Abstract
MicroRNAs are essential regulators of cancer-associated genes at the posttranscriptional level, and their expression is altered in cancer tissues. Herein we sought to identify the regulation of miR-615-3p in NSCLC progression and its mechanism. miR-615-3p expression was significantly downregulated in NSCLC tissue compared to control normal tissue. Exogenous overexpression of miR-615-3p inhibited the growth and metastasis of NSCLC cells. In addition, the in vivo mouse xenograft model showed that overexpression of miR-615-3p inhibited NSCLC growth and lung metastasis, whereas decreased expression of miR-615-3p caused an opposite outcome. Furthermore, we revealed that insulin-like growth factor 2 (IGF2) expression was negatively correlated with the miR-615-3p level in NSCLC specimens, and IGF2 knockdown mimicked the effect of miR-615-3p inhibition on NSCLC cell proliferation, migration, and invasion. In addition, overexpression of IGF2 rescued the inhibition of miR-615-3p in NSCLC cells. Together, our results indicated that miR-615-3p played important roles in the regulation of NSCLC growth and metastasis by targeting IGF2.
Key words: Non-small cell lung cancer (NSCLC), miR-615-3p, IGF2, Metastasis
INTRODUCTION
Non-small cell lung cancer (NSCLC) is one of the most deadly cancers, and its incidence has drastically increased worldwide1. Once NSCLC cells have disseminated to distant and internal organs, NSCLC will be almost resistant to the current clinical treatment and become an incurable disease, with a median survival time of only 6–9 months2,3. Hence, it is noteworthy that the occurrence of cancer cell metastasis is a key promoter of NSCLC, making it aggressive and life-threatening, and the identification of metastasis-related biomarkers and therapeutic targets is therefore urgent for the treatment of NSCLC.
MicroRNAs (miRNAs) are small noncoding RNAs and posttranscriptionally control their downstream target gene expression4,5. Substantive studies have shown that altered expression of miRNAs is associated with human cancer progression, including growth, metastasis and angiogenesis, and chemotherapy resistance6–8. miRNAs commonly function as a tumor suppressor oncogene and play an important role in the diagnosis and treatment of cancer8,9. In NSCLC, several miRNAs such as miR-145, miR-9, and miR-182 have been demonstrated to influence NSCLC cell invasion and metastasis, indicating a great potential for targeting miRNAs in NSCLC treatment10–12. However, abnormal miRNAs that regulate NSCLC invasion and metastasis have not yet been fully investigated. Specifically, previous studies identified five miRNAs to be closely associated with the prognosis of patients with NSCLC: miR-615-3p, miR-142-5p, miR-150-5p, miR-342-3p, and miR-146b-5p13–17. miR-615-3p was shown to be dysregulated in different types of malignancies such as gastric cancer, colorectal carcinoma, hepatocellular carcinoma, and breast cancer18,19. However, the role of miR-615-3p in NSCLC growth and metastasis has not been studied.
Insulin-like growth factor 2 (IGF2) is considered to play a crucial role in cancer progression and metastasis; it shows a high level of expression in epithelial cancers, including prostate cancer, hepatocellular carcinoma, lung cancer, and pancreatic cancer, and its expression is correlated with a poor prognosis20. Through driving epithelial–mesenchymal transition (EMT), IGF2 contributes to the metastasis of carcinoma cells, and prior studies demonstrated that IGF2 conferred stemness and resistance21–23. Inhibition of IGF2 reversed EMT and chemoresistance in chemoresistant human lung cancer cells24. In this study, we found that miR-615-3p was significantly downregulated in NSCLC samples, including clinical specimens and cell lines. With overexpression of miR-615-3p, cell growth, invasion, and metastasis were significantly inhibited, and tumor growth was inhibited, whereas reduced expression of miR-615-3p exhibited the opposite outcomes. In addition, we demonstrated that IGF2, the miR-615-3p target gene, plays very important roles in this process.
MATERIALS AND METHODS
NSCLC Specimens and Cell Culture
The frozen human NSCLC samples and paratumor tissue were obtained from Binzhou Central Hospital (Shandong, P.R. China). This research was approved by the Institutional Research Committee of Binzhou Central Hospital. Written informed consent was obtained from all patients before participation in this study. The NSCLC cell lines, including H1975, A549, HCC827, and PLC/PRF/5, were obtained from the Chinese Academy of Sciences Cell Bank of Type Culture Collection (CBTCCCAS; Shanghai, P.R. China). The lung cancer cell lines were cultured as monolayers in RPMI-1640 or DMEM culture media supplemented with 10% FBS (Wisent, Quebec, Canada) and 100 μg/ml streptomycin/penicillin and maintained in an incubator with a humidified atmosphere of 95% air and 5% CO2 at 37°C.
Transfection of miR-615-3p Precursors and Inhibitor
miR-615-3p and its positive control (miR-NC), as well as miR-615-3p inhibitor and its negative control (miR-NC inhibitor), were purchased from Ambion (GeneCopoeia, Guangzhou, P.R. China). Transfection of miR-615-3p or inhibitor was carried out using Lipofectamine RNAiMAX transfection reagent (Invitrogen, Carlsbad CA, USA) according to the manufacturer’s protocol.
Transient Transfection and Lentiviral Constructs
Transfection of vectors or short hairpin RNA (shRNA) into cells was performed with Lipofectamine 2000 reagent (Invitrogen). The shRNAs against the human IGF2 gene were designed and purchased commercially from Genepharma Co., Ltd. (Shanghai, P.R. China). For establishment of stable IGF2-overexpressing cells, lentiviral vector encoding human IGF2 cDNA was constructed by Genepharma Co., Ltd. and designated as pLV-IGF2. The empty vector was used as control (designated as pLV-vector).
Luciferase Reporter Assay
293T cells were seeded in a 96-well plate at 70% to 80% confluence. After 24 h, cells were cotransfected with miR-615-3p and the wild-type (WT) or mutant (MUT) 3′-UTR of IGF2. Forty-eight hours after transfection, 293T cells were collected, and Renilla luciferase activity was assessed by a dual-luciferase reporter assay system (Promega, Madison, WI, USA).
MTT Assay
Cells were seeded in 96-well culture plates and cultured for 1, 2, or 3 days. MTT (5 mg/ml) was added into the 96-well culture plates, and the cells were incubated for 4 h. Then the cell supernatant was removed, and 200 μl of DMSO was added; the optical density (OD) value was measured at 490 nm25.
Wound Healing Assay
Cells were seeded into a six-well plate and cultured for 24 h. The monolayer cells were scratched by a 100-μl tip, and cells were cultured with serum-free medium. The images of wound closure at 0 and 24 h were recorded using a common microscope, and the width of the wound was obtained using ImageJ software4.
