Graphical abstract
Keywords: Hepatocellular carcinoma, Microvascular invasion, Deep learning, Interpretation of machine learning, Blood test
Abstract
Microvascular invasion (MVI) is one of the most important factors leading to poor prognosis for hepatocellular carcinoma (HCC) patients, and detection of MVI prior to surgical operation could great benefit patient’s prognosis and survival. Since it is still lacking effective non-invasive strategy for MVI detection before surgery, novel MVI determination approaches were in urgent need. In this study, complete blood count, blood test and AFP test results are utilized to perform preoperative prediction of MVI based on a novel interpretable deep learning method to quantify the risk of MVI. The proposed method termed as “Interpretation based Risk Prediction” can estimate the MVI risk precisely and achieve better performance compared with the state-of-art MVI risk estimation methods with concordance indexes of 0.9341 and 0.9052 on the training cohort and the independent validation cohort, respectively. Moreover, further analyses of the model outputs demonstrate that the quantified risk of MVI from our model could serve as an independent preoperative risk factor for both recurrence-free survival and overall survival of HCC patients. Thus, our model showed great potential in quantification of MVI risk and prediction of prognosis for HCC patients.
1. Introduction
Hepatocellular Carcinoma (HCC), with its mortality ranking the 3rd among all cancer-related death worldwide, has become one major challenge for human health. The prognosis of HCC patients remained poor, mainly resulted from the high potential of recurrence and metastasis. Microvascular invasion (MVI), a marker representing aggressive tumor behavior, has been reported to correlate with worse prognostic outcomes, especially after potentially curative surgery [1], [2], [3]. The accurate determination of MVI presence in HCC could great assist clinical decision and thus effectively benefit patients’ prognosis with prolonged survival.
Currently, numerous studies have dedicated their efforts to the precise diagnosis and risk evaluation of MVI. The traditional strategy of histologic examination provides the gold standard for MVI detection [4], while the fact that its diagnosis was based on biopsy or surgical samples greatly limited its value in preoperative evaluation [5]. In recent days, novel approaches integrating imaging features have become one of the noteworthy directions of MVI detection and achieved excellent performance [6], [7]. Meanwhile, some predictive models based on clinical features were also proposed [8]. On the other hand, liquid biopsy, a quick and non-invasive strategy to acquire tumor information with greatly reduced risk of medical complication, also draw more and more attention. Liquid biopsy provided a potent tool to facilitate MVI detection without requiring tissue samples information, making it highly suitable in preoperative MVI evaluation scenario. Detection of MVI ahead of surgical operation could be invaluable, as the MVI itself is a key factor affecting surgery strategy making for commonly applied options including hepatectomy and liver transplantation [9]. Complete blood count, blood test for liver damage and AFP test are three most common liquid biopsy tests that HCC patients received regularly to obtain information regarding blood cells, specific types of enzymes and metabolites, all of which could reflect patients’ disease status. Previous studies have linked the blood counts as a serum biomarker for tumor progression [10], while liver function abnormality has also been considered as an indication for HCC burden [11], [12]. Meanwhile, AFP were considered as one of the most noteworthy circulating biomarker for MVI presence [13]. Considering the close relationships between the circulating system and tumor metastasis, comprehensive utilization of these three tests might provide a precise evaluation of MVI risks. However, the large number of parameters presented great difficulty for building an accurate predictive model for MVI based on traditional modeling strategies.
Over the last few years, we have seen the fast development of deep learning, which has become one of the most promising research focuses on the medical field [14], [15], [16], [17]. Under a relatively fixed structure, deep learning algorithms can learn features from big data through continuously iterating their weights, which could overcome the difficulty of non-linear mapping between the input and output data, making it suitable for constructing a predictive model for clinical outcomes influenced by numerous of different factors, such as MVI. To our best knowledge, there are still very few studies focusing on predicting MVI presence with deep learning methods. Cucchetti et al. [18] firstly used a 3-layer fully-connected neural network, i.e., artificial neural network (ANN) to predict MVI with four variables including serum AFP; Men et al. [19] utilized contrast-enhanced MR images to predict MVI by a method combining 3D convolutional neural network (CNN) and long-short term memory (LSTM). Despite the accomplished progress, the “black box” issue stands between the theories and real-world applications. The black box issue of deep learning means that there is so little information about “What does the model exactly depend on to give this prediction”, leading to the doubt about the reliability of the models. The explanation of machine learning models has now attracted increasing attention [20], [21], and it is an obstacle that needs to be overcome in practical applications.
To explore the possibility for MVI risk evaluation based on liquid biopsy, we utilize deep learning methods to fully investigate the relationship between MVI and blood parameters. An Interpretation based Risk Prediction method, abbreviated to the IRP method, was proposed to predict MVI risk based on complete blood count and blood test results. A deep learning explanation method termed as “Local Interpretable Model-agnostic Explanations (LIME)”, proposed by Ribeiro et al. [22], was used to explain the features learnt by the deep learning models and build a scoring model to predict MVI risk of HCC patients. Our results revealed several parameters significantly correlated with MVI risk including Lactate dehydrogenase (LDH), Gamma glutamyl transpeptidase (GGTP) and Aspartate amino transferase (AST). The proposed method could precisely predict the MVI risk of HCC patients with superior performance compared to other state-of-art scoring models, while also provide quantification for the impact of each parameter on the prediction results. Further validation of the model using an independent cohort also demonstrated excellent performance in MVI prediction. Considering that our model was built based on the routine blood test, it could greatly benefit future clinical evaluation of patients’ prognosis and clinical outcomes.
2. Materials and methods
2.1. Patients of the training cohort
A total of 1007 patients received liver resection surgery at Mengchao Hepatobiliary Hospital of Fujian Medical University from 2014 to 2019 were enrolled as the training cohort for this study. The results of complete blood count, blood tests, and AFP tests were acquired within 2 days before surgery, and clinical information as well as pathological examination results were also obtained (Supplementary Tables S1 and S2). The pathological identification of MVI is determined by the presence of cancer cell nests in vessels lined by endothelial cells under a microscope. For each tumor sample, 4 Paraffin blocks corresponding to 4 different junction region of tumor and adjacent liver tissues were used to identify MVI by at least 3 experienced pathologists according to Clinical guideline [23]. In this study, the collection and usage of all patients’ information were approved by the Ethics Committee of Mengchao Hepatobiliary Hospital of Fujian Medical University and followed principles of the Declaration of Helsinki. The results of complete blood count, blood test, and AFP tests contained 18 blood parameters including Lactate dehydrogenase (LDH), Gamma glutamyl transpeptidase (GGTP), Plateletcrit (PCT), AST/ALT ratio (AST/ALT), Platelet count (PLT), Alkaline phosphatase (ALP), Aspartate amino transferase (AST), Prealbumin (PAB), Neutrophil count (NC), Direct bilirubin (DBIL), Mean corpuscular hemoglobin (MCH), Total bilirubin (TBIL), Leukocyte count (WBC), Lymphocyte count (TLC), Mean corpuscular hemoglobin concentration (MCHC), Serum creatinine (SCR), Erythrocyte count (RBC) and Alpha Fetoprotein (AFP), which were used as inputs for the following deep learning algorithms.
