Abstract
Cancer-associated fibroblasts (CAFs) play a predominant role in regulating tumor progression. Understanding how CAFs communicate with osteosarcoma is crucial for developing novel approaches for osteosarcoma therapy. Exosomes are able to transmit messages between cells. In this study, we demonstrated that CAFs transfer exosomes to osteosarcoma cells, which promotes osteosarcoma cell migration and invasion. Using a miRNA microarray analysis, we identified 13 miRNAs that are significantly increased in exosomes derived from cancer-associated fibroblasts (CAFs) and corresponding paracancer fibroblasts (PAFs). In vitro studies further validated that the levels of microRNA-1228 (miR-1228) were increased in CAFs, its secreted exosomes, and in recipient osteosarcoma cells, which can downregulate endogenous SCAI mRNA and protein level in osteosarcoma. Furthermore, our findings demonstrate that SCAI was downregulated in osteosarcoma tissues. Taken together, this study provides evidence that CAF exosomal miR-1228 is able to promote osteosarcoma invasion and migration by targeting SCAI, which may represent a critical therapeutic target for osteosarcoma treatment.
Key words: Osteosarcoma, Exosome, miRNAs, miR-1228, Cancer-associated fibroblasts (CAFs), Suppressor of cancer cell invasion (SCAI)
INTRODUCTION
Osteosarcoma (OS) is the eighth most common form of childhood cancer, comprising 2.4% of all malignancies in pediatric patients and about 20% of all primary bone cancers1,2. Tumor metastasis is known to be the major factor affecting OS patient survival. More than 20% of OS patients are diagnosed at an advanced stage, which is accompanied by metastasis into the lung, and the 5-year survival of these patients is extremely poor1,3. Therefore, a thorough investigation into the mechanisms of OS tumor invasion and metastasis may facilitate the identification of potential molecular targets for OS therapy.
The tumor microenvironment consists of various components such as stromal fibroblasts, endothelial cells, and extracellular matrix4. Previous studies have demonstrated that cancer-associated stromal fibroblasts (CAFs) are functionally and phenotypically different from fibroblasts in normal tissues5. Several studies have also demonstrated that CAFs can directly affect tumor cell malignancy abilities, such as proliferation, motility, drug resistance, and the epithelial-to-mesenchymal transition (EMT)6,7.
Intercellular communication between tumors and CAFs may involve direct cell-to-cell communication or the release of soluble mediators, such as cytokines, chemokines, or exosomes. Exosomes are nano-sized (30–150 nm) vesicles shed from living cells and then released into the extracellular environment. It is now well established that these vesicles can transfer their contents to and modify the phenotype of recipient cells, thereby acting as critical intracellular communication devices8–10. Exosomes can also contribute to cancer growth by enhancing antiapoptotic and other oncogenic pathways such as angiogenesis, invasion, and metastasis11–13.
MicroRNAs (miRNAs) are small, noncoding RNA molecules that negatively regulate gene expression at the posttranscriptional level in a sequence-specific manner, primarily via base pairing to the 3′-untranslated region of the target messenger RNA transcripts14,15. Mounting evidence implicates miRNAs as regulators of the tumor phenotype through their ability to modulate the expression of critical genes and signaling networks involved in tumor genesis and downstream malignant processes16–18.
In the present study, we demonstrate that exosomes derived from CAFs are able to promote the migration and invasion of osteosarcoma cells through the shuttling of miR-1228, subsequently leading to depression of the expression of suppressor of cancer cell invasion (SCAI) in osteosarcoma cells.
MATERIALS AND METHODS
Clinical Samples
The human study was approved by the Institutional Ethics Committee of Kunshan Hospital (Shanghai, P.R. China). For the evaluation of SCAI expression using tumor specimens, 10 patients with diagnosed OS were enrolled in this study. Ten pairs of matched primary osteosarcoma adjacent tumor-free tissues (5 cm from the cut edge of the tumor edge) were also obtained.
