Abstract
RAS-responsive element-binding protein 1 (RREB1) is a transcription factor that is implicated in RAS signaling and multiple tumors. However, the role of RREB1 in acute myeloid leukemia has not been studied. We found that RREB1 is overexpressed in AML patients and myeloid leukemia cell lines (NB4 and HL-60), and RREB1 expression was significantly decreased during granulocytic differentiation of myeloid leukemia cells induced by all-trans retinoic acid (ATRA). Then we performed a RREB1 knockdown assay in NB4 and HL-60 cells; the results showed that knockdown of RREB1 upregulated expression of CD11b, CEBPβ, and microRNA-145 (miR-145), which hinted that knockdown of RREB1 enhanced granulocytic differentiation of myeloid leukemia cells. In addition, inhibitor of miR-145 can offset the enhanced effect on granulocytic differentiation mediated by downregulation of RREB1. These collective findings demonstrated that RREB1 blocks granulocytic differentiation of myeloid leukemia cells by inhibiting the expression of miR-145 and downstream targets of the RAS signal pathway. These may provide a promising therapeutic target for AML patients.
Key words: RAS-responsive element-binding protein 1 (RREB1), Differentiation, NB4, HL-60, Acute myeloid leukemia (AML)
INTRODUCTION
Acute myeloid leukemia (AML) is a common hematopoietic malignancy, accounting for 60% to 70% of adult acute leukemia in China each year. It is a highly heterogeneous clonal disease caused by mutations in acquired myeloid progenitor cells1,2. Kinds of genetic alterations occur in AML, which cause uncontrollable proliferation, inhibition of differentiation, and apoptosis3. Studies have found genetic mechanism involved in AML including inhibition of tumor suppressor genes, activation of oncogenes, gene mutation, gene deletion, and gene translocation4. Therefore, to explore the roles of new genes in AML will not only help to elucidate the pathogenesis but also may discover new effective therapeutic targets.
RAS-responsive element-binding protein 1 (RREB1 gene) was found initially in human medullary thyroid carcinomas5. It has been demonstrated that RREB1 is implicated in RAS signaling and multiple malignancies, which is involved in cell differentiation, proliferation, apoptosis, adhesion, and migration6–9. Previous studies have identified roles of RREB1 in cell differentiation in multiple cell backgrounds6,7,10,11. In medullary thyroid carcinomas, enhanced expression of RREB1 confers the ability to mediate increased transactivation of the CT gene promoter during Ras (or Raf)-induced differentiation5. Another study in the adult midgut of Drosophila showed that RREB1 (Hindsight homolog) is required for EC differentiation in the context of undifferentiated intestinal stem cell (ISC)-to-plus enterocyte (EC) differentiation, but not in the context of adult midgut precursors (AMP)-to-EC differentiation of stem cells12. It hints that RREB1 plays a very complicated role even under the same cell background. In addition, RREB1 has been identified as a dysregulated gene in APL screened by Hu133A GeneChips13, indicating that RREB1 may be involved in AML development.
In addition, a lack of miR-145 expression has been found in APL cells and samples, and inhibition of miR-145 attenuated neutrophil differentiation of APL cells14. Interestingly, a reverse relationship has been demonstrated between miR-145 and RREB1 in many tumors, such as colorectal tumors and pancreatic cancers15,16. However, the relationship of RREB1 gene and miR-145 in the differentiation of AML cells has not been reported. In our current study, we aimed to explore the underlying mechanism of RREB1 and miR-145 in the differentiation of AML cell lines.
NB4 cells and HL-60 cells were chosen to confirm our hypothesis. NB4 is an APL/AML-M3 cell line that harbors the t(15;17) chromosomal translocation. HL-60 cells have also been considered as an APL cell line, but it lacks the specific t(15;17) chromosomal translocation of APL17. Thus, in this context, we describe HL-60 cells with the more general term of AML cell line18–20.
MATERIALS AND METHODS
Cell Lines and Cell Culture
NB4 and HL-60 cells were maintained in our own laboratory by culture in RPMI-1640 medium (Gibco-Life Technologies, Carlsbad, CA, USA) containing 10% fetal bovine serum (FBS; Gemini, Yi Meng Science and Technology Co., Ltd, Chengdu, P.R. China) and 1% penicillin–streptomycin (100 U/ml penicillin and 100 μg/ml streptomycin) at 37°C in a humidified atmosphere containing 5% CO2.
Quantitative Real-Time PCR (qRT-PCR)
Total RNA was extracted from the cells with TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s protocol, followed by phenol–chloroform extraction and isopropyl alcohol precipitation. The RT-PCR analysis was conducted by application of SYBR Premix ex tag II (Takara Bio, Dalian, P.R. China). Reactions were run using a real-time system (Bio-Rad Laboratories, Inc., Hercules, CA, USA). All primers were synthesized and purchased from TSINGKE Biotechnology Co., Ltd. (Shanghai, P.R. China). The sequences of primers used in this study were: β-actin 5′-TGACGTGGACATCCGCAAAG-3′ (forward) and 5′-CTGGAAGGTGGACAGCGAGG-3′ (reverse); RREB1 5′-GCTGGCGGTCCCAATCTACT-3′ (forward) and 5′-AAGCTGTCTGAAGCCGTGGT-3′ (reverse); CD11b 5′-ACTGGTGAAGCCAATAACGCA-3′ (forward) and 5′-TCCGTGATGACAACTAGGATCTT-3′ (reverse); CEBPβ 5′-ATGTTCCTACGGGCTTGTTG-3′ (forward) and 5′-CCCAAAGGCTTTGTAACCA-3′ (reverse); miR-145 5′-ACGCGGTCCAGTTTCCCAGGA-3′ (forward) and 5′-ATCCAGTGCAGGGTCCGAGG-3′ (reverse); RT-primer 5′-GTCGTATCCGTGCAGGTCCGAGGTATTCGCACTGGATACGACAGGGAT-3′; U6 5′-CTCGCTTCGGCAGCACA-3′ (forward) and 5′-AACGCTTCACGAATTTGCGT-3′ (reverse); RT-primer 5′-CGCTTCACGAATTTGCGT-3′. β-Actin and U6 were used as housekeeping genes.
