Abstract
Non-small cell lung cancer (NSCLC) represents the leading cause of cancer-related mortality worldwide. More and more reports have identified important roles for long noncoding RNAs (lncRNAs) in cancer development. ENST457720 expression was upregulated in lung adenocarcinoma in a microarray-based lncRNA screen. We determined the expression levels of ENST457720 in NSCLC tissues with quantitative real-time PCR and then studied their clinical significance. We explored the biological significance of ENST457720 with gain- and loss-of-function analyses in vitro and in vivo. In this study, ENST457720 was expressed at higher levels in NSCLC tissues than in paired normal tissues. Higher ENST457720 expression was associated with larger tumor sizes, lymph node metastasis, and advanced TNM stage. ENST457720 silencing suppressed NSCLC cell proliferation in vitro and in vivo. Moreover, ENST457720 knockdown inhibited NSCLC invasion and reversed the epithelial-to-mesenchymal transition. ENST457720 promoted NSCLC proliferation and invasion, which may be a novel potential therapeutic target for NSCLC.
Key words: Non-small cell lung cancer (NSCLC), Long noncoding RNAs (lncRNAs), ENST457720, Proliferation, Apoptosis
INTRODUCTION
Non-small cell lung cancer (NSCLC), mainly consisting of adenocarcinoma and squamous cell carcinoma, is the main cause of cancer-related death worldwide1. Despite the great advances in the treatment of NSCLC, the 5-year survival rate is still less than 15%2. The failure of NSCLC treatments is mainly attributed to the high incidence of cancer recurrence and metastasis1,2. Thus, studies identifying novel biomarkers and revealing the underlying molecular mechanisms of NSCLC progression are important.
Long noncoding RNAs (lncRNAs) are a class of RNAs that are longer than 200 nucleotides (nt) in length with little protein coding capacity. lncRNAs participate in various biological processes, including cell differentiation and development3, tumorigenesis4, inflammation5, and angiogenesis6. Based on accumulating evidence, lncRNAs regulate multiple hallmarks of cancer, such as proliferation4, cell metabolism7, apoptosis8, and chemoresistance9.
Expression profiles of lncRNAs in early stage human NSCLC tissues and adjacent nontumor tissues identified a number of differentially expressed lncRNAs10. According to the microarray data, ENST457720 expression was significantly upregulated in lung adenocarcinoma. ENST457720 is included in multiple public ncRNA databases, also known as lnc-ABCA1-1 (LNCipedia and Ensembl). It is located on human chromosome 9 (chr9:107753077-107754062) and contains two exons. The ENST457720 transcript is approximately 606 nt. However, the role of ENST457720 in the progression of NSCLC and its underlying mechanism remain elusive.
In this study, the level of the ENST457720 transcript was markedly increased in NSCLC tissues compared to paired adjacent normal lung tissues. Knockdown of ENST457720 attenuated cell proliferation, induced cancer cell apoptosis, and suppressed invasion by reversing the epithelial-to-mesenchymal transition (EMT) in NSCLC.
MATERIALS AND METHODS
Patient Samples
Written consent was obtained from all enrolled patients. This study was performed under the guidance of the Human Ethics Committee of Pulmonary Hospital at Tongji University (Shanghai, P.R. China) with approval number A145. A cohort of 60 NSCLC [stage IA–IIIA, adenocarcinoma (n = 32), squamous cell carcinoma (n = 28)] and pair-matched adjacent normal lung tissues (>3 cm away from the tumor) were obtained from patients who underwent surgery at Shanghai Pulmonary Hospital, Tongji University School of Medicine, between 2008 and 2011. The final diagnosis was made by two experienced pathologists. All samples were stored at −80°C before RNA isolation and quantitative real-time polymerase chain reaction (RT-qPCR) analysis. None of these patients received any preoperative treatments, including chemotherapy and radiotherapy.
Cell Culture
The normal human bronchial epithelial cell line (16HBE) and three NSCLC cell lines [NCI-H1975 (EGFR mutations: L858R + T790M), A549 (wild-type EGFR), and SK-MES-1 (exon 19 deletion + T790M)] were obtained from the Chinese Academy of Sciences (Shanghai, P.R. China). Cell culture was performed using previously described methods11.