Transwell Invasion Assay
Cells were seeded into a chamber with a pore size of 8 μm coated with Matrigel and placed in a 24-well culture plate. After 24 h, the lower chamber cells were fixed and stained with 0.1% crystal violet. The invaded cells were counted26.
Colony Formation Assay
Indicated cells were seeded in 25-mm3 culture dish and cultured in an incubator for 3 weeks. The colonies were stained with 0.1% crystal violet, and the number of colonies was counted27.
qRT-PCR Assay
The total RNA was extracted from cells using TRIzol (TakaraBio, Tokyo, Japan). One microgram of RNA was reverse transcripted to cDNA using a PrimeScript RT Reagent Kit (TakaraBio). qRT-PCR was performed to detect the mRNA level of miR-615-3p or other genes using IQ™ SYBR Green Supermix and the iQ5 real-time detection system28. The formula of relative expression value was determined through the 2(−ΔΔCt) method. We used the reference gene GAPDH when detecting the mRNA level of miR-615-3p in NSCLC tissues and paratumor tissues. The primers used for the PCR were as follows: GAPDH, 5′-CTGGGCTACACTGAGCACC-3′ (forward) and 5′-AAGTGGTCGTTGAGGGCAATG-3′ (reverse); IGF2, 5′-TATATCGGAAACCTCAGCGAGA-3′ (forward) and 5′-GGACCGAGTGCTCAACTTCT-3′ (reverse).
Xenograft Model of NSCLC and NSCLC Cell Metastasis In Vivo
All nude mice were bred and housed in AAALAC-accredited SPF rodent facilities at Binzhou Central Hospital. All experiments were conducted in accordance with standard operating procedures approved by the University Committee on the Use and Care of Animals at Binzhou Central Hospital. miR-615-3p and its positive control (miR-NC) as well as miR-615-3p inhibitor and its negative control (miR-NC inhibitor)-transfected cells were subcutaneously implanted into the flanks of nude mice (n = 6 for each group). The tumor sizes were measured once every week with a caliper and calculated as tumor volume = length × width2/2. For NSCLC cell metastasis in vivo, the cells were transfected with miR-615-3p or miR-615-3p inhibitor and injected into the tail vein of nude mice. After 14 days, the mice were sacrificed, and the lungs were excised. Lungs were stained with Bouin’s solution for 24 h and then paraffin embedded, sectioned, and stained with H&E29.
Statistical Analysis
Results are presented as the mean ± SD for the three repeated experiments in each group, and the difference of two samples was compared using the Student’s t-test. A value of p < 0.05 was considered as significant.
RESULTS
The Expression of miR-615-3p Is Reduced in NSCLC
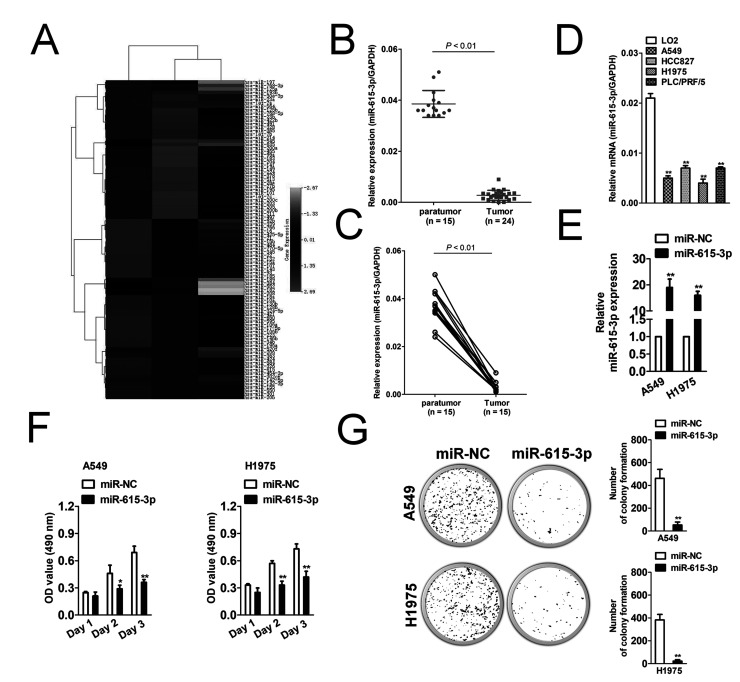
Gene expression data sets used for statistical analysis were acquired from the GEO database with the accession code GSE53882. The screening was performed in GEO data sets, which contained both the NSCLC samples and the matched normal lung samples. As shown in Figure 1A, miR-615-3p was most reduced in NSCLC tissues. In order to confirm the pattern of miR-615-3p expression in NSCLC and control normal tissue, miR-615-3p levels were detected in 24 NSCLC tissues and 15 paratumor tissues. As shown in Figure 1B, the mRNA level of miR-615-3p in NSCLC was much lower than that in the control group. In addition, miR-615-3p was significantly downregulated in NSCLC tissue compared to the paratumor tissue from the same patient (Fig. 1C). miR-615-3p levels were also detected in a panel of NSCLC cell lines by qRT-PCR. Similar results were obtained in four NSCLC cell lines in which the level of miR-615-3p was lowly expressed in all NSCLC cell lines (Fig. 1D). In order to investigate the role of miR-615-3p in the growth of NSCLC, we constructed the miR-615-3p overexpression system. miR-615-3p was significantly overregulated in miR-615-3p-transfected NSCLC A549 and H1975 cells (Fig. 1E). The MTT assay results showed that the induction of miR-615-3p significantly inhibited the proliferation of NSCLC cells (Fig. 1F). Colony formation growth analysis revealed that miR-615-3p overexpression inhibited the colony growth of tumor cells (Fig. 1G). These results indicate that miR-615-3p is a tumor promoter in NSCLC in vitro.
Figure 1.
