Abstract
Tripartite motif-containing 14 (TRIM14) is abnormally expressed in several human cancers. However, the function and expression of TRIM14 in human breast cancer are still largely unknown. To understand the biological function of TRIM14 in breast cancer, we measured the expression level of TRIM14. Cell proliferation and cell apoptosis were measured after TRIM14 overexpression or knockdown. Upregulation of TRIM14 was found in human breast cancer specimens and cell lines. Reduction of TRIM14 inhibited cell proliferation but increased cell apoptosis in the BT474 and MDA-MB-231 cell lines. Further study showed that knockdown of TRIM14 upregulated the expression of BAX while downregulating the expression of BCL2. In addition, the expression of SHP-1 was increased, and the phosphorylation of STAT3 (p-STAT3) was inhibited. Conversely, overexpression of TRIM14 had the opposite effects. Additionally, cryptotanshinone, a STAT3 inhibitor, inhibited cell proliferation but increased cell apoptosis in the BT474 and MDA-MB-231 cell lines. In conclusion, TRIM14 may act as an oncogene in human breast cancer and may be a novel strategy for human breast cancer.
Key words: Apoptosis, Breast cancer, Src-homology protein tyrosine phosphatase-1 (SHP-1), STAT3, Tripartite motif-containing 14 (TRIM14)
INTRODUCTION
Breast cancer is a widespread cancer in women, and it is both the most prevalent form of cancer and the greatest killer. Each year there are almost a million new cases of breast cancer, and 458,000 deaths in both the developed and developing world1. Despite that, the survival rate of breast cancer has increased significantly over the last few years; the major breast cancer treatment is chemotherapy, which has some limitations. In recent years, several molecularly targeted agents have been demonstrated to be effective for cancer treatments, such as trastuzumab, a monoclonal antibody targeting the human epidermal growth factor receptor 2 (HER2), which has a great effect in treating HER2+ breast cancer2. So there is still a need to find new biomarkers to predict tumor progression, which can be potential therapeutic targets.
Signal transducer and activator of transcription 3 (STAT3) plays a key role in many cancer cell lines and cancer tissues, including cell proliferation, invasion, migration, apoptosis resistance, tumor angiogenesis, and epithelial–mesenchymal transition (EMT)3–5. In addition, STAT3 is associated with oncogene addiction in several types of cancers, including human breast cancer6. STAT3 is necessary for normal breast and is activated in all classes of breast cancer7. Further, in about 60% of breast tumors, STAT3 is phosphorylated8. Growth factors induce tyrosine phosphorylation of STAT3 that directly activates gene expression, which is associated with tumor cell survival and proliferation9. Recent evidence suggests that phosphorylated STAT3 can promote the survival of breast cancer cells10. There are therapeutic benefits from targeting STAT3 in cancer treatment. Currently, inhibition of STAT3 transcription and inhibition of upstream receptors are regular strategies for inhibiting STAT3 activation11. Thus, STAT3 is a strategy for human anticancer treatment, including human breast cancer.
Src-homology protein tyrosine phosphatase-1 (SHP-1), containing protein tyrosine phosphatase (PTP), expresses most abundantly in hematopoietic cells. SHP-1 negatively regulates the JAK/STAT pathway in leukemias and lymphomas12. It has been reported that SHP-1 acts as a cancer suppressor in human breast cancer13. SHP-1 inhibits the phosphorylation of p-STAT3 at the tyrosine 705 residue14. An aberrant SHP-1/p-STAT3 signaling pathway was found in many human cancers15,16, including breast cancer17. Importantly, regulating the SHP-1/p-STAT3 signaling pathway may be a new treatment for human cancer.
The tripartite motif (TRIM) family of proteins consist of a RING domain (R), two B-box domains (B1 and B2), and a coiled-coil (CC) region18. Recent studies indicate that TRIM family members may serve as oncogenes or tumor suppressors19–22. In breast cancer, TRIM44 could promote cell proliferation and migration by enhancing NF-κB signaling23. TRIM28 enhances cell migration and invasion in human breast cancer by regulating TWIST1 and EMT24. On the other hand, TRIM29 acts as a hypoxia-induced tumor suppressor gene in human breast cancer25. Moreover, TRIM24 and TRIM28 have been found to inhibit STAT1 signaling26,27. TRIM14 is a member of the TRIM family and is located at chromosome 9q22. Previously, Su et al. found that overexpression of TRIM14 promotes the aggressiveness of tongue squamous cell carcinoma28. Upregulation of TRIM14 was found in human osteosarcoma specimens and cell lines29. However, the function and expression of TRIM14 in human breast cancer are still unclear.
Here we found the upregulation of TRIM14 in human breast cancer specimens and cell lines. Knockdown of TRIM14 inhibited cell proliferation but increased cell apoptosis. Our findings suggest that TRIM14 may act as an oncogene in breast cancer and could be a potential therapeutic target.
MATERIALS AND METHODS
Patient Samples
A total of 30 human breast cancer tissues and 30 matched normal adjacent tissues were collected from Shanghai Traditional Chinese edicine-Integrated Hospital (Shanghai, P.R. China) with informed consent. Both cancer and normal specimens were frozen immediately in liquid nitrogen until RNA extraction. All the experiments were approved by Shanghai Traditional Chinese Medicine-Integrated Hospital.
Cell Culture
The normal epithelial breast cell line, MCF-10A, was obtained from the American Type Culture Collection (ATCC; Manassas, VA, USA) and grown in EMBM (ATCC) with 10% fetal bovine serum (FBS; Gibco, Carlsbad, CA, USA) and 1% penicillin/streptomycin (Gibco), plus 100 ng/ml cholera toxin (ATCC). Two human breast cancer cell lines, BT474 and MDA-MB-231, were obtained from Cell Bank of the Chinese Academy of Sciences (Shanghai, P.R. China) and grown in RPMI-1640 medium (Hyclone, Logan, UT, USA) with 10% FBS and antibiotics. All cells were cultivated at 37°C in a 5% CO2 incubator.
