Abstract
Acquired resistance remains a key challenge in epidermal growth factor receptor (EGFR)–tyrosine kinase inhibitors (TKIs) therapy in lung adenocarcinoma (LUAD). Recent studies have shown that Notch signaling is associated with drug resistance. However, its role and possible mechanisms in EGFR-TKI resistance are not yet clear. In our study, we found that among four members of NOTCH1–4, only NOTCH3 was upregulated in LUAD tissues and TKI-resistant cell line (HCC827GR6). Knockdown of NOTCH3 by siRNA significantly inhibited proliferative ability, and decreased colony and sphere formation in HCC827GR6 cells. Then miR-150 was identified as a posttranscriptional regulator of NOTCH3. Its expression was downregulated in LUAD tissues and negatively correlated with NOTCH3 mRNA. The cell proliferation and IC50 of gefitinib were decreased in HCC827GR6 cells transfected with miR-150 mimic, but was reversed when cotransfected with NOTCH3 overexpressed vector. Moreover, we also enrolled 20 patients with advanced LUAD who have taken TKIs as first-line therapy in this study. We found that collagen 1A1 (COL1A1) expression was increased significantly in LUAD tissues both at mRNA and protein levels, and positively correlated with NOTCH3 expression verified in our data and TCGA data. Univariate survival analysis showed that patients with high protein expression of NOTCH3 or COL1A1 were associated with shorter overall survival (OS). Taken together, these results suggest that miR-150/NOTCH3/COL1A1 axis contributed to EGFR–TKI resistance in LUAD, which provide a potential therapeutic target for LUAD treatment.
Key words: Lung adenocarcinoma, EGFR–TKIs resistance, NOTCH3, miR-150, COL1A1
INTRODUCTION
Lung cancer remains the leading cause of cancer-associated death worldwide1. Lung adenocarcinoma (LUAD) is the fastest growing subtype in recent years and is often diagnosed at advanced stages of the disease. Epidermal growth factor receptor (EGFR)–tyrosine kinase inhibitors (TKIs) have been widely used in advanced LUAD patients harboring EGFR mutations2. However, most patients will acquire resistance to EGFR–TKI, and disease progression usually occurs. Several major mechanisms of acquired resistance have been revealed, including T790M mutations, epithelial–mesenchymal transition (EMT) and proapoptotic protein Bcl-2-like 11 (BIM) deletion polymorphism3,4. Further study is still needed to deepen the understanding of the molecular mechanism of EGFR-TKI resistance.
Notch pathway has been implicated in cell fate determination5. The mammalian Notch family consists of four receptors, NOTCH1–4. Notch signaling is activated by ligand binding that results in production of the NOTCH intracellular domain (NICD). Translocation of NICD into the nucleus induces target gene transcription6,7. Overexpression of NOTCH3 has been reported to be associated with poor prognosis for non-small cell lung cancer (NSCLC)8,9. Using inhibitors to prevent NOTCH3 activation could reduce the proliferation in lung cancer cells10. Moreover, elevated NOTCH3 expression is associated with resistance to chemotherapy in many solid tumors including lung cancer11–14. However, the role of NOTCH3 in EGFR-TKI resistance in NSCLC is not fully elucidated.
MicroRNA (miRNAs), small noncoding RNAs of ∼22 nucleotides, play an important role in development and progression of tumors15. miRNAs bind to the 3′-untranslated regions (3′-UTRs) of target mRNAs, thereby regulating several biological processes16. Several NOTCH-associated miRNAs have been identified in cancers. miR-491-5p suppresses cell growth and invasion by targeting NOTCH3 in nasopharyngeal carcinoma17. There were reciprocal regulation loops between NOTCH2 pathway and miR-23b in controlling gastric carcinogenesis18.
In this study, we investigated the role of NOTCH3 in acquired resistance to EGFR–TKI and identified posttranscriptional regulation of NOTCH3 by candidate miRNA in LUAD.
MATERIALS AND METHODS
Patients
From the year 2008 to 2010, 20 patients with advanced LUAD who were diagnosed by bronchoscopy or percutaneous biopsy in Xuzhou Central Hospital affiliated to Xuzhou Medical University were included. All patients had EGFR-sensitive mutations and no brain metastases, and had taken oral TKIs as first-line therapy. The formalin-fixed tissues and fresh tissues frozen in liquid nitrogen were studied in this study. Information on survival was obtained through active follow-up based on the verification of patients’ vital status. The overall survival (OS) was defined as the time between the initiation of treatment to the date of death or last follow-up by January 1, 2018. The study was carried out in accordance with the approved ethical standards of the ethics committee in our hospital, and informed consent was obtained from all participants.
Cell Culture and Cell Treatment
Human gefitinib-sensitive HCC827 LUAD cell line and gefitinib-resistant cell line (HCC827GR6) were gifted by Dr. Pasi A. Jänne (Dana-Farber Cancer Institute, Boston, MA, USA). The cells were cultured in RPMI-1640 medium (Hyclone, Logan, UT, USA) containing 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin in a humidified 5% CO2 incubator at 37°C. HCC827GR6 cells were cultured in gefitinib (LC Laboratories, Woburn, MA, USA) with a final concentration at 1 μM.
Quantitation Real-Time Polymerase Chain Reaction (qRT-PCR)
Total RNA was isolated from lung tissues and cell lines using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. Complementary DNA was synthesized by TaqMan MicroRNA RT Kit (Applied Biosystems, Foster City, CA, USA) and PrimeScript™ RT reagent Kit (TaKaRa, Shiga, Japan). qRT-PCR was performed using TaqMan MicroRNA Assays (Applied Biosystems) and SYBR Premix Ex Taq™ Kit (TaKaRa). The primer sequences used are listed in Table 1. For amplification, cDNA was initially denatured at 95°C for 10 min, 40 cycles of 95°C for 15 s, 60°C for 35 s, and 72°C for 40 s (ABI PRISM 7300 Sequence Detection system). Standard curve with gene expression amplification was plotted to difference in gene expression and standard deviation in the replicate groups. Ct values for each well and for each replicate group were calculated. Relative RNA expression is represented by 2−ΔΔCt.
Table 1.