MicroRNA-615-3p (miR-615-3p) inhibits non-small cell lung cancer (NSCLC) cell growth. (A) Microarray analysis of miRNA expression in NSCLC tissues and corresponding normal tissues. (B) Total RNA was extracted from the paratumor and tumor tissues from patients with NSCLC, and then reverse transcribed into cDNA. The mRNA level of miR-615-3p was determined by qRT-PCR assay. N represents the total number of patients. (C) The two connected points represent the level of miR-615-3p in the tumor and paratumor tissue from the same patient with NSCLC. (D) miR-615-3p levels were measured in five different NSCLC cell lines by qRT-PCR. The data are presented as mean ± SD. **p < 0.01, compared to LO2. (E) A549 and SK-MEL-2 cells were transfected with miR-NC or miR-615-3p. The total RNA was isolated and the level of miR-615-3p was determined by qRT-PCR. qPCR values were normalized to the levels of GAPDH. Data are presented as the mean ± SD from three independent measurements. (F) Cells were plated into 96-well plates and cultured for 1, 2, or 3 days. MTT assay was performed, and the optical density (OD) value was measured at 490 nm. (G) Cells were subjected to colony formation assay in a six-well culture plate. The cell colonies were counted after 3 weeks. *p < 0.05, **p < 0.01, compared to miR-NC.
miR-615-3p Overregulation Inhibits Cell Migration and Invasion
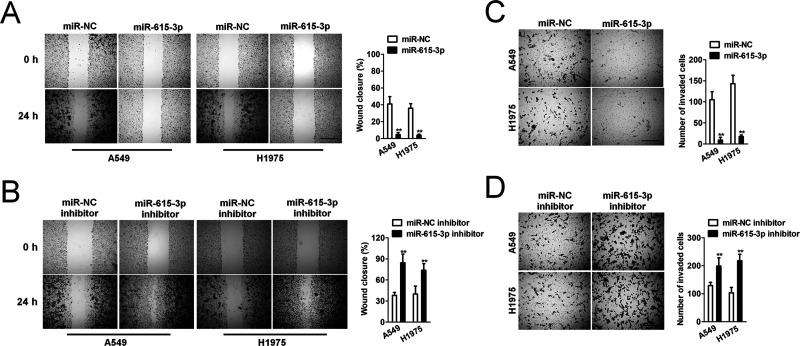
To investigate the effect of miR-615-3p on movement ability and wound healing, we performed a wound healing assay, in which we observed that the distance between the wound edges of miR-615-3p-transfected A549 and H1975 cells was markedly longer than those of control cells (Fig. 2A). Additionally, we observed shorter distance in wound healing of miR-615-3p inhibitor-transfected A549 and H1975 cells compared with control cells (Fig. 2B). To investigate the effect of altering the expression of miR-615-3p on NSCLC cell invasion in vitro, we performed a Transwell invasion assay, in which we observed that miR-615-3p overexpression remarkably suppressed both A549 and H1975 cells invasion (Fig. 2C). As expected, we observed that miR-615-3p inhibitor significantly accelerated NSCLC cell invasion in vitro (Fig. 2D).
Figure 2.
Effects of miR-615-3p on migration and invasion in NSCLC cells (A) Wound healing assay with NSCLC A549 and H1975 cells. Microscopic observations were recorded 0 and 24 h after scratching the cell surface. A representative image is shown (left). Histograms show the percentage of wound closure (right). (B) Transwell assay. Photographs show NSCLC cells that invaded through the Transwell membrane with Matrigel (left). Histograms show the numbers of invasion cells (right). **p < 0.01, compared to miR-NC. (C) Wound healing assay with NSCLC A549 and H1975 cells. Microscopic observations were recorded 0 and 24 h after scratching the cell surface. A representative image is shown (left). Histograms show the percentage of wound closure (right). (D) Transwell assay. Photographs show cells that invaded through the membrane (left). Histograms show the numbers of invasion cells (right). **p < 0.01, compared to miR-NC inhibitor.
miR-615-3p Overexpression Inhibits NSCLC Cell Metastasis In Vivo
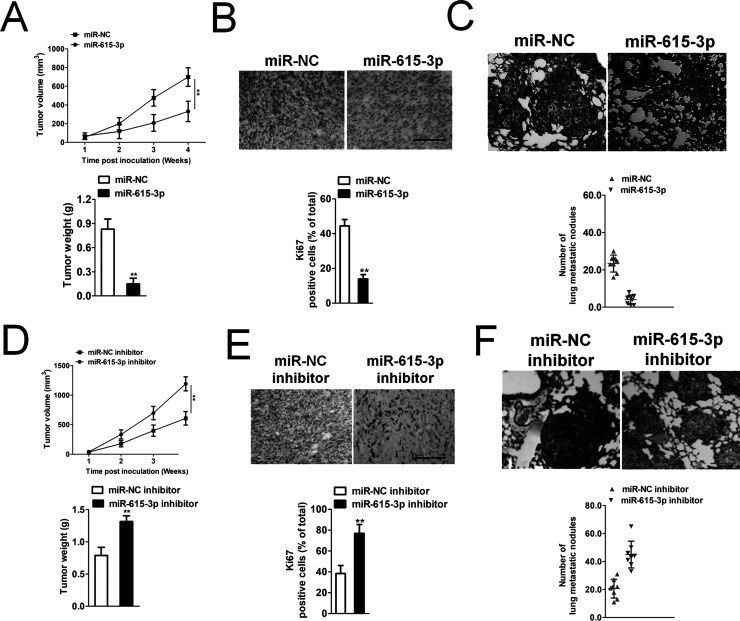
In vitro, miR-615-3p significantly inhibited NSCLC cell proliferation, migration, and invasion. Whether miR-615-3p could regulate tumor growth and metastasis in vivo needed to be further investigated. A549 cells transfected with miR-615-3p were subcutaneously implanted into nude mice, and the tumors were monitored and measured every week for 4 weeks. miR-615-3p overexpression remarkably inhibited the growth and the tumor volume of the NSCLC cells (Fig. 3A). We dissected the mice to harvest the tumor, noting that the Ki-67 staining was significantly lower in the miR-615-3p group than in the miR-NC group (Fig. 3B), which confirmed that miR-615-3p inhibited NSCLC growth in vivo. In addition, the incidence of lung metastasis of NSCLC cells in the miR-615-3p transfection group was remarkably suppressed, compared with the control group (Fig. 3C). To further determine the effects of miR-615-3p on in vivo tumor growth and metastasis, parental or miR-615-3p downregulated A549 cells were implanted into the nude mice to establish xenograft models. Downregulation of miR-615-3p markedly accelerated the tumor growth of A549 cells in vivo (Fig. 3D). Then we analyzed the level of Ki-67 in the paraffin sections of the tumor by immunohistochemical staining. The results of the immunohistochemical assay showed that the Ki-67 level in the tumors of the miR-615-3p downregulated group was double that of the controls (Fig. 3E). Finally, parental or miR-615-3p downregulated A549 cells were injected into nude mice via the tail vein. As expected, when the mice were dissected, there was a significant increase in the lung tumor lesions between the miR-615-3p downregulated group and the control group (Fig. 3F). All these results demonstrated that miR-615-3p inhibits the growth and metastasis of NSCLC cells in vivo.
Figure 3.
