Abstract
Hepatocellular carcinoma (HCC) has high morbidity and mortality rates, and the number of new cases and deaths from liver cancer are increasing. However, the details of the regulation in HCC remain largely unknown. Plant homeodomain finger protein 8 (PHF8) is a JmjC domain-containing protein. Recently, PHF8 was reported to participate in several types of cancer. However, the biological function and clinical significance of PHF8 in HCC remain unknown. In this study, we investigate the role of PHF8 in HCC growth and metastasis. We used bioinformatics analysis and identified the differentially expressed PHF8 in primary HCC and metastasis HCC. Immunohistochemistry analysis demonstrated that PHF8 was expressed higher in human HCC tissues than in corresponding adjacent noncancerous tissues. Silencing PHF8 in HCC cells significantly decreased the cells’ ability of proliferation, migration, invasion, and sphere formation. On the contrary, overexpression of PHF8 promoted these properties. In addition, the analysis in vivo showed that PHF8 overexpression promoted tumor formation and metastasis in nude mice. In the end, the RNA-sequence assay showed that CUL4A is upregulated by the PHF8. Taken together, these results demonstrated that PHF8 was a novel oncogene in HCC, which may contribute to therapeutic approaches aimed at targeting components of the PHF8 and provide new insights into the mechanisms governing the developmental programs in HCC.
Key words: Hepatocellular carcinoma (HCC), PHF8, Proliferation, Metastasis
INTRODUCTION
Hepatocellular carcinoma (HCC) has become the most common leading cause of cancer-associated mortality globally1, which has high morbidity and mortality rates, and the number of new cases and deaths from liver cancer are increasing2. However, the details of the regulation in HCC remain largely unknown.
Histone modification pattern change is involved in the development and metastasis of cancer. Histone demethylases have been linked to various cancers. For instance, LSD1 inhibits breast cancer’s metastasis, but promotes proliferation, migration, and invasion of non-small cell lung cancer (NSCLC)3. KDM5B promotes bladder and lung tumor progression but serves as a tumor suppressor in melanoma4. Histone demethylases play important roles during HCC initiation and metastasis as well5. Previously, investigators also considered targeting histone demethylases as a new weapon in the fight against cancer6. Thus, illustrating the role of histone demethylases in HCC is critical for the treatment of this disease. The KDM7 family of histone demethylases, which consists of three members, KDM7A, KDM7B [plant homeodomain finger protein 8 (PHF8)], and KDM7C, is characterized by the presence of a C-terminal JmjC domain and a PHD finger domain in the N-terminal portion7. The members of this family have been associated with tumor formation processes7. PHF8 catalyzes the removal of H3K9me2/me1 and monomethylated histone H4 lysine 20 (H4K20me1); its PHD finger binds H3K4me3 and contributes to the demethylase activity of the protein at appropriate target sites8. Recently, PHF8 was reported to participate in several types of cancer, including prostate cancer, leukemia, and esophageal squamous cell carcinoma9. Previous data identified that PHF8 was overexpressed in prostate cancer with an impact on cell proliferation, migration, and invasion. However, the biological function and clinical significance of PHF8 in HCC remain to be investigated.
In the present study, we found that the PHF8 was upregulated in HCC and correlates with poor prognosis of HCC. PHF8 enhanced metastasis and progression of HCC cells in vitro and in vivo. Thus, we identified PHF8 as an oncogenic protein in HCC and suggested that PHF8 might be a potential therapeutic target for preventing HCC progression.
MATERIALS AND METHODS
Cell Lines
The HCC cell lines were purchased from ATCC. The HCC cell lines were cultured in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% fetal bovine serum (FBS) at 37°C in an atmosphere containing 5% CO2.
Western Blot
Cells were lysed with cell lysis buffer (Beyotime, Zhejiang, P.R. China). Total proteins were applied to SDS-polyacrylamide gel. After electrophoresis, the proteins were transferred to PVDF membranes, followed by blocking in the buffer containing 5% fat-free dry milk. The membranes were then probed with indicated antibodies overnight, and then washed and incubated with conjugated secondary antibodies for 1.5 h and finally visualized using Chemiluminescent ECL reagent (Vigorous Biotechnology, Beijing, P.R. China).
Quantitative Real-Time PCR (qPCR)
Total RNA samples were extracted from lung cancer tissues or cells with TRIzol regent (TaKaRa, Dalian, P.R. China). cDNA was synthesized from 1 μg of total RNA with One Step RT-PCR Kit (TaKaRa). qPCR was performed with the SYBR Green (TaKaRa) detection method on an ABI-7500 RT-PCR system (Applied Biosystems, Foster City, CA, USA).
Immunohistochemical Analysis
Immunohistochemistry analysis was performed on samples of liver adenocarcinoma tissues and corresponding adjacent noncancerous tissues. The samples were fixed with 10% formalin, embedded in paraffin, and sliced into 5-μm sections. The sections were then stained. Staining was observed in five randomly selected high-power fields. The staining intensity was based on the average percentage of positive cells. The scoring results were analyzed by two investigators.