Cell Lines and Cell Culture
The human osteosarcoma cell lines MG-63 and HOS were purchased from the ATCC (Manassas, VA, USA). The cells were cultured in high-glucose Dulbecco’s modified Eagle’s medium (DMEM; Gibco, Brooklyn, NY, USA), supplemented with 10% fetal bovine serum (FBS; Gibco), at 37°C with 5% CO2. CAFs and corresponding paracancer fibroblasts (PAFs) were prepared as previously described19. In brief, six pairs of matched primary osteosarcoma adjacent tumor-free tissues (5 cm from the cut edge of the tumor edge) were obtained. The tissues were immersed into serum-free DMEM. Tissues were cut into 1-mm3 fragments and were collected into C-type tubes with 5 ml of serum-free DMEM containing 0.5% collagenase I for 1 h of digestion. After gentle treatment using a MACS Dissociator (Miltenyi Biotec, Germany) for 2 min, a single-cell suspension was then collected and centrifuged at 1,000 rpm/min for 5 min. The cell pellets were suspended with 15% fetal bovine serum (FBS)-containing DMEM and were seeded into T25 tissue culture flasks. Nonadherent cells and tissues were removed by washing twice with PBS after 48 h. The adherent stromal fibroblasts were then further incubated for 4–7 days.
Exosome Purification
Exosomes were prepared from the supernatant of CAFs and PAFs by differential centrifugations as previously described20. Briefly, CAFs were cultured in high-glucose DMEM (Gibco) supplement with 10% exosome-depleted FBS (SBI, Palo Alto, CA, USA) for 2 days, after which the medium was collected, centrifuged at 3,500 × g for 10 min to eliminate cells, followed by filtration through a 0.22-μm filter to remove cell debris. Exosomes were then pelleted by ultracentrifugation (Beckman SW-34 rotor, Brea, CA, USA) at 120,000 × g for 70 min.
Electron Microscopy
Exosomes to be examined by transmission electron microscope (TEM) were isolated and loaded onto a carbon-coated electron microscopy grid as described previously20. The samples were fixed with 2% glutaraldehyde and 2% paraformaldehyde in 0.1 mol/L sodium cacodylate buffer at pH 7.3 for 3 h at room temperature. Samples were critical-point dried, mounted on specimen stubs, sputter coated, and visualized using a TEM.
Western Blot Analysis
Protein was separated on a 10% SDS-PAGE gel and transferred to polyvinylidene difluoride membranes (Millipore, Boston, MA, USA). Membranes were blocked and then incubated overnight with mouse anti-CD9 (1:2,000; Sigma-Aldrich, St. Louis, MO, USA), mouse anti-CD63 (1:1,000; Sigma-Aldrich), mouse anti-Alix (1:1,000; Sigma-Aldrich), rabbit anti-calnexin (1:1,500; Abcam, Cambridge, MA, USA), mouse anti-SCAI (1:1,000; Sigma-Aldrich), rabbit anti-fibronectin (1:2,000; Sigma-Aldrich), and rabbit anti-α-SMA (1:1,500; Sigma-Aldrich). Horseradish peroxidase-conjugated anti-mouse or anti-rabbit IgG (1:5,000; Sigma-Aldrich) was used as a secondary antibody (diluted 1:5,000 in TBST).
Scratch Assay
The scratch assay was performed to measure cell migration in vitro as described previously21. Briefly, cells were reseeded onto 60-mm dishes to create a confluent monolayer. The cell monolayer was scraped in a straight line to create a “scratch” with a p200 pipette tip and then incubated with tumor-derived exosomes (10 mg/ml). The first image of the scratch was acquired, and cells were cultured in the incubator at 37°C for 24 h prior to acquisition of the second image. The percent wound closure (%) = migrated cell surface area/total surface area × 100.
Invasion Assay
A cell invasion assay was conducted with BioCoat Matrigel Invasion Chambers (Corning, Corning, NY, USA) according to the manufacturer’s instructions. Briefly, cells were seeded into the extracellular matrix layer. Exosomes (10 mg/ml) were added to the bottom chambers as chemoattractant. After 24 h the cells that migrated through the membrane were stained with 0.4% crystal violet, and the number of cells was counted under 200× magnification. Data represent at least three experiments performed in triplicate.
Transfection
Cells were transfected with miR-1228 mimic using RNA-fectin reagent or with plasmid DNA using DNA-fectin (both from Applied Biological Materials, Vancouver, Canada) following the manufacturer’s protocol. The Lenti-X shRNA expression system (Clontech/TaKaRa, Mountain View, CA, USA) was used for the shRNA-mediated inhibition of SCAI expression, and the knocking down efficiency of SCAI lentiviral shRNAs has been demonstrated.
