Abstract
Background
Culture‐based assessment of the fecal microbiome using fecal culture profiles frequently is performed in dogs with chronic diarrhea, but the diagnostic value of this approach has not been determined.
Objectives
To compare the reported results of fecal culture profiles and the polymerase chain reaction‐based dysbiosis index (DI) between dogs with chronic diarrhea and healthy dogs; to assess interlaboratory variability in bacterial and fungal cultures among 3 veterinary diagnostic laboratories (diagnostic laboratory 1 [L1], diagnostic laboratory 2 [L2], diagnostic laboratory 3 [L3]); and to compare the reported interpretation of culture profiles (normobiosis versus dysbiosis) with those of the DI.
Animals
Eighteen dogs with chronic diarrhea (CDG) and 18 healthy control dogs (HG).
Methods
In this prospective, case‐control study, fecal samples were submitted to 3 commercial laboratories for fecal culture. The microbiota was assessed using PCR assays. Dogs receiving antimicrobials were excluded.
Results
Dysbiosis index was significantly increased in CDG (mean, 0.9; SD, 3.8; 95% confidence interval [CI], −1.0; 2.8) compared to HG (mean, −3.0; SD, 2.8; CI, −4.3; −1.6; P = .0002), whereas cultures from all laboratories failed to detect significant differences (P = .66, .18, and .66, respectively). Hemolytic Escherichia coli was the only potential enteropathogen on culture, but no significant difference was found between CDG and HG. For diagnosis of dysbiosis, culture showed no agreement with DI (L1, κ = −0.21; CI, −0.44; −0.02; L2, κ = −0.33; CI, −0.58; −0.08; L3, κ = −0.25; CI, −0.39; −0.11). Furthermore, variability among the 3 laboratories was high (L1/L2, κ = 0.15; CI, −0.05; 0.35; L1/L3, κ = −0.08; CI, −0.01; −0.16; L2/L3, κ = −0.06; CI, −0.33; −0.20).
Conclusions and clinical importance
Fecal cultures failed to distinguish between diseased and healthy dogs, and a high level of interlaboratory variation for culture was found.
Keywords: antibiotic, canine, chronic enteropathyEscherichia coli, interlaboratory
Abbreviations
- CDG
chronic diarrhea group
- DI
dysbiosis index
- HG
healthy group
- L1
diagnostic laboratory 1
- L2
diagnostic laboratory 2
- L3
diagnostic laboratory 3
1. INTRODUCTION
Culture‐based assessment of feces is a diagnostic tool that should be used to identify specific or opportunistic enterpathogenic bacteria (eg, Salmonella spp., Campylobacter jejuni, specific enteropathogenic Escherichia coli strains, Yersinia spp., Clostridium perfringens, Clostridium difficile) and fungi in animals showing clinical signs associated with infectious acute or chronic diarrhea. 1 , 2 , 3 Several commercial veterinary diagnostic laboratories offer fecal culture as a tool to assess microbial composition (ie, growth of gram‐negative and gram‐positive flora) and, furthermore, to provide treatment recommendations based on their own interpretation of normobiosis and dysbiosis. Doing so is problematic, because aerobic culture‐based methods do not adequately represent the mostly anaerobic intestinal microbiota. Several limitations are associated using fecal culture to diagnose the cause of diarrhea, such as lack of standardization with regard to sampling technique (eg, amount of feces), shipping (eg, chilled vs. room temperature), and methodology (eg, culture media, subsampling, dilution error, method used to count colonies) among different laboratories. However, it should be implicit that submission of a fecal sample to a diagnostic laboratory is a request to confirm the diagnosis of an infectious disease. Fecal cultures should not be submitted to infer what constitutes normal versus abnormal feces, especially in dogs with chronic diarrhea. In addition, the diagnostic value of fecal cultures in dogs with chronic diarrhea without signs of systemic inflammation is questionable, 4 especially because putative bacterial enteropathogens frequently are isolated from healthy dogs. 4 , 5 , 6 , 7 Thus, the clinical utility of this method for identifying potentially enteropathogenic bacteria in dogs with chronic diarrhea is unclear.
Novel molecular genetic‐based tests have been developed to assess the microbiota and have identified complex bacterial communities in the intestine of dogs. Compositional changes in the microbiota have been documented in dogs with chronic enteropathies and may play a role in the pathogenesis of the disease. 8 , 9 Some of these molecular genetic‐based tests for dysbiosis are rapid PCR‐based methods and represent promising tools for assessment of dysbiosis in dogs with chronic diarrhea. 10 , 11
Chronic diarrhea in dogs has been defined as having a duration of ≥3 weeks. 12 The first stage of the diagnostic evaluation usually aims to rule out extraintestinal diseases and parasites. 12 , 13 Although imaging and histopathological evaluations frequently are restricted to patients with severe clinical signs, gastrointestinal protein loss, or suspicion of neoplastic infiltration or invasive infectious agents, fecal cultures sometimes are included in the first routine evaluation of these cases. More specifically, clinicians frequently submit fecal samples for cost‐intensive “fecal culture profiles”, although studies evaluating the diagnostic utility of fecal cultures in dogs with chronic diarrhea are lacking. 4 , 14
We hypothesize that the diagnostic value of fecal culture profiles in dogs with chronic diarrhea is questionable. Thus, we aimed to (1) compare the results of fecal cultures and the interpretations provided by 3 diagnostic laboratories in dogs with chronic diarrhea and healthy control dogs; (2) compare these results to the interpretation of the PCR‐based dysbiosis index (DI) to verify dysbiosis; and (3) to assess interlaboratory variability in bacterial and fungal cultures among 3 commercial laboratories (diagnostic laboratory 1 [L1], diagnostic laboratory 2 [L2], diagnostic laboratory 3 [L3]).