Colony Formation Assay
As for colony formation assays, the cells were seeded (100/35-mm dish) in triplicate and cultured for 14 days. Cell colonies were fixed, stained with 0.25% crystal violet in 50% ethanol for 20–30 min, air dried, and counted. A region containing > 50 cells was considered one colony. The number of colonies was recorded.
Transwell Assay
The invasion ability of HCC cells was performed using 24-well Transwells (8-mm pore size; Corning Life Sciences, Corning, NY, USA) coated with 1 mg/ml of Matrigel (BD Sciences, San Jose, CA, USA). The cells were seeded in the upper chamber of the wells in 100 μl of FBS-free medium, and 500 μl of 20% FBS medium was added to the lower chambers. Following incubation for 24 h, the cells remaining on the upper membrane were removed with cotton wool. Cells that had invaded through the membrane were stained with methanol and 0.1% crystal violet, and photographed with a phase-contrast inverted microscope (Olympus, Tokyo, Japan). The cells from at least five random microscopic felids (×40) were counted.
Soft Colony Formation Assay
Cells were suspended in 1.5 ml of complete medium supplemented with 0.45% low-melting point agarose. The cells were placed in 35-mm tissue culture plates containing 1.5 ml of complete medium and agarose (0.75%) on the bottom layer. The plates were incubated at 37°C with 5% CO2 for 2 weeks. Cell colonies were stained with 0.005% crystal violet and analyzed using a microscope.
Tumor Metastasis Model
Nude mice were maintained in a specific pathogen-free facility. Cells were injected into the tail veins of mice. After 2 months, the mice were sacrificed by anesthesia with chloral hydrate. The lung tissues were dissected out for hematoxylin and eosin (H&E) staining.
TCGA RNASeqV2 Expression Correlation and Outcome Analysis
The Kaplan–Meier overall survival plot and the log-rank p value for The Cancer Genome Atlas (TCGA) HCC were generated using R with HCC samples (TCGA Network, 2015) that had RNASeqV2 data and clinical information with at least 6 months of follow-up information or a death event. The samples were split according to whether the expression measured by RNASeqV2 fell into the lower one-third quantile or the upper two-thirds quantile of expression.
TCGA Genetic Alteration Analysis
Analysis of genetic alterations in HCC TCGA data was performed on cBioPortal (http://www.cbioportal.org). A Fisher’s exact test was performed on the confusion matrix of co-occurrence of alterations in PHF8 and CUL4A. CBioPortal (http://www.cbioportal.org) containing samples of HCC and its GISTIC 2.0 changes in copy number, all exon group sequencing mutation data, and RNASeqV2 RSEM expression data were used in this study. The p values were calculated using a pairwise t-test in R, performing pairwise comparisons between group levels with a Holm correction for multiple testing.
Statistical Analysis
SPSS 20.0 software (IBM, Armonk, NY, USA) was used for statistical analysis. The significance of differences between groups was estimated by Student’s t-test and chi-square test as appropriate. The survival curves were calculated by the Kaplan–Meier method with the log-rank test applied for comparison. Survival data were evaluated using univariate and multivariate Cox proportional hazards model. Variables with a value of p < 0.05 in univariate analysis were used in subsequent multivariate analysis on the basis of Cox regression analyses.
RESULTS
High Expression of PHF8 Predicts Poor Survival
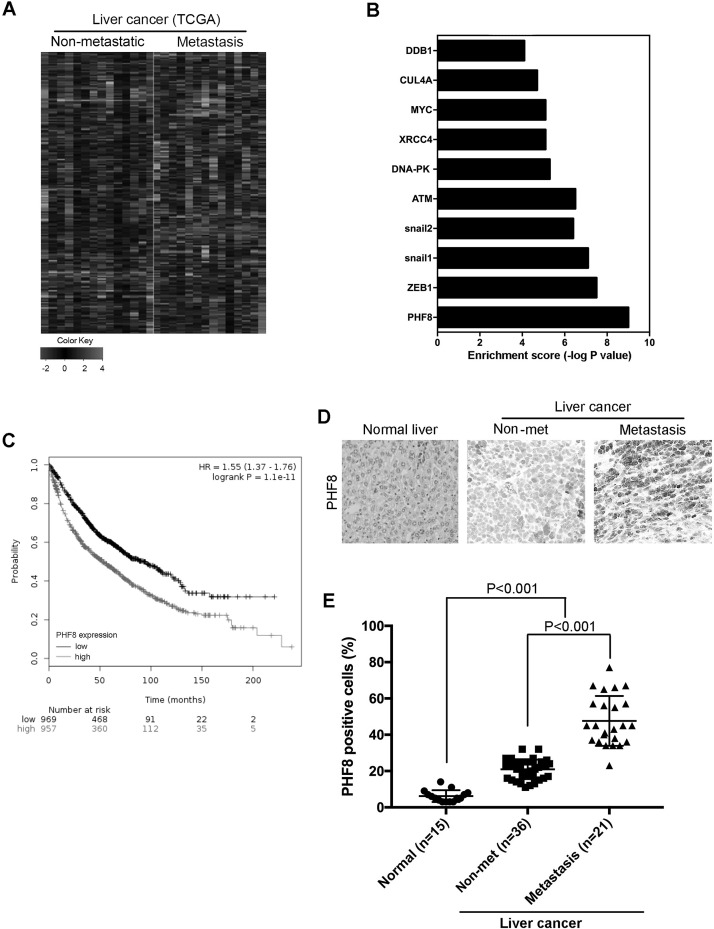
We used datasets from TCGA databases; genes that are dysregulated in primary liver cancer and metastasis liver cancer are outlined (Fig. 1A). Moreover, bioinformatics analysis identified 10 differentially expressed genes including PHF8 (Fig. 1B). These data analyses indicate that PHF8 may play an important role in human liver cancer metastasis. To further explore whether PHF8 participates in human liver cancer, we analyzed the correlation between PHF8 mRNA level and survival using the Kaplan–Meier method. Significantly, high PHF8 level predicted poor survival (Fig. 1C). The finding indicates that PHF8 is involved in human HCC. Then we examined the expression of PHF8 in paired human HCC specimens by immunohistochemistry (IHC). The expression level of PHF8 protein in HCC tissues was significantly higher than those in peritumor tissues (Fig. 1D and E). More importantly, we found that PHF8 overexpression was significantly correlated with distant metastasis (Fig. 1E).
