Abstract
This study aimed to explore the effects of lncRNA BCAR4 on the viability and aggressiveness of non-small cell lung cancer (NSCLC) cells. qRT-PCR was used to determine the expression of BCAR4 and GLI2 downstream genes in NSCLC tissues and cell lines. Chromatin isolation by RNA purification (CHIRP) and Western blot were employed to measure the expression of the GLI2 downstream proteins. Ki-67 expression in nude mice tumors was tested by immunohistochemistry. MTT assay, wound healing assay, and Transwell assay were used to assess NSCLC cell viability and aggressiveness, respectively. Tumor xenograft was conducted to determine the effects of BCAR4 and GLI2 on NSCLC tumorigenesis in vivo. The expression of BCAR4 in NSCLC tissues and cells was significantly higher than the normal level. The overexpression of BCAR4 promoted NSCLC cell viability, migration, and invasion. The suppression of BCAR4 and GLI2 showed the opposite effects. The overexpression of BCAR4 led to an increase in the expression of GLI2 downstream proteins, while the suppression of BCAR4 and GLI2 reduced their expression. In a tumor xenograft assay, the tumors in mice of the BCAR4 group showed the biggest volume, while those in mice of the si-GLI2 group showed the smallest volume. Ki-67 showed much higher levels in the BCAR4 overexpression group but much lower levels in the si-GLI2 group. In summary, the cooperative mechanism of lncRNA BCAR4 and GLI2 might provide a new opportunity for treating NSCLC.
Key words: GLI2, Long noncoding RNA BCAR4, Non-small cell lung cancer (NSCLC)
INTRODUCTION
Approximately 222,500 new cases of lung and bronchus cancers were diagnosed and around 155,870 cases of related deaths were confirmed in the US recently1. Non-small cell lung cancer (NSCLC) is a major subtype of lung cancer. Nearly 70%–80% of lung cancer cases are NSCLC2, indicating its significant prevalence. Despite advances in early diagnosis and therapeutic methods for lung cancer, the 5-year survival rate of lung cancer still remains poor at about 18%1,3,4. The high incidence of death is mainly due to the occurrence of invasion and metastasis in NSCLC5. Therefore, a detailed understanding of the potential invasion and metastasis mechanism of NSCLC progression is urgently needed.
A plethora of long noncoding RNAs (lncRNAs) are associated with tumorigenesis. For instance, lncRNA PANDAR was identified as a pancreatic ductal adenocarcinoma facilitator6, whereas lncRNA CASC2 acted as a gastric cancer suppressor7. lncRNA breast cancer antiestrogen resistance 4 (BCAR4) was initially identified as antiestrogen resistant in breast cancer cells8. Recent studies have suggested that BCAR4 could act as a promoter of various human malignancies. For instance, BCAR4 promoted cell proliferation and migration of chondrosarcoma9. The upregulation of BCAR4 predicted a poor prognosis of patients with colorectal cancer10. Li et al. reported that BCAR4 promoted NSCLC cell proliferation and invasion through affecting epithelial–mesenchymal transition5. Increased expression of BCAR4 was significantly associated with a poorer 5-year overall survival rate for NSCLC patients11. Although previous studies have looked at the functions of BCAR4 in NSCLC, the detailed mechanism through which BCAR4 regulates NSCLC cell progression and prognosis still remains unclear and requires further exploration.
Glioma-associated oncogene 2 (GLI2) is a primary transcriptional activator of the sonic hedgehog (Shh) signaling pathway, which plays a crucial role during embryonic development and tumor formation12,13. Recent studies have demonstrated that aberrant expression of GLI2 is involved in many neoplasms. For instance, Xu et al. reported that GLI2 enhanced glioma cell proliferation via the miR-124/AURKA axis14. Nagao-Kitamoto et al. revealed that the knockdown of GLI2 inhibited the metastasis of osteosarcomas15. Also, networking of the lncRNA BCAR4 and GLI2 in several malignancies has been reported. For example, it was found that BCAR4 promoted osteosarcoma advancement by inducing gene transcription dependent on GLI2 16. Xing et al. found that BCAR4 contributed to breast cancer cell aggressiveness by regulating the Shh pathway17. However, the biological significance of GLI2 in NSCLC still remains to be investigated.
We aimed to study the effects of lncRNA BCAR4 on NSCLC cells. The expression of BCAR4 and GLI2-related proteins in NSCLC tissues and cell lines was verified. The correlation between BCAR4 and clinicopathological characteristics of patients was also analyzed. Meanwhile, we explored the biological roles of BCAR4 in NSCLC cells. We elucidated the relationship between lncRNA BCAR4 and GLI2 downstream proteins and revealed how the interaction between BCAR4 and GLI2 influences NSCLC cell activities.
MATERIALS AND METHODS
Tissue Samples Collection
A total of 64 cases of NSCLC tissues and the adjacent tissues were obtained from patients who underwent surgeries at The Second Affiliated Hospital of Mudanjiang Medical University from February 2015 to June 2016. The patients who had received radiotherapy or chemotherapy before the operation were excluded from our study. We obtained the approval of the medical ethics committee of The Second Affiliated Hospital of Mudanjiang Medical University before the study. Informed consent was obtained from all patients.
Cell Culture
The normal human bronchial epithelium cell line BEAS-2B and NSCLC cell lines A549 and HCC827 were purchased from BeNa Culture Collection (BNCC; Suzhou, P.R. China). All cell lines were cultured in high-glucose Dulbecco’s modified Eagle’s medium (H-DMEM) with 10% fetal bovine serum (FBS; Sigma-Aldrich, Cambridge, MA, USA).
qRT-PCR
The TRIzol (Invitrogen, Carlsbad, CA, USA) extraction method was used to obtain total RNA from the tissue samples and cells. After overall quality of RNA was assessed by the agarose gel electrophoresis (AGE) method, total RNA was reverse transcribed into cDNA by utilizing TaKaRa PrimeScript RT reagent Kit (TaKaRa, Beijing, P.R. China), followed by qRT-PCR using SYBR Premix Ex Taq (TaKaRa). GAPDH was used as the internal control. The relative expression of mRNA and lncRNA was calculated using the 2−ΔΔCT method. The primers used for qRT-PCR are shown in Table 1.