2. MATERIAL AND METHODS
2.1. Animals
Our study was designed as a prospective, case‐control (1:1) trial and was approved by the animal care and use committee (Ethikkommission) of the Centre for Clinical Veterinary Medicine LMU, Munich (reference 156‐07‐02‐2019).Two groups of 18 dogs each were included in the study, 1 consisted of dogs with chronic diarrhea (CDG) and 1 of dogs without clinical signs serving as controls (HG). All dogs were recruited between February and November 2019 by the same clinician (MW). Dogs of either sex, neuter status, body weight, and at least 1 year of age were included. The HG included dogs presented for vaccination or annual health checkups and these dogs had no clinically relevant history or findings on physical examination. Exclusion criteria for HG were gastrointestinal signs or administration of antimicrobials or probiotics during the last 4 weeks before presentation. Only dogs with diarrhea of a minimum duration of 3 weeks were enrolled into the CDG. Exclusion criteria for CDG were administration of antimicrobials or probiotics during the 4 weeks before to presentation, clinical, or laboratory findings suggesting the necessity of antimicrobial treatment (rectal temperature > 39.0°C [102.2 °F], white blood cell count <5 × 109/L or >20 × 109/L, and band neutrophils >1.5 × 109/L), suspicion or documentation of neoplastic infiltration of the intestine, and moderate to severe hypoalbuminemia (<2.3 g/dL). The clinical history of all dogs (HG and CDG) was recorded by using a standardized protocol with specific questions regarding the onset of diarrhea (CDG), fecal quality, number of defecations per day, vomiting, appetite, current treatment and previous treatment, other concurrent diseases, diet, and current dietary changes. The diagnostic evaluation in the CDG consisted of a CBC (in all dogs), serum biochemistry profile (ie, alanine aminotransferase, alkaline phosphatase, creatinine, urea nitrogen, total protein, albumin, glucose, sodium, potassium, chloride, phosphate, total calcium; all dogs), serum cobalamin and folate concentrations (14/18 dogs), basal serum cortisol concentration or adrenocorticotropin hormone stimulation test (14/18 dogs), fecal flotation (17/18 dogs), Giardia spp. ELISA (16/18 dogs), and abdominal ultrasound examination (16/18 dogs).
2.2. Sample collection
Naturally passed feces from each dog were collected by the owner on the day of presentation. Fecal samples were mixed and immediately divided into 4 equally sized aliquots for fecal culture at 3 different commercial reference laboratories as well as microbiota analysis by quantitative PCR (qPCR), respectively. Fecal samples for culture were submitted to the 3 laboratories according to their instructions: samples were sent on the same day to L1 and L2 by courier, and to L3 by regular mail. The aliquots for molecular genetics‐based microbiota analysis were frozen at −80°C and were sent as a batch on dry ice to the Gastrointestinal Laboratory at Texas A&M University at the end of the study period. No information about the dogs' history was provided to any of the laboratories at the time of submission.
2.3. Fecal culture
Each laboratory offered its unique test panel. Thus, the included tests and microbiological methods varied among the laboratories and were not standardized. The following diagnostic tests were offered at all 3 laboratories as part of their routinely offered “fecal culture profile”: bacteriology (aerobically incubated), mycology, and specific testing for obligate and facultative pathogenic bacteria (Salmonella, Campylobacter spp., and Yersinia spp.). Laboratory 1 additionally performed a clostridial culture as part of its fecal profile. All 3 laboratories provided their own interpretation of test results (eg, “presence of abnormal flora” or “detection of pathogenic isolates” or both). Laboratory 1 and L2 subdivided the interpretation of the results into gram + and gram ‐ spectrum of bacteria based on general microbiological nomenclature. Table S2 summarizes all results of tests performed for each individual dog.
2.4. Microbiota analysis
2.4.1. DNA extraction
The DNA was extracted from an aliquot of 100 mg of each fecal sample using a MoBio Power soil DNA isolation kit (MoBio Laboratories, Carlsbad, California) according to the manufacturer's instructions. The bead‐beating step was performed on a homogenizer (FastPrep‐24; MP Biomedicals, Santa Ana, California) at a speed of 4 m/s for 60 seconds. Fecal DNA was frozen at −80°C until further analysis.
2.4.2. Quantitative PCR
The abundances of total bacteria and 7 bacterial taxa (ie, Faecalibacterium spp., Turicibacter spp., Streptococcus spp., E. coli, Blautia spp., Fusobacterium spp., and Clostridium hiranonis), which had been identified as being altered in dogs with gastrointestinal disease in previous studies, were quantified by specific qPCR assays. The results were used to calculate the previously described DI using a mathematical algorithm. 10 The technique, containing the oligonucleotide sequence of the primers and the annealing temperatures, has already been described in detail elsewhere. 10 A DI < 0 indicates normobiosis, whereas a DI ≥2 indicates dysbiosis, and values between 0 and 2 are considered equivocal. The abundances of C. perfringens 16S rRNA and C. perfringens enterotoxin genes in feces were quantified by qPCR assays using previously reported oligonucleotide primers and assay conditions. 15 The PCR conditions were 95°C for 20 seconds, 40 cycles at 95°C for 5 seconds, and 10 seconds at the optimized annealing temperature. For probe‐based assays, the master mix consisted of 10 μL of TaqMan reaction mixtures, consisting of 5 μL of TaqMan Fast Universal PCR master mix (2×), No AmpErase UNG (Applied Biosystems, Foster City, California), 0.4 μL of each primer (concentration, 400 nM), 0.2 μL of the probe (concentration, 200 nM), 1 μL of 1% bovine serum albumin (BSA; concentration, 0.1%), 1 μL of water, and 2 μL of DNA (1:10 or 1:100 dilution). For N′,N′‐dimethyl‐N‐[4‐[(E)‐(3‐methyl‐1,3‐benzothiazol‐2‐ylidene)methyl]‐1‐phenylquinolin‐1‐ium‐2‐yl]‐N‐propylpropane‐1,3‐diamine‐based (SYBR) assays, PCR procedures ran at 95°C for 2 minutes, 40 cycles at 95°C for 5 seconds, and 10 seconds at the optimized annealing temperature with 10 μL of SYBR‐based reaction mixtures consisting of 5 μL of SsoFast EvaGreen supermix (Biorad Laboratories, Hercules, California), 0.4 μL of each primer (concentration, 400 nM), 1 μL of 1% BSA (concentration, 0.1%), 1.6 μL of water, and 2 μL of DNA (1:10 or 1:100 dilution). The oligonucleotide sequences of the primers, probes, and the annealing temperatures are presented in Table S1.
2.5. Statistical analyses
Statistical analyses were performed using GraphPad Prism (GraphPad Prism c7.0, GraphPad Software, San Diego, California) and a web‐based program for calculating Cohen's kappa and weighted kappa values (https://www.graphpad.com/quickcalcs/kappa1/). The distribution of data was tested using the D'Agonisto‐Pearson omnibus normality test. The association of categorical variables (ie, sex, gram‐positive or gram‐negative bacteria, or hemolytic or mucoid growing E. coli, Proteus mirabilis, Klebsiella spp., Acinetobacter johnsonii, alpha‐hemolytic Streptococcus spp., aerobic spore‐forming bacteria, Enterococcus spp., fungal, or Clostridia spp. between CDG and HG) and group (CDG or HG) were assessed using Fisher's exact test. Differences in continuous variables (ie, age, weight, DI, abundances of Faecalibacterium spp., Turicibacter spp., Streptococcus spp., E. coli, Blautia spp., Fusobacterium spp., C. hiranonis, total bacteria, C. perfringens, and C. perfringens enterotoxin gene) between CDG and HG were evaluated using an unpaired t test or the Mann‐Whitney U test depending on their distribution.
