Abstract
This study aimed to investigate the effect and underlying mechanism of lncRNA CASC2 in malignant melanoma (MM). Expression of CASC2 in MM tissues and cells was detected. A375 cells were transfected with pc-CASC2, si-CASC2, miR-18a-5p inhibitor, or corresponding controls, and then cell proliferation, migration, and invasion were detected using MTT assay, colony formation assay, and Transwell analysis, respectively. The relationship of miR-18a-5p and CASC2 or RUNX1 was detected by luciferase reporter assay. The levels of CASC2 and RUNX1 were significantly reduced in MM tissues compared with normal skin tissues or cells, while the miR-18a-5p level was obviously increased (all p < 0.01). Cell viability, colony number, migration, and invasion were significantly decreased in cells with pc-CASC2 compared with cells transfected with pcDNA3.1 (all p < 0.05). These effects were consistent with the cells transfected with miR-18a-5p inhibitor. The luciferase reporter assay revealed that CASC2 acted as a molecular sponge for miR-18a-5p, and RUNX1 was a target gene of miR-18a-5p. Moreover, CASC2 overexpression promoted the expression of RUNX1, while upregulated miR-18a-5p significantly reversed the effect of CASC2 on the RUNX1 level (all p < 0.05). Upregulated CASC2 may inhibit cell proliferation, migration, and invasion through regulating miR-18a-5p and its target gene RUNX1 in MM.
Key words: Malignant melanoma, lncRNA CASC2, miR-18a-5p, Cell proliferation, Migration and invasion
INTRODUCTION
Malignant melanoma (MM) is a common skin cancer, with characteristics of high invasiveness and lethality1. In 2009, an increasing incidence rate of 3.1% per year was reported in the US2. Although considerable progress has been made in the treatment strategies for MM, such as surgical excision, radiation, and chemotherapy, there is only 6–9 months of median survival in patients with metastatic MM. Therefore, it is imperative to further understand the underlying mechanism and search for effective therapeutic targets in MM.
Long noncoding RNAs (lncRNAs) are commonly defined as noncoding RNA molecules with length greater than 200 nucleotides. Multiple studies have indicated the regulating roles of lncRNAs in various diseases3, including autoimmune diseases4,5, cardiovascular diseases6, neurodegenerative diseases7, and cancers8, by mediating target gene transcription. Thus, understanding the mechanisms of lncRNAs in rheumatoid arthritis fibroblast-like synoviocytes (RA-FLSs) may contribute to the search for an effective treatment for MM. Previous studies have shown that lncRNA cancer susceptibility candidate 2 (CASC2) is downregulated in several cancers, such as renal cell carcinoma9, glioma10, and bladder cancer11, and it plays an important role as a tumor inhibitor. However, the expression and role of CASC2 in MM are still unclear.
In addition, microRNAs (miRNAs), small noncoding RNAs, play a significant regulatory role in oncogenesis through mediating target gene expression and pathways related to cancer12. Evidence suggests that the functions of miRNAs are involved in the immune response, inflammation reaction, infection, and cell metabolism, growth, and migration13,14. Accumulating studies have focused on the potential roles of miRNAs in various types of cancers, such as colorectal cancer15, hepatocellular carcinoma16, chronic lymphocytic leukemia17, and breast cancer18. It has been demonstrated that the role of lncRNAs can be regulated by miRNAs in cancer development19. A recent study has found that miR-18a-5p is upregulated in MM20. However, few studies have investigated the role and mechanism of miR-18a-5p in MM.
A study revealed that CASC2 functions as a competing endogenous RNA by sponging miR-18a in colorectal cancer21, so we proposed that CASC2 and miR-18a-5p are coinvolved in the development of MM. The present study first analyzed the levels of CASC2 and miR-18a-5p in MM tissues and normal skin tissues. We then up- or downregulated the level of CASC2 or miR-18a-5p in the human MM cell line A375 and evaluated the effects of the aberrant expression of CASC2 or miR-18a-5p on cell proliferation, migration, and invasion, aiming to investigate the regulatory role of CASC2 and miR-18a-5p in MM.
