Abstract
Profilin 2 (PFN2) was found to be mainly expressed in neurons and involved in the development of the brain. In recent years, emerging evidence indicated that PFN2 is also significantly upregulated in various cancers including head and neck cancer (HNSC) and influences cancer cell proliferation, migration, and invasion. However, the role of PFN2 in HNSC development and progression remains unclear. The aim of our study was to investigate the role of PFN2 in the development of HNSC and its possible molecular mechanisms. Bioinformatics showed that increased expression of PFN2 in tumors correlated highly with poor prognosis of HNSC patients. Our results indicated that PFN2 was highly expressed in HNSC tissues and in HNSC cell lines. Knockdown of PFN2 inhibited proliferation, invasion, and migration of HNSC cells, while PFN2 overexpression produced the opposite effects. Using a nude mouse xenograft model, we substantiated the tumor-promoting effect of PFN2 on HNSC in vivo. Furthermore, we found that PFN2 downregulation reduced the phosphorylation of Akt and GSK-3β and reduced the expression of β-catenin in HNSC cells. The opposite was observed when PFN2 was overexpressed. Collectively, these results suggest that PFN2 promotes the proliferation and metastasis of HNSC by activating the PI3K/Akt/β-catenin signaling pathway. Although further validation is needed, we speculate that PFN2 plays a crucial role in HNSC and may be a promising therapeutic target and prognostic biomarker.
Key words: Head and neck cancer (HNSC), Profilin 2 (PFN2), Proliferation, Migration, Invasion, PI3K/Akt/β-catenin pathway
INTRODUCTION
Head and neck cancer (HNSC) arises from the oral and nasal cavities, pharynx, and larynx1. It is the sixth most common cancer worldwide with more than 650,000 patients diagnosed each year and a mortality rate of 300,0002. Unlike many other epithelial cancers, most HNSCs are locally advanced with some that metastasize to nearby cervical lymph nodes3. Tobacco and alcohol consumption and human papillomavirus (HPV) infection are the major known causes of HNSC4. Despite medical advancements in HNSC detection and treatment, the 5-year survival rate of patients has not been significantly improved5. At present, even the most aggressive treatment yields a mere 30% response rate, and the median progression-free survival and median overall survival are not significantly extended6,7. HNSC patients often have difficulty in breathing and swallowing after radiotherapy and chemotherapy, which further reduce their quality of life8. Therefore, research into the molecular mechanism behind HNSC tumorigenesis is both necessary and beneficial to the discovery of novel therapeutic strategies.
Profilins (PFNs) are a family of actin-binding proteins that contains four isoforms encoded by PFN1, PFN2, PFN3, and PFN49. They are involved in cell motility and cell shape changes through regulating the dynamics of actin polymerization and reorganization of the actin cytoskeleton9. In mammalian cells, PFN1 and PFN2 are the two most widely expressed isoforms of the PFN family10. Recently, the role of PFNs in regulating tumor progression and modulating Akt signaling pathway has been proposed11. Originally, PFN2 was thought to be expressed exclusively in the developing nervous system and participate in neurotransmitter exocytosis12. In recent years, studies have shown that PFN2 dysregulation is correlated with the occurrence of multiple tumors13–17. It was found that PFN2 promoted tumor cell migration and invasion by inducing an epithelial-to-mesenchymal transition (EMT) phenotype in lung cancer, colorectal cancer (CRC), and esophageal squamous cell carcinoma (ESCC)16–18. PFN2 was also shown to positively correlate with invasion depth and lymph node metastasis and acts as an independent prognostic factor for poor overall survival in ESCC16. However, one study found that the expression of PFN2 was downregulated in metastatic colon cancer, and low PFN2 expression was correlated with enhanced EMT in CRC cells19. While PFN2 expression may be cancer type and stage specific, how its level changes in tumors remains controversial. In particular, the general function of PFN2 in HNSC is poorly understood.
In our study, we surveyed the expression of PFN2 in HNSC tissues from online database and further verified the results in multiple HNSC cell lines. PFN2 was then overexpressed and knocked down in vitro, and changes in HNSC cell proliferation, migration, and invasion were examined. Results indicated that PFN2 was significantly upregulated in HNSC tissues and cell lines. Knockdown of PFN2 strikingly restrained proliferation, migration, and invasion of HNSC cells. In the nude mouse xenograft model, we found that PFN2 knockdown inhibited the proliferation of HNSC cells, while overexpression of PFN2 did the opposite. Furthermore, we discovered that PFN2 facilitated the oncogenic ability of HNSC cells by activating the phosphatidylinositol-4,5-bisphosphate 3-kinase (PI3K)/AKT serine/threonine kinase (Akt)/β-catenin pathway.
MATERIALS AND METHODS
Cell Lines and Cell Culture
The human HNSC cell lines Fadu, SSC-9, SSC-25, OSC-19, and Cal-27, and normal oral mucosal epithelial cells, HOK, were purchased from American Type Culture Collection (ATCC, Manassas, VA, USA) and cultured in DMEM/F12 (Gibco, Thermo Fisher Scientific, Waltham, MA, USA) supplemented with 10% fetal bovine serum (Gibco, Thermo Fisher Scientific), 100 U/ml penicillin, and 100 μg/ml streptomycin (Gibco, Thermo Fisher Scientific). The cells were incubated at 37°C in a humidified atmosphere of 5% CO2.
Lentivirus Construction and Cell Transduction
The cDNA and short hairpin RNA for PFN2 were generated (Synbio Technologies, Suzhou, Jiangsu, P.R. China) and confirmed by sequencing, followed by insertion into the overexpression vector pLVX-IRES-ZsGreen1 and knockdown vector PLKO.1, respectively. Recombinant plasmids were transfected into 293T cells together with packaging plasmid psPAX2 and G protein of the vesicular stomatitis virus (VSV-G) envelope plasmid pMD2.G to generate the lentivirus.