RNA Extraction and Quantitative Real-Time Polymerase Chain Reaction
RNA extraction and RT-qPCR were performed as previously described11. The primer sequences were β-actin, 5′-GCTTCGGCAGCACATATACTAA-3′ (forward) and 5′-AACGCTTCACGAATTTGCGT-3′ (reverse); glyceraldehyde-3-phosphate dehydrogenase (GAPDH), 5′-TGTGTCCGTCGTGGATCTGA-3′ (forward) and 5′-CCTGCTTCACCACCTTCTTGA-3′ (reverse); and ENST457720, 5′-GCAAGTGTTGAAGGTACGACAA-3′ (forward) and 5′-AGATGCAATGGAGGCATACTAAA-3′ (reverse).
Cell Proliferation Assay, Transwell Assay, and Western Blot Analysis
The cell proliferation assays, Transwell assays, and Western blot analyses were performed using previously described methods11. The following primary antibodies were used: E-cadherin (1:400; Abcam, Cambridge, UK), vimentin (1:600; Abcam), and GAPDH (1:2,000; Abcam).
Absolute Quantitative PCR
Briefly, we first chemically synthesized ENST457720 (GeneChem, Shanghai, P.R. China), introduced it into Escherichia coli, and sequenced it to verify that no variations occurred. After the expansion of Escherichia coli, we extracted the plasmids. We then constructed the standard curve with serially diluted RT-qPCR products of ENST457720 used as templates.
Plasmid and Transfection
The synthesized full-length ENST457720 sequence (GeneChem) was subcloned into pcDNA3.1 vectors (Invitrogen, Grand Island, NY, USA) for overexpression. For ENST457720 silencing, siRNAs targeting ENST457720 were synthesized with the following nucleotide sequences: siRNA1, GGAAGGUUGACAUAAU; siRNA2, GAGAGACAUAGAAAU (GeneChem). When the cells reached 80% confluence, transfections were performed using Lipofectamine 3000 (Invitrogen).
Cell Cycle Analysis
Cells transiently transfected with the desired vector were harvested 48 h after transfection. The cells were washed three times with cold PBS and were then fixed in 70% ethanol in PBS at −20°C for 24 h. After fixation, cells were washed with cold PBS and stained with 0.5 ml of propidium iodide (PI) staining buffer, which contains 200 mg/ml RNase A and 50 μg/ml PI, at 37°C for 30 min at 4°C in the dark. The cell cycle was assessed using a flow cytometer (FACSCalibur, BD Biosciences, San Jose, CA, USA) after PI staining. Data are expressed as a percent distribution of cells in the G0/G1, S, and G2/M phases of the cell cycle.
Nude Mouse Xenograft Model
The animal experiments were approved by the Animal Experiment Center of Shanghai Pulmonary Hospital, Tongji University School of Medicine. The experiments were carried out in accordance with the National Institutes of Health guide for the care and use of Laboratory animals (NIH Publications No. 8023, revised 1978). NCI-H1975 cells in which ENST457720 was stably knocked down or control cells (1 × 106/0.1 ml) were subcutaneously injected into 5-week-old nude mice (female, n = 4 each group). The tumor volume was determined weekly using the following formula: 0.5 × length × width2. Four weeks after the cell inoculation, the nude mice were euthanized and photographed.
Statistical Analysis
All statistical analyses were performed using SPSS 17.0 (SPSS, Chicago, IL, USA). Data are presented as the means ± standard deviation (SD). Student’s t-test was used to compare differences unless indicated otherwise. The Mann–Whitney test was employed to determine the association between ENST457720 expression and clinical characteristics. A value of p < 0.05 was considered significant.
