Abstract
Cholangiocarcinoma (CCA) is the second most common primary hepatobiliary carcinoma. The long noncoding RNA (lncRNA) small nucleolar RNA host gene 1 (SNHG1) has been reported to contribute to the progression of multiple cancers. Nonetheless, the functions and hidden mechanism of SNHG1 remain unclear in CCA. In this study, the SNHG1 levels were boosted in CCA cell lines, and knockdown of SNHG1 repressed CCA cell proliferation and invasion in vitro. The data also demonstrated that miR-140 could act as a target of SNHG1 in CCA and inhibited CCA cell proliferation and invasion, whereas the inhibition effects were relieved by overexpression of SNHG1. In addition, Toll-like receptor 4 (TLR4), an NF-κB-activating signal, was identified to be a target of miR-140. SNHG1, as a competing endogenous RNA (ceRNA) for miR-140, enhanced TLR4 expression and activated NF-κB signaling, thereby regulating growth and tumorigenesis in CCA. Animal experiments further confirmed this conclusion. Collectively, these findings not only uncovered a key role of SNHG1/miR-140/TLR4/NF-κB signaling axis in CCA tumorigenesis and progression but also denoted the probable utilization of SNHG1 as a therapeutic target for CCA.
Key words: Cholangiocarcinoma (CCA), Long noncoding RNAs (lncRNAs) SNHG1, miR-140, Toll-like receptor 4 (TLR4), NF-κB signaling
INTRODUCTION
Cholangiocarcinoma (CCA) originates from malignant transformation of biliary epithelial cells located in the intrahepatic and extrahepatic bile ducts and is the second most common primary hepatobiliary carcinoma1–3. Over the past decades, there has been a growing number of CCA around the world, especially in Asia and Latin America, and currently surgical resection remains the most effective treatment for CCA4,5. However, surgical resection is advocated only for CCA patients in its early stage and is not a viable option for patients in its advanced stage. Additionally, excision surgery is related to a high relapse rate of patients with biliary tract cancer, and the 5-year survival rate is only 10%6. Unfortunately, being short of applicable approaches for early diagnosis, most CCA patients are not diagnosed until they are at the advanced stages7. Furthermore, CCA is also insensitive to conventional chemotherapy and radiotherapy. Hence, there is great need to explore new therapeutic targets and biomarkers associated with diagnosis/prognosis by elucidating the vital molecular mechanisms of CCA initiation and progression to improve patient survival time.
Long noncoding RNAs (lncRNAs), a type of endogenous noncoding RNAs with more than 200 nucleotides in length, play crucial roles in the regulation of multiple biological processes. Increasing evidence points out that lncRNAs can function by a competing endogenous RNA (ceRNA) mechanism. In this mechanism, lncRNAs compete with mRNAs for shared microRNAs, leading to the derepression of the mRNAs8,9. lncRNAs have been shown to be involved in various disease pathogenesis, including, but not limited to, cancers. In fact, many studies show that lncRNAs have been identified as oncogenes or tumor suppressors, and thus inhibited/promoted the initiation and progression of cancers. In CCA, lncRNAs H19 and HULC are activated by oxidative stress and then promote cell migration as well as invasion through a ceRNA manner1. lncRNAs AFAP1-AS1 and NRAT1 promote cell growth and metastasis; lncRNA CCAT1 promotes cell migration, invasion, and epithelial–mesenchymal transition (EMT) through suppressing miR-152; and lncRNA PANDAR is observed to be upregulated and promotes tumorigenesis in CCA10–13. lncRNA SNHG1 (small nucleolar RNA host gene 1) has been reported to be upregulated and contribute to malignancy, metastasis, and poor prognosis of lung cancer, hepatocellular carcinoma, prostate cancer, glioma, colorectal carcinoma, gastric cancer, and ovarian carcinoma14–20. Thus, SNHG1 is a potential therapeutic target as well as a diagnostic and prognostic biomarker in these cancers. However, the expression and detailed function of SNHG1 in CCA remain unclear.
We confirmed the expression of SNHG1 in CCA cell lines and explored the functional roles of SNHG1 in CCA through performing in vitro and in vivo experiments. We also investigated the molecular mechanism by which SNHG1 functioned in CCA.
MATERIALS AND METHODS
Cell Culture
Four CCA cell lines (HCCC-9810, SSP25, RBE, and HuCCT-1) were purchased from the Cell Bank of Chinese Academy of Sciences (Shanghai, P.R. China). Human intrahepatic biliary epithelial cells (HIBECs) were preserved in our laboratory. Cells were cultivated in RPMI-1640 (Gibco, Grand Island, NY, USA) supplemented with 10% fetal bovine serum (FBS; Invitrogen, Carlsbad, CA, USA) in a 37°C humidified atmosphere with 5% CO2.
Cell Transfection
The overexpressing sequence for SNHG1 was constructed into the adenoviral vector and named as AdSNHG1, and the adenoviral vector expressing a control scrambled sequence (AdCtr) acted as the negative control. The shRNA sequence for SNHG1 was ligated into lentiviral vector and named as shSNHG1. The lentiviral vector expressing scrambled shRNA (shCtr) was used as the negative control. The negative control mimic (miR-Ctr) was used as the control of miR-140 mimics (miR-140). All these sequences or vectors were acquired from GenePharma Corporation (Shanghai, P.R. China) and transfected into HCCC-9810 and RBE cells using Lipofectamine 2000 (Invitrogen).