Fadu and Cal-27 cells were infected with lentivirus containing pLVX-PFN2-IRES-ZsGreen1 to establish stable PFN2 high-expression cell lines. OSC-19 cells were infected with lentivirus containing short hairpin RNA for PFN2 to establish a stable PFN2 low-expression cell line. Cells with either empty pLVX-IRES-ZsGreen1 (Vector) or empty pLKO.1 (Sh-Ctrl) were used as transduction control. In addition, uninfected cells were used as the overall baseline control. Lentivirus-infected cells were screened by puromycin (2 μg/ml). The efficiencies of overexpression and knockdown were determined by Western blot.
Western Blot Analysis
Cells were lysed in radioimmunoprecipitation assay buffer (Beyotime Biotechnology, Shanghai, P.R. China). Protein concentration was quantified by bicinchoninic acid protein assay kit (Beyotime Biotechnology). Equal amounts of protein were separated with 8–12% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and transferred onto polyvinylidene fluoride membrane (Millipore, Boston, MA, USA). The membranes were incubated with primary antibodies: PFN2 (Abcam, San Francisco, CA, USA), tubulin (Santa Cruz Biotechnology, Santa Cruz, CA, USA), p-Akt (Ser473; Cell Signaling Technology, Beverly, MA, USA), p-Akt (Thr308; Cell Signaling Technology), Akt (Cell Signaling Technology), pGSK3β (Ser9; Cell Signaling Technology), GSK3β (Cell Signaling Technology), and β-catenin (Cell Signaling Technology). After sufficient incubation and wash, the corresponding secondary antibody was added, and bands were detected with ECL chemiluminescence kit (Beyotime Biotechnology).
Cell Viability Assay
HNSC cells with PFN2 overexpression and knockdown were seeded into 96-well plates at 1,000 cells per well. The Cell Counting Kit-8 (CCK-8) reagent was added on 0, 1, 2, 3, and 4 days. The absorbance was measured with a microplate reader at 450 nm.
Colony Formation Assay
HNSC cells with PFN2 overexpression or knockdown were seeded into six-well plates at a density of 400 cells per well and cultured in DMEM/F12 containing 10% fetal bovine serum for 2 weeks. The colonies were then fixed with methanol for 15 min and stained with crystal violet for 10 min.
Apoptosis Assays
Apoptosis of HNSC cells after PFN2 transduction was assessed by Annexin-V–phycoerythrin (PE) and 7-amino-actinomycin D (7-AAD) apoptosis detection kit (BD, Franklin Lakes, NJ, USA) according to manufacturer’s instructions. Briefly, cells were treated with 10 μM cisplatin for 24 h, and then the cells were collected and washed twice with cold PBS. After that, the cells were stained with 5 μl of annexin V and 5 μl of 7-AAD for 15 min in the dark. Cell apoptosis was analyzed by CytoFLEX flow cytometer.
Scratch Wound Assay and Transwell Invasion Assay
For the scratch wound assay, confluent HNSC cells were scratched with a plastic tip across the center. Five images were randomly taken at 0 h and 24 h after scratching, and the distances of migrated cells were measured under a light microscope.
For the Transwell invasion assay, HNSC cells were seeded in the upper chamber of the Matrigel-coated Transwell insert. Cells that did not invade were removed using a cotton swab, stained by crystal violet, and counted under an inverted microscope in five randomly selected views.
Nude Mouse Xenograft Model
HNSC cells with PFN2 overexpression or knockdown and the appropriate control cells were digested and suspended in cold PBS at a concentration of 5 × 107 cells/ml. Subsequently, the cells were subcutaneously inoculated into the back right forearm of 6-week-old nude mice (100 μl per mouse). Two weeks after inoculation, tumors were observed. The length and width of the tumor were measured every 3 days. The tumor volume was calculated as follows: V = length × width2/2. Tumors were harvested 33 days after inoculation, weighed, and imaged. All applicable international, national, and/or institutional guidelines for the care and use of animals were followed.
Statistical Analysis
Significance was determined using two-tailed unpaired Student’s t-test or one-way ANOVA. Differences in tumor growth were analyzed by two-way ANOVA. Results were expressed as mean ± SEM. A value of p < 0.05 was considered to be statistically significant. All statistical analyses were performed with Graph Pad Prism 6.0 software.