The agreement in classifying the fecal microbiota as normobiotic and equivocal or dysbiotic by the DI and the 3 laboratories each (ie, abnormal vs normal microbiota, presence of growing gram‐negative/gram‐positive microbiota, hemolytic E. coli) was evaluated using Cohen's kappa (κ) coefficient. 16 For culture results with >2 categories (eg, mycology) a weighted kappa was calculated. The interpretation of Cohen's kappa coefficient can be found in the legend of Table 2.
TABLE 2.
Comparison of agreement of microbiota analysis and fecal cultures between individual laboratories
| Laboratory | Laboratory | Level of agreement | CI | Level of agreement |
|---|---|---|---|---|
| DI | L1 | κ = −0.21 | −0.44; −0.02 | Disagreement |
| DI | L2 | κ = −0.33 | −0.58; −0.08 | Disagreement |
| DI | L3 | κ = −0.25 | −0.39; −0.11 | Disagreement |
| L1 | L2 | κ = 0.15 | −0.05; 0.35 | Poor agreement |
| L2 | L3 | κ = −0.06 | −0.33; 0.20 | Disagreement |
| L1 | L3 | κ = 0.08 | −0.01; 0.16 | Poor agreement |
Note: Interpretation of Cohen's kappa (κ) value: κ < 0: disagreement; 0 ≤ κ ≥ 0.4: poor agreement; 0.4 < κ > 0.75: fair to good agreement; κ ≥ 0.75: strong agreement.
Abbreviations: CI, confidence interval; DI, dysbiosis index; L1, diagnostic laboratory 1; L2, diagnostic laboratory 2; L3, diagnostic laboratory 3.
3. RESULTS
3.1. Study population
Thirty‐six dogs (CDG, n = 18; HG, n = 18) were included in the study. Frequency of breed, sex, body weight, and age did not differ between CDG and HG (Table 1). In CDG, the median duration of diarrhea was 13.5 (range, 1‐96) months. Owners described soft stool quality in 9/18 (50%), watery diarrhea in 9/18 (50%), and intermittent hematochezia in 6/18 (33%) dogs. The number of defecations per day was increased (>3 times per day) in 11/18 (61%) dogs. Seven of 18 (39%) dogs had additional chronic vomiting and 4/18 (22%) had decreased appetite. Six of 18 (33%) dogs received a commercial diet, 5/18 (28%) a hydrolyzed protein diet, 4/18 (22%) a commercial single protein/single carbohydrate diet, and 3/18 (17%) a home‐cooked diet. Six of 18 dogs (33%) received medication at the date of presentation (levothyroxine PO [n = 2], pancreatic enzymes PO [n = 1], pantoprazole PO [n = 1], prednisolone PO [n = 1], oclacitinib PO [n = 1], omeprazole PO [n = 1], metamizole PO [n = 1], cobalamin supplement PO [n = 1]). Drugs and dietary supplements (excluding antimicrobials) that the patients received in the 2 years before presentation included: fenbendazole PO, toltrazuril PO, probiotic PO, metamizole PO, omeprazole PO, maropitant PO, prednisolone PO, pantoprazole PO, tramadol PO, meloxicam PO, and cyclosporin PO. Antimicrobials that were administered to the dogs at least 2 months before presentation were: metronidazole PO (9/18 in total: 1/18, 2 months; 2/18, 3 months; 3/18, 4 months; 1/18, 7 months; 1/18, 12 months; and 1/18 24 months before sample collection), amoxicillin‐clavulanic acid PO (2/18; 2 and 10 months previously), and trimethoprim‐sulfonamide PO (1/18; 3 months previously). Concurrent diseases in CDG included dermatological disorders (2/18), hypothyroidism (2/18), and orthopedic problems (1/18).
TABLE 1.
Demographics of dogs with chronic diarrhea and healthy control dogs
| CDG (n = 18) | HG (n = 18) | P value | |||
|---|---|---|---|---|---|
| Sex | 10 male, 8 females | 6 male, 12 females | .31 | ||
| Neutered/intact | 5/13 | 9/9 | .31 | ||
| Breeds | Mixed breed (4), Australian Shepard dog (1), Bracke (1), Cavalier King Charles Spaniel (1), Chow‐Chow (1), Dalmatian (1), German Shepard dog (1), Magyar Vizsla (1), Maltese (1), Miniature Pinscher (1), Pug (1), Small Muensterlaender (1), Standard Poodle (1), Standard Schnauzer (1), Whippet (1) | Mixed breed (4), Labrador Retriever (3), Border Collie (2), Dachshund (2), Dobermann Pinscher (2), Beagle (1), Bearded Collie (1), Chihuahua (1), German Shepard Dog (1), Goldendoodle (1) | |||
| Median | Range | Median | Range | ||
| Body weight (kg) | 14.5 | 2.7‐30 | 20.6 | 3.5‐33.3 | .27 |
| Age (years) | 5.0 | 1.0‐12.0 | 3.0 | 1.0‐10.0 | .16 |
Abbreviations: CDG, chronic diarrhea group; HG, healthy group; n, number of dogs.
3.2. Comparison of microbiota analysis by qPCR between CDG and HG
The DI was significantly higher (P = .0002) in CDG (mean, 0.9; SD, 3.8; 95% CI, −1.0; 2.8) in comparison to HG (mean, −3.0; SD, 2.8; CI, −4.3; −1.6; Figure 1). An increased DI (> 2) was found in 44% (8/18) of the dogs of the CDG and in 6% (1/18) of the HG. The abundance of Faecalibacterium (P = .01) and Fusobacterium (P = .03) was significantly decreased in the CDG in comparison to the HG (Figure 1), whereas abundances of Turicibacter, Streptococcus, E. coli, Blautia, and C. hiranonis were not significantly different between CDG and HG. All dogs with a decreased abundance of C. hiranonis in both groups had a DI > 2. Only 1 of the CDG dogs with DI > 2 had a normal abundance of C. hiranonis, but in this dog Streptococcus was increased (Figure 1). Polymerase chain reaction was performed for C. perfringens and C. perfringens enterotoxins genes, but no difference between CDG and HG could be identified. Clostridium perfringens was found in 16/18 (89%) dogs of CDG and in all dogs (100%) of the HG.
FIGURE 1.