Reverse Transcription and Real-Time PCR
Real-time PCR was used to measure the mRNA levels of target genes. Cellular and tissue RNA were extracted using TRIzol reagent (Invitrogen, Carlsbad, CA, USA), and a cDNA synthesis kit was used to obtain cDNA. The following mixture was prepared to give a 25-μl reverse transcriptase (RT) solution: 12 μl of RNA-Primer Mix, 5 μl of 5× RT reaction buffer, 1 μl of 25 mM dNTPs, 1 μl of 25 U/μl RNase inhibitor, 1 μl of 200 U/μl M-MLV Rtase (Promega, Madison, WI, USA), 1 μl of oligo(dt)18, and 4 μl of RNase free water (ddH2O). The reaction was completed on an ABI 7300 system (Applied Biosystems, Foster City, CA, USA) using the following steps: 60 min at 37°C, 5 min at 85°C, 5 min at 4°C, then stored at −20°C. The primers were designed using Primer 3 online Software. Real-time PCR experiments were performed with SYBR Green qPCR Mixes (Thermo Fisher Scientific Inc., Grand Island, NY, USA). In a 25-μl reaction solution, 12.5 μl of SYBR Green Mix, 0.5 μl of former primer, 0.5 μl of reverse primer, 2 μl of cDNA, and 9.5 μl of ddH2O were added. The reaction was completed on an ABI 7300 system using the following steps: 10 min at 95°C, then followed by 40 cycles of 15 s at 95°C, 45 s at 60°C. Expression of target genes was normalized with GAPDH and calculated using the 2−ΔΔct method. All data represented the average of three replicates. Primers used were as follows: TRIM14 (NM_014788.3), 5′-GGATTTGTGTCTCCGTTCTG-3′ and 5′-TCTGTCTGCCTGGTATTCTG-3′; STAT3 (NM_139276), 5′-TGTCTAAAGGTCCCTCATC-3′ and 5′-CCATAGTGTGCATCATGTC-3′; GAPDH (NM_001256799.1), 5′-CACCCACTCCTCCACCTTTG-3′ and 5′-CCACCACCCTGTTGCTGTAG-3′.
Western Blotting
Whole cells were rinsed twice with PBS, then lysed in RIPA lysis buffer (Solarbio, Beijing, P.R. China) containing protease inhibitors. For tissues, 1 g of each tissue was cut down by scalpel on ice as quickly as possible. Then the tissues were washed twice with iced PBS and then lysed with a homogenizer in RIPA lysis buffer containing protease inhibitors. After 30 min on ice, the harvested cell lysis was centrifuged at 12,000 × g at 4°C for 10 min. Protein concentration was measured with a bicinchoninic acid (BCA) protein assay kit (Thermo Fisher Scientific Inc.). An equal amount of protein (20 μg) was loaded onto 10% polyacrylamide gels for SDS-PAGE, and then transferred to a nitrocellulose membrane (Millipore Corp., Bedford, MA, USA). After 1 h, the membrane was blocked with PBST containing 5% BSA and immunoblotted with respective antibodies for 4–6 h: TRIM14 (ab185349; 1:800 dilution; Abcam, Cambridge, MA, USA); SHP-1 (ab32559; 1:1,000 dilution; Abcam); BCL2 (ab32124; 1:1,000 dilution; Abcam); BAX (ab32503; 1:800 dilution; Abcam); STAT3 (ab50761; 1:800 dilution; Abcam); p-STAT3 (ab76315; 1:500 dilution; Abcam); and GAPDH (#5174; 1:3,000 dilution; Cell Signaling Technology, Danvers, MA, USA). The blots were incubated for 1 h at room temperature with secondary antibody (all from Abcam) tagged with horseradish peroxidase (HRP). Then the blots were washed twice with TBST. The membranes were developed by an enhanced chemiluminescence (ECL) kit (Millipore). Bands were recorded with ImageJ and normalized to GAPDH.
Lentivirus Infection and Inhibitor
To construct the TRIM14 overexpression lentivirus, full-length human TRIM14 cDNA was inserted into pLVX-Puro vector (Clontech Laboratories, Inc., Mountain View, CA, USA). DNA sequencing was used to verify the vector sequence. HEK-293T cells were plated into 10-cm dishes. After 24 h of culturing, the plasmids including pLVX-puro vector or pLVX-puro-TRIM14, psPAX2, and pMD2G were transfected into HEK293T cells according to the manufacturer’s instructions. After 48 h posttransfection, vector and TRIM14 overexpression viral supernatants were harvest and used to transduce the BT474 cell line.
Three lentivirus shRNAs targeting human TRIM14 (5′-ACAGGCCCTAAACAAGTCT-3′; 5′-GTAGGCACACAGTAAACGT-3′; 5′- TAGACTTTCAGTGCATTAG-3′) and a negative control (NC) shRNA were obtained from Genechem (Shanghai, P.R. China). shRNAs were inserted into PLKO.1 (Addgene, Cambridge, MA, USA) to construct the TRIM14-silencing lentivirus and verified by sequencing. The same method as described above was used to obtain NC and shTRIM14 viral supernatant. NC and shTRIM14 viral supernatants were harvested and used to transduce BT474 and MDA-MB-231 cells. These two cell lines were infected with recombinant lentiviruses expressing TRIM14 or shTRIM14 using vector or NC lentiviruses as control. After 48 h, the cells were collected to confirm transfection efficiency.