Table 1.
Primers Used in qPCR
| Primer Name | Sequence (5′–3′) |
|---|---|
| BCAR4 forward | 5′-TACAACCACTGCACTACCTG-3′ |
| BCAR4 reverse | 5′-TGGAATGCTTGAAGGCTGCT-3′ |
| GLI2 forward | 5′-GTGAGGCAACTCCAAGGT-3′ |
| GLI2 reverse | 5′-CAGATGGACGTGCGATGT-3′ |
| RPS3 forward | 5′-AACTGGTAAGATTGGCCCTAAGAAG-3′ |
| RPS3 reverse | 5′-TGTTATGCTGTGGGGACTGG-3′ |
| IL-6 forward | 5′-AGACAGCCACTCACCTCTTCAG-3′ |
| IL-6 reverse | 5′-TTCTGCCAGTGCCTCTTTGCTG-3′ |
| MUC5AC forward | 5′-CCACTGGTTCTATGGCAACACC-3′ |
| MUC5AC reverse | 5′-GCCGAAGTCCAGGCTGTGCG-3′ |
| TGF-β1 forward | 5′-TACCTGAACCCGTGTTGCTCTC-3′ |
| TGF-β1 reverse | 5′-GTTGCTGAGGTATCGCCAGGAA-3′ |
| GAPDH forward | 5′-ACCTGACCTGCCGTCTAGAA-3′ |
| GAPDH reverse | 5′-TCCACCACCCTGTTGCTGTA-3′ |
Western Blot
The protein concentration of protein lysate was quantified using the BCA Protein Assay Kit (BioTek Corporation, Beijing, P.R. China). Proteins were separated on an SDS-PAGE gel. Subsequently, the separated protein blots were blotted to a PVDF membrane. The membrane was then blocked in 5% skimmed milk (milk powder dissolved in TBST buffer) for 5 min. Thereafter, primary antibodies were incubated with the membrane overnight at 4°C. The primary antibodies were as follows: mouse anti-human RPS3, IL-6, MUC5AC, TGF-β1, and GAPDH (1:500; Cell Signaling Technology Inc., Beverly, MA, USA). The membranes were cleaned and then incubated with Peroxidase AffiniPure rabbit anti-mouse IgG (1:2,000; Cell Signaling Technology). The densitometry of the bands was determined by the ImageJ2X software.
Cell Transfection
The pcDNA3.1 plasmid vectors, BCAR4 siRNA, and GLI2 siRNA were obtained from Sangon Biotech Co., Ltd (Shanghai, P.R. China). The plasmid constructs were transfected into A549 and HCC827 cells using Lipofectamine 2000 (Invitrogen). The experimental groups were as follows: lncRNA BCAR4 group (pcDNA3.1-lncRNA BCAR4 plasmid-transfected group), negative control (NC) group (pcDNA3.1 empty plasmid-transfected group), si-BCAR4 group (BCAR4 siRNA-transfected group), si-GLI2 group (GLI2 siRNA-transfected group), and si-GLI2 + BCAR4 group.
MTT Assay
One day after transfection, cells were seeded into a 96-well plate (1 × 104 cells/well). At 0, 24, 48, and 72 h after the inoculation, 20 μ1 of MTT solution (5 mg/ml; Sigma-Aldrich, P.R. China-Mainland) was added to each well. Four hours after the addition of the MTT solution, the medium was discarded, and 150 μ1 of DMSO was added into each well. Thereafter, the optical density (OD at 490 nm) of the cells was measured.
Wound Healing Assay
A549 cells grew in a 24-well plate (5 × 105 cells/well) for 1 day to reach 70–80% confluence as a monolayer. The monolayers were scratched using new 1-ml pipette tips. The wells were cleaned using PBS several times to remove the detached cells. Subsequently A549 cells were grown for an additional 18 h. After that, the cells were fixed in 3.7% paraformaldehyde for 0.5 hour. The monolayers were photographed under a microscope.
Transwell Assay
Matrigel (50 mg/L; Sigma-Aldrich) was diluted in serum-free media in a proportion of 1:6. Sixty microliters of the mixture was added to each Transwell chamber and stored in a 37°C incubator overnight. Residual fluid in the chambers was discarded before 200 μ1 of serum-free medium was added.
Five hundred microliters of complete media containing 10% FBS was added to each well of the 24-well plate. Two hundred microliters of cell suspension (1 × 106/ml) was then added into the Matrigel-coated inserts. The inserts were taken out after 48 h and stained with crystal violet. The invaded cells were observed under a microscope (200×).
Dual-Luciferase Reporter Gene Assay
Cells were seeded into 96-well plates (100 μl/well) and incubated at 37°C overnight. Plasmid vector pcDNA3.1 (0.2 μg) and FuGENE HD (0.3 μl) were purchased from Promega (Beijing, P.R. China). Wild-type (WT) and mutated (Mut)-type BCAR4 3′-untranslated region (3′-UTR) were, respectively, inserted into the XhoI/NotI sites of pcDNA3.1 to construct recombinant sequences. Afterward, the A549 and HCC827 cells were cotransfected. At 48 h after transfection, the relative luciferase activity was confirmed following the Dual-Luciferase Reporter Assay Kit instructions (Promega). Firefly luciferase activity was normalized relative to Renilla luciferase.
Chromatin Isolation by RNA Purification (CHIRP)
CHIRP assay was conducted as previously described18 with minor modifications using the Magna CHIRP RNA Interactome Kit (Millipore, Frankfurt, Germany). The sequence of BCAR4 probes from a previous study was used19. The isolated DNA was quantitated by qRT-PCR. LacZ was used as the NC. The primers used for CHIRP are shown in Table 2.
Table 2.