MATERIALS AND METHODS
Tissue Samples and Cell Line
A total of 12 cutaneous MM tissues were collected from patients with cutaneous MM who underwent surgical resection of their tumors. The patients did not receive preoperative chemotherapy. In addition, 12 normal skin tissues were collected from healthy unaffected skin of the same patients. For this study, the approval of the ethics committee of our hospital and informed consent from each patient were obtained. The MM cell lines A375, A2058, and HTB63 and normal human epidermal melanocytes HEMa-LP cells were all purchased from Shanghai Cell Bank of the Chinese Academy of Sciences (CAS; Shanghai, Beijing). These cells were maintained in RPMI-1640 medium (Gibco, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (FBS; Gibco) and antibiotics.
Cell Treatment
A375 cells were seeded onto a 60-mm dish and cultured for 24 h. To evaluate the roles of CASC2 and miR-18a-5p on MM, cells were transiently transfected with pc-CASC2, pcDNA3.1, si-CASC2, si-control, miR-18a-5p inhibitor, inhibitor control, miR-18a-5p mimic, mimic control, or pc-RUNX1, respectively (RiboBio Corp., Guangzhou, P.R. China) using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA). To further evaluate the relationship of CASC2 and miR-18a-5p, cells were transfected with pc-CASC2, pc-CASC2 + miR-18a-5p mimic, or pc-CASC2 + mimic control, respectively. After treatment for 48 h, cells with various treatments were subjected to the following experiments.
Luciferase Reporter Assay
The potential target gene of miR-18a-5p was predicted using sequence analysis. The wild-type (wt) 3′-untranslated region (3′-UTR) fragment of CASC2 or RUNX1 that can bind to miR-18a-5p or the mutant 3′-UTR fragments were amplified from the genomic DNA and cloned into the pGL3 vector. A375 cells were cotransfected with miR-18a-5p mimic and luciferase reporter comprising wt or mutant CASC2 or RUNX1 for 48 h using Lipofectamine 2000. The Dual Luciferase Assay kit (Promega, Madison, WI, USA) was used to measure the activity of luciferase.
MTT Assay
MTT assay was used to detect cell viability. A375 cells (1 × 104) were incubated in 96-well plates. On the following day, cells were treated with the above-mentioned transfections for 24, 48, or 72 h. Then 10 μl of MTT (5 mg/ml; Sigma-Aldrich, St. Louis, MO, USA) was added into each well for 4 h at 37°C. Next, 100 μl of dimethyl sulfoxide (DMSO; Sigma-Aldrich) was used to dissolve the formazan crystals. The absorbances at 495 nm were measured using a microplate reader (Molecular Devices, Sunnyvale, CA, USA).
Colony Formation Assay
A375 cells (0.5 × 103) were seeded into each well of six-well plates and cultured for 10 days in Dulbecco’s modified Eagle’s medium (DMEM; Gibco) containing 10% FBS. Subsequently, cells were stained with 0.5% (w/v) crystal violet dissolved in ethanol (Sigma-Aldrich) for 5 min. The mean number of colonies was calculated from 10 different fields of vision. All experiments were performed in triplicate.
Transwell Analysis
Three hundred microliters of A375 cell suspension with 5 × 105 cells/ml was seeded into the upper chamber of a Transwell chamber (for invasion assay, precoated with Matrigel; Millipore, Bedford, MA, USA) in serum-free medium. Then 500 μl of DMEM containing 10% FBS was added to the lower chamber. After incubation for 24 h, the cells that did not migrate through the pores were carefully wiped away using a cotton-tipped swab. The filter membrane was stained with crystal violet. For the migration assay, the chamber was not treated with Matrigel. Ultimately, five fields were randomly selected to count the cell number under an inverted microscope (Olympus Corp., Tokyo, Japan).