Cryptotanshinone (Selleck, Houston, TX, USA) is a common inhibitor of STAT3. Cells were seeded in 96-well or 6-well plates and cultured for 24 h with 5 μM cryptotanshinone. The treated cells were used to detect cell proliferation and apoptosis.
Cell Proliferation Assay
Transducted and untransducted cells were seeded into 96-well plates. After 24, 48, and 72 h of transduction, 90 μl of culture medium and 10 μl of CCK-8 solution were added to each well and incubated for 1 h. After incubation, a microplate reader (Bio-Rad Laboratories, Inc., Hercules, CA, USA) was used to detect the absorbance at 450 nm. Experiments were performed in triplicate.
Cell Apoptosis Assay
Apoptosis was measured using the Annexin-V Apoptosis Detection Kit (BD Biosciences, San Jose, CA, USA). All the cells were harvested, then incubated with annexin V and PI at room temperature for 15 min in the dark. Annexin V or PI staining alone was used as a control. Cell apoptosis was assessed by flow cytometry (BD Biosciences). Experiments were performed in triplicate.
Statistical Analysis
All statistical analyses were performed as mean ± SD. Statistical significance between groups was determined using ANOVA. A value of p < 0.05 was considered statistically significant.
RESULTS
TRIM14 Was Overexpressed in Human Breast Cancer Tissues and Cell Lines
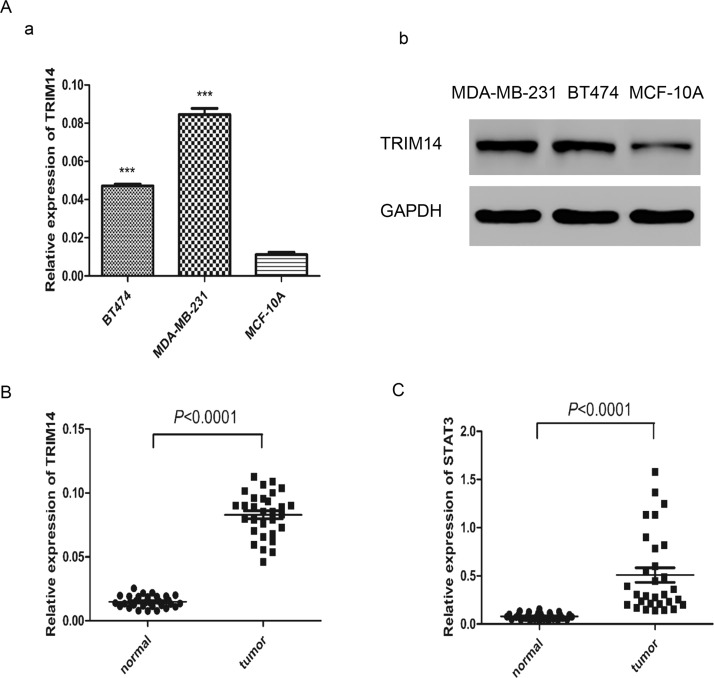
To determine the baseline of TRIM14 in vitro, the expression level of TRIM14 was detected in two human breast cancer cell lines (BT474 and MDA-MB-231) and a normal breast cell line (MCF-10A) by real-time PCR and Western blot. Our results showed that a higher level of TRIM14 was found in these two cell lines than in MCF-10A (Fig. 1A). We further detected TRIM14 and STAT3 expression in breast cancer and normal specimens using real-time PCR. The mRNA levels of TRIM14 and STAT3 were both higher in cancer tissues than in normal tissues (Fig. 1B and C). These results suggested that TRIM14 was upregulated in human breast cancer tissues and cell lines. STAT3 was upregulated in human breast cancer, which indicated that the STAT3 pathway may be activated in human breast cancer.
Figure 1.
Tripartite motif-containing 14 (TRIM14) was enhanced in human breast cancer tissues and cell lines. (A) TRIM14 expression level in different human breast cancer cell lines (BT474 and MDA-MB-231) and the normal breast cell line (MCF-10A) was analyzed by real-time PCR (a) and Western blot (b). (B) mRNA level of TRIM14 in cancer samples (n = 30) and normal samples (n = 30). (C) mRNA level of signal transducer and activator of transcription 3 (STAT3) in breast cancer samples (n = 30) and normal samples (n = 30). ***p < 0.0001 versus normal cell line.
Lentivirus Infection Reduced the Expression of TRIM14
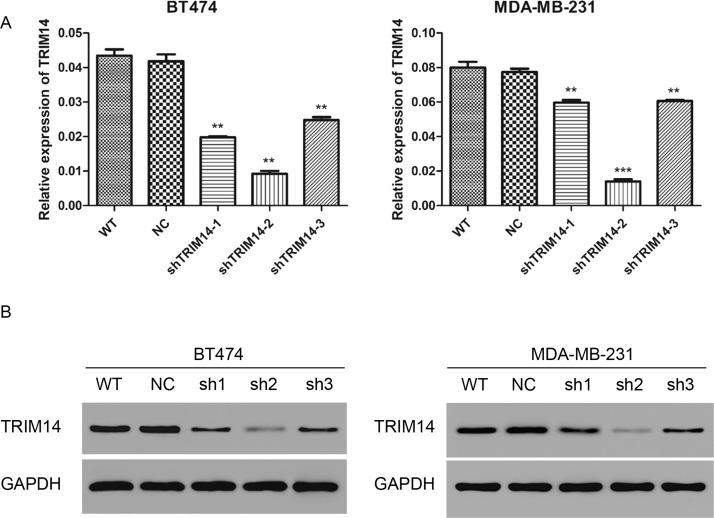
Three lentivirus shRNAs targeting human TRIM14 (shTRIM14-1, shTRIM14-2, and shTRIM14-3) and NC were transduced into BT474 and MDA-MB-231 cells. To measure the knockdown efficiency, TRIM14 was detected by real-time PCR and Western blot. As shown in Figure 2A and B, after 48 h of transduction, TRIM14 transcription and expression level were decreased notably compared with NC (knockdown efficiency: in the BT474 cell line, shTRIM14-1, 52.6 ± 0.5%; shTRIM14-2, 77.9 ± 0.1%; shTRIM14-3, 40.6 ± 0.1%; in the MDA-MB-231 cell line, shTRIM14-1, 22.8 ± 0.2%; shTRIM14-2, 81.8 ± 0.2%; shTRIM14-3, 21.6 ± 0.1%). ShTRIM14-2 was chosen for the following assays, as it had the best knockdown efficiency.