Figure 1.
High PHF8 expression level predicted poor survival. (A) Heat map of differentially expressed genes in primary liver cancer and metastasis liver cancer in The Cancer Genome Atlas (TCGA) dataset. (B) Bioinformatics analysis identified 10 differentially expressed genes in primary liver cancer and metastasis liver cancer. (C) Kaplan–Meier curve comparing time to survival between hepatocellular carcinomas (HCCs) with the plant homeodomain finger protein 8 (PHF8). (D) Patients with higher expression of PHF8. PHF8 protein levels were analyzed by immunohistochemistry (IHC) staining. (E) The graph showed quantitative analysis. All experiments were repeated three times. Error bars indicated standard deviation.
Stable Construction of Cell Lines
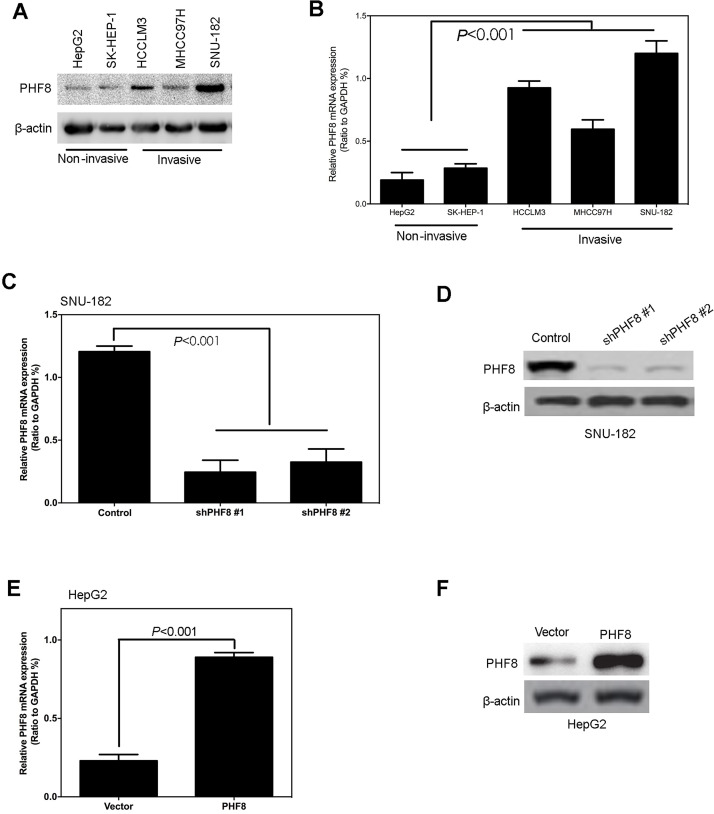
To study the potential oncogenic mechanism of PHF8 in hepatoma cells, we surveyed the level of endogenous PHF8 expression among various hepatoma cell lines. As shown in Figure 2A and B, PHF8 was highly expressed in invasive cancer cells (HCCLM3, SNU-182, and MHCC97H), compared with noninvasive liver cell line (HepG2, SK-HEP-1). To further assess the potential function of PHF8 in hepatocellular carcinoma, we next employed a loss-of-function strategy in PHF8 high-expressing human liver cancer cell lines SUN-182 using lentivirus-mediated short hairpin RNAs (shRNAs). qPCR and Western blot analysis showed that the knockdown of PHF8 by shRNAs (shPHF8 #1, #2) significantly decreased both the mRNA and protein expression of PHF8 (Fig. 2C and D). On the contrary, stably overexpressed PHF8 significantly elevated PHF8 mRNA and protein levels compared with the control (Fig. 2E and F). When infected with retrovirus carrying shPHF8, PHF8 protein level was significantly decreased in SUN-182 HCC cell lines.