Immunoblotting
Cells were lysed with ice-cold radioimmunoprecipitation assay (RIPA) lysis buffer (Cell Signaling Biological Reagent Co., Ltd., Shanghai, P.R. China) supplemented with protease inhibitor phenylmethane sulfonyl fluoride (PMSF; Cell Signaling Biological Reagent Co., Ltd.). A BCA protein assay kit (Beyotime Biotechnology, Shanghai, P.R. China) was used to measure the concentration of protein samples. Protein (50 μg) from NB4 and HL-60 cells was loaded onto every lane. The following primary antibodies were used for immunoblotting: β-actin (1:1,000; Beijing Zhongshan Golden Bridge Biotechnology, Co., Ltd, Beijing, P.R. China), RREB1 (AP19046b; 1:1,000; Abgent, San Diego, CA, USA), CEBPβ (WL0056a; 1:1,000; Wanleibio, Co., Ltd., Beijing, P.R. China), CD11b (ab133357; 1:1,000; Abcam, Cambridge, UK), ERK (ab184699; 1:1,000; Abcam), p-ERK (ab76299; 1:1,000; Abcam), AKT (ab8805; 1:1,000; Abcam), pAKT (ab81283; 1:1,000; Abcam). The chemiluminescence reaction was performed using Pierce ECL West Femto substrate (Thermo Fisher Scientific Co., Ltd, Shanghai, P.R. China), and images were captured using an Image Lab system (Bio-Rad Laboratories, Inc.).
Cell Morphological Analysis
Cells were collected and then washed twice with cold phosphate-buffered saline (PBS), resuspended with 100 μl of PBS. Ten microliters of the cell suspension was used to prepare a smear that was allowed to dry naturally. Wright’s stain was added dropwise according to the instructions. After drying naturally, cell morphological changes were observed by a BX51 polarizing microscope (Olympus, Tokyo, Japan).
Cell Transfection
To perform the RREB1 knockdown assay, the lentiviral vectors short hairpin (sh) RREB1-homo-1257, shRREB1-homo-1777, shRREB1-v1092, and LV-NC were purchased from Shanghai GenePharma Co., Ltd. (Shanghai, P.R. China). NB4 and HL-60 cells were transfected following the manufacturer’s recommendations. After culture for 3 days, puromycin (Sigma-Aldrich, St. Louis, MO, USA) was added for 7 days to ensure the transfection rate. Successful transfection was confirmed by RT-qPCR and immunoblotting. For overexpression (downregulation) of miR-145, NB4 and HL-60 cells were transfected with scramble or mimics (inhibitors), respectively. Lipofectamine™ 2000 reagent (Invitrogen) was used to enhance the efficiency of transfection according to the manufacturer’s instructions.
Indirect Immunofluorescence Assay
Cells treated with all-trans retinoic acid (ATRA; 10 nmol/L) or dimethyl sufoxide (DMSO; 0.1%) were washed using fresh PBS then fixed with 4% paraformaldehyde for 20 min. Triton X-100 (0.1% in PBS) was used to permeabilize the cells for another 15 min. The cells were blocked with 10% goat serum (in PBS) for 30 min at room temperature and washed three times with PBS. The cells on slides were incubated overnight with the corresponding primary antibodies. Secondary goat antibody against rabbit IgG fluorescein isothiocyanate (FITC; 1:200; Beijing Zhongshan Golden Bridge Biotechnology) was used to detect rabbit IgG for 1 h at room temperature. Nuclei were stained with 4′6-diamidino-2-phenylindole (DAPI) at room temperature for 30 min. Finally, 70% glycerol was used to immobilize the coverslips, and the cells were observed with a fluorescence microscope (Nikon, Tokyo, Japan).
Flow Cytometry Assay
Cells were collected and washed twice using cold PBS (3,000 rpm, 2 min). Five microliters of phycoerythrin (PE)-conjugated CD11b antibody (12011342; eBioscience, Inc., San Diego, CA, USA) was added and incubated at room temperature for 30 min in darkness. Then cells were analyzed using flow cytometry (BD influx; BD Biosciences, San Jose, CA, USA).
Statistical Analysis
GraphPad Prism 5 software (GraphPad, La Jolla, CA, USA) was utilized for statistical analysis. Data are shown as mean ± SD. Student’s t-test or one-way ANOVA was used to evaluate the statistical significance. A value of p < 0.05 was considered significant.