Primers Used in Chromatin Isolation by RNA Precipitation (CHIRP)
| Primer Name | Sequence (5′–3′) |
|---|---|
| RPS3 forward | 5′-AATGAGTTTCACTGAGGGAT-3′ |
| RPS3 reverse | 5′-CAAAACAAGCAATGTGAGGT-3′ |
| IL-6 forward | 5′-TCTGCA AGATGCCACAAGGT-3′ |
| IL-6 reverse | 5′-TGAAGCCCACTTGGTTCAGG-3′ |
| MUC5AC forward | 5′-GAAACTGGGCTCTACCCGGCAGACA-3′ |
| MUC5AC reverse | 5′-CCTGTCCCTCAGCAGCCTCTGA-3′ |
| TGF-β1 forward | 5′-TCCCCAGCTAGCTAGAACAGA-3′ |
| TGF-β1 reverse | 5′-CTCCTTCCATAGCTTTGCCCA-3′ |
| LacZ forward | 5′-GGTCTATATAAGCAGAGCTG-3′ |
| LacZ reverse | 5′-GTGGTATGGCTGATTATGATCAG-3′ |
Tumor Xenograft in Nude Mice
Sixteen female BALB/c nude mice (4 to 6 weeks old) that were bred with specific pathogen-free (SPF) conditions were divided into four groups. Mice in different groups were injected with A549 cells from the NC group, BCAR4 group, si-GLI2 group, and si-GLI2 + BCAR4 group, respectively. Briefly, 100 μ1 of cell suspension (1.5 × 106 cells) was subcutaneously injected into the back of each nude mouse. The tumor growth was recorded every 3 days. The volume of the tumor was calculated by the formula: volume = [(length + width)/2] × 3 × 0.5236.
Immunohistochemistry
The 4-μm-thick paraffin-embedded mice tumor tissues were dewaxed by xylene and washed with PBS. Antigen retrieval was thermally induced (microwave heating, 96°C, for 10 min, repeated twice) in PBS, and 0.3% hydrogen peroxide–methanol was used to block endogenous peroxidase activity. Bovine serum albumin (BSA; 5%) was incubated with the tissues for 30 min to block nonspecific antibody fixation. The primary antibody (mouse anti-human Ki-67 monoclonal antibody; 1:200; Sigma-Aldrich) was incubated with tissues at 4°C overnight. On the next day, the tissues were washed and incubated with biotin-labeled rabbit anti-mouse IgG (1:200; Sigma-Aldrich). 3,3′-Diaminobenzidine (DAB; ab64238; Abcam) was used to stain the tissues. Hematoxylin was used for counterstaining. The results were observed using a microscope (OLYMPUS BX-41; Tokyo, Japan).
Statistical Analysis
Data are presented as mean ± standard deviation. Data analyses were performed using the statistical software SPSS 19.0. Diagrams were produced using the GraphPad Prism 6.0 software. The t-test and ANOVA were used to analyze the significance of difference between groups. A value of p < 0.05 was considered as statistically significant.
RESULTS
The Expression of BCAR4 in NSCLC Tissues and Cell Lines
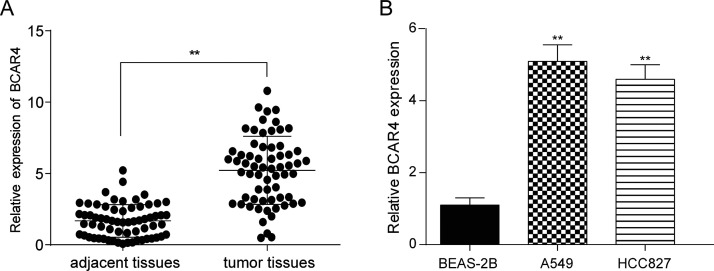
Results of the qRT-PCR showed that the expression of BCAR4 was significantly higher in NSCLC tissues than in adjacent normal tissues (p < 0.01) (Fig. 1A). In addition, the level of BCAR4 in the A549 and HCC827 cells was significantly higher than that in the BEAS-2B cells (p < 0.01) (Fig. 1B). In addition, the relationship between the expression of BCAR4 and clinical pathology was tested, and the results showed that there was no significant correlation with sex, age, cancer type, and smoking history (p > 0.05), but was significantly correlated with lymph metastasis (p < 0.05) (Table 3).
Figure 1.
Long noncoding RNA breast cancer antiestrogen resistance 4 (lncRNA BCAR4) was highly expressed in non-small cell lung cancer (NSCLC) tissues and cells. (A) The expression level of lncRNA BCAR4 was significantly higher in NSCLC tissues than in adjacent tissues. **p < 0.01, compared with adjacent tissues. (B) The expression level of lncRNA BCAR4 was significantly higher in NSCLC cell lines A549 and HCC827 than in normal human bronchial epithelium cells BEAS-2B. **p < 0.01, compared with human bronchial epithelium cells.
Table 3.
Relationship Between Expression of lncRNA BCAR4 and Clinical Pathology
| Pathological Feature | Sample No. | lncRNA BCAR4 [n (%)] | p Value | |
|---|---|---|---|---|
| Low Expression | High Expression | |||
| Sex | 0.4887 | |||
| Male | 35 | 6 (40.3%) | 29 (59.7%) | |
| Female | 29 | 7 (36.4%) | 22 (63.6%) | |
| Age (years) | 0.2256 | |||
| <60 | 24 | 8 (62.5%) | 16 (37.5%) | |
| ≥60 | 40 | 5 (65.5%) | 35 (37.5%) | |
| Cancer type | 0.8856 | |||
| SCC | 30 | 6 (61.1%) | 24 (38.9%) | |
| AC | 34 | 7 (63.1%) | 37 (36.9%) | |
| Smoking history | 0.9003 | |||
| Positive | 40 | 9 (63.4%) | 31 (36.6%) | |
| Negative | 24 | 4 (60.9%) | 20 (39.1%) | |
| Lymph metastasis | 0.0312* | |||
| Positive | 41 | 5 (63.4%) | 36 (36.6%) | |
| Negative | 23 | 8 (60.9%) | 15 (39.1%) | |
SCC, squamous cell carcinoma; AC, adenomatous carcinoma.
p < 0.05, chi-square test.