Figure 2.
Stable construction of cell lines. PHF8 expression was analyzed by Western blotting (A) and quantitative qPCR (B) in liver cancer cell lines. PHF8 expression was analyzed by qPCR (C) and Western blotting (D) in SUN-182 cells. PHF8 expression was analyzed by qPCR (E) and Western blotting (F) in HepG2. All experiments were repeated three times. Error bars indicated standard deviation.
PHF8 Promotes the Malignant Tumor Characteristics of HCC Cells
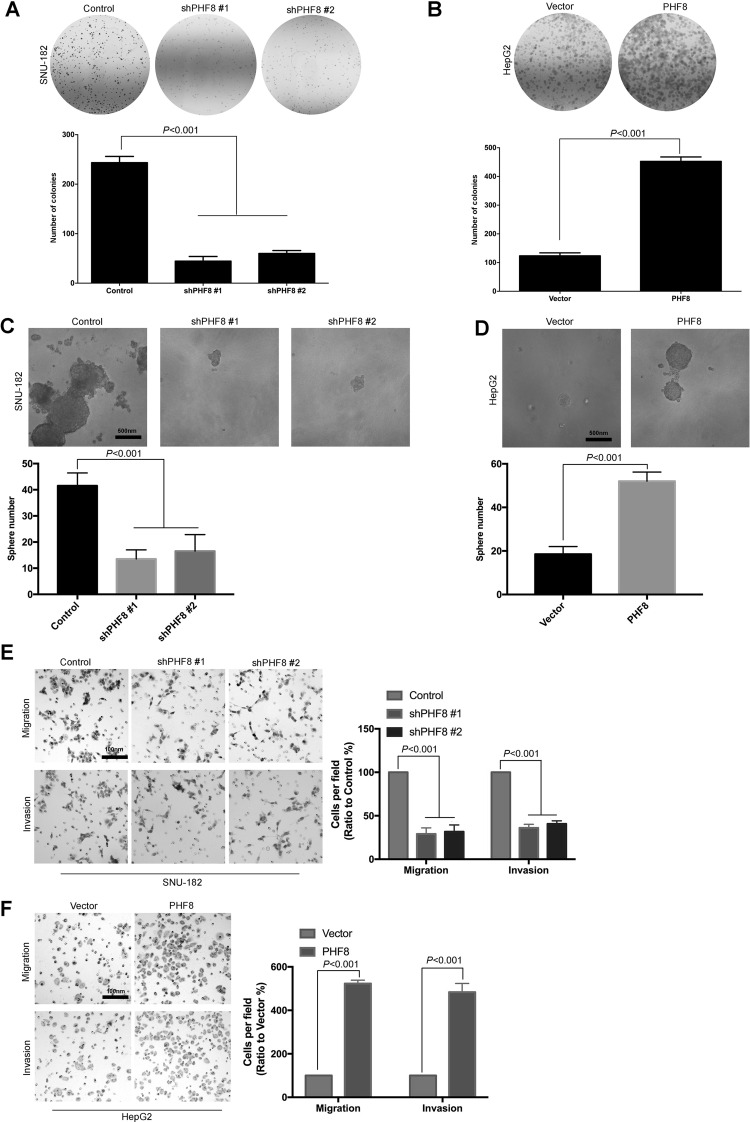
Colony formation assay showed that knockdown of PHF8 led to a slight decrease in colony number (Fig. 3A), whereas overexpression of PHF8 showed a significant increase in the number of colonies of HepG2 cells (Fig. 3B). The result suggested that PHF8 could promote proliferative capacity of HCC cells. Recent research showed that PHF8 was associated with cancer stem cell-enriched populations of several epithelial tumor cells. We next investigated whether PHF8 was associated with cancer stem cell-like traits of human liver cancer cell lines. We observed that PHF8-overexpressed cells have a higher capacity for self-renewal. Compared with control cells, significantly fewer and smaller hepatospheres were observed in PHF8 knockdown cells (Fig. 3C). PHF8-overexpressed cells formed much larger and a greater number of tumor spheres compared with the control (Fig. 3D). The results suggested that PHF8 could promote the cancer stem-like capacity of HCC cells. Previous research has found that PHF8 could promote epithelial-to-mesenchymal transition in breast cancer cells10. We confirmed that PHF8 also can promote HCC cell migration and invasion. Silencing PHF8 significantly decreased the migration and invasion of HCC cells (Fig. 3E), whereas overexpression of PHF8 led to increase in migration and invasion compared with the control (Fig. 3F).
Figure 3.