Effect of miR-615-3p on A549 cell tumor growth and metastasis in vivo. (A) A549 cells were transfected with miR-NC or miR-615-3p and then were implanted into the nude mice. The tumor volume was measured every 3 days. The tumors of the two groups were separated at the end of treatment. (B) The tumors were removed and embedded in paraffin for immunohistochemical staining with Ki-67. (C) Metastatic lesions in the lungs of the NSCLC cell metastasis models are shown at 6 weeks after tail vein injection. The total number of lung metastatic lesions in the miR-615-3p transfection groups was much less than those in controls. (D) A549 cells were transfected with miR-NC inhibitor or miR-615-3p inhibitor and then were implanted into the nude mice. The tumor volume was measured every 3 days. (E) The tumors were removed and embedded in paraffin for immunohistochemical staining with Ki-67. (F) Metastatic lesions in the lungs of the models are shown at 6 weeks after tail vein injection. The total number of lung metastatic lesions in the miR-615-3p downregulated group was much higher than those in controls. **p < 0.01, compared to miR-NC inhibitor.
miR-615-3p Directly Binds to the 3′-UTR of IGF2
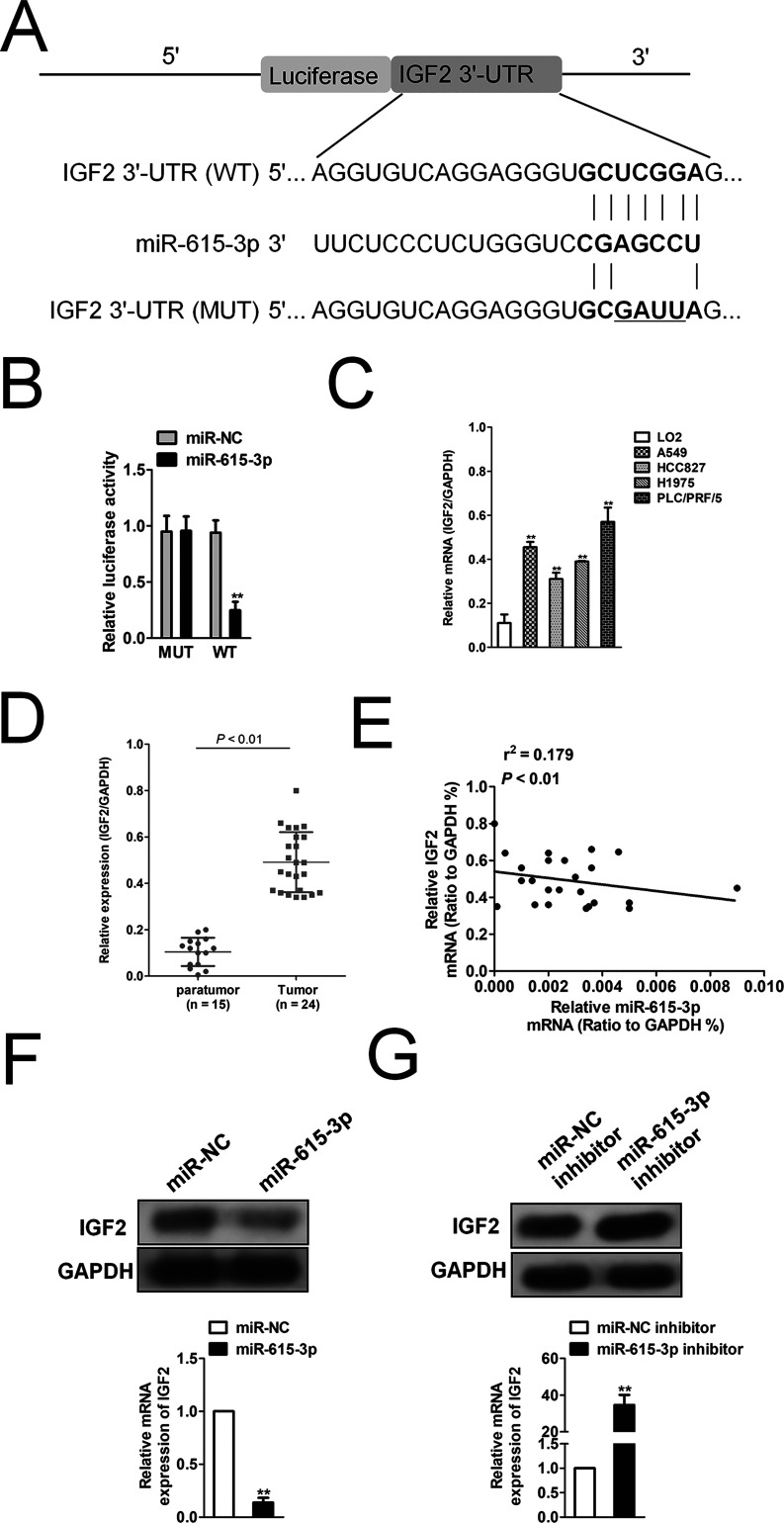
Generally, miRNAs regulate their target genes by binding to the 3′-UTR of genes. miRBase and TargetScan were used to predict the target genes of miR-615-3p, and we found that IGF2 may be a target gene of miR-615-3p (Fig. 4A). To investigate whether miR-615-3p binds directly to the 3′-UTR of IGF2 and modulates IGF2 expression, the 3′-UTR of IGF2 was cloned into the downstream of the luciferase reporter gene. Luciferase activity analysis revealed that miR-615-3p could significantly reduce the activity of luciferase (Fig. 4B). Taken together, the results indicated that miR-615-3p regulated IGF2 by binding to the 3′-UTR region directly. qRT-PCR assay analysis showed that NSCLC cell lines exhibited high IGF2 expression (Fig. 4C). In addition, IGF2 was highly expressed in the NSCLC samples by analyzing the level of IGF2 in NSCLC samples compared to normal tissue samples (Fig. 4D). We also investigated the relationship between miR-615-3p and IGF2 in NSCLC specimens. As shown in Figure 4E, there was a negative correlation between the miR-615-3p levels and IGF2 levels. Additionally, IGF2 was significantly downregulated after cells were transfected with miR-615-3p (Fig. 4F), whereas IGF2 was remarkably upregulated after cells were transfected with miR-615-3p inhibitor (Fig. 4G). These results indicate that IGF2 may be the target gene of miR-615-3p.
Figure 4.
