Abstract
Artemis is a key protein of NHEJ (nonhomologous end joining), which is the major pathway for the repair of IR-induced DSBs in mammalian cells. However, the expression of Artemis in tumors and the influence of silencing Artemis on tumor sensitivity to radiation have not been investigated fully. In this study, we investigated how the expression levels of Artemis may affect the treatment outcome of radiotherapy and chemotherapy in colorectal cancer cells. First, we found that the expression of Artemis is strong in some human rectal cancer samples, being higher than in adjacent normal tissues using immunohistochemical staining. We then knocked down Artemis gene in a human colorectal cancer cell line (RKO) using lentivirus-mediated siRNAs. Compared to the control RKO cells, the Artemis knockdown cells showed significantly increased sensitivity to bleomycin, etoposide, camptothecin, and IR. Induced by DNA-damaging agents, delayed DNA repair kinetics was found by the γ-H2AX foci assay, and a significantly increased cell apoptosis occurred in the Artemis knockdown RKO cells through apoptosis detection methods and Western blot. We also found that the p53/p21 signaling pathway may be involved in the apoptosis process. Taken together, our study indicates that manipulating Artemis can enhance colorectal cancer cell sensitivity to DNA-damaging agents. Therefore, Artemis can serve as a therapeutic target in rectal cancer therapy.
Key words: Colorectal cancer, Artemis, Small interfering RNA (siRNA), DNA repair, Ionizing radiation
INTRODUCTION
Radiotherapy is becoming a standard treatment in clinical cancer therapy nowadays. About 60%–70% cancer patients receive radiotherapy combined with or without chemotherapy in their treatment regimen, especially for nonmetastatic tumors. Considerable evidence has demonstrated that the intrinsic radiosensitivity of tumor cells is a primary determinant for the treatment outcome of radiation therapy1. Indeed, DNA repair in cancer cells plays a key role in innate and acquired cellular resistance to DNA-damaging agents including irradiation (IR). Defects in the DNA repair process in cells can result in cellular hypersensitivity to such agents2.
IR induces two major forms of DNA damage: single-strand breaks (SSBs) and double-strand breaks (DSBs). The SSBs are detected and repaired by base excision repair (BER) and SSB repair pathways, respectively. Two major pathways, namely, nonhomologous end joining (NHEJ) and homologous recombination or homology-directed repair (HDR), are involved in the repair of IR-induced DSBs in mammalian cells. In particular, NHEJ is active throughout the cell cycle and plays a major role in IR-induced DSB repair in human cells3,4. However, complex or clustered DNA lesions may occur in the presence of DSBs. Such lesions can result in cell death without appropriate repair.
Artemis is a key molecule in the NHEJ system. The Ku proteins, a heterodimer of Ku70 and Ku80, recruit the catalytic subunit of DNA protein kinase (DNA-PKcs) into DNA strand break sites, thereby activating Artemis, which is responsible for processing DNA ends before X-ray repair cross-complementing 4 (XRCC4), and DNA ligase IV along with the newly identified component XRCC4-like factor (XLF)/Cernunnos facilitate the final ligation step4. Certain mutations in the Artemis protein in humans have been linked to hypersensitivity to DSB-inducing agents, as well as severe deficiency of B and T lymphocytes [radiosensitive severe combined immune deficiency (RS-SCID)]5. Based on the previous studies, Artemis is an enzyme belonging to the metallo-β-lactamase (β-Lact) superfamily and an important factor in the variable-diversity-joining [V(D)J] recombination/DNA repair6,7. Artemis functions in V(D)J recombination and NHEJ as a hairpin and 5′ and 3′ overhang endonuclease. The kinase activity of the DNA-PKcs is required for Artemis activation as an endonuclease8,9. Artemis can form a complex with the DNA-PKcs in the presence of DNA. Upon complex formation, the DNA-PKcs phosphorylates Artemis and activates its endonucleolytic activity on 5′ and 3′ overhangs and hairpins generated by the recombination-activating gene (RAG) complex. It appears that the DNA-PKcs regulates Artemis’s activity by inducing phosphorylation and complex formation, which is critical for the hairpin-opening step of V(D)J recombination and for the 5′ and 3′ overhang processing in nonhomologous DNA end joining10–12. Artemis also has 5′ exonuclease activity, which would permit Artemis to act on 5′ overhangs more efficiently13. More evidence confirmed that Artemis can remove lesions or secondary structures, thereby inhibiting end resection and precluding the completion of NHEJ14–16.
Besides its effects on classical DNA-PKcs-dependent nonhomologous end joining (C-NHEJ), Artemis is also involved in homologous recombination repair (HRR). Artemis and ataxia telangiectasia mutated (ATM) together promote homologous recombination of radiation-induced DSBs during the G2 phase17. A recent study showed that Artemis is participating in DSB repair by all major repair pathways, including HRR, C-NHEJ, and an alternative form of end joining (A-EJ)18. Beyond that, the role of Artemis in DNA repair also includes cell cycle regulation and maintaining normal telomere function19–21.
The critical role of Artemis in radiation-induced damage has been supported by the evidence that the cells lacking or possessing a mutation of Artemis were hypersensitive to radiation as revealed by persistent cell cycle arrest following radiation22–25. In contrast, overexpression of Artemis showed a marked radioprotection for both high and low linear energy transfer (LET) radiation26. Taken together, the expression level of Artemis is a key factor for radiosensitivity. However, the expression of Artemis in tumors and the influence of silencing Artemis on the tumor’s sensitivity to radiation have not really been extensively explored. In this study, we first found that the expression of Artemis is higher in some human rectal cancer samples compared to adjacent normal tissues. Then using human colorectal cancer cell lines, we performed some experiments to explore the influence of silencing Artemis on tumor radiosensitivity. Our results indicate that Artemis can serve as a therapeutic target to increase cancer cell sensitivity to DNA-damaging agents including radiation in rectal cancer therapy.