RESULTS
ENST457720 Is Expressed at Higher Levels in NSCLC Tissues
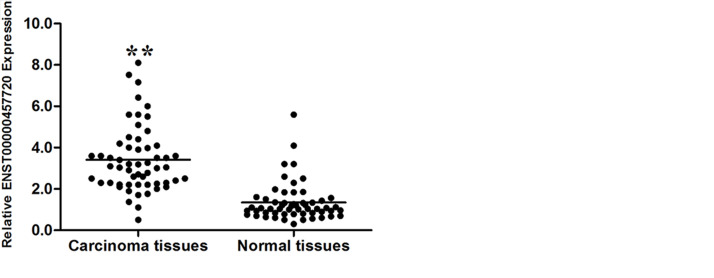
The expression pattern of ENST457720 in NSCLC cell lines (including the adenocarcinoma and squamous carcinoma subtypes) was measured to confirm that ENST457720 expression was upregulated in NSCLC. ENST457720 expression was upregulated in NSCLC cells, including A549, NCI-H1975, and SK-MES-1 cells, compared to 16HBE cells, the normal bronchial epithelial cell line (Fig. 1A). Afterward, ENST457720 expression was determined in 60 pairs of NSCLC tissues and matched normal tissues using RT-qPCR. ENST457720 expression was significantly upregulated in NSCLC tissues compared to paired normal tissues (p < 0.05) (Fig. 1B). Furthermore, the correlation between ENST457720 expression and clinicopathological factors was analyzed. Higher levels of ENST457720 in NSCLC tissues were associated with larger tumor sizes (p = 0.020, p < 0.05), lymph node metastasis (p = 0.001, p < 0.05), and advanced TNM stage (p = 0.035, p < 0.05); however, no significant correlations with age, gender, and smoking history were observed.
Figure 1.
ENST457720 expression in non-small cell lung cancer (NSCLC) cell lines and cancer tissues. (A) Quantitative real-time polymerase chain reaction (RT-qPCR) analysis of ENST457720 expression levels in NSCLC cell lines (SK-MES-1, NCI-H1975, and A549) compared with the normal bronchial epithelial cell line (16HBE). (B) ENST457720 expression was analyzed by RT-qPCR in NSCLC samples and adjacent nontumor liver tissues (n = 60). ENST457720 expression level was normalized to that of β-actin. **p < 0.01.
ENST457720 Knockdown Suppressed NSCLC Cell Proliferation and Induced Apoptosis
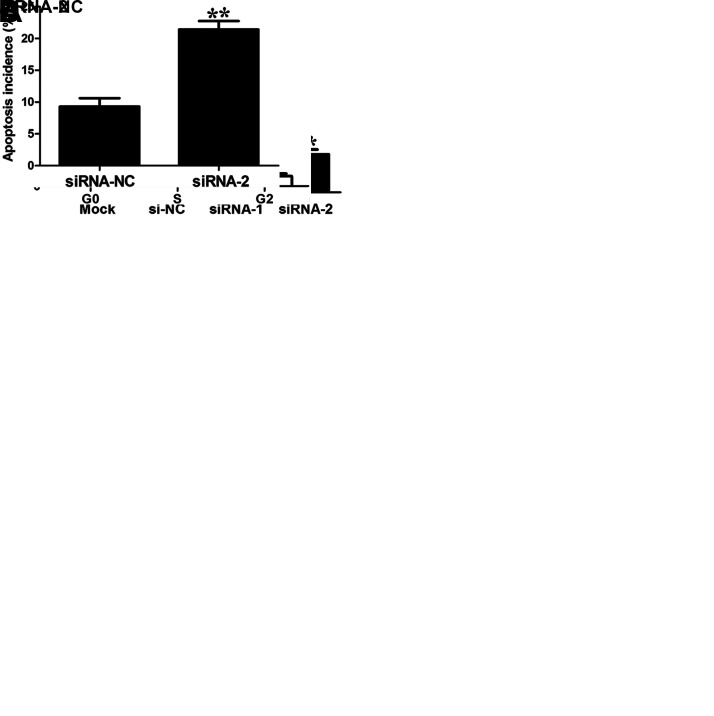
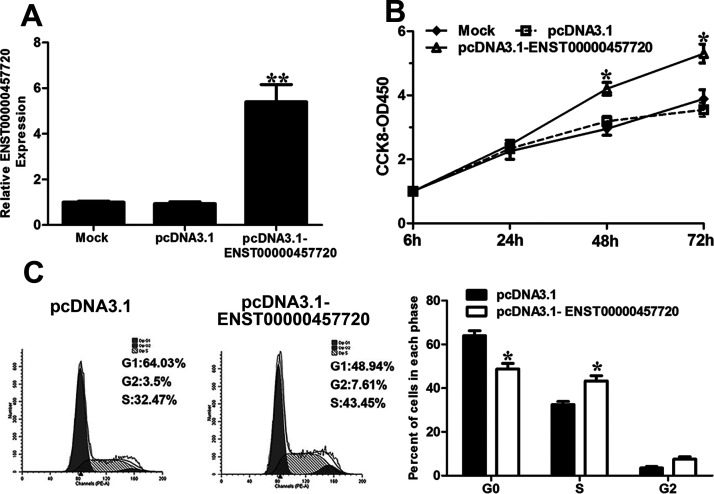
We chose the NCI-H1975 cell line for the ENST457720 knockdown experiments. Forty-eight hours after transfection, the RT-qPCR analysis of ENST457720 confirmed that the ENST457720-specific siRNA was effective (Fig. 2A), and we determined the effect of ENST457720 knockdown on NSCLC cell proliferation. ENST457720 silencing significantly decreased the viability of NCI-H1975 cells (Fig. 2B). Furthermore, a significant decrease in the percentage of cells in the S phase was observed following ENST457720 knockdown (Fig. 2C). The annexin V/PI assay revealed an obvious increase in the percentage of apoptotic cells following ENST457720 knockdown (Fig. 2D). We selected A549 cells for the ENST457720 overexpression experiments (Fig. 3A). ENST457720 overexpression obviously increased cell viability (Fig. 3B) and the percentage of cells in the S phase (Fig. 3C).
