Abstract
ROS1 rearrangements define a distinct molecular subset of non-small-cell lung cancer (NSCLC), which can be treated effectively with crizotinib, a tyrosine kinase inhibitor (TKI) targeting ROS1/MET/ALK rearrangements. Diverse efficacy was observed in ROS1-rearranged NSCLC patients. Because of its rareness, very limited studies have investigated the correlation between different fusion partners and response to crizotinib. In this study, we retrospectively screened 6,235 advanced NSCLC patients (stage IIIB to IV) from five hospitals and identified 106 patients with ROS1 rearrangements based on either plasma or tumor tissue testing using capture-based targeted sequencing. The most frequently occurring fusion partners included cluster of differentiation 74 (CD74), ezrin (EZR), syndecan 4 (SDC4), and tropomyosin 3 (TPM3), occurring in 49.1%, 17%, 14.2%, and 4.7% of patients, respectively. Among them, 38 patients were treated with crizotinib. Seventeen patients were treatment naive, and the remaining were previously treated with pemetrexed-based chemotherapy. Collectively, there was no significant difference among patients with various types of ROS1 fusion partners in overall survival (OS) and progression-free survival (PFS). Patients who were treated with crizotinib as first-line therapy showed comparable PFS (p = 0.26) to patients who were previously treated with pemetrexed-based chemotherapy. For treatment-naive patients, patients with low baseline ROS1 allelic fraction (AF) had a statistically significant longer OS than those with high ROS1 AF (184 vs. 110 days, p = 0.048). Collectively, our study demonstrates that ROS1 + patients with various fusion partners show comparable efficacy to crizotinib.
Key words: ROS1 fusion, Crizotinib, Non-small-cell lung cancer (NSCLC)
INTRODUCTION
The c-ros oncogene 1 (ROS1), structurally similar to anaplastic lymphoma kinase (ALK), encodes for an orphan tyrosine receptor kinase belonging to the insulin receptor family1. Chromosomal rearrangements involving ROS1 was first identified as an oncogene in non-small-cell lung cancer (NSCLC) in 20072. ROS1 fusion involves a portion of ROS1 that contains the kinase activity with several partners, including, but not limited to, CD74, SLC34A2, EZR, and SDC4, resulting in the constitutive activation of ROS1 and subsequently triggering prosurvival pathways3. Such oncogenic property has been described in a number of cancers, including NSCLC4. ROS1 rearrangements, defining a molecular subset of NSCLC with distinct clinical features, are clinically associated with younger age, no history of smoking, Asian ethnicity, advanced stage, and adenocarcinoma histologic type5.
ROS1 rearrangements, occurring in 1%–2% of NSCLC patients, have become a validated therapeutic target5–7. Both preclinical and clinical studies have reported the efficacy of ROS1 TKIs to cell lines and patients harboring such mutations8. Crizotinib is a first-in-class, orally available, small-molecule tyrosine kinase inhibitor targeting ALK, MET proto-oncogene, receptor tyrosine kinase (MET), and ROS1 8,9. In a phase I crizotinib trial (PROFILE 1001), the objective response rate (ORR) was 72%, and the median progression-free survival (PFS) was 19.2 months in advanced ROS1-rearranged NSCLC patients10. Furthermore, two phase II studies also reported notable efficacy to crizotinib in ROS1-rearranged NSCLC patients with an ORR ranging from 71% to 80% and a PFS of 9–10 months11,12. A similar finding was also observed in the largest ROS1+ NSCLC cohort (OO-1201) reported to date, consisting of 127 East Asian ROS1+ patients. It reported a median PFS of 15.9 months13. Crizotinib, granted breakthrough therapy for the ROS1 indication, was approved by the US Food and Drug Administration (FDA) in March 2016 based on an expansion of phase I trial.
Numerous studies have reported differential efficacies observed in patients harboring various types of specific mutation, such as EGFR 14–16. In addition, previous studies have reported differential response magnitude and duration to crizotinib among patients harboring different ALK fusion partners17. Diverse responses to crizotinib were also observed in patients harboring ROS1-rearranged NSCLC10–13. Very few studies have examined the effect of different ROS1 fusion partners on the efficacy of ROS1 inhibitors. A recent study with 36 ROS1-rearranged NSCLC patients showed that patients carrying CD74-ROS1 had a statistically significantly shorter median PFS (12.63 months) than patients harboring other partners (17.63 months) (p = 0.048)18. More investigation is needed to elucidate whether different fusion partners can affect the therapeutic efficacy of ROS1 inhibitors. In this study, we further explored the relationship between ROS1 fusion partners and the therapeutic efficacy of crizotinib by retrospectively investigating PFS and OS of 38 advanced ROS1-rearranged NSCLC patients treated with crizotinib.
MATERIALS AND METHODS
Patients
We retrospectively screened 6,235 patients with advanced NSCLC who were treated in any of the following five participating hospitals between December 2015 and June 2017. Participating hospitals included Shenzhen People’s Hospital, Xiangya Hospital, First Affiliated Hospital of Fujian Medical School Quanzhou Branch, Renmin Hospital of Wuhan University, and Xiamen Cancer Hospital. We identified 106 with ROS1 rearrangements, and 38 of them were treated with crizotinib and had survival data. The study was approved by the ethical committee of each participating hospital. All patients provided written informed consent. The inclusion criteria were as follows: 1) histopathological diagnosed as stage IIIB/IV NSCLC; 2) NGS analysis showed a translocation or inversion event of ROS1; 3) oral crizotinib therapy was administered at any time throughout the course of the disease. Patients with ALK and/or MET mutations were excluded with mutations on ALK from the study.