PHF8 promoted HCC cell proliferation, migration, and invasion in vitro. Colony formation assays in SNU-182 cells with PHF8 knockdown (A) and in HepG2 cells with PHF8 overexpression (B) were performed. The graph showed quantitative analysis. PHF8 overexpressed HepG2 cells (C) formed much larger and a greater number of tumor spheres than SNU-182 cells with PHF8 knockdown (D). PHF8 inhibition decreased cells’ ability of migration and invasion in SNU-182 cells (E). PHF8 overexpression increased cells’ ability of migration and invasion in HepG2 cells (F). All experiments were repeated three or four times. Error bars indicated standard deviation.
PHF8 Promoted HCC Tumor Formation In Vivo
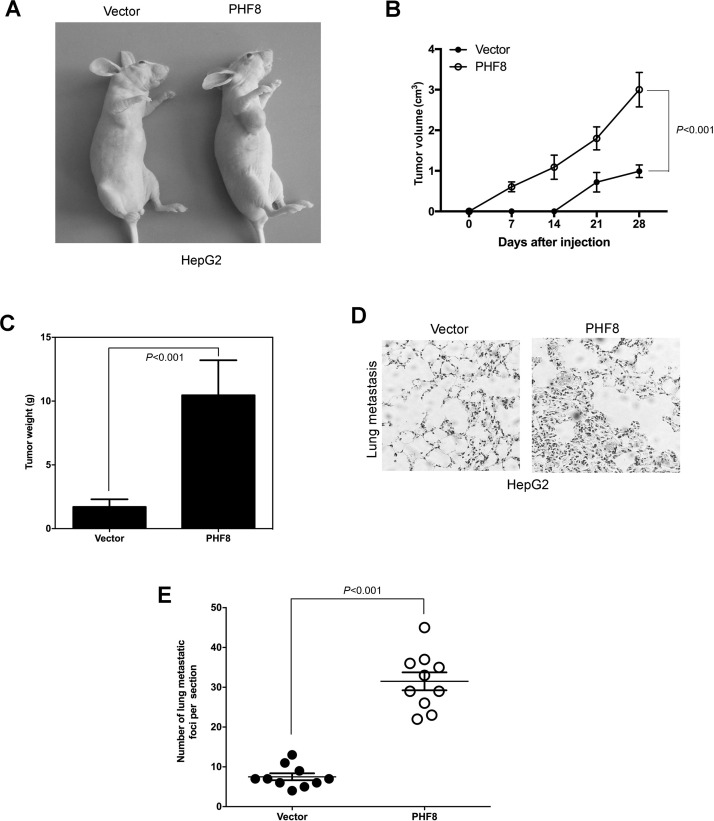
To extend our in vitro observations, we investigated whether PHF8 could regulate tumorigenic capacity of HCC cells in vivo. HepG2–PHF8 cells and their control cells were subcutaneously injected into nude mice. Tumor size was measured up to 28 days after injection. As expected, the tumors from HepG2–PHF8 cells grew more rapidly at the implantation site than the control cells (Fig. 4A–C). These results suggested that PHF8 might be an important regulator of proliferation in HCC cells in vivo.
Figure 4.
PHF8 overexpression promoted HCC cell tumor formation and metastasis in vivo. (A) Representative images of tumor from PHF8 overexpression and its control HepG2 cells. (B) Growth curve of tumors formed by PHF8 overexpression and its control HepG2 cells. (C) The weight of tumors formed by PHF8 overexpression and its control HepG2 cells. (D) Representative images of lung tissues measured by hematoxylin and eosin (H&E) stain with injection of PHF8 overexpression and its control HepG2 cells into the tail vein. (E) The numbers of metastatic foci per section in the lung of individual mouse with injection of PHF8 overexpression and its control HepG2 cells into the tail vein. All experiments were repeated three or four times. Error bars indicated standard deviation.
Overexpression of PHF8 Promoted HCC Cell Metastasis In Vivo
Due to PHF8’s contribution to migration and invasion in human liver cancer cells in vitro, we wondered whether PHF8 could participate in the metastasis in vivo. Cells were injected in tail veins of nude mice. Two months later, the mice were sacrificed by anesthesia with chloral hydrate. The lung was dissected out for H&E staining. As shown in Figure 4D and E, PHF8-overexpressed cells enhanced distant metastasis abilities compared with the control.