Figure 2.
Effect of ENST457720 silencing on cell proliferation. (A) RT-qPCR analysis of ENST457720 expression following treatment of H1975 cells with siRNA-ENST457720. (B) Cell counting kit 8 (CCK-8) assays were performed to determine the proliferation of H1975 cells. Data represent the mean ± standard deviation (SD) from three independent experiments. (C) Cell cycle analysis determined the relative cell numbers in each cell cycle phase after propidium iodide (PI) staining of ENST457720-downregulated H1975 cells. (D) Annexin V/PI staining and flow cytometry analysis assessing apoptosis in ENST457720-downregulated H1975 cells. Data represent the mean ± SD from three independent experiments. *p < 0.05; **p < 0.01.
Figure 3.
Effect of ENST457720 overexpression on cell proliferation. (A) RT-qPCR analysis of ENST457720 expression following treatment of A549 cells with pcDNA3.1-ENST457720. (B) CCK-8 assays were performed to determine the proliferation of A549 cells after transfection with pcDNA3.1-NC or pcDNA3.1-ENST457720. (C) Cell cycle analysis determined the relative cell numbers in each cell cycle phase after PI staining of ENST457720-upregulated A549 cells. *p < 0.05; **p < 0.01.
ENST457720 Knockdown Suppressed NSCLC Cell Invasion by Reversing the Epithelial-to-Mesenchymal Transition
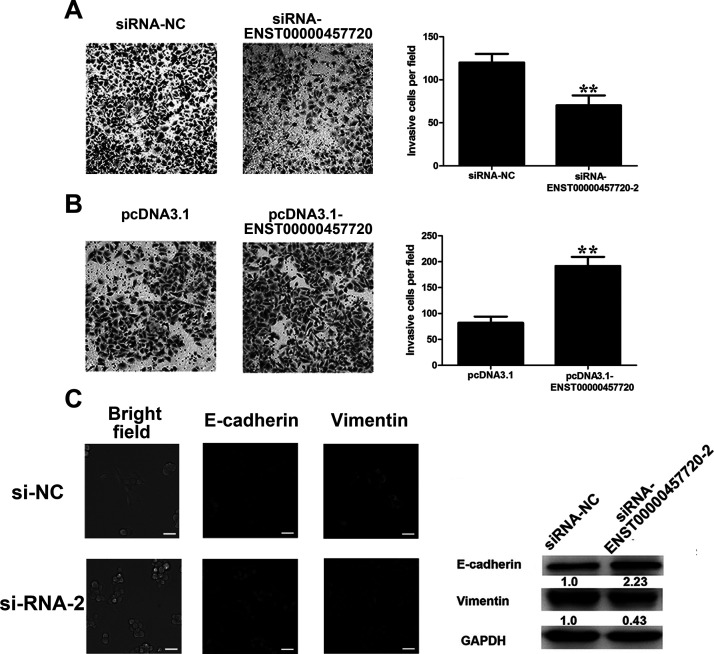
We also determined the effect of ENST457720 on cell invasion. As shown in the Transwell invasion assay, ENST457720 silencing markedly suppressed the invasion of H1975 cells (Fig. 4A), whereas ENST457720 overexpression promoted the invasion of A549 cells (Fig. 4B). A link between cancer cell invasion and the EMT has been established12,13. During this process, cells adopt a mesenchymal phenotype, as illustrated by decreased expression of epithelial markers (E-cadherin) and increased expression of mesenchymal markers (vimentin). After ENST457720 silencing, cells shifted from a spindled-shaped morphology to a more rounded, epithelial morphology. A significant upregulation of E-cadherin expression and downregulation of vimentin expression were observed in the Western blot analysis and immunofluorescence staining (Fig. 4C). Thus, ENST457720 promoted the EMT.