2.2. Preprocessing
The following three steps were applied as the preprocessing of data to deal with the missing values. First, the blood parameters with more than 20% missing values were removed. Second, the patients’ data with more than 20% missing variables were removed. Finally, the remaining missing values were filled through the MI method using SPSS. A total of 916 patients were finally included to build the model.
2.3. Study design
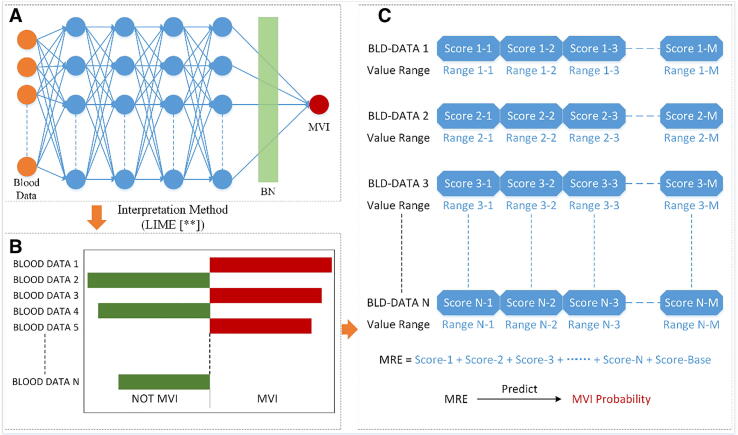
Fig. 1 shows the pipeline of the IRP method for MVI risk prediction. We first trained a deep learning model by letting the neural network learn the relation between the blood data and MVI. After the network was well trained, an explainer was applied to explain the features learned by the learner. To further assess the explained features and build the MVI prediction model, we constructed a scoring system by the explained features to quantify the risk of MVI. To perform internal validation of the model performance, we deploy both 5-fold cross validation and bootstrap procedure, and the whole pipeline was repeated for each validation process. After that, a final model was trained using all patients in the training cohort.
Fig. 1.
The pipeline for the proposed IRP method. (A) Is the fully-connected neural network used for learning the features between the input blood test data and MVI. Each of four blue hidden layers has 32 neurons. Before the last fully-connected layer, batch normalization was applied to accelerate training and avoid overfitting. (B) Shows the result of the explanation of the learner obtained by the interpretation method “LIME”. The length of each column represents the impact of the corresponding variable on MVI (or NOT MVI). (C) Is the scoring model formed from (B). Each variable has an independent score according to its value. The sum of all the scores predicts the risk of MVI. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
2.4. Data augmentation
Recently, the deep learning method has been considered as a data-hungry technology [24], [25], [26]. Lack of training data could lead to over-fitting and poor performance in the real-world applications. Therefore, we performed data augmentation to better train our model. Considering that the concentration of various parameters in the blood usually fluctuates with time, we proposed a data augmentation method of blood data based on Gaussian distribution. For each blood parameter , the largest 10% of the samples and the smallest 10% of the samples are excluded to prevent the impact of extreme values. Then the difference between the minimum and maximum values is taken to calculate the perturbation of each dimension. For a given sample, the generated new data array is:
| (1) |
where is the original data array of the sample and G is the random number array generated under Gaussian distribution with a mean value of 0 and a variance of 0.052. We added a small disturbance on the original data by this function to simulate the fluctuation of blood data in a certain range.
2.5. Learn the features using a DL model
Considering the input blood parameters may have inner relation, we constructed a fully connected neural network to learn the features between the 18 blood parameters and MVI using Keras with Tensorflow as backend. The constructed network structure contained 6 layers, which is shown in Fig. 1A. Within the network, each hidden layer contained 32 neurons. The binary cross-entropy loss was applied as the loss function. A batch normalization layer was applied to avoid overfitting and accelerate the training process.
2.6. Explain the features learned by the learner
We used Local Interpretable Model-Agnostic Explanations (LIME) [22], proposed by Ribeiro et al., to explain our well-trained deep learning model. The LIME algorithm in processing our numerical blood data can be divided into the following six steps:
-
1.
Set the coordinate system of the original blood data as O. Divide the data of each dimension equally into M groups according to its value, then re-encode it into 1 to M to form a new data coordinate system P.
-
2.
Randomly generate K points in coordinate system P, and inverse these K points to coordinate system O.
-
3.
Use the well-trained machine learning model to calculate the prediction values of these K data points in O. Then bring each calculated predicted value into the coordinate system P as the value of each data point.
-
4.
Encode the patient data to be analyzed into the coordinate system P according to the method in 1. Calculate the Euclidean distances from K generated points to the patient data point in P.
-
5.
Do ridge regression using these K data points in P. This regression is weighted by a function of Euclidean distances calculated in 4, the greater the distance, the less the weight.
-
6.
Taking the regression equation as following:
| (2) |
where N is the number of input data dimension X , and b is the intercept. is the impact of each data dimension on MVI. In our experiment, considering the training samples we have, M was set to 4, and K was set to 5000.
Through the LIME algorithm, we could know which data dimension and which value range the machine learning model depends on to give the current judgment. To get the overall impacts of all blood parameters on MVI, all original patient data in the training set were analyzed using LIME, and the average of their results was calculated weighted by the patient’s categories (MVI or NOT MVI).
2.7. Construct MVI scoring model
To further assess the explained result and give the prediction of MVI risk for HCC patients, the overall average impacts of each blood parameter were recombined into a scoring model (Fig. 1C). In the scoring model, every blood parameter for each patient was scored separately. The sum of scores of all dimensions and a basic score, which is acquired by linear transformation, was taken as the MVI Risk Estimation (MRE). By correlating the MRE gained from the scoring model with the ground-truth MVI ratios, we finally obtained a risk prediction model for MVI.
