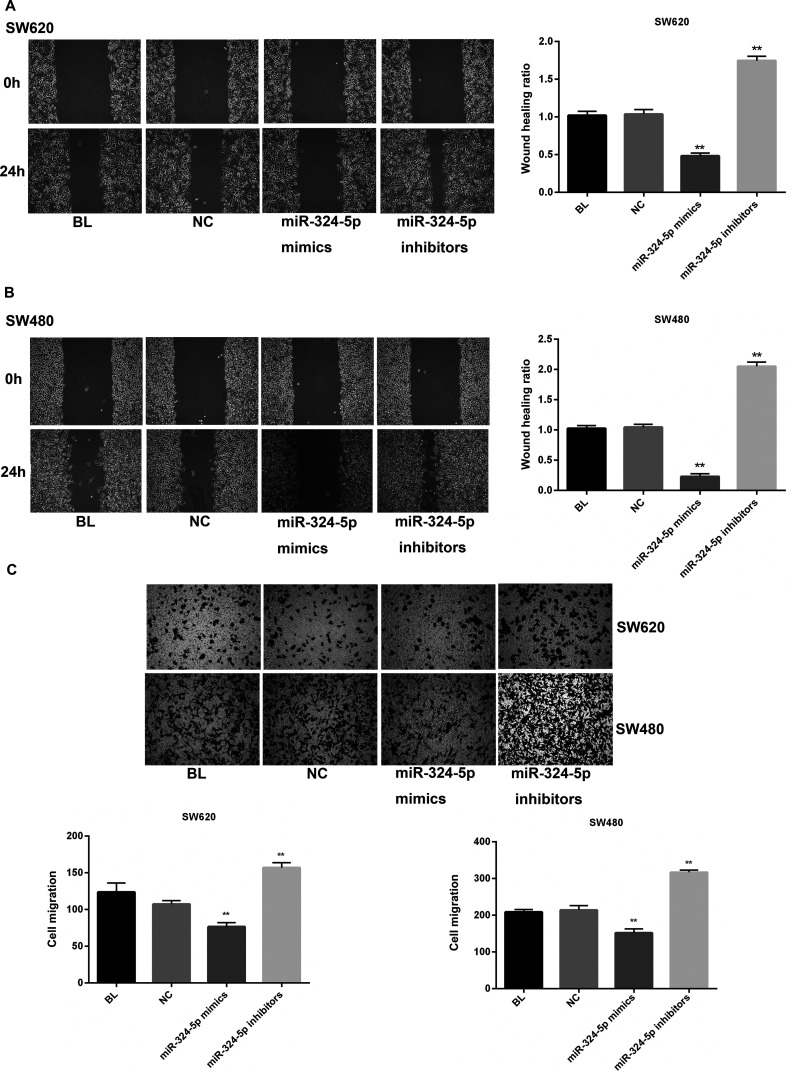
miR-324-5p Transfection Suppresses the Migration and Invasion of CRC Cells In Vitro
To further observe the effect of miR-324-5p on cell motility in CRC, the wound healing assay and Transwell invasion assay were performed. As exhibited in Figure 3A, wound closure of miR-324-5p-transfected SW620 cells was slower than cells in the blank (BL) or NC group after scratching, while miR-324-5p inhibition significantly promoted wound closure. Likewise, SW480 cells showed similar results (Fig. 3B). Invasive ability was also observed in a parallel invasive assay (Fig. 3C). In both SW620 and SW480 cells, the number of miR-324-5p mimic-transfected cells that penetrated the membrane largely decreased compared with the BL and NC groups. Also, the miR-324-5p inhibitor group showed markedly more penetrating cells than the BL and NC groups. Taken together, our findings demonstrated that miR-324-5p could suppress the migration and invasion of CRC cells.
Figure 3.
miR-324-5p suppresses migration and invasion of colorectal cells. Wound healing assay verified the effect of miR-324-5p on cell migration in SW620 (A) and SW480 (B) cells. (C) Transwell invasion assay was used to measure the invasion of SW620 and SW480 cells. **p < 0.01 versus NC group.