Figure 2.
Lentivirus infection reduced the expression of TRIM14. (A) TRIM14 expression level was analyzed by real-time PCR in BT474 and MDA-MB-231 cells 48 h after transduction with shTRIM14 (shTRIM14-1, -2, and -3) and NC lentivirus. (B) TRIM14 expression level was analyzed by Western blot in BT474 and MDA-MB-231 cells. WT, wild-type cells; NC, negative control cells; shTRIM14, shTRIM14-transducted cells. **p < 0.01; ***p < 0.001 versus NC.
TRIM14 Knockdown Inhibited Cell Proliferation but Increased Cell Apoptosis
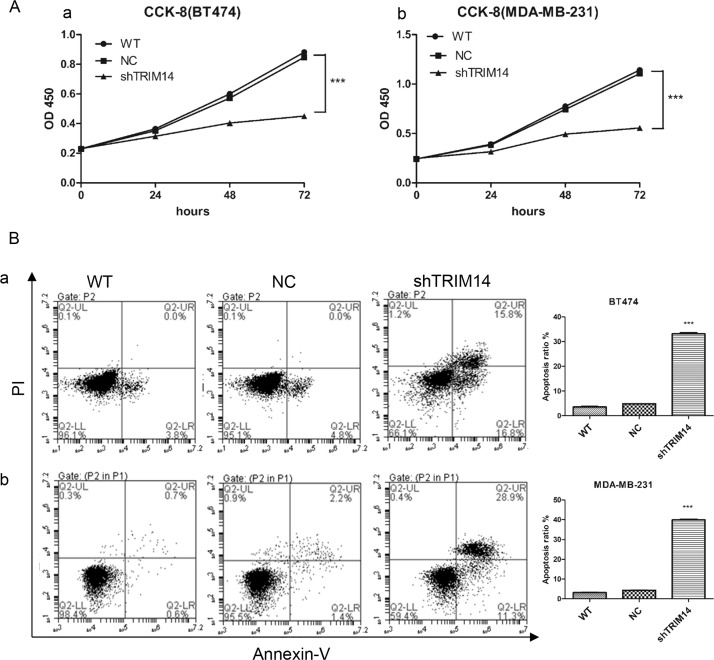
Cell proliferation was measured by CCK-8. As shown in Figure 3A, reduction of TRIM14 inhibited cell proliferation in BT474 and MDA-MB-231 cells, compared with NC. Furthermore, more apoptosis was observed in groups with TRIM14 knockdown than in the NC group [apoptosis ratio: knockdown group in BT474, 33.13 ± 0.75%; in MDA-MB-231, 39.93 ± 0.59%; NC group in BT474, 4.87 ± 0.15%; in MDA-MB-231, 4.37 ± 0.15%; wild-type (WT) group in BT474, 3.6 ± 0.3%; in MDA-MB-231, 3.17 ± 0.15%] (Fig. 3B). These results suggested that knockdown of TRIM14 inhibited cell proliferation but increased cell apoptosis.
Figure 3.
TRIM14 knockdown inhibited cell proliferation but increased cell apoptosis. (A) Cell proliferation was measured after transduction with shTRIM14-2 and NC lentivirus by CCK-8 in BT474 (a) and MDA-MB-231 (b). (B) Cell apoptosis was measured after transduction with shTRIM14-2 and NC lentivirus by flow cytometry in BT474 (a) and MDA-MB-231 (b). ***p < 0.001 versus NC.
Downregulation of TRIM14 Activated the Apoptosis Pathway in Breast Cancer Cells
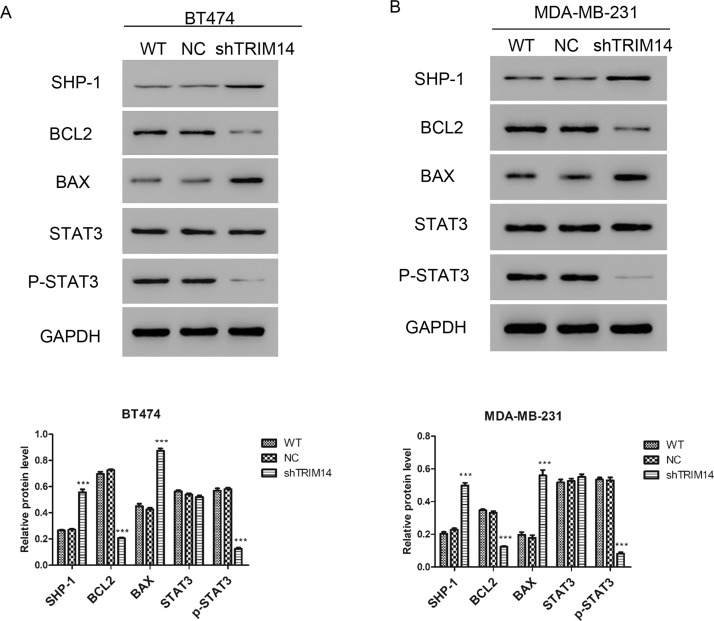
For further studying the mechanism of TRIM14, we proposed that TRIM14 may play a role in apoptosis based on previous data29. To verify our hypothesis, we detected the expression of related protein by Western blot in BT474 and MDA-MB-231. The results shown in Figure 4 reveal that the expression of BAX was upregulated, while the expression of BCL2 was downregulated after transduction with shTRIM14 lentivirus. As shown in Figure 1C, STAT3 was overexpressed in breast cancer tissues, so we detected whether the suppression of TRIM14 by shTRIM14 lentivirus could regulate the expression of SHP-1. We found that transduction with shTRIM14 lentivirus could promote the expression of SHP-1. SHP-1 can prevent the activation of STAT3, so we detected the phosphorylation level of STAT3. We found that transduction with shTRIM14 lentivirus inhibited the expression of p-STAT3. These results indicated that downregulation of TRIM14 may inhibit STAT3 pathway by upregulating SHP-1.