2.8. Concordance index
Concordance index (C-index) was used to assess the accuracy in the prediction of MVI risk of the scoring model. As one of the most frequently used metrics in survival analysis and morbidity analysis, C-index is the proportion of the paired individuals whose survival time meet the prediction. When it comes to morbidity, C-index is the proportion of the paired patient groups whose incidences of disease meet the prediction. It is usually considered as a good model if the C-index is around 0.70, and below 0.50 indicates an invalid model [27]. In our study, patients were grouped according to their MRE. To assess the model more precisely, different numbers of groups were applied, and the average value of the C-index was used as the final result.
2.9. 5-fold cross validation
To evaluate the performance of our model, we deployed 5-fold cross validation. To perform 5-fold cross validation, we first randomly divide all 916 patients in the training cohort into 5 non-overlapping subsets with each subset containing approximately same number of patients. Then, one of the subsets were selected as test set while the other four combined were selected as the training set. After conducting above mentioned modeling process, C-index for the used training set and test set were obtained. These steps were repeated for 5 times and all divided subsets were used as test set for once, generating 5 models and corresponding C-index on their respective training set and test set. The average C-index of 5 models is used to evaluate the prediction performance of the model under the current network structure and parameter settings. After the internal validation, final learning process was performed using all patients in the training cohorts.
2.10. Independent validation
To further evaluate MRE’s performance, an independent validation cohort of HCC patients was enrolled. This cohort included 1085 additional HCC patients received standard HCC management at Eastern Hepatobiliary Surgery Hospital of Second Military Medical University (n = 535) or Mengchao Hepatobiliary Hospital of Fujian Medical University (n = 550), with all 18 blood parameters for deep learning model available (Supplementary Table S1). The clinical information for this cohort was also collected (Supplementary Table S3). The collection and usage of all patients’ information were approved by the Ethics Committee of the two centers, and the requirement of written informed consent was waived, and all procedures were performed in accordance with the Declaration of Helsinki.
2.11. Survival analysis
To evaluate the relationship between MRE and patients’ survival, univariate and multi-variable Cox regression analysis were performed. We further divided patients based on the cut-off of median for MRE in each cohort and use Kaplan-Meier survival analysis to evaluate the differences between the survival of the two groups.
3. Results
3.1. Construction of the deep learning model and the scoring model
A total of 1007 patients received surgical operation were enrolled as the training cohorts for our study. After filtering based on data availability of complete blood count and blood test results, 916 patients remained for downstream analysis. Extracting data from complete blood count and blood test results generated 17 blood related parameters. These 17 blood parameters, along with AFP, were used for downstream learning process. To provide an accurate evaluation of MVI risk, we first build a deep fully connected neural network to learn the features between the 18 blood parameters and MVI. The constructed neural network contained 4 hidden layers, and each hidden layer contains 32 neurons. The detailed structure of the neural network is shown in Fig. 1A.
Although the nonlinear mapping between layers in our models can sufficiently deal with the nonlinear relationship between input and output, it should be also noticed that it causes so called “black box” issue [21], [28] for deep learning. How to convert this nonlinear relation into understandable and quantified representation is the key to the breakthrough of the “black box” issue. To address this, an explainer, i.e., LIME, was applied to provide an understandable interpretation of the nonlinear relation between 18 blood parameters and MVI occurrence after the deep learning model was well trained. Based on the idea of perturbing inputs for explanations, LIME is capable to handle almost any data type, such as image, natural language, and numerical data type, and it is very suitable for interpreting our model. For each sample in the training set, LIME output the impact of every blood parameter on MVI occurrence.
To better quantify the risk of MVI, we constructed a scoring model according to the explained results obtained by LIME. The four intervals of each blood parameters have their corresponding scores, and these scores would be assigned to patients according to the value of each feature. Then, the sum of scores for all 18 features and a base score were calculated to present the overall MVI risk, or to say, MRE of patients. The relevance between the MRE and the ground-truth MVI probability was quantified by C-index.
To validate the performance of our built-up model, we performed 5-fold cross validation and bootstrap procedure. 5-fold cross validation result showed that our proposed process achieved high performance, with an average c-index of 0.9176 in training set and an average c-index of 0.83242 in test set. The detailed results of each internal validation were available in Supplementary Table S4. After internal validation, all patients within the training cohort were used to generate the final deep learning model and corresponding scoring model.
3.2. Interpretation results showed the most important factors related to MVI occurrence
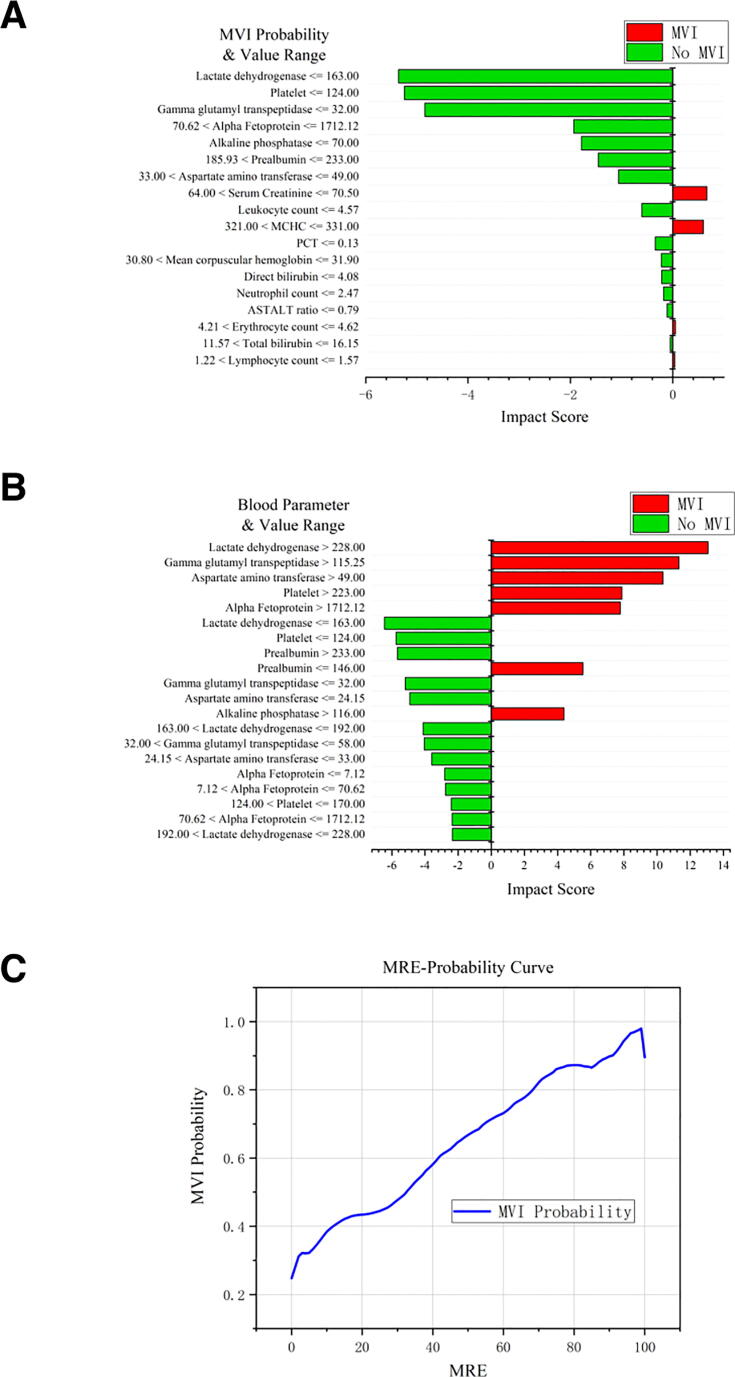