Insulin-like growth factor 2 (IGF2) is a direct target of miR-615-3p. (A) Predicted miR-615-3p target sequences in the 3′-UTR of IGF2. (B) Luciferase activity of IGF2 (wild type or mutation) was evaluated after miR-615-3p transfection into 293T cells. **p < 0.01, compared to miR-NC. (C) The expression level of IGF2 was detected by qRT-PCR assay in several NSCLC cell lines. **p < 0.01, compared to LO2 cells. (D) Total RNA was extracted from tumor tissues of patients with NSCLC, and IGF2 expression level was measured by qRT-PCR. N stands for total numbers of patients. (E) The correlation analysis of miR-615-3p and IGF2 expressions. (F) The level of IGF2 in A549 cells after cells transfected with miR-615-3p was measured by qRT-PCR and Western blotting assays. (G) The expression level of IGF2 in A549 cells after cells transfected with miR-615-3p inhibitor was measured by qRT-PCR and Western blot analysis. **p < 0.01, compared to miR-NC inhibitor.
The Effect of miR-615-3p on NSCLC Is Rescued by IGF2 Overexpression
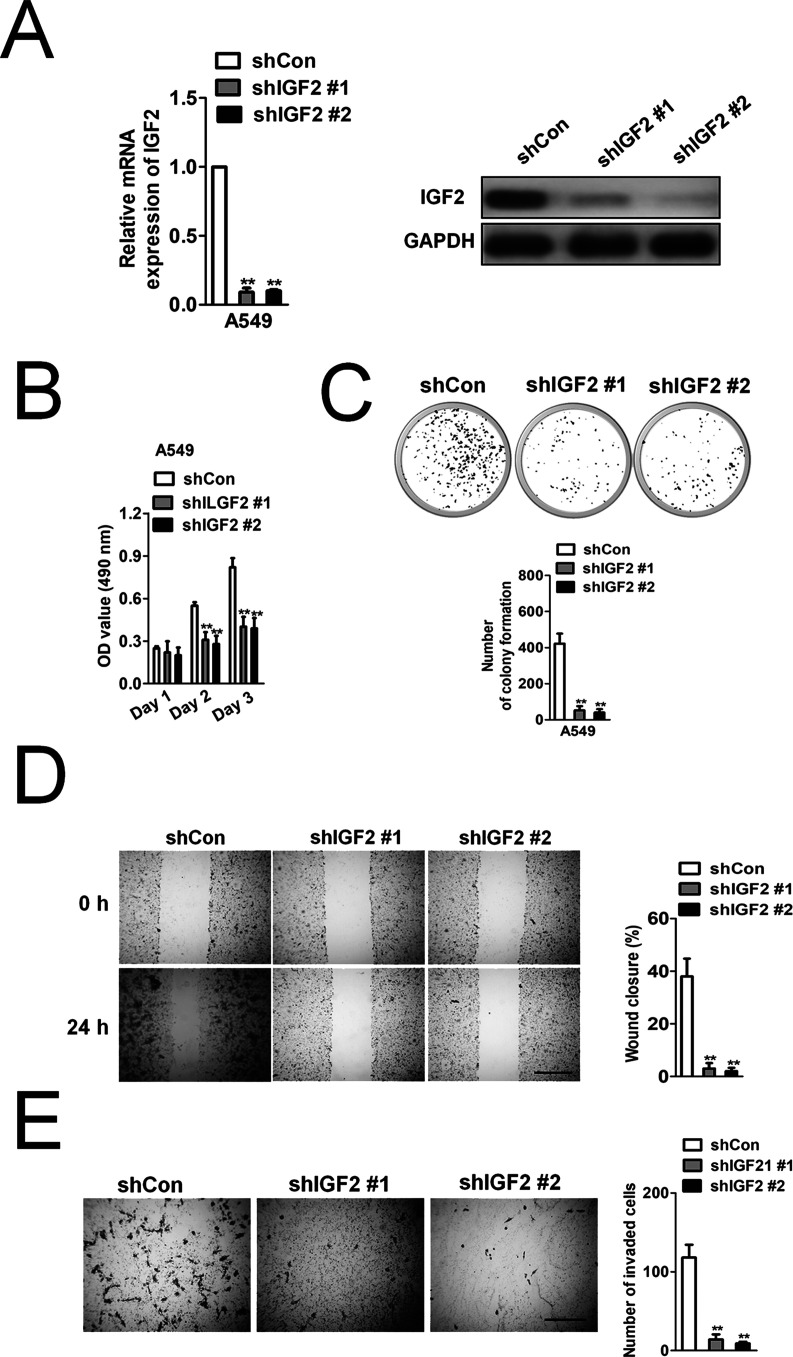
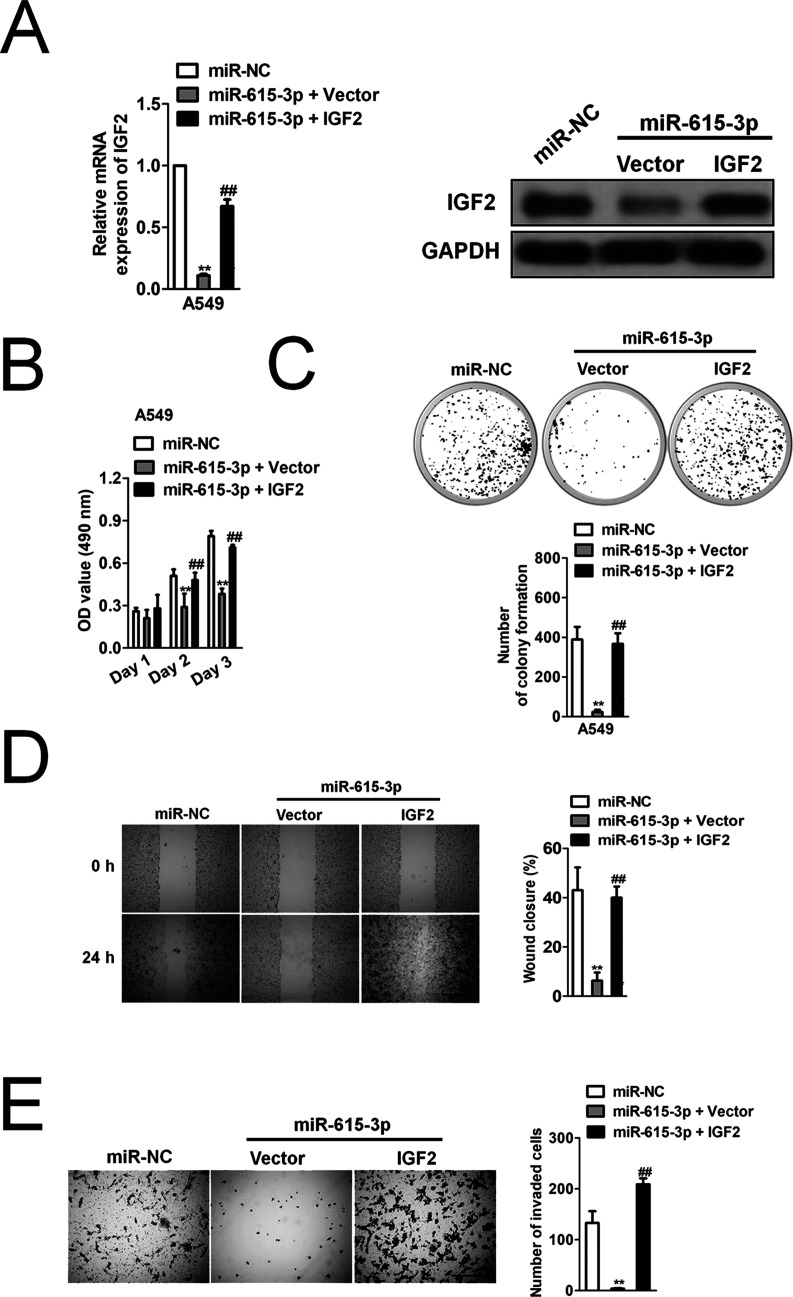
Nevertheless, little is known about the role of IGF2 in NSCLC progression and whether IGF2 knockdown could mimic miR-615-3p on NSCLC. Two different shRNAs, which specifically target IGF2, were used to stably knock down the expression of IGF2 in the A549 cells (Fig. 5A). To reveal the effect of IGF2 on NSCLC growth and metastasis, the effect of IGF2 knockdown on A549 cell proliferation, colony formation, migration, and invasion was examined (Fig. 5B–E). The stable knockdown of IGF2 in A549 had a similar effect to that of miR-615-3p overexpression in A549 cells on growth, migration, and invasion in vitro, which suggested that IGF2 might exert a carcinogenic effect on NSCLC growth and metastasis. To investigate whether miR-615-3p inhibited the proliferation and invasion of NSCLC by inhibiting IGF2, we constructed A549 cell lines that were cotransfected with miR-615-3p and IGF2 overexpression. Western blot and qRT-PCR analysis showed that ectopic expression of IGF2 rescued its expression after A549 cells were transfected with miR-615-3p (Fig. 6A). MTT assay, colony formation, wound healing, and Transwell invasion assay showed that IGF2 overexpression restored the inhibitory effect of miR-615-3p in NSCLC cell proliferation and metastasis (Fig. 6B–E).
Figure 5.
IGF2 knockdown mimics the inhibitory effects of miR-615-3p on cell invasion and proliferation in A549 cells. (A) To knock down IGF2, shIGF2 or shCon was transfected into A549 cells. The IGF2 expression level was detected by qRT-PCR and Western blot. (B) A549 cell proliferation activity was measured by MTT. (C) Colony formation ability of A549 cells was measured by colony formation assay. The image and quantitative analysis of colonies are shown. (D) A549 cells were transfected with shIGF2 or shCon, and then the migration ability of the cells was investigated with the wound healing assay. Quantitative analysis of the percentage of wound closure from three independent experiments was shown. (E) The invasion capacity of A549 cells was investigated with the Transwell invasion assay. Quantitative analysis of the invasive cells is shown. The data are presented as mean ± SD. **p < 0.01, compared to shCon.
Figure 6.