Tissue DNA Extraction
Tumor tissue samples obtained by either diagnostic or surgical procedures were used for ROS1 fusion detection. DNA was extracted using QIAamp DNA FFPE tissue kit (Qiagen, Manchester, UK) according to the manufacturer’s instructions. DNA concentration was measured using Qubit dsDNA assay (Life Technologies, Carlsbad, CA, USA).
Preparation of Plasma Cell-Free DNA
For each eligible patient, plasma was collected during routine care either prior to first-line therapy or at specified time points throughout the trial. Whole blood was collected into one 10-ml EDTA-containing vacutainer (BD Vacutainer® Venous Blood Collection) and was spun into plasma within 4 h of collection. Cell-free DNA (cfDNA) was extracted from 4 to 5 ml of plasma using the QIAamp Circulating Nucleic Acid kit (Qiagen)19 and quantified by the Qubit 2.0 Fluorimeter with the dsDNA HS assay kits (Life Technologies). A minimum of 50 ng of cfDNA was required for NGS library construction.
NGS Library Preparation
DNA shearing was performed on tissue DNA using the M220 Focused-ultrasonicator (Covaris, Woburn, MA, USA), followed by end repair, phosphorylation, and adaptor ligation. Fragments in the range of 200–400 bp were size selected by Agencourt AMPure XP beads (Beckman Coulter, Brea, CA, USA) followed by hybridization with capture probe baits, hybrid selection with magnetic beads, and PCR amplification. Finally, a high-sensitivity DNA assay was performed to assess the quality and size of the fragments, and indexed samples were sequenced on Illumina NextSeq™ 500 paired-end system (Illumina, Inc., Hayward, CA, USA).
Capture-Based Targeted DNA Sequencing
DNA was profiled using a commercially available capture-based sequencing panel (Burning Rock Biotech Ltd, Guangzhou, P.R. China), targeting 168 genes and spanning 160 K of human genomic regions. DNA was hybridized with the capture probes baits, selected with magnetic beads, and PCR amplified. A bioanalyzer high-sensitivity DNA assay was then performed to assess the quality and size of the fragments, and indexed samples were sequenced on Nextseq500 sequencer (Illumina, Inc) with pair-end reads.
Sequence Data Analysis
Burrows–Wheeler aligner 0.7.1020 was used for mapping the pair-end reads to the human genome (hg19). Local alignment optimization, variant calling, and annotation were performed using GATK 3.2, MuTect, and VarScan. DNA translocation analysis was performed using both Tophat2 and Factera 1.4.3. Variants were filtered using the VarScan filter pipeline. Loci with depth less than 100 were filtered out. White blood cells were sequenced to filter out germline mutations. At least two and five supporting reads were needed for INDELs in plasma and CSF samples, respectively, whereas eight supporting reads were needed for SNVs to be called in both plasma and CSF samples. According to the ExAC, 1,000 genomes, dbSNP, ESP6500SI-V2 database, variants with population frequency over 0.1% were grouped as SNP and excluded from further analysis. Remaining variants were annotated with ANNOVAR and SnpEff v3.6. DNA translocation analysis was performed using both Tophat2 and Factera 1.4.3.
Copy number variation was detected by in-house analysis scripts based on the depth of coverage data of capture intervals. Coverage data were corrected against sequencing bias resulting from GC content and probe design. The average coverage of all captured regions was utilized to normalize the coverage of different samples to comparable scales. Copy number was calculated based on the ratio between the depth of coverage in tumor samples and average coverage of an adequate number (n > 50) of samples without copy number variation as references as to each capture interval. Copy number variation is called if the coverage data of the gene region was quantitatively and statistically significantly different from its reference control. The limit of detection for CNVs is 1.5 for deletion and 2.64 for amplification.
Statistical Analysis
Progression-free survival (PFS) was defined as the time from commencement of crizotinib treatment to progressive disease (PD) according to RECIST criteria21. OS was calculated as the period from the date of first-line systemic treatment to the date of any cause of death or the last follow-up visit. PFS and OS were analyzed using the Kaplan–Meier method and compared between different groups using the log-rank test.
Continuous variables were summarized as either means ± standard deviations (SD) or medians with interquartile ranges and categorical variables by frequencies. Unpaired Wilcoxon signed-rank test was used for continuous variable comparison and two-sided Fisher’s exact test was used to compare categorical data, as appropriate. A value of p < 0.05 was considered statistically significant. All bioinformatics analyses were performed with R (version 3.3.3, the R Foundation for Statistical Computing, Vienna, Austria) and RStudio (version 1.1.383).
RESULTS
Patient Characteristics
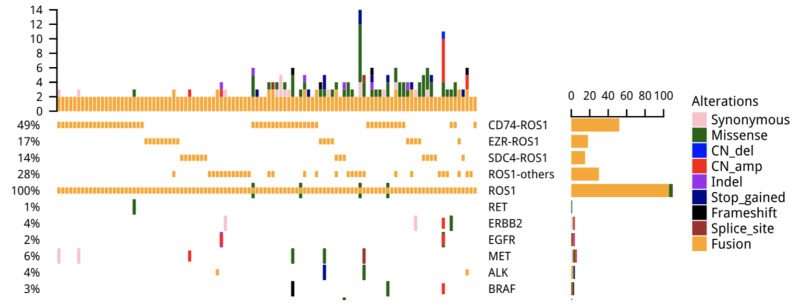
We screened 6,235 advanced NSCLC patients and identified 106 patients with ROS1 rearrangements. The median age of the ROS1+ cohort was 54 years (ranged from 26 to 79 years). Sixty-five of them were males (61.32%) and the remaining 41 were females. Except for three cases of squamous cell carcinoma, all of them were adenocarcinoma (103). Except for eight patients, who were diagnosed with stage IIIB, all others were diagnosed with stage IV disease. Interestingly, five patients had concurrent driver mutation in addition to ROS1 rearrangement: two patients had concurrent EGFR sensitizing mutation, two patients had ALK fusion, and one patient had MET amplification. One patient with EGFR sensitizing mutation also had ERBB2 amplification (Fig. 1). Patients’ characteristics are summarized in Table 1. Among the 106 ROS1-rearranged patients, 38 were treated with crizotinib, with a median age of 55 (range from 26 to 79 years). Sixty-one percent (23/38) of them were females, and the remaining 39% (15/38) were males. Patient characteristics of this crizotinib-treated cohort are shown in Table 1.