Figure 4.
Downregulation of TRIM14 activated apoptosis pathway. Expressions of SHP-1, BAX, BCL2, and p-STAT3 were detected by Western blot after transduction with shTRIM14 and NC lentivirus in BT474 (A) and MDA-MB-231 (B). ***p < 0.001 versus NC.
An Inhibitor of STAT3 Inhibited Cell Proliferation but Increased Cell Apoptosis
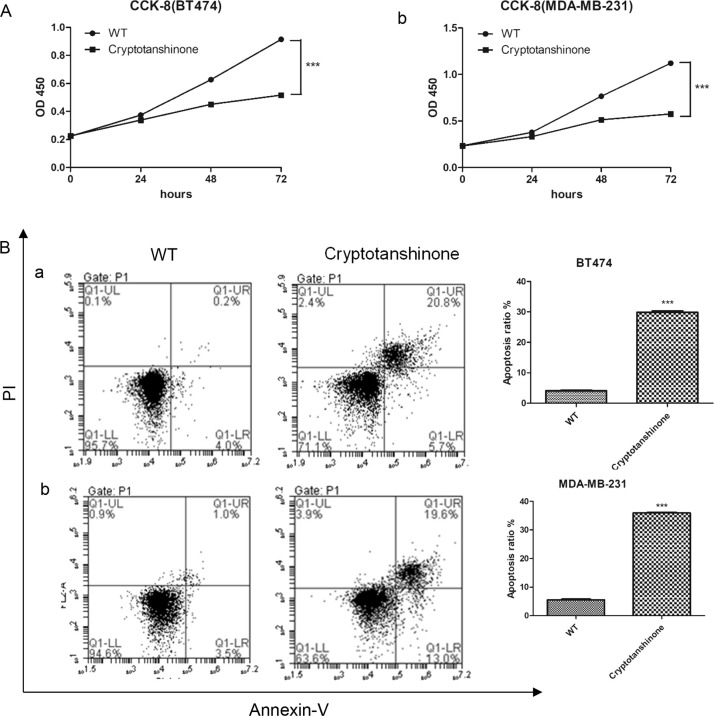
Based on previous data, reduction of TRIM14 inhibited cell proliferation and inhibited the phosphorylation of STAT3. Cells were treated with cryptotanshinone, a common inhibitor of STAT3, and then cell proliferation and apoptosis were measured by CCK-8 and annexin V assay, respectively. As shown in Figure 5A, cryptotanshinone-treated groups inhibited cell proliferation in BT474 and MDA-MB-231 cells, compared with WT. Furthermore, more apoptosis was observed in the groups with cryptotanshinone (apoptosis ratio: cryptotanshinone group in BT474, 29.87 ± 0.91%; in MDA-MB-231, 35.93 ± 0.42%; WT group in BT474, 4.1 ± 0.26%; in MDA-MB-231, 5.53 ± 0.42%) (Fig. 5B). These results suggested that treatment with a STAT3 inhibitor inhibited cell proliferation but increased cell apoptosis.
Figure 5.
Inhibitor of STAT3 inhibited cell proliferation but increased cell apoptosis. (A) Cell proliferation was measured after treatment with cryptotanshinone by CCK-8 in BT474 (a) and MDA-MB-231 (b). (B) Cell apoptosis was measured after treatment with cryptotanshinone by flow cytometry in BT474 (a) and MDA-MB-231 (b). cryptotanshinone: cells treated with cryptotanshinone. ***p < 0.001 versus WT.