Effects of BCAR4 Modulation on NSCLC Cell Viability and Aggressiveness
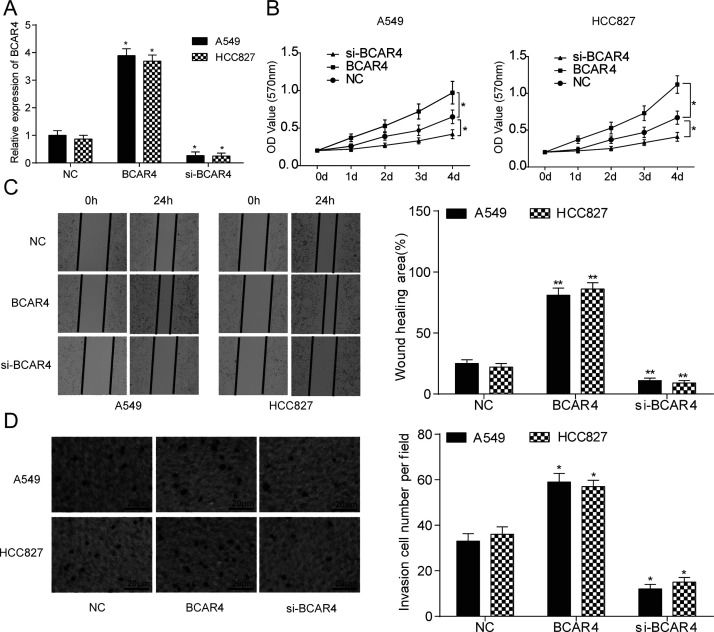
BCAR4 in the overexpression group was significantly upregulated, while in the si-BCAR4 group, it was significantly underexpressed compared with the NC group in both cell lines (p < 0.05) (Fig. 2A). The PCR results suggested that BCAR4 overexpression or knockdown cell lines had been successfully established. Results of the MTT assay demonstrated that cell viability in the BCAR4 overexpression group was significantly stronger, while that in the si-BCAR4 group was significantly weaker compared with the NC group (p < 0.05) (Fig. 2B). In both cell lines, a bigger wound healing area and a larger invaded cell number were seen in the BCAR4 overexpression group. In addition, a smaller wound healing area and a smaller invaded cell number were seen in the BCAR4 knockdown group, compared with the NC group (p < 0.05) (Fig. 2C and D).
Figure 2.
lncRNA BCAR4 promoted NSCLC cell proliferation, migration, and invasion. (A) The PCR results showed that NSCLC cell lines A549 and HCC827 were successfully transfected with BCAR4 overexpression plasmid and BCAR4 siRNA. *p < 0.05, compared with the NC group. (B) The viability of A549 and HCC827 cells was stronger in the BCAR4 group but weaker in the si-BCAR4 group. *p < 0.05, compared with the NC group. (C) The wound healing area was remarkably larger in the BCAR4 group, while the wound healing area was much smaller in the si-BCAR4 group. **p < 0.01, compared with the NC group. (D) There were significantly more invaded cells in the BCAR4 group, whereas there were less invaded cells in the si-BCAR4 group. *p < 0.05, compared with the NC group.
Effects of BCAR4 Modulation on GLI2
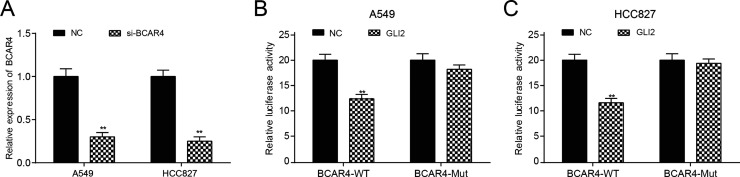
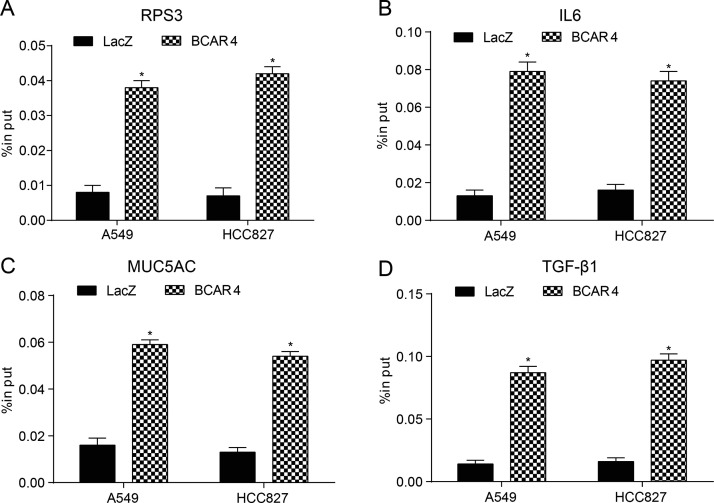
The transfection efficiency of BCAR4 siRNA was determined by qRT-PCR, as shown in Figure 3A. Moreover, overexpression of GLI2 significantly repressed the luciferase activity of the luciferase reporter containing BCAR4 3′-UTR-WT but not the reporter containing BCAR4 3′-UTR-Mut both in the A549 and HCC827 cells, as shown in the dual-luciferase reporter assay (p < 0.05) (Fig. 3B and C). According to the results of the CHIRP assay (Fig. 4A–D), GLI2 downstream protein levels of RPS3, IL-6, MUC5AC, and TGF-β1 were dramatically increased after BCAR4 was upregulated (p < 0.05), thus manifesting the regulation of BCAR4 on GLI2 downstream proteins.
Figure 3.
BCAR4 regulated glioma-associated oncogene 2 (GLI2). (A) The PCR results showed that BCAR4 knockdown was successfully established in NSCLC cell lines. **p < 0.01, compared with the NC group. The luciferase activity of BCAR4-wild type (WT) was decreased, whereas BCAR4 3′-untranslated region (3′-UTR)-Mut displayed no obvious difference in the (B) A549 and (C) HCC827 cells. **p < 0.01, compared with the NC group.
Figure 4.
BCAR4 regulated the downstream proteins of GLI2. Results of chromatin isolation by RNA precipitation (CHIRP) assay showed that levels of the GLI2 downstream proteins (A) RPS3, (B) IL-6, (C) MUC5AC, and (D) TGF-β1 were dramatically higher in the BCAR4 group. *p < 0.05, compared with the LacZ [negative control (NC)] group.