Figure 1.
Oncoprint outcome of concomitant mutations in 106 c-ros oncogene 1 (ROS1) fusion-positive patients.
Table 1.
Clinicopathologic Features of Patients With C-Ros Oncogene 1 (ROS1) Rearrangement (n = 106)
| Characteristic | ROS1 Fusion-Positive Patients (n = 106) | ROS1 Fusion-Positive Patients Treated With Crizotinib (n = 38) |
|---|---|---|
| Age (years) | ||
| Median | 54 | 55 |
| Range | 26–79 | 26–79 |
| Sex | ||
| Male | 65 (61.32%) | 15 (39.47%) |
| Female | 41 (38.68%) | 23 (60.53%) |
| Histologic type [n (%)] | ||
| Adenocarcinoma | 100 (94.34%) | 35 (92.11%) |
| Squamous cell carcinoma | 3 (2.83%) | 1 (2.63%) |
| Adenosquamous carcinoma | 3 (2.83%) | 2 (5.26%) |
| Disease stage [n (%)] | ||
| IIIB | 8 (7.55%) | 5 (13.16%) |
| ≥ IV | 98 (92.45%) | 33 (86.84%) |
| Brain metastasis | ||
| Yes | 9 (8.49%) | 8 (21.05%) |
| No | 32 (30.19%) | 22 (57.89%) |
| Unknown | 65 (61.32%) | 8 (21.05%) |
| Concurrent driven mutation [n (%)] | ||
| EGFR mutation | 2 (1.88%) | 0 (0%) |
| ALK fusion | 2 (1.88%) | 0 (0%) |
| MET amplification | 1 (0.94%) | 0 (0%) |
| ERBB2 amplification | 1 (0.94%) | 0 (0%) |
ALK, anaplastic lymphoma kinase; MET, MET proto-oncogene, receptor tyrosine kinase.
ROS1 Fusion Partner and Variant Frequencies
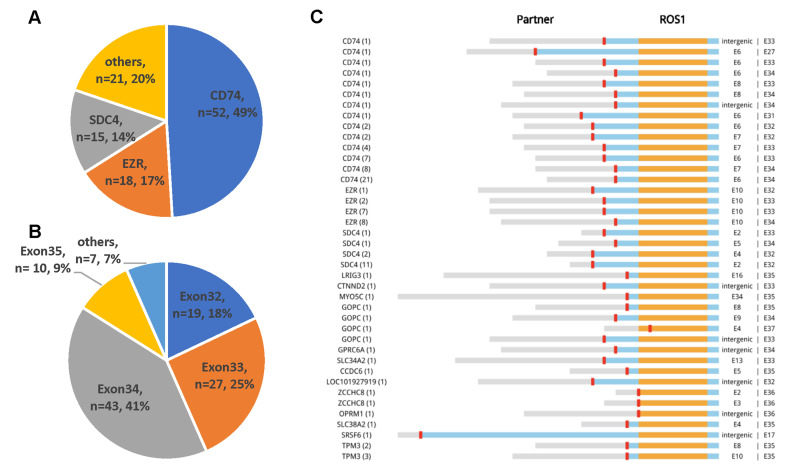
From 106 ROS1-rearranged patients, we identified 14 partners: 10 known and 4 novel partners. The frequencies of common ROS1 fusion partners are shown in Figure 2A. The most common partner was CD74, occurring in 49.1% (52/106) of patients; followed by EZR and SDC4, occurring in 17% (18/106) and 14.2% (15/106) of patients, respectively. Other partners included TPM3 (5/106), GOPC (4/106), ZCCHC8 (2/106), SLC34A2 (2/106), CCDC6 (1/106), MYO5C (1/106), and GPRC6A (1/106). All of the abovementioned partners have been reported7,22. In addition, we also have revealed four novel partners: LRIG3, CTNND2, OPRM1, and SRSF6.
Figure 2.
Overall landscape of ROS1 fusion partners detected by capture-based next-generation sequencing (n = 106). (A) Frequency of ROS1 fusions partners. (B) Distribution of ROS1 breakpoints. (C) Schematic representations of ROS1 fusion partners. Orange, tyrosine kinase domain; blue, transmembrane domain; red, coiled-coil domain.
Next, we investigated the distribution of ROS1 breakpoints. A majority of patients had a breakpoint between exons 32 and 34 (Fig. 2B and C), with minimal effect in the transmembrane and tyrosine kinase domains of the ROS1 protein22–24. Ten patients had a breakpoint at exon 35, which disrupts the structures of the transmembrane domain24. Detailed breakpoint information is summarized in Table 2.
Table 2.