Real-Time PCR (RT-PCR)
Total RNA was extracted from tissues or cells using the TRIzol reagent (Invitrogen). The cDNA was synthesized using High Capacity cDNA Reverse Transcription kit (Applied Biosystems, Foster City, CA, USA) for lncRNAs and protein-encoding genes, and TaqMan MicroRNA Reverse Transcription kit (Applied Biosystems) for miRNA. Then the synthesized cDNA was subjected to PCR as previously described21. The PCR products were separated by electrophoresis and visualized by ethidium bromide staining. β-Actin was used as the reference for SNHG1 and Toll-like receptor 4 (TLR4), and U6 snRNA served as the loading control for miR-140. The relative expression levels were calculated using the 2−ΔΔCt method.
Cell Proliferation Assays
MTT assays were employed to assess cell proliferation. The transfected cells with 1.5 × 103 cells/well were seeded in 96-well plates and maintained in RPMI-1640 including 10% of FBS for different times (0, 24, 48, or 72 h), and then 20 μl of 5 mg/ml MTT solution was added to each well for 4 h of continuous incubation at 37°C. Following the incubation, the supernatant was then discarded, and 200 μl of dimethyl sulfoxide (DMSO; Sigma-Aldrich, St. Louis, MO, USA) was added into wells. Optical density (OD) value at 490 nm was recorded by microplate reader (Molecular Devices, Sunnyvale, CA, USA), and cell growth curve was plotted based on the OD values.
Cell Invasion Assays
Cell invasion was detected by Transwell chambers with 8.0-μm pore size (Millipore, Bedford, MA, USA). A total of 1 × 105 cells in 100 μl of FBS-free RPMI-1640 were seeded on the upper chamber precoated with Matrigel (BD Biosciences, San Jose, CA, USA). RPMI-1640 (200 μl) with 20% FBS was added into the lower chamber. After 48 h of incubation, the cells on the upper surface were carefully removed, and the invaded cells on the lower surface were fixed with 4% paraformaldehyde and stained with 0.1 mg/ml crystal violet (Sigma-Aldrich). Finally, the invaded cells were counted manually under a microscope.
Western Blot
The cellular proteins were extracted from tissues or cells using the lysis buffer (pH 7.6) with the protease inhibitors (Beyotime Institute of Biotechnology, Nanjing, China). The protein concentration was measured by the BCA protein assay kit (Pierce, Rockford, IL, USA). The proteins were separated in 12% of sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to Immobilon-PSQ membranes (Millipore). The membranes were blocked by incubation in Tris-buffered saline (TBS) supplemented with 5% skim milk at room temperature for 1 h and then probed with the specific primary antibodies at 4°C overnight. After washing three times with Tween-20/Tris-buffered saline (TTBS), the membranes were probed with HRP-conjugated secondary antibodies (Boster, Wuhan, P.R. China) at room temperature for 2 h. The bound antibodies were detected by an enhanced chemiluminescence detection reagent (Amersham, Piscataway, NJ, USA). The following primary antibodies were used: anti-PCNA (Abcam, Cambridge, UK), anti-NF-κB-p65 (Abcam), anti-TLR4 (Abcam), and anti-β-actin (Abcam) that was used as the internal reference.
Luciferase Reporter Assays
The SNHG1 and TLR4 3′-UTR cDNAs comprising the miR-140 recognition site were amplified by PCR, cloned into the pMIR-luciferase reporter vector, and named as SNHG1-wt and TLR4-wt, respectively. In addition, the luciferase reporters SNHG1-mut and TLR4-mut that encompassed the mutated sequence at miR-140 binding site were generated. HCCC-9810 and RBE cells were cotransfected with miR-Ctr or miR-140 and pMIR-luciferase reporter vectors containing the gene fragments of SNHG1 or TLR4 3′UTR with wt or mut binding sites of miR-140 by Lipofectamine 2000. Forty-eight hours after transfection, the luciferase activities were assayed by Dual-Luciferase Reporter Gene Assay kit (BioVision, Milpitas, CA, USA).
NF-κB Activity Assays
The NF-κB activity was detected in a whole cell extract using the ELISA-based kit (Active Motif, Carlsbad, CA, USA) according to manufacturer’s instructions.
Animal Experiments
All animal experiments were performed in accordance with the Guide for NIH and the institutional ethical guidelines. The athymic female BALB/c mice (5 weeks) were obtained from Slac Laboratory Animal Co. Ltd. (Shanghai, P.R. China) and were randomly assigned to the shCtr group or the shSNHG1 group with five mice in each group. HCCC-9810 cells (2 × 106) transfected with shCtr or shSNHG1 were suspended in 100 μl of PBS and subcutaneously injected into a single side of the posterior flank of each mouse. Tumor size was measured every 5 days. After 5 weeks of injection, all mice were sacrificed and weighed. Then the tumor tissues were excised, weighed, imaged, and used for RT-PCR, Western blot, and NF-κB activity assays.
Statistical Analyses
All statistical tests were performed by SPSS 17.0 statistical software (IBM, Chicago, IL, USA) and GraphPad Prism 5.01 software (GraphPad Software, Inc., La Jolla, CA, USA). The results were presented as mean ± SD of at least three independent experiments and analyzed by Student’s t-test. Statistical significance was set at a value of p < 0.05.
RESULTS
SNHG1 Is Upregulated in CCA Cell Lines
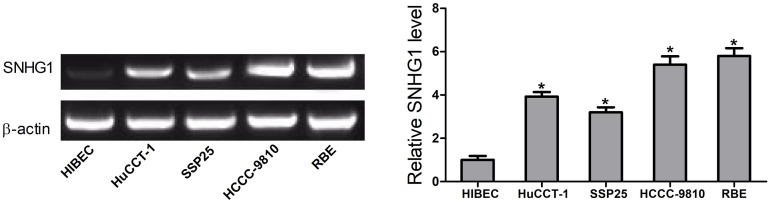
SNHG1 expression was determined in CCA cell lines HCCC-9810, SSP25, RBE, and HuCCT-1, and human intrahepatic biliary epithelial cells (HIBECs) by RT-PCR. These results revealed that there was a marked rise in SNHG1 expression in CCA cell lines compared with HIBECs (Fig. 1).