Association Between BCAR4 Expression and GLI2 Downstream Protein Expression in NSCLC Tissues
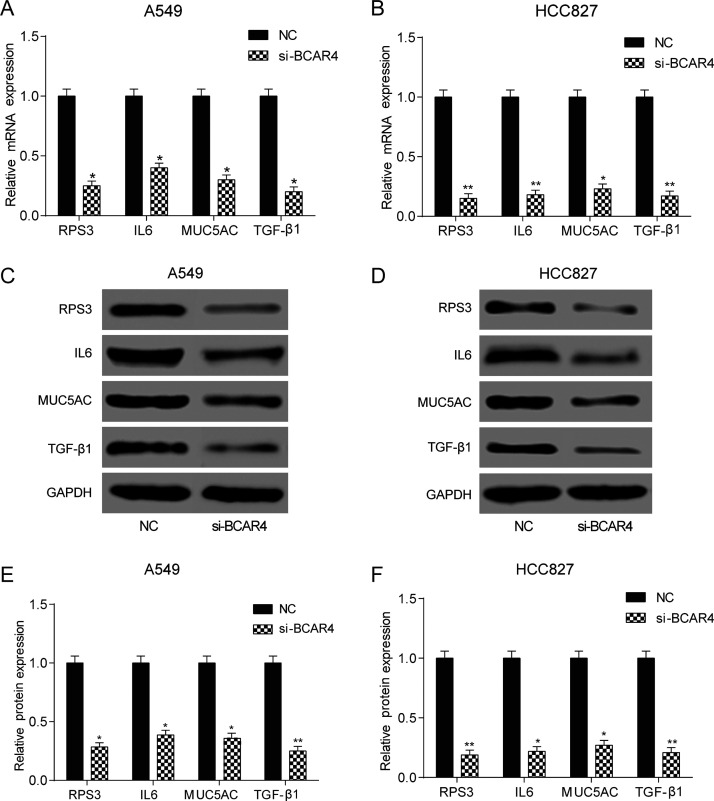
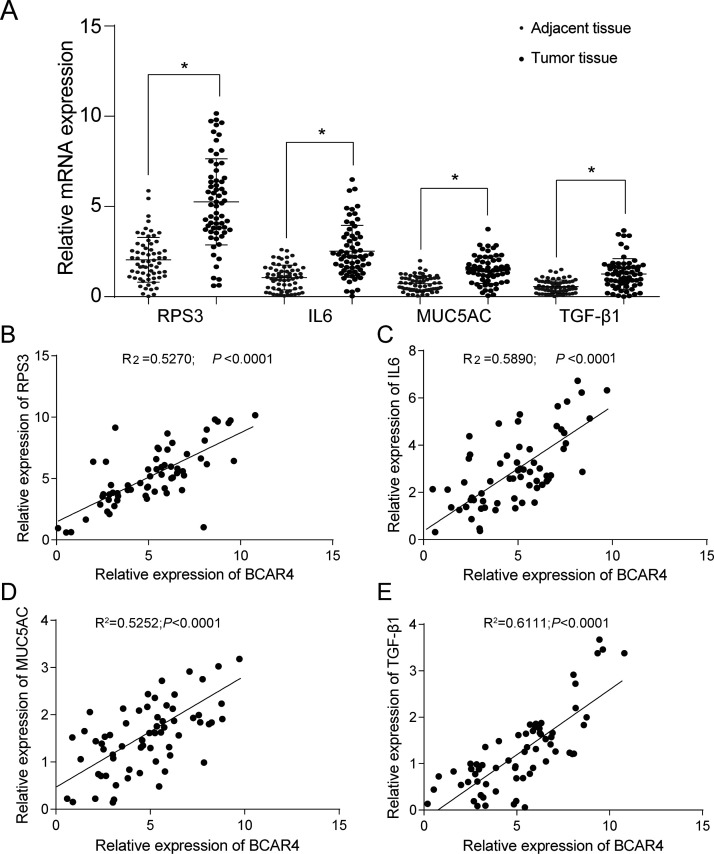
According to qRT-PCR and Western blotting, both mRNA and protein expressions of RPS3, IL-6, MUC5AC, and TGF-β1 significantly declined after the silencing of BCAR4 in both cell lines (p < 0.05) (Fig. 5A–F). Meanwhile, the RPS3, IL-6, MUC5AC, and TGF-β1 levels were significantly higher in the NSCLC tissues compared with the adjacent tissues (Fig. 6A). In addition, their expression levels were positively correlated with BCAR4 expression levels (Fig. 6B–E).
Figure 5.
BCAR4 regulated GLI2 expression. The mRNA expression of RPS3, IL-6, MUC5AC, and TGF-β1 was significantly lower in the si-BCAR4 group of (A) A549 and (B) HCC827 cells. *p < 0.05, **p < 0.01, compared with the NC group. (C–F) In both cell lines, the protein expression of RPS3, IL-6, MUC5AC, and TGF-β1 was significantly lower in the si-BCAR4 group. *p < 0.05, **p < 0.01, compared with the NC group.
Figure 6.
GLI2 downstream protein expression was positively correlated with BCAR4 expression. (A) Results of the qRT-PCR showed that the expression levels of RPS3, IL-6, MUC5AC, and TGF-β1 were much higher in NSCLC tissues than in adjacent tissues. *p < 0.05, compared with adjacent tissues. The expression levels of (B) RPS3, (C) IL-6, (D) MUC5AC, and (E) TGF-β1 were positively correlated with BCAR4 expression.