ROS1 Fusion Variants Described in Non-Small-Cell Lung Cancer
| Exon 32 | Exon 33 | Exon 34 | Exon 35 | Others | |
|---|---|---|---|---|---|
| CD74 | 4 | 14 | 32 | 0 | 2 |
| EZR | 1 | 9 | 8 | 0 | 0 |
| SDC4 | 13 | 1 | 1 | 0 | 0 |
| Others | 1 | 3 | 2 | 10 | 5 |
CD74, cluster of differentiation 74; EZR, ezrin; SDC4, syndecan 4.
Association Between ROS1 Fusion Partners and Crizotinib Response
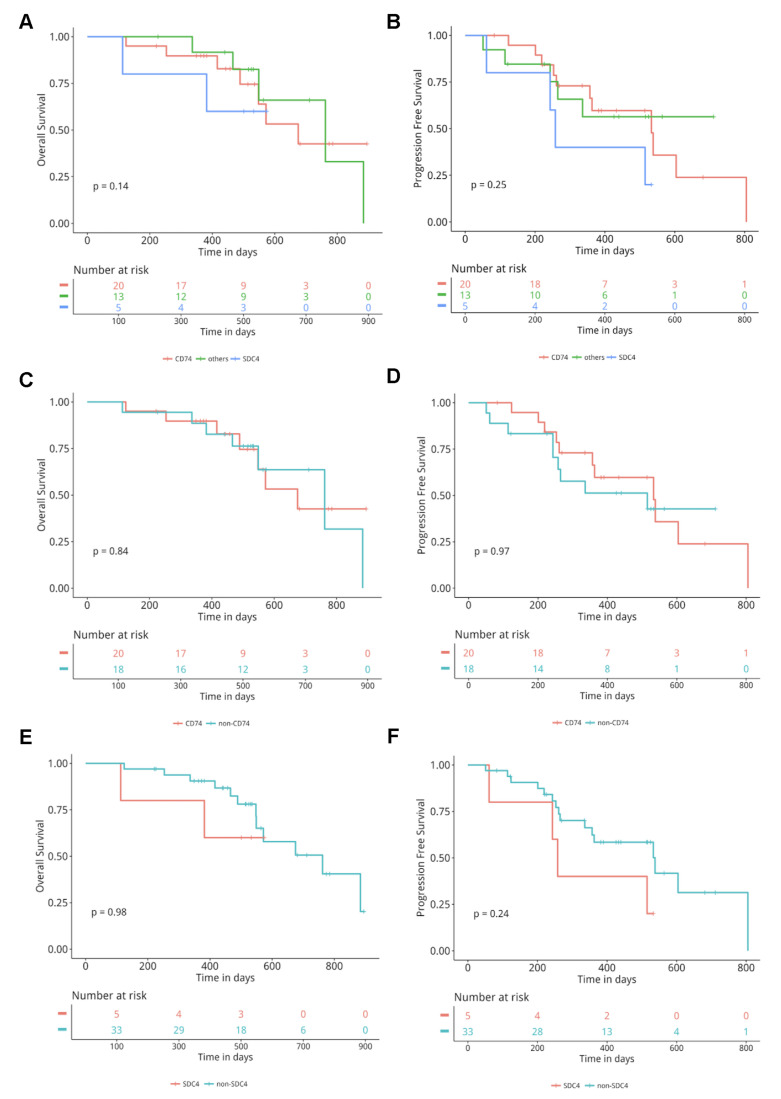
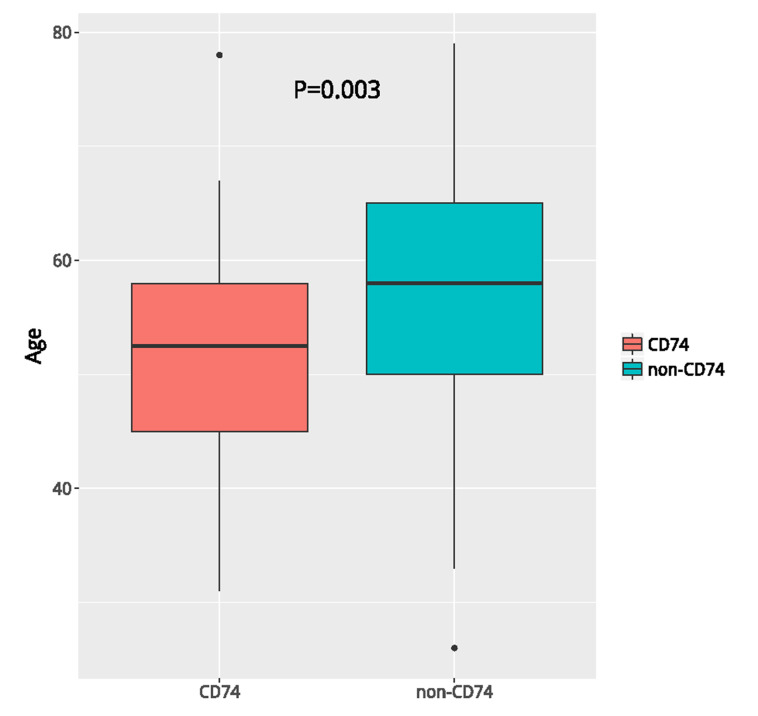
Next, we evaluated the efficacy of crizotinib in terms of PFS and OS in 38 patients. According to the frequency of ROS1 fusion partners, we classified these patients into three subgroups: patients with CD74-ROS1, SDC4-ROS1, and others. All three groups of patients exhibited comparable OS (p = 0.14) and PFS (p = 0.25) (Fig. 3A–B), consistent with the result that no difference was observed in the clinical outcomes in response to TKIs among the various ROS1 fusion partners24. Next, we classified patients based on the most common fusion partners into CD74 versus non-CD74 and SDC4 versus non-SDC4. No difference in OS and PFS was observed between the CD74 versus non-CD74 (p = 0.84 for OS; p = 0.97 for PFS) group (Fig. 3C and D). The median OS and PFS for patients with SDC4-ROS1 were 501 and 258 days, respectively, compared with 516 and 357 days for those who harbored other fusion partners (p = 0.98 for OS; p = 0.24 for PFS) (Fig. 3E and F). The basic clinical parameters such as age, gender, and stage were comparable between the SDC4 and non-SDC4 groups. Patients with CD74 partner were significantly younger than patients harboring non-CD74 partner (p = 0.003) (Fig. 4). Taken together, these results showed that the efficacy of crizotinib was comparable among patients with various types of ROS1 fusion partners.