Figure 1.
Detection of small nucleolar RNA host gene 1 (SNHG1) expression in cholangiocarcinoma (CCA) cell lines HCCC-9810, SSP25, RBE, and HuCCT-1, as well as human intrahepatic biliary epithelial cells (HIBECs) by real-time (RT)-PCR. *p < 0.05.
Knockdown of SNHG1 Inhibits CCA Cell Proliferation and Invasion
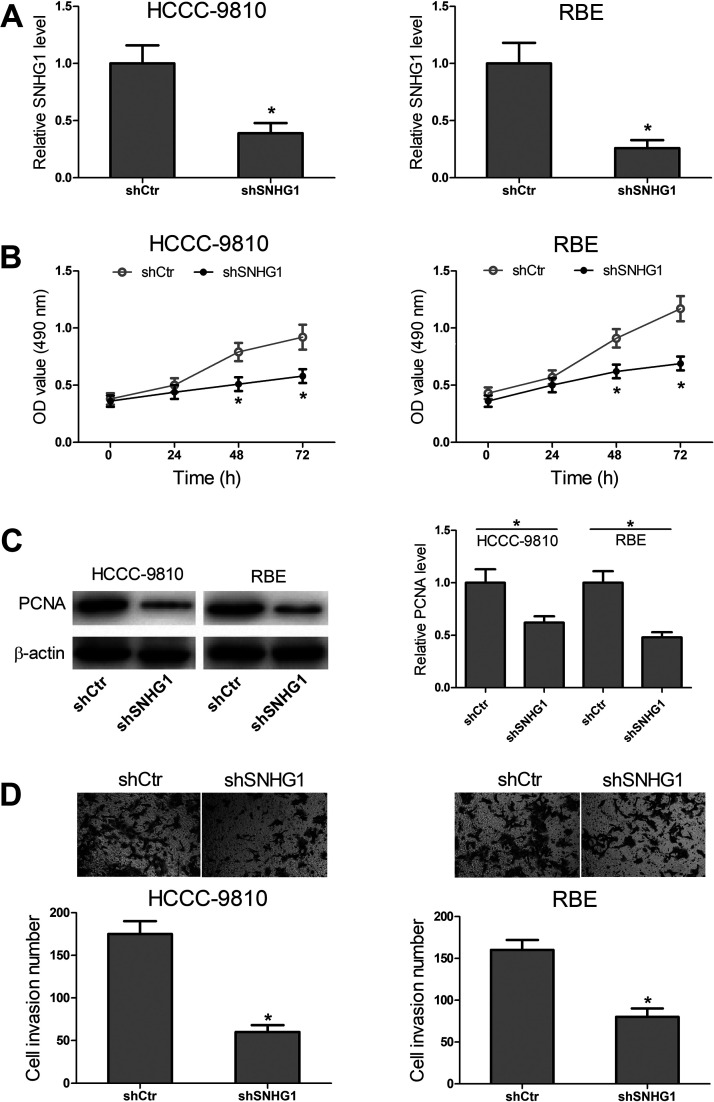
To determine the functional roles of SNHG1 in CCA cells, HCCC-9810 and RBE cells were transfected with shSNHG1 or shCtr, and RT-PCR was performed to confirm the transfection efficiency (Fig. 2A). Then MTT and Western blot assays were carried out to measure the effect that SNHG1 had on cell proliferation. As shown in Figure 2B, the results from MTT assays showed that SNHG1 knockdown obviously inhibited cell growth of HCCC-9810 and RBE cells. Consistently, the Western blot assays showed that the proliferation marker PCNA expression in the shSNHG1 group was less than that in the shCtr group (Fig. 2C). Transwell assays were used to assess the effect of SNHG1 expression on HCCC-9810 and RBE cell invasion. The results showed that decreased cell invasion was observed in the shSNHG1 group (Fig. 2D). These data indicated that downregulation of SNHG1 possessed tumor-inhibitory effects that SNHG1 knockdown inhibited cell proliferation and invasion in vitro.
Figure 2.
Effects of knockdown of SNHG1 on CCA cell proliferation and invasion. (A) SNHG1 expression was determined by RT-PCR in HCCC-9810 and RBE cells transfected with scrambled shRNA (shCtr) or shSNHG1. (B) MTT assays were used to detect cell proliferation of the transfected HCCC-9810 and RBE cells. (C) After transfection, Western blot assays were carried out to analyze proliferation marker PCNA expression. (D) Transwell assays were performed to detect cell invasion of the transfected HCCC-9810 and RBE cells. *p < 0.05.