The explainer of the deep learning model, e.g., LIME, provided a quantified evaluation of the impact for each blood parameter on MVI occurrence. Fig. 2A shows how LIME works on a single patient (patient ID: 2014005480). For this patient, 14 blood parameters, including LDH, PLT and GGTP, indicated this patient has a low risk of having MVI, while 4 blood parameters such as SCR and MCHC indicated a high risk of getting MVI. Integration of all related parameters and their impacts showed that this patient has an over-all low risk of MVI, which is consistent with the pathological reports showing no MVI in this patient. To obtain the overall average impact of each input variable on MVI, all the samples in the training set were analyzed by LIME. The average of the explanation results was calculated and weighted by samples’ categories. The value of each blood parameter dimension is divided into four intervals in our experiment. Therefore, the impact scores of all 18 × 4 parameter dimensions and value range pairs, which covers the whole input data space, were obtained. The detailed impact scores were available in Supplementary Table S5. Fig. 2B shows the top twenty blood parameter and value range pairs that have the greatest impact on MVI occurrence. In our model, LDH plays the most important role with a mean absolute score of 6.49 in the prediction of MVI, followed by GGTP and AST with the mean absolute score of 5.65 and 5.13, respectively. Noteworthily, AFP, with its well-known correlation with MVI, ranked 5th among all blood parameters and had a mean absolute score of 3.93. The patient’s risk of MVI increases significantly if variables such as LDH, GGTP, AST reached certain higher levels, while the risk reduces if these variables dropped down to lower than a certain value. This result could help us to determine which blood parameters should be paid more attention to.
Fig. 2.
(A) The explanation results of a single sample. 11 variables contribute to no MVI and 6 variables contribute to MVI. The pathological report showed this patient did not have MVI. The length of each column represents the impact of the corresponding variable on MVI (or NOT MVI). (B) Top-10 high impact feature and value range pairs. (C) MRE-Probability Curve of the proposed model. The MRE are mostly located between 0 and 100. With the increase of the MRE, the risk of MVI rises. The linear correlation coefficient R2 achieves 0.9415.
3.3. The scoring model achieved better performance on C-index compared with the state-of-art method and independent validation proved its applicability
The MRE scoring model based on LIME explaining results provided quantifiable evaluation of MVI risk for HCC patients. Fig. 2C shows the correlation between the MRE and MVI probability. Most MRE were located between 0 and 100. With the increase of MRE, the probability of MVI increased. C-index was used to quantify the relevance between the MRE and the ground-truth MVI probability. Our proposed method achieved good accuracy in estimating the risk of MVI, with a C-index of 0.9341 in the whole training cohorts. To better validation this scoring model’s performance, we further collect another cohort, constituted of a total of 1085 HCC patients, all with 18 blood parameters utilized by our model available. MRE showed excellent consistence with patients’ MVI risk, achieving a C-index of 0.9052. As shown in Table 1, compared with the state-of-art method of scoring the MVI risk [7], our proposed method achieved better performance with less data for modeling judging by C-index. It’s also well known that MVI is correlated with tumor size. To explore the application of MRE in tumor with relatively small size, we calculated C-index for small HCCs (nodule < 2 cm) additionally, achieving 0.85 and 0.822 respectively in the training cohort and independent validation cohort. Considered that our model was built based solely on results of blood tests and did not require any additional information from surgical operation or imaging, it could great benefit non-invasive evaluation of MVI risk for HCC patients.
Table 1.
Comparison of concordance index of different scoring methods.
| Method | Training Set Samples | Testing Set Samples | Training Set C-index | Testing Set C-index |
|---|---|---|---|---|
| Nomogram * | 707 | 297 | 0.81 | 0.80 |
| IRP Method | 916 | 1085 | 0.9341 | 0.9052 |
3.4. Relationship between MRE and clinical features
With MRE accurately predict MVI risk in HCC patients, we further explored if the model’s score might be related to other clinical features. We first divided patients in the training cohort into two groups, one group with MRE above median and the other with MRE below median, defined as high-risk group and low-risk group. Comparison of the clinical features between the two groups showed that, higher MRE was related with larger tumor (p < 0.0001), more possibility to have satellite nodules (p < 0.0001), and lower Edmondson-Steiner grade (p = 0.0025), all indicating tumor progression. These results are not surprising since MVI is also an indicator of cancer progression. Meanwhile, we also noticed significant more probability of having less complete tumor envelope (p = 0.027) and tumor thrombus (p < 0.0001) for the high-risk group, indicating that tumors have greater potential of metastasize. The more sever condition of tumor also lead to rising difficulty in surgical resection, reflected by significant more blood loss during surgery (p < 0.0001). Noteworthily, significant differences in alcohol assumption history (p < 0.05) were also revealed. However, we did not observe any differences in Child–Pugh score between the two groups. We also divided the training cohort into groups according to MVI presence, and observe similar correlations between MVI and tumor size, satellite nodules Edmondson-Steiner grade, tumor envelope as well tumor thrombus (all p < 0.0001). However, we did not see significant change in alcohol assumption between the two groups, suggesting that previous correlation between alcohol assumption history and MRE might reflect its influence on circulating system rather than MVI risk. Similar analysis was conducted for the independent validation cohort and yield accordant results (Supplementary Table S6), further supporting that the MRE could indeed reflect the risk of MVI by providing quantification of tumor progression status.