List of the Primer Sequences
| Gene | Sequences (5′–3′) |
|---|---|
| NOTCH1 | F: TGCCAGACCAACATCAAC |
| R: CTCATAGTCCTCGGATTGC | |
| NOTCH2 | F: GTGTTGACTTCTGCTCTC |
| R: ACTTCTTCCTCCTACTACC | |
| NOTCH3 | F: GTCTTCCAGATTCTCATCC |
| R: ATCC ACAGCATTGACATC | |
| NOTCH4 | F: GATGTGGATGAGTGTGAGAC |
| R: CAGGTGG CAGCAATACAG | |
| COL1A1 | F: ACGTCCTGGTGAAGTTGGTC |
| R: ACGCTGTCCAGCAATACCTT | |
| GAPDH | F: CAATGACCCCTTCATTGACC |
| R: TGGAAGATGGTGATGGGATT | |
| miR-150 | F: TCTCCCAACCCTTGTACCAGTG |
| R: CAGTGCGTGTCGTGGAGT | |
| U6 | F: GTGCTCGCTTCGGCAGCACAT |
| R: GTTTAAGCACTTCGCAAGGTA |
Western Blotting
Cell protein lysates were separated by 10% SDS-PAGE and blotted onto a polyvinylidene fluoride (PVDF) membrane (Roche Diagnostics, Mannheim, Germany). After soaking in 10 ml of 5% nonfat milk in Tris-buffered saline with Tween 20 (TBST) solution for 1 h, the membrane was incubated with human polyclonal NOTCH3 (1:500 dilution; Abcam, Cambridge, UK), monoclonal NICD3 (1:1,000 dilution; Abcam), monoclonal HES1 (1:1,000 dilution; Cell Signaling Technology, Danvers, MA, USA), and polyclonal antibody against GAPDH (1:2,000 dilution; Sigma-Aldrich, St. Louis, MO, USA). Horseradish peroxidase-conjugated goat anti-rabbit IgG was used as secondary antibody (1:5,000 dilution; Sigma-Aldrich). Results were visualized following treatment with enhanced chemiluminescent (ECL) substrate.
Transient Transfection
The lentivirus-mediated small interfering RNA siNOTCH3-1 and siNOTCH3-2 (GenePharma Corp., Suzhou, P.R. China) were designed and manufactured to knock down NOTCH3. The open reading frame of NOTCH3 that was generated by PCR was then inserted into the pEGFP-C1 expression vector, which was named pEGFP-NOTCH3. A chemically modified RNA-based miRNA precursor, miR-150 mimic, was purchased from GenePharma Corp. These recombinant vectors and miR-150 mimic were transfected to HCC827GR6 cells by Lipofectamine 3000 (Invitrogen) according to the manufacturer’s instructions. After 48 h, the cells were collected for further experiments. qRT-PCR and Western blot were used to validate the efficiency of transfection.
Cell Viability and Proliferation Analysis
Cell viability was assessed using the Cell Counting Kit 8 (CCK-8). In brief, cells were seeded into 96-well plates at an initial density of 5 × 103 cells/well for 1–4 days. Afterward, 90 μl of fresh serum-free medium and 10 μl of CCK-8 reagent (Beyotime, Shanghai, P.R. China) were added to each well after decanting the old medium, and the culture was continued at 37°C for 1 h. The optical density was determined using scanning with a microplate reader (Promega, Madison, WI, USA) at 450 nm. IC50 values were calculated by gefitinib concentration–response curve using a GraphPad Prism 5.0 software (concentration gradient: 0.001, 0.01, 0.1, 1, 10, and 100 μM).
Colony Formation Assay
Cells were plated in 96-well plates at a density of 5,000 cells/well for 10 days. The medium was changed every 3 days. At the end of the experiment the cells were fixed with 4% formaldehyde for 15 min followed by PBS washing. After that, the cells were stained with crystal violet for 15 min, and the cells were washed with PBS three times. The colonies were evaluated by inverse microscopy.
Spheroid Formation Assay
The single cells were seeded in ultra-low attachment surface 96-well culture plates in serum-free DMEM/F-12 medium (Invitrogen) supplemented with 20 ng/ml epidermal growth factor (Invitrogen), 20 ng/ml basic fibroblast growth factor (Peprotech, Rocky Hill, NJ, USA), 0.4% bovine serum albumin (Sigma-Aldrich), and 5 mg/ml insulin (Sigma-Aldrich). The formed spheres were counted 10 days after seeding by inverse microscopy.
Immunohistochemistry (IHC)
Formalin-fixed, paraffin-embedded tissues were sliced consecutively into 4-μm sections and then subjected to IHC analysis. The sections were incubated with NOTCH3 (1:400 dilution; Abcam) and COL1A1 antibody (1:400 dilution; Abcam) at 4°C overnight. After washing in PBS, sections were further incubated with HRP-conjugated secondary antibody for 30 min at 37°C. Then substrate-chromogen (DAB) solution was employed to incubate the tumor tissues for 10 min. Finally, automated hematoxylin was used to counterstain the slides for 5 min. Cells positive for NOTCH3 and COL1A1 appeared yellow or yellowish brown. The immunostaining was microscopically evaluated by two independent pathologists. A semiquantitative scoring system was based on the staining intensity and proportion of positive cells19.
Bioinformatics Analysis
Nine independent microarray datasets from the Oncomine Cancer Microarray database (https://www.oncomine.org) were used for evaluating the expression status of NOTCH3 in LUAD20. Candidate miRNAs targeting NOTCH3 were predicted using the TargetScan algorithm (http://www.targetscan.org/). Publicly available data of lung adenocarcinoma were downloaded and analyzed from the Cancer Genome Atlas Project (TCGA) Project (https://cancergenome.nih.gov/).
Luciferase Reporter Assay
Human embryonic kidney HEK293T cells were seeded in 96-well plates at 1 × 104 cells per well. When the cells reached 60% confluence, they were cotransfected with wild-type pGL3-NOTCH3 3′-UTR, mutated pGL3-NOTCH3 3′-UTR plasmids, and either scramble miRNA or miR-150 mimics using Lipofectamine 3000 (Invitrogen). Luciferase activity was measured with the Dual-Luciferase Reporter Assay System (Promega) after 48 h and expressed as the ratio between firefly and Renilla luciferase activity (Fluc/Rluc).
Statistical Analysis
Data are expressed as mean ± standard deviation from three independent experiments. The relationship between the expression of miR-150 and NOTCH3, COL1A1, and NOTCH3 was examined by Pearson’s correlation analysis. Comparisons among groups were performed with the variance analysis. Overall survival (OS) was calculated by the Kaplan–Meier method and compared using log-rank testing. A value of p < 0.05 was considered to be a statistically significant result. Statistical tests were performed using SPSS 16.0 (SPSS, Inc., Chicago, IL, USA).