RESULTS
Expression of RREB1 in AML Cells
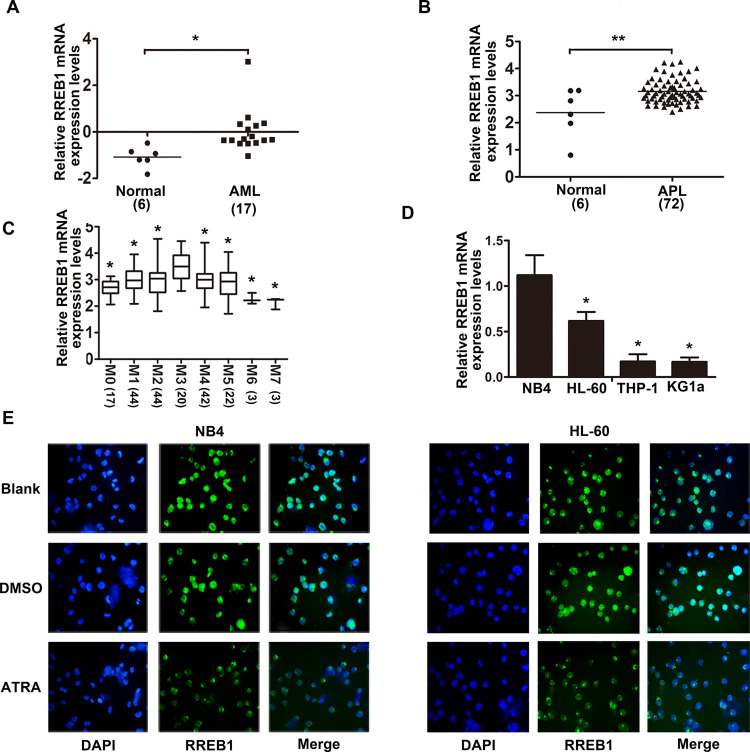
Results from Oncomine dataset showed that RREB1 is highly expressed in AML and APL patients compared with normal people (Fig. 1A and B), and the expression of RREB1 in all AML subtypes was also analyzed by Oncomine dataset (Fig. 1C). Then AML cell lines (NB4, HL-60, THP-1, and KG1a) were chosen to analyze the mRNA level of RREB1 (Fig. 1D), which showed that RREB1 expression in NB4 cells was higher than in HL-60, THP-1, and KG1a cells. Furthermore, the expression and localization of RREB1 protein after dealing with ATRA in NB4 and HL-60 cells were observed by immunofluorescence assay, which displayed that RREB1 is mainly distributed in the nucleus, and ATRA treatment only decreased the expression of RREB1, but did not change the localization (Fig. 1E). These data hinted that RREB1 may be involved in the progression of AML.
Figure 1.
RAS-responsive element-binding protein 1 (RREB1) is highly expressed in acute myeloid leukemia (AML). (A, B) The mRNA levels of RREB1 in AML and APL patients versus normal people were analyzed using Oncomine datasets. (C) The mRNA levels of RREB1 in AML subtypes were analyzed using Oncomine datasets. *p < 0.05 compared with M3, **p < 0.01 compared with normal. (D) Expression of RREB1 mRNA in NB4, HL-60, THP-1, and KG1a cells was detected by quantitative real-time PCR (qRT-PCR). *p < 0.05 compared with NB4. (E) Expression and localization of RREB1 in NB4 and HL-60 cells after ATRA treatment were detected by immunofluorescence assay.
ATRA Suppresses RREB1 Expression in NB4 and HL-60 Cells
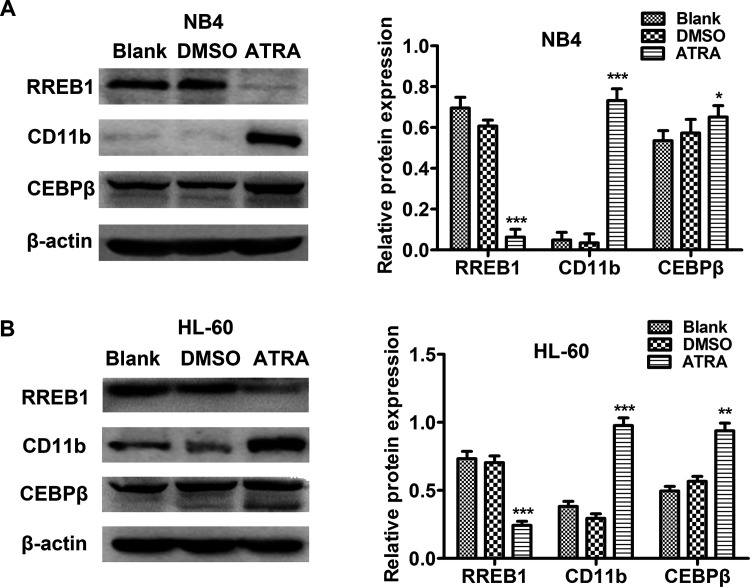
Next we examined the effects of ATRA on the expression of RREB1 in AML cell lines. NB4 and HL-60 cells were treated with ATRA (1 μmol/L) for 48 h; we found that the expression of RREB1 decreased significantly in the ATRA-treated group compared with the Blank group. At the same time, successful granulocytic differentiation of NB4 (Fig. 2A) and HL-60 (Fig. 2B) cells was confirmed by the significant increase in CD11b and CEBPβ protein expression. These results showed that RREB1 expression was dramatically decreased during granulocytic differentiation of leukemia cells induced by ATRA.
Figure 2.
All-trans retinoic acid (ATRA) suppresses RREB1 expression in AML cell lines. (A, B) Expression levels of RREB1, CD11b, and CEBPβ protein in NB4 and HL-60 cells were measured by Western blot after ATRA treatment. *p < 0.05, **p < 0.01, ***p < 0.001, compared with Blank.
Downregulation of RREB1 Enhances Granulocytic Differentiation in NB4 and HL-60 Cells
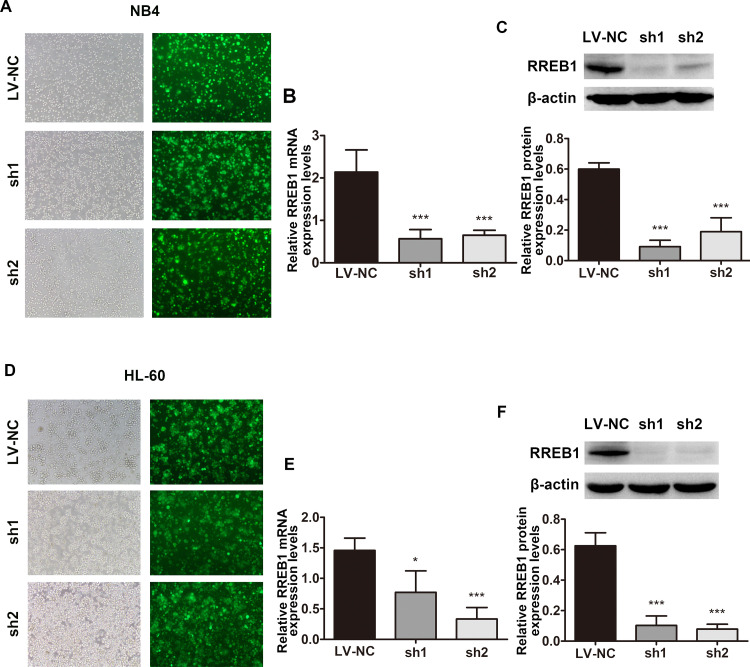
Based on our findings that RREB1 is overexpressed in AML cells and can be downregulated by ATRA, we hypothesized that RREB1 plays a role in differentiation arrest of myeloid leukemia cells. To assess this, a lentivirus target RREB1 was constructed to perform a loss-of-function experiment and then transfected into NB4 and HL-60 cells. Positive clones were screened by purinomycin (7 days), and the transfection efficiency of NB4 (Fig. 3A) and HL-60 (Fig. 3D) cells was identified under fluorescence microscope, which showed that green fluorescence protein+ (GFP+) cells were over 90%. In addition, successful knockdown of RREB1 expression was confirmed at both the mRNA level (Fig. 3B and E) and protein level (Fig. 3C and F).