Figure 4.
Effect of ENST457720 on cell invasion. (A) Transwell assays were performed to investigate the migratory ability of H1975 cells after transfection with si-NC or si-ENST457720. (B) Transwell assays were performed to investigate the migratory ability of A549 cells after transfection with pcDNA3.1 or pcDNA3.1-ENST457720. Data represent the mean ± SD from three independent experiments. **p < 0.01. (C) Western blot analysis, bright field, and immunofluorescence images of phenotypic markers after ENST457720 silencing in H1975 cells.
ENST457720 Suppressed Cell Proliferation In Vivo
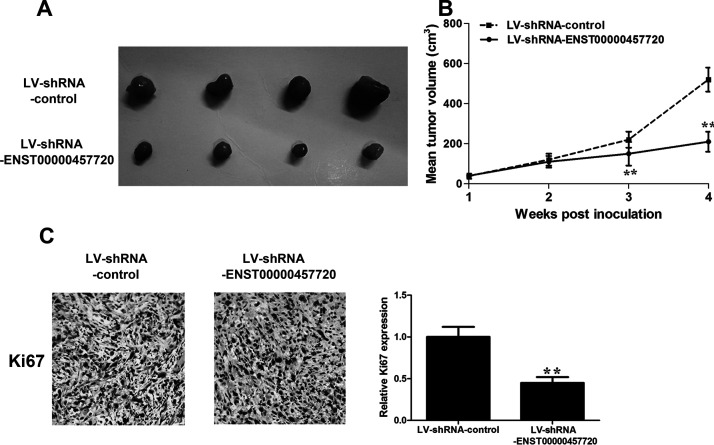
H1975 cells stably expressing sh-ENST457720 or the control vector were subcutaneously injected into either flank of nude mice to provide additional evidence for the oncogenic activity of ENST457720 in tumor progression. We measured tumor volumes weekly, and the mice were sacrificed 4 weeks after the cell inoculation. The tumor volume and tumor growth curve were significantly suppressed in the ENST457720-silenced group (Fig. 5A and B). According to the immunohistochemical analysis, the expression of the proliferation marker Ki-67 was obviously reduced in the ENST457720-silenced group (Fig. 5C). In general, ENST457720 promotes NSCLC tumor growth both in vitro and in vivo.
Figure 5.
Effect of ENST457720 on cell growth in vivo. (A) Total number of tumors removed from the mice 28 days after injection of H1975 cells transfected with shRNA-ENST457720 or shRNA-NC. (B) Tumor growth curve. Points indicate mean (n = 4), and bars indicate SD. (C) Representative images (×200) of immunohistochemistry (IHC) staining of the tumor. The IHC staining showed that ENST457720 silencing decreased the proliferation index marker Ki-67. **p < 0.01.