Quantitative Reverse Transcription Polymerase Chain Reaction (qRT-PCR)
After various treatments, the total RNA was extracted using TRIzol (Invitrogen), and complementary DNA synthesis was carried out using miRNA-specific primers (Invitrogen). The PCR primers for miR-18a-5p and U6 were commercially obtained from Applied Biosystems (Foster City, CA, USA). The CASC2 sense primer was 5′-GCACATTGGACGGTGTTTCC-3′ and the antisense primer was 5′-CCCAGTCCTTCACAGGTCAC-3′; the RUNX1 sense primer was 5′-CTGCCCATCGCTTTCAAGGT-3′ and the antisense primer was 5′-GCCGAGTAGTTTTCATCATTGCC-3′; and the glyceraldehyde-3-phosphate dehydrogenase (GAPDH) sense primer was 5′-GCACCGTCAAGGCTGAGAAC-3′ and the antisense primer was 5′-TGGTGAAGACGCCAGTGGA-3′. The PCR parameters were set as follows: 95°C for 10 min, 40 cycles of 94°C for 30 s, 58°C for 30 s, and 72°C for 15 s. U6 or GAPDH was used as the reference gene, and relative gene expression level was calculated using the comparative threshold (Ct) cycle method (2−ΔΔ Ct). All experiments were performed in triplicate.
Western Blotting
Protein from cells was extracted using radioimmunoprecipitation assay (RIPA) lysis buffer (Beyotime Institute of Biotechnology, Shanghai, P.R. China), and the concentration was measured using the BCA Protein Quantitative Assay (Beyotime Institute of Biotechnology). A total of 50 μg of protein sample (per lane) was separated on sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gel, blotted onto polyvinylidene difluoride (PVDF) membranes, and blocked in 5% nonfat milk for 1 h. The membranes were probed with rabbit anti-RUNX1 polyclonal antibodies (1:500; Santa Cruz Biotechnology, Santa Cruz, CA, USA) and mouse anti-GAPDH monoclonal antibody (1:2,000; Sigma-Aldrich) overnight at 4°C, respectively. After washing three times with phosphate-buffered saline (PBS), the membranes were incubated with appropriate IgG (H + L)-HRP (1:5,000; Santa Cruz Biotechnology) second antibody for 2 h at room temperature. Ultimately, the proteins were detected with enhanced chemiluminescence (Millipore).
Statistical Analysis
Statistical analysis was carried out using a statistical analysis software (SPSS 19.0; SPSS Inc., Chicago, IL, USA). Data were expressed as the mean ± standard deviation and analyzed by one-way analysis of variance. A value of p < 0.05 was considered to indicate a statistically significant result.
RESULTS
Effect of CASC2 on MM Cell Proliferation, Migration, and Invasion
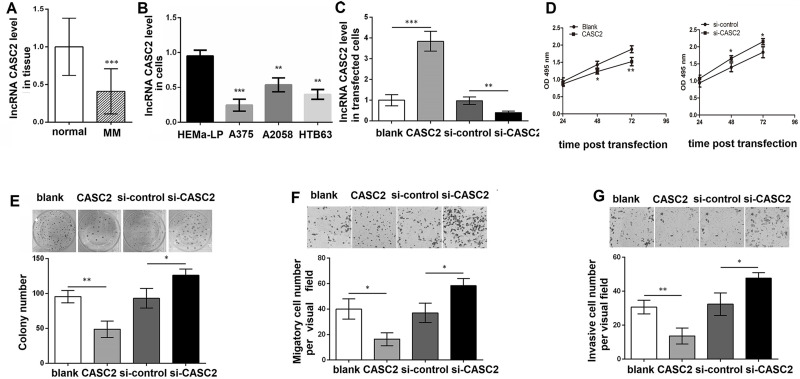
qRT-PCR analysis revealed that the CASC2 level was decreased in MM tissues compared with normal skin tissues (p < 0.001) (Fig. 1A). In addition, we analyzed the expression of CASC2 in MM cell lines. The results showed that the CASC2 level was also significantly decreased in A375, A2058, and HTB63 compared to epidermal melanocytes HEMa-LP cells (p < 0.01 or p < 0.001) (Fig. 1B). To evaluate the effect of CASC2 on MM, the CASC2 level was altered in A375 cells. As shown in Figure 1C, the CASC2 level was significantly increased in cells with pc-CASC2 compared with cells with pcDNA3.1 (p < 0.001), and the transfection with si-CASC2 obviously inhibited the CASC2 level compared with the transfection with si-control (p < 0.01). The MTT assay revealed that cell viability was significantly inhibited in cells with pc-CASC2 compared with pcDNA3.1-treated cells, while cell viability was obviously increased in cells treated with si-CASC2 compared with that in cells treated with si-control (p < 0.05) (Fig. 1D). Consistent with the results of the MTT assay, the colony formation assay also found that the number of colonies was significantly decreased in CASC2 upregulated cells, while obviously increased in CASC2 downregulated cells compared with control cells (p < 0.05) (Fig. 1E). In addition, the Transwell assay showed that both migratory and invasive cell numbers were remarkably lower in cells with pc-CASC2 than those in cells with pcDNA3.1, but were higher in si-CASC2-treated cells compared with si-control (p < 0.05) (Fig. 1F and G).