Figure 3.
RREB1 was successfully knocked down in AML cell lines. (A, D) Transfection efficiency of shRNA in NB4 and HL-60 cells was observed under fluorescence microscope. (B, E) The mRNA levels of RREB1 in NB4 and HL-60 cells were detected by qRT-PCR. (C, F) The protein levels of RREB1 in NB4 and HL-60 cells were analyzed by Western blot. *p < 0.05, ***p < 0.001, compared with LV-NC.
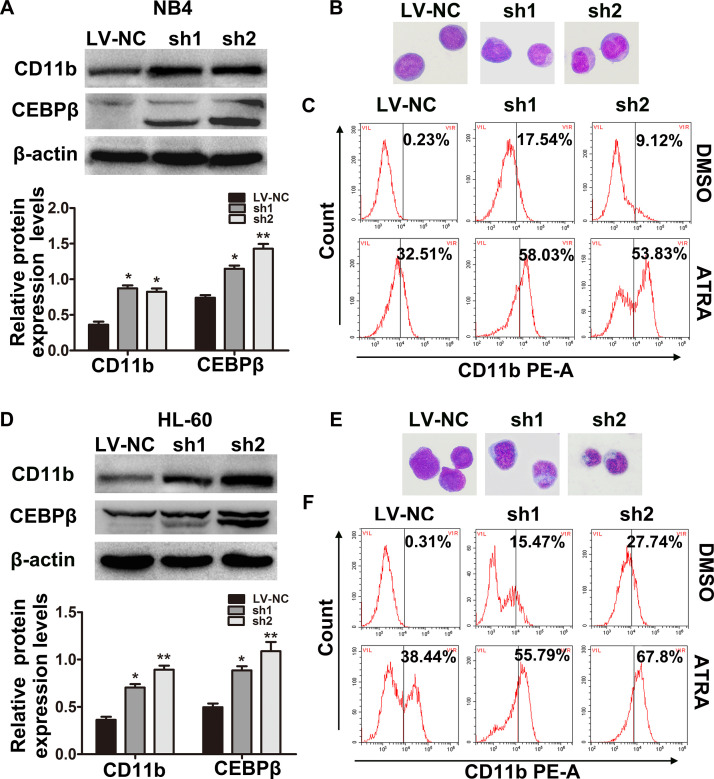
Furthermore, the expression of granulocyte differentiation marker protein CD11b and CEBPβ in the RREB1 knockdown groups was elevated compared with the LV-NC group (Fig. 4A and D). At the same time, representative Wright’s staining displayed that the differentiation of NB4 (Fig. 4B) and HL-60 (Fig. 4E) cells was enhanced in two shRREB1 groups, showing as the deviation and lobulation of the nucleus. On the other hand, we wondered if the downregulation of RREB1 would have an effect on the differentiation-inducing ability of ATRA. Therefore, ATRA (10 nmol/L) was added to the culture with NB4 and HL-60 cells after lentivirus transfection, and CD11b+ cells were analyzed by flow cytometry (Fig. 4C and F). Results showed that knockdown of RREB1 enhanced the expression of CD11b induced by ATRA. These experiments suggest that knockdown of RREB1 enhances the granulocytic differentiation of AML cells.
Figure 4.
Downregulation of RREB1 enhances differentiation of NB4 and HL-60 cells. (A) The protein levels of CD11b and CEBPβ in NB4 (A) and HL-60 (D) cells were examined by Western blot. Representative Wright–Giemsa staining images showed the change in cell morphology in NB4 (B) and HL-60 (E) cells. Original magnification: 100×. The CD11b+ cells in NB4 (C) and HL-60 (F) cells were analyzed by flow cytometry. *p < 0.05, **p < 0.01, compared with LV-NC.