BCAR4 Promoted HCC827 Cell Viability, Migration, and Invasion by Regulating GLI2
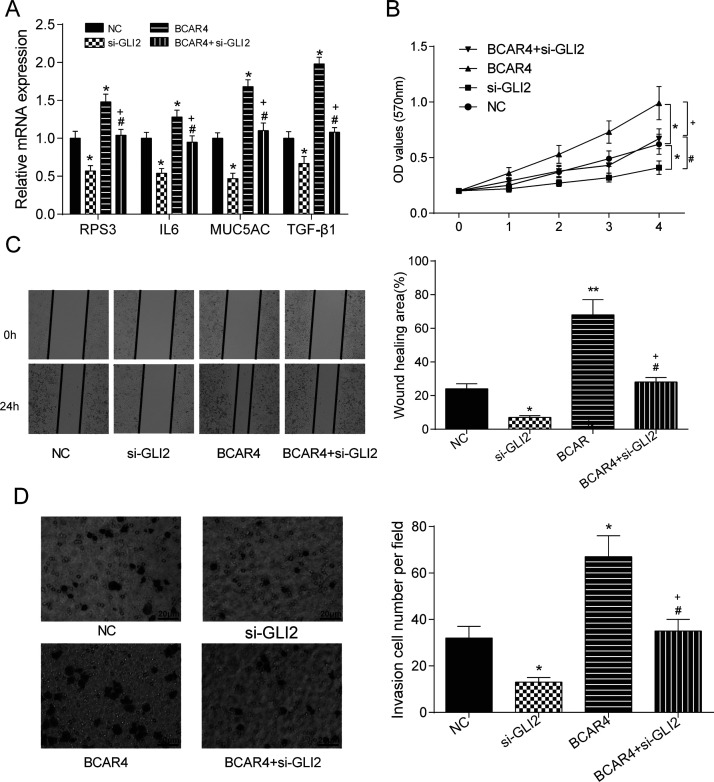
The transfection efficiency in HCC827 cells was better than that in A549 cells; therefore, HCC827 cells were selected for the following experiments. The expression levels of GLI2 downstream proteins were remarkably higher in the BCAR4 overexpression group, while the silencing of GLI2 led to the reduced levels of GLI2 downstream proteins compared with the NC group. The expression of the proteins in the BCAR4 + si-GLI2 group had no significant difference with that in the NC group, but showed significantly higher levels than that in the si-GLI2 group and lower levels than that in the BCAR4 group (p < 0.05) (Fig. 7A). Results of the MTT assay demonstrated that overexpression of BCAR4 manifestly improved cell viability, while the silencing of GLI2 inhibited cell viability, and the cell viability in the BCAR4 + si-GLI2 group was not significantly different from that in the NC group (p < 0.05) (Fig. 7B). Similarly, the overexpression of BCAR4 promoted cell aggressiveness; the knockdown of GLI2 inhibited cell aggressiveness. Yet, the aggressiveness of cells in the BCAR4 + si-GLI2 group was not significantly different from those in the NC group (p < 0.05) (Fig. 7C and D).
Figure 7.
BCAR4 promoted NSCLC cell proliferation, migration, and invasion by regulating GLI2. (A) The expression levels of GLI2 downstream proteins were remarkably higher in the BCAR4 overexpression group, while the levels of GLI2 downstream proteins were much lower in the si-GLI2 group. The expression levels of the proteins in the BCAR4 + si-GLI2 group showed no significant difference from those in the NC group. *p < 0.05, compared with the NC group; #p < 0.05, compared with the si-GLI2 group; +p < 0.05, compared with the BCAR4 group. (B) Cell viability in the BCAR4 overexpression group was dramatically improved, while cell viability in the si-GLI2 group was suppressed. *p < 0.05, compared with the NC group; #p < 0.05, compared with the si-GLI2 group; +p < 0.05, compared with the BCAR4 group. (C) The wound healing area in the BCAR4 overexpression group was manifestly larger, whereas the wound healing area in the si-GLI2 group was much smaller. *p < 0.05, **p < 0.01 compared with the NC group; #p < 0.05, compared with the si-GLI2 group; +p < 0.05, compared with the BCAR4 group. (D) There were notably more invaded cells in BCAR4 overexpression group, while much fewer invaded cells were observed in the si-GLI2 group. *p < 0.05, compared with the NC group; #p < 0.05, compared with the si-GLI2 group; +p < 0.05, compared with the BCAR4 group.
BCAR4 Promoted Tumor Growth by Regulating GLI2 In Vivo
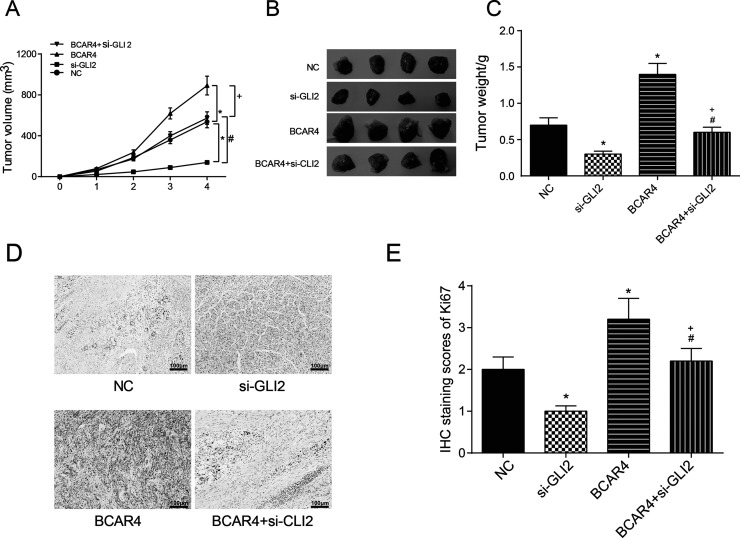
Tumor volumes and weights were more significant in the BCAR4 overexpression group but were less significant in the si-GLI2 group compared with the NC group. The tumor volumes and weights of mice in the si-GLI2 + BCAR4 group were not different from those of the NC group (Fig. 8A–C). The expression of Ki-67 was highest in the BCAR4 group and lowest in the si-GLI2 group. No significant difference was seen in the tumors of mice in the si-GLI2 + BCAR4 group compared with those in the NC group (Fig. 8D and E), suggesting that BCAR4 promoted tumor growth in vivo. The overexpression of BCAR4 led to the increase in the expression of GLI2 downstream proteins, while the suppression of BCAR4 and GLI2 reduced their expression. In tumor xenograft assay, tumors in mice of the BCAR4 group showed the biggest volume, while those in mice of the si-GLI2 group showed the smallest volume. Ki-67 showed much higher levels in the BCAR4 overexpression group but much lower levels in the si-GLI2 group.