MATERIALS AND METHODS
Tumor Samples
Tumor samples were obtained from 25 colorectal carcinoma patients undergoing primary tumor biopsy by colonoscopy at the Sir Run Run Shaw Hospital (Hangzhou, P.R. China) during the period between January 2012 and November 2014. They included 9 women and 16 men with ages ranging from 42 to 75 (median, 62) years. None of the patients received any chemotherapy or radiotherapy prior to biopsy. All patients provided signed, informed consent for their tissues to be used for scientific research. The ethics committee of the Sir Run Run Shaw Hospital approved the study.
Cells
Human colorectal cancer cell lines RKO and HCT116 were obtained from the American Type Culture Collection (ATCC; Manassas, VA, USA). RKO cells were cultured in RPMI-1640, and HCT116 cells were cultured in McCoy’s 5A. All culture media were supplemented with 10% fetal bovine serum (FBS; Gibco, Carlsbad, CA, USA), 100 U/ml penicillin, and 100 U/ml streptomycin. All cells were cultured in a humidified incubator (37°C, 5% CO2). The cells in the logarithmic phase were used for all experiments.
Reagents
Artemis antibody was provided by Hangzhou AuaAn Biotechnology Co. (Hangzhou, P.R. China). Anti-p53, anti-phospho-p53 (S46), anti-p21, and anti-β-actin antibodies were purchased from Cell Signaling Technology, Inc. (Danvers, MA, USA). Anti-phospho-γ-H2AX (Ser139) and anti-cleaved caspase 3 antibodies were purchased from Epitomics, Inc. (Burlingame, CA, USA).
Construction of Lentiviral Vectors and Lentivirus Infection
Three predesigned small interfering RNA (siRNA) sequences targeting Artemis (GenBank accession number NM 005702) were designed by Innovation Biotech Co. (Shanghai, P.R. China). The specificity for Artemis silencing was determined by transfecting the three siRNAs into HEK293 cell lines using Lipofectamine 2000 according to the manufacturer’s instructions (Invitrogen, Carlsbad, CA, USA). The Artemis siRNA target sequence (5′-GCATTAAGCCATCCACCATGT-3′) was selected for the construction with lentiviral vector pLenO-THM. A nonsilencing sequence (5′-TTCTCCGAACGTGTCACGT-3′) was used as a negative control (NC). Construction of lentiviral vectors and vector packaging were carried out by Innovation Biotech Co. The final titer of recombinant lentivirus was adjusted to 3.5 × 108 TU/ml.
The RKO cells (2 × 104) were seeded in 24-well plates overnight before transfection. The virus was added to each well containing an enhanced infection solution and incubated for 8–12 h at 37°C, followed by incubation for 96 h in complete RPMI-1640 medium. The cells were then harvested for subsequent studies.
Real-Time RT-PCR
Total RNA of RKO cells was extracted for reverse transcription (RT) reaction. The Moloney murine leukemia virus (M-MLV) reverse transcriptase (Promega, Madison, WI, USA) was used to synthesize the cDNA of Artemis. Quantitative real-time PCR was performed using TaqMan® Gene Expression Master Mix and the Applied Biosystems StepOneplus™ Real-Time PCR System (Applied Biosystems, Foster City, CA, USA). Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) served as an internal control. Specific PCR primers and TaqMan probes were used as previously described27. All PCR data were normalized. The relative fold change was calculated using the ΔΔCt method based on the results from three independent experiments.
Cytotoxicity and Proliferation Assay
A standard MTT assay was performed to compare tumor cell sensitivities to DNA damage agents. Cells (4 × 103 cells per well) were cultured in 96-well plates for 24 h, then incubated with different concentrations of bleomycin, etoposide, camptothecin, or cisplatin for another 72 h. Cell viability was measured by relative absorbance of MTT. IC50 value was calculated using GraphPad Prism 5.0 software.
For proliferation assay, cells (2 × 104 cells/well) were seeded in 24-well plates initially (37°C, 5% CO2 incubator overnight). The numbers of living cells were counted daily for 7 days using trypan blue staining. The growth curve was done using GraphPad Prism 5.0 software.
Ionizing Radiation and Colony Formation Assay
Irradiation was carried out using six MV X-rays generated by a linear accelerator at a dose rate of 2 Gy/min (PRIMUS-M; Siemens, Erlangen, Germany). Cells cultured in six-well plates were irradiated with X-rays (0–6 Gy) and continuously cultured for another 10 to 14 days to allow colony formation. Cells were washed and fixed with 75% methanol, then stained with 0.5% crystal violet in methanol. The colonies consisting of >50 cells were counted under a dissecting microscope. The surviving fraction was calculated by the GraphPad Prism 5.0 software based on the multitarget/single-hit model [SF = 1 − (1 − e − D/D0)N]. The sensitizing enhancement ratio (SER) was calculated based on the formula: SER = D0 (control cells)/D0 (testing cells). D0 represents the dose that can reduce cell survival to 37%.
Immunohistochemistry
Immunohistochemistry was performed as described previously. Briefly, formalin-fixed paraffin-embedded tissue was mounted onto poly-l-lysine-coated slides, deparaffinized, and rehydrated. After quenching the endogenous peroxidase activity with 0.3% H2O2 (in absolute methanol) for 30 min, the slides were treated with 5% bovine serum albumin to block nonspecific staining and incubated overnight with primary antibodies detecting Artemis. Slides were then incubated for 1 h with biotinylated anti-rabbit IgG (Vector Laboratories Inc., Burlingame, CA, USA) for Artemis. The slides were incubated with the avidin–biotin–peroxidase complex (Vector Laboratories Inc.) for 1 h, and antibody binding was visualized with 3,3′-diaminobenzidine tetrahydrochloride. Last, the sections were lightly counterstained with Mayer’s hematoxylin.