Overexpression of TRIM14 Promoted Cell Proliferation but Inhibited Cell Apoptosis
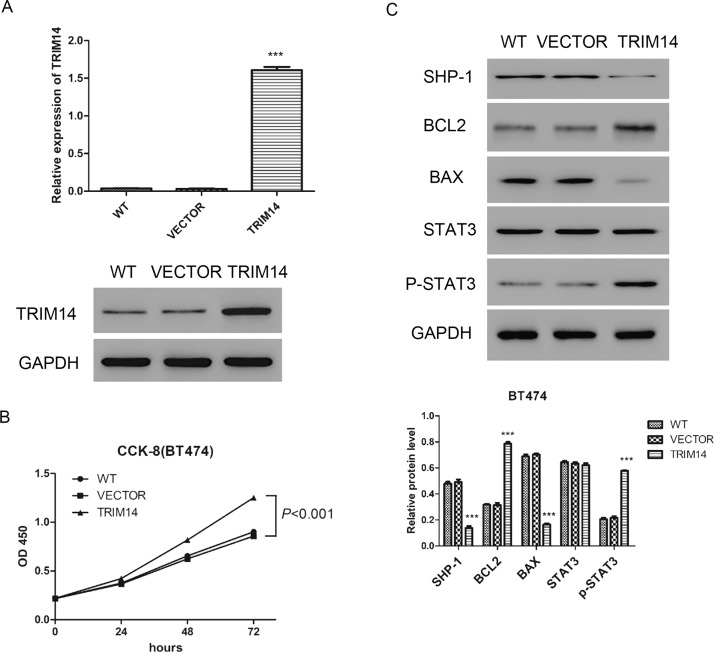
TRIM14 overexpression or vector lentivirus was transduced into BT474 for 48 h. Expression of TRIM14 was highly increased in the TRIM14 overexpression group compared with the vector group (Fig. 6A). Overexpression of TRIM14 promoted the proliferation of BT474 cells, compared with cells transduced with vector (Fig. 6B). Furthermore, the expression of BAX was downregulated while the expression of BCL2 was upregulated after TRIM14 overexpression. Interestingly, TRIM14 overexpression could inhibit SHP-1 and promote p-STAT3 (Fig. 6C). These results indicated that overexpression of TRIM14 promoted cell proliferation but inhibited cell apoptosis. TRIM14 overexpression could upregulate the expression of p-STAT3.
Figure 6.
Overexpression of TRIM14 enhanced cell proliferation but inhibited cell apoptosis. (A) Expression of TRIM14 was detected by real-time PCR and Western blot. (B) Cell proliferation was measured after transduction with TRIM14 overexpression and vector lentivirus by CCK-8 in BT474. (C) Expressions of SHP-1, BAX, BCL2, and p-STAT3 were detected by Western blot after transduction in BT474. VECTOR, vector lentivirus transduced cells; TRIM14, TRIM14 overexpression lentivirus transduced cells. ***p < 0.001 versus VECTOR.
DISCUSSION
TRIM14 is a member of the TRIM family. Some TRIM family proteins play a key role in tumor progression. TRIM family members participate in a range of biological processes, such as cell growth, development, apoptosis, transcriptional regulation, and cellular differentiation30. Recent studies have reported the upregulation of TRIM14 in many tumor cell lines28,29,31. For example, overexpression of TRIM14 enhances proliferation, cell cycle progression, clone formation, migration, and invasion in osteosarcoma cells29. However, the function and expression of TRIM14 in breast cancer remain unclear. Our data showed that upregulation of TRIM14 in breast cancer tissues and cell lines (Fig. 1). TRIM14 knockdown inhibited cell proliferation but increased cell apoptosis in BT474 and MDA-MB-231 cells. Conversely, overexpression of TRIM14 promoted cell proliferation but inhibited cell apoptosis (Fig. 6).
To explore the mechanisms, we detected the expression levels of some tumor-related proteins. As reported, BCL2 family members include cell apoptosis suppressors (such as BCL2 and BCL-w) and proapoptotic inducers (such as BAX)32. BCL2 is an antiapoptotic molecule promoting cancer cell survival through the mitochondrial apoptotic pathway33,34. BAX is a molecule needed for cell apoptosis. In particular, the balance between BAX and BCL2 is important for the regulation of apoptosis35. In this study, knockdown of the expression of TRIM14 inhibited the expression of BCL2 while increasing the expression of BAX. Conversely, overexpression of TRIM14 increased the expression of BCL2 while inhibiting the expression of BAX.
STAT3 is an oncogenic transcription factor, and hyperactivation of STAT3 has been found in many human cancers. STAT3 activation plays a key role in several cellular processes such as cell survival, proliferation, invasion, migration, and cell apoptosis36. Not only is SHP-1 an SH2-containing protein, but it also is a nonreceptor phosphatase12. SHP-1 could silence the JAK/STAT pathway by dephosphorylating STAT314. Targeting the SHP-1/p-STAT3 signaling pathway may be a novel strategy for breast cancer37,38. Here we found that knockdown of the expression of TRIM14 promoted SHP-1 while inhibiting p-STAT3 (Fig. 4). Conversely, overexpression of TRIM14 downregulated SHP-1 while upregulating p-STAT3 (Fig. 6).
In conclusion, TRIM14 was upregulated in both tissues and cell lines of human breast cancer. Knockdown of TRIM14 inhibited cell proliferation but increased cell apoptosis. Furthermore, the balance between BAX and BCL2 and the SHP-1/p-STAT3 signaling pathway was disturbed by TRIM14, which induced breast cancer initiation and progression. Our results suggest that TRIM14 acts as an oncogene in human breast cancer and thus may be a potential target for treatment.
ACKNOWLEDGMENT
The authors declare no conflicts of interest.
Footnotes
The authors declare no conflicts of interest.