RESULTS
PFN2 Expression Is Upregulated and Associated With Poor Outcomes in HNSC
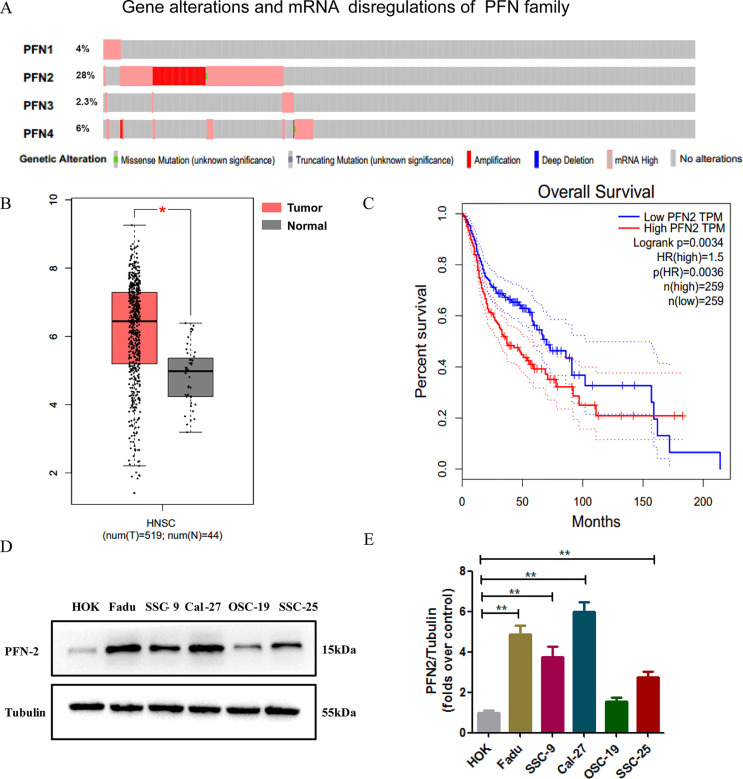
We analyzed 530 sequenced HNSC samples from The Cancer Genome Atlas (TCGA) by cBioPortal, and mRNA alterations in the four PFN isoforms were evaluated. Alteration frequencies of PFN1, PFN2, PFN3, and PFN4 were found to be 4%, 28%, 2.3%, and 6%, respectively (Fig. 1A). In particular, PFN2 displayed the highest alteration frequency in HNSC, in which the most common alterations were gene amplification and mRNA upregulation. We further searched the PFN2 expression in the Gene Expression Profiling Interactive Analysis (GEPIA) database and found the expression of PFN2 in 519 HNSC samples to be much higher than the 44 noncancerous head and neck tissues (Fig. 1B). Further analysis on GEPIA revealed a highly positive correlation between increased PFN2 expression and shorter overall survival (Fig. 1C). Next, we investigated the expression of PFN2 in HNSC cell lines and found that PFN2 was significantly upregulated in HNSC cell lines compared to control human oral keratinocytes (HOK) (Fig. 1D and E). The above results indicate that PFN2 may play a vital role in the development and progression of HNSC.
Figure 1.
Profilin 2 (PFN2) is highly expressed in head and neck cancer (HNSC) and correlates with poor prognosis. (A) Gene alterations and mRNA dysregulations of the PFN gene family in HNSC. Data were obtained from the cBioPortal online database. (B) Data from the Gene Expression Profiling Interactive Analysis (GEPIA) database showed a significant upregulation of PFN2 mRNA in HNSC tissues when compared with noncancerous tissues. (C) Survival analysis using the GEPIA database showed high PFN2 expression was correlated with shorter overall survival. HR, hazard ratio. (D) The expression of PFN2 in five different HNSC cell lines and control oral epithelial cells were detected by Western blot. (E) Quantitative analysis of the Western blot results in (D). Data were expressed as mean ± SEM from five independent experiments, n = 5. *p < 0.05, **p < 0.01.
PFN2 Regulates Cell Proliferation and Apoptosis in HNSC Cells
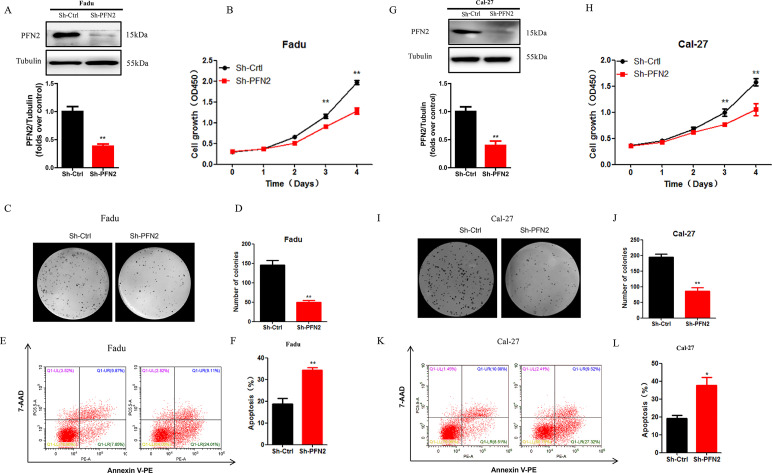
To investigate the function of PFN2 in HNSC, PFN2 was knocked down in HNSC cell lines, Fadu, and Cal27. Knockdown efficiency was validated by Western blot (Fig. 2A and G). CCK-8 assay revealed that PFN2 knockdown significantly slowed cell proliferation (Fig. 2B and H). This result was complemented by the clonal formation experiment, which showed significantly lower colony formation in the PFN2 knockdown cells (Fig. 2C and I). We next examined whether PFN2 is involved in the process of apoptosis. Annexin V/7-AAD staining demonstrated that the percentage of apoptotic cells was significantly increased in the PFN2 knockdown of Fadu and Cal-27 cells (Fig. 2E, F, K, and L).
Figure 2.
PFN2 knockdown suppressed tumor cell proliferation and promoted cell apoptosis. (A, G) Stable knockdown of PFN2 in Fadu and Cal-27 cells was verified by Western blot. (B, H) Cell viability under PFN2 knockdown was examined by CCK-8 assay in Fadu and Cal-27 cells. (C, I) Colony formation of HNSC cells was significantly inhibited by PFN2 knockdown in Fadu and Cal-27 cells. (D, J) The number of colonies in PFN2 knockdown group and control group in Fadu and Cal-27 cells. (E, K) The proportion of apoptotic cells increased after PFN2 knockdown, as assayed by annexin V/7-AAD staining. (F, L) The percentage of apoptotic cells in PFN2 knockdown group and control group in Fadu and Cal-27 cells. Data were expressed as mean ± SEM from five independent experiments, n = 5. *p < 0.05, **p < 0.01.