SNHG1 Targets miR-140
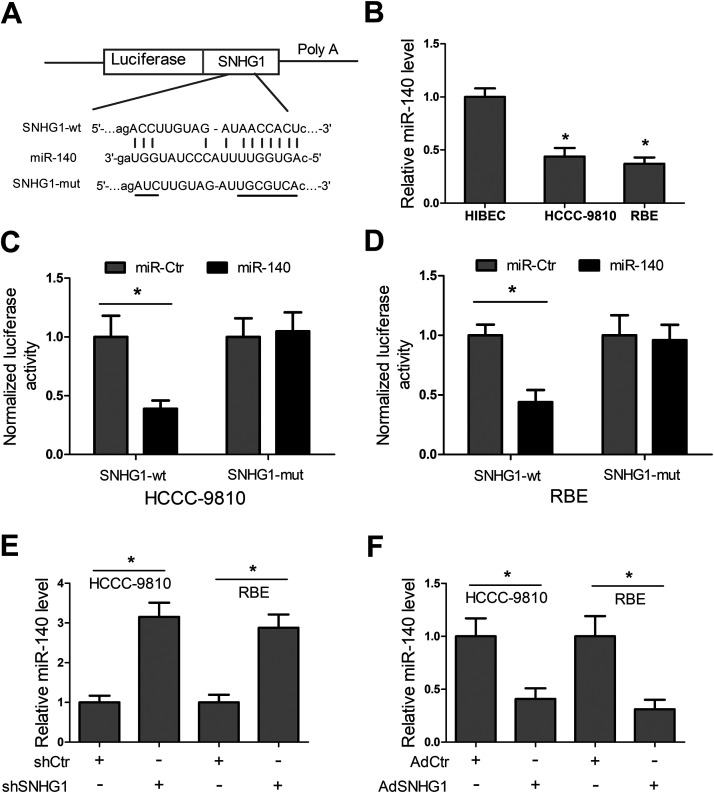
Recently, lncRNAs have been widely reported to function by a ceRNA mechanism. To investigate whether SNHG1 functioned via a similar mechanism in CCA, the online software starBase v2.0 for miRNA recognition sequences in SNHG1 was employed to predict the putative binding site, and the results showed that there existed a putative binding site for miR-140 (Fig. 3A). The RT-PCR results showed that miR-140 expression was significantly reduced in HCCC-9810 and RBE cells compared with HIBECs (Fig. 3B). To verify the direct binding between SNHG1 and miRNA-140, luciferase reporter constructs were generated. We observed that miR-140 significantly declined the luciferase activity of reporter vector comprising SNHG1-wt, while it had no significant effect on the luciferase activity of reporter vector containing SNHG1-mut (Fig. 3C and D). Additionally, our results exhibited that SNHG1 knockdown significantly enhanced miR-140 level, and SNHG1 overexpression markedly reduced miR-140 level (Fig. 3E and F). Collectively, these data indicated that SNHG1 targeted miR-140 and inhibited miR-140 expression in HCCC-9810 and RBE cells.
Figure 3.
SNHG1 targets miR-140 and represses its expression in CCA cells. (A) Diagram of the miR-140 putative binding sites and corresponding mutant sites in SNHG1 sequences. (B) Detection of miR-140 expression in CCA cell lines HCCC-9810 and RBE, and HIBECs by RT-PCR. (C, D) Luciferase reporter assays were performed to verify the direct binding between miR-140 and SNHG1 in HCCC-9810 and RBE cells. (E, F) miR-140 expression was determined in HCCC-9810 and RBE cells after transfection with shCtr, shSNHG1, adenoviral vector expressing a control scrambled sequence (AdCtr), or AdSNHG1. *p < 0.05.
SNHG1 Functions by Targeting miR-140
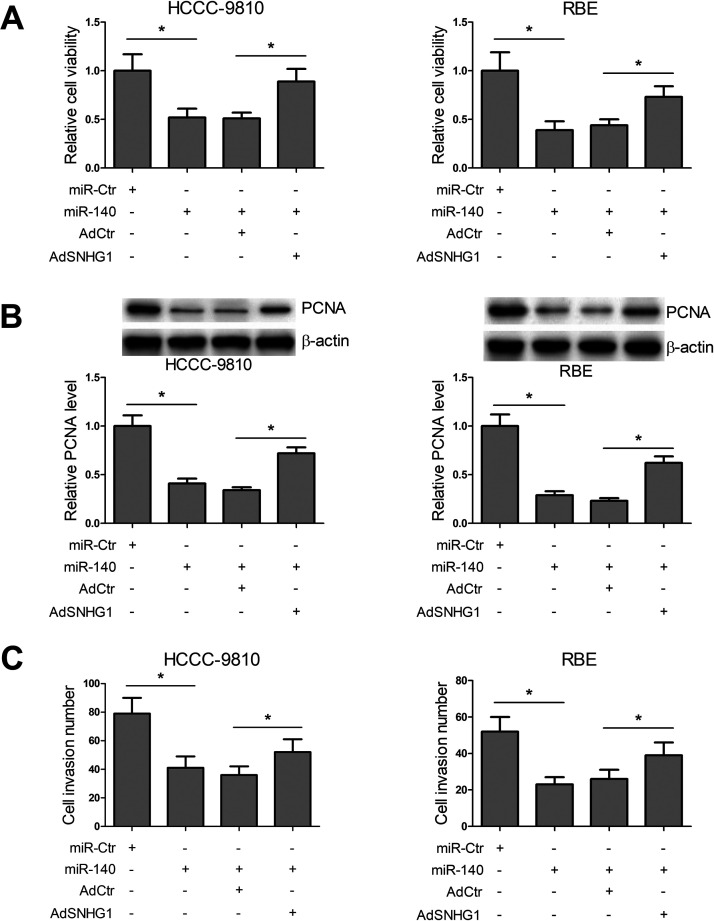
In view of the suppressive effect of SNHG1 on expression of miR-140, we further investigated whether SNHG1 functioned by targeting miR-140. HCCC-9810 and RBE cells were transfected with miR-Ctr, miR-140, AdCtr, or AdSNHG1, alone or in combination, and then MTT, Western blot, and Transwell assays were conducted. MTT and Western blot assays revealed that miR-140 significantly inhibited cell viability and PCNA expression, whereas overexpression of SNHG1 dramatically relieved the inhibitory effects caused by miR-140 (Fig. 4A and B). Transwell assays showed that cell invasion number in miR-140-transfected cells was much less than that in the control group, whereas the amount of invaded cells was largely recovered through upregulation of SNHG1 (Fig. 4C). These data indicated that SNHG1 functioned by targeting miR-140 in CCA cells.
Figure 4.