Dysbiosis index. This figure shows the dysbiosis index, A, and the abundances of Faecalibacterium, B, Fusobacterium, C, Streptococcus, D, Blautia, E, Escherichia coli, F, Turicibacter, G, and Clostridium hiranonis, H, in dogs with chronic diarrhea (CDG) and healthy control dogs (HG). Dots show individual dogs, bars show the means for each group. The reference intervals are shaded in grey. In A, green dots represent dogs with a decreased abundance of Clostridium hiranonis. A dysbiosis index <0 indicates normobiosis. A dysbiosis index above 2 indicates dysbiosis. The interval between 0 and 2 is defined as equivocal
3.3. Comparison of fecal cultures between CDG and HG
Fecal culture results from all 3 laboratories were not significantly different between CDG and HG. The tests offered, the results, and their interpretation differed among the 3 laboratories and, therefore, main findings are presented for each laboratory separately. The term “normal or abnormal microbiota” was used as interpretation by the laboratories themselves when the results of cultures were reported.
Laboratory 1 (L1) reported “abnormal microbiota” in 14/18 dogs of CDG and in 16/18 dogs of HG (P = .66; Figure 2). Gram‐negative bacteria were not found in 2 dogs of each group. Gram‐positive bacteria could not be cultured in 3 dogs from each group. The only identified bacteria considered as potential enteropathogen by the laboratory was hemolytic E. coli in 4/18 CDG and 8/18 HG dogs (P = .29; Figure 3). Only L1 tested for anaerobic bacterial growth. Clostridium spp. (>1 million colony forming units/g) was found in 8/18 of CDG and in 10/18 of HG. All reported bacterial groups are summarized in Table 3. Unrequested susceptibility testing for antimicrobials was routinely provided for 6 different bacterial isolates (ie, Acinetobacter spp., Buttiauxella ferragutiae, hemolytic E. coli [not in cases with a low growth rate], mucoid growing E. coli, Enterobacter cloacae, and Klebsiella variicola).
FIGURE 2.

Fecal culture interpretation. Interpretations of the fecal culture results for dysbiosis given by the 3 commercial laboratories in dogs with chronic diarrhea (CDG) and healthy control dogs (HG). There was no significant association between the interpretation provided by each laboratory and group (CDG or HG)
FIGURE 3.
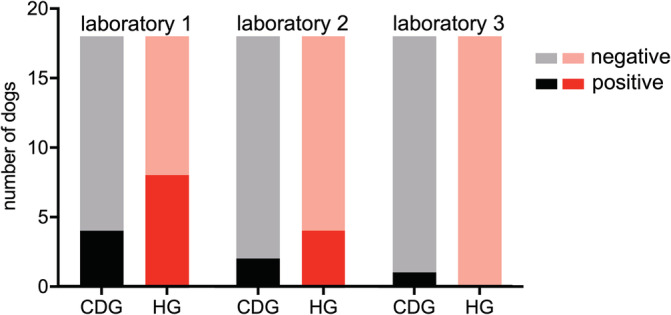
Growth of hemolytic Escherichia coli. Comparison of growth of fecal hemolytic E. coli reported by 3 different commercial laboratories in dogs with chronic diarrhea (CDG) and healthy control dogs (HG). There was no significant association between the growth of hemolytic E. coli as reported by each laboratory and group (CDG or HG)
TABLE 3.
Bacterial isolates reported by laboratory 1 in dogs with chronic diarrhea and healthy control dogs
| n (CDG) | n (HG) | Bacterial isolate | P value |
|---|---|---|---|
| 0 | 2 | Acinetobacter spp. | .49 |
| 5 | 6 | Aerobic, spore‐forming bacteria | >.99 |
| 6 | 7 | Alpha‐hemolytic Streptococcus spp. | >.99 |
| 1 | 0 | Bacillus cereus | >.99 |
| 1 | 0 | Buttiauxella ferraguti | >.99 |
| 8 | 10 | Clostridium spp. | .74 |
| 1 | 0 | Enterobacter cloacae | >.99 |
| 8 | 7 | Enterococcus spp. | >.99 |
| 16 | 16 | Escherichia coli | >.99 |
| 4 | 8 | Hemolytic E. coli | .29 |
| 1 | 1 | Mucoid growing E. coli | >.99 |
| 1 | 1 | Klebsiella spp. | >.99 |
| 2 | 1 | Proteus mirabilis | >.99 |
| 1 | 0 | Pseudomonas spp. | >.99 |
| 0 | 1 | Staphylococcus epidermidis | >.99 |
Abbreviations: CDG, chronic diarrhea group; HG, healthy group.
Laboratory 2 (L2) interpreted the bacterial microbiota as “abnormal” in 5/18 dogs of CDG and 10/18 dogs of HG (P = .18; Figure 2). Gram‐negative bacteria were present in 16/18 dogs of each group. Gram‐positive bacteria were cultured in 17/18 dogs of CDG and 12/18 dogs of HG. Hemolytic growing E. coli were documented in 2/18 dogs of CDG and 4/18 dogs of HG (P = .66; Figure 3). No other potential enteropathogens were isolated from any of the samples.
Laboratory 3 (L3) reported “abnormal microbiota” in 4/18 dogs of CDG and 2/18 dogs of HG (Figure 2; P = .66) Hemolytic E. coli were found in none of the HG and only in 1 dog of CDG (P > .99) and for this strain a sensitivity testing for antimicrobials was provided (Figure 3). As was the case for L2, no other enteropathogens were found by L3.
Fungal culture showed no significant difference in any variable between HG and CDG for any of the 3 laboratories. Fungal organisms were cultured in 4/18 (CDG) and 1/18 (HG) by L1 (P = .34), none of the samples by L2 (P > .99), and 3/18 (CDG) and 1/18 (HG) by L3 (P = .60; Table 4).
TABLE 4.
Results of fungal cultures of dogs with chronic diarrhea and healthy control dogs
| Isolate | Laboratory 1 | Laboratory 2 | Laboratory 3 | |||
|---|---|---|---|---|---|---|
| n (CDG) | n (HG) | n (CDG) | n (HG) | n (CDG) | n (HG) | |
| No growth | 14 | 17 | 18 | 18 | 15 | 17 |
| Candida spp. (mild) | 2 | 1 | ‐ | ‐ | ‐ | ‐ |
| Candida spp. (moderate) | 1 | ‐ | ‐ | ‐ | 1 | 1 |
| Candida spp. (severe) | 1 | ‐ | ‐ | ‐ | 1 | ‐ |
| Geotrichophythum sp. | ‐ | ‐ | ‐ | ‐ | 1 | ‐ |
Abbreviations: CDG, chronic diarrhea group; HG, healthy group; n, number of dogs.
3.4. Comparison of interpretation of microbiota analysis and fecal cultures among individual laboratories
For the following evaluation, all dogs in both groups were analyzed collectively. The overall assessment of “abnormal intestinal microbiota” for any of the 3 laboratories did not agree with the DI. Overall levels of agreement between each laboratory and the DI are shown in Table 2.