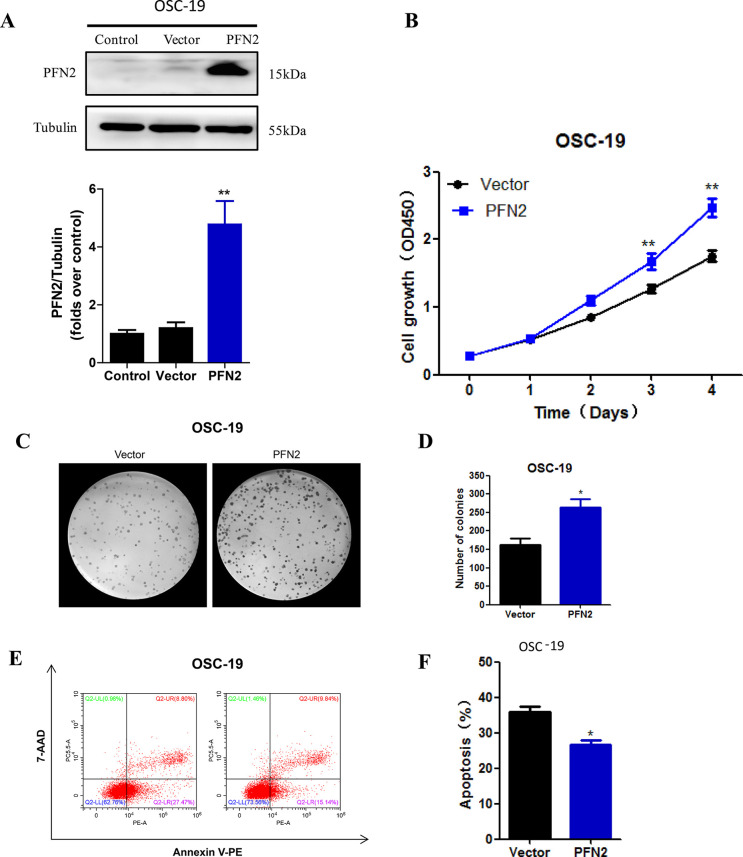
To validate the tumor-promoting ability of PFN2 in HNSC, we overexpressed PFN2 in OSC-19 cells and confirmed the overexpression by Western blot (Fig. 3A). As expected, overexpression of PFN2 accelerated OSC-19 cell proliferation as evidenced by CCK-8 and colony formation assays (Fig. 3B–D). Moreover, PFN2 overexpression significantly inhibited cisplatin-induced apoptosis, which is consistent with its ability to regulate cell proliferation in HNSC cells (Fig. 3E and F).
Figure 3.
PFN2 overexpression promoted tumor cell proliferation and inhibited cell apoptosis. (A) Stable overexpression of PFN2 in OSC-19 cells was verified by Western blot. (B) Cell viability under PFN2 overexpression was examined by CCK-8 assay. (C, D) Colony formation of HNSC cells was significantly enhanced by PFN2 overexpression. (E, F) The proportion of apoptotic cells decreased after PFN2 overexpression, as assayed by annexin V/7-AAD staining. Data were expressed as mean ± SEM from five independent experiments, n = 5. *p < 0.05, **p < 0.01.
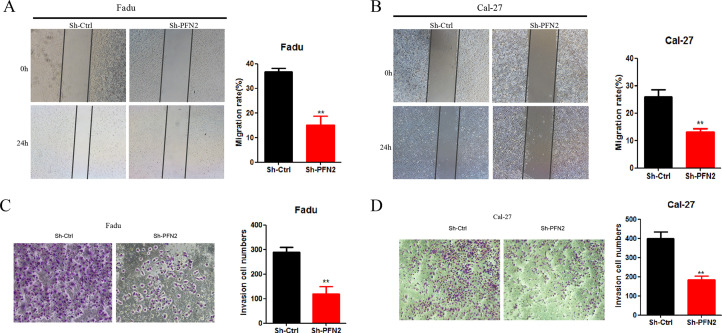
PFN2 Regulates Migration and Invasion of HNSC Cells
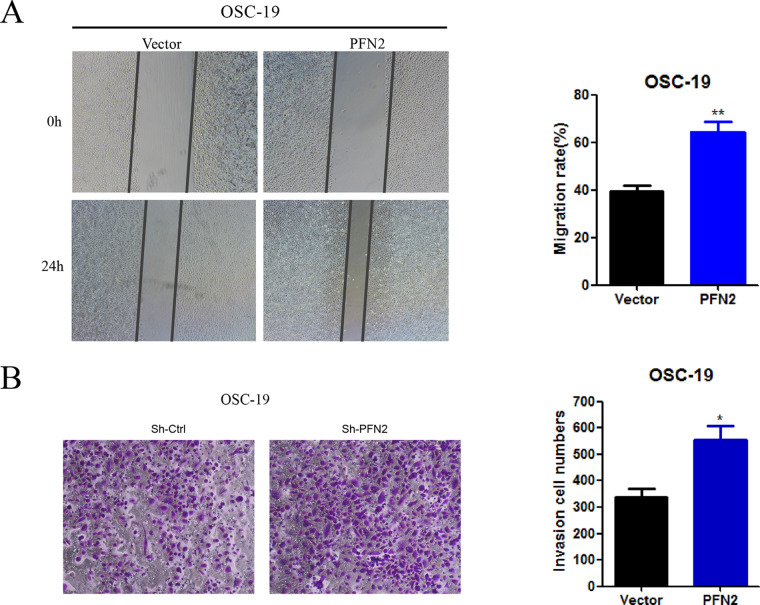
We further investigated the role of PFN2 in tumor migration and invasion using the scratch wound healing and Transwell invasion assays. PFN2 knockdown reduced Fadu and Cal27 cell migration toward the scratch wound as well as reduced invasive cell number (Fig. 4A–D). On the other hand, PFN2 overexpression generated the opposite effect where cell migration and invasion were enhanced in PFN2 overexpressed OCS-19 cells (Fig. 5A and B). Our findings reveal a critical role of PFN2 in promoting cell migration and invasion in HNSC.
Figure 4.
PFN2 knockdown suppressed cell migration and invasion. (A, B) Cell migration was inhibited under PFN2 knockdown in Fadu and Cal-27 cells. (C, D) The relative cell invasion ratio was reduced in Fadu and Cal-27 cells with PFN2 knockdown. Data were expressed as mean ± SEM from five independent experiments, n = 5. **p < 0.01.