SNHG1 functions by targeting miR-140 in CCA cells. (A) HCCC-9810 and RBE cells were transfected with negative control mimic (miR-Ctr), miR-140, miR-140 + AdCtr, or miR-140 + AdSNHG1, and MTT assays were used to detect cell activity. (B) After transfection, PCNA expression was determined by Western blot. (C) Transwell assays were conducted to detect invasion of the transfected cells. *p < 0.05.
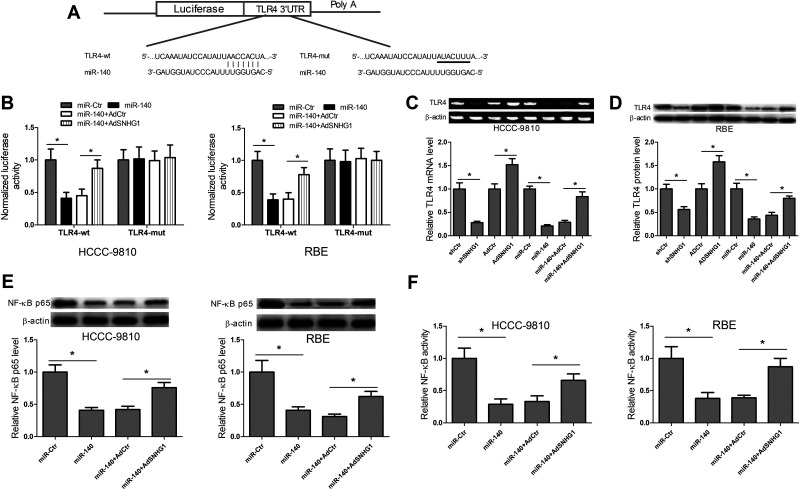
SNHG1-miR-140 Axis Controls TLR4 and the NF-κB Pathway
To further explore the underlying mechanisms by which SNHG1-miR-140 axis regulated CCA cell proliferation and invasion, TargetScan database prediction was carried out to confirm the miR-140 recognition sequence on TLR4 3′-UTR (Fig. 5A). Luciferase reporter assays were performed to verify the direct binding between miR-140 and TLR4. The results disclosed that miR-140 transfection obviously diminished the luciferase activity of reporter vector containing TLR4-wt compared with miR-Ctr transfection, while cotransfection with miR-140 and AdSNHG1 alleviated the inhibitory roles caused by miR-140 transfection (Fig. 5B). Then the expression levels of TLR4 mRNA and protein were also determined in HCCC-9810 and RBE cells transfected with shCtr, shSNHG1, AdCtr, AdSNHG1, miR-Ctr, miR-140, miR-140 + AdCtr, or miR-140 + AdSNHG1. The results showed that levels of TLR4 mRNA and protein expression were inhibited by shSNHG1 and miR-140 transfection compared with their respective control groups, and were enhanced by AdSNHG1 transfection compared with AdCtr transfection. However, the cotransfection with AdSNHG1 and miR-140 relieved the inhibitory effects of miR-140 transfection on TLR4 mRNA and protein expression (Fig. 5C and D). These data suggested that SNHG1 regulated TLR4 expression via ceRNA crosstalk. Activation of the NF-κB pathway is strongly implicated in a variety of hematologic and solid tumor malignancies, and is involved in the promotion of cell proliferation and angiogenesis, inhibition of apoptosis, and stimulation of invasion and metastasis. TLR4 can activate NF-κB protein through myeloid differentiation factor 88 dependent or independent pathway. Thus, the effects of SNHG1-miR-140 axis on the NF-κB pathway were explored. The NF-κB p65 protein expression and NF-κB activity were determined in HCCC-9810 and RBE cells transfected with miR-Ctr, miR-140, miR-140 + AdCtr, or miR-140 + AdSNHG1 by performing Western blot and ELISA, respectively. What stands out in Figure 5E and F is that miR-140 transfection significantly inhibited NF-κB p65 protein expression and NF-κB activity, while the cotransfection with AdSNHG1 and miR-140 relieved the inhibitory effects caused by miR-140 transfection. All in all, these data indicated that SNHG1, as a ceRNA for miR-140, boosted the expression of TLR4 and activated the NF-κB pathway.
Figure 5.
SNHG1–miR-140 axis controls Toll-like receptor 4 (TLR4) and the NF-κB pathway in CCA cells. (A) Diagram of the miR-140 putative binding sites and corresponding mutant sites in TLR4 3′-UTR region. (B) Dual-luciferase reporter assays were carried out to determine the direct binding between miR-140 and AdSNHG1 in HCCC-9810 and RBE cells. (C, D) TLR4 mRNA and protein expression were determined in HCCC-9810 and RBE cells after transfection with shCtr, shSNHG1, AdCtr, AdSNHG1, miR-Ctr, miR-140, miR-140 + AdCtr, or miR-140 + AdSNHG1 by RT-PCR and Western blot, respectively. (E, F) Detection of NF-κB p65 protein expression and NF-κB activity in HCCC-9810 and RBE cells after transfection with miR-Ctr, miR-140, miR-140 + AdCtr, or miR-140 + AdSNHG1 by Western blot and ELISA, respectively. *p < 0.05.