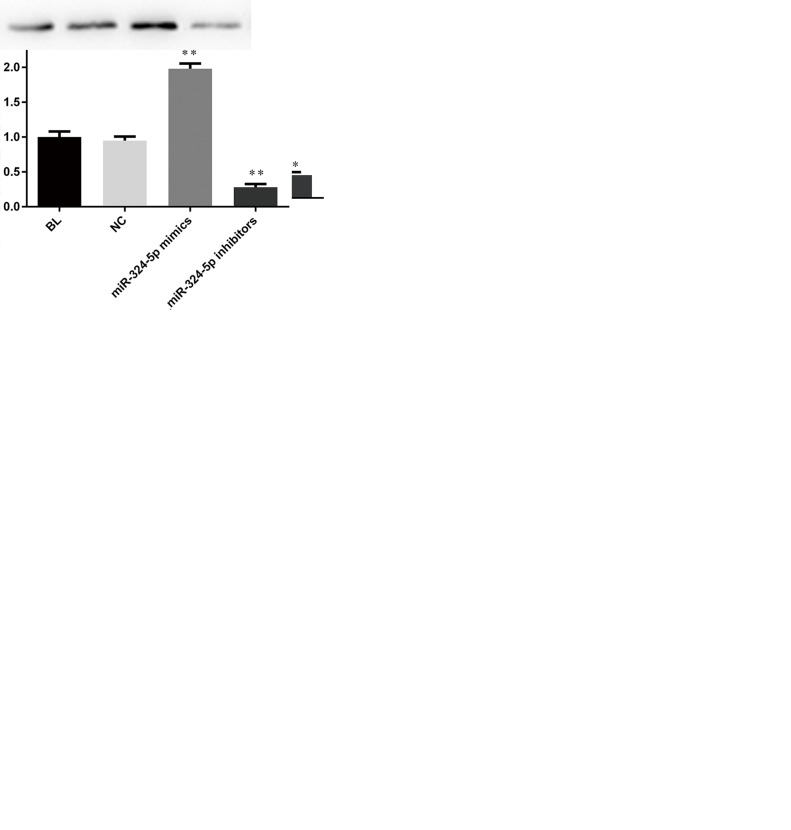
ELAVL1 Expression in SW620 and SW480 Cells Was Significantly Inhibited by miR-324-5p
We then tried to determine the influence of miR-324-5p on ELAVL1 expression in CRC cells. Figure 4A shows that in SW620 and SW480 cells, ELAVL1 protein had a much lower expression in the miR-324-5p-transfected group, while miR-324-5p inhibition significantly promoted ELAVL1 expression to a level even higher than in the BL and NC groups. In addition, RT-qPCR was applied to evaluate the mRNA level of ELAVL1. As indicated in Figure 4B, the mRNA level of ELAVL1 in CRC cells showed the same tendency as with protein. miR-324-5p overexpression decreased ELAVL1 expression, and the corresponding inhibitors markedly reversed the low expression of ELAVL1. Results showed that miR-324-5p may negatively regulate ELAVL1 in CRC.
Figure 4.
miR-324-5p overexpression suppressed ELAVL1. In SW620 and SW480 cells, ELAVL1 expressions in protein (A) and mRNA (B) were measured by Western blotting. **p < 0.01 versus NC group.