REFERENCES
- 1. Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61(2):69–90. [DOI] [PubMed] [Google Scholar]
- 2. Vogel CL, Cobleigh MA, Tripathy D, Gutheil JC, Harris LN, Fehrenbacher L, Slamon DJ, Murphy M, Novotny WF, Burchmore M, Shak S, Stewart SJ, Press M. Efficacy and safety of trastuzumab as a single agent in first-line treatment of HER2-overexpressing metastatic breast cancer. J Clin Oncol. 2002;20(3):719–26. [DOI] [PubMed] [Google Scholar]
- 3. Grandis JR, Drenning SD, Chakraborty A, Zhou MY, Zeng Q, Pitt AS, Tweardy DJ. Requirement of Stat3 but not Stat1 activation for epidermal growth factor receptor-mediated cell growth in vitro. J Clin Invest. 1998;102(7):1385–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gritsko T, Williams A, Turkson J, Kaneko S, Bowman T, Huang M, Nam S, Eweis I, Diaz N, Sullivan D, Yoder S, Enkemann S, Eschrich S, Lee JH, Beam CA, Cheng J, Minton S, Muro-Cacho CA, Jove R. Persistent activation of stat3 signaling induces survivin gene expression and confers resistance to apoptosis in human breast cancer cells. Clin Cancer Res. 2006;12(1):11–9. [DOI] [PubMed] [Google Scholar]
- 5. Doucette TA, Kong LY, Yang Y, Ferguson SD, Yang J, Wei J, Qiao W, Fuller GN, Bhat KP, Aldape K, Priebe W, Bogler O, Heimberger AB, Rao G. Signal transducer and activator of transcription 3 promotes angiogenesis and drives malignant progression in glioma. Neuro Oncol. 2012;14(9):1136–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sharma SV, Gajowniczek P, Way IP, Lee DY, Jiang J, Yuza Y, Classon M, Haber DA, Settleman J. A common signaling cascade may underlie “addiction” to the Src, BCR-ABL, and EGF receptor oncogenes. Cancer Cell 2006;10(5):425–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Alvarez JV, Febbo PG, Ramaswamy S, Loda M, Richardson A, Frank DA. Identification of a genetic signature of activated signal transducer and activator of transcription 3 in human tumors. Cancer Res. 2005;65(12):5054–62. [DOI] [PubMed] [Google Scholar]
- 8. Yeh YT, Ou-Yang F, Chen IF, Yang SF, Wang YY, Chuang HY, Su JH, Hou MF, Yuan SS. STAT3 ser727 phosphorylation and its association with negative estrogen receptor status in breast infiltrating ductal carcinoma. Int J Cancer 2006;118(12):2943–7. [DOI] [PubMed] [Google Scholar]
- 9. Yu CL, Meyer DJ, Campbell GS, Larner AC, Carter-Su C, Schwartz J, Jove R. Enhanced DNA-binding activity of a Stat3-related protein in cells transformed by the Src oncoprotein. Science 1995;269(5220):81–3. [DOI] [PubMed] [Google Scholar]
- 10. Gough DJ, Corlett A, Schlessinger K, Wegrzyn J, Larner AC, Levy DE. Mitochondrial STAT3 supports Ras-dependent oncogenic transformation. Science 2009;324(5935):1713–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wang X, Crowe PJ, Goldstein D, Yang JL. STAT3 inhibition, a novel approach to enhancing targeted therapy in human cancers (review). Int J Oncol. 2012;41(4):1181–91. [DOI] [PubMed] [Google Scholar]
- 12. Oka T, Ouchida M, Koyama M, Ogama Y, Takada S, Nakatani Y, Tanaka T, Yoshino T, Hayashi K, Ohara N, Kondo E, Takahashi K, Tsuchiyama J, Tanimoto M, Shimizu K, Akagi T. Gene silencing of the tyrosine phosphatase SHP1 gene by aberrant methylation in leukemias/lymphomas. Cancer Res. 2002;62(22):6390–4. [PubMed] [Google Scholar]
- 13. Thangaraju M, Sharma K, Liu D, Shen SH, Srikant CB. Interdependent regulation of intracellular acidification and SHP-1 in apoptosis. Cancer Res. 1999;59(7):1649–54. [PubMed] [Google Scholar]
- 14. Tai WT, Cheng AL, Shiau CW, Huang HP, Huang JW, Chen PJ, Chen KF. Signal transducer and activator of transcription 3 is a major kinase-independent target of sorafenib in hepatocellular carcinoma. J Hepatol. 2011;55(5):1041–8. [DOI] [PubMed] [Google Scholar]
- 15. Beldi-Ferchiou A, Skouri N, Ben Ali C, Safra I, Abdelkefi A, Ladeb S, Mrad K, Ben Othman T, Ben Ahmed M. Abnormal repression of SHP-1, SHP-2 and SOCS-1 transcription sustains the activation of the JAK/STAT3 pathway and the progression of the disease in multiple myeloma. PLoS One 2017;12(4):e0174835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fan LC, Shiau CW, Tai WT, Hung MH, Chu PY, Hsieh FS, Lin H, Yu HC, Chen KF. SHP-1 is a negative regulator of epithelial-mesenchymal transition in hepatocellular carcinoma. Oncogene 2015;34(41):5252–63. [DOI] [PubMed] [Google Scholar]
- 17. Liu CY, Su JC, Ni MH, Tseng LM, Chu PY, Wang DS, Tai WT, Kao YP, Hung MH, Shiau CW, Chen KF. Obatoclax analog SC-2001 inhibits STAT3 phosphorylation through enhancing SHP-1 expression and induces apoptosis in human breast cancer cells. Breast Cancer Res Treat. 2014;146(1):71–84. [DOI] [PubMed] [Google Scholar]