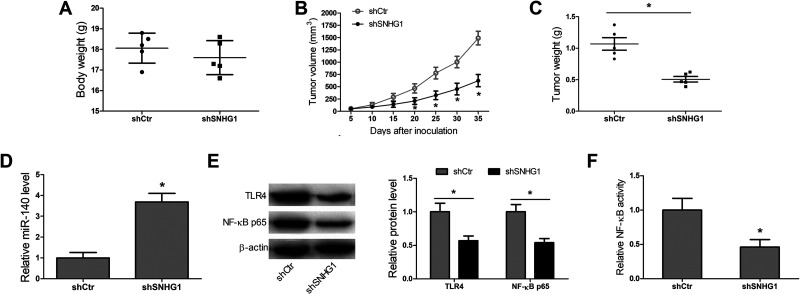
Knockdown of SNHG1 Inhibits CCA Growth In Vivo
To further confirm that knockdown of SNHG1 played a tumor-inhibitory role in CCA, the HCCC-9810 cells transfected with shCtr or shSNHG1 were inoculated into a single side of the posterior flank of all mice. The tumor size was monitored once every 5 days. These observations showed that there were no obvious differences in body weight between shCtr group and shSNHG1 group (Fig. 6A). What can be clearly seen in Figure 6B and C is that the shSNHG1 group had less tumor volume and tumor weight than the shCtr group. Additionally, the xenografts in the shSNHG1 group had a significant increase in miR-140 level, and marked decreases in TLR4 and NF-κB p65 protein expression, and NF-κB activity (Fig. 6D–F). Taken together, these data indicated that lncRNA SNHG1 regulated growth and tumorigenesis via inhibition of miR-140 and activation of the TLR4/NF-κB pathway in CCA.
Figure 6.
Knockdown of SNHG1 inhibits CCA growth in vivo. (A) The body weight of mice in the shCtr group and shSNHG1 group. (B, C) Tumor volume and tumor weight in the shCtr group and shSNHG1 group. (D–F) miR-140, TLR4, and NF-κB p65 expression, and NF-κB activity in xenografts of the shCtr group and the shSNHG1 group. *p < 0.05.
DISCUSSION
Increasing evidence has pointed out that lncRNAs are associated with the tumorigenesis and progression, and are thought of as therapeutic targets as well as diagnostic and prognostic markers for a variety of cancers, including CCA. In CCA, lncRNAs such as H19, HULC, AFAP1-AS1, NRAT1, CCAT1, PANDAR, PCAT1, CRNDE, and CCAT2 have been shown to be involved in the initiation and progression, including preserving proliferative signaling, evading growth suppressors, inhibiting cell death, and activating invasion and metastasis1,10–13,22–24. Recently, SNHG1 has been reported to play an oncogenic role in several cancers. For example, Cui et al.14 found that the SNHG1 is upregulated in non-small cell lung cancer and promotes progression of cancer via repression of miR-101-3p and activation of the Wnt/β-catenin pathway; Zhang et al.15 reported that SNHG1 exacerbates hepatocellular carcinoma progression via suppression of miR-195; Li et al.16 found that SNHG1/miR-199a-3p/CDK7 axis contributes to the progression of prostate cancer; Wang et al.17 reported that SNHG1 positively modulates the expression levels of NOB1 through sponging miR-326 and boosts tumorigenesis in osteosarcoma; and Qi et al.18 have found that in colorectal cancer SNHG1 enhances cell proliferation and metastasis via activating the Wnt/β-catenin signaling pathway, and what stands out is that the upregulated SNHG1 indicates a poor prognosis. Yet, the expression profile and biological functions of SNHG1 in CCA are still unclear.
In this study, our results showed that SNHG1 was significantly increased in CCA cell lines compared to HIBECs, and what was interesting was that SNHG1 knockdown markedly repressed proliferation and invasion of CCA cells. These data suggested that SNHG1 acted as a tumor oncogene in CCA.
Recently, the ceRNA mechanism has been extensively reported, and evidence has confirmed the interaction among lncRNA, miRNA, and mRNA (miRNA target) in cancers. lncRNA MALAT1 has been confirmed to work as a ceRNA to promote the expression of ZEB2 via sponging miR-200s in clear cell kidney carcinoma; lncRNA HOTAIR has been reported to derepress HER2 expression through competitively binding to miR-331-3p in gastric cancer; and lncRNA H19 has been found to promote β-catenin expression by sponging miR-200a in colorectal cancer25–27. In this study, we hypothesized that SNHG1 may work as a ceRNA in CCA. To verify that, we made use of starBase v2.0 software to predict the potential miRNAs interacting with SNHG1. Our data exhibited that some possible miRNAs including miR-140 could bind to SNHG1. Furthermore, many reports have pointed out that miR-140 may perform the function of tumor inhibition in cancers. For example, miR-140 inhibits tumor growth and metastasis by targeting insulin-like growth factor 1 receptor in non-small cell lung cancer; downregulation of miR-140 induces epithelial–mesenchymal transition (EMT) and promotes invasion by targeting Slug in esophageal cancer; and miR-140 suppresses TGFB receptor 1 (TGFBR1) and fibroblast growth factor 9 (FGF9) and inhibits tumor growth and metastasis of hepatocellular carcinoma28–30. In addition, recently, miR-140 is reported to inhibit CCA progression31. Based on these findings, the miR-140 was selected and further investigated. RT-PCR analyses showed that miR-140 was decreased in CCA cell lines compared with HIBECs. Luciferase reporter assays confirmed the direct binding of miR-140 to SNHG1. SNHG1 knockdown dramatically increased miR-140 expression, while SNHG1 overexpression significantly reduced miR-140 expression. In addition, our results revealed that miR-140 significantly constrained cell proliferation and invasion, whereas SNHG1 overexpression dramatically relieved the effects caused by miR-140.