miR-324-5p Directly Binds and Regulates ELAVL1
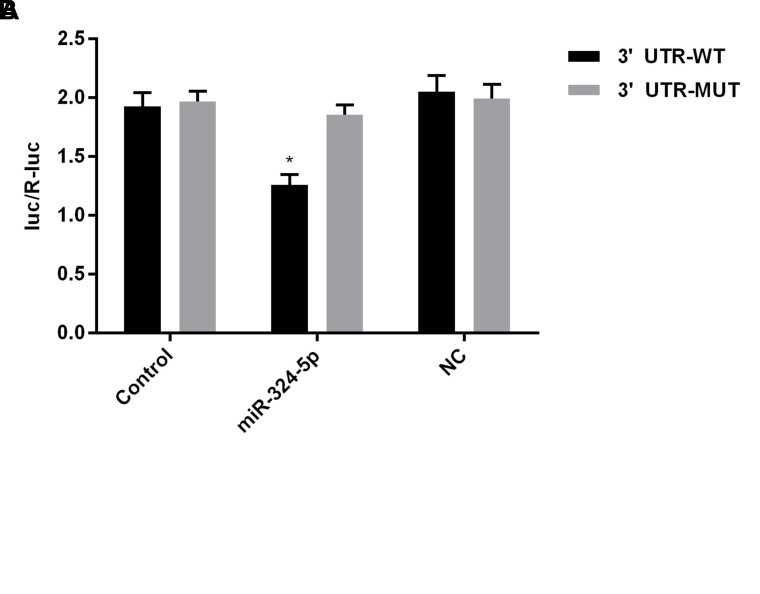
To further understand the connection of miR-324-5p and ELAVL1 in CRC, the potential target genes of miR-324-5p were analyzed by TargetScan databases. As shown in Figure 5A, ELAVL1 was one of the candidate targets. Bioinformatic analysis indicated that the 3′-UTR of ELAVL1 contains a predicted binding site for miR-324-5p. Luciferase reporter assays were then used to confirm that ELAVL1 is regulated by miR-324-5p. In Figure 5B, the relative activity of luciferase in the reporter containing a wild-type ELAVL1 3′-UTR was significantly decreased after miR-324-5p cotransfection in SW620 cells, thus confirming that ELVAL1 serves as a direct target of miR-324-5p.
Figure 5.
ELAVL1 was a direct target of miR-324-5p. (A) Predicted miR-324-5p target sequences in the 3′-untranslated region (3′-UTR) of ELAVL1. (B) The miR-324-5p target sequence from ELAVL1 was cloned into the 3′-UTR of a luciferase reporter gene. Seed site mut was used to control for binding specificity. Luciferase activity was determined by the Dual-Luciferase Reporter Assay System. *p < 0.05 versus NC group.
Proinvasion/Metastasis Protein Expression Downstream of ELAVL1 Was Inhibited by miR-324-5p in SW620 and SW480 Cells
To explore deeper the mechanism of miR-324-5p-induced proliferation and invasion inhibition, classic proinvasion/metastasis proteins MMP-9, uPA, and uPAR21 were tested by Western blotting. As shown in Figure 6, it was revealed that expression of the above proteins significantly dropped in miR-324-5p-transfected cells compared with BL and NC cells. On the other hand, miR-324-5p inhibition markedly increased the levels of MMP-9, uPA, and uPAR. On this basis, we suspect that miR-324-5p regulates cell invasion and proliferation via targeting ELAVL1 and affecting its downstream proinvasion/metastasis proteins.
Figure 6.
miR-324-5p overexpression suppressed matrix metalloproteinase 9 (MMP-9) and urokinase-type plasminogen activator (uPA) system. In SW620 (A) and SW480 (B) cells, the proinvasion factors MMP-9, uPA, and uPA receptor (uPAR) were tested by Western blotting. **p < 0.01 versus NC group.
DISCUSSION
Taken together, major findings in our current study may be listed as follows: 1) in CRC cell lines SW620 and SW480, miR-324-5p was downregulated in parallel with an overexpression of ELAVL1. 2) miR-324-5p transfection significantly suppressed proliferation ability in both SW620 and SW480 cells. Migration and invasion were also inhibited markedly by miR-324-5p overexpression. 3) miR-324-5p transfection decreased the expression of ELAVL1. 4) ELAVL1 was a direct target of miR-324-5p, and downstream proinvasion factors of ELAVL1, MMP-9, uPA, and uPAR were inhibited by miR-324-5p overexpression.