Figure 5.
PFN2 overexpression promoted cell migration and invasion. (A) Cell migration was promoted under PFN2 overexpression in OSC-19 cells. (B) The relative cell invasion ratio was improved under PFN2 overexpression in OSC-19 cells. Data were expressed as mean ± SEM from five independent experiments, n = 5. *p < 0.05, **p < 0.01.
PFN2 Promotes Tumor Growth Through the Upregulation of PI3K/Akt/β-Catenin Signaling Pathway
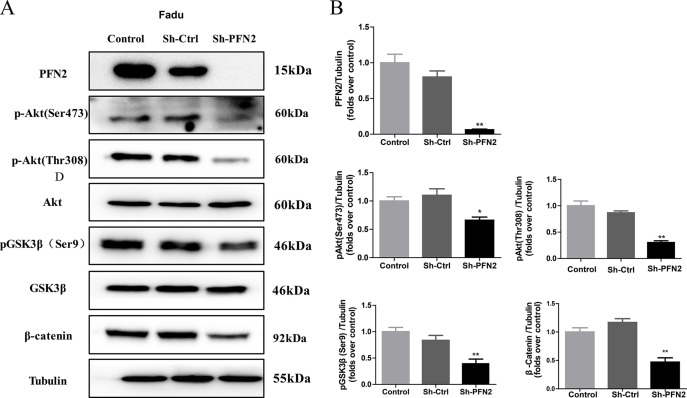
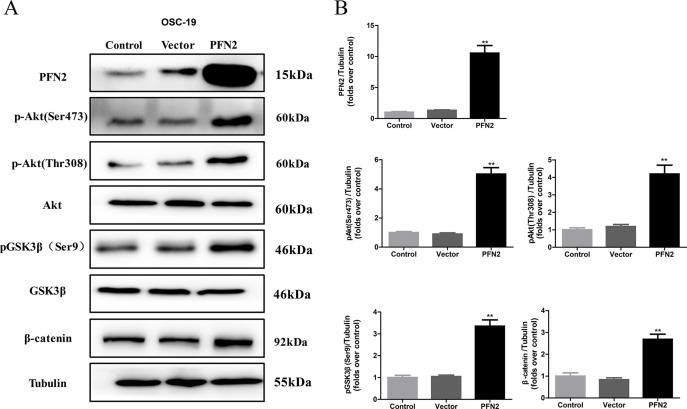
PI3K/Akt signaling pathway, the most frequently altered pathway in human tumors, regulates the cell cycle and plays a critical role in inhibiting apoptosis and promoting cell growth and proliferation20. One study based on 151 cases of head and neck cancer found that PI3K/Akt pathway is the most frequently mutated oncogenic pathway21. To understand the function of PFN2 in HNSC growth, we first examined the phosphorylation state of Akt and its downstream effector, glycogen synthase kinase 3 β (GSK3β), in HNSC cells upon PFN2 alteration. As shown in Figure 6A, phosphorylation of Akt at Ser473 and Thr308 were dramatically inhibited in PFN2 knockdown Fadu cells. On the other hand, overexpression of PFN2 in OSC-19 cells substantially increased the phosphorylation of Akt at Ser473 and Thr308 (Fig. 7A and B). Further, we searched for the effectors downstream of PI3K/Akt, and the results showed that the levels of glycogen synthase kinase-3 β phosphorylated at serine9 (pGSK3β Ser9) and β-catenin were also decreased in the PFN2 knockdown Fadu cells, but were increased in the OSC-19 cells with PFN2 overexpression (Figs. 6A and 7A). Collectively, our results indicate that PFN2 may potentially regulate HNSC proliferation and migration through the PI3K/Akt/β-catenin signaling pathway.
Figure 6.
PFN2 knockdown inhibited the PI3K/Akt/β-catenin signaling pathway activation. (A) Phosphorylation of Akt at Ser473 and Thr308, phosphorylation of GSK3β at Ser9, and the expression of β-catenin were significantly suppressed in PFN2 knockdown Fadu cells. (B) Statistical analysis of each protein expression. Data were expressed as mean ± SEM from five independent experiments, n = 5. *p < 0.05, **p < 0.01.
Figure 7.
PFN2 overexpression activated the PI3K/Akt/β-catenin signaling pathway. (A) Phosphorylation of Akt at Ser473 and Thr308, phosphorylation of GSK3β at Ser9, and the expression of β-catenin were significantly upregulated in OSC-19 cells with PFN2 overexpression. (B) Statistical analysis of each protein expression. Data were expressed as mean ± SEM from five independent experiments, n = 5. **p < 0.01.
PFN2 Manipulation Affects HNSC Growth In Vivo
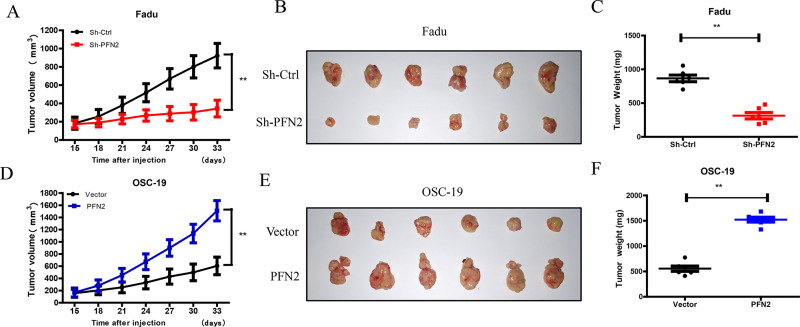
In line of the tumor-promoting effect of PFN2 in vitro, we next examined the role of PFN2 in HNSC in vivo. Fadu cells with PFN2 knockdown or OSC-19 cells with PFN2 overexpression were subcutaneously inoculated into the back right forelimb of nude mice, respectively. Fadu xenografts with PFN2 knockdown showed significantly slower tumor growth than controls. In contrast, xenografts from PFN2-overexpressed OSC-19 cells grew much faster than the control group (Fig. 8A and D). Furthermore, the tumor weight of PFN2 knockdown group was lighter than that of the control group (Fig. 8C). The opposite was true for tumor weight after PFN2 overexpression (Fig. 8F). These results support our in vitro findings that PFN2 promotes tumor cell proliferation.