miR-145 Is Associated With Granulocytic Differentiation Blockade of Myeloid Leukemia Cells Mediated by RREB1
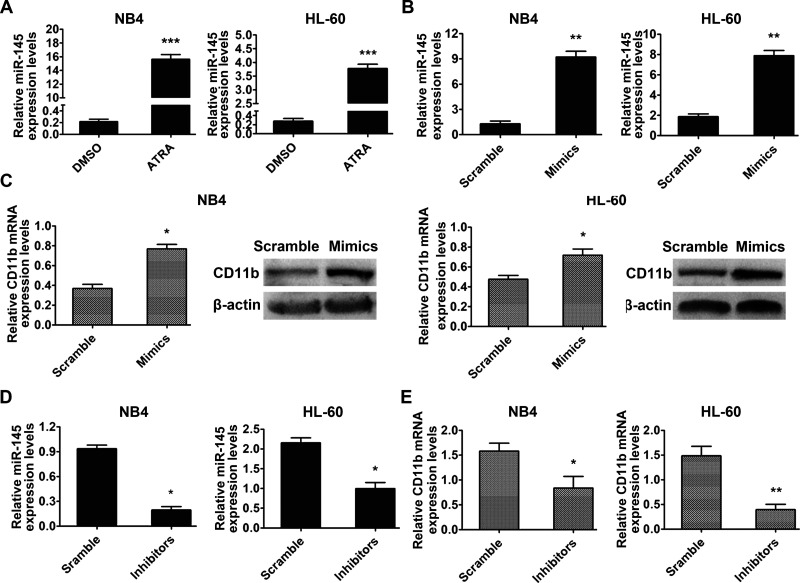
We examined the involvement of miR-145 in granulocytic differentiation in NB4 and HL-60 cells. The effects of ATRA on miR-145 in NB4 and HL-60 cells were measured, and the results showed that miR-145 expression increased significantly after ATRA treatment (Fig. 5A). Then miR-145 mimics (Fig. 5B) or inhibitors (Fig. 5D) were transfected into NB4 and HL-60 cells separately. CD11b expression was upregulated in the miR-145 mimics group at both the mRNA and protein levels compared with the scramble group (Fig. 5C), whereas the inhibition of miR-145 decreased the CD11b mRNA level (Fig. 5E). These results hint that enhanced miR-145 expression could induce granulocyte differentiation of NB4 and HL-60 cells.
Figure 5.
MicroRNA-145 (miR-145) is associated with granulocytic differentiation of myeloid leukemia cells. (A) miR-145 expression in NB4 and HL-60 cells treated with ATRA was analyzed by qRT-PCR. ***p < 0.001 compared with dimethyl sulfoxide (DMSO). (B) Levels of miR-145 in NB4 and HL-60 cells transfected with miR-145 negative control (scramble) and mimics were examined by qRT-PCR; U6 was used as an internal control. (C) The expression of CD11b was detected at both the mRNA level and protein level. β-Actin was used as an internal control. (D) Downregulation of miR-145 induced by inhibitors was measured by qRT-PCR. (E) Inhibition of miR-145 attenuated the mRNA level of CD11b measured by qRT-PCR. *p < 0.05, **p < 0.01.
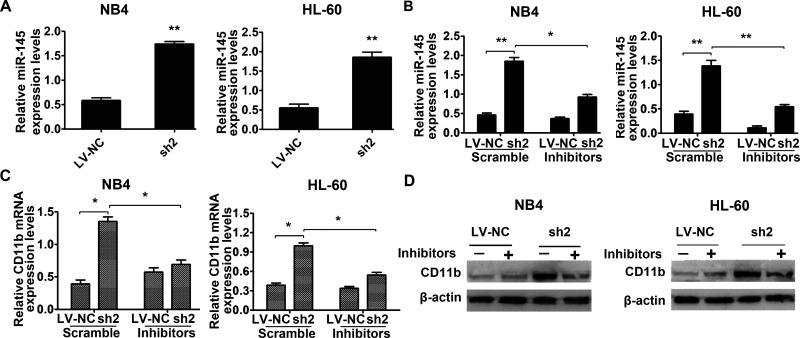
Subsequently, we detected a notable increase in miR-145 expression in the sh-RREB1 group in both NB4 and HL-60 cells (Fig. 6A). Hence, miR-145 inhibitors or scramble were transfected into shRREB1-NB4 and HL-60 cells. The results showed that expression of CD11b decreased at both the mRNA level (Fig. 6C) and protein level (Fig. 6D) in the shRREB1 group, indicating that inhibition of miR-145 abolished the enhancement of granulocyte differentiation induced by downregulation of RREB1. These results suggest that RREB1 weakens the granulocytic differentiation of AML cells by downregulation of miR-145.
Figure 6.
miR-145 is involved in the granulocytic differentiation blockade of NB4 and HL-60 cells mediated by RREB1. (A) The expression of miR-145 in the RREB1 knockdown group and LV-NC group was evaluated by qRT-PCR. (B) Successful miR-145 inhibition of NB4 and HL-60 cells was confirmed by qRT-PCR. *p < 0.05, **p < 0.01 compared with the LV-NC group. Expression of CD11b in NB4 and HL-60 cells transfected with miR-145 scramble or inhibitors after knockdown of RREB1 was measured by qRT-PCR (C) and Western blot (D). U6 and β-actin were used as negative control. *p < 0.05 compared with scramble.
The Roles of RREB1 in HL-60 and NB4 Cells Are Associated With the RAS/MEK/ERK Pathway and PML-RARα Degradation Separately
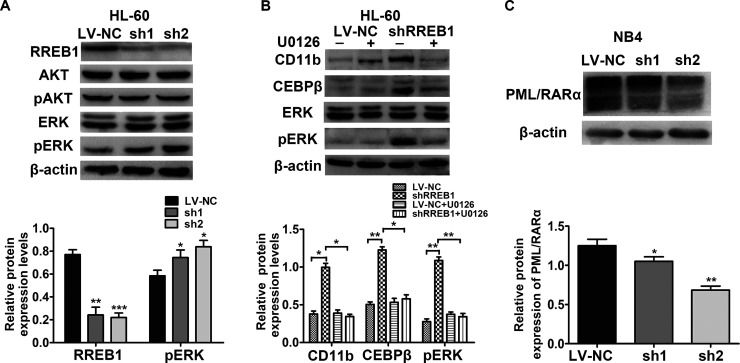
To further clarify the role of RREB1 in the blockade of granulocytic differentiation, we measured the expression of AKT, pAKT, ERK, and pERK protein in NB4 and HL-60 cells after RREB1 was knocked down (Fig. 7A). The results showed that only the expression of pERK in HL-60 cells increased, while other proteins did not obviously change. Proteins in NB4 cells did not show significant changes (data not shown). Then small molecule inhibitor of phosphorylated ERK (U0126) was used to culture with HL-60 cells after transfection by shRREB1 lentivirus. Decreased protein levels of pERK, CD11b, and CEBPβ were observed (Fig. 7B). The results showed that the RAS/MEK/ERK pathway is associated with the negative role of RREB1 in the differentiation of HL-60 cells. In addition, we detected the expression of PML-RARα protein in NB4 cells after knockdown of RREB1; the results showed that downregulation of RREB1 leads to a portion of PML-RARα fusion protein degradation (Fig. 7C). These results showed that the enhanced differentiation of NB4 cells after downregulation of RREB1 is associated with degradation of PML-RARα.