DISCUSSION
Although great efforts have been dedicated to clarifying the pathophysiological mechanism of NSCLC progression, including mutations in and amplifications of protein coding genes14–16 and miRNAs17–19, the underlying mechanisms remain to be explored. In recent years, more and more studies have underscored the significance of lncRNAs in the progression of NSCLC20–22. For example, LINC00511 is upregulated and associated with oncogenic activity and a poor prognosis in patients with NSCLC23. MEG3 regulates the EMT in lung cancer cell lines by repressing the recruitment of JARID2 and EZH2 and histone H3 methylation in the regulatory regions of the CDH1 and microRNA-200 family genes24. As shown in the study by Wan et al20, PVT1 promotes NSCLC cell proliferation by epigenetically regulating LATS2 expression. With the development of high-throughput screening technologies for lncRNAs, hundreds of lncRNAs have been shown to be differentially expressed in NSCLC to date10,25; however, further studies are required to uncover their functions in the development of NSCLC.
ENST457720 was shown to be upregulated in early lung adenocarcinoma by Wang et al10. The biological role of ENST457720 in NSCLC is unknown. First, ENST457720 is included in multiple public ncRNA databases, and the ENST457720 transcript is approximately 606 nt. According to the coding potential calculator (CPC) algorithm26, ENST457720 has a CPC score of −1.019, which significantly discriminates ENST457720 from protein coding genes (positive score). The codon substitution frequency (CSF) score27 for ENST457720 is −131.9962. Thus, ENST457720 is a noncoding RNA. The copy number of ENST457720 was approximately 50% of the HOTAIR copy number in NCI-H1975 cells. The expression levels of lncRNAs are much lower than that of protein coding genes9, so we determined the precise expression level of ENST457720. ENST457720 expression was comparable to a well-characterized lncRNA, HOTAIR28,29, in NSCLC cells, providing evidence that ENST457720 might play a role in the development of NSCLC.
In addition, ENST457720 expression was upregulated in NSCLC tissues compared to adjacent normal tissues. Correlations were observed between ENST457720 expression and a larger tumor diameter and lymph node metastasis. Moreover, we investigated the function of ENST457720 using gain- and loss-of-function assays. ENST457720 knockdown significantly suppressed cell viability and induced cell apoptosis, whereas ENST457720 overexpression increased cell viability and the percentage of cells in the S phase. ENST457720 silencing also inhibited NSCLC cell invasion. We examined the effect of ENST457720 on the EMT to elucidate its underlying mechanism. The EMT contributes to cancer invasion and metastasis13,14. ENST457720 knockdown reversed the EMT process. Thus, the oncogenic effect of ENST457720 on cancer invasion may be attributed to the EMT.
We examined ENST457720 expression in a relatively small cohort of patients, with 60 subjects enrolled. Further studies should be conducted in a large cohort of patients, and more mechanistic investigations should be performed to clarify the mechanism underlying the oncogenic activity of ENST457720 in NSCLC.
In summary, ENST457720 expression was upregulated in NSCLC tissues. ENST457720 increased NSCLC cell proliferation and invasion. Our study provides new evidence describing the important roles of lncRNAs in NSCLC progression.
ACKNOWLEDGMENT
This work was supported by Shanghai Municipal Health Bureau Project (Grant No. 201540097).
Footnotes
The authors declare no conflicts of interest.
REFERENCES
- 1. Smith RA, Manassaram-Baptiste D, Brooks D, Cokkinides V, Doroshenk M, Saslow D, Wender RC, Brawley OW. Cancer screening in the United States, 2014: A review of current American Cancer Society guidelines and current issues in cancer screening. CA Cancer J Clin. 2014;64(1):30–51. [DOI] [PubMed] [Google Scholar]