miR-324-5p is widely reported to be involved in human diseases. A previous study demonstrated that miR-324-5p attenuates mitochondrial fission and cardiomyocyte apoptosis, thus playing a part in cardiac diseases16. miR-324-5p was also observed to inhibit glioma cell proliferation by targeting glioma-associated oncogene 115. More importantly, this miRNA shows an aberrant expression in almost all human cancers, including breast cancer, esophageal carcinoma, and CRC9. In the present study, we mainly explore the role of miR-324-5p in CRC and associated mechanisms. First, this miRNA was confirmed to be low in CRC cells (Fig. 1A). Moreover, existence of miR-324-5p was found to significantly restrain proliferation (Fig. 2C and D), migration, and invasion (Fig. 3) of CRC cells, indicating that miR-324-5p may inhibit CRC development and progression. Earlier evidence provided support that miR-324-5p might function as a therapeutic molecule in CRC22,23, which is consistent with our findings, even though the mechanisms mentioned are quite different.
The RBP ELAVL1 is mainly located in the nucleus but is translocated into the cytoplasm upon cellular stress24. It is generally regarded as an mRNA-stabilizing factor to increase gene expression by binding to the 3′-UTRs of mRNAs. Previous studies show ELAVL1 takes part in cell cycle regulation, cell migration, tumorigenesis, and inflammation25. Furthermore, ELAVL1 aberrant expression is observed in various cancers, like breast, colon, and brain cancers26 and is demonstrated to correlate with poor prognosis27. However, articles focusing on the potential effect of ELAVL1 and relative molecular mechanisms in CRC were not found. In our study, for the first time, an miR-324-5p–ELAVL1 connection was proposed in CRC. We demonstrated that ELAVL1 has an abnormally high expression in CRC (Fig. 1B and C), while miR-324-5p transfection dramatically reduces ELAVL1 (Fig. 4). To confirm that miR-324-5p targets ELAVL1, a luciferase reporter assay was carried out (Fig. 5), which indicated that in CRC cells, ELAVL1 serves as a direct target of miR-324-5p. Therefore, we came to the conclusion that ELAVL1 is directly regulated by miR-324-5p and that miR-324-5p might inhibit cell proliferation and invasion in CRC, at least partially via ELAVL1 targeting.
To confirm our suspicions above and gain a deeper understanding of how miR-324-5p affects CRC, some downstream molecules of ELAVL1 were investigated (Fig. 6). The invasive growth of tumor cells is strongly dependent on ECM reconstruction28. The uPA system (uPA and its receptor uPAR) and matrix metalloproteinases (MMP-2/MMP-9) are well established as being responsible for ECM reconstruction29. Strong clinical and experimental evidence has demonstrated a correlation of elevated uPA and MMP levels with cancer progression and metastasis30,31 in CRC. In addition, ELAVL1 was believed to regulate uPA and MMPs21. In the current study, we found that miR-324-5p overexpression significantly reduced the level of uPA, uPAR, and MMP-9, which may happen to explain the invasion- and migration-inhibiting effects of miR-324-5p. However, to be more rigorous, follow-up studies are needed, and ELAVL1 inhibition or knockout should be explored.
To sum up, this study suggested that miR-324-5p inhibits cell proliferation and invasion in CRC at least in part via negatively regulating ELAVL1. Therefore, miR-324-5p might function as an effective tumor-suppressing miRNA in CRC. Our study is the first to describe miR-324-5p–ELAVL1 regulation and provides a novel and promising therapeutic target in CRC.