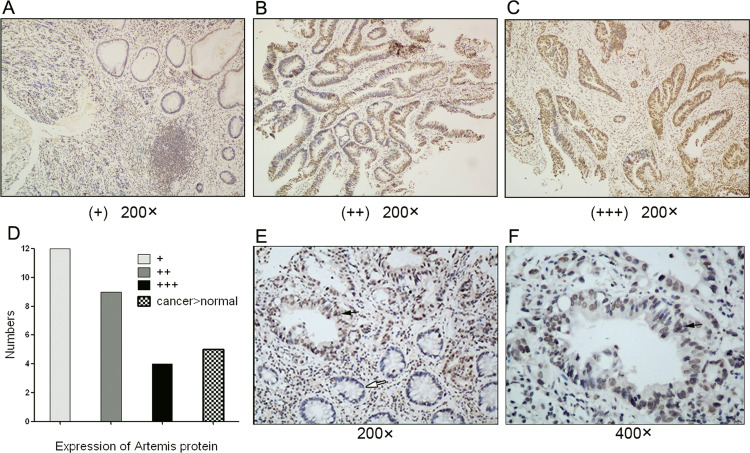
The expression of Artemis protein in rectal cancer was evaluated according to the positive proportion and the staining degree of Artemis protein. The following are scores according to the proportion of positive cells: <5% (0); ≧5% (1); ≧25% (2); ≧50% (3); ≧75% (4). The following are scores according to the staining degree: negative (0), no brown particles; weakly positive (1), scattered in shallow or small brown granules; moderately positive (2), big brown granules; strong positive (3), brown granules were densely distributed. Finally, the above two scores were multiplied to get the final score of each sample: 1–4 is weakly positive (+), 5–8 is moderately positive (++), >8 is strongly positive (+++).
Immunoblotting and Immunofluorescence Staining
Western blotting was performed as described previously27. For immunofluorescence staining, cells were cultured on a coverslip (12 mm × 12 mm) overnight, then irradiated with X-rays (2 Gy). At the indicated time point, cells were fixed and then stained with anti-γ-H2AX antibody (Ser139) and subsequently with a fluorescence-activated cell sorting (FITC)-conjugated secondary antibody. Cell nuclei were counterstained with 4,6-diamidino-2-phenylindole (DAPI; Sigma-Aldrich, Merck KGaA, Darmstadt, Germany). The slides were observed and photographed under an LSM710 inverted confocal microscope (Zeiss, Oberkochen, Germany). γ-H2AX foci in the nuclei were observed and counted. More than 50 cells in each treatment were counted by two observers. The average number of foci per cell is shown.
Assays for Apoptosis
Caspase 3/7 assay was purchased from Promega Co. and performed as per the manual. Briefly, cells were planted in 96-well plates and treated with etoposide for different times. One hundred microliters of Caspase-Glo 3/7 Reagent was added to each well of a 96-well plate containing 100 μl of blank or cells. The well contents were mixed and incubated at room temperature for 60 min. The luminescence of each sample was measured in a plate-reading luminometer.
DNA DAPI staining was used to distinguish apoptotic cells. Cells were plated in 24-well plates and treated with etoposide. Media were removed and replaced with 0.5 ml of 75% ethanol to fix the cells for 10 min. The cells were then washed with phosphate-buffered saline (PBS) two times, and 0.5 ml of (5 μg/ml) DAPI solution was added for 5 min. The cell nucleus was then detected using a fluorescence microscope.
Statistical Analysis
All data were presented as means ± standard deviation (SD). Statistical significance (p < 0.05) was determined by the Student’s t-test.
RESULTS
The Expression of Artemis Is Different in Human Rectal Cancer Samples and Colorectal Cancer Cell Lines
We first assessed the expression of Artemis in 25 cases of human rectal carcinoma specimens by immunochemical staining as a preliminary experiment. Artemis protein staining was weakly positive (+) in 12 cases, moderately positive (++) in 9 cases, and strongly positive (+++) in 4 cases of rectal cancer tissues (Fig. 1A–C). The analysis results are shown in Figure 1D. These results indicated that more than half of the patients with rectal cancer had moderate or high expression of Artemis protein. Further, a stronger staining of Artemis protein is seen in cancer tissues compared with adjacent normal tissues in five cases (Fig. 1D–F). So Artemis protein expression is increased in human rectal cancer. Taken together, it is necessary to study whether Artemis expression affects the radiosensitivity of colorectal cancer cells.
Figure 1.
The expression of Artemis in human rectal cancer samples and adjacent normal tissues. Representative image of Artemis protein immunochemical staining in rectal cancer samples: weakly positive (A), moderately positive (B), strongly positive (C). (D) The analysis result of Artemis protein expression in 25 cases of rectal cancer. (E, F) A stronger staining of Artemis protein is seen in rectal cancer tissues compared with adjacent normal tissues. The black arrow denotes Artemis expression in adenocarcinoma cells. The white arrow denotes Artemis expression in normal gland cells. Magnification: 200×, 400×.