RESULTS
The Expression of NOTCH3 Was Upregulated in LUAD Tissues and TKI-Resistant Cells
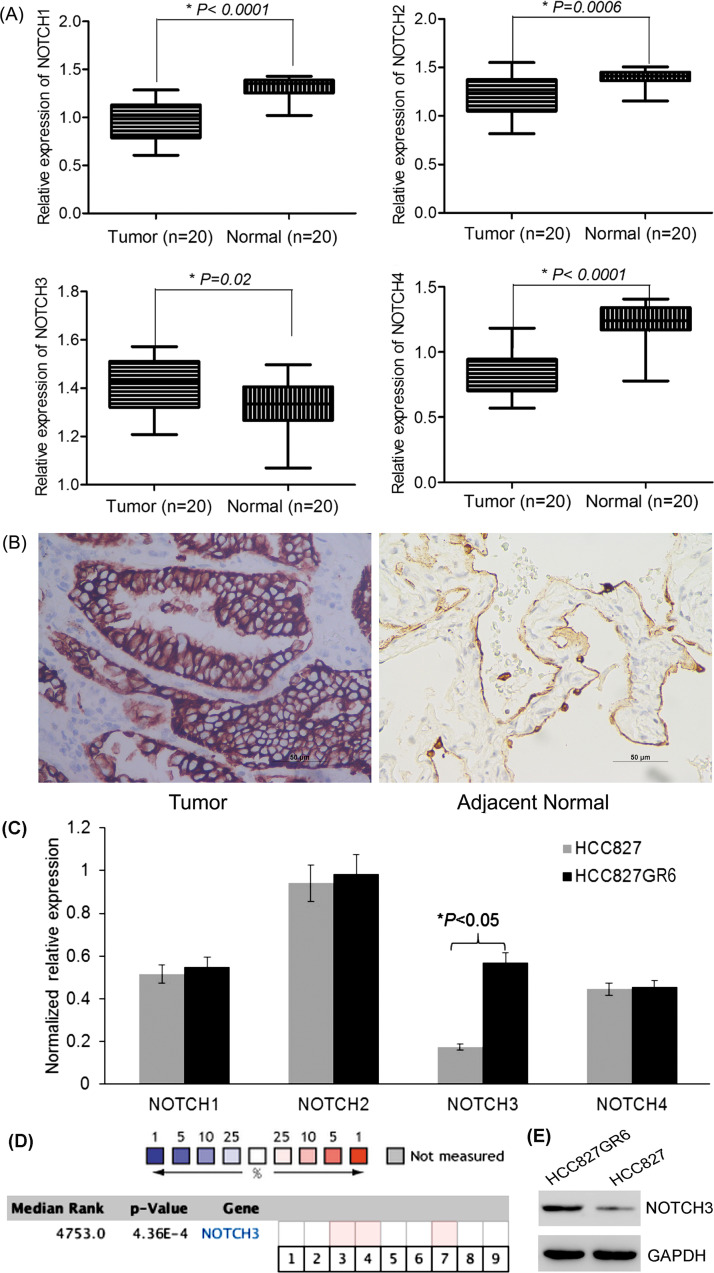
To reveal the role of NOTCH family in LUAD, qRT-PCR was performed to measure NOTCH1-4 levels in 20 pairs of LUAD tissues and adjacent normal tissues. NOTCH3 expression was significantly higher in LUAD tissues compared with matched normal tissues, while NOTCH1, 2, 4 levels were decreased (Fig. 1A). Further, NOTCH3 protein expression was upregulated in LUAD tissues as verified by IHC (Fig. 1B). To clarify whether NOTCH3 was related to TKI resistance, NOTCH levels were determined in HCC827 and HCC827GR6 cell lines. The results showed that only NOTCH3 mRNA was overexpressed in HCC827GR6 cells, while other NOTCH levels showed no significant difference (Fig. 1C).
Figure 1.
Expression of NOTCH3 in lung adenocarcinoma (LUAD) tissues and tyrosine kinase inhibitor (TKI)-resistant cell line. (A) Expression levels of NOTCH3 mRNA were significantly higher in LUAD tissues, compared to matched normal tissues, while NOTCH1,2,4 levels were significantly lower in LUAD tissues than normal tissues (20 pairs). (B) Immunohistochemical staining of NOTCH3 in LUAD and matched normal tissues. Scale bar: 50 μm. Positive staining is located in the cell membrane. (C) NOTCH3 mRNA was upregulated in gefitinib-resistant HCC827GR6 cells compared to parental HCC827 cells, while other NOTCH members showed no significant difference. (D) Meta-analysis of NOTCH3 gene expression from nine Oncomine databases. Colored squares indicate the median rank for NOTCH3 (lung adenocarcinoma vs. normal tissue) across nine analyses. Beer lung (1), Bhattacharjee lung (2), Garber lung (3), Hou lung (4), Landi lung (5), Okayama lung (6), Selamat lung (7), Stearman lung (8), Su lung (9). The p value is given for the median rank analysis. (E) NOTCH3 protein expression was upregulated in HCC827GR6 cells compared to parental HCC827 cells.
Oncomine database also showed that NOTCH3 was overexpressed in the majority of LUAD tissues compared to normal controls, based on a meta-analysis, which included nine analyses from the Oncomine algorithms (887 LUAD samples, p = 0.000436) (Fig. 1D). Consistent with mRNA level, NOTCH3 protein was significantly upregulated in HCC827GR6 cells (Fig. 1E). These results suggested that NOTCH3 expression might be correlated with TKI resistance in LUAD.