RNA Extraction and miRNA Profiling by RT-PCR
RNA from exosomes was isolated and enriched using a miRNeasy Mini Kit (Qiagen, Gaithersburg, MD, USA) according to the manufacturer’s instructions, and the total RNA from osteosarcoma treated with the exosomes (100 mg/ml) after 24 h was extracted using TRIzol (Invitrogen, Carlsbad, CA, USA). Mature miRNA-1228 was reverse transcribed with specific RT primers and was then quantified with a TaqMan probe using TaqMan miRNA assays (Applied Biosystems, Foster City, CA, USA). The human snRNA RNU6B (U6) was used for data normalization of miRNA expression. The U6 was used for normalization of exosomal miRNA expression.
miRNA Microarray Assays
Microarray assays were performed on exosomes from CAFs and PAFs using Human miR Microarray Kits (Agilent, Santa Clara, CA, USA). A total of 100 ng of total RNA from each sample was phosphatase treated and then labeled with cyanine 3-pCp. The labeled RNA was purified using Micro Bio-spin columns (Bio-Rad, Hercules, CA, USA) and was subsequently hybridized to a human miRNA microarray slide at 55°C for 20 h. After hybridization, the slides were washed with Gene Expression Wash Buffer (Agilent) and scanned on an Agilent Microarray Scanner using Agilent’s Scan Control version A.7.0.1 software. Raw hybridization intensities were obtained using Agilent’s feature extraction software, and a total of 2,006 miRNAs were probed.
Statistical Analysis
The comparisons of means among groups were analyzed by one-way ANOVA, and the Dunn multiple comparison test was further used to determine significant differences between groups. All statistical analyses were performed using the SPSS package (version 13.0). A value of p < 0.05 was considered statistically significant.
RESULTS
Exosomes Derived From CAFs Are Internalized by Osteosarcoma Cells
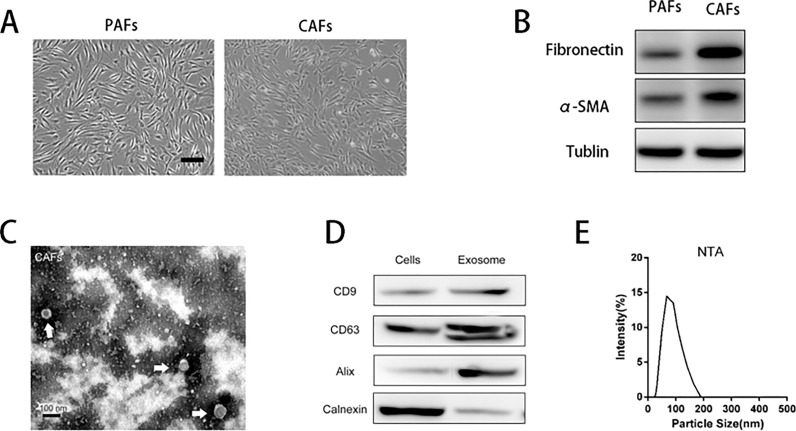
Primary CAFs and PAFs were isolated from HCC tumor tissues using 0.5% type I collagenase. We next characterized the phenotypes of PAFs and CAFs (Fig. 1A). We performed Western blotting to analyze the expression of the well-recognized markers fibronectin and α-SMA (Fig. 1B), indicating the expression of fibronectin and α-SMA in CAFs. Sequential centrifugations were used to collect exosomes from the supernatant of CAFs. The purified exosomes were characterized by TEM, Western blot, and nanoparticle tracking analysis (NTA). Exosomes purified from conditioned media by ultracentrifugation exhibited typical cup-shaped morphology by electron microscopy (Fig. 1C), similar to previously described exosomes20. The studied vesicles were further confirmed as exosomes by Western blot analysis, which show the presence of the surface proteins CD63, CD9, Alix (commonly used marker of exosomes), and absence of calnexin (commonly used marker of endoplasmic reticulum) (Fig. 1D). NTA result also shows that the size of purified exosome ranges from 30 to 150 nm (Fig. 1E).
Figure 1.
Identification of exosomes derived from cancer-associated fibroblasts (CAFs). (A) Representative morphology of para-cancer fibroblasts (PAFs) and CAFs derived from osteosarcoma (OS) patients. Scale bar: 100 μm. (B) Western blot analysis of the expression of CAF markers (α-SMA, fibronectin) in isolated fibroblasts. (C) Electron microscopy images of exosomes purified from the conditioned medium of CAFs. Scale bar: 100 nm. (D) Western blot analysis showing the presence of CD9, CD63, and Alix, and the absence of calnexin in CAF-secreted exosomes. (E) An exosome size range of 30–100 nm was measured by nanoparticle tracking analysis.