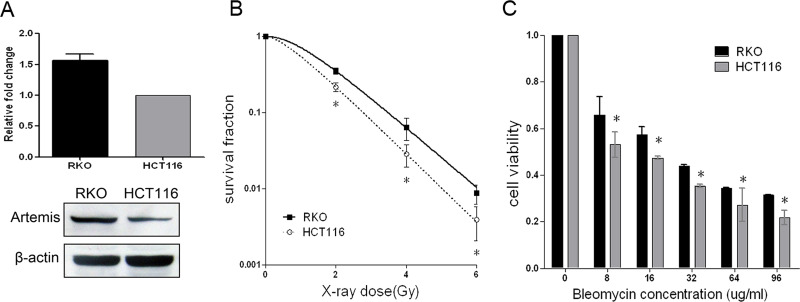
Then we studied the correlation of Artemis expression with intrinsic radiosensitivity in two different human colorectal cancer cell lines RKO and HCT116. Both RKO cells and HCT116 cells are mutLhomolog 1 (MLH1)-deficient cells. Using Western blotting and qPCR, the translation and transcription levels of Artemis in RKO and HCT116 cells were determined, respectively (Fig. 2A). We found that the expression level of Artemis was higher in RKO cells than in HCT116 cells. We also compared their radiosensitivity by colony-forming assays to X-ray and MTT assay to bleomycin (Fig. 2B and C). As a result, the RKO cells were more radioresistant than HCT116 cells.
Figure 2.
Correlation of Artemis expression and radiosensitivity in human colorectal cancer cells. Artemis mRNA and protein expression in RKO and HCT116 cells (A). Cancer cell sensitivity to X-ray (B) and bleomycin (C) was assessed in RKO and HCT116 cells. Data represent the results from three independent experiments. Error bars represent (SD) of three independent experiments. *p < 0.05.
Artemis Was Knocked Down in RKO Cells Using Lentivirus-Mediated siRNA
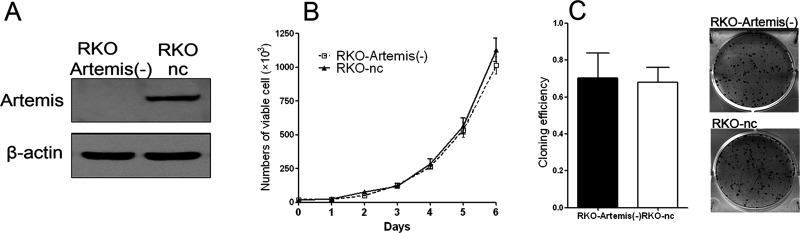
To study how the expression levels of Artemis may affect cancer cell radiosensitivity, we knocked down Artemis in RKO cells using lentivirus-mediated siRNA technology. The expression levels of Artemis protein were determined in the selected cell clones by Western blotting. As shown in Figure 3A, the expression level of Artemis in the Artemis-silencing RKO cells was markedly decreased, indicating a successful knockdown by specific siRNA. The established cell line was then used as the in vitro cell model for the following experiments.
Figure 3.
Knockdown of Artemis in RKO cell by lentivirus-mediated small interfering RNA (siRNA). The expression levels of Artemis protein (A), cell proliferation (B), and cell colony formation (C) were assessed in the RKO-Artemis (−) cells and control RKO cells (RKO-nc).
We next measured cell proliferation after Artemis silencing in RKO cells and found a similar proliferation pattern and rate in the Artemis-silencing RKO cells [RKO-Artemis (−) cells] and control RKO cells (RKO-nc cells). There was no statistical significance in cell numbers on day 6 (p = 0.083) (Fig. 3B). The colony formation experiment was also done to evaluate the effects of Artemis silencing on cellular capacity in colony formation. The clone formation rates of the RKO-Artemis (−) cells and RKO-nc cells were 70% and 67.8%, respectively. There was no statistically significant difference between them (p = 0.86) (Fig. 3C).
The Sensitivity of the RKO-Artemis (−) Cell Was Increased to DNA-Damaging Agents Including Irradiation
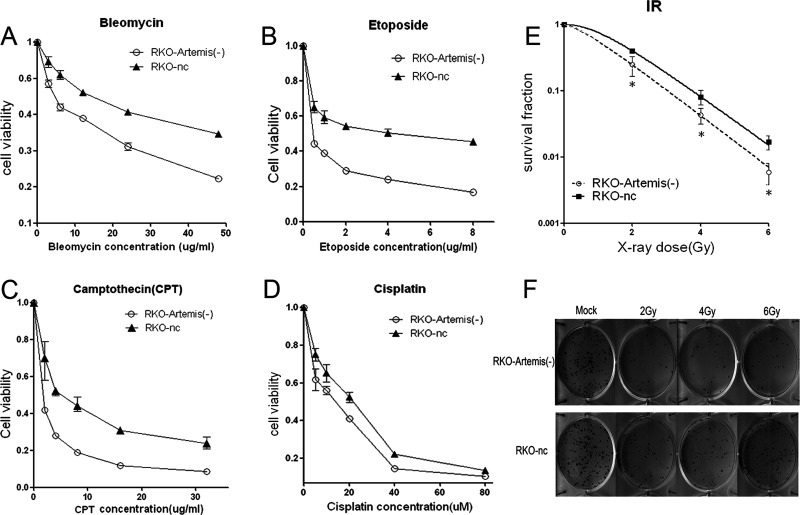
To analyze if Artemis silencing may affect cell sensitivity to DNA-damaging agents, the RKO-Artemis (−) cells and RKO-nc cells were exposed to a variety of DNA-damaging agents and followed by standard MTT analysis. As shown in Figure 4, compared with control RKO-nc cells, the RKO-Artemis (−) cells showed significantly increased sensitivity to bleomycin (Fig. 4A), etoposide (Fig. 4B), and camptothecin (Fig. 4C). We also examined the sensitivity of the RKO-Artemis (−) cells to a DNA cross-linking agent, cisplatin. We found that the RKO-Artemis (−) cells only showed a mild increase in sensitivity to cisplatin (Fig. 4D). Our results indicate that the Artemis-silencing RKO cells showed a similar phenotype to the previously reported Artemis-deficient cells28.