Knockdown of NOTCH3 Inhibits Growth and Stemness of TKI-Resistant Cells
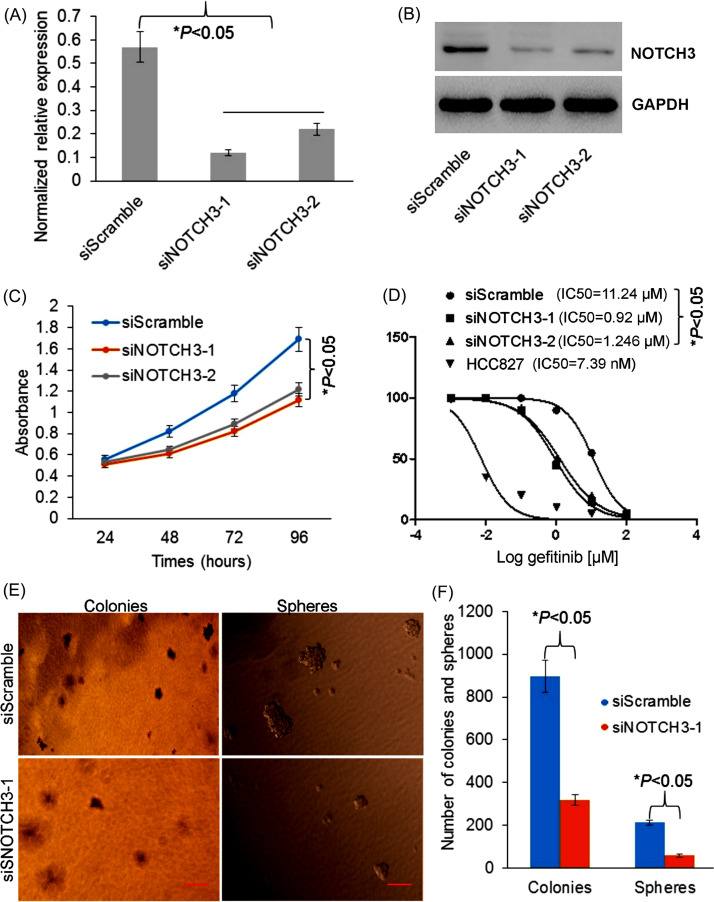
To study the role of NOTCH3 in regulating proliferation of TKI-resistant cells, we used specific siRNAs to knock down NOTCH3 expression. First, the siNOTCH3-1 and siNOTCH3-2 were used to knock down NOTCH3 expression in HCC827GR6 cell line, and downexpression of NOTCH3 was verified in mRNA and protein levels (Fig. 2A and B). Next, CCK-8 assay was performed in HCC827GR6 cells transfected with siScramble, siNOTCH3-1, and siNOTCH3-2. As we expected, the cell proliferation was decreased in the cells transfected with siNOTCH3-1 and siNOTCH3-2 compared to the controls (Fig. 2C). Also, IC50 values of gefitinib in the cells transfected with siNOTCH3-1 and siNOTCH3-2 were decreased (Fig. 2D). Moreover, colony and sphere formation of stem-like property were decreased in HCC827GR6 cells transfected with siNOTCH3-1 compared to the cells with siScramble control (Fig. 2E and F). Altogether, our data indicated that NOTCH3 knockdown was able to inhibit cell growth and stemness in TKI-resistant cells, gefitinib sensitivity increased.
Figure 2.
Effects of NOTCH3 siRNA on the growth and stemness of TKI-resistant cells. (A) Quantitation real-time polymerase chain reaction (qRT-PCR) and (B) Western blot showed NOTCH3 expression was decreased in HCC827GR6 cells transfected with siNOTCH3-1 and siNOTCH3-2. (C) The cell proliferation and (D) IC50 of gefitinib were decreased in HCC827GR6 cells transfected with siNOTCH3-1 and siNOTCH3-2. (E) Representative photographs for the colony formation and the sphere formation in HCC827GR6 cells transfected with siScramble and siNOTCH3-1. (F) Knockdown of NOTCH3 reduced colony number and sphere formation efficiency with significant difference.
miR-150 Was Identified as a Posttranscriptional Regulator of NOTCH3 in LUAD
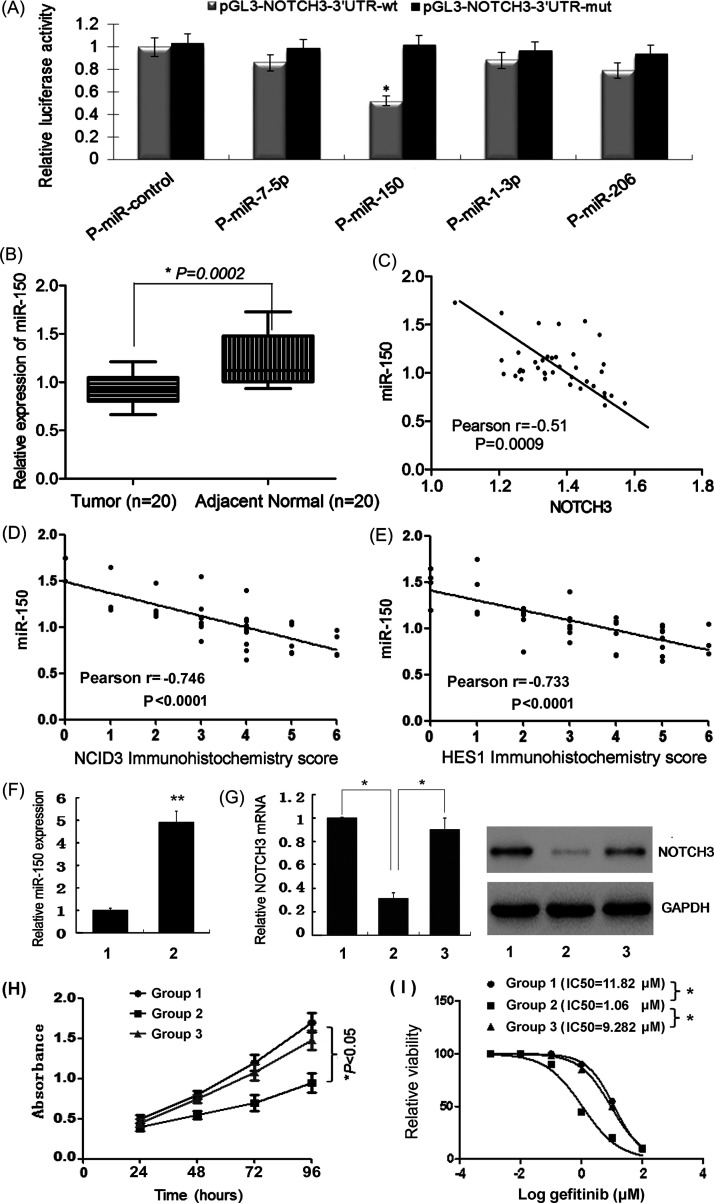
To identify novel miRNAs that might contribute to chemoresistance and carcinogenesis by targeting NOTCH3, we searched for computationally predicted candidate miRNAs using a web-based miRNA database (www.targetscan.org). Among multiple candidate miRNAs with potentially high binding capacity to the 3′-UTR of NOTCH3, four putative miRNAs (miR-7, miR-150, miR-1, and miR-206) were selected because of their previously reported differential expression in human cancers. Only the miR-150 mimic decreased the luciferase activity of the wild-type 3′-UTR reporter (51 ± 10%, p < 0.05), while that of the mutated reporter was not significantly affected, as shown by luciferase assays (Fig. 3A). Then the expression of miR-150 in LUAD tissues was quantified by qRT-PCR. miR-150 levels were significantly lower in LUAD tissues compared with adjacent normal tissues (Fig. 3B), and negatively correlated with NOTCH3 (Fig. 3C), NCID3 (Fig. 3D), and HES1 (Fig. 3E) expression, both at cancerous and normal tissues. These data indicate that miR-150 directly targets the 3′-UTR of NOTCH3, thus inhibiting NOTCH3 expression and Notch signal pathway.