REFERENCES
- 1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA-Cancer J Clin. 2016;66(1):7–30. [DOI] [PubMed] [Google Scholar]
- 2. Goldberg RM, Sargent DJ, Morton RF, Fuchs CS, Ramanathan RK, Williamson SK, Findlay B, Pitot HC, Alberts SR. A randomized controlled trial of fluorouracil plus leucovorin, irinotecan, and oxaliplatin combinations in patients with previously untreated metastatic colorectal cancer. J Clin Oncol. 2004;22(1):23–30. [DOI] [PubMed] [Google Scholar]
- 3. Grothey A, Sargent DJ, Goldberg RM, Schmoll H. Survival of patients with advanced colorectal cancer improves with the availability of fluorouracil-leucovorin, irinotecan, and oxaliplatin in the course of treatment. J Clin Oncol. 2004;22(7):1209–14. [DOI] [PubMed] [Google Scholar]
- 4. Cassidy J, Twelves C, Van Cutsem E, Hoff PM, Bajetta E, Boyer MJ, Bugat R, Burger U, Garin A, Graeven U. First-line oral capecitabine therapy in metastatic colorectal cancer: A favorable safety profile compared with intravenous 5-fluorouracil/leucovorin. Ann Oncol. 2002;13(4):566–75. [DOI] [PubMed] [Google Scholar]
- 5. Leva GD, Garofalo M, Croce CM. MicroRNAs in cancer. Annu Rev Pathol-Mech. 2014;9(1):287–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ambros V. The functions of animal microRNAs. Nature 2004;431(7006):350–5. [DOI] [PubMed] [Google Scholar]
- 7. John B, Enright AJ, Aravin AA, Tuschl T, Sander C, Marks DS. Human microRNA targets. PLoS Biol. 2004;2(11):1862–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cao L, Xie B, Yang X, Liang H, Jiang X, Zhang D, Xue P, Chen D, Shao Z. miR-324-5p suppresses hepatocellular carcinoma cell invasion by counteracting ECM degradation through post-transcriptionally downregulating ETS1 and SP1. PLoS One 2015;10(7):e0133074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kuo W, Yu S, Li S, Lam H, Chang H, Chen W, Yeh C, Hung S, Liu T, Wu T. microRNA-324 in human cancer: miR-324-5p and miR-324-3p have distinct biological functions in human cancer. Anticancer Res. 2016;36(10):5189–96. [DOI] [PubMed] [Google Scholar]
- 10. Abdelmohsen K, Gorospe M. Posttranscriptional regulation of cancer traits by HuR. Wiley Interdiscip Rev RNA 2010;1(2):214–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Simone LE, Keene JD. Mechanisms coordinating ELAV/Hu mRNA regulons. Curr Opin Genet Dev. 2013;23(1):35–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Young LE, Sanduja S, Standoli KB, Pena EA, Price RL, Dixon DA. The mRNA binding proteins HuR and tristetraprolin regulate cyclooxygenase 2 expression during colon carcinogenesis. Gastroenterology 2009;136(5):1669–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Blanco FF, Preet R, Aguado A, Vishwakarma V, Stevens LE, Vyas A, Padhye S, Xu L, Weir S, Anant S. Impact of HuR inhibition by the small molecule MS-444 on colorectal cancer cell tumorigenesis. Oncotarget 2016;7(45):74043–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ahmed D, Eide PW, Eilertsen IA, Danielsen SA, Eknæs M, Hektoen M, Lind GE, Lothe RA. Epigenetic and genetic features of 24 colon cancer cell lines. Oncogenesis 2013;2(9):1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Xu H, Zong H, Shang M, Ming X, Zhao J, Ma C, Cao L. MiR-324-5p inhibits proliferation of glioma by target regulation of GLI1. Eur Rev Med Pharmacol Sci. 2014;18(6):828–32. [PubMed] [Google Scholar]
- 16. Wang K, Zhang D, Long B, An T, Zhang J, Zhou L, Liu C, Li P. NFAT4-dependent miR-324-5p regulates mitochondrial morphology and cardiomyocyte cell death by targeting Mtfr1. Cell Death Dis. 2015;6(12):1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kwan HY, Yang Z, Fong WF, Hu YM, Yu ZL, Hsiao WL. The anticancer effect of oridonin is mediated by fatty acid synthase suppression in human colorectal cancer cells. J Gastroenterol. 2013;48(2):182–92. [DOI] [PubMed] [Google Scholar]