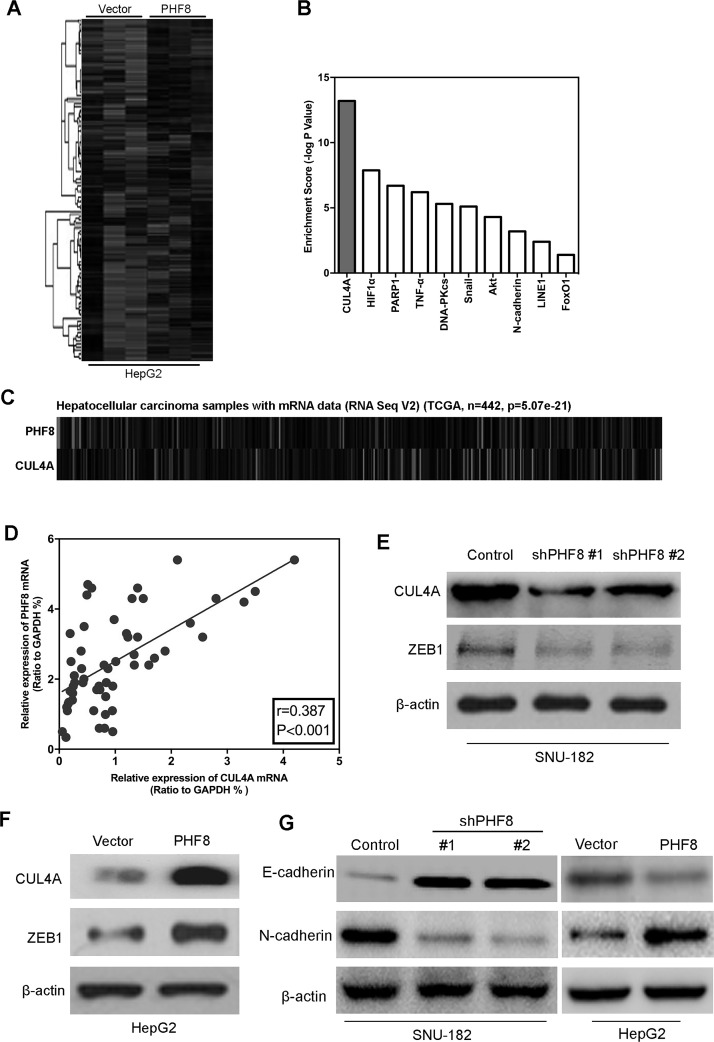
PHF8 Induced the Expression of CUL4A
To better understand the mechanisms of how PHF8 is engaged in proliferation, migration, and invasion functionality, we performed gene expression profiling on HepG2–PHF8 and its control cells. RNA-sequence analysis identified a list of significantly differentially expressed genes after PHF8’s overexpression, including the downregulation of CUL4A (Fig. 5A and B). The data from TCGA showed that the expression of PHF8 was positively correlated with CUL4A expression (Fig. 5C). The same correlation was also observed in our HCC tissues by measuring the mRNA expressions of PHF8 and CUL4A using qPCR (Fig. 5D). Together with previous reports linking CUL4A with tumorigenesis and metastasis in carcinomas11–13, we examined whether CUL4A could be a downstream target of PHF8. Expression of CUL4A in the cells with altered PHF8 expression was evaluated by Western blot. Silencing PHF8 in SNU-182 cells dramatically decreased CUL4A expression (Fig. 5E). Moreover, overexpression of PHF8 in HepG2 cells greatly increased CUL4A expression (Fig. 5F). It has been demonstrated that CUL4A targets many proteins for degradation, such as ZEB114. Thus, we measured the expression level of ZEB1 in PHF8-altered cells. The results showed that silencing PHF8 significantly inhibited the expression of ZEB1 (Fig. 5E), while ectopic expression of PHF8 drastically upregulated its expression (Fig. 5F). CUL4A has been reported to regulate EMT14, so we examined whether PHF8 can alter EMT marker expression in liver cancer cells. As shown in Figure 5G, the epithelial cell marker E-cadherin dramatically decreased after PHF8 overexpression in HepG2 cells, and the mesenchymal cell marker N-cadherin was significantly upregulated. On the contrary, silencing PHF8 in SNU-182 cells has reversed the results. All of these results indicated that CUL4A was the downstream target of PHF8.
Figure 5.
PHF8 induced the expression of CUL4A. (A) Gene expression profiling on PHF8 overexpression and its control HepG2 cells was assayed by RNA-sequence analyses. (B) Gene set enrichment analysis showed the top 10 regulated by PHF8 overexpression in HepG2 cells. (C) The data from TCGA showed that the expression of PHF8 was positively correlated with CUL4A expression in HCC. (D) CUL4A mRNA expression was positively correlated withPHF8 mRNA in HCC tissues. (E) The expression of CUL4A and its target gene ZEB1 was assayed by Western blot in PHF8 knockdown and its control SNU-182 cells. (F) The expression of CUL4A and its target gene ZEB1 was assayed by Western blot in PHF8 overexpression and its control HepG2 cells. (G) The expression of E-cadherin and N-cadherin was assayed by Western blot in indicated cells.
DISCUSSION
Emerging evidence has suggested that overexpression of PHF8 was associated with several different types of cancers, including prostate cancer15, esophageal squamous cell carcinoma16, lung cancer9, and breast cancer17. However, the role of PHF8 in HCC is still unclear.
In this study, we performed a study to investigate the role of PHF8 in HCC and its potential prognostic value. We found that the expression of PHF8 was high in HCC cell lines and tissues compared with the control, which was associated with poor patient prognosis further. We also found that decreased expression of PHF8 in HCC cells inhibited the cells’ ability of migration and invasion significantly compared with the control. Furthermore, we also found that the expression of CUL4A was affected with PHF8 synchronously, indicating that PHF8 might modulate HCC’s progression through CUL4A and its target genes, such as ZEB1. These results suggest that PHF8 is an important oncogene that is involved in initiation, growth, and metastasis of hepatocellular carcinoma.