Exosomes Derived From CAFs Promote the Migration and Invasion of Osteosarcoma Cells
Once secreted, exosomes deliver biologic information by internalization by neighboring or distant cells8,22. We next investigated the effects of CAF-derived exosomes on the migration and invasion of osteosarcoma cells. The results are presented in Figure 2. Wound healing assays showed that CAF exosomes significantly increased the migration of MG-63 and HOS (Fig. 2A and B), compared with PBS and PAF exosomes. In accordance with the migration results, invasion was also significantly increased by CAF-derived exosomes compared with PBS and PAF exosomes (Fig. 2C and D). These results indicate that CAF-derived exosomes play a pivotal role in migration and invasion ability.
Figure 2.
Exosomes derived from CAFs promote the migration and invasion of OS cells. (A) OS cells treated as indicated were subjected to Transwell migration. Images were acquired at 0 h and 24 h (magnification 200×). (B) Quantitative analysis of scratch wound closure. (C) OS cells treated as indicated were subjected to invasion assay. Images were acquired at 24 h (magnification 200×). (D) Quantitative analysis of the invaded cells. Data represent at least three experiments performed in triplicate. **p < 0.01.
miR-1228 Is Significantly Increased in the Exosomes of CAFs
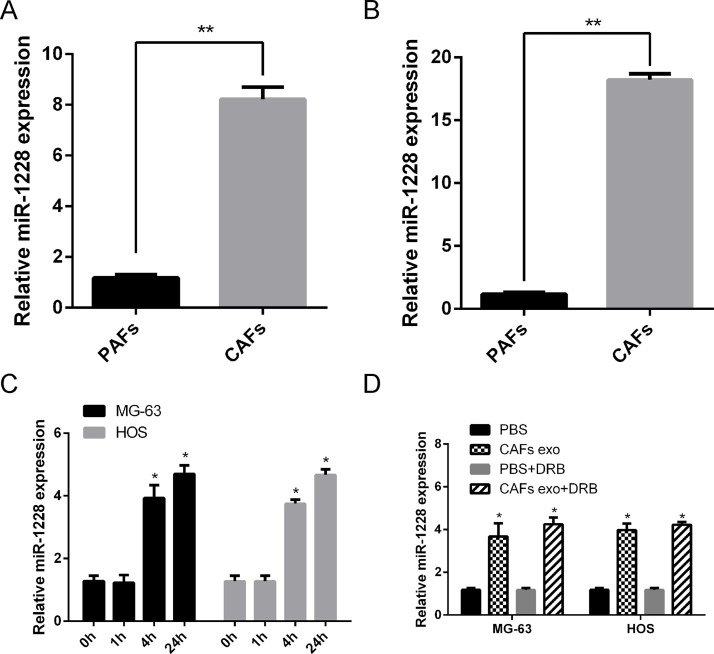
The exosomal transfer of miRNAs has been regarded as a novel and important mechanism of genetic exchange between cells7. To identify differentially expressed miRNAs in exosomes from CAFs and PAFs, miRNA microarray technology was applied and identified changes in 18 functional miRNAs (13 upregulated and 5 downregulated) in exosomes (Table 1). miR-1228 has been characterized as an oncogenic driver in many human cancers, including ovarian cancer, and we further focused on this miRNA. qRT-PCR analyses showed a higher level of miR-1228 in CAFs (p < 0.05) (Fig. 3A) and in exosomes purified from CAFs (Fig. 3B), compared with those derived from PAFs. Furthermore, increased levels of miR-1228 were observed in both osteosarcoma cell lines after 24-h incubation with CAF-derived exosomes (Fig. 3C). To further confirm that CAF-secreted miR-1228 can be transferred to osteosarcoma cells via exosomes, we measured the miR-1228 levels in MG-63 and HOS cells treated with exosomes derived from CAFs. An increased expression of the cellular level of mature miR-1228 was observed in recipient osteosarcoma cells with kinetics starting at 4 h and peaking at 24 h. We conclude that this increase in miR-1228 expression reflects the exosome-mediated miRNA transfer but is not an induction of miR-1228’s endogenous expression in the recipient cells, as its level in exosome-treated cells was not significantly affected by an RNA polymerase II inhibitor (Fig. 3D). These results indicate that an enrichment in the level of miR-1228 was observed in CAFs and also in exosomes secreted by CAFs and the recipient osteosarcoma cells.
Table 1.