3.5. MRE could serve as an independent pre-surgical risk factor for RFS and OS
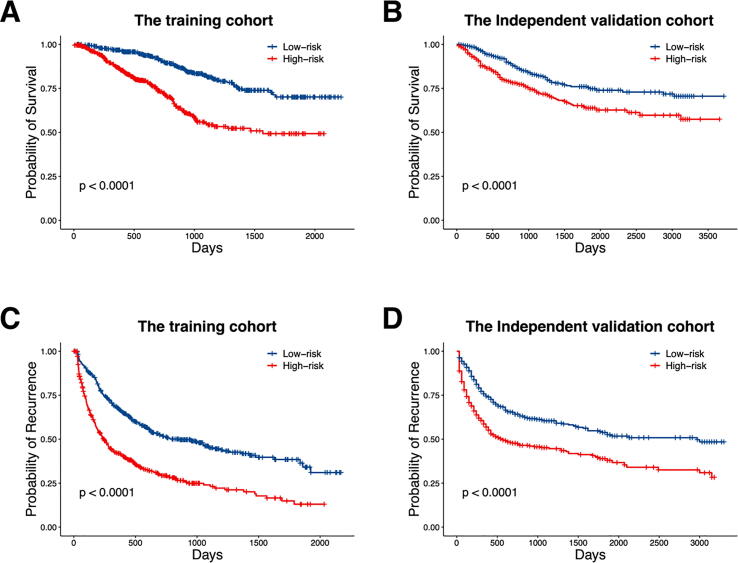
The occurrence of MVI has long been considered as an important factor related to HCC prognosis. Since our model was built based on the correlation between the predicted scores and MVI risk of HCC patients, it is natural to investigate whether these scores showed relevance with HCC survival and recurrence. Fig. 3 shows the Kaplan-Meier curves of overall survival (OS) and recurrence-free survival (RFS) for the two patient groups in both training cohort and independent validation cohort. For the overall survival, the log-rank P-value of 5.36E-13 was achieved for the training cohort (Fig. 3A), and P-value of 8.7E-5 was achieved in the independent validation cohort (Fig. 3B). Considering the relatively short follow-up time for some patients, the differences might be more significant when conducting longer term of follow-up. Meanwhile, survival analysis also shows that the recurrence-free survivals in the two risk groups are drastically different, with log-rank P values of 2.72E-17 (Fig. 3C) and 2.49E-09 (Fig. 3D) in the training cohort and the independent validation cohort, respectively. These associations pointed out that the predicted MRE from our model was a significant factor affecting the recurrence of HCC, and further had a great impact on patients’ prognosis.
Fig. 3.
Survival and recurrence-free survival differences of training and testing set under the MRE threshold of 50: survival of training set (A), survival of testing set (B), recurrence-free survival of training set (C), recurrence-free survival of testing set (D).
When performing univariate Cox regression analysis for the MRE as well as other clinical parameters in the training cohort, we found that the derived MRE served as one of the most significant factors affecting both RFS and OS, which is shown in Table 2. Other clinical parameters correlated with patients’ prognosis include age, weight, parameters reflecting tumor progression including tumor maximum diameter, Edmondson-Steiner grade, number of tumor and parameters reflecting how resection goes include the blood lose and intraoperative blood transfusion (all p < 0.05). To further evaluate the application of our model in pre-surgery prognosis evaluation, we performed multi-variable Cox analysis including the MRE and all clinical parameters (weight and BMI were removed since they were unavailable in the independent testing cohort) that could be acquired before the surgical operation. Noteworthily, the MRE derived by our model remained as one independent risk factor for both RFS and OS (Table 3), with the complete formula shown as follows:
Table 2.
Univariate Cox Regression Analysis for MRE in included HCC patients.
| B | SE | Wald | Sig. | Exp(B) | 95.0% CI for Exp(B) |
||
|---|---|---|---|---|---|---|---|
| Lower | Upper | ||||||
| RFS | 0.021 | 0.002 | 122.230 | 0.000 | 1.021 | 1.017 | 1.025 |
| OS | 0.024 | 0.003 | 72.640 | 0.000 | 1.025 | 1.019 | 1.031 |
Table 3.
Multivariate Cox Regression analysis of pre-surgery parameters in included HCC patients.
| Parameter | B | SE | Wald | Sig. | Exp(B) | 95.0% CI for Exp(B) |
||
|---|---|---|---|---|---|---|---|---|
| Lower | Upper | |||||||
| RFS | Sex | 0.273 | 0.131 | 4.330 | 0.037 | 1.314 | 1.016 | 1.699 |
| Imaging tumor maximum diameter | 0.012 | 0.003 | 23.983 | 0.000 | 1.012 | 1.007 | 1.017 | |
| MRE | 0.088 | 0.015 | 35.891 | 0.000 | 1.092 | 1.061 | 1.124 | |
| OS | Imaging tumor maximum diameter | 0.068 | 0.023 | 8.860 | 0.003 | 1.070 | 1.023 | 1.119 |
| MRE | 0.018 | 0.004 | 19.865 | 0.000 | 1.018 | 1.010 | 1.026 | |
Using the above formulas, we calculated the prognostic risk score for the independent validation cohort and divided them using median of the score as cut-off. Kaplan-Meier curves of the divided two groups showed that they have dramatically different prognosis (Supplementary Fig. S1), further supporting the value of MRE in prognosis evaluation. Thus, our model could also be utilized to evaluate patients’ status and this evaluation could be achieved even before the surgical operation, providing an important strategy for better prognosis management.
4. Discussion
Microvascular invasion is one of the most crucial factors leading to HCC recurrence after liver hepatectomy and transplantation. Quantifying the risk of MVI occurrence before surgery would help to determine the therapeutic schedule and is conducive to the achievement of precision medicine. It also helped surgical decision making between liver resection and liver transplantation, with MVI being a strong contra indication to the second option. An accurate evaluation strategy that minimum the collateral risk to patients is in demand and our method meets the urgent need by utilizing the blood test results to predict the risk of MVI for HCC patients based on the interpretation of the deep learning model. This approach, which could be termed as a liquid-biopsy for MVI, achieved high performance, and might further aid HCC patients’ clinical management.