Figure 8.
BCAR4 promoted NSCLC development in vivo by regulating GLI2. (A–C) The tumors in nude mice were significantly larger and heavier in the BCAR4 overexpression group, while the tumors were much smaller and lighter in the si-GLI2 group compared with the NC group. *p < 0.05, compared with the NC group; #p < 0.05, compared with the si-GLI2 group; +p < 0.05, compared with the BCAR4 group. (D, E) The expression of Ki-67 in nude mice tumor samples was significantly higher in the BCAR4 overexpression group, while it was much lower in the si-GLI2 group. *p < 0.05, compared with the NC group. #p < 0.05, compared with the si-GLI2 group; +p < 0.05, compared with the BCAR4 group.
DISCUSSION
BCAR4 was overexpressed in NSCLC tissues and cell lines compared with adjacent tissues and a normal bronchial epithelium cell line, respectively. In addition, the overexpression of BCAR4 promoted NSCLC cell proliferation and aggressiveness. BCAR4 was positively correlated with GLI2 downstream proteins in NSCLC cell lines and tissues. BCAR4 promoted NSCLC cell progression and aggressiveness through GLI2.
Aberrant expression of various lncRNAs in NSCLC has been previously reported. For instance, lncRNA 00312 was downregulated in NSCLC tissues and impeded cell proliferation in vitro and in vivo20, whereas lncRNA BLACAT1 was upregulated in NSCLC and promoted cell metastasis through sponging miR-14421. BCAR4 was first discovered in breast cancer and might be resistant to the anticancer drug tamoxifen5. Later, van Agthoven et al. found that BCAR4 was associated with high proliferation in primary breast cancers and promoted the proliferation of IPH-926 lobular carcinoma cells22. Elevated expression of BCAR4 had also been found in NSCLC, chondrosarcoma, colorectal cancer, and osteosarcoma5,9,10,16. Our study focused on the function of BCAR4 in NSCLC cell progression. The experimental results demonstrated the upregulated expression of BCAR4 in NSCLC tissues and cell lines. The overexpression of BCAR4 encouraged NSCLC cell viability and aggressiveness, which was analogous with the study of Li et al5. Taken together, we speculated that BCAR4 could be a potential NSCLC facilitator.
To further investigate the molecular mechanism of BCAR4, we identified the role of GLI2, a transcriptional factor of the Shh signaling pathway, in the process of tumorigenesis of NSCLC. The impact of GLI2 on tumorigenesis and metastasis had been explored in various tumors including hepatocellular carcinoma, glioma, renal cell carcinoma, etc.13,14,23. However, only a few studies focused on the GLI2 involvement in NSCLC. For instance, Giroux Leprieur et al. found that GLI2 was expressed in NSCLC patients and not expressed in non-NSCLC patients, which indicated that GLI2 overexpression was closely correlated with prognosis in advanced NSCLC24. Ji et al. found the evidence to connect GLI2 and the growth of lung squamous cell cancer (LSCC) stem cell-like cells, that is, GLI2 was found to cooperate with FGFR1 to promote LSCC stem cell-like cell growth25. Based on the exploration of the interaction between BCAR4 and GLI2 in other tumors, we attempted to investigate the interaction between BCAR4 and GLI2 in NSCLC. We herein found the positive relationship between BCAR4 and GLI2 downstream proteins RPS3, IL-6, MUC5AC, and TGF-β1. In addition, we detected that upregulation of BCAR4 promoted NSCLC cell proliferation and metastasis, which was neutralized by GLI2 knockdown. Therefore, we deduced that BCAR4 might synergize with GLI2 to promote the development of NSCLC.
We have confirmed the effects of BCAR4 and GLI2 interaction on NSCLC cell lines. Yet, our study still had several limitations. For example, the sample size was small. We would like to conduct further studies with a larger cohort to support our findings. In addition, BCAR4 may interact with other factors when regulating NSCLC cell progression. Thus, further studies are warranted to reach an exhaustive understanding of BCAR4’s regulation mechanism in NSCLC.
In conclusion, our study unraveled the overexpression of lncRNA BCAR4 in NSCLC tissues and cell lines. We demonstrated the positive correlation between the expression of BCAR4 and GLI2 downstream proteins. Furthermore, we explored the cooperative mechanism of BCAR4 and GLI2 in promoting NSCLC cell viability and aggressiveness. These results may provide a rational strategy for the treatment of NSCLC.
CONCLUSION
BCAR4 was overexpressed in NSCLC and promoted NSCLC cell viability and aggressiveness by targeting GLI2.
ACKNOWLEDGMENT
This study was supported by the Natural Science Foundation of Heilongjiang Province, P.R. China (No. H2015078).
Footnotes
The authors declare no conflicts of interest.