Agreement between L1 and L2 for the growth of gram‐negative bacteria was fair to good (κ = 0.44; CI, −0.02; 0.89), but there was a disagreement on the growth of gram‐positive bacteria (κ = −0.22; CI, −0.34; −0.10). Laboratory 3 provided no results on the growth of gram‐positive or gram‐negative bacteria.
Agreement on growth of hemolytic E. coli results was fair to good between L1 and L2 (κ = 0.57; CI, 0.29; 0.85), poor between L1 and L3 (κ = 0.11; CI, −0.09; 0.31), and there was overall disagreement between L2 and L3 (κ = −0.05; CI, −0.14; −0.04).
Assessment of fungal cultures showed disagreement or poor agreement among the 3 laboratories (L1 and L2: κ = 0.00; CI, 0.00; 0.00; L1 and L3: κ = 0.28; CI, −0.07; 0.63; L2 and L3: κ = 0.00; CI, 0.00; 0.00), and there was also poor agreement for the presence of Candida spp. (L1 and L2: weighted κ = 0.00; L1 and L3: weighted κ = 0.33; L2 and L3: weighted κ = 0.00).
4. DISCUSSION
The objective of our prospective clinical trial was to compare the results of fecal cultures with the DI in dogs with chronic diarrhea. An important aspect is that the DI assesses the composition of the microbiota, whereas culture as performed and interpreted by the 3 laboratories attempted to both assess the microbiota as well as document the presence of enteropathogenic bacteria. Assessing microbiota composition based on culture should, however, not be the diagnostic method of choice to assess dysbiosis because of the limitations stated in the introduction. Nevertheless, offering fecal culture for this purpose is still common practice in several veterinary diagnostic laboratories. Our study did not detect any significant differences in commercial fecal culture results in dogs with chronic diarrhea compared to healthy dogs. Furthermore, agreement on the interpretation of normobiosis vs dysbiosis was found to be poor among the 3 laboratories. No putative bacterial pathogens could be detected in any dog except hemolytic E. coli and the clinical relevance of this finding must be questioned because of the similar and often higher isolation rates in dogs of the HG. These findings raise the question as to whether routine fecal culture testing is of any use in dogs with chronic diarrhea.
The DI is a qPCR‐based tool and has been used to assess fecal dysbiosis in dogs with chronic enteropathy, 10 , 17 dogs with acute diarrhea, 18 , 19 and healthy dogs after antimicrobial administration. 10 , 20 , 21 As shown in previous studies, a significant difference in the occurrence of dysbiosis was found between dogs in the CDG and the HG, with 8/18 dogs in the CDG having a DI > 2.
The etiology of chronic diarrhea in dogs is poorly understood but is presumed to be multifactorial. Several studies suggest a critical role of the intestinal microbiota, 8 , 22 and dysbiosis has been described in human and canine patients with chronic enteropathies (CE). 23 , 24 Documentation of changes in the intestinal microbiota is important, because dysbiosis is considered a factor in the pathogenesis of CE. 25 Alterations in the intestinal microbiota can lead to functional changes, such as a decrease in short‐chain fatty acids 17 and abnormal bile acid metabolism. 18 , 20 , 26 For example, secondary bile acids have local and systemic anti‐inflammatory properties and are an important driver of a healthy gut metabolism. 27 In the colon, primary bile acids are converted to secondary bile acids. The main converter of bile acids in dogs is C. hiranonis. 18 , 20 , 26 Decreased abundance of C. hiranonis leads to lack of conversion of primary to secondary bile acids, which is associated with an increased DI. 10 , 18 , 20 , 26 Decreased numbers of C. hiranonis also were associated with a lower fecal concentration of secondary bile acids and with an increased abundance of E. coli. 27 In our study, all dogs with decreased abundance of C. hiranonis had a DI > 2, including 8/18 in CDG and 1/18 in the HG. The latter dog had no signs of gastrointestinal disease and no medication history before sample collection. This finding suggests that a small subset of clinically healthy dogs can have subclinical dysbiosis. Interestingly, after 1 year this dog developed chronic diarrhea. Therefore, long‐term studies are warranted to evaluate the effect of dysbiosis on developing chronic gastrointestinal signs and the potential role of DI as an early marker for chronic gastrointestinal disease.
Abundances of Faecalibacterium and Fusobacterium were decreased in some of the dogs of CDG. Decreased Faecalibacterium is a consistent finding in dogs and people with CE and gained attention because of their ability to secrete anti‐inflammatory peptides in in vitro studies. 28 , 29 Although, most of the time, the causal relationship between dysbiosis and disease remains unclear, it seems important to recognize intestinal dysbiosis so as to incorporate this information into individual treatment strategies. Besides treatment of the underlying disease process in dogs with CE (eg, food‐responsive disease, immune‐mediated inflammation), restoration of the normal microbiota might be useful as an adjunctive treatment.
The culture results of all 3 laboratories failed to detect any difference between CDG and HG. The definition of an “abnormal microbiota” varied broadly among the laboratories and none of the laboratories provided information about their diagnostic criteria, thus emphasizing that a clear consensus is lacking when using fecal culture. Laboratory 1 routinely gave unrequested detailed information about the isolates identified and suggested a dysbiotic state in all but 2 of the HG dogs and in all but 4 of the CDG dogs. In comparison, L2 and L3 concluded dysbiosis less frequently in both groups of dogs. Surprisingly, more dogs with an interpretation of “abnormal microbiota” were reported in the HG compared to the CDG by both L1 and L2. This finding is in contrast to the results of the DI and to observations from studies showing that changes in the microbiota are associated with CE but usually not present in healthy individuals. Hemolytic E. coli were the only identified bacteria considered as facultative pathogens. Historical data suggest that the ability of E. coli to hemolyze erythrocytes is linked to different virulence factors, and these isolates can be the causative agent in extraintestinal diseases (eg, urinary tract infections, wound infections). 30 , 31 However, hemolytic E. coli are part of the normal intestinal microbiota of healthy individuals and the pathogenic role of hemolytic E. coli strains in CE is not clear. 32 Moreover, none of the laboratories provided information on what specific hemolytic E. coli strain was present and whether it was a pathogenic isolate or not. Other classic enteropathogens, such as Salmonella spp., thermophilic Campylobacter spp., and Yersinia enterocolica were not found in any of the samples, which support the idea that enteropathogens do not play a major role in dogs with CE.