Exosomal miRNAs Overexpressed in Cancer-Associated Fibroblasts (CAFs) Compared to Paracancer Fibroblasts (PAFs)
| No. | miRNA | Fold Expression |
|---|---|---|
| 1 | miR-1228 | 33.21 ± 2.14 |
| 2 | miR-10a | 32.38 ± 1.74 |
| 3 | miR-6126 | 30.29 ± 0.68 |
| 4 | miR-675 | 19.07 ± 1.06 |
| 5 | miR-375 | 19.03 ± 1.01 |
| 6 | miR-552 | 15.02 ± 0.04 |
| 7 | miR-99a | 15.00 ± 0.22 |
| 8 | miR-7e | 14.28 ± 2.71 |
| 9 | miR-106b | 12.92 ± 0.57 |
| 10 | miR-142 | 12.25 ± 0.47 |
| 11 | miR-218 | 12.02 ± 0.11 |
| 12 | miR-32 | 11.99 ± 0.24 |
| 13 | miR-139 | 11.87 ± 0.27 |
Figure 3.
miR-1228 enriches in CAFs, its secreted exosomes, and the recipient OS cells. (A) Cellular RNA was extracted from PAFs and CAFs and subjected to miR-1228 qRT-PCR. Data were normalized to levels of U6. (B) Exosomal RNA was extracted from PAFs and CAFs and subjected to miR-1228 qRT-PCR. Data were normalized to levels of miR-16. (C) qRT-PCR analyses were used to detect the levels of miR-1228 in OS cells after incubation with exosomes from CAFs (p < 0.05). (D) CAF-secreted exosomes were fed to MG-63 and HOS cells in the presence or absence of 5,6-dichloro-1-β-d-ribofuranosylbenzimidazole (20 μM). After 24 h, RNA extracted from the recipient cells was analyzed for miR-1228 level. *p < 0.05. **p < 0.01.
Exosomal miR-1228 Transferred From CAFs Modulates Migration of Osteosarcoma Cells
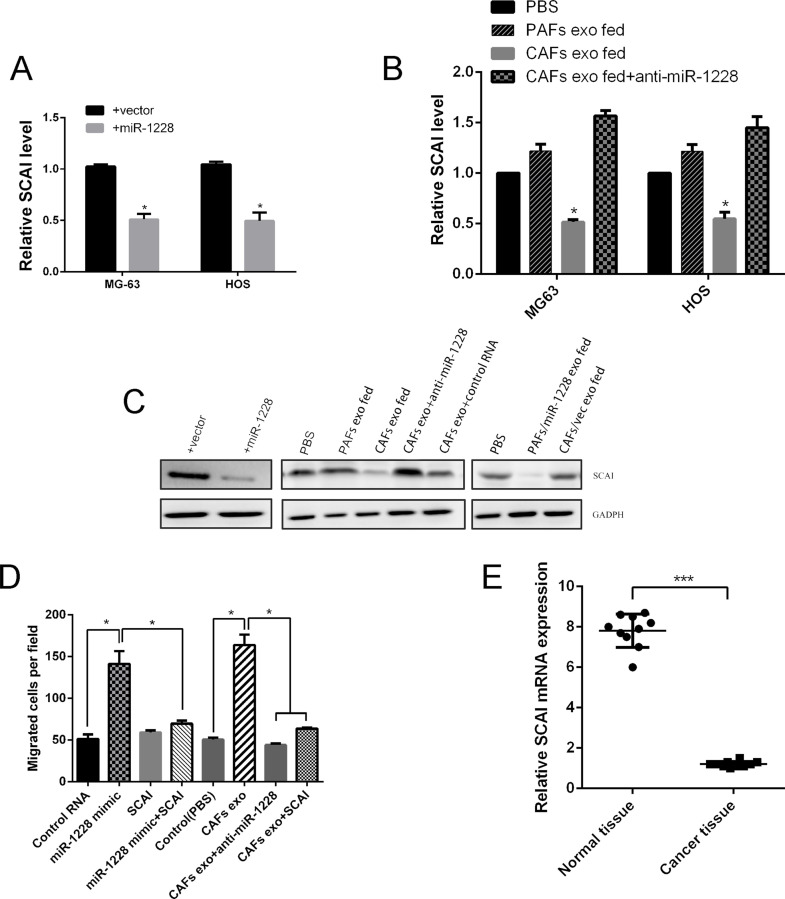
We next investigated the potential mechanism of the function of exosomal miR-1228 on cell invasion and migration. To assess whether miR-1228 effects on cellular invasion and migration are also due to the suppression of SCAI expression in osteosarcoma, we performed a rescue experiment. Ectopic expression of miR-1228, or treatment with exosomes derived from the CAFs (high miR-1228), but not the PAFs (low miR-1228), result in a significant reduction in SCAI expression at both the mRNA and protein levels in MG-63 and HOS osteosarcoma cells (Fig. 4A–C).The effect of CAFs exosomes could be abolished by transfecting the recipient cells with miR-1228 inhibitor (Fig. 4B and C).We also overexpressed SCAI in osteosarcoma cells and the migration of MG63 was decreased, compared with miR-1228 mimics treatment (Fig. 4D). Consistent with these results, ectopic expression of miR-1228 or treatment with CAF exosomes significantly induced migration in MG-63 through the miR-1228/SCAI-mediated mechanism. To further evaluate the expression of SCAI in osteosarcoma tissues, qRT-PCR was used to measure the SCAI mRNA expression levels in osteosarcoma, and we observed that SCAI mRNA expression was significantly decreased in osteosarcoma tissue compared with adjacent normal tissues (Fig. 4E). These findings suggested that CAF-derived miR-1228-induced osteosarcoma cell invasion and migration were mediated by SCAI.