REFERENCES
- 1. Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin. 2017;67(1):7–30. [DOI] [PubMed] [Google Scholar]
- 2. Sun SJ, Lin Q, Ma JX, Shi WW, Yang B, Li F. Long non-coding RNA NEAT1 acts as oncogene in NSCLC by regulating the Wnt signaling pathway. Eur Rev Med Pharmacol Sci. 2017;21(3):504–10. [PubMed] [Google Scholar]
- 3. Wang H, Shen Q, Zhang X, Yang C, Cui S, Sun Y, Wang L, Fan X, Xu S. The long non-coding RNA XIST controls non-small cell lung cancer proliferation and invasion by modulating miR-186-5p. Cell Physiol Biochem. 2017;41(6):2221–9. [DOI] [PubMed] [Google Scholar]
- 4. Zhao Z, Wang J, Wang S, Chang H, Zhang T, Qu J. LncRNA CCAT2 promotes tumorigenesis by over-expressed Pokemon in non-small cell lung cancer. Biomed Pharmacother. 2017;87:692–7. [DOI] [PubMed] [Google Scholar]
- 5. Li N, Gao WJ, Liu NS. LncRNA BCAR4 promotes proliferation, invasion and metastasis of non-small cell lung cancer cells by affecting epithelial-mesenchymal transition. Eur Rev Med Pharmacol Sci. 2017;21(9):2075–86. [PubMed] [Google Scholar]
- 6. Jiang Y, Feng E, Sun L, Jin W, You Y, Yao Y, Xu Y. An increased expression of long non-coding RNA PANDAR promotes cell proliferation and inhibits cell apoptosis in pancreatic ductal adenocarcinoma. Biomed Pharmacother. 2017;95:685–91. [DOI] [PubMed] [Google Scholar]
- 7. Zhou J, Huang H, Tong S, Huo R. Overexpression of long non-coding RNA cancer susceptibility 2 inhibits cell invasion and angiogenesis in gastric cancer. Mol Med Rep. 2017;16(4):5235–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Weidle UH, Birzele F, Kollmorgen G, Ruger R. Long non-coding RNAs and their role in metastasis. Cancer Genomics Proteomics 2017;14(3):143–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Shui X, Zhou C, Lin W, Yu Y, Feng Y, Kong J. Long non-coding RNA BCAR4 promotes chondrosarcoma cell proliferation and migration through activation of mTOR signaling pathway. Exp Biol Med. (Maywood) 2017;242(10):1044–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Li Q, Dai Y, Wang F, Hou S. Differentially expressed long non-coding RNAs and the prognostic potential in colorectal cancer. Neoplasma 2016;63(6):977–83. [DOI] [PubMed] [Google Scholar]
- 11. Gong J, Zhang H, He L, Wang L, Wang J. Increased expression of long non-coding RNA BCAR4 is predictive of poor prognosis in patients with non-small cell lung cancer. Tohoku J Exp Med. 2017;241(1):29–34. [DOI] [PubMed] [Google Scholar]
- 12. Fu L, Wu H, Cheng SY, Gao D, Zhang L, Zhao Y. Set7 mediated Gli3 methylation plays a positive role in the activation of sonic hedgehog pathway in mammals. eLife 2016;5:e15690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Shi C, Huang D, Lu N, Chen D, Zhang M, Yan Y, Deng L, Lu Q, Lu H, Luo S. Aberrantly activated Gli2-KIF20A axis is crucial for growth of hepatocellular carcinoma and predicts poor prognosis. Oncotarget 2016;7(18):26206–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Xu L, Liu H, Yan Z, Sun Z, Luo S, Lu Q. Inhibition of the Hedgehog signaling pathway suppresses cell proliferation by regulating the Gli2/miR-124/AURKA axis in human glioma cells. Int J Oncol. 2017;50(5):1868–78. [DOI] [PubMed] [Google Scholar]
- 15. Nagao-Kitamoto H, Nagata M, Nagano S, Kitamoto S, Ishidou Y, Yamamoto T, Nakamura S, Tsuru A, Abematsu M, Fujimoto Y, Yokouchi M, Kitajima S, Yoshioka T, Maeda S, Yonezawa S, Komiya S, Setoguchi T. GLI2 is a novel therapeutic target for metastasis of osteosarcoma. Int J Cancer 2015;136(6):1276–84. [DOI] [PubMed] [Google Scholar]
- 16. Chen F, Mo J, Zhang L. Long noncoding RNA BCAR4 promotes osteosarcoma progression through activating GLI2-dependent gene transcription. Tumour Biol. 2016;37(10):13403–12. [DOI] [PubMed] [Google Scholar]
- 17. Xing Z, Park PK, Lin C, Yang L. LncRNA BCAR4 wires up signaling transduction in breast cancer. RNA Biol. 2015;12(7):681–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yang L, Lin C, Jin C, Yang JC, Tanasa B, Li W, Merkurjev D, Ohgi KA, Meng D, Zhang J, Evans CP, Rosenfeld MG. lncRNA-dependent mechanisms of androgen-receptor-regulated gene activation programs. Nature 2013;500(7464):598–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Xing Z, Lin A, Li C, Liang K, Wang S, Liu Y, Park PK, Qin L, Wei Y, Hawke DH, Hung MC, Lin C, Yang L. lncRNA directs cooperative epigenetic regulation downstream of chemokine signals. Cell 2014;159(5):1110–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhu Q, Lv T, Wu Y, Shi X, Liu H, Song Y. Long non-coding RNA 00312 regulated by HOXA5 inhibits tumour proliferation and promotes apoptosis in non-small cell lung cancer. J Cell Mol Med. 2017;21(9):2184–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ye JR, Liu L, Zheng F. Long noncoding RNA bladder cancer associated transcript 1 promotes the proliferation, migration, and invasion of nonsmall cell lung cancer through sponging miR-144. DNA Cell Biol. 2017;36(10):845–52. [DOI] [PubMed] [Google Scholar]
- 22. van Agthoven T, Dorssers LC, Lehmann U, Kreipe H, Looijenga LH, Christgen M. Breast cancer anti-estrogen resistance 4 (BCAR4) drives proliferation of IPH-926 lobular carcinoma cells. PLoS One 2015;10(8):e0136845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhou J, Zhu G, Huang J, Li L, Du Y, Gao Y, Wu D, Wang X, Hsieh JT, He D, Wu K. Non-canonical GLI1/2 activation by PI3K/AKT signaling in renal cell carcinoma: A novel potential therapeutic target. Cancer Lett. 2016;370(2):313–23. [DOI] [PubMed] [Google Scholar]
- 24. Giroux Leprieur E, Vieira T, Antoine M, Rozensztajn N, Rabbe N, Ruppert AM, Lavole A, Cadranel J, Wislez M. Sonic hedgehog pathway activation is associated with resistance to platinum-based chemotherapy in advanced non-small-cell lung carcinoma. Clin Lung Cancer 2016;17(4):301–8. [DOI] [PubMed] [Google Scholar]
- 25. Ji W, Yu Y, Li Z, Wang G, Li F, Xia W, Lu S. FGFR1 promotes the stem cell-like phenotype of FGFR1-amplified non-small cell lung cancer cells through the Hedgehog pathway. Oncotarget 2016;7(12):15118–34. [DOI] [PMC free article] [PubMed] [Google Scholar]