Laboratory 1 and L3 provided unrequested antimicrobial susceptibility testing for hemolytic E. coli as part of the fecal panel. Sensitivity testing was based only on selected isolates found on the agar plates. Thus, it does not reflect the resistance pattern of all (hemolytic) E. coli in the intestines. Unjustified antibiotic usage can lead to a higher proportion of resistant E. coli isolates in canine feces. 19 By providing sensitivity testing of facultative enteropathogens in dogs with chronic diarrhea, veterinarians might get the impression that antibiotics are indicated in these cases. However, antibiotic treatment should not be based on fecal culture findings. Specialists in veterinary gastroenterology strongly suggested in a recent proposal for rational antibacterial use in dogs with chronic diarrhea that antibiotics should be reserved for those dogs with evidence of true infection (ie, signs of systemic inflammatory response syndrome or evidence of adherent‐invasive bacteria in intestinal biopsy samples). 33 Untargeted use of antibiotics based on fecal culture results likely contributes to the spreading of resistant bacteria and stands in contrast to principles of responsible antibiotic stewardship. 34 , 35
Laboratory 1 and L2 separated between gram‐positive and gram‐negative bacteria based on general microbiological nomenclature in their results. Interestingly, both laboratories found no gram‐negative bacteria in 8 dogs, but were in agreement only in 2 of these dogs. Laboratory 1 failed to detect any gram‐positive bacteria in 7 dogs, and L2 in 6 dogs. Both laboratories only agreed for 1 dog. Two dogs in L2 and 1 dog in L1 had neither a gram‐positive nor a gram‐negative cultivatable microbiota (all belonged to the HG). Primarily aerobically growing bacteria (eg, Enterococcus as part of the gram‐positive microbiota and Enterobacteriaceae, such as E. coli, as part of the gram‐negative microbiota) are cultivated in routine fecal cultures. 36 , 37 , 38 However because of sequencing techniques, it is known that strictly anaerobic bacteria predominate in the intestinal microbiota. Thus, failing to culture anaerobic bacteria from fecal cultures leads to an inadequate representation of the overall composition of the intestinal microbiota. 39 , 40 , 41 Our results emphasize that the absence or presence of cultivable microbiota impedes assessment of the composition of the intestinal microbiota.
Laboratory 1 identified increased growth of Clostridium spp. in 50% of the samples, which was defined as “abnormal” by the laboratory itself. According to literature, this percentage is considered low, because with appropriate sample handling and culture techniques C. perfringens can be identified in feces of 80% healthy dogs. 42 , 43 Clostridium spp. are characterized by anaerobic growth. 43 Under aerobic conditions, which are usually present during transportation and shipping of fecal samples, certain clostridial strains can form spores within minutes, which can require specialized culture methods to induce germination. Thus, the presence of Clostridia spp. might be underestimated by culture. 44 , 45 There was no difference in Clostridium‐positive samples between CDG and HG. The relevance of clostridial growth in dogs with intestinal disease is questionable, because it is not the presence of clostridial species, but the presence of certain enterotoxins that likely plays a pathogenic role (eg, in acute hemorrhagic diarrhea syndrome), and is associated with clinical signs. 46 , 47 , 48
In comparison, PCR detected the C. perfringens 16S rRNA gene in all except 2 samples. Samples from dogs of the CDG did not have a higher abundance of the gene compared to HG. Thus, our findings further support the notion that clostridial strains, in general, and C. perfringens, in particular, are considered unlikely to have played a role in the pathogenesis of chronic diarrhea in the present study population.
Aerobic fungi were only isolated from a few samples, with the majority consisting of Candida spp, and no difference was found between CGD and HG. This finding is consistent with recent molecular genetic‐based investigations that documented a similar abundance of Candida spp. in fecal samples from dogs with acute diarrhea and healthy dogs. 49 Studies in humans showed that the fungal microbiota might play a role in CE 50 and thus the inability of fungal culture to discern a difference between groups in our study is concerning.
Significant disagreement was found among laboratories in the interpretation of abnormal microbiota. Factors that explain differences could include random errors, a systematic bias of the analytical procedure, application errors within the laboratories, interpretation errors, and preanalytical (including transport) errors. 51 , 52 Transportation of samples differed and was based on the instructions provided by the individual laboratories. Moreover, bias could be caused by different subsampling methods, which can either take place in the clinic by dividing samples or in the laboratory.55 Furthermore, differences in culture methods among laboratories potentially could have a substantial impact on culture results.
Establishing guidelines for adequate sample handling and shipping conditions would be essential for reproducible results. However, even if these procedures were to be standardized, definition of dysbiosis based on culture methods is not defined and primarily based on individual subjective interpretation. Consequently systematic bias could occur among different microbiologists.
Agreement between fecal culture results and DI generally was poor. The DI was significantly different between healthy and diseased individuals, whereas in contrast, fecal cultures did not show such a difference.
Our study had some important limitations. First, it was not possible to assess the laboratories' agreement on the assessment of the presence of enteropathogenic bacteria other than hemolytic E. coli, because these organisms were not found in any of the samples. However, the results do support that classic enteropathogens do not play an important role in dogs with CE. Second, information about the microbiological methods of the 3 commercial laboratories was not available. It can be assumed that methodological differences among the laboratories existed, which limits the direct comparability of results. However, an aim of the study was to assess the agreement of culture results among different laboratories from a clinical perspective, independent of their methods. Our findings show that clinicians might receive different results depending on the laboratory they choose. Third, it is difficult to define a gold standard for the evaluation of dysbiosis. However, recent studies have indicated that the results of the DI agreed well with more comprehensive sequencing methods. 18 , 20 Moreover, our results suggest that the DI is a relevant tool to assess dysbiosis in dogs with CE and healthy dogs, comparable to findings of previous studies.
Our results indicate that fecal cultures are not useful for identifying dysbiosis of dogs with CE. In fact, interpretation of culture results and routinely provided sensitivity testing for antibiotics can even be misleading and result in unnecessary antibiotic treatment. Fecal culture should be reserved for detecting enteropathogens without giving any recommendations on their treatment.
CONFLICT OF INTEREST
Drs Suchodolski, Lidbury, and Steiner are employees of the Gastrointestinal Laboratory, which performs diagnostic testing, including the dysbiosis index, on a fee‐for‐service basis.
OFF‐LABEL ANTIMICROBIAL DECLARATION
Authors declare no off‐label use of antimicrobials.
INSTITUTIONAL ANIMAL CARE AND USE COMMITTEE (IACUC) OR OTHER APPROVAL DECLARATION
Prospective collection and analysis of canine fecal samples was approved by the Ethics Committee of the Centre of Veterinary Medicine, LMU, Germany (reference 156‐07‐02‐2019).
HUMAN ETHICS APPROVAL DECLARATION
Authors declare human ethics approval was not needed for this study.
Supporting information
Table S1 Oligonucleotides primers/probes used in this study.
Table S2 Fecal culture profiles of three commercial laboratories in dogs with chronic diarrhea and healthy individuals.