- 2. Verdecchia A, Francisci S, Brenner H, Gatta G, Micheli A, Mangone L, Kunkler I; EUROCARE-4 Working Group. Recent cancer survival in Europe: A 2000-02 period analysis of EUROCARE-4 data. Lancet Oncol. 2007;8(9):784–96. [DOI] [PubMed] [Google Scholar]
- 3. Yu CY, Kuo HC. The trans-spliced long noncoding RNA tsRMST impedes human embryonic stem cell differentiation through WNT5A-mediated inhibition of the epithelial-to-mesenchymal transition. Stem Cells 2016;34(8):2052–62. [DOI] [PubMed] [Google Scholar]
- 4. Ma MZ, Zhang Y, Weng MZ, Wang SH, Hu Y, Hou ZY, Qin YY, Gong W, Zhang YJ, Kong X, Wang JD, Quan ZW. Long noncoding RNA GCASPC, a target of miR-17-3p, negatively regulates pyruvate carboxylase-dependent cell proliferation in gallbladder cancer. Cancer Res. 2016;76(18):5361–71. [DOI] [PubMed] [Google Scholar]
- 5. Atianand MK, Hu W, Satpathy AT, Shen Y, Ricci EP, Alvarez-Dominguez JR, Bhatta A, Schattgen SA, McGowan JD, Blin J, Braun JE, Gandhi P, Moore MJ, Chang HY, Lodish HF, Caffrey DR, Fitzgerald KA. A long noncoding RNA lincRNA-EPS acts as a transcriptional brake to restrain inflammation. Cell 2016;165(7):1672–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Jia P, Cai H, Liu X, Chen J, Ma J, Wang P, Liu Y, Zheng J, Xue Y. Long non-coding RNA H19 regulates glioma angiogenesis and the biological behavior of glioma-associated endothelial cells by inhibiting microRNA-29a. Cancer Lett. 2016;381(2):359–69. [DOI] [PubMed] [Google Scholar]
- 7. Zhang P, Cao L, Fan P, Mei Y, Wu M. LncRNA-MIF, a c-Myc-activated long non-coding RNA, suppresses glycolysis by promoting Fbxw7-mediated c-Myc degradation. EMBO Rep. 2016;17(8):1204–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zhang Y, He Q, Hu Z, Feng Y, Fan L, Tang Z, Yuan J, Shan W, Li C, Hu X, Tanyi JL, Fan Y, Huang Q, Montone K, Dang CV, Zhang L. Long noncoding RNA LINP1 regulates repair of DNA double-strand breaks in triple-negative breast cancer. Nat Struct Mol Biol. 2016;23(6):522–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Qu L, Ding J, Chen C, Wu ZJ, Liu B, Gao Y, Chen W, Liu F, Sun W, Li XF, Wang X, Wang Y, Xu ZY, Gao L, Yang Q, Xu B, Li YM, Fang ZY, Xu ZP, Bao Y, Wu DS, Miao X, Sun HY, Sun YH, Wang HY, Wang LH. Exosome-transmitted lncARSR promotes sunitinib resistance in renal cancer by acting as a competing endogenous RNA. Cancer Cell 2016;29(5):653–68. [DOI] [PubMed] [Google Scholar]
- 10. Wang P, Lu S, Mao H, Bai Y, Ma T, Cheng Z, Zhang H, Jin Q, Zhao J, Mao H. Identification of biomarkers for the detection of early stage lung adenocarcinoma by microarray profiling of long noncoding RNAs. Lung Cancer 2015;88(2):147–53. [DOI] [PubMed] [Google Scholar]
- 11. Luo J, Tang L, Zhang J, Ni J, Zhang HP, Zhang L, Xu JF, Zheng D. Long non-coding RNA CARLo-5 is a negative prognostic factor and exhibits tumor pro-oncogenic activity in non-small cell lung cancer. Tumour Biol. 2014;35(11):11541–9. [DOI] [PubMed] [Google Scholar]
- 12. Bogachek MV, De Andrade JP, Weigel RJ. Regulation of epithelial-mesenchymal transition through SUMOylation of transcription factors. Cancer Res. 2015;75(1):11–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Li L, Li W. Epithelial-mesenchymal transition in human cancer: Comprehensive reprogramming of metabolism, epigenetics, and differentiation. Pharmacol Ther. 2015;150:33–46. [DOI] [PubMed] [Google Scholar]