Figure 4.
Cancer cell sensitivity to DNA-damaging agents in the RKO-Artemis (−) cells and control RKO cells. MTT assays were used. (A) Bleomycin, (B) etoposide, (C) camptothecin, and (D) cisplatin. Colony formation assays were performed to measure radiosensitivity (E). (F) Representative colony formation on day 14 after irradiation (IR). Data represent the results from three independent experiments. Error bars represent SD of three independent experiments. *p < 0.05.
We performed a colony formation assay to measure the radiosensitivity of the Artemis-silencing RKO cells. A significantly higher sensitivity to IR was observed in the RKO-Artemis (−) cells than in the RKO-nc cells (Fig. 4E and F). In particular, the cell survival fraction was significantly decreased in the RKO-Artemis (−) cells versus the RKO-nc cells (0.5% vs. 1.9%) after high-dose IR (6 Gy). The calculated D0 values of the RKO-Artemis (−) cells and RKO-nc cells were 1.022 and 1.245, respectively. The calculated sensitizing enhancement ratio (SER) was 1.218.
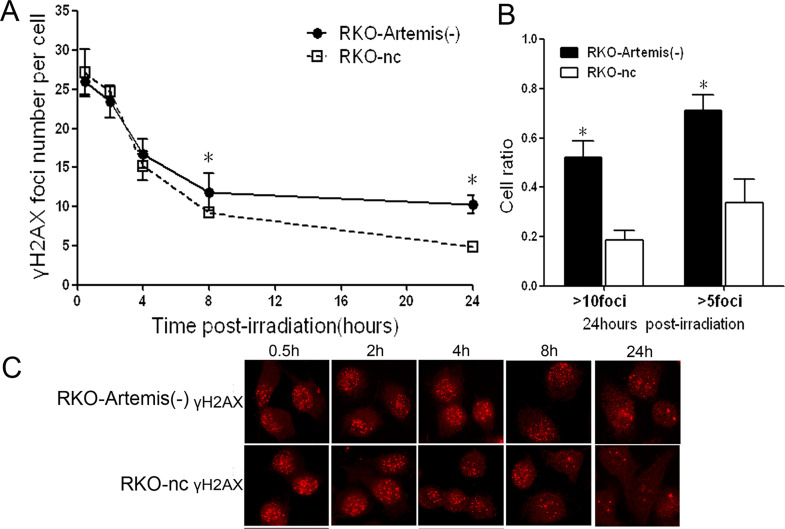
The DNA DSB Repair Time Was Prolonged in the RKO-Artemis (−) Cells After Irradiation
The γ-H2AX foci assay was employed to monitor the kinetics of the DSB rejoining process. After 2 Gy of IR, γ-H2AX foci were seen in both the RKO-Artemis (−) cells and RKO-nc cells, and gradually decreased along with the DNA repair process (Fig. 5A and C). We counted the numbers of foci per nucleus and calculated the mean (Fig. 5A). Within 4 h postradiation, the average number of γ-H2AX foci of the two groups was compatible without statistical difference (p > 0.05). However, from 8 to 24 h, the repair kinetics were obviously different between the RKO-Artemis (−) cells and RKO-nc cells. After 24 h, the residual number of foci in the RKO-Artemis (−) cell was about twofold of that in RKO-nc cells. Furthermore, the percentage of cells with residual foci (≥10 or ≥5) in the RKO-Artemis (−) cells were 52.2% and 71.1%, which were significantly higher than in the RKO-nc cells (18.4% and 33.8%, respectively) (p < 0.05) (Fig. 5B). These data indicate an impaired DNA repair in the RKO-Artemis (−) cells.
Figure 5.
Impaired double-strand break (DSB) rejoining in RKO-Artemis (−) cells after IR. (A) The average number of γ-H2AX foci per cell was counted and shown. Each bar represents mean ± SD. At least 100 nuclei were evaluated per sample. (B) Cell ratio with residual foci more than 10 or 5 at 24 h after IR. Data represent the results from three independent experiments. Error bars represent SD of three independent experiments. *Statistical significance (p < 0.05). (C) Representative image of γ-H2AX foci under an immunofluorescence microscope.
The Cell Apoptosis Increased in the RKO-Artemis (−) Cells Under the DNA-Damaging Agents
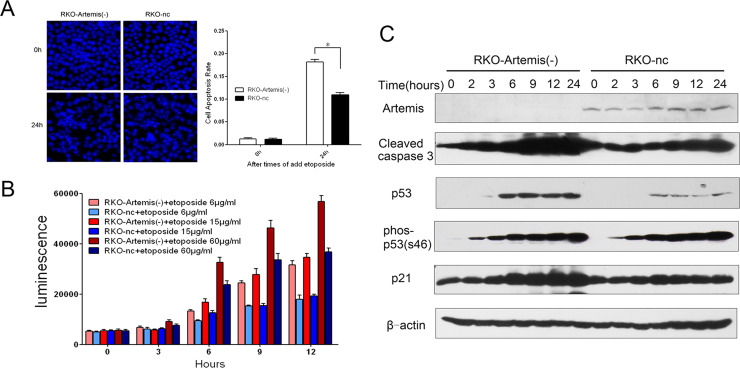
We studied how knockdown of Artemis affects DNA-damaging agent-induced cell apoptosis in RKO cells. DNA DAPI staining was used to distinguish apoptotic cells with nuclear pyknosis, fragmentation, and nucleosome. After 24 h of treatment with the DNA-damaging agent etoposide (6 μg/ml), cells were fixed and stained with DAPI. The apoptotic cells were counted under a fluorescence microscope. A greater amount of apoptotic cells was seen in the RKO-Artemis (−) cells compared to RKO-nc cells (Fig. 6A).