Figure 8.
Effects of PFN2 expression on tumor growth in vivo. Fadu cells with PFN2 knockdown and OSC-19 cells with PFN2 overexpression were subcutaneously injected to construct the xenograft tumor model, and tumor growth was monitored. Two weeks after transplantation, the length (L) and width (W) of the tumor were measured every 3 days with vernier calipers. (A, D) Tumor volume was calculated according to the formula: (L × W 2)/2, and the growth curves of xenografts in nude mouse were drawn. (B, E) Tumors isolated from mice 33 days after transplantation. (C, F) The tumor nodules were weighed after they were harvested. Data were expressed as mean ± SEM from six independent experiments, n = 6. **p < 0.01.
DISCUSSION
Profilins are a family of actin-binding proteins that are known for regulating the dynamics of actin polymerization22. PFN1 and PFN2 are the two major isoforms of PFN. PFN1 is widely expressed in many tissues, whereas PFN2 is mainly expressed in the brain23. In recent years, increasing evidence has revealed the important functions of PFN2 in various cancers. For example in lung cancer, PFN2 epigenetically promotes the transcription of Smad2 and Smad3 and thereby accelerates cancer growth and metastasis17. In addition, increased expression of PFN2 is associated with poor survival of lung cancer patients17. Downregulation of PFN2 hinders cell invasion and migration in ESCC via promoting expression of epithelial markers and decreasing expression of mesenchymal markers16. Furthermore, regression analysis revealed that high PFN2 expression was related to lymph node metastases and shorter overall survival of ESCC patients16. A recent bioinformatic study showed that PFN2 was dramatically upregulated in HNSC tissues, and its overexpression was significantly associated with poor prognosis of HNSC patients24. However, the biological function of PFN2 specifically in HNSC has not been reported. In this study, genetic alterations of the PFN gene family in HNSC were evaluated using the cBioPortal online database. We found that dysregulation in the PFN2 isoform (28%) was the most common, which was mainly made up of gene amplification and mRNA upregulation. Furthermore, we observed that HNSC was remarkably upregulated in HNSC tissues and cell lines. The overall survival of patients with high PFN2 expression is significantly lower than patients with low PFN2 expression, demonstrating that PFN2 is closely related to the prognosis of HNSC patients.
It is clear that PFN2 plays a role in HNSC. We then established stable PFN2 overexpression and PFN2 knockdown cell lines to investigate the functional significance of PFN2 on tumor progression. We found that PFN2 depletion significantly inhibited HNSC cell proliferation, migration, and invasion, and induced cell apoptosis. Conversely, the opposite effects were observed when PFN2 was overexpressed in HNSC cells. Moreover, these observations were confirmed in the in vivo tumor model. Our study, for the first time, reveals that PFN2 functions as an oncogene by promoting tumor cell proliferation and metastasis in HNSC.
The PI3K/Akt signaling pathway is involved in the regulation of various cellular biological processes such as proliferation, cell cycle, differentiation, metabolism, migration, and angiogenesis, and is often overactivated in many cancers25. Previous research indicates that the PI3K pathway is the most frequently overactivated pathway in HNSC21. In the present study, we explored the potential regulatory role of PFN2 on the PI3K/Akt pathway in HNSC and found that PFN2 knockdown reduced the phosphorylation of Akt at Ser473 and Thr308. Studies have shown that the PI3K/Akt pathway is regulated by multiple interconnecting pathways in tumors. For example, the epidermal growth factor receptor (EGFR) can directly activate PI3K through recruiting p85 and promoting its binding with the activated receptor26. Upon EGFR dimerization, the complex can also stimulate Ras, which leads to a phosphorylation cascade that eventually activates the PI3K/Akt signaling pathway27. G-protein-coupled receptors (GPCRs) promote the production of phosphatidylinositol 3,4,5-triphosphate (PIP3) and can activate a unique PI3K isoform28. In addition, PI3K is negatively regulated by many other proteins such as the phosphatase and tensin homolog (PTEN), which turn off the activation of the PI3K/Akt signaling pathway29. Therefore, PFN2 may likely promote the activation of PI3K/Akt indirectly through both positive and negative means in HNSC.
As one of the downstream effectors, GSK-3β is directly phosphorylated by Akt, which leads to stabilization of β-catenin in the canonical Wnt signaling pathway30. The stability of β-catenin is controlled by the adenomatous polyposis coli (APC)/Axin destruction complex, which consists of adenomatous polyposis coli, Axin, and two kinases, GSK3 and casein kinase α/β31. GSK-3β has three major phosphorylation sites, among which phosphorylation at Ser9 leads to the inhibition of GSK-3β and APC/Axin destruction complex32. The phosphorylation and degradation of β-catenin is inhibited as a result of the inactivation of the destruction complex32. Therefore, we also examined the phosphorylation of GSK-3β and found that phosphorylation of GSK-3β at Ser9 decreased after PFN2 knockdown, and as a result, the expression of β-catenin was also inhibited. Opposite results were obtained in PFN2-overexpressed OSC-19 cells. The accumulation of β-catenin in the cytoplasm promotes transcription of downstream target genes that are vital in cell proliferation, migration, and invasion33. Multiple studies have revealed that the PI3K/Akt/GSK-3β signaling pathway is involved in many aspects of cancer pathophysiology in various tissues34–38. Our results also indicated that PFN2 takes advantage of this pathway by enhancing phosphorylation of Akt and GSK-3β and promoting the stability and intracellular levels of β-catenin in HNSC, which leads to tumor cell proliferation and metastasis.