Figure 3.
Validation of candidate miRNAs targeting NOTCH3. (A) Among four putative miRNAs, cotransfection of miR-150 reduced NOTCH3 luciferase activity in HEK293T cells. (B) miR-150 levels were significantly lower in lung adenocarcinoma tissues compared with adjacent normal tissues. miR-150 levels were negatively correlated with NOTCH3 mRNA expression (C), NCID3 protein (D), and HES1 protein (E) expression, both at lung adenocarcinoma tissues and normal tissues (n = 40). (F) qRT-PCR detection of miR-150 expression in HCC827GR6 cells transfected with miR-NC (group 1) or miR-150 mimic (group 2). (G) qRT-PCR and Western blot detection of NOTCH3 expression in HCC827GR6 cells transfected with miR-NC (group 1), miR-150 mimic (group 2), or cotransfected with miR-150 mimic and pEGFP-NOTCH3 (group 3). (H) The cell proliferation and (I) IC50 of gefitinib were decreased in HCC827GR6 cells transfected with miR-150 mimic, but was reversed when cotransfected with pEGFP-NOTCH3.
To study miR-150 function, miR-150 mimic was successfully transfected into HCC827GR6 cells (Fig. 3F), which caused downregulation of NOTCH3 (Fig. 3G), inhibition of cell proliferation (Fig. 3H), and decrease in gefitinib IC50 values (Fig. 3I). However, these changes were reversed when HCC827GR6 cells were cotransfected with miR-150 mimic and pEGFP-NOTCH3, which indicated that miR-150 could improve TKI sensitivity by directly downregulating NOTCH3.
Overexpression of NOTCH3 Was Correlated With the Expression of COL1A1 and Unfavorable Prognosis of LUAD Patients
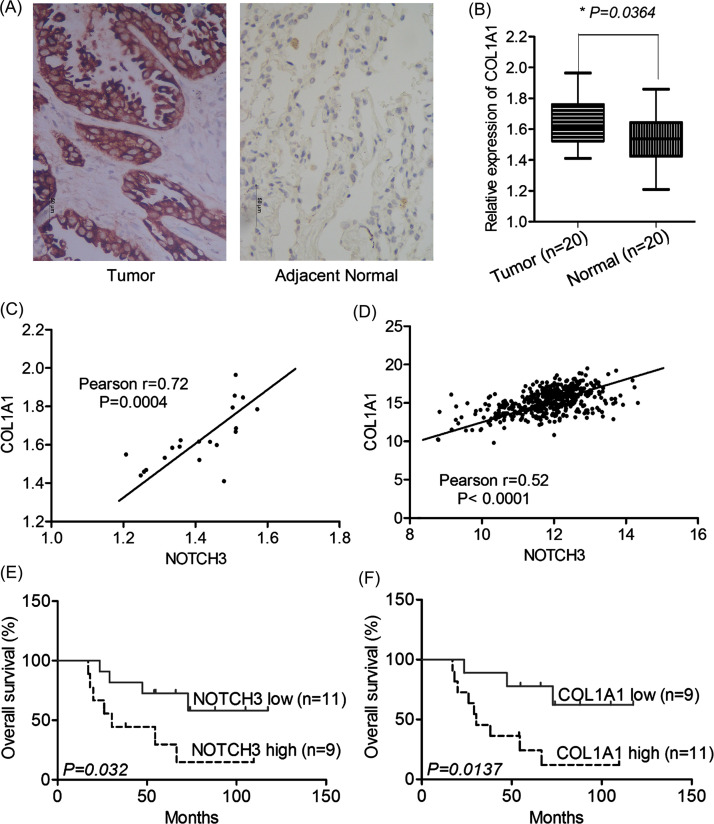
Collagen 1A1 (COL1A1) is a novel player implicated in the complex network of hypoxia response in NSCLC. We detected COL1A1 expression by qRT-PCR and IHC in 20 pairs of LUAD tissues. COL1A1 expression was increased significantly in LUAD tissues compared with matched normal tissues both in mRNA and protein levels (Fig. 4A and B). Then the relationship between COL1A1 and NOTCH3 expression was evaluated. The level of COL1A1 mRNA in LUAD tissues was positively correlated with NOTCH3 expression in our data (n = 20) (Fig. 4C), which was further verified in TCGA data (n = 445) (Fig. 4D). According to the IHC semiquantitative score, NOTCH3 expression levels were divided into low expression (n = 11) and high expression (n = 9) in these LUAD cases. Univariate survival analysis showed that patients with high expression of NOTCH3 were associated with shorter OS than that of low expression [39.9 months (95% CI 27.2–52.7) vs. 60.1 months (95% CI 48.5–71.6), p = 0.032] (Fig. 4E). Also, COL1A1 expression levels were divided into low expression (n = 9) and high expression (n = 11) in these LUAD cases, and high expression of COL1A1 was associated with shorter OS [38.8 months (95% CI 28.4–49.3) vs. 62.4 months (95% CI 50.4–74.5), p = 0.0137] (Fig. 4F). These results suggested that NOTCH3 may affect LUAD prognosis and TKI resistance by upregulating COL1A1.
Figure 4.
NOTCH3 correlated with COL1A1 as predictors for poor outcome of lung adenocarcinoma (LUAD) patients treated with TKIs. (A) Immunohistochemical staining of COL1A1 in LUAD and matched normal tissues. Scale bar: 50 μm. Positive staining is located in the cytoplasm. (B) COL1A1 mRNA expression was increased significantly in LUAD tissues compared with normal tissues (20 pairs). The level of COL1A1 mRNA expression in LUAD tissues was positively correlated with NOTCH3 in our cohort (n = 20; C) and TCGA data (n = 445; D). Patients with high expression of NOTCH3 protein (E) or COL1A1 protein (F) were associated with shorter overall survival.