So far, the golden standard for MVI determination is via pathological examination on tumor tissue samples collected during surgery. Other methods usually have disadvantages preventing them from clinical applications. For example, biopsy for confirmation of MVI has been proven unreliable [5], which is largely due to intratumoral heterogeneity, and could increase the risk of the tumor metastasis [29], [30]. Meanwhile, medical imaging often fails to detect MVI resulted from the limitation of the imaging approaches [31]. Blood test served as a non-invasive strategy to acquire tumor information, can relieve the distress of patients compared with the needle biopsy. Also, Studies have reported that blood tests presented parameters correlated with MVI of HCC patients [32], [33], [34], [13], [35], [36], while it is hard to precisely quantify the differences of their impacts on the prediction of MVI. In the present study, the proposed IRP method could not only offer the quantified impact on MVI for each involved blood parameter but also construct a prediction model for preoperative estimation of MVI risk for HCC patients. Interestingly, although the model in the present study was constructed by the blood parameters and MVI, the MRE of HCC patients showed significant relevance to the survival and recurrence-free survival with a log-rank P-value of 1.8E-4 and 8.6E-12 in the enrolled cohort, respectively. The finding is not unexpected since MVI itself is one of the most critical factors leading to HCC recurrence and a key marker that could help guide treatment selection [5], [37], [38], [39]. The revealed correlation between MRE and patients’ prognosis therefore offers a novel solution to provide quantifiable evaluation for HCC clinical outcome based on easy-to-obtain variables. Furthermore, the MRE might represent an integrated status reflecting the influence tumor burden laid on the circulating system. However, we must notice that the correlation between these blood parameters and MVI was previously rarely reported, which is accordant with the scientific consensus that Machine learning solutions are usually difficult to directly relate to existing biological knowledge. Our model might reflect a rather complicated dynamical status at which these blood parameters let us get a glimpse on, and further investigation is still required to fully understand how they are connected.
It is worth noting that the whole modeling process was carried out by the machine learning method automatically, without prior knowledge, making the proposed method also suitable to other similar risk prediction tasks. In the present study, the deep learning model learned the features between 18 pre-selected blood parameter and MVI occurrence, and automatically converted the well-learned features into the form we can understand by the explanation method. The explained results demonstrated that which variables are the most important factors leading to MVI occurrence and should be paid more attention to, such as LDH, GGTP, and AST. Meanwhile, the evaluation of the MVI risk for each HCC patient is also of importance. In this study, the explanation results of all the blood parameters were combined to form a scoring model to give the estimate of MVI risk for HCC patients, and the assessment of scoring model validated the accuracy of the interpretation results. Since the deep learning model supports input data with any length, any variable considered to have potential value in predicting MVI can be used as the input data for modeling. In scientific research, the effectiveness of newly discovered indices can be pre-validated by adding them to existing models. Compared with the traditional machine learning method, the prediction model built on the explanations of machine learning models offers more transparency, users can find the basis for the predicted results, making the model more reliable. Moreover, there are some studies applied the deep learning method to predict MVI for HCC [17], [19], while these deep learning models involved numerous computations of the neurons in the artificial neural network, making it use the high-performance computer as the basic hardware equipment, limiting its wide use. The computation process of the scoring model in this study is not only easier to be understood than the “black box” model but also convenient to be computed. It is suitable to be used in most clinical scenarios.
There are some more issues needed to be declared in this section. A new data augmentation method was proposed in this paper to generate more training data. Although there is no research declares that how much training data is enough to train a model, it is generally believed that as the data dimension of the input layer increases, the amount of training data required to achieve the same accuracy also increases. The study has shown that data augmentation methods can indeed improve the performance of the deep learning model, but the improvement is limited by the equivalent amount of real data [40] so that real data should be added as much as possible.
It’s also important to point out that while building our model solely based on blood test results make it one of the most convenient and cost-efficient approach for MVI risk evaluation, the integration of MRE and other strategies might still possess potential for a more comprehensive and more accurate determination for MVI presence. Many other clinical features have also been reported to related with MVI such as large tumor size [41] non-smooth tumor margin [42], and Circular RNAs such as ciRS-7 [43]. The combination of different strategies of multi-omics has shown great promise and become one of the most important research focus recently. In future multi-omics integration, our model will sure provide irreplaceable insights and help build a more comprehensive view for MVI evaluation.
CRediT authorship contribution statement
Geng Chen: Conceptualization, Methodology, Formal analysis, Investigation, Data curation, Writing - original draft, Writing - review & editing. Rendong Wang: Conceptualization, Methodology, Software, Validation, Writing - original draft, Writing - review & editing. Chen Zhang: Methodology, Software, Visualization, Writing - review & editing. Lijia Gui: Software, Investigation, Writing - original draft. Yuan Xue: Software, Investigation, Visualization. Xianlin Ren: Data curation, Formal analysis, Resources. Zhenli Li: Data curation, Formal analysis, Resources. Sijia Wang: Software, Writing - review & editing. Zhenxi Zhang: Methodology, Writing - review & editing. Jing Zhao: Data curation. Huqing Zhang: Methodology, Writing - review & editing. Cuiping Yao: Conceptualization, Methodology, Supervision, Writing - review & editing. Jing Wang: Conceptualization, Methodology, Supervision, Writing - review & editing. Jingfeng Liu: Conceptualization, Methodology, Supervision, Resources, Writing - review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work is supported by the Natural Science Foundation of China (Grant No. 61727823 and No. 61775178), National Science and Technology Major Project of China (Grant No. 2018ZX10302205), Regional development project of Fujian province (Grant No. 2019Y3001), the Science and Technology development project of central government guiding local government (Grant No. 2019L3027).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2021.01.014.
Contributor Information
Cuiping Yao, Email: zsycp@mail.xjtu.edu.cn.
Jing Wang, Email: wangjing@mail.xjtu.edu.cn.