- 18. Sui Y, Sun M, Wu F, Yang L, Di W, Zhang G, Zhong L, Ma Z, Zheng J, Fang X. Inhibition of TMEM16A expression suppresses growth and invasion in human colorectal cancer cells. PLoS One 2014;9(12):e115443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lin H, Liu W, Fang Z, Liang X, Li J, Bai Y, Lin L, You H, Pei Y, Wang F. Overexpression of DHX32 contributes to the growth and metastasis of colorectal cancer. Sci Rep. 2015;5:9247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Han Q, Zhang HY, Zhong BL, Wang XJ, Zhang B, Chen H. microRNA-145 inhibits cell migration and invasion and regulates epithelial-mesenchymal transition (EMT) by targeting connective tissue growth factor (CTGF) in esophageal squamous cell carcinoma. Med Sci Monit. 2016;22:3925–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Alahmadi W, Alghamdi M, Alsouhibani N, Khabar KSA. miR-29a inhibition normalizes HuR over-expression and aberrant AU-rich mRNA stability in invasive cancer. J Pathol. 2013;230(1):28–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chen Y, Wang S, Mu R, Luo X, Liu Z, Liang B, Zhuo H, Hao X, Wang Q, Fang D. Dysregulation of the miR-324-5p-CUEDC2 axis leads to macrophage dysfunction and is associated with colon cancer. Cell Rep. 2014;7(6):1982–93. [DOI] [PubMed] [Google Scholar]
- 23. Sun LN, Xing C, Zhi Z, Liu Y, Chen LY, Shen T, Zhou Q, Liu YH, Gan WJ, Wang JR. Dicer suppresses cytoskeleton remodeling and tumorigenesis of colorectal epithelium by miR-324-5p mediated suppression of HMGXB3 and WASF-2. Oncotarget 2017;8(34):55776–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lu Y, Chang S, Hafner M, Li X, Tuschl T, Elemento O, Hla T. ELAVL1 modulates transcriptome-wide miRNA binding in murine macrophages. Cell Rep. 2014;9(6):2330–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gorospe M, Tominaga K, Wu X, Fahling M, Ivan M. Post-transcriptional control of the hypoxic response by RNA-binding proteins and microRNAs. Front Mol Neurosci. 2011;4(4):7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Vo DT, Abdelmohsen K, Martindale JL, Qiao M, Tominaga K, Burton TL, Gelfond JAL, Brenner A, Patel V, Trageser D. The oncogenic RNA-binding protein musashi1 is regulated by HuR via mRNA translation and stability in glioblastoma cells. Mol Cancer Res. 2012;10(1):143–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wang J, Guo Y, Chu H, Guan Y, Bi J, Wang B. Multiple functions of the RNA-binding protein HuR in cancer progression, treatment responses and prognosis. Int J Mol Sci. 2013;14(5):10015–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Schuler PJ, Bendszus M, Kuehnel S, Wagner S, Hoffmann TK, Goldbrunner R, Vince GH. Urokinase plasminogen activator, uPAR, MMP-2, and MMP-9 in the C6-glioblastoma rat model. In Vivo 2012;26(4):571–6. [PubMed] [Google Scholar]
- 29. Ny T, Wahlberg P, Brandstrom IJM. Matrix remodeling in the ovary: Regulation and functional role of the plasminogen activator and matrix metalloproteinase systems. Mol Cell Endocrinol. 2002;187(1–2):29–38. [DOI] [PubMed] [Google Scholar]
- 30. Kim T, Song K, Li G, Choi H, Park H, Lim K, Hwang B, Yoon W. Activity and expression of urokinase-type plasminogen activator and matrix metalloproteinases in human colorectal cancer. BMC Cancer 2006;6(1):211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Roomi MW, Kalinovsky T, Niedzwiecki A, Rath M. Modulation of uPA, MMPs and their inhibitors by a novel nutrient mixture in human colorectal, pancreatic and hepatic carcinoma cell lines. Int J Oncol. 2015;47(1):370–6. [DOI] [PubMed] [Google Scholar]