Figure 3.
Comparison of overall survival (OS) and progression-free survival (PFS) among different ROS1 fusion pattern-positive patients treated with crizotinib. Kaplan–Meier curves for OS (A) and PFS (B) in the cluster of differentiation 74 (CD74)-ROS1, SDC4-ROS1, and others groups. Kaplan–Meier curves for OS (C) and PFS (D) in the CD74-ROS1 versus the non-CD74-ROS1 group. Kaplan–Meier curves for OS (E) and PFS (F) in the SDC4-ROS1 versus the non-SDC4-ROS1 group.
Figure 4.
Comparison of the median age between patients in the CD74-ROS1 versus the non-CD74-ROS1 group.
OS and PFS of Crizotinib in Patients With Different Treatment History
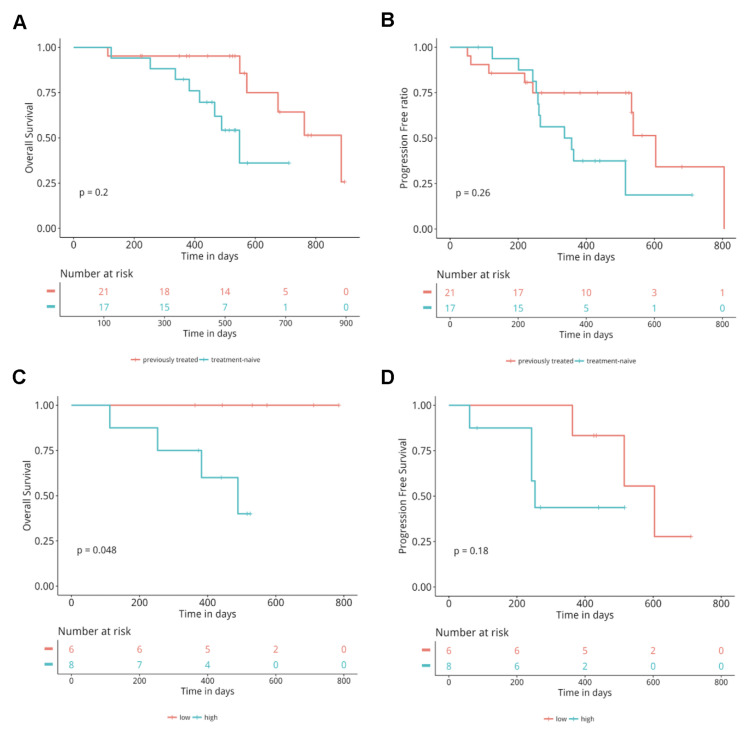
In this cohort, 17 patients were treatment naive, and 21 patients were previously treated. Next, we explored whether there is a difference in efficacy between treatment-naive patients compared to previously treated patients. Our data revealed comparable PFS (p = 0.26) and OS (p = 0.2) between the two groups.
Baseline ROS1 AF Correlates With OS
Next, we correlated baseline characteristics of 15 treatment-naive patients with clinical responses. We used the median baseline allelic fraction (AF) of ROS1 (1.6%) in blood as the binary classifier to group the patients into two groups. Our analysis revealed that patients with high baseline ROS1 AF with a median OS of 411 days were associated with a significantly shorter OS compared to patients with low ROS1 AF with a median OS of 552 days (p = 0.048) (Fig. 5C). Both groups of patients had comparable PFS (248 days vs. 474 days, p = 0.18) (Fig. 5D). Taken together, these data demonstrate that baseline ROS1 AF might be a good prognostic biomarker but not a predictive biomarker.
Figure 5.
Comparison of OS and PFS between different groups regarding the treatment lines of crizotinib or baseline ROS1 allelic fraction. Kaplan–Meier curves for OS (A) and PFS (B) in the crizotinib treatment-naive versus previously treated group. Kaplan–Meier curves for OS (C) and PFS (D) in the high baseline ROS1 AF versus high baseline ROS1 AF group. The analysis was adjusted for age, sex, histology, stage, presence or absence of brain metastases.
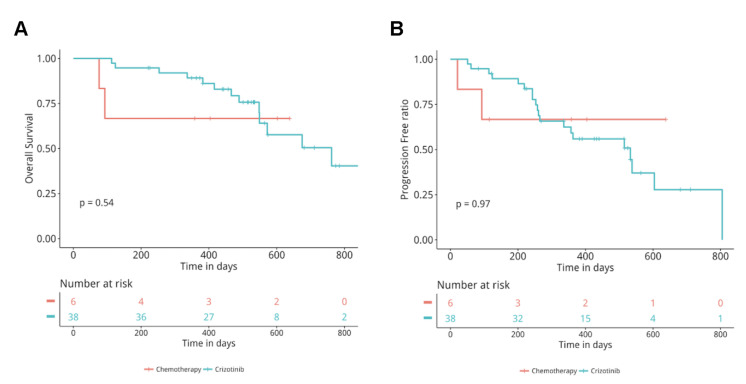
Treatment Responses of Crizotinib Versus Pemetrexed-Based Therapy in ROS1-Rearranged Patients
Studies have shown a durable response to pemetrexed-based therapy in ROS1 + patients with a median PFS of 9 months25,26. We included 6 ROS1-rearranged patients treated with pemetrexed and platinum doublet therapy from the 106 patients with ROS1 rearrangements and compared PFS and OS to ROS1-rearranged patients treated with crizotinib. Our analysis revealed comparable PFS (p = 0.3) and OS (p = 0.54) between the two groups (Fig. 6A and B).