The initiation and progression of tumor have been recognized as a complex process, which is relevant to both genetic and epigenetic alterations18. Furthermore, global changes in histone modification patterns are functionally associated with cancer development and recurrence19–21. Epigenetic mechanisms have been proven to be important in cancer development22. Histone methylation, a common form of epigenetic regulation controlled by both methyltransferases and demethylases, has been reported to play fundamental roles in many cellular processes22.
As a histone demethylase, PHF8 could activate its target genes through demethylation of the transcriptionally repressive marks, H3K9me2/1 and H4K20me123. PHF8 could bind to H3K4me3 via its PHD finger, which is typically enriched at the transcription start sites (TSSs) and may play a role in their recruitment to target promoters24. Histone demethylase PHF8 acts on mH4K20me1, monomethylated and dimethylated H3 lysine 9 (H3K9me1/2), and dimethylated H3 lysine 27 (H3K27me2), serving as a transcription coactivator25–27. Bioinformatics analysis has shown that PHF8 binding sites overlap with the consensus sequences of several transcription factors significantly: E2F1, ETS-1 SP1, FOXO1, TCF, and MYC-MAX28. These data indicate that PHF8 may exert distinct functions in different types of cancer10.
In the present study, we show that PHF8 also participates in the regulation of liver cancer as a histone demethylase. Thus, PHF8 might be a potential therapeutic target for preventing HCC progression.
ACKNOWLEDGMENT
The authors declare no conflicts of interest.
Footnotes
The authors declare no conflicts of interest.
REFERENCES
- 1. Braillon A. Hepatocellular carcinoma. Lancet 2012;380(9840):469; author reply 470–61. [DOI] [PubMed] [Google Scholar]
- 2. Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012;62(1):10–29. [DOI] [PubMed] [Google Scholar]
- 3. Ambrosio S, Sacca CD, Majello B. Epigenetic regulation of epithelial to mesenchymal transition by the lysine-specific demethylase LSD1/KDM1A. Biochim Biophys Acta Gene Regul Mech. 2017;1860(9):905–10. [DOI] [PubMed] [Google Scholar]
- 4. Han M, Xu W, Cheng P, Jin H, Wang X. Histone demethylase lysine demethylase 5B in development and cancer. Oncotarget 2017;8(5):8980–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ji X, Jin S, Qu X, Li K, Wang H, He H, Guo F, Dong L. Lysine-specific demethylase 5C promotes hepatocellular carcinoma cell invasion through inhibition BMP7 expression. BMC Cancer 2015;15:801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Tang B, Qi G, Tang F, Yuan S, Wang Z, Liang X, Li B, Yu S, Liu J, Huang Q, Wei Y, Zhai R, Lei B, Yu H, Tomlinson S, He S. Aberrant JMJD3 expression upregulates Slug to promote migration, invasion, and stem cell-like behaviors in hepatocellular carcinoma. Cancer Res. 2016;76(22):6520–32. [DOI] [PubMed] [Google Scholar]
- 7. Tsukada Y, Ishitani T, Nakayama KI. KDM7 is a dual demethylase for histone H3 Lys 9 and Lys 27 and functions in brain development. Genes Dev. 2010;24(5):432–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Walsh RM, Shen EY, Bagot RC, Anselmo A, Jiang Y, Javidfar B, Wojtkiewicz GJ, Cloutier J, Chen JW, Sadreyev R, Nestler EJ, Akbarian S, Hochedlinger K. Phf8 loss confers resistance to depression-like and anxiety-like behaviors in mice. Nat Commun. 2017;8:15142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Shen Y, Pan X, Zhao H. The histone demethylase PHF8 is an oncogenic protein in human non-small cell lung cancer. Biochem Biophys Res Commun. 2014;451(1):119–25. [DOI] [PubMed] [Google Scholar]
- 10. Shao P, Liu Q, Maina PK, Cui J, Bair TB, Li T, Umesalma S, Zhang W, Qi HH. Histone demethylase PHF8 promotes epithelial to mesenchymal transition and breast tumorigenesis. Nucleic Acids Res. 2017;45(4):1687–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wang Y, Zhang P, Liu Z, Wang Q, Wen M, Wang Y, Yuan H, Mao JH, Wei G. CUL4A overexpression enhances lung tumor growth and sensitizes lung cancer cells to erlotinib via transcriptional regulation of EGFR. Mol Cancer 2014;13:252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Xu Y, Wang Y, Ma G, Wang Q, Wei G. CUL4A is overexpressed in human pituitary adenomas and regulates pituitary tumor cell proliferation. J Neurooncol. 2014;116(3):625–32. [DOI] [PubMed] [Google Scholar]