Figure 7.
The roles of RREB1 in HL-60 and NB4 cells are related to the RAS/MEK/ERK pathway and PML-RARα degradation separately. (A) Protein levels of RREB1, AKT, pAKT, ERK, and pERK of the LV-NC and shRNA groups in HL-60 cells were analyzed by Western blot. (B) The expression of CD11b, CEBPβ, ERK, and pERK protein in LV-NC and shRREB1 group treated with U0126 or not were detected by Western blot. *p < 0.05, **p < 0.01, ***p < 0.001, compared with LV-NC. (C) The expression of PML–RARα fusion protein was analyzed in NB4 cells. *p < 0.05, **p < 0.01.
DISCUSSION
In this study, we found that RREB1 was significantly upregulated in AML patients compared with normal people. What is more, RREB1 expression in AML cell lines (NB4 and HL-60) was remarkably suppressed after ATRA treatment. Subsequent RREB1 loss of function showed that knockdown of RREB1 enhanced granulocytic differentiation of NB4 and HL-60 cells. Further studies showed that the negative role of RREB1 in differentiation of AML cells was associated with the RAS/MEK/ERK pathway and miR-145 in vitro. In summary, inhibition of RREB1 is a promising strategy to induce granulocytic differentiation in AML for potential therapy.
RAS proteins play vital roles in regulating cell growth and proliferation, which are considered as a molecular switch that controls extracellular signal21. RAS proteins have complex downstream cascade and can be activated by a complex network, which leads to its wide range of influence on tumor development22. The mechanism of AML involves gene mutations that promote proliferation and impair differentiation, and studies have found that the RAS pathway was considered as a key component of leukemogenesis3,23. Previously, studies have shown that a large proportion of AML harbor mutations in NRAS, KRAS, or genes that activate Ras signaling, which makes Ras signaling an attractive therapeutic target for AML24. The abnormally high expression of the RREB1 gene in AML is likely the consequence of genetic mutation in the RAS pathway.
RREB1, the protein encoded by this gene, is a zinc finger transcription factor that binds to RAS-responsive elements (RREs) of gene promoters25. It was originally identified as a target of the RAS/RAF signaling cascade and has been shown to both activate and inhibit transcription of genes in response to the RAS pathway16. RREB1 has emerged as a likely human oncogene from multiple screenings16,26,27. A study has identified that RREB1 is a repressor of the mammalian ζ-globin genes during erythroid differentiation development, where RREB1 can decrease the expression of ζ-globin genes by binding to gene promoters through the zinc finger, and depletion of RREB1 upregulated the ζ-globin expression, which may be involved in the progression of α-thalassemia28. Another study of adult Drosophila midgut showed that RREB1/Hindsight (Hnt) is a key factor in the regulation of development and differentiation of adult midgut12. In addition, RREB1 was also found abundantly expressed in colorectal cancer, and suppressing RREB1 mediated by circular RNA circITGA7 can inhibit tumor cell proliferation and metastasis via upregulation of ITGA7 gene29, which is in line with our results that knockdown of RREB1 inhibits proliferation of NB4 and HL-60 cells (data not shown).
miR-145 has been found to be involved in multiple cancers and always shows low expression30,31. The same results were found in APL cell lines, and it was proven that low expression of miR-145 has a close relationship with the differentiation arrest of APL, since lentivirus target miR-145 tends to attenuate the differentiation of APL cells14. Furthermore, there is a strong relationship between miR-145 and RREB1. For example, in colorectal tumors and pancreatic cancer, RREB1 represses miR-145 expression through two RREs in the miR-145 promoters32. In addition, downregulation of RREB1 also induces miR-145 expression in chronic constriction injury in rats33. In our RREB1 knockdown assay, the upregulation of miR-145 was detected in the shRREB1-2 group, and the inhibitor of miR-145 could offset the effect of RREB1 depletion, implying that miR-145 is a likely downstream target of RREB1 in AML cells. However, this effect was only detected in the shRREB1-2 group but not in the shRREB1-1 group. The possible reason may be because RREB1 is an alternatively spliced transcription factor, since different RREB1 alternative isoforms have been found in urologic cancer26. So we measured the mRNA level of RREB1α, RREB1β, RREB1δ, and RREB1ε in AML cells after transfection (data not shown), which showed that two interference groups had different knockdown efficiencies on different subtypes of RREB1. Protein level was not examined because no available antibodies exist. These indicate that variants of RREB1 may play different roles in the same cell background.
In addition, considering that RREB1 was identified as a downstream target of KRAS in pancreatic adenocarcinomas34, we measured two main effectors of the KRAS (MAPK and PI3K) signal pathway. However, only increased expression of pERK in HL-60 cells was detected. A previous study identified that the RAF/MEK/ERK pathway was involved in ATRA-induced differentiation in NB4 and HL-60 cells by enhancing the expression of CEBPβ, CEBPε, and PU.135, which is consistent with our RREB1 knockdown assay, where the expression of CEBPβ and pERK proteins increased in the shRREB1 group. Furthermore, inhibitor of pERK (U0126) abrogated the effect of enhanced differentiation in HL-60 cells mediated by knockdown of RREB1, which demonstrated that the role of RREB1 in HL-60 cells is involved in the RAS/MEK/ERK signal pathway. We believed the different effects in NB4 cells were caused by different cell backgrounds from HL-60 cells. NB4 cells were considered derived from human APL cells, which harbors the PML–RARα fusion protein, while HL-60 cells do not. Lee et al. have found different gene expression in ATRA-induced differentiation of NB4 and HL-60 cells36. We also identified the expression of the PML–RARα fusion protein in NB4 cells when RREB1 was knocked down; the results indicate that knockdown of RREB1 degrades part of the fusion protein (Fig. 7C). This may be an important reason why knocking down RREB1 can promote the differentiation of NB4 cells.