Figure 6.
Comparison of OS and PFS between chemotherapy and crizotinib-treated patients. Kaplan–Meier curves for OS (A) and PFS (B) in the chemotherapy versus crizotinib group. The analysis was adjusted for age, sex, histology, stage, presence or absence of brain metastases.
DISCUSSION
This retrospective study comprehensively investigated the relationship between the efficacy of crizotinib and different fusion partners in advanced ROS1-rearranged Chinese NSCLC patients. The incidence of ROS1 rearrangement was 1.7% in our study, in an agreement with previous studies5,27,28. Consistent with previous reports, patients with ROS1 fusion were younger and had a nonsmoker and adenocarcinoma predominance29–34. Our study demonstrated that there was no difference in PFS and OS among ROS1-rearranged patients harboring various fusion partners. Furthermore, we also revealed comparable PFS in treatment-naive and previously treated patients (p = 0.26). Treatment-naive patients with low ROS1 AF at baseline had a significantly longer OS compared to patients with a high ROS1 AF (p = 0.048).
At present, various methods are available for analyzing ROS1 status, including IHC, FISH, PCR-based, and NGS-based methods. Each detection method is associated with its own distinct advantages and challenges. Among these, NGS-based methods offer an alternative diagnostic option with the advantage of providing detailed information regarding fusion partners, thus allowing for the detection of novel partners. In this study, the most common fusion partner was CD74, consistent with published studies22,35. In addition, we also identified four novel ROS1 fusion partners that have not been reported previously: LRIG3, CTNND2, OPRM1, and SRSF6.
Several studies have investigated the efficacy of crizotinib in NSCLC patients with ROS1 rearrangement30,31,36. We revealed a median PFS of 12 months, comparable with published studies. The EUROS1 Cohort study reported that ROS1 fusion patients treated with crizotinib had a median PFS of 9.1 months30. Another retrospective study consisting of entirely Chinese patients reported a median PFS of 9.8 months36. Furthermore, a latest study reported a median PFS of 12.7 months for TKIs (crizotinib, ceritinib, TPX-0005, and entrectinib) in a Korean population24. Several studies also evaluated whether there is a difference in efficacy of crizotinib in patients harboring diverse fusion partners but with conflicting results. A current study reported that patients with non-CD74 fusion partners had a significantly longer PFS (p = 0.048)35. Another study revealed no difference in PFS among patients with SCL34A2, EZR, or CD7436. We did not reveal any difference in PFS and OS among crizotinib-treated ROS1-rearranged patients harboring various fusion partners. Further investigations conducted in larger cohorts involving multicenters are necessary to address this issue more comprehensively.
Studies have shown a durable response to pemetrexed-based treatment in ROS1 fusion-positive patients with a PFS of 6∼8 months24. In our study, pemetrexed-treated patients had a median PFS of 8 months, comparable to published studies24. Interestingly, we observed that patients treated with pemetrexed-based treatment had a comparable PFS (p = 0.97) and OS (p = 0.54) to patients treated with crizotinib. In addition, our study also revealed comparable PFS in treatment-naive patients and previously treated patients (p = 0.26), suggesting equivalent efficacy. Larger prospective studies are needed to elucidate the most optimal treatment strategy or sequence of regiments for ROS-rearranged patients. Our study also showed that an inverse correlation between baseline ROS1 AF and OS, suggesting the prognostic value of baseline ROS1 AF in treatment-naive patients. This phenomenon has not been reported in previous studies, which can provide useful evidence for clinical practice.
In conclusion, our study revealed a prevalence rate of 1.7% for ROS1 rearrangements in Chinese NSCLC patients and evaluated associations between various ROS1 fusion partners and the efficacy of crizotinib. We found no difference in PFS and OS in patients with various fusion partners. Furthermore, we also determined that ROS1 AF can potentially act as a prognostic marker for crizotinib efficacy. Due to the limited sample sizes, larger multicenter, prospective studies are necessary to validate some of these findings.
ACKNOWLEDGMENT
The authors declare no conflicts of interest.
Footnotes
The authors declare no conflicts of interest.