Figure 6.
Cell apoptosis in RKO-Artemis (−) cells after treatment with the DNA-damaging agent etoposide. (A) Apoptotic cells were counted under a fluorescence microscope 24 h after treatment with etoposide (6 μg/ml). (B) Caspase 3/7 enzyme activity was measured after treatment with different doses of etoposide (6, 15, and 60 μg/ml) in the RKO-Artemis (−) cells and control RKO-nc cells. Data represent the results from three independent experiments. Error bars represent SD of three independent experiments. *p < 0.05. (C) Cells were pretreated with etoposide (6 μg/ml for 0–24 h). Total cell lysate was harvested at indicated time points after IR for Western blot analysis. Cleaved caspase 3, total p53, phosphorylated p53 (phos-p53), and total p21 were determined by Western blotting. β-Actin served as loading control.
We used Caspase-Glo™ 3/7, a biological luminescence detection method, to measure the activity of caspase 3/7. We treated the RKO-Artemis (−) cells and RKO-nc cells with different concentrations of etoposide (6, 15, and 60 μg/ml), and then measured the activity of caspase 3/7. Significantly increased activities with time- and dose-dependent patterns were noted (Fig. 6B). Compared with RKO-nc cells, the RKO-Artemis (−) cells’ caspase 3/7 activity started increasing earlier and faster. In Western blot analysis, the RKO-Artemis (−) cells showed significantly higher levels of apoptosis-related caspase activities, and the p53/p21 signaling pathway was also stimulated after treatment with 6 μg/ml etoposide (Fig. 6C). Of note, greater caspase 3 activities were detected in the RKO-Artemis (−) cells in a time-dependent manner compared to RKO-nc cells. In the meantime, the medication-induced p53 and p21 expressions were significantly higher in the RKO-Artemis (−) cells. These results demonstrate an increased cell apoptosis in the RKO-Artemis (−) cells, and the p53/p21 signaling pathway is likely involved.
DISCUSSION
The role of Artemis in radiation-induced DNA damage and repair has been well studied and documented in the literature as previously described17,19–25. Artemis serves as a multifunctional protein in DSB repair by all major repair pathways18. However, there are few studies focusing on whether regulating the expression of Artemis can improve the radiosensitivity of tumors. In this study, we found that knockdown of Artemis in human colorectal cancer cells can enhance cancer cell sensitivity to DNA-damaging agents including radiation and indicated that Artemis has the potential to be a therapeutic target in rectal cancer therapy.
In a preliminary study, we found that Artemis is moderately or highly expressed in more than half of human rectal cancer samples, even being higher than in adjacent normal tissues in some cases. This finding suggests that it is necessary to study the relationship between Artemis expression and sensitivity to radiotherapy in rectal cancer. In a following work, we will collect more samples to study the expression of Artemis in rectal cancer and the correlation between the Artemis expression and the radiosensitivity of tumors. In the present study, in vitro, we mainly used the human colorectal cancer cell lines to study this question. Our results indicate that RKO cells with relatively higher Artemis expression are more radioresistant than HCT116 cells with lower Artemis expression. In order to observe the effect of the Artemis expression level on the radiosensitivity of colorectal cancer cells, we established Artemis knockdown RKO cells using lentiviral-mediated RNAi technology. Further experiments revealed that inhibition of Artemis in RKO cells enhanced tumor cell sensitivity to other DNA-damaging agents (bleomycin, camptothecin, and etoposide) in addition to IR. Our result is consistent with previous reports on the increased sensitivities in Artemis-defective phenotypes22,24,28,29. The results generated from the RKO-Artemis (−) cells indicated dysfunctional DNA repair, resulting in more DSBs, may eventually lead to chromosomal mutation and cell death. In fact, higher levels of γ-H2AX foci were observed in the RKO-Artemis (−) cells 8 h later after exposure, highly supporting the impaired reconnection of DSBs (rejoining). This is also consistent with a role of Artemis in end processing prior to DSB rejoining30,31. These data were further supported by the results from MTT assays for short-term toxicity after IR and the data from clonogenic survival assay for long-term survival.
DNA-damaging agents especially IR induce cell apoptosis, and its role in chemotherapy and radiotherapy has been widely investigated and well documented32. Previous studies also focused on enhancing cancer cell apoptosis by regulating apoptosis-related gene expression and signaling pathways33,34. We found that certain DNA-damaging agents including IR induced more cell apoptosis in the Artemis-silencing RKO cells. In addition, our data showed that the p53/p21 pathway was critically involved in DNA-damaging agent-induced cell apoptosis in the Artemis-silencing RKO cells. The p53/p21 system is known to be critical in the response of human colon carcinoma cells to DNA-damaging agents35. We hypothesize that Artemis silencing causes DSB repair defects, which lead to DNA damage and chromosome mutations, and then p53 protein and relevant pathways will be activated in those damaged cells. It has been reported that Artemis inhibits the expression of p53 protein and that Artemis silencing induces p53 upregulation in a variety of cells36. Similarly, the RKO-Artemis (−) cells expressed significantly higher levels of p53 than the control RKO cells. In addition, it is also possible that Artemis silencing abolished the DNA-PKcs inhibition of p53 phosphorylation since the phosphorylation levels of p53 were higher in the earlier time period in the RKO-Artemis (−) cells. Moreover, a p53-independent G1 arrest was observed in RKO cells expressing wild-type p53, suggesting an unknown mechanism may also have been involved37. Further studies are needed to clarify this issue.