In conclusion, we propose a new gene involved in granulocytic differentiation of AML cells, which is highly expressed in NB4 and HL-60 cells, and can be downregulated by ATRA. RREB1 plays a negative role in granulocyte differentiation by depressing the downstream targets of the RAS/MEK/ERK pathway and partially inhibiting the expression of tumor suppressor miR-145. Our findings propose that RREB1 could be a promising therapeutic target for patients with AML. Due to the fact that RREB1 has different isoforms in AML, the interaction network is extremely complex. Therefore, the specific molecular mechanism still needs to be clarified.
ACKNOWLEDGMENTS
This work was supported by the National Natural Science Foundation of China (Grant No. 81772280).
Footnotes
The authors declare no conflicts of interest.
REFERENCES
- 1. Li S, Mason CE, Melnick A. Genetic and epigenetic heterogeneity in acute myeloid leukemia. Curr Opin Genet Dev. 2016;36:100–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Rubio P, Campos B, Digiorge JA, Gallego MS, Medina A, Rossi JG, Felice MS, Alonso CN. NPM1, FLT3 and CEBPA mutations in pediatric patients with AML from Argentina: Incidence and prognostic value. Int J Hematol. 2016;104(5):582–90. [DOI] [PubMed] [Google Scholar]
- 3. Medinger M, Passweg JR. Acute myeloid leukaemia genomics. Br J Haematol. 2017;179(4):530–42. [DOI] [PubMed] [Google Scholar]
- 4. Kavianpour M, Ahmadzadeh A, Shahrabi S, Saki N. Significance of oncogenes and tumor suppressor genes in AML prognosis. Tumour Biol. 2016;37(8):10041–52. [DOI] [PubMed] [Google Scholar]
- 5. Thiagalingam A, De Bustros A, Borges M, Jasti R, Compton D, Diamond L, Mabry M, Ball DW, Baylin SB, Nelkin BD. RREB-1, a novel zinc finger protein, is involved in the differentiation response to Ras in human medullary thyroid carcinomas. Mol Cell Biol. 1996;16(10):5335–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lee DH, Ko JJ, Ji YG, Chung HM, Hwang T. Proteomic identification of RREB1, PDE6B, and CD209 up-regulated in primitive gut tube differentiated from human embryonic stem cells. Pancreas 2012;41(1):65–73. [DOI] [PubMed] [Google Scholar]
- 7. Terriente-Felix A, Li J, Collins S, Mulligan A, Reekie I, Bernard F, Krejci A, Bray S. Notch cooperates with Lozenge/Runx to lock haemocytes into a differentiation programme. Development 2013;140(4):926–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Liu H, Hew HC, Lu ZG, Yamaguchi T, Miki Y, Yoshida K. DNA damage signalling recruits RREB-1 to the p53 tumour suppressor promoter. Biochem J. 2009;422(3):543–51. [DOI] [PubMed] [Google Scholar]
- 9. Melani M, Simpson KJ, Brugge JS, Montell D. Regulation of cell adhesion and collective cell migration by hindsight and its human homolog RREB1. Curr Biol. 2008;18(7):532–7. [DOI] [PubMed] [Google Scholar]
- 10. Siegfried A, Romary C, Escudie F, Nicaise Y, Grand D, Rochaix P, Barres B, Vergez S, Chevreau C, Coindre JM, Uro-Coste E, Le Guellec S. RREB1-MKL2 fusion in biphenotypic “oropharyngeal” sarcoma: New entity or part of the spectrum of biphenotypic sinonasal sarcomas? Genes Chromosomes Cancer 2018;57(4):203–10. [DOI] [PubMed] [Google Scholar]
- 11. Ray SK, Nishitani J, Petry MW, Fessing MY, Leiter AB. Novel transcriptional potentiation of BETA2/NeuroD on the secretin gene promoter by the DNA-binding protein Finb/RREB-1. Mol Cell Biol. 2003;23(1):259–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Baechler BL, McKnight C, Pruchnicki PC, Biro NA, Reed BH. Hindsight/RREB-1 functions in both the specification and differentiation of stem cells in the adult midgut of Drosophila. Biol Open 2015;5(1):1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Marstrand TT, Borup R, Willer A, Borregaard N, Sandelin A, Porse BT, Theilgaard-Monch K. A conceptual framework for the identification of candidate drugs and drug targets in acute promyelocytic leukemia. Leukemia 2010;24(7):1265–75. [DOI] [PubMed] [Google Scholar]
- 14. Batliner J, Buehrer E, Fey MF, Tschan MP. Inhibition of the miR-143/145 cluster attenuated neutrophil differentiation of APL cells. Leuk Res. 2012;36(2):237–40. [DOI] [PubMed] [Google Scholar]
- 15. Kent OA, Chivukula RR, Mullendore M, Wentzel EA, Feldmann G, Lee KH, Liu S, Leach SD, Maitra A, Mendell JT. Repression of the miR-143/145 cluster by oncogenic Ras initiates a tumor-promoting feed-forward pathway. Genes Dev. 2010;24(24):2754–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kent OA, Fox-Talbot K, Halushka MK. RREB1 repressed miR-143/145 modulates KRAS signaling through downregulation of multiple targets. Oncogene 2013;32(20):2576–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lee KH, Chang MY, Ahn JI, Yu DH, Jung SS, Choi JH, Noh YH, Lee YS, Ahn MJ. Differential gene expression in retinoic acid-induced differentiation of acute promyelocytic leukemia cells, NB4 and HL-60 cells. Biochem Biophys Res Commun. 2002;296(5):1125–33. [DOI] [PubMed] [Google Scholar]