DISCUSSION
The emergence of EGFR-TKIs, small molecules that bind to the tyrosine kinase domain of EGFR to inhibit its phosphorylation and the subsequent receptor activation and signal transduction, is a breakthrough in LUAD treatment. However, the resistance to EGFR-TKIs acquired by most patients during the treatment is a major obstacle that needs to be overcome21. Although our previous studies have revealed some preliminary results22,23, the mechanisms of EGFR–TKI resistance have not been well elucidated.
Active Notch signal has been documented to be involved in the development, prognosis, and TKI-acquired resistance of NSCLC8,9,24. Currently, we demonstrated that NOTCH3 was overexpressed in LUAD tissues compared to normal tissues, among family members NOTCH1–4. Also, in the TKI-resistant cell line, only NOTCH3 expression was upregulated in HCC827GR6 cells. Knockdown of NOTCH3 could reverse TKI resistance in vitro; moreover, overexpression of NOTCH3 protein in patients treated with EGFR–TKIs was associated with shorter OS, which suggested that NOTCH3 expression contributes to TKI resistance in LUAD. Consistent with our results, Diluvio et al. found that NOTCH3-specific inhibition increases triple-negative breast cancer (TNBC) sensitivity to TKI–gefitinib in TNBC-resistant cells25. Mechanistically, NOTCH3 is able to regulate the activated EGFR membrane localization; NOTCH3 inhibition induces the EGFR internalization and its intracellular arrest, without involving receptor degradation. Previous studies have also shown that overexpression of NOTCH3 was correlated with higher possibility of chemoresistance in NSCLC patients, and NOTCH3-specific inhibition could reduce the resistance to paclitaxel treatment13,14. The supposed mechanism includes downregulating of apoptosis-associated protein expression by NOTCH3. Another study confirmed a crucial role of NOTCH3 in the increase of stem-like property in NSCLC cells via upregulation of cancer stem cells (CSC) markers ALDH1A1 and CD44 and activation of autophagy26. But how NOTCH3 is involved in EGFR-TKI resistance needs to be further uncovered.
Dysregulation of miRNAs has been shown to contribute to the initiation, progression, and drug resistance of cancer. The Notch signaling pathway is also regulated by miRNAs. A previous study has identified miR-223 associated with resistance to erlotinib through regulating the Notch signaling pathway27. Jeong et al. report that microRNA-136 inhibits cancer stem cell activity and enhances the antitumor effect of paclitaxel against chemoresistant ovarian cancer cells by targeting NOTCH328. We showed here that miR-150 is involved in TKI resistance in LUAD by directly targeting NOTCH3. The miR-150, located on chromosome 19q13, may function as either a tumor suppressor or oncogene in different tumor types, which is dependent on its expression level and the action of its target genes in certain tumor types29,30. For example, miR-150 promotes the proliferation and migration of lung cancer cells by targeting SRC kinase signaling inhibitor 131, whereas it inhibits the aggressiveness of lung squamous cell carcinoma cells by downregulating matrix metalloproteinase 1432. Furthermore, it has been reported that NOTCH3 is a direct target of miR-150. miR-150 enhances apoptotic and antitumor effects of paclitaxel in paclitaxel-resistant ovarian cancer cells by targeting NOTCH333. Ghisi et al. found that miR-150 is significantly upregulated during T-cell maturation progresses. miR-150 targets NOTCH3 and plays important roles both in T-cell differentiation and leukemogenesis, and forced expression of miR-150 reduces NOTCH3 levels in T-cell lines and has adverse effects on their proliferation and survival34. However, the role of miR-150 in TKI resistance in lung cancer has not yet been clarified. In our study, miR-150 expression is significantly downregulated in LUAD tissues compared with matched normal tissues, and the level of miR-150 is negatively correlated with NOTCH3, as well as Notch signaling components NCID3 and HES1 protein expression. The cell proliferation and IC50 of gefitinib are decreased in HCC827GR6 cells transfected with miR-150 mimic, but reversed when cotransfected with NOTCH3 overexpressed vector. Therefore, miR-150 may play an important role in overcoming TKI resistance by directly inhibiting NOTCH3 expression and Notch signal pathway.
Tumor microenvironment is described as key regulators of resistance to anticancer drugs35,36. The cancer stroma is comprised of various types of immune cells, endothelial cells, fibroblasts, and extracellular matrix (ECM)37. COL1A1, an ECM protein, is regarded as the most important collagen in the tumor microenvironment. Increasing COL1A1 by cytokine stimulation could induce EMT phenotype, apoptotic resistance, and cell adhesion-mediated drug resistance38,39. A recent study showed that COL1A1 was correlated with hypoxia markers in NSCLC40. Solid tumors are often characterized by regions with reduced oxygen levels, and COL1A1 could assist in the adaptive responses to hypoxia events, thus promoting tumor aggressiveness and drug resistance41. Furthermore, collagen type I has been shown to induce EGFR–TKI resistance in EGFR-mutated cancer cells via mTOR activation42. Sincere Notch signaling can directly upregulate COL1A1 and COL1A2 promoter activity43. We are interested in studying the relationship between NOTCH3 and COL1A1 in EGFR–TKI resistance. As expected, our study showed that COL1A1 expression was upregulated in LUAD tissues compared with matched normal tissues, and its level was positively correlated with NOTCH3. We also found that high expression of either NOTCH3 or COL1A1 protein in patients treated with EGFR–TKIs was associated with shorter OS. Therefore, we propose that the effect of NOTCH3 on the prognosis and TKI resistance of LUAD may be carried out by upregulating COL1A1.
In summary, our study identified that NOTCH3 upregulation was associated with EGFR–TKI resistance in LUAD. Moreover, miR-150 was identified to directly target NOTCH3, and NOTCH3 expression was positively correlated with COL1A1 and poor outcome of LUAD patients, suggesting that the miR-150/NOTCH3/COL1A1 axis might be a potential therapeutic target for LUAD treatment.
ACKNOWLEDGMENT
This work was supported by the National Natural Science Foundation of China (No. 81472615) and Health and Family Planning Commission of Nanjing (No. ZKX15014). This work is dedicated in memory of Dr. Xiexin Yin, a professor in Chinese Pharmaceutical University.
Footnotes
The authors declare no conflicts of interest.