- 14. Chen Y, Tang Q, Xiao Q, Yang L, Hann SS. Targeting EP4 downstream c-Jun through ERK1/2-mediated reduction of DNMT1 reveals novel mechanism of solamargine-inhibited growth of lung cancer cells. J Cell Mol Med. 2017;21(2):222–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wan J, Xu W, Zhan J, Ma J, Li X, Xie Y, Wang J, Zhu WG, Luo J, Zhang H. PCAF-mediated acetylation of transcriptional factor HOXB9 suppresses lung adenocarcinoma progression by targeting oncogenic protein JMJD6. Nucleic Acids Res. 2016;44(22):10662–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Klingbeil O, Lesche R, Gelato KA, Haendler B, Lejeune P. Inhibition of BET bromodomain-dependent XIAP and FLIP expression sensitizes KRAS-mutated NSCLC to pro-apoptotic agents. Cell Death Dis. 2016;7:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rothschild SI, Gautschi O, Batliner J, Gugger M, Fey MF, Tschan MP. MicroRNA-106a targets autophagy and enhances sensitivity of lung cancer cells to Src inhibitors. Lung Cancer 2017;107:73–83. [DOI] [PubMed] [Google Scholar]
- 18. Deng Z, Rong Y, Teng Y, Zhuang X, Samykutty A, Mu J, Zhang L, Cao P, Yan J, Miller D, Zhang HG. Exosomes miR-126a released from MDSC induced by DOX treatment promotes lung metastasis. Oncogene 2017;36(5):639–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhang WC, Chin TM, Yang H, Nga ME, Lunny DP, Lim EK, Sun LL, Pang YH, Leow YN, Malusay SR, Lim PX, Lee JZ, Tan BJ, Shyh-Chang N, Lim EH, Lim WT, Tan DS, Tan EH, Tai BC, Soo RA, Tam WL, Lim B. Tumour-initiating cell-specific miR-1246 and miR-1290 expression converge to promote non-small cell lung cancer progression. Nat Commun. 2016;7:11702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wan L, Sun M, Liu GJ, Wei CC, Zhang EB, Kong R, Xu TP, Huang MD, Wang ZX. Long noncoding RNA PVT1 promotes non-small cell lung cancer cell proliferation through epigenetically regulating LATS2 expression. Mol Cancer Ther. 2016;15(5):1082–94. [DOI] [PubMed] [Google Scholar]
- 21. Han Y, Wu Z, Wu T, Huang Y, Cheng Z, Li X, Sun T, Xie X, Zhou Y, Du Z. Tumor-suppressive function of long noncoding RNA MALAT1 in glioma cells by downregulation of MMP2 and inactivation of ERK/MAPK signaling. Cell Death Dis. 2016;7:e2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Qiu M, Xu Y, Wang J, Zhang E, Sun M, Zheng Y, Li M, Xia W, Feng D, Yin R, Xu L. A novel lncRNA, LUADT1, promotes lung adenocarcinoma proliferation via the epigenetic suppression of p27. Cell Death Dis. 2015;6:e1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sun CC, Li SJ, Li G, Hua RX, Zhou XH, Li DJ. Long intergenic noncoding RNA 00511 acts as an oncogene in non-small-cell lung cancer by binding to EZH2 and suppressing p57. Mol Ther Nucleic Acids 2016;5(11):1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Terashima M, Tange S, Ishimura A, Suzuki T. MEG3 Long noncoding RNA contributes to the epigenetic regulation of epithelial-mesenchymal transition in lung cancer cell lines. J Biol Chem. 2017;292(1):82–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wang Y, Qian CY, Li XP, Zhang Y, He H, Wang J, Chen J, Cui JJ, Liu R, Zhou H, Xiao L, Xu XJ, Zheng Y, Fu YL, Chen ZY, Chen X, Zhang W, Ye CC, Zhou HH, Yin JY, Liu ZQ. Genome-scale long noncoding RNA expression pattern in squamous cell lung cancer. Sci Rep. 2015;5:11671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kong L, Zhang Y, Ye ZQ, Liu XQ, Zhao SQ, Wei L, Gao G. CPC: Assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res. 2007;35(Web Server issue):W345–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lin MF, Jungreis I, Kellis M. PhyloCSF: A comparative genomics method to distinguish protein coding and non-coding regions. Bioinformatics 2011;27(13):i275–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhuang Y, Wang X, Nguyen HT, Zhuo Y, Cui X, Fewell C, Flemington EK, Shan B. Induction of long intergenic non-coding RNA HOTAIR in lung cancer cells by type I collagen. J Hematol Oncol. 2013;6(35):1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wang R, Shi Y, Chen L, Jiang Y, Mao C, Yan B, Liu S, Shan B, Tao Y, Wang X. The ratio of FoxA1 to FoxA2 in lung adenocarcinoma is regulated by LncRNA HOTAIR and chromatin remodeling factor LSH. Sci Rep. 2015;5:17826. [DOI] [PMC free article] [PubMed] [Google Scholar]