Jingfeng Liu, Email: drjingfeng@126.com.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Lim K.-C., Chow P.-K.-H., Allen J.C., Chia G.-S., Lim M., Cheow P.-C. Microvascular invasion is a better predictor of tumor recurrence and overall survival following surgical resection for hepatocellular carcinoma compared to the Milan criteria. Ann Surg. 2011;254(1):108–113. doi: 10.1097/SLA.0b013e31821ad884. [DOI] [PubMed] [Google Scholar]
- 2.Choi K.K., Kim S.H., Choi S.B., Lim J.H., Choi G.H., Choi J.S. Portal venous invasion: the single most independent risk factor for immediate postoperative recurrence of hepatocellular carcinoma. J Gastroenterol Hepatol. 2011;26(11):1646–1651. doi: 10.1111/j.1440-1746.2011.06780.x. [DOI] [PubMed] [Google Scholar]
- 3.Du M., Chen L., Zhao J., Tian F., Zeng H., Tan Y. Microvascular invasion (MVI) is a poorer prognostic predictor for small hepatocellular carcinoma. BMC cancer. 2014;14(1):38. doi: 10.1186/1471-2407-14-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cong W.-M., Bu H., Chen J., Dong H., Zhu Y.-Y., Feng L.-H. Practice guidelines for the pathological diagnosis of primary liver cancer: 2015 update. World J Gastroenterol. 2016;22(42):9279. doi: 10.3748/wjg.v22.i42.9279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rodríguez-Perálvarez M., Luong T.V., Andreana L., Meyer T., Dhillon A.P., Burroughs A.K. A systematic review of microvascular invasion in hepatocellular carcinoma: diagnostic and prognostic variability. Ann Surg Oncol. 2013;20(1):325–339. doi: 10.1245/s10434-012-2513-1. [DOI] [PubMed] [Google Scholar]
- 6.Banerjee S., Wang D.S., Kim H.J., Sirlin C.B., Chan M.G., Korn R.L. A computed tomography radiogenomic biomarker predicts microvascular invasion and clinical outcomes in hepatocellular carcinoma. Hepatology. 2015;62(3):792–800. doi: 10.1002/hep.27877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lei Z., Li J., Wu D., Xia Y., Wang Q., Si A. Nomogram for preoperative estimation of microvascular invasion risk in hepatitis B virus–related hepatocellular carcinoma within the milan criteria. JAMA Surg. 2016;151(4):356–363. doi: 10.1001/jamasurg.2015.4257. [DOI] [PubMed] [Google Scholar]
- 8.Schlichtemeier S.M., Pang T.C., Williams N.E., Gill A.J., Smith R.C., Samra J.S. A pre-operative clinical model to predict microvascular invasion and long-term outcome after resection of hepatocellular cancer: the Australian experience. Eur J Surg Oncol. 2016;42(10):1576–1583. doi: 10.1016/j.ejso.2016.05.032. [DOI] [PubMed] [Google Scholar]
- 9.Dhir M., Melin A.A., Douaiher J., Lin C., Zhen W.K., Hussain S.M. A review and update of treatment options and controversies in the management of hepatocellular carcinoma. Ann Surg. 2016;263(6):1112–1125. doi: 10.1097/SLA.0000000000001556. [DOI] [PubMed] [Google Scholar]
- 10.Rana A.P.S., Kaur M., Zonunsanga B., Puri A., Kuka A.S. Preoperative peripheral blood count in breast carcinoma: predictor of prognosis or a routine test. Int. J Breast Cancer. 2015;2015 doi: 10.1155/2015/964392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lopez J., Balasegaram M., Thambyrajah V., Timor J. The value of liver function tests in hepatocellular carcinoma. Malays J Pathol. 1996;18:95–100. [PubMed] [Google Scholar]
- 12.Johnson P.J., Berhane S., Kagebayashi C., Satomura S., Teng M., Reeves H.L. Assessment of liver function in patients with hepatocellular carcinoma: a new evidence-based approach—the ALBI grade. J Clin Oncol. 2015;33(6):550. doi: 10.1200/JCO.2014.57.9151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McHugh P.P., Gilbert J., Vera S., Koch A., Ranjan D., Gedaly R. Alpha-fetoprotein and tumour size are associated with microvascular invasion in explanted livers of patients undergoing transplantation with hepatocellular carcinoma. Hpb. 2010;12(1):56–61. doi: 10.1111/j.1477-2574.2009.00128.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shen D., Wu G., Suk H.-I. Deep learning in medical image analysis. Annu Rev Biomed Eng. 2017;19:221–248. doi: 10.1146/annurev-bioeng-071516-044442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Litjens G., Kooi T., Bejnordi B.E., Setio A.A.A., Ciompi F., Ghafoorian M. A survey on deep learning in medical image analysis. Med Image Anal. 2017;42:60–88. doi: 10.1016/j.media.2017.07.005. [DOI] [PubMed] [Google Scholar]
- 16.Lee J.-G., Jun S., Cho Y.-W., Lee H., Kim G.B., Seo J.B. Deep learning in medical imaging: general overview. Korean J Radiol. 2017;18(4):570–584. doi: 10.3348/kjr.2017.18.4.570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Greenspan H., Van Ginneken B., Summers R.M. Guest editorial deep learning in medical imaging: overview and future promise of an exciting new technique. IEEE Trans Med Imaging. 2016;35(5):1153–1159. [Google Scholar]
- 18.Cucchetti A., Piscaglia F., Grigioni A.D.E., Ravaioli M., Cescon M., Zanello M. Preoperative prediction of hepatocellular carcinoma tumour grade and micro-vascular invasion by means of artificial neural network: a pilot study. J Hepatol. 2010;52(6):880–888. doi: 10.1016/j.jhep.2009.12.037. [DOI] [PubMed] [Google Scholar]
- 19.Men S, Ju H, Zhang L, Zhou W, editors. Prediction of Microvascular Invasion of Hepatocellar Carcinoma With Contrast-Enhanced MR Using 3D CNN And LSTM. 2019 IEEE 16th International Symposium on Biomedical Imaging (ISBI 2019); 2019: IEEE.
- 20.Samek W, Wiegand T, Müller K-R. Explainable artificial intelligence: Understanding, visualizing and interpreting deep learning models. arXiv preprint arXiv:170808296. 2017.
- 21.Koh PW, Liang P, editors. Understanding black-box predictions via influence functions. Proceedings of the 34th International Conference on Machine Learning-Volume 70; 2017: JMLR. org.
- 22.Ribeiro MT, Singh S, Guestrin C, editors. Why should i trust you?: Explaining the predictions of any classifier. Proceedings of the 22nd ACM SIGKDD international conference on knowledge discovery and data mining; 2016: ACM.