In summary, our results indicate that PFN2 is highly expressed in HNSC tumor tissues and correlates with poor prognosis in patients. PFN2 promotes tumor cell proliferation and metastasis by activating the PI3K/Akt/β-catenin signaling pathway in HNSC. These results support PFN2 as an important oncogenic protein and potential therapeutic target in HNSC.
ACKNOWLEDGMENTS
This research work was supported by the Basic Research Program of Wenzhou City (Y20180207) and National Natural Science Foundation of China (Nos. 81873376, 81574074).
Footnotes
The authors declare no conflicts of interest.
REFERENCES
- 1. Argiris A, Karamouzis MV, Raben D, Ferris RL. Head and neck cancer. Lancet 2008;371(9625):1695–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer 2015;136(5):E359–86. [DOI] [PubMed] [Google Scholar]
- 3. Hammerman PS, Hayes DN, Grandis JR. Therapeutic insights from genomic studies of head and neck squamous cell carcinomas. Cancer Discov. 2015;5(3):239–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Leemans CR, Braakhuis BJ, Brakenhoff RH. The molecular biology of head and neck cancer. Nat Rev Cancer 2011;11(1):9–22. [DOI] [PubMed] [Google Scholar]
- 5. Mao L, Hong WK, Papadimitrakopoulou VA. Focus on head and neck cancer. Cancer Cell 2004;5(4):311–6. [DOI] [PubMed] [Google Scholar]
- 6. Colevas AD. Chemotherapy options for patients with metastatic or recurrent squamous cell carcinoma of the head and neck. J Clin Oncol. 2006;24(17):2644–52. [DOI] [PubMed] [Google Scholar]
- 7. Gibson MK, Li Y, Murphy B, Hussain MH, DeConti RC, Ensley J, Forastiere AA, Eastern Cooperative Oncology G. Randomized phase III evaluation of cisplatin plus fluorouracil versus cisplatin plus paclitaxel in advanced head and neck cancer (E1395): An intergroup trial of the Eastern Cooperative Oncology Group. J Clin Oncol. 2005;23(15):3562–7. [DOI] [PubMed] [Google Scholar]
- 8. Zhu G, Peng F, Gong W, She L, Wei M, Tan H, Chen C, Zhang D, Li G, Huang D, Zhang X, Liu Y. Hypoxia promotes migration/invasion and glycolysis in head and neck squamous cell carcinoma via an HIF-1alpha-MTDH loop. Oncol Rep. 2017;38(5):2893–900. [DOI] [PubMed] [Google Scholar]
- 9. Li Y, Grenklo S, Higgins T, Karlsson R. The profilin:actin complex localizes to sites of dynamic actin polymerization at the leading edge of migrating cells and pathogen-induced actin tails. Eur J Cell Biol. 2008;87(11):893–904. [DOI] [PubMed] [Google Scholar]
- 10. Witke W, Podtelejnikov AV, Di Nardo A, Sutherland JD, Gurniak CB, Dotti C, Mann M. In mouse brain profilin I and profilin II associate with regulators of the endocytic pathway and actin assembly. EMBO J. 1998;17(4):967–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Das T, Bae YH, Wells A, Roy P. Profilin-1 overexpression upregulates PTEN and suppresses AKT activation in breast cancer cells. J Cell Physiol. 2009;218(2):436–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pilo Boyl P, Di Nardo A, Mulle C, Sassoe-Pognetto M, Panzanelli P, Mele A, Kneussel M, Costantini V, Perlas E, Massimi M, Vara H, Giustetto M, Witke W. Profilin2 contributes to synaptic vesicle exocytosis, neuronal excitability, and novelty-seeking behavior. EMBO J. 2007;26(12):2991–3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ma CY, Zhang CP, Zhong LP, Pan HY, Chen WT, Wang LZ, Andrew OW, Ji T, Han W. Decreased expression of profilin 2 in oral squamous cell carcinoma and its clinicopathological implications. Oncol Rep. 2011;26(4):813–23. [DOI] [PubMed] [Google Scholar]
- 14. Mouneimne G, Hansen SD, Selfors LM, Petrak L, Hickey MM, Gallegos LL, Simpson KJ, Lim J, Gertler FB, Hartwig JH, Mullins RD, Brugge JS. Differential remodeling of actin cytoskeleton architecture by profilin isoforms leads to distinct effects on cell migration and invasion. Cancer Cell 2012;22(5):615–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yan J, Ma C, Gao Y. MicroRNA-30a-5p suppresses epithelial-mesenchymal transition by targeting profilin-2 in high invasive non-small cell lung cancer cell lines. Oncol Rep. 2017;37(5):3146–54. [DOI] [PubMed] [Google Scholar]
- 16. Cui XB, Zhang SM, Xu YX, Dang HW, Liu CX, Wang LH, Yang L, Hu JM, Liang WH, Jiang JF, Li N, Li Y, Chen YZ, Li F. PFN2, a novel marker of unfavorable prognosis, is a potential therapeutic target involved in esophageal squamous cell carcinoma. J Transl Med. 2016;14(1):137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tang YN, Ding WQ, Guo XJ, Yuan XW, Wang DM, Song JG. Epigenetic regulation of Smad2 and Smad3 by profilin-2 promotes lung cancer growth and metastasis. Nat Commun. 2015;6:8230. [DOI] [PubMed] [Google Scholar]
- 18. Kim MJ, Lee YS, Han GY, Lee HN, Ahn C, Kim CW. Profilin 2 promotes migration, invasion, and stemness of HT29 human colorectal cancer stem cells. Biosci Biotechnol Biochem. 2015;79(9):1438–46. [DOI] [PubMed] [Google Scholar]