The effect of miR-615-3p on NSCLC cells can be rescued by IGF2 overexpression. (A) IGF2 overexpression lentivirus was transfected into A549 cells. The expression level of IGF2 was detected by qRT-PCR analysis and Western blot. (B) Cell proliferation activity was measured by MTT assay. (C) Growth ability was measured by the colony formation assay in vitro. The image and quantitative analysis of colonies are shown. (D) A549 cells were cotransfected with IGF2 or miR-615-3p, and then the migration ability of the cells was investigated with the wound healing assay. Quantitative analysis of the percentage of wound closure from three independent experiments is shown. (E). The invasion ability of the cells was investigated with the Transwell invasion assay. Quantitative analysis of the invading cells from three independent experiments is shown. The data are presented as mean ± SD. **p < 0.01, compared to miR-NC. ##p < 0.01, compared to cells cotransfected with miR-615-3p and vector.
DISCUSSION
NSCLC is one of the most aggressive tumors, and the occurrence of metastasis leads to a sharp decline in the survival rate of patients30–32. Thus, identification of metastatic-related biomarkers and therapeutic targets will help improve NSCLC33,34. Recently, miRNAs have been reported to be involved in the regulation of cancer invasion and metastasis, and have been shown to be potentially noninvasive biomarkers in various cancers35,36. Here we report that miR-615-3p is a novel metastasis-associated miRNA that plays a significant role in regulating NSCLC growth and metastasis. We found a significant decrease in miR-615-3p levels in NSCLC tissues and NSCLC cell lines. In addition, overexpression of miR-615-3p inhibits the aggressiveness and migration of NSCLC cells by direct targeting of IGF2. Finally, overexpression of miR-615-3p suppresses the growth and metastasis of NSCLC in vivo. In conclusion, our results suggest that metastatic-associated miR-615-3p is not only a potential biomarker but also a valuable therapeutic target for NSCLC.
It has been reported that miR-615-3p directly targets certain oncogenes and that overexpression of miR-615-3p in tumors leads to upregulation of these target genes37. In recent years, several studies on the role of miR-615-3p in tumor progression have been performed38,39. In our study, we found that miR-615-3p was downregulated in NSCLC tissues and cell lines. More importantly, miR-615-3p overexpression inhibited NSCLC metastasis, with a low degree of migration and invasive abilities, indicating the great potential of miR-615-3p for controlling NSCLC metastasis. Our study also demonstrated the role of miR-615-3p in inhibiting the growth of NSCLC cells in vitro and in vivo, further enhancing the regulation of miR-615-3p in NSCLC. In summary, these results demonstrate the great value of miR-615-3p in the growth and metastasis of NSCLC.
It is well known that miRNAs regulate gene expression by binding to the 3′-UTR of the target gene40,41. In this study, bioinformatics was used to predict the target gene, and IGF2 was selected as the candidate target. The findings were consistent with IGF2 levels being downregulated and upregulated in A549 cells after transfection with miR-615-3p and miR-615-3p inhibitor, respectively. The luciferase reporter assay is a direct method for target validation and confirmed that IGF2 is a direct target gene for miR-615-3p. In addition, the use of shRNA knockdown of IGF2 inhibits A549 cell growth, migration, and invasion, suggesting that IGF2 also plays an important role in regulating NSCLC cell proliferation and metastasis. Importantly, the overexpression of IGF2 rescued the miR-615-3p inhibitory effect on the proliferation, migration, and invasion of NSCLC cells, thus confirming that IGF2 is a direct functional target for miR-615-3p in NSCLC.