CONCLUSION
We found that Artemis can be overexpressed in human rectal cancer and Artemis silencing can enhance the sensitivity of colorectal cancer cells to a series of DNA-damaging agents including IR. Certain DNA-damaging agents such as topoisomerase I inhibitor CPT-11 has become a second-line treatment for advanced colorectal cancer. A combination of radiotherapy and chemotherapy may hold the key for a better treatment outcome. Our study strongly indicates that Artemis can be used as a target to enhance colorectal cancer cell sensitivity to DNA-damaging agents including IR. This is clinically important in improving the efficacy of radiotherapy and chemotherapy in colorectal cancer patients.
ACKNOWLEDGMENTS
This work was supported in part by grants from the Chinese National Natural Science Foundation (Nos. 81441086 and 81672976) and Zhejiang Provincial Natural Science Foundation of China (Nos. LY14H160016 and LY14H110002).
Footnotes
The authors declare no conflicts of interest.
REFERENCES
- 1. Gerweck LE, Vijayappa S, Kurimasa A, Ogawa K, Chen DJ. Tumor cell radiosensitivity is a major determinant of tumor response to radiation. Cancer Res. 2006;66(17):8352–5. [DOI] [PubMed] [Google Scholar]
- 2. Ding J, Miao ZH, Meng LH, Geng MY. Emerging cancer therapeutic opportunities target DNA-repair systems. Trends Pharmacol Sci. 2006;27(6):338–44. [DOI] [PubMed] [Google Scholar]
- 3. Mahaney BL, Meek K, Lees-Miller SP. Repair of ionizing radiation-induced DNA double-strand breaks by non-homologous end-joining. Biochem J. 2009;417(3):639–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wang C, Lees-Miller SP. Detection and repair of ionizing radiation-induced DNA double strand breaks: New developments in nonhomologous end joining. Int J Radiat Oncol Biol Phys. 2013;86(3):440–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Moshous D, Callebaut I, de Chasseval R, Corneo B, Cavazzana-Calvo M, Le Deist F, Tezcan I, Sanal O, Bertrand Y, Philippe N, Fischer A, de Villartay JP. Artemis, a novel DNA double-strand break repair/V(D)J recombination protein, is mutated in human severe combined immune deficiency. Cell 2001;105(2):177–86. [DOI] [PubMed] [Google Scholar]
- 6. Pannicke U, Ma Y, Hopfner KP, Niewolik D, Lieber MR, Schwarz K. Functional and biochemical dissection of the structure-specific nuclease ARTEMIS. EMBO J. 2004;23(9):1987–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Poinsignon C, Moshous D, Callebaut I, de Chasseval R, Villey I, de Villartay JP. The metallo-beta-lactamase/beta-CASP domain of Artemis constitutes the catalytic core for V(D)J recombination. J Exp Med. 2004;199(3):315–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ma Y, Pannicke U, Lu H, Niewolik D, Schwarz K, Lieber MR. The DNA-dependent protein kinase catalytic subunit phosphorylation sites in human Artemis. J Biol Chem. 2005;280(40):33839–46. [DOI] [PubMed] [Google Scholar]
- 9. Niewolik D, Pannicke U, Lu H, Ma Y, Wang LC, Kulesza P, Zandi E, Lieber MR, Schwarz K. DNA-PKcs dependence of Artemis endonucleolytic activity, differences between hairpins and 5’ or 3’ overhangs. J Biol Chem. 2006;281(45):33900–9. [DOI] [PubMed] [Google Scholar]
- 10. Ma Y, Pannicke U, Schwarz K, Lieber MR. Hairpin opening and overhang processing by an Artemis/DNA-dependent protein kinase complex in nonhomologous end joining and V(D)J recombination. Cell 2002;108(6):781–94. [DOI] [PubMed] [Google Scholar]
- 11. Ma Y, Schwarz K, Lieber MR. The Artemis:DNA-PKcs endonuclease cleaves DNA loops, flaps, and gaps. DNA Repair 2005;4(7):845–51. [DOI] [PubMed] [Google Scholar]
- 12. Goodarzi AA, Yu Y, Riballo E, Douglas P, Walker SA, Ye R, Harer C, Marchetti C, Morrice N, Jeggo PA, Lees-Miller SP. DNA-PK autophosphorylation facilitates Artemis endonuclease activity. EMBO J. 2006;25(16):3880–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Li S, Chang HH, Niewolik D, Hedrick MP, Pinkerton AB, Hassig C, Schwarz K, Lieber MR. Evidence that the DNA endonuclease ARTEMIS also has intrinsic 5’ exonuclease activity. J Biol Chem. 2014;289(11):7825–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Povirk LF, Zhou T, Zhou R, Cowan MJ, Yannone SM. Processing of 3’-phosphoglycolate-terminated DNA double strand breaks by Artemis nuclease. J Biol Chem. 2007;282(6):3547–58. [DOI] [PubMed] [Google Scholar]
- 15. Malyarchuk S, Castore R, Shi R, Harrison L. Artemis is required to improve the accuracy of repair of double-strand breaks with 5’-blocked termini generated from non-DSB-clustered lesions. Mutagenesis 2013;28(3):357–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mohapatra S, Yannone SM, Lee SH, Hromas RA, Akopiants K, Menon V, Ramsden DA, Povirk LF. Trimming of damaged 3’ overhangs of DNA double-strand breaks by the Metnase and Artemis endonucleases. DNA Repair 2013;12(6):422–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Beucher A, Birraux J, Tchouandong L, Barton O, Shibata A, Conrad S, Goodarzi AA, Krempler A, Jeggo PA, Lobrich M. ATM and Artemis promote homologous recombination of radiation-induced DNA double-strand breaks in G2. EMBO J. 2009;28(21):3413–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Moscariello M, Wieloch R, Kurosawa A, Li F, Adachi N, Mladenov E, Iliakis G. Role for Artemis nuclease in the repair of radiation-induced DNA double strand breaks by alternative end joining. DNA Repair 2015;31:29–40. [DOI] [PubMed] [Google Scholar]