- 13. Wang Y, Ma G, Wang Q, Wen M, Xu Y, He X, Zhang P, Wang Y, Yang T, Zhan P, Wei G. Involvement of CUL4A in regulation of multidrug resistance to P-gp substrate drugs in breast cancer cells. Molecules 2013;19(1):159–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wang Y, Wen M, Kwon Y, Xu Y, Liu Y, Zhang P, He X, Wang Q, Huang Y, Jen KY, LaBarge MA, You L, Kogan SC, Gray JW, Mao JH, Wei G. CUL4A induces epithelial-mesenchymal transition and promotes cancer metastasis by regulating ZEB1 expression. Cancer Res. 2014;74(2):520–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bjorkman M, Ostling P, Harma V, Virtanen J, Mpindi JP, Rantala J, Mirtti T, Vesterinen T, Lundin M, Sankila A, Rannikko A, Kaivanto E, Kohonen P, Kallioniemi O, Nees M. Systematic knockdown of epigenetic enzymes identifies a novel histone demethylase PHF8 overexpressed in prostate cancer with an impact on cell proliferation, migration and invasion. Oncogene 2012;31(29):3444–56. [DOI] [PubMed] [Google Scholar]
- 16. Sun X, Qiu JJ, Zhu S, Cao B, Sun L, Li S, Li P, Zhang S, Dong S. Oncogenic features of PHF8 histone demethylase in esophageal squamous cell carcinoma. PLoS One 2013;8(10):e77353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wang Q, Ma S, Song N, Li X, Liu L, Yang S, Ding X, Shan L, Zhou X, Su D, Wang Y, Zhang Q, Liu X, Yu N, Zhang K, Shang Y, Yao Z, Shi L. Stabilization of histone demethylase PHF8 by USP7 promotes breast carcinogenesis. J Clin Invest. 2016;126(6):2205–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sharma S, Kelly TK, Jones PA. Epigenetics in cancer. Carcinogenesis 2010;31(1):27–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fraga MF, Ballestar E, Villar-Garea A, Boix-Chornet M, Espada J, Schotta G, Bonaldi T, Haydon C, Ropero S, Petrie K, Iyer NG, Perez-Rosado A, Calvo E, Lopez JA, Cano A, Calasanz MJ, Colomer D, Piris MA, Ahn N, Imhof A, Caldas C, Jenuwein T, Esteller M. Loss of acetylation at Lys16 and trimethylation at Lys20 of histone H4 is a common hallmark of human cancer. Nat Genet. 2005;37(4):391–400. [DOI] [PubMed] [Google Scholar]
- 20. Seligson DB, Horvath S, McBrian MA, Mah V, Yu H, Tze S, Wang Q, Chia D, Goodglick L, Kurdistani SK. Global levels of histone modifications predict prognosis in different cancers. Am J Pathol. 2009;174(5):1619–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Seligson DB, Horvath S, Shi T, Yu H, Tze S, Grunstein M, Kurdistani SK. Global histone modification patterns predict risk of prostate cancer recurrence. Nature 2005;435(7046):1262–6. [DOI] [PubMed] [Google Scholar]
- 22. Greer EL, Shi Y. Histone methylation: A dynamic mark in health, disease and inheritance. Nat Rev Genet. 2012;13(5):343–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lim HJ, Dimova NV, Tan MK, Sigoillot FD, King RW, Shi Y. The G2/M regulator histone demethylase PHF8 is targeted for degradation by the anaphase-promoting complex containing CDC20. Mol Cell Biol. 2013;33(21):4166–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Horton JR, Upadhyay AK, Qi HH, Zhang X, Shi Y, Cheng X. Enzymatic and structural insights for substrate specificity of a family of jumonji histone lysine demethylases. Nat Struct Mol Biol. 2010;17(1):38–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kleine-Kohlbrecher D, Christensen J, Vandamme J, Abarrategui I, Bak M, Tommerup N, Shi X, Gozani O, Rappsilber J, Salcini AE, Helin K. A functional link between the histone demethylase PHF8 and the transcription factor ZNF711 in X-linked mental retardation. Mol Cell 2010;38(2):165–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Liu W, Tanasa B, Tyurina OV, Zhou TY, Gassmann R, Liu WT, Ohgi KA, Benner C, Garcia-Bassets I, Aggarwal AK, Desai A, Dorrestein PC, Glass CK, Rosenfeld MG. PHF8 mediates histone H4 lysine 20 demethylation events involved in cell cycle progression. Nature 2010;466(7305):508–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Qi HH, Sarkissian M, Hu GQ, Wang Z, Bhattacharjee A, Gordon DB, Gonzales M, Lan F, Ongusaha PP, Huarte M, Yaghi NK, Lim H, Garcia BA, Brizuela L, Zhao K, Roberts TM, Shi Y. Histone H4K20/H3K9 demethylase PHF8 regulates zebrafish brain and craniofacial development. Nature 2010;466(7305):503–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Asensio-Juan E, Gallego C, Martinez-Balbas MA. The histone demethylase PHF8 is essential for cytoskeleton dynamics. Nucleic Acids Res. 2012;40(19):9429–40. [DOI] [PMC free article] [PubMed] [Google Scholar]