In conclusion, we describe the downregulation of miR-615-3p in NSCLC. Overexpression of miR-615-3p inhibits the growth, invasion, and metastasis of NSCLC, whereas downregulation of IGF2 resulted in the opposite outcomes. IGF2, a direct target gene for miR-615-3p, plays a crucial role in this process. These results may help us to better understand the mechanism of miR-615-3p regulation of NSCLC and might provide a novel therapeutic target for the clinical treatment of NSCLC metastasis.
ACKNOWLEDGMENT
The authors declare no conflicts of interest.
Footnotes
The authors declare no conflicts of interest.
REFERENCES
- 1. Ma R, Yang Y, Tu Q, Hu K. Overexpression of T-box transcription factor 5 (TBX5) inhibits proliferation and invasion in non-small cell lung carcinoma cells. Oncol Res. 2017;25(9):1495–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zhao Y, Adjei AA. New strategies to develop new medications for lung cancer and metastasis. Cancer Metastasis Rev. 2015;34(2):265–75. [DOI] [PubMed] [Google Scholar]
- 3. Reck M, Heigener DF, Mok T, Soria JC, Rabe KF. Management of non-small-cell lung cancer: Recent developments. Lancet 2013;382(9893):709–19. [DOI] [PubMed] [Google Scholar]
- 4. Sun J, Tian X, Lu SQ, Hu HB. MicroRNA-4465 suppresses tumor proliferation and metastasis in non-small cell lung cancer by directly targeting the oncogene EZH2. Biomed Pharmacother. 2017;96:1358–62. [DOI] [PubMed] [Google Scholar]
- 5. Li Y, Liang Y, Sang Y, Song X, Zhang H, Liu Y, Jiang L, Yang Q. MiR-770 suppresses the chemo-resistance and metastasis of triple negative breast cancer via direct targeting of STMN1. Cell Death Dis. 2018;9(1):14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zhu Y, Li T, Chen G, Yan G, Zhang X, Wan Y, Li Q, Zhu B, Zhuo W. Identification of a serum microRNA expression signature for detection of lung cancer, involving miR-23b, miR-221, miR-148b and miR-423-3p. Lung Cancer 2017;114:6–11. [DOI] [PubMed] [Google Scholar]
- 7. Ng L, Wan TM, Man JH, Chow AK, Iyer D, Chen G, Yau TC, Lo OS, Foo DC, Poon JT, Leung WK, Pang RW, Law WL. Identification of serum miR-139-3p as a non-invasive biomarker for colorectal cancer. Oncotarget 2017;8(16):27393–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Huang P, Xi J, Liu S. MiR-139-3p induces cell apoptosis and inhibits metastasis of cervical cancer by targeting NOB1. Biomed Pharmacother. 2016;83:850–6. [DOI] [PubMed] [Google Scholar]
- 9. Zhang X, He X, Liu Y, Zhang H, Chen H, Guo S, Liang Y. MiR-101-3p inhibits the growth and metastasis of non-small cell lung cancer through blocking PI3K/AKT signal pathway by targeting MALAT-1. Biomed Pharmacother. 2017;93:1065–73. [DOI] [PubMed] [Google Scholar]
- 10. Li Y, Li Y, Liu J, Fan Y, Li X, Dong M, Liu H, Chen J. Expression levels of microRNA-145 and microRNA-10b are associated with metastasis in non-small cell lung cancer. Cancer Biol Ther. 2016;17(3):272–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Li G, Wu F, Yang H, Deng X, Yuan Y. MiR-9-5p promotes cell growth and metastasis in non-small cell lung cancer through the repression of TGFBR2. Biomed Pharmacother. 2017;96:1170–8. [DOI] [PubMed] [Google Scholar]
- 12. Li Y, Zhang H, Li Y, Zhao C, Fan Y, Liu J, Li X, Liu H, Chen J. MiR-182 inhibits the epithelial to mesenchymal transition and metastasis of lung cancer cells by targeting the Met gene. Mol Carcinog. 2018;57(1):125–36. [DOI] [PubMed] [Google Scholar]
- 13. Li J, Yang H, Li Y, Liu Y, Chen S, Qi C, Zhang Q, Lan T, He X, Guan XY, Wang L. MicroRNA-146 up-regulation predicts the prognosis of non-small cell lung cancer by miRNA in situ hybridization. Exp Mol Pathol. 2014;96(2):195–9. [DOI] [PubMed] [Google Scholar]
- 14. Lin Q, Chen T, Lin Q, Lin G, Lin J, Chen G, Guo L. Serum miR-19a expression correlates with worse prognosis of patients with non-small cell lung cancer. J Surg Oncol. 2013;107(7):767–71. [DOI] [PubMed] [Google Scholar]
- 15. Jiang LP, Zhu ZT, He CY. Expression of miRNA-26b in the diagnosis and prognosis of patients with non-small-cell lung cancer. Future Oncol. 2016;12(9):1105–15. [DOI] [PubMed] [Google Scholar]
- 16. Zhang J, Xu L, Yang Z, Lu H, Hu D, Li W, Zhang Z, Liu B, Ma S. MicroRNA-10b indicates a poor prognosis of non-small cell lung cancer and targets E-cadherin. Clin Transl Oncol. 2015;17(3):209–14. [DOI] [PubMed] [Google Scholar]
- 17. Eilertsen M, Andersen S, Al-Saad S, Richardsen E, Stenvold H, Hald SM, Al-Shibli K, Donnem T, Busund LT, Bremnes RM. Positive prognostic impact of miR-210 in non-small cell lung cancer. Lung Cancer 2014;83(2):272–8. [DOI] [PubMed] [Google Scholar]
- 18. Rahmoon MA, Youness RA, Gomaa AI, Hamza MT, Waked I, El Tayebi HM, Abdelaziz AI. MiR-615-5p depresses natural killer cells cytotoxicity through repressing IGF-1R in hepatocellular carcinoma patients. Growth Factors 2017;35(2–3):76–87. [DOI] [PubMed] [Google Scholar]
- 19. Wang J, Liu L, Sun Y, Xue Y, Qu J, Pan S, Li H, Qu H, Wang J, Zhang J. miR-615-3p promotes proliferation and migration and inhibits apoptosis through its potential target CELF2 in gastric cancer. Biomed Pharmacother. 2018;101:406–13. [DOI] [PubMed] [Google Scholar]
- 20. Cui H. Loss of imprinting of IGF2 as an epigenetic marker for the risk of human cancer. Dis Markers 2007;23(1–2):105–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Brouwer-Visser J, Huang GS. IGF2 signaling and regulation in cancer. Cytokine Growth Factor Rev. 2015;26(3):371–7. [DOI] [PubMed] [Google Scholar]