Figure 4.
Exosomal miR-1228 transferred from CAFs modulates the migration of OS cells. (A) MG-63 and HOS cells were transduced with miR-1228 or vector and analyzed for suppressor of cancer cell invasion (SCAI) expression by qRT-PCR. (B) MG-63 and HOS cells were treated as indicated and were analyzed for SCAI mRNA expression. (C) MG-63 treated as indicated was analyzed by Western blot. (D) MG-63 treated as indicated was subjected to transwell migration. Cells that had migrated within 8 h were quantified from triplicate wells. (E) Relative mRNA expression of SCAI in OS tissue (n = 10) and adjacent normal tissue (n = 10). *p < 0.05. ***p < 0.001.
DISCUSSION
Previous studies have demonstrated that CAFs play a critical role in affecting tumor cell malignancy abilities, such as proliferation, motility, drug resistance, and EMT7,19. However, the underlying mechanism(s) has not been well characterized. In recent years, a large number of studies have recognized that the exosomal transfer of factors, such as miRNAs or proteins, may play an important role in communication between cancer cells and the CAFs23–25. However, there are few reports about the specific effect of the transfer of exosomes from CAFs to osteosarcoma cells.
In the present study, we purified exosomes from CAFs and PAFs media, and characterized the structure of exosomes by TEM, Western blot, and NTA (Fig. 1B–D). We then demonstrated a novel function where CAF-derived exosomes could promote the migration and invasion of osteosarcoma (Fig. 2A and D). Understanding the role of CAF-derived exosomes in osteosarcoma provides important new insights into the mechanisms of osteosarcoma progression and recurrence.
To further validate the mechanism, we compared the miRNA profiles of exosomes from six paired CAFs and PAFs of osteosarcoma patients using miRNA array. miRNAs were confirmed to be contained in exosomes and could be transferred between cells26. We identified a total of 18 miRNAs that showed a significant difference between CAF and PAF exosomes. After screening the miRNAs with the criterion that miRNAs should be detected in all of the samples with at least a twofold change, we chose miR-1228 for further experiments. Although miR-1228 has been shown to be overexpressed in many different cancer types and to promote cancer cell proliferation, migration, and survival, the function of exosomal miR-1228 in osteosarcoma has not been characterized27. Our in vitro studies showed that the levels of miR-1228 not only increased in CAFs and exosomes from CAFs (Fig. 3A and B) but also increased in the recipient osteosarcoma cells (Fig. 3C). We also observed that pretransfection of antagomiR-1228 decreased the level of miR-1228 in secreted exosomes and the recipient osteosarcoma cells, which then resulted in reduction in migration and invasion. This experiment with the antagomiR-1228 provided further evidence that exosomal miR-1228 transferred from CAFs was able to modulate the migration of osteosarcoma cells (Fig. 4A and D).
We next investigated the possible downstream signaling pathways that may participate in the effect of miR-1228 in osteosarcoma. Previous studies have demonstrated that miR-1228 promotes tumor migration and invasion by targeting SCAI in breast cancer28–30. In this study, we demonstrated that the exosomal miR-1228 could promote invasion and migration of osteosarcoma by downregulating SCAI mRNA expression of SCAI in osteosarcoma (Fig. 4B and C).
In conclusion, the present study provides the first evidence that CAFs are able to transfer miR-1228 via exosomes to human osteosarcoma cells, eventually leading to promote migration and invasion of the osteosarcoma cells in vitro. Enforced miR-1228 expression can then lead to a reduced expression of SCAI, which is a key mediator that controls tumor migration and invasion. The findings from our study are important as they provide new insights into the pathogenesis of osteosarcoma and identify potential new targets that can then be used in the treatment of osteosarcoma.