- 18. Reymond A, Meroni G, Fantozzi A, Merla G, Cairo S, Luzi L, Riganelli D, Zanaria E, Messali S, Cainarca S, Guffanti A, Minucci S, Pelicci PG, Ballabio A. The tripartite motif family identifies cell compartments. EMBO J. 2001;20(9):2140–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chen Y, Guo Y, Yang H, Shi G, Xu G, Shi J, Yin N, Chen D. TRIM66 overexpresssion contributes to osteosarcoma carcinogenesis and indicates poor survival outcome. Oncotarget 2015;6(27):23708–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sutton SK, Koach J, Tan O, Liu B, Carter DR, Wilmott JS, Yosufi B, Haydu LE, Mann GJ, Thompson JF, Long GV, Liu T, McArthur G, Zhang XD, Scolyer RA, Cheung BB, Marshall GM. TRIM16 inhibits proliferation and migration through regulation of interferon beta 1 in melanoma cells. Oncotarget 2014;5(20):10127–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhu Z, Wang Y, Zhang C, Yu S, Zhu Q, Hou K, Yan B. TRIM25 blockade by RNA interference inhibited migration and invasion of gastric cancer cells through TGF-beta signaling. Sci Rep. 2016;6:19070. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 22. Wang Y, He D, Yang L, Wen B, Dai J, Zhang Q, Kang J, He W, Ding Q, He D. TRIM26 functions as a novel tumor suppressor of hepatocellular carcinoma and its downregulation contributes to worse prognosis. Biochem Biophys Res Commun. 2015;463(3):458–65. [DOI] [PubMed] [Google Scholar]
- 23. Kawabata H, Azuma K, Ikeda K, Sugitani I, Kinowaki K, Fujii T, Osaki A, Saeki T, Horie-Inoue K, Inoue S. TRIM44 is a poor prognostic factor for breast cancer patients as a modulator of NF-kappaB signaling. Int J Mol Sci. 2017;18(9):E1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wei C, Cheng J, Zhou B, Zhu L, Khan MA, He T, Zhou S, He J, Lu X, Chen H, Zhang D, Zhao Y, Fu J. Tripartite motif containing 28 (TRIM28) promotes breast cancer metastasis by stabilizing TWIST1 protein. Sci Rep. 2016;6:29822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Dukel M, Streitfeld WS, Tang TC, Backman LR, Ai L, May WS, Brown KD. The breast cancer tumor suppressor TRIM29 is expressed via ATM-dependent signaling in response to hypoxia. J Biol Chem. 2016;291(41):21541–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tisserand J, Khetchoumian K, Thibault C, Dembele D, Chambon P, Losson R. Tripartite motif 24 (Trim24/Tif1alpha) tumor suppressor protein is a novel negative regulator of interferon (IFN)/signal transducers and activators of transcription (STAT) signaling pathway acting through retinoic acid receptor alpha (Raralpha) inhibition. J Biol Chem. 2011;286(38):33369–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kamitani S, Ohbayashi N, Ikeda O, Togi S, Muromoto R, Sekine Y, Ohta K, Ishiyama H, Matsuda T. KAP1 regulates type I interferon/STAT1-mediated IRF-1 gene expression. Biochem Biophys Res Commun. 2008;370(2):366–70. [DOI] [PubMed] [Google Scholar]
- 28. Su X, Wang J, Chen W, Li Z, Fu X, Yang A. Overexpression of TRIM14 promotes tongue squamous cell carcinoma aggressiveness by activating the NF-kappaB signaling pathway. Oncotarget 2016;7(9):9939–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Xu G, Guo Y, Xu D, Wang Y, Shen Y, Wang F, Lv Y, Song F, Jiang D, Zhang Y, Lou Y, Meng Y, Yang Y, Kang Y. TRIM14 regulates cell proliferation and invasion in osteosarcoma via promotion of the AKT signaling pathway. Sci Rep. 2017;7:42411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cambiaghi V, Giuliani V, Lombardi S, Marinelli C, Toffalorio F, Pelicci PG. TRIM proteins in cancer. Adv Exp Med Biol. 2012;770:77–91. [DOI] [PubMed] [Google Scholar]
- 31. Zhan W, Han T, Zhang C, Xie C, Gan M, Deng K, Fu M, Wang JB. TRIM59 promotes the proliferation and migration of non-small cell lung cancer cells by upregulating cell cycle related proteins. PLoS One 2015;10(11):e0142596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Goping IS, Gross A, Lavoie JN, Nguyen M, Jemmerson R, Roth K, Korsmeyer SJ, Shore GC. Regulated targeting of BAX to mitochondria. J Cell Biol. 1998;143(1):207–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Juin P, Geneste O, Gautier F, Depil S, Campone M. Decoding and unlocking the BCL-2 dependency of cancer cells. Nat Rev Cancer 2013;13(7):455–65. [DOI] [PubMed] [Google Scholar]
- 34. Braso-Maristany F, Filosto S, Catchpole S, Marlow R, Quist J, Francesch-Domenech E, Plumb DA, Zakka L, Gazinska P, Liccardi G, Meier P, Gris-Oliver A, Cheang MCU, Perdrix-Rosell A, Shafat M, Noel E, Patel N, McEachern K, Scaltriti M, Castel P, Noor F, Buus R, Mathew S, Watkins J, Serra V, Marra P, Grigoriadis A, Tutt AN. PIM1 kinase regulates cell death, tumor growth and chemotherapy response in triple-negative breast cancer. Nat Med. 2016;22(11):1303–13. [Erratum Nat Med. 2017;23(4):526] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. van Delft MF, Huang DC. How the Bcl-2 family of proteins interact to regulate apoptosis. Cell Res. 2006;16(2):203–13. [DOI] [PubMed] [Google Scholar]
- 36. Lim CP, Cao X. Structure, function, and regulation of STAT proteins. Mol Biosyst. 2006;2(11):536–50. [DOI] [PubMed] [Google Scholar]
- 37. Liu CY, Tseng LM, Su JC, Chang KC, Chu PY, Tai WT, Shiau CW, Chen KF. Novel sorafenib analogues induce apoptosis through SHP-1 dependent STAT3 inactivation in human breast cancer cells. Breast Cancer Res. 2013;15(4):R63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Huang TT, Su JC, Liu CY, Shiau CW, Chen KF. Alteration of SHP-1/p-STAT3 signaling: A potential target for anticancer therapy. Int J Mol Sci. 2017;18(6):E1234. [DOI] [PMC free article] [PubMed] [Google Scholar]