- 19. Zhang H, Yang W, Yan J, Zhou K, Wan B, Shi P, Chen Y, He S, Li D. Loss of profilin 2 contributes to enhanced epithelial–mesenchymal transition and metastasis of colorectal cancer. Int J Oncol. 2018;53(3):1118–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhou K, Chen J, Wu J, Wu Q, Jia C, Xu YXZ, Chen L, Tu W, Yang G, Kong J, Kou J, Jiang S. Atractylenolide III ameliorates cerebral ischemic injury and neuroinflammation associated with inhibiting JAK2/STAT3/Drp1-dependent mitochondrial fission in microglia. Phytomedicine 2019;59:152922. [DOI] [PubMed] [Google Scholar]
- 21. Zhou K, Wu J, Chen J, Zhou Y, Chen X, Wu Q, Xu Y, Tu W, Lou X, Yang G, Jiang S. Schaftoside ameliorates oxygen glucose deprivation-induced inflammation associated with the TLR4/Myd88/Drp1-related mitochondrial fission in BV2 microglia cells. J Pharmacol Sci. 2019;139(1):15–22. [DOI] [PubMed] [Google Scholar]
- 22. Fruman DA, Rommel C. PI3K and cancer: Lessons, challenges and opportunities. Nat Rev Drug Discov. 2014;13(2):140–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lui VW, Hedberg ML, Li H, Vangara BS, Pendleton K, Zeng Y, Lu Y, Zhang Q, Du Y, Gilbert BR, Freilino M, Sauerwein S, Peyser ND, Xiao D, Diergaarde B, Wang L, Chiosea S, Seethala R, Johnson JT, Kim S, Duvvuri U, Ferris RL, Romkes M, Nukui T, Kwok-Shing Ng P, Garraway LA, Hammerman PS, Mills GB, Grandis JR. Frequent mutation of the PI3K pathway in head and neck cancer defines predictive biomarkers. Cancer Discov. 2013;3(7):761–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lambrechts A, Kwiatkowski AV, Lanier LM, Bear JE, Vandekerckhove J, Ampe C, Gertler FB. cAMP-dependent protein kinase phosphorylation of EVL, a Mena/VASP relative, regulates its interaction with actin and SH3 domains. J Biol Chem. 2000;275(46):36143–51. [DOI] [PubMed] [Google Scholar]
- 25. Witke W, Sutherland JD, Sharpe A, Arai M, Kwiatkowski DJ. Profilin I is essential for cell survival and cell division in early mouse development. Proc Natl Acad Sci USA 2001;98(7):3832–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Liu J, Wu Y, Wang Q, Liu X, Liao X, Pan J. Bioinformatic analysis of PFN2 dysregulation and its prognostic value in head and neck squamous carcinoma. Future Oncol. 2018;14(5):449–59. [DOI] [PubMed] [Google Scholar]
- 27. Hanahan D, Weinberg RA. Hallmarks of cancer: The next generation. Cell 2011;144(5):646–74. [DOI] [PubMed] [Google Scholar]
- 28. Engelman JA. Targeting PI3K signalling in cancer: Opportunities, challenges and limitations. Nat Rev Cancer 2009;9(8):550–62. [DOI] [PubMed] [Google Scholar]
- 29. Bussink J, van der Kogel AJ, Kaanders JH. Activation of the PI3-K/AKT pathway and implications for radioresistance mechanisms in head and neck cancer. Lancet Oncol. 2008;9(3):288–96. [DOI] [PubMed] [Google Scholar]
- 30. Arcaro A, Guerreiro AS. The phosphoinositide 3-kinase pathway in human cancer: Genetic alterations and therapeutic implications. Curr Genomics 2007;8(5):271–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Psyrri A, Seiwert TY, Jimeno A. Molecular pathways in head and neck cancer: EGFR, PI3K, and more. Am Soc Clin Oncol Educ Book 2013:246–55. [DOI] [PubMed] [Google Scholar]
- 32. Zhang X, Jiang D, Jiang W, Zhao M, Gan J. Role of TLR4-mediated PI3K/AKT/GSK-3beta signaling pathway in apoptosis of rat hepatocytes. Biomed Res Int. 2015;2015:631326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Clevers H. Wnt/beta-catenin signaling in development and disease. Cell 2006;127(3):469–80. [DOI] [PubMed] [Google Scholar]
- 34. Tejeda-Munoz N, Robles-Flores M. Glycogen synthase kinase 3 in Wnt signaling pathway and cancer. IUBMB Life 2015;67(12):914–22. [DOI] [PubMed] [Google Scholar]
- 35. Nusse R, Clevers H. Wnt/beta-catenin signaling, disease, and emerging therapeutic modalities. Cell 2017;169(6):985–99. [DOI] [PubMed] [Google Scholar]
- 36. Martini M, De Santis MC, Braccini L, Gulluni F, Hirsch E. PI3K/AKT signaling pathway and cancer: An updated review. Ann Med. 2014;46(6):372–83. [DOI] [PubMed] [Google Scholar]
- 37. He F, Chen H, Yang P, Wu Q, Zhang T, Wang C, Wei J, Chen Z, Hu H, Li W, Cao J. Gankyrin sustains PI3K/GSK-3beta/beta-catenin signal activation and promotes colorectal cancer aggressiveness and progression. Oncotarget 2016;7(49):81156–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wu K, Fan J, Zhang L, Ning Z, Zeng J, Zhou J, Li L, Chen Y, Zhang T, Wang X, Hsieh JT, He D. PI3K/Akt to GSK3beta/beta-catenin signaling cascade coordinates cell colonization for bladder cancer bone metastasis through regulating ZEB1 transcription. Cell Signal 2012;24(12):2273–82. [DOI] [PubMed] [Google Scholar]