ACKNOWLEDGMENT
The authors declare no conflicts of interest.
Footnotes
The authors declare no conflicts of interest.
REFERENCES
- 1. Kansara M, Teng MW, Smyth MJ, Thomas DM. Translational biology of osteosarcoma. Nat Rev Cancer 2014;14(11):722–35. [DOI] [PubMed] [Google Scholar]
- 2. Gianferante DM, Mirabello L, Savage SA. Germline and somatic genetics of osteosarcoma—Connecting aetiology, biology and therapy. Nat Rev Endocrinol. 2017;13(8):480–91. [DOI] [PubMed] [Google Scholar]
- 3. Michaelis J. Osteosarcoma. Lancet 1988;1(8595):1174. [DOI] [PubMed] [Google Scholar]
- 4. Hu Y, Li D, Wu A, Qiu X, Di W, Huang L, Qiu L. TWEAK-stimulated macrophages inhibit metastasis of epithelial ovarian cancer via exosomal shuttling of microRNA. Cancer Lett. 2017;393:60–7. [DOI] [PubMed] [Google Scholar]
- 5. Yang L, Achreja A, Yeung TL, Mangala LS, Jiang D, Han C, Baddour J, Marini JC, Ni J, Nakahara R, Wahlig S, Chiba L, Kim SH, Morse J, Pradeep S, Nagaraja AS, Haemmerle M, Kyunghee N, Derichsweiler M, Plackemeier T, Mercado-Uribe I, Lopez-Berestein G, Moss T, Ram PT, Liu J, Lu X, Mok SC, Sood AK, Nagrath D. Targeting stromal glutamine synthetase in tumors disrupts tumor microenvironment-regulated cancer cell growth. Cell Metab. 2016;24(5):685–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Labernadie A, Kato T, Brugues A, Serra-Picamal X, Derzsi S, Arwert E, Weston A, Gonzalez-Tarrago V, Elosegui-Artola A, Albertazzi L, Alcaraz J, Roca-Cusachs P, Sahai E, Trepat X. A mechanically active heterotypic E-cadherin/N-cadherin adhesion enables fibroblasts to drive cancer cell invasion. Nat Cell Biol. 2017;19(3):224–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Garcia-Silva S, Peinado H. Melanosomes foster a tumour niche by activating CAFs. Nat Cell Biol. 2016;18(9):911–3. [DOI] [PubMed] [Google Scholar]
- 8. Becker A, Thakur BK, Weiss JM, Kim HS, Peinado H, Lyden D. Extracellular vesicles in cancer: Cell-to-cell mediators of metastasis. Cancer Cell 2016;30(6):836–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hoshino A, Costa-Silva B, Shen TL, Rodrigues G, Hashimoto A, Tesic Mark M, Molina H, Kohsaka S, Di Giannatale A, Ceder S, Singh S, Williams C, Soplop N, Uryu K, Pharmer L, King T, Bojmar L, Davies AE, Ararso Y, Zhang T, Zhang H, Hernandez J, Weiss JM, Dumont-Cole VD, Kramer K, Wexler LH, Narendran A, Schwartz GK, Healey JH, Sandstrom P, Labori KJ, Kure EH, Grandgenett PM, Hollingsworth MA, de Sousa M, Kaur S, Jain M, Mallya K, Batra SK, Jarnagin WR, Brady MS, Fodstad O, Muller V, Pantel K, Minn AJ, Bissell MJ, Garcia BA, Kang Y, Rajasekhar VK, Ghajar CM, Matei I, Peinado H, Bromberg J, Lyden D. Tumour exosome integrins determine organotropic metastasis. Nature 2015;527(7578):329–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Huang Z, Feng Y. Exosomes derived from hypoxic colorectal cancer cells promote angiogenesis through Wnt4-induced beta-catenin signaling in endothelial cells. Oncol Res. 2017;25(5):651–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Skog J, Wurdinger T, van Rijn S, Meijer DH, Gainche L, Sena-Esteves M, Curry WT Jr, Carter BS, Krichevsky AM, Breakefield XO. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol. 2008;10(12):1470–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yu Z, Zhao S, Ren L, Wang L, Chen Z, Hoffman RM, Zhou J. Pancreatic cancer-derived exosomes promote tumor metastasis and liver pre-metastatic niche formation. Oncotarget 2017;8(38):63461–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lang HL, Hu GW, Zhang B, Kuang W, Chen Y, Wu L, Xu GH. Glioma cells enhance angiogenesis and inhibit endothelial cell apoptosis through the release of exosomes that contain long non-coding RNA CCAT2. Oncol Rep. 2017;38(2):785–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rupaimoole R, Calin GA, Lopez-Berestein G, Sood AK. miRNA deregulation in cancer cells and the tumor microenvironment. Cancer Discov. 2016;6(3):235–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chen LL. The biogenesis and emerging roles of circular RNAs. Nat Rev Mol Cell Biol. 2016;17(4):205–11. [DOI] [PubMed] [Google Scholar]