ACKNOWLEDGMENTS
No funding was received for this study. Preliminary results were presented at the 2020 ACVIM Forum on Demand and at the virtual 2020 ECVIM congress. Open access funding enabled and organized by Projekt DEAL.
Werner M, Suchodolski JS, Lidbury JA, Steiner JM, Hartmann K, Unterer S. Diagnostic value of fecal cultures in dogs with chronic diarrhea. J Vet Intern Med. 2021;35:199–208. 10.1111/jvim.15982
REFERENCES
- 1. Batt RM, Rutgers HC, Sancak AA. Enteric bacteria: friend or foe? J Small Anim Pract. 1996;37:261‐267. [DOI] [PubMed] [Google Scholar]
- 2. McDonough PL, Simpson KW. Diagnosing emerging bacterial infections: salmonellosis, campylobacteriosis, clostridial toxicosis, and helicobacteriosis. Semin Vet Med Surg (Small Anim). 1996;11:187‐197. [DOI] [PubMed] [Google Scholar]
- 3. Marks SL, Rankin SC, Byrne BA, Weese JS. Enteropathogenic bacteria in dogs and cats: diagnosis, epidemiology, treatment, and control. J Vet Intern Med. 2011;25:1195‐1208. [DOI] [PubMed] [Google Scholar]
- 4. Marks SL, Kather EJ. Bacterial‐associated diarrhea in the dog: a critical appraisal. Vet Clin North Am Small Anim Pract. 2003;33:1029‐1060. [DOI] [PubMed] [Google Scholar]
- 5. Burnens AP, Angéloz‐Wick B, Nicolet J. Comparison of Campylobacter carriage rates in diarrheic and healthy pet animals. Zoonoses Public Health. 1992;39:175‐180. [DOI] [PubMed] [Google Scholar]
- 6. Rossi M, Hanninen ML, Revez J, et al. Occurrence and species level diagnostics of Campylobacter spp., enteric Helicobacter spp. and Anaerobiospirillum spp. in healthy and diarrheic dogs and cats. Vet Microbiol. 2008;129:304‐314. [DOI] [PubMed] [Google Scholar]
- 7. Tupler T, Levy JK, Sabshin SJ, Tucker SJ, Greiner EC, Leutenegger CM. Enteropathogens identified in dogs entering a Florida animal shelter with normal feces or diarrhea. J Am Vet Med Assoc. 2012;241:338‐343. [DOI] [PubMed] [Google Scholar]
- 8. Suchodolski JS, Markel ME, Garcia‐Mazcorro JF, et al. The fecal microbiome in dogs with acute diarrhea and idiopathic inflammatory bowel disease. PloS One. 2012;7:e51907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Xenoulis PG, Palculict B, Allenspach K, Steiner JM, van House AM, Suchodolski JS. Molecular‐phylogenetic characterization of microbial communities imbalances in the small intestine of dogs with inflammatory bowel disease. FEMS Microbiol Ecol. 2008;66:579‐589. [DOI] [PubMed] [Google Scholar]
- 10. AlShawaqfeh MK, Wajid B, Minamoto Y, et al. A dysbiosis index to assess microbial changes in fecal samples of dogs with chronic inflammatory enteropathy. FEMS Microbiol Ecol. 2017;93:136. [DOI] [PubMed] [Google Scholar]
- 11. Heilmann RM, Steiner JM. Clinical utility of currently available biomarkers in inflammatory enteropathies of dogs. J Vet Intern Med. 2018;32:1495‐1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Marks SL. Diarrhea In: Washabau RJ, Day MJ, eds. Canine and Feline Gastroenterology. St. Louis, MO: Elsevier Health Sciences; 2012:99‐108. [Google Scholar]
- 13. Hall EJ, Day MJ. Chronic small intestinal disease In: Ettinger SJ, Feldman EC, Cote E, eds. Textbook of Veterinary Internal Medicine. 8th ed. St. Louis, MO: Elsevier Health Sciences; 2017:1516‐1564. [Google Scholar]
- 14. Jergens AE. Dyschezia and tenesmus In: Washabau RJ, Day MJ, eds. Canine and Feline Gastroenterology. St. Louis, MO: Elsevier Health Sciences; 2012:109‐113. [Google Scholar]
- 15. Minamoto Y, Dhanani N, Markel ME, Steiner JM, Suchodolski JS. Prevalence of Clostridium perfringens, Clostridium perfringens enterotoxin and dysbiosis in fecal samples of dogs with diarrhea. Vet Microbiol. 2014;174:463‐473. [DOI] [PubMed] [Google Scholar]
- 16. Fleiss JL. Statistical methods for rates and proportions. 2nd ed. New York, NY: John Wiley & Sons; 1981:217‐225. [Google Scholar]
- 17. Minamoto Y, Minamoto T, Isaiah A, et al. Fecal short‐chain fatty acid concentrations and dysbiosis in dogs with chronic enteropathy. J Vet Intern Med. 2019;33:1608‐1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chaitman J, Ziese A‐L, Pilla R, et al. Fecal microbial and metabolic profiles in dogs with acute diarrhea receiving either fecal microbiota transplantation or oral metronidazole. Front Vet Sci. 2020;7:192‐192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Werner M, Suchodolski JS, Straubinger RK, et al. Effect of amoxicillin‐clavulanic acid on clinical scores, intestinal microbiome, and amoxicillin‐resistant Escherichia coli in dogs with uncomplicated acute diarrhea. J Vet Intern Med. 2020;34(3):1166‐1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Pilla R, Gaschen F, Barr JW, et al. Effects of metronidazole on the fecal microbiome and metabolome in healthy dogs. J Vet Intern Med. 2020;34(5):1853‐1866. online print ahead. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Manchester AC, Webb CB, Blake AB, et al. Long‐term impact of tylosin on fecal microbiota and fecal bile acids of healthy dogs. J Vet Intern Med. 2019;33:2605‐2617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Minamoto Y, Otoni CC, Steelman SM, et al. Alteration of the fecal microbiota and serum metabolite profiles in dogs with idiopathic inflammatory bowel disease. Gut Microbes. 2015;6:33‐47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Duboc H, Rajca S, Rainteau D, et al. Connecting dysbiosis, bile‐acid dysmetabolism and gut inflammation in inflammatory bowel diseases. Gut. 2013;62:531‐539. [DOI] [PubMed] [Google Scholar]