- 19. Zhang X, Succi J, Feng Z, Prithivirajsingh S, Story MD, Legerski RJ. Artemis is a phosphorylation target of ATM and ATR and is involved in the G2/M DNA damage checkpoint response. Mol Cell Biol. 2004;24(20):9207–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Geng L, Zhang X, Zheng S, Legerski RJ. Artemis links ATM to G2/M checkpoint recovery via regulation of Cdk1-cyclin B. Mol Cell Biol. 2007;27(7):2625–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yasaei H, Slijepcevic P. Defective Artemis causes mild telomere dysfunction. Genome Integr. 2010;1(1):3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wang J, Pluth JM, Cooper PK, Cowan MJ, Chen DJ, Yannone SM. Artemis deficiency confers a DNA double-strand break repair defect and Artemis phosphorylation status is altered by DNA damage and cell cycle progression. DNA Repair 2005;4(5):556–70. [DOI] [PubMed] [Google Scholar]
- 23. Evans PM, Woodbine L, Riballo E, Gennery AR, Hubank M, Jeggo PA. Radiation-induced delayed cell death in a hypomorphic Artemis cell line. Hum Mol Genet. 2006;15(8):1303–11. [DOI] [PubMed] [Google Scholar]
- 24. Darroudi F, Wiegant W, Meijers M, Friedl AA, van der Burg M, Fomina J, van Dongen JJ, van Gent DC, Zdzienicka MZ. Role of Artemis in DSB repair and guarding chromosomal stability following exposure to ionizing radiation at different stages of cell cycle. Mutat Res. 2007;615(1–2):111–24. [DOI] [PubMed] [Google Scholar]
- 25. Mohapatra S, Kawahara M, Khan IS, Yannone SM, Povirk LF. Restoration of G1 chemo/radioresistance and double-strand-break repair proficiency by wild-type but not endonuclease-deficient Artemis. Nucleic Acids Res. 2011;39(15):6500–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sridharan DM, Whalen MK, Almendrala D, Cucinotta FA, Kawahara M, Yannone SM, Pluth JM. Increased Artemis levels confer radioresistance to both high and low LET radiation exposures. Radiat Oncol. 2012;7(1):96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Liu H, Sun X, Zhang S, Ge W, Zhu Y, Zhang J, Zheng S. The dominant negative mutant Artemis enhances tumor cell radiosensitivity. Radiother Oncol. 2011;101(1):66–72. [DOI] [PubMed] [Google Scholar]
- 28. Musio A, Marrella V, Sobacchi C, Rucci F, Fariselli L, Giliani S, Lanzi G, Notarangelo LD, Delia D, Colombo R, Vezzoni P, Villa A. Damaging-agent sensitivity of Artemis-deficient cell lines. Eur J Immunol. 2005;35(4):1250–6. [DOI] [PubMed] [Google Scholar]
- 29. Kurosawa A, Koyama H, Takayama S, Miki K, Ayusawa D, Fujii M, Iiizumi S, Adachi N. The requirement of Artemis in double-strand break repair depends on the type of DNA damage. DNA Cell Biol. 2008;27(1):55–61. [DOI] [PubMed] [Google Scholar]
- 30. Riballo E, Kuhne M, Rief N, Doherty A, Smith GC, Recio MJ, Reis C, Dahm K, Fricke A, Krempler A, Parker AR, Jackson SP, Gennery A, Jeggo PA, Lobrich M. A pathway of double-strand break rejoining dependent upon ATM, Artemis, and proteins locating to gamma-H2AX foci. Mol Cell 2004;16(5):715–24. [DOI] [PubMed] [Google Scholar]
- 31. Kocher S, Rieckmann T, Rohaly G, Mansour WY, Dikomey E, Dornreiter I, Dahm-Daphi J. Radiation-induced double-strand breaks require ATM but not Artemis for homologous recombination during S-phase. Nucleic Acids Res. 2012;40(17):8336–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Rupnow BA, Knox SJ. The role of radiation-induced apoptosis as a determinant of tumor responses to radiation therapy. Apoptosis 1999;4(2):115–43. [DOI] [PubMed] [Google Scholar]
- 33. Krystal GW, Sulanke G, Litz J. Inhibition of phosphatidylinositol 3-kinase-Akt signaling blocks growth, promotes apoptosis, and enhances sensitivity of small cell lung cancer cells to chemotherapy. Mol Cancer Ther. 2002;1(11):913–22. [PubMed] [Google Scholar]
- 34. Ohtsuka T, Liu XF, Koga Y, Kitajima Y, Nakafusa Y, Ha CW, Lee SW, Miyazaki K. Methylation-induced silencing of ASC and the effect of expressed ASC on p53-mediated chemosensitivity in colorectal cancer. Oncogene 2006;25(12):1807-11. [DOI] [PubMed] [Google Scholar]
- 35. Ravizza R, Gariboldi MB, Passarelli L, Monti E. Role of the p53/p21 system in the response of human colon carcinoma cells to doxorubicin. BMC Cancer 2004;4:92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zhang X, Zhu Y, Geng L, Wang H, Legerski RJ. Artemis is a negative regulator of p53 in response to oxidative stress. Oncogene 2009;28(22):2196–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Slichenmyer WJ, Nelson WG, Slebos RJ, Kastan MB. Loss of a p53-associated G1 checkpoint does not decrease cell survival following DNA damage. Cancer Res. 1993;53(18):4164–8. [PubMed] [Google Scholar]