- 16. Cao Z, Zheng X, Cao L, Liang N. MicroRNA-539 inhibits the epithelial-mesenchymal transition of esophageal cancer cells by twist-related protein 1-mediated modulation of melanoma associated antigen A4 (MAGEA4). Oncol Res. 2018;26(4):529–36. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 17. Xiao J, Li G, Zhou J, Wang S, Liu D, Shu G, Zhou J, Ren F. MicroRNA-520b functions as a tumor suppressor in colorectal cancer by inhibiting DCUN1D1. Oncol Res. 2018;26(4):593–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Li Y, Chen M, Liu J, Li L, Yang X, Zhao J, Wu M, Ye M. Upregulation of microRNA-18b contributes to the development of colorectal cancer through inhibiting CDKN2B. Mol Cell Biol. 2017;37(22). pii: e00391–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhang Z, Li X, Sun W, Yue S, Yang J, Li J, Ma B, Wang J, Yang X, Pu M, Ruan B, Zhao G, Huang Q, Wang L, Tao K, Dou K. Loss of exosomal miR-320a from cancer-associated fibroblasts contributes to HCC proliferation and metastasis. Cancer Lett. 2017;397:33–42. [DOI] [PubMed] [Google Scholar]
- 20. Thery C, Amigorena S, Raposo G, Clayton A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr Protoc Cell Biol. 2006;Chapter 3:Unit 3.22. [DOI] [PubMed] [Google Scholar]
- 21. Wang G, Fu Y, Liu G, Ye Y, Zhang X. miR-218 inhibits proliferation, migration, and EMT of gastric cancer cells by targeting WASF3. Oncol Res. 2017;25(3):355–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Akishiba M, Takeuchi T, Kawaguchi Y, Sakamoto K, Yu HH, Nakase I, Takatani-Nakase T, Madani F, Graslund A, Futaki S. Cytosolic antibody delivery by lipid-sensitive endosomolytic peptide. Nat Chem. 2017;9(8):751–61. [DOI] [PubMed] [Google Scholar]
- 23. Aran D, Butte AJ. Digitally deconvolving the tumor microenvironment. Genome Biol. 2016;17(1):175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Quail DF, Joyce JA. The microenvironmental landscape of brain tumors. Cancer Cell 2017;31(3):326–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tonry C, Armstrong J, Pennington S. Probing the prostate tumour microenvironment II: Impact of hypoxia on a cell model of prostate cancer progression. Oncotarget 2017;8(9):15307–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007,9(6):654–9. [DOI] [PubMed] [Google Scholar]
- 27. Lin L, Liu D, Liang H, Xue L, Su C, Liu M. MiR-1228 promotes breast cancer cell growth and metastasis through targeting SCAI protein. Int J Clin Exp Pathol. 2015;8(6):6646–55. [PMC free article] [PubMed] [Google Scholar]
- 28. Lin KC, Moroishi T, Meng Z, Jeong HS, Plouffe SW, Sekido Y, Han J, Park HW, Guan KL. Regulation of Hippo pathway transcription factor TEAD by p38 MAPK-induced cytoplasmic translocation. Nat Cell Biol. 2017;19(8):996–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wada M, Canals D, Adada M, Coant N, Salama MF, Helke KL, Arthur JS, Shroyer KR, Kitatani K, Obeid LM, Hannun YA. P38 delta MAPK promotes breast cancer progression and lung metastasis by enhancing cell proliferation and cell detachment. Oncogene 2017;36(47):6649–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zheng HL, Wang LH, Sun BS, Li Y, Yang JY, Wu CF. Oligomer procyanidins (F2) repress HIF-1alpha expression in human U87 glioma cells by inhibiting the EGFR/AKT/mTOR and MAPK/ERK1/2 signaling pathways in vitro and in vivo. Oncotarget 2017;8(49):85252–62. [DOI] [PMC free article] [PubMed] [Google Scholar]