NF-κB plays a central role in the regulation of innate and adaptive immunity. In recent years, many studies have reported that the NF-κB pathway is frequently constitutively activated in multiple cancers, which contributes to cancer initiation and maintenance of established cancer via upregulation of multiple antiapoptotic and other oncogenic genes32. NF-κB-activating signals classically originate from members of the interleukin (IL)-1, tumor-necrosis factor (TNF), or Toll-like receptor (TLR) superfamily, which facilitate the release and nuclear translocation of NF-κB via induction of the phosphorylation, ubiquitination, and degradation of IκBα and IκBβ. Among them, TLR4 is one of the most common NF-κB-activating signals, and TLR4/NF-κB pathway has been widely reported to participate in the pathogenesis of many diseases, including cancers. Woods et al.33 reported that TLR4 activates NF-κB in human ovarian granulosa tumor cells; He et al.34 found that TLR4/NF-κB signaling induces apoptosis resistance and boosts the immune escape of human lung cancer cells; and Duan et al.35 reported that Mycoplasma hyorhinis induces EMT in gastric cancer cell MGC803 through the TLR4/NF-κB signaling pathway. In addition, previous studies also have shown that caffeic acid phenethyl ester and cepharanthine exert antitumor activity on CCA by the inhibition of NF-κB36,37. In this study, our results showed that TLR4 was a target of miR-140, and the TLR4/NF-κB signaling could be regulated by SNHG1-miR-140 axis. These data indicated that SNHG1, as a ceRNA for miR-140, promoted TLR4 expression and activated the NF-κB signaling, thereby regulating growth and tumorigenesis of CCA. Animal experiments further confirmed this conclusion.
To conclude, our study first uncovered that SNHG1 functioned as an oncogenic lncRNA that enhanced tumorigenesis and progression of CCA through the miR-140/TLR4/NF-κB signaling axis. Our findings suggested that SNHG1 might act as a therapeutic target as well as a diagnostic and prognostic marker for CCA.
ACKNOWLEDGMENT
The authors declare no conflicts of interest.
Footnotes
The authors declare no conflicts of interest.
REFERENCES
- 1. Wang W-T, Ye H, Wei P-P, Han B-W, He B, Chen Z-H, Chen Y-Q. LncRNAs H19 and HULC, activated by oxidative stress, promote cell migration and invasion in cholangiocarcinoma through a ceRNA manner. J Hematol Oncol. 2016;9(1):1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Rizvi S, Gores GJ. Pathogenesis, diagnosis, and management of cholangiocarcinoma. Gastroenterology 2013;145(6):1215–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–32. [DOI] [PubMed] [Google Scholar]
- 4. Sirica AE, Nathanson MH, Gores GJ, LaRusso NF. Pathobiology of biliary epithelia and cholangiocarcinoma: Proceedings of the Henry M. and Lillian Stratton basic research single-topic conference. Hepatology 2008;48(6):2040–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wang Y, Meng R, Duan J, Liu G, Chen J, Li S, Ji X. Nephrotic syndrome may be one of the important etiologies of cerebral venous sinus thrombosis. J Stroke Cerebrovasc Dis. 2016;25(10):2415–22. [DOI] [PubMed] [Google Scholar]
- 6. Skipworth J, Olde Damink S, Imber C, Bridgewater J, Pereira S, Malagó M. Surgical, neo-adjuvant and adjuvant management strategies in biliary tract cancer. Aliment Pharmacol Ther. 2011;34(9):1063–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lau SHY, Lau WY. Current therapy of hilar cholangiocarcinoma. HBPD INT. 2012;11(1):12–7. [DOI] [PubMed] [Google Scholar]
- 8. Sen R, Ghosal S, Das S, Balti S, Chakrabarti J. Competing endogenous RNA: The key to posttranscriptional regulation. Sci World J. 2014;2014:1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kartha RV, Subramanian S. Competing endogenous RNAs (ceRNAs): New entrants to the intricacies of gene regulation. Front Genet. 2014;5:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Shi X, Zhang H, Wang M, Xu X, Zhao Y, He R, Zhang M, Zhou M, Li X, Peng F. LncRNA AFAP1-AS1 promotes growth and metastasis of cholangiocarcinoma cells. Oncotarget 2017;8(35):58394–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhang C, Li J-Y, Tian F-Z, Zhao G, Hu H, Ma Y-F, Yang Y-L. lncRNA NEAT1 promotes growth and metastasis of cholangiocarcinoma cells. Oncol Res. 2018;26(6):879–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhang S, Xiao J, Chai Y, Du YY, Liu Z, Huang K, Zhou X, Zhou W. LncRNA-CCAT1 Promotes migration, invasion, and EMT in intrahepatic cholangiocarcinoma through suppressing miR-152. Dig Dis Sci. 2017;62(11):3050–58. [DOI] [PubMed] [Google Scholar]
- 13. Xu Y, Jiang X, Cui Y. Upregulated long noncoding RNA PANDAR predicts an unfavorable prognosis and promotes tumorigenesis in cholangiocarcinoma. Onco Targets Ther. 2017;10:2873–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cui Y, Zhang F, Zhu C, Geng L, Tian T, Liu H. Upregulated lncRNA SNHG1 contributes to progression of non-small cell lung cancer through inhibition of miR-101-3p and activation of Wnt/β-catenin signaling pathway. Oncotarget 2017;8(11):17785–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhang H, Zhou D, Ying M, Chen M, Chen P, Chen Z, Zhang F. Expression of long non-coding RNA (lncRNA) small nucleolar RNA host gene 1 (SNHG1) exacerbates hepatocellular carcinoma through suppressing miR-195. Med Sci Monit. 2016;22:4820–29. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 16. Li J, Zhang Z, Xiong L, Guo C, Jiang T, Zeng L, Li G, Wang J. SNHG1 lncRNA negatively regulates miR-199a-3p to enhance CDK7 expression and promote cell proliferation in prostate cancer. Biochem Biophys Res Commun. 2017;487(1):146–52. [DOI] [PubMed] [Google Scholar]