- 18. Pelicci PG, Lanfrancone L, Brathwaite MD, Wolman SR, Dalla-Favera R. Amplification of the c-myb oncogene in a case of human acute myelogenous leukemia. Science 1984;224(4653):1117–21. [DOI] [PubMed] [Google Scholar]
- 19. Ishizawa J, Kojima K, Chachad D, Ruvolo P, Ruvolo V, Jacamo RO, Borthakur G, Mu H, Zeng Z, Tabe Y, Allen JE, Wang Z, Ma W, Lee HC, Orlowski R, Sarbassov dos D, Lorenzi PL, Huang X, Neelapu SS, McDonnell T, Miranda RN, Wang M, Kantarjian H, Konopleva M, Davis RE, Andreeff M. ATF4 induction through an atypical integrated stress response to ONC201 triggers p53-independent apoptosis in hematological malignancies. Sci Signal. 2016;9(415):ra17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yu SH, Zhu KY, Chen J, Liu XZ, Xu PF, Zhang W, Yan L, Guo HZ, Zhu J. JMJD3 facilitates C/EBPbeta-centered transcriptional program to exert oncorepressor activity in AML. Nat Commun. 2018;9(1):3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Simanshu DK, Nissley DV, McCormick F. RAS proteins and their regulators in human disease. Cell 2017;170(1):17–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Brock EJ, Ji K, Reiners JJ, Mattingly RR. How to target activated Ras proteins: Direct inhibition vs. induced mislocalization. Mini Rev Med Chem. 2016;16(5):358–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bowen DT, Frew ME, Hills R, Gale RE, Wheatley K, Groves MJ, Langabeer SE, Kottaridis PD, Moorman AV, Burnett AK, Linch DC. RAS mutation in acute myeloid leukemia is associated with distinct cytogenetic subgroups but does not influence outcome in patients younger than 60 years. Blood 2005;106(6):2113–9. [DOI] [PubMed] [Google Scholar]
- 24. Pomeroy EJ, Eckfeldt CE. Targeting Ras signaling in AML: RALB is a small GTPase with big potential. Small GTPases 2017:1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Liang H, Ding X, Zhou C, Zhang Y, Xu M, Zhang C, Xu L. Knockdown of eukaryotic translation initiation factors 3B (EIF3B) inhibits proliferation and promotes apoptosis in glioblastoma cells. Neurol Sci. 2012;33(5):1057–62. [DOI] [PubMed] [Google Scholar]
- 26. Nitz MD, Harding MA, Smith SC, Thomas S, Theodorescu D. RREB1 transcription factor splice variants in urologic cancer. Am J Pathol. 2011;179(1):477–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Milon BC, Agyapong A, Bautista R, Costello LC, Franklin RB. Ras responsive element binding protein-1 (RREB-1) down-regulates hZIP1 expression in prostate cancer cells. Prostate 2010;70(3):288–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chen RL, Chou YC, Lan YJ, Huang TS, Shen CK. Developmental silencing of human zeta-globin gene expression is mediated by the transcriptional repressor RREB1. J Biol Chem. 2010;285(14):10189–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Li X, Wang J, Zhang C, Lin C, Zhang J, Zhang W, Zhang W, Lu Y, Zheng L, Li X. Circular RNA circITGA7 inhibits colorectal cancer growth and metastasis by modulating the Ras pathway and upregulating transcription of its host gene ITGA7. J Pathol. 2018;246(2):166–79. [DOI] [PubMed] [Google Scholar]
- 30. Shen H, Shen J, Wang L, Shi Z, Wang M, Jiang BH, Shu Y. Low miR-145 expression level is associated with poor pathological differentiation and poor prognosis in non-small cell lung cancer. Biomed Pharmacother. 2015;69:301–5. [DOI] [PubMed] [Google Scholar]
- 31. Campayo M, Navarro A, Vinolas N, Diaz T, Tejero R, Gimferrer JM, Molins L, Cabanas ML, Ramirez J, Monzo M, Marrades R. Low miR-145 and high miR-367 are associated with unfavourable prognosis in resected nonsmall cell lung cancer. Eur Respir J. 2013;41(5):1172–8. [DOI] [PubMed] [Google Scholar]
- 32. Yu Y, Nangia-Makker P, Farhana L, G Rajendra S, Levi E, Majumdar AP. miR-21 and miR-145 cooperation in regulation of colon cancer stem cells. Mol Cancer 2015;14:98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pang X, Tang Y, Zhang D. Role of miR-145 in chronic constriction injury in rats. Exp Ther Med. 2016;12(6):4121–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kent OA, Sandi MJ, Burston HE, Brown KR, Rottapel R. An oncogenic KRAS transcription program activates the RHOGEF ARHGEF2 to mediate transformed phenotypes in pancreatic cancer. Oncotarget 2017;8(3):4484–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Weng XQ, Sheng Y, Ge DZ, Wu J, Shi L, Cai X. RAF-1/MEK/ERK pathway regulates ATRA-induced differentiation in acute promyelocytic leukemia cells through C/EBPbeta, C/EBPepsilon and PU.1. Leuk Res. 2016;45:68–74. [DOI] [PubMed] [Google Scholar]
- 36. Lee KH, Chang MY, Ahn JI, Yu DH, Jung SS, Choi JH, Noh YH, Lee YS, Ahn MJ. Differential gene expression in retinoic acid-induced differentiation of acute promyelocytic leukemia cells, NB4 and HL-60 cells. Biochem Biophys Res Commun. 2002;296(5):1125–33. [DOI] [PubMed] [Google Scholar]