REFERENCES
- 1. Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin. 2017;67:7–30. [DOI] [PubMed] [Google Scholar]
- 2. Lee DH. Treatments for EGFR-mutant non-small cell lung cancer (NSCLC): The road to a success, paved with failures. Pharmacol Ther. 2017;174:1–21. [DOI] [PubMed] [Google Scholar]
- 3. Yi Y, Zeng S, Wang Z, Wu M, Ma Y, Ye X, Zhang B, Liu H. Cancer-associated fibroblasts promote epithelial-mesenchymal transition and EGFR-TKI resistance of non-small cell lung cancers via HGF/IGF-1/ANXA2 signaling. Biochim Biophys Acta 2018;1864:793–803. [DOI] [PubMed] [Google Scholar]
- 4. Nakagawa T, Takeuchi S, Yamada T, Ebi H, Sano T, Nanjo S, Ishikawa D, Sato M, Hasegawa Y, Sekido Y, Yano S. EGFR-TKI resistance due to BIM polymorphism can be circumvented in combination with HDAC inhibition. Cancer Res. 2013;73:2428–34. [DOI] [PubMed] [Google Scholar]
- 5. Pannuti A, Foreman K, Rizzo P, Osipo C, Golde T, Osborne B, Miele L. Targeting Notch to target cancer stem cells. Clin Cancer Res. 2010;16:3141–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Yin L, Velazquez OC, Liu ZJ. Notch signaling: Emerging molecular targets for cancer therapy. Biochem Pharmacol. 2010;80:690–701. [DOI] [PubMed] [Google Scholar]
- 7. Andersson ER, Lendahl U. Therapeutic modulation of Notch signalling—Are we there yet? Nat Rev Drug Discov. 2014;13:357–78. [DOI] [PubMed] [Google Scholar]
- 8. Ye YZ, Zhang ZH, Fan XY. Notch3 overexpression associates with poor prognosis in human non-small-cell lung cancer. Med Oncol. 2013;30:595. [DOI] [PubMed] [Google Scholar]
- 9. Yuan X, Wu H, Xu H, Han N, Chu Q, Yu S, Chen Y, Wu K. Meta-analysis reveals the correlation of Notch signaling with non-small cell lung cancer progression and prognosis. Sci Rep. 2015;5:10338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Konishi J, Kawaguchi KS, Vo H, Haruki N, Gonzalez A, Carbone DP, Dang TP. Gamma-secretase inhibitor prevents Notch3 activation and reduces proliferation in human lung cancers. Cancer Res. 2007;67:8051–7. [DOI] [PubMed] [Google Scholar]
- 11. Kang H, Jeong JY, Song JY, Kim TH, Kim G, Huh JH, Kwon AY, Jung SG, An HJ. Notch3-specific inhibition using siRNA knockdown or GSI sensitizes paclitaxel-resistant ovarian cancer cells. Mol Carcinog. 2016;55:1196–209. [DOI] [PubMed] [Google Scholar]
- 12. Rahman MT, Nakayama K, Rahman M, Katagiri H, Katagiri A, Ishibashi T, Ishikawa M, Iida K, Nakayama S, Otsuki Y, Miyazaki K. Notch3 overexpression as potential therapeutic target in advanced stage chemoresistant ovarian cancer. Am J Clin Pathol. 2012;138:535–44. [DOI] [PubMed] [Google Scholar]
- 13. Shi C, Qian J, Ma M, Zhang Y, Han B. Notch 3 protein, not its gene polymorphism, is associated with the chemotherapy response and prognosis of advanced NSCLC patients. Cell Physiol Biochem. 2014;34:743–52. [DOI] [PubMed] [Google Scholar]
- 14. He F, Du T, Jiang Q, Zhang Y. Synergistic effect of Notch-3-specific inhibition and paclitaxel in non-small cell lung cancer (NSCLC) cells via activation of the intrinsic apoptosis pathway. Med Sci Monit. 2017;23:3760–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Garzon R, Marcucci G, Croce CM. Targeting microRNAs in cancer: Rationale, strategies and challenges. Nat Rev Drug Discov. 2010;9:775–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010;11:597–610. [DOI] [PubMed] [Google Scholar]
- 17. Zhang Q, Li Q, Xu T, Jiang H, Xu LG. miR-491-5p suppresses cell growth and invasion by targeting Notch3 in nasopharyngeal carcinoma. Oncol Rep. 2016;35:3541–7. [DOI] [PubMed] [Google Scholar]
- 18. Huang TT, Ping YH, Wang AM, Ke CC, Fang WL, Huang KH, Lee HC, Chi CW, Yeh TS. The reciprocal regulation loop of Notch2 pathway and miR-23b in controlling gastric carcinogenesis. Oncotarget 2015;6:18012–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Deng W, Jiao N, Li N, Wan X, Luo S, Zhang Y. Decreased expression of PinX1 protein predicts poor prognosis of colorectal cancer patients receiving 5-FU adjuvant chemotherapy. Biomed Pharmacother. 2015;73:1–5. [DOI] [PubMed] [Google Scholar]
- 20. Yuan Y, Sun S, Jiao N, Shu Y, Zhang Y. Upregulation of HOXA10 protein expression predicts poor prognosis for colorectal cancer. Genet Test Mol Biomarkers 2018;22:390–7. [DOI] [PubMed] [Google Scholar]
- 21. Jackman D, Pao W, Riely GJ, Engelman JA, Kris MG, Jänne PA, Lynch T, Johnson BE, Miller VA. Clinical definition of acquired resistance to epidermal growth factor receptor tyrosine kinase inhibitors in non-small-cell lung cancer. J Clin Oncol. 2010;28:357–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wang YS, Wang YH, Xia HP, Zhou SW, Schmid-Bindert G, Zhou CC. MicroRNA-214 regulates the acquired resistance to gefitinib via the PTEN/AKT pathway in EGFR-mutant cell lines. Asian Pac J Cancer Prev. 2012;13:255–60. [DOI] [PubMed] [Google Scholar]
- 23. Tian Y, Zhang Z, Miao L, Yang Z, Yang J, Wang Y, Qian D, Cai H, Wang Y. Anexelekto (AXL) increases resistance to EGFR-TKI and activation of AKT and ERK1/2 in non-small cell lung cancer cells. Oncol Res. 2016;24:295–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Xie M, He CS, Wei SH, Zhang L. Notch-1 contributes to epidermal growth factor receptor tyrosine kinase inhibitor acquired resistance in non-small cell lung cancer in vitro and in vivo. Eur J Cancer 2013;49:3559–72. [DOI] [PubMed] [Google Scholar]