- 17. Wang J, Cao L, Wu J, Wang Q. Long non-coding RNA SNHG1 regulates NOB1 expression by sponging miR-326 and promotes tumorigenesis in osteosarcoma. Int J Oncol. 2018;52(1):77–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Qi H, Wang J, Wang F, Ma H. Long non-coding RNA SNHG1 promotes cell proliferation and tumorigenesis in colorectal cancer via Wnt/β-catenin signaling. Pharmazie 2017;72(7):395–401. [DOI] [PubMed] [Google Scholar]
- 19. Hu Y, Ma Z, He Y, Liu W, Su Y, Tang Z. LncRNA-SNHG1 contributes to gastric cancer cell proliferation by regulating DNMT1. Biochem Biophys Res Commun. 2017;491(4):926–31. [DOI] [PubMed] [Google Scholar]
- 20. Ge J, Wu X, Yang X, Gao J, Wang F, Ye K. Role of long non-coding RNA SNHG1 in occurrence and progression of ovarian carcinoma. Eur Rev Med Pharmacol Sci. 2018;22:329–35. [DOI] [PubMed] [Google Scholar]
- 21. Shiraki K, Tsuji N, Shioda T, Isselbacher KJ, Takahashi H. Expression of Fas ligand in liver metastases of human colonic adenocarcinomas. Proc Natl Acad Sci USA 1997;94(12):6420–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhang F, Wan M, Xu Y, Li Z, Leng K, Kang P, Cui Y, Jiang X. Long noncoding RNA PCAT1 regulates extrahepatic cholangiocarcinoma progression via the Wnt/β-catenin-signaling pathway. Biomed Pharmacother. 2017;94:55–62. [DOI] [PubMed] [Google Scholar]
- 23. Xia XL, Xue D, Xiang TH, Xu HY, Song DK, Cheng PG, Wang JQ. Overexpression of long non-coding RNA CRNDE facilitates epithelial-mesenchymal transition and correlates with poor prognosis in intrahepatic cholangiocarcinoma. Oncol Lett. 2018;15(4):4105–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Xu Y, Yao Y, Qin W, Zhong X, Jiang X, Cui Y. Long non-coding RNA CCAT2 promotes cholangiocarcinoma cells migration and invasion by induction of epithelial-to-mesenchymal transition. Biomed Pharmacother. 2018;99:121–27. [DOI] [PubMed] [Google Scholar]
- 25. Xiao H, Tang K, Liu P, Chen K, Hu J, Zeng J, Xiao W, Yu G, Yao W, Zhou H. LncRNA MALAT1 functions as a competing endogenous RNA to regulate ZEB2 expression by sponging miR-200s in clear cell kidney carcinoma. Oncotarget 2015;6(35):38005–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Liu X-h, Sun M, Nie F-q, Ge Y-b, Zhang E-b, Yin D-d, Kong R, Xia R, Lu K-h, Li J-h. Lnc RNA HOTAIR functions as a competing endogenous RNA to regulate HER2 expression by sponging miR-331-3p in gastric cancer. Mol Cancer 2014;13(1):1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yang W, Ning N, Jin X. The lncRNA H19 promotes cell proliferation by competitively binding to miR-200a and derepressing β-catenin expression in colorectal cancer. Biomed Res Int. 2017;2017:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yuan Y, Shen Y, Xue L, Fan H. miR-140 suppresses tumor growth and metastasis of non-small cell lung cancer by targeting insulin-like growth factor 1 receptor. PloS One 2013;8(9):e73604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Li W, Jiang G, Zhou J, Wang H, Gong Z, Zhang Z, Min K, Zhu H, Tan Y. Down-regulation of miR-140 induces EMT and promotes invasion by targeting Slug in esophageal cancer. Cell Physiol Biochem. 2014;34(5):1466–76. [DOI] [PubMed] [Google Scholar]
- 30. Yang H, Fang F, Chang R, Yang L. MicroRNA-140-5p suppresses tumor growth and metastasis by targeting transforming growth factor β receptor 1 and fibroblast growth factor 9 in hepatocellular carcinoma. Hepatology 2013;58(1):205–17. [DOI] [PubMed] [Google Scholar]
- 31. Yu J, Zhang W, Tang H, Qian H, Yang J, Zhu Z, Ren P, Lu B. Septin 2 accelerates the progression of biliary tract cancer and is negatively regulated by mir-140-5p. Gene 2016;589(1):20–6. [DOI] [PubMed] [Google Scholar]
- 32. Lee H, Herrmann A, Deng J-H, Kujawski M, Niu G, Li Z, Forman S, Jove R, Pardoll DM, Yu H. Persistently activated Stat3 maintains constitutive NF-κB activity in tumors. Cancer Cell 2009;15(4):283–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Woods DC, White YA, Dau C, Johnson A. TLR4 activates NF-κB in human ovarian granulosa tumor cells. Biochem Biophys Res Commun. 2011;409(4):675–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. He W, Liu Q, Wang L, Chen W, Li N, Cao X. TLR4 signaling promotes immune escape of human lung cancer cells by inducing immunosuppressive cytokines and apoptosis resistance. Mol Immunol. 2007;44(11):2850–59. [DOI] [PubMed] [Google Scholar]
- 35. Duan H, Qu L, Shou C. Mycoplasma hyorhinis induces epithelial-mesenchymal transition in gastric cancer cell MGC803 via TLR4-NF-κB signaling. Cancer Lett. 2014;354(2):447–54. [DOI] [PubMed] [Google Scholar]
- 36. Seubwai W, Vaeteewoottacharn K, Hiyoshi M, Suzu S, Puapairoj A, Wongkham C, Okada S, Wongkham S. Cepharanthine exerts antitumor activity on cholangiocarcinoma by inhibiting NF-κB. Cancer Sci. 2010;101(7):1590–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Onori P, DeMorrow S, Gaudio E, Franchitto A, Mancinelli R, Venter J, Kopriva S, Ueno Y, Alvaro D, Savage J. Caffeic acid phenethyl ester decreases cholangiocarcinoma growth by inhibition of NF-κB and induction of apoptosis. Int J Cancer 2009;125(3):565–76. [DOI] [PMC free article] [PubMed] [Google Scholar]