REFERENCES
- 1. Acquaviva J, Wong R, Charest A. The multifaceted roles of the receptor tyrosine kinase ROS in development and cancer. Biochim Biophys Acta 2009;1795(1):37–52. [DOI] [PubMed] [Google Scholar]
- 2. Rikova K, Guo A, Zeng Q, Possemato A, Yu J, Haack H, Nardone J, Lee K, Reeves C, Li Y. Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell 2007;131(6):1190–203. [DOI] [PubMed] [Google Scholar]
- 3. Davies KD, Doebele RC. Molecular pathways: ROS1 fusion proteins in cancer. Clin Cancer Res. 2013;19(15):4040–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Rikova K, Guo A, Zeng Q, Possemato A, Yu J, Haack H, Nardone J, Lee K, Reeves C, Li Y, Hu Y, Tan Z, Stokes M, Sullivan L, Mitchell J, Wetzel R, Macneill J, Ren JM, Yuan J, Bakalarski CE, Villen J, Kornhauser JM, Smith B, Li D, Zhou X, Gygi SP, Gu TL, Polakiewicz RD, Rush J, Comb MJ. Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell 2007;131(6):1190–203. [DOI] [PubMed] [Google Scholar]
- 5. Bergethon K, Shaw AT, Ou SH, Katayama R, Lovly CM, Mcdonald NT, Massion PP, Siwaktapp C, Gonzalez A, Fang R. ROS1 rearrangements define a unique molecular class of lung cancers. J Clin Oncol. 2012;30(8):863–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hirsch FR, Suda K, Wiens J, Bunn PA Jr. New and emerging targeted treatments in advanced non-small-cell lung cancer. Lancet 2016;388(10048):1012–24. [DOI] [PubMed] [Google Scholar]
- 7. Takeuchi K, Soda M, Togashi Y, Suzuki R, Sakata S, Hatano S, Asaka R, Hamanaka W, Ninomiya H, Uehara H. RET, ROS1 and ALK fusions in lung cancer. Nat Med. 2012;18(3):378–81. [DOI] [PubMed] [Google Scholar]
- 8. Yasuda H, de Figueiredo-Pontes LL, Kobayashi S, Costa DB. Preclinical rationale for use of the clinically available multitargeted tyrosine kinase inhibitor crizotinib in ROS1-translocated lung cancer. J Thorac Oncol. 2012;7(7):1086–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Christensen JG, Zou HY, Arango ME, Li Q, Lee JH, McDonnell SR, Yamazaki S, Alton GR, Mroczkowski B, Los G. Cytoreductive antitumor activity of PF-2341066, a novel inhibitor of anaplastic lymphoma kinase and c-Met, in experimental models of anaplastic large-cell lymphoma. Mol Cancer Ther. 2007;6(12 Pt 1):3314–22. [DOI] [PubMed] [Google Scholar]
- 10. Shaw AT, Ou SH, Bang YJ, Camidge DR, Solomon BJ, Salgia R, Riely GJ, Varellagarcia M, Shapiro GI, Costa DB. Crizotinib in ROS1-rearranged non-small-cell lung cancer. N Engl J Med. 2015;371(21):1189–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mazières J, Zalcman G, Crinò L, Biondani P, Barlesi F, Filleron T, Dingemans AM, Léna H, Monnet I, Rothschild SI. Crizotinib therapy for advanced lung adenocarcinoma and a ROS1 rearrangement: Results from the EUROS1 cohort. J Clin Oncol. 2015;33(9):992–9. [DOI] [PubMed] [Google Scholar]
- 12. Moro-Sibilot D, Faivre L, Zalcman G, Pérol M, Barlesi F, Otto J, Monnet I, Cortot AB, Wislez M, Lena H. Crizotinib in patients with advanced ROS1-rearranged non-small cell lung cancer (NSCLC). Preliminary results of the ACSé phase II trial. J Clin Oncol. 2015;33(Suppl):8065. [Google Scholar]
- 13. Wu Y-L, Yang JC-H, Kim D-W, Lu S, Zhou J, Seto T, Yang J-J, Yamamoto N, Ahn M-J, Takahashi T. Phase II study of crizotinib in East Asian patients with ROS1-positive advanced non-small-cell lung cancer. J Clin Oncol. 2018;36(14):1405–11. [DOI] [PubMed] [Google Scholar]
- 14. Zofia P, Niederst MJ, Karlovich CA, Wakelee HA, Neal JW, Mari MK, Linnea F, Hata AN, Lockerman EL, Anuj K. Heterogeneity underlies the emergence of EGFRT790 wild-type clones following treatment of T790M-positive cancers with a third-generation EGFR inhibitor. Cancer Discov. 2015;5(7):713–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kobayashi Y, Togashi Y, Yatabe Y, Mizuuchi H, Jangchul P, Kondo C, Shimoji M, Sato K, Suda K, Tomizawa K. EGFR exon 18 mutations in lung cancer: Molecular predictors of augmented sensitivity to afatinib or neratinib as compared with first- or third-generation TKIs. Clin Cancer Res. 2015;21(23):5305–13. [DOI] [PubMed] [Google Scholar]
- 16. Finlay MRV, Mark A, Susan A, Peter B, Bethel PA, Box MR, Bradbury RH, Brown SJ, Sam B, Andrew C. Discovery of a potent and selective EGFR inhibitor (AZD9291) of both sensitizing and T790M resistance mutations that spares the wild type form of the receptor. J Med Chem. 2014;57(20):8249–67. [DOI] [PubMed] [Google Scholar]
- 17. Yoshida T, Oya Y, Tanaka K, Shimizu J, Horio Y, Kuroda H, Sakao Y, Hida T, Yatabe Y. Differential crizotinib response duration among ALK fusion variants in ALK-positive non-small-cell lung cancer. J Clin Oncol. 2016;34(28):3383–9. [DOI] [PubMed] [Google Scholar]
- 18. Li Z, Shen L, Ding D, Huang J, Zhang J, Chen Z, Lu S. Efficacy of crizotinib among different types of ROS1 fusion partners in patients with ROS1-rearranged non-small cell lung cancer. J Thorac Oncol. 2018;13(7):987–95. [DOI] [PubMed] [Google Scholar]