- 25. Diluvio G, Del Gaudio F, Giuli MV, Franciosa G, Giuliani E, Palermo R, Besharat ZM, Pignataro MG, Vacca A, d’Amati G, Maroder M, Talora C, Capalbo C, Bellavia D, Checquolo S. NOTCH3 inactivation increases triple negative breast cancer sensitivity to gefitinib by promoting EGFR tyrosine dephosphorylation and its intracellular arrest. Oncogenesis 2018;7:42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ma Y, Li M, Si J, Xiong Y, Lu F, Zhang J, Zhang L, Zhang P, Yang Y. Blockade of Notch3 inhibits the stem-like property and is associated with ALDH1A1 and CD44 via autophagy in non-small lung cancer. Int J Oncol. 2016;48:2349–58. [DOI] [PubMed] [Google Scholar]
- 27. Zhang H, Chen F, He Y, Yi L, Ge C, Shi X, Tang C, Wang D, Wu Y, Nian W. Sensitivity of non-small cell lung cancer to erlotinib is regulated by the Notch/miR-223/FBXW7 pathway. Biosci Rep. 2017;37:BSR20160478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jeong JY, Kang H, Kim TH, Kim G, Heo JH, Kwon AY, Kim S, Jung SG, An HJ. MicroRNA-136 inhibits cancer stem cell activity and enhances the anti-tumor effect of paclitaxel against chemoresistant ovarian cancer cells by targeting Notch3. Cancer Lett. 2017;386:168–78. [DOI] [PubMed] [Google Scholar]
- 29. Xu DD, Zhou PJ, Wang Y, Zhang Y, Zhang R, Zhang L, Chen SH, Fu WY, Ruan BB, Xu HP, Hu CZ, Tian L, Qin JH, Wang S, Wang X, Liu QY, Ren Z, Gu XK, Li YH, Liu Z, Wang YF. miR-150 suppresses the proliferation and tumorigenicity of leukemia stem cells by targeting the Nanog signaling pathway. Front Pharmacol. 2016;7:439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Li H, Ouyang R, Wang Z, Zhou W, Chen H, Jiang Y, Zhang Y, Li H, Liao M, Wang W, Ye M, Ding Z, Feng X, Liu J, Zhang B. MiR-150 promotes cellular metastasis in non-small cell lung cancer by targeting FOXO4. Sci Rep. 2016;6:39001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Cao M, Hou D, Liang H, Gong F, Wang Y, Yan X, Jiang X, Wang C, Zhang J, Zen K, Zhang CY, Chen X. miR-150 promotes the proliferation and migration of lung cancer cells by targeting SRC kinase signalling inhibitor 1. Eur J Cancer 2014;50:1013–24. [DOI] [PubMed] [Google Scholar]
- 32. Suetsugu T, Koshizuka K, Seki N, Mizuno K, Okato A, Arai T, Misono S, Uchida A, Kumamoto T, Inoue H. Downregulation of matrix metalloproteinase 14 by the antitumor miRNA, miR-150-5p, inhibits the aggressiveness of lung squamous cell carcinoma cells. Int J Oncol. 2018;52:913–24. [DOI] [PubMed] [Google Scholar]
- 33. Kim TH, Jeong JY, Park JY, Kim SW, Heo JH, Kang H, Kim G, An HJ. miR-150 enhances apoptotic and anti-tumor effects of paclitaxel in paclitaxel-resistant ovarian cancer cells by targeting Notch3. Oncotarget 2017;8:72788–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ghisi M, Corradin A, Basso K, Frasson C, Serafin V, Mukherjee S, Mussolin L, Ruggero K, Bonanno L, Guffanti A, De Bellis G, Gerosa G, Stellin G, D’Agostino DM, Basso G, Bronte V, Indraccolo S, Amadori A, Zanovello P. Modulation of microRNA expression in human T-cell development: Targeting of NOTCH3 by miR-150. Blood 2011;117:7053–62. [DOI] [PubMed] [Google Scholar]
- 35. Straussman R, Morikawa T, Shee K, Barzily-Rokni M, Qian ZR, Du J, Davis A, Mongare MM, Gould J, Frederick DT, Cooper ZA, Chapman PB, Solit DB, Ribas A, Lo RS, Flaherty KT, Ogino S, Wargo JA, Golub TR. Tumour micro-environment elicits innate resistance to RAF inhibitors through HGF secretion. Nature 2012;487:500–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yoshida T, Ishii G, Goto K, Neri S, Hashimoto H, Yoh K, Niho S, Umemura S, Matsumoto S, Ohmatsu H, Iida S, Niimi A, Nagai K, Ohe Y, Ochiai A. Podoplanin-positive cancer-associated fibroblasts in the tumor microenvironment induce primary resistance to EGFR-TKIs in lung adenocarcinoma with EGFR mutation. Clin Cancer Res. 2015;21:642–51. [DOI] [PubMed] [Google Scholar]
- 37. Lu P, Weaver VM, Werb Z. The extracellular matrix: A dynamic niche in cancer progression. J Cell Biol. 2012;196:395–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Liu J, Eischeid AN, Chen XM. Col1A1 production and apoptotic resistance in TGF-β1-induced epithelial-to-mesenchymal transition-like phenotype of 603B cells. PLoS One 2012;7:e51371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Song Y, Kim JS, Choi EK, Kim J, Kim KM, Seo HR. TGF-β-independent CTGF induction regulates cell adhesion mediated drug resistance by increasing collagen I in HCC. Oncotarget 2017;8:21650–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Oleksiewicz U, Liloglou T, Tasopoulou KM, Daskoulidou N, Gosney JR, Field JK, Xinarianos G. COL1A1, PRPF40A, and UCP2 correlate with hypoxia markers in non-small cell lung cancer. J Cancer Res Clin Oncol. 2017;143:1133–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Januchowski R, Świerczewska M, Sterzyńska K, Wojtowicz K, Nowicki M, Zabel M. Increased expression of several collagen genes is associated with drug resistance in ovarian cancer cell lines. J Cancer 2016;7:1295–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yamazaki S, Higuchi Y, Ishibashi M, Hashimoto H, Yasunaga M, Matsumura Y, Tsuchihara K, Tsuboi M, Goto K, Ochiai A, Ishii G. Collagen type I induces EGFR-TKI resistance in EGFR-mutated cancer cells via mTOR activation through Akt-independent pathway. Cancer Sci. 2018;109:2063–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hu M, Ou-Yang HF, Wu CG, Qu SY, Xu XT, Wang P. Notch signaling regulates col1α1 and col1α2 expression in airway fibroblasts. Exp Biol Med. (Maywood) 2014;239:1589–96. [DOI] [PubMed] [Google Scholar]