- 23.Bureau of Medical Administration, National Health. Diagnosis, Management, and Treatment of Hepatocellular Carcinoma (V2017). Zhonghua gan zang bing za zhi= Zhonghua ganzangbing zazhi= Chinese journal of hepatology. 2017; 25(12): 886. [DOI] [PMC free article] [PubMed]
- 24.Oliver A, Odena A, Raffel CA, Cubuk ED, Goodfellow I, editors. Realistic evaluation of deep semi-supervised learning algorithms. Advances in Neural Information Processing Systems; 2018.
- 25.Kuznietsov Y, Stuckler J, Leibe B, editors. Semi-supervised deep learning for monocular depth map prediction. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition; 2017.
- 26.Maheshwari R, Moudgil K, Parekh H, Sawant R, editors. A Machine Learning Based Medical Data Analytics and Visualization Research Platform. 2018 International Conference on Current Trends towards Converging Technologies (ICCTCT); 2018: IEEE.
- 27.Chaudhary K., Poirion O.B., Lu L., Garmire L.X. Deep learning–based multi-omics integration robustly predicts survival in liver cancer. Clin Cancer Res. 2018;24(6):1248–1259. doi: 10.1158/1078-0432.CCR-17-0853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fong RC, Vedaldi A, editors. Interpretable explanations of black boxes by meaningful perturbation. Proceedings of the IEEE International Conference on Computer Vision; 2017.
- 29.Stigliano R., Marelli L., Yu D., Davies N., Patch D., Burroughs A. Seeding following percutaneous diagnostic and therapeutic approaches for hepatocellular carcinoma. What is the risk and the outcome?: seeding risk for percutaneous approach of HCC. Cancer Treat Rev. 2007;33(5):437–447. doi: 10.1016/j.ctrv.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 30.Silva M.A., Hegab B., Hyde C., Guo B., Buckels J.A., Mirza D.F. Needle track seeding following biopsy of liver lesions in the diagnosis of hepatocellular cancer: a systematic review and meta-analysis. Gut. 2008;57(11):1592–1596. doi: 10.1136/gut.2008.149062. [DOI] [PubMed] [Google Scholar]
- 31.Gouw A.S., Balabaud C., Kusano H., Todo S., Ichida T., Kojiro M. Markers for microvascular invasion in hepatocellular carcinoma: where do we stand? Liver Transpl. 2011;17(S2):S72–S80. doi: 10.1002/lt.22368. [DOI] [PubMed] [Google Scholar]
- 32.Bertuzzo V.R., Cescon M., Ravaioli M., Grazi G.L., Ercolani G., Del Gaudio M. Analysis of factors affecting recurrence of hepatocellular carcinoma after liver transplantation with a special focus on inflammation markers. Transplantation. 2011;91(11):1279–1285. doi: 10.1097/TP.0b013e3182187cf0. [DOI] [PubMed] [Google Scholar]
- 33.Gan W., Zhang M.-X., Wang J.-X., Fu Y.-P., Huang J.-L., Yi Y. Prognostic impact of lactic dehydrogenase to albumin ratio in hepatocellular carcinoma patients with Child-Pugh i who underwent curative resection: a prognostic nomogram study. Cancer Manage Res. 2018;10:5383. doi: 10.2147/CMAR.S176317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao W.-C., Fan L.-F., Yang N., Zhang H.-B., Chen B.-D., Yang G.-S. Preoperative predictors of microvascular invasion in multinodular hepatocellular carcinoma. Eur J Surg Oncol. 2013;39(8):858–864. doi: 10.1016/j.ejso.2013.04.003. [DOI] [PubMed] [Google Scholar]
- 35.Poté N., Cauchy F., Albuquerque M., Voitot H., Belghiti J., Castera L. Performance of PIVKA-II for early hepatocellular carcinoma diagnosis and prediction of microvascular invasion. J Hepatol. 2015;62(4):848–854. doi: 10.1016/j.jhep.2014.11.005. [DOI] [PubMed] [Google Scholar]
- 36.Hirokawa F., Hayashi M., Miyamoto Y., Asakuma M., Shimizu T., Komeda K. Outcomes and predictors of microvascular invasion of solitary hepatocellular carcinoma. Hepatol Res. 2014;44(8):846–853. doi: 10.1111/hepr.12196. [DOI] [PubMed] [Google Scholar]
- 37.Roayaie S., Blume I.N., Thung S.N., Guido M., Fiel M.I., Hiotis S. A system of classifying microvascular invasion to predict outcome after resection in patients with hepatocellular carcinoma. Gastroenterology. 2009;137(3):850–855. doi: 10.1053/j.gastro.2009.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hou Y.-F., Li B., Wei Y.-G., Yang J.-Y., Wen T.-F., Xu M.-Q. Second hepatectomy improves survival in patients with microvascular invasive hepatocellular carcinoma meeting the Milan criteria. Medicine. 2015;94(48) doi: 10.1097/MD.0000000000002070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Meniconi R.L., Komatsu S., Perdigao F., Boëlle P.-Y., Soubrane O., Scatton O. Recurrent hepatocellular carcinoma: a Western strategy that emphasizes the impact of pathologic profile of the first resection. Surgery. 2015;157(3):454–462. doi: 10.1016/j.surg.2014.10.011. [DOI] [PubMed] [Google Scholar]
- 40.Wong SC, Gatt A, Stamatescu V, McDonnell MD, editors. Understanding data augmentation for classification: when to warp? 2016 international conference on digital image computing: techniques and applications (DICTA); 2016: IEEE.
- 41.Schlichtemeier S., Pang T., Williams N., Gill A., Smith R., Samra J. A pre-operative clinical model to predict microvascular invasion and long-term outcome after resection of hepatocellular cancer: The Australian experience. Eur J Surg Oncol. 2016;42(10):1576–1583. doi: 10.1016/j.ejso.2016.05.032. [DOI] [PubMed] [Google Scholar]
- 42.Hu H., Zheng Q., Huang Y., Huang X.W., Lai Z.C., Liu J. A non-smooth tumor margin on preoperative imaging assesses microvascular invasion of hepatocellular carcinoma: a systematic review and meta-analysis. Sci Rep. 2017;7(1):15375. doi: 10.1038/s41598-017-15491-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu L., Zhang M., Zheng X., Yi P., Lan C., Xu M. The circular RNA ciRS-7 (Cdr1as) acts as a risk factor of hepatic microvascular invasion in hepatocellular carcinoma. J Cancer Res Clin Oncol. 2017;143(1):17–27. doi: 10.1007/s00432-016-2256-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.