- 22. Murphy R, Ibanez L, Hattersley A, Tost J. IGF2/H19 hypomethylation in a patient with very low birthweight, preocious pubarche and insulin resistance. BMC Med Genet. 2012;13:42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ogawa T, Ogawa K, Shiga K, Furukawa T, Nagase H, Hashimoto S, Kobayashi T, Horii A. Upregulation of IGF2 is associated with an acquired resistance for cis-diamminedichloroplatinum in human head and neck squamous cell carcinoma. Eur Arch Otorhinolaryngol. 2010;267(10):1599–606. [DOI] [PubMed] [Google Scholar]
- 24. Kondo M, Takahashi T. [Altered genomic imprinting in the IGF2 and H19 genes in human lung cancer]. Nihon Rinsho 1996;54(2):492–6. [PubMed] [Google Scholar]
- 25. Li D, He C, Wang J, Wang Y, Bu J, Kong X, Sun D. MicroRNA-138 inhibits cell growth, invasion, and EMT of non-small cell lung cancer via SOX4/p53 feedback loop. Oncol Res. 2018;26(3):385–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Fan N, Zhang J, Cheng C, Zhang X, Feng J, Kong R. MicroRNA-384 represses the growth and invasion of non-small-cell lung cancer by targeting astrocyte elevated gene-1/Wnt signaling. Biomed Pharmacother. 2017;95:1331–7. [DOI] [PubMed] [Google Scholar]
- 27. He J, Tian N, Yang Y, Jin L, Feng X, Hua J, Lin S, Wang B, Li H, Wang J. miR-185 enhances the inhibition of proliferation and migration induced by ionizing radiation in melanoma. Oncol Lett. 2017;13(4):2442–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Xu G, Cai J, Wang L, Jiang L, Huang J, Hu R, Ding F. MicroRNA-30e-5p suppresses non-small cell lung cancer tumorigenesis by regulating USP22-mediated Sirt1/JAK/STAT3 signaling. Exp Cell Res. 2018;362(2):268–78. [DOI] [PubMed] [Google Scholar]
- 29. Zeng Y, Zhu J, Shen D, Qin H, Lei Z, Li W, Liu Z, Huang JA. MicroRNA-205 targets SMAD4 in non-small cell lung cancer and promotes lung cancer cell growth in vitro and in vivo. Oncotarget 2017;8(19):30817–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Garber ME, Troyanskaya OG, Schluens K, Petersen S, Thaesler Z, Pacyna-Gengelbach M, van de Rijn M, Rosen GD, Perou CM, Whyte RI, Altman RB, Brown PO, Botstein D, Petersen I. Diversity of gene expression in adenocarcinoma of the lung. Proc Natl Acad Sci USA 2001;98(24):13784–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Talbot SG, Estilo C, Maghami E, Sarkaria IS, Pham DK, O-charoenrat P, Socci ND, Ngai I, Carlson D, Ghossein R, Viale A, Park BJ, Rusch VW, Singh B. Gene expression profiling allows distinction between primary and metastatic squamous cell carcinomas in the lung. Cancer Res. 2005;65(8):3063–71. [DOI] [PubMed] [Google Scholar]
- 32. Huang L, Li F, Deng P, Hu C. MicroRNA-223 promotes tumor progression in lung cancer A549 cells via activation of the NF-kappaB signaling pathway. Oncol Res. 2016;24(6):405–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tsoulos N, Papadopoulou E, Metaxa-Mariatou V, Tsaousis G, Efstathiadou C, Tounta G, Scapeti A, Bourkoula E, Zarogoulidis P, Pentheroudakis G, Kakolyris S, Boukovinas I, Papakotoulas P, Athanasiadis E, Floros T, Koumarianou A, Barbounis V, Dinischiotu A, Nasioulas G. Tumor molecular profiling of NSCLC patients using next generation sequencing. Oncol Rep. 2017;38(6):3419–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Beer DG, Kardia SL, Huang CC, Giordano TJ, Levin AM, Misek DE, Lin L, Chen G, Gharib TG, Thomas DG, Lizyness ML, Kuick R, Hayasaka S, Taylor JM, Iannettoni MD, Orringer MB, Hanash S. Gene-expression profiles predict survival of patients with lung adenocarcinoma. Nat Med. 2002;8(8):816–24. [DOI] [PubMed] [Google Scholar]
- 35. Hao C, Xu X, Ma J, Xia J, Dai B, Liu L, Ma Y. MicroRNA-124 regulates the radiosensitivity of non-small cell lung cancer cells by targeting TXNRD1. Oncol Lett. 2017;13(4):2071–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Liu C, Li G, Ren S, Su Z, Wang Y, Tian Y, Liu Y, Qiu Y. miR-185-3p regulates the invasion and metastasis of nasopharyngeal carcinoma by targeting WNT2B in vitro. Oncol Lett. 2017;13(4):2631–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Pu HY, Xu R, Zhang MY, Yuan LJ, Hu JY, Huang GL, Wang HY. Identification of microRNA-615-3p as a novel tumor suppressor in non-small cell lung cancer. Oncol Lett. 2017;13(4):2403–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bai Y, Li J, Li J, Liu Y, Zhang B. MiR-615 inhibited cell proliferation and cell cycle of human breast cancer cells by suppressing of AKT2 expression. Int J Clin Exp Med. 2015;8(3):3801–8. [PMC free article] [PubMed] [Google Scholar]
- 39. Mukai R, Tomimaru Y, Nagano H, Eguchi H, Mimori K, Tomokuni A, Asaoka T, Wada H, Kawamoto K, Marubashi S, Doki Y, Mori M. miR-615-3p expression level in bone marrow is associated with tumor recurrence in hepatocellular carcinoma. Mol Clin Oncol. 2015;3(3):487–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kojima S, Enokida H, Yoshino H, Itesako T, Chiyomaru T, Kinoshita T, Fuse M, Nishikawa R, Goto Y, Naya Y, Nakagawa M, Seki N. The tumor-suppressive microRNA-143/145 cluster inhibits cell migration and invasion by targeting GOLM1 in prostate cancer. J Hum Genet. 2014;59(2):78–87. [DOI] [PubMed] [Google Scholar]
- 41. Gallach S, Jantus-Lewintre E, Calabuig-Farinas S, Montaner D, Alonso S, Sirera R, Blasco A, Uso M, Guijarro R, Martorell M, Camps C. MicroRNA profiling associated with non-small cell lung cancer: Next generation sequencing detection, experimental validation, and prognostic value. Oncotarget 2017;8(34):56143–57. [DOI] [PMC free article] [PubMed] [Google Scholar]