- 24. Honneffer J, Minamoto Y, Suchodolski J. Microbiota alterations in acute and chronic gastrointestinal inflammation of cats and dogs. World J Gastroenterol. 2014;20:16489‐16497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Dandrieux JRS, Mansfield CS. Chronic enteropathy in canines: prevalence, impact and management strategies. Vet Med (Auckl). 2019;10:203‐214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Giaretta PR, Rech RR, Guard BC, et al. Comparison of intestinal expression of the apical sodium‐dependent bile acid transporter between dogs with and without chronic inflammatory enteropathy. J Vet Intern Med. 2018;32:1918‐1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wang S, Martins R, Sullivan MC, et al. Diet‐induced remission in chronic enteropathy is associated with altered microbial community structure and synthesis of secondary bile acids. Microbiome. 2019;7:126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sokol H, Pigneur B, Watterlot L, et al. Faecalibacterium prausnitzii is an anti‐inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci USA. 2008;105:16731‐16736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rossi G, Pengo G, Caldin M, et al. Comparison of microbiological, histological, and immunomodulatory parameters in response to treatment with either combination therapy with prednisone and metronidazole or probiotic VSL# 3 strains in dogs with idiopathic inflammatory bowel disease. PloS One. 2014;9:e94699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hacker J, Schroter G, Schrettenbrunner A, et al. Hemolytic Escherichia coli strains in the human fecal flora as potential urinary pathogens. Zentralbl Bakteriol Mikrobiol Hyg A. 1983;254:370‐378. [PubMed] [Google Scholar]
- 31. Beutin L. The different hemolysins of Escherichia coli . Med Microbiol Immunol. 1991;180:167‐182. [DOI] [PubMed] [Google Scholar]
- 32. Beutin L. Escherichia coli as a pathogen in dogs and cats. Vet Res. 1999;30(2–3):285‐298. [PubMed] [Google Scholar]
- 33. Cerquetella M, Rossi G, Suchodolski JS, et al. Proposal for rational antibacterial use in the diagnosis and treatment of dogs with chronic diarrhoea. J Small Anim Pract. 2020;61:211‐215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Borg MA, Zarb P, Scicluna EA, et al. Antibiotic consumption as a driver for resistance in Staphylococcus aureus and Escherichia coli within a developing region. Am J Infect Control. 2010;38:212‐216. [DOI] [PubMed] [Google Scholar]
- 35. Sørum H, Sunde M. Resistance to antibiotics in the normal flora of animals. Vet Res. 2001;32:227‐241. [DOI] [PubMed] [Google Scholar]
- 36. Selbitz HJ, Truyen U, Valentin‐Weigand P, Wieler LH, Ewer C, Selbitz HJ. Enterobactericeae Tiermedizinische Mikrobiologie, Infektions‐und Seuchenlehre. Stuttgart, Germany: Georg Thieme Verlag; 2015. [Google Scholar]
- 37. Selbitz HJ, Truyen U, Valentin‐Weigand P. Grampositive Kokken Tiermedizinische Mikrobiologie, Infektions‐und Seuchenlehre. Stuttgart, Germany: Georg Thieme Verlag; 2015. [Google Scholar]
- 38. Mentula S, Harmoinen J, Heikkilä M, et al. Comparison between cultured small‐intestinal and fecal microbiotas in beagle dogs. Appl Environ Microbiol. 2005;71:4169‐4175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hayashi H, Sakamoto M, Benno Y. Phylogenetic analysis of the human gut microbiota using 16S rDNA clone libraries and strictly anaerobic culture‐based methods. Microbiol Immunol. 2002;46:535‐548. [DOI] [PubMed] [Google Scholar]
- 40. Greetham HL, Giffard C, Hutson RA, Collins MD, Gibson GR. Bacteriology of the Labrador dog gut: a cultural and genotypic approach. J Appl Microbiol. 2002;93:640‐646. [DOI] [PubMed] [Google Scholar]
- 41. Wang RF, Cao WW, Cerniglia CE. PCR detection and quantitation of predominant anaerobic bacteria in human and animal fecal samples. Appl Environ Microbiol. 1996;62:1242‐1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Marks SL, Kather EJ, Kass PH, Melli AC. Genotypic and phenotypic characterization of Clostridium perfringens and Clostridium difficile in diarrheic and healthy dogs. J Vet Intern Med. 2002;16:533‐540. [DOI] [PubMed] [Google Scholar]
- 43. Marks SL, Melli A, Kass PH, Jang SS, Barkhoodarian A, Hirsh DC. Evaluation of methods to diagnose Clostridium perfringens‐associated diarrhea in dogs. J Am Vet Med Assoc. 1999;214:357‐360. [PubMed] [Google Scholar]
- 44. Paredes‐Sabja D, Shen A, Sorg JA. Clostridium difficile spore biology: sporulation, germination, and spore structural proteins. Trends Microbiol. 2014;22:406‐416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Jump RL, Pultz MJ, Donskey CJ. Vegetative Clostridium difficile survives in room air on moist surfaces and in gastric contents with reduced acidity: a potential mechanism to explain the association between proton pump inhibitors and C. difficile‐associated diarrhea? Antimicrob Agents Chemother. 2007;51:2883‐2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Mehdizadeh Gohari I, Unterer S, Whitehead AE, Prescott JF. NetF‐producing Clostridium perfringens and its associated diseases in dogs and foals. J Vet Diagn Invest. 2020;32:230‐238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sindern N, Suchodolski JS, Leutenegger CM, et al. Prevalence of Clostridium perfringens netE and netF toxin genes in the feces of dogs with acute hemorrhagic diarrhea syndrome. J Vet Intern Med. 2019;33:100‐105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ziese AL, Suchodolski JS, Hartmann K, et al. Effect of probiotic treatment on the clinical course, intestinal microbiome, and toxigenic Clostridium perfringens in dogs with acute hemorrhagic diarrhea. PloS One. 2018;13:e0204691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Foster ML, Dowd SE, Stephenson C, et al. Characterization of the fungal microbiome (mycobiome) in fecal samples from dogs. Vet Med Int. 2013;2013:658373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ott SJ, Kühbacher T, Musfeldt M, et al. Fungi and inflammatory bowel diseases: alterations of composition and diversity. Scand J Gastroenterol. 2008;43:831‐841. [DOI] [PubMed] [Google Scholar]
- 51. Corry JE, Jarvis B, Passmore S, et al. A critical review of measurement uncertainty in the enumeration of food microorganisms. Food Microbiol. 2007;24:230‐253. [DOI] [PubMed] [Google Scholar]
- 52. Niemi RM, Niemelä SI. Measurement uncertainty in microbiological cultivation methods. Accred Qual Assur. 2001;6:372‐375. [Google Scholar]
- 53. O'Toole G, Kaplan HB, Kolter R. Biofilm formation as microbial development. Annu Rev Microbiol. 2000;54:49–79. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1 Oligonucleotides primers/probes used in this study.
Table S2 Fecal culture profiles of three commercial laboratories in dogs with chronic diarrhea and healthy individuals.