- 19. Qiagen. 2010 09/05/2016 QIAamp(r) MiniElute(r) virus spin handbook: For simultaneous purification of viral RNA and DNA from plasma, serum, and cell-free body fluids. 2010. [accessed 2016 September 5]. https://www.qiagen.com/gb/resources/resourcedetail?id=3b3ea70c-a15d-4032-9cb0-0a5d762c9313&lang=en
- 20. Li H, Durbin R. Fast and accurate long-read alignment with Burrows–Wheeler transform. Bioinformatics 2010;26(5):589–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Eisenhauer EA, Therasse P, Bogaerts J, Schwartz LH, Sargent D, Ford R, Dancey J, Arbuck S, Gwyther S, Mooney M, Rubinstein L, Shankar L, Dodd L, Kaplan R, Lacombe D, Verweij J. New response evaluation criteria in solid tumours: Revised RECIST guideline (version 1.1). Eur J Cancer 2009;45(2):228–47. [DOI] [PubMed] [Google Scholar]
- 22. Lin JJ, Shaw AT. Recent advances in targeting ROS1 in lung cancer. J Thora Oncol. 2017;12(11):1611–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Uguen A, De Braekeleer M. ROS1 fusions in cancer: A review. Future Oncology 2016;12(16):1911–28. [DOI] [PubMed] [Google Scholar]
- 24. Park S, Ahn B-C, Lim SW, Sun J-M, Kim HR, Hong MH, Lee SH, Ahn JS, Park K, La Choi Y. Characteristics and outcome of ROS1-positive non-small cell lung cancer patients in routine clinical practice. J Thorac Oncol. 2018;13(9):1373–82. [DOI] [PubMed] [Google Scholar]
- 25. Chen YF, Hsieh MS, Wu SG, Chang YL, Yu CJ, Yang JC, Yang PC, Shih JY. Efficacy of pemetrexed-based chemotherapy in patients with ROS1 fusion-positive lung adenocarcinoma compared with in patients harboring other driver mutations in East Asian populations. J Thorac Oncol. 2016;11(7):1140–52. [DOI] [PubMed] [Google Scholar]
- 26. Song Z, Su H, Zhang Y. Patients with ROS1 rearrangement-positive non-small-cell lung cancer benefit from pemetrexed-based chemotherapy. Cancer Med. 2016;5(10):2688–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Davies KD, Le AT, Theodoro MF, Skokan MC, Aisner DL, Berge EM, Terracciano LM, Incarbone M, Roncalli M, Cappuzzo F. Identifying and targeting ROS1 gene fusions in non-small cell lung cancer. Clin Cancer Res. 2011;18(17):4570–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Scheffler M, Schultheis A, Teixido C, Michels S, Morales-Espinosa D, Viteri S, Hartmann W, Merkelbach-Bruse S, Fischer R, Schildhaus HU. ROS1 rearrangements in lung adenocarcinoma: Prognostic impact, therapeutic options and genetic variability. Oncotarget 2015;6(12):10577–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chen Y-F, Hsieh M-S, Wu S-G, Chang Y-L, Shih J-Y, Liu Y-N, Tsai M-F, Tsai T-H, Yu C-J, Yang JC-H. Clinical and the prognostic characteristics of lung adenocarcinoma patients with ROS1 fusion in comparison with other driver mutations in East Asian populations. J Thorac Oncol. 2014;9(8):1171–9. [DOI] [PubMed] [Google Scholar]
- 30. Mazières J, Zalcman G, Crinò L, Biondani P, Barlesi F, Filleron T, Dingemans A-MC, Léna H, Monnet I, Rothschild SI. Crizotinib therapy for advanced lung adenocarcinoma and a ROS1 rearrangement: Results from the EUROS1 cohort. J Clin Oncol. 2015;33(9):992–9. [DOI] [PubMed] [Google Scholar]
- 31. Shaw AT, Ou S-HI, Bang Y-J, Camidge DR, Solomon BJ, Salgia R, Riely GJ, Varella-Garcia M, Shapiro GI, Costa DB. Crizotinib in ROS1-rearranged non-small-cell lung cancer. N Engl J Med. 2014;371(21):1963–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhu Q, Zhan P, Zhang X, Lv T, Song Y. Clinicopathologic characteristics of patients with ROS1 fusion gene in non-small cell lung cancer: A meta-analysis. Transl Lung Cancer Res. 2015;4(3):300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cai W, Li X, Su C, Fan L, Zheng L, Fei K, Zhou C, Manegold C, Schmid-Bindert G. ROS1 fusions in Chinese patients with non-small-cell lung cancer. Ann Oncol. 2013;24(7):1822–7. [DOI] [PubMed] [Google Scholar]
- 34. Lee HJ, Seol HS, Kim JY, Chun S-M, Suh Y-A, Park Y-S, Kim S-W, Choi C-M, Park S-I, Kim DK. ROS1 receptor tyrosine kinase, a druggable target, is frequently overexpressed in non-small cell lung carcinomas via genetic and epigenetic mechanisms. Ann Surg Oncol. 2013;20(1):200–8. [DOI] [PubMed] [Google Scholar]
- 35. Li Z, Shen L, Ding D, Huang J, Zhang J, Chen Z, Lu S. Efficacy of crizotinib among different types of ROS1 fusion partners in patients with ROS1-rearranged non-small-cell lung cancer. J Thorac Oncol. 2018;13(7):987–95. [DOI] [PubMed] [Google Scholar]
- 36. Zhang L, Jiang T, Zhao C, Li W, Li X, Zhao S, Liu X, Jia Y, Yang H, Ren S. Efficacy of crizotinib and pemetrexed-based chemotherapy in Chinese NSCLC patients with ROS1 rearrangement. Oncotarget 2016;7(46):75145–54. [DOI] [PMC free article] [PubMed] [Google Scholar]