Figure 1.
Long noncoding RNA (lncRNA) cancer susceptibility candidate 2 (CASC2) inhibits malignant melanoma (MM) cell proliferation, migration, and invasion. (A) The CASC2 level in MM tissues and normal skin tissues using quantitative reverse transcription polymerase chain reaction (qRT-PCR). (B) The CASC2 level in A375, A2058, HTB63, and epidermal melanocytes HEMa-LP cells. (C) The CASC2 level in A375 cells with pc-CASC2, pcDNA3.1 (blank), si-CASC2, or si-control using qRT-PCR. (D) The cell viability in A375 cells with pc-CASC2, pcDNA3.1 (blank), si-CASC2, or si-control for 24, 48, and 72 h, respectively, using the MTT assay. (E) The colony number in A375 cells with pc-CASC2, pcDNA3.1 (blank), si-CASC2, or si-control using colony formation assay. (F) The migratory cell number in A375 cells with pc-CASC2, pcDNA3.1 (blank), si-CASC2, or si-control using Transwell analysis. (G) The invasive cell number in A375 cells with pc-CASC2, pcDNA3.1 (blank), si-CASC2, or si-control using Transwell analysis. *p < 0.05, **p < 0.01, and ***p < 0.001 compared with normal.
Effect of miR-18a-5p on MM Cell Proliferation, Migration, and Invasion
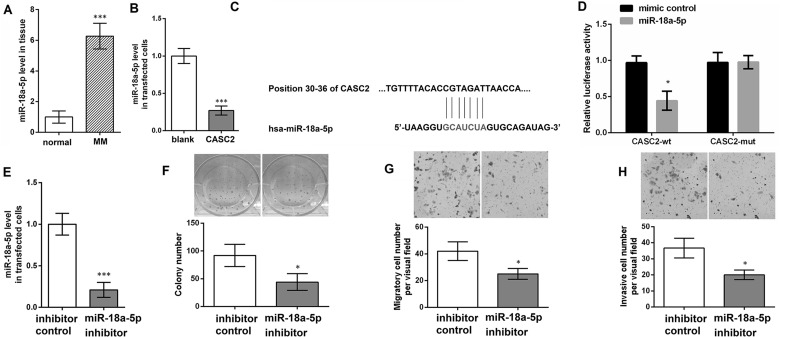
qRT-PCR analysis revealed that the miR-18a-5p level was significantly increased in MM tissues compared with normal skin tissues (p < 0.001) (Fig. 2A). In addition, compared with cells transfected with pcDNA3.1, the miR-18a-5p level was obviously reduced in cells with pc-CASC2 (p < 0.001) (Fig. 2B). To examine whether CACS2 is involved in MM by functioning as a competitive endogenous RNA (ceRNA) to miR-18a-5p, we predicted miRNA target sites and found that there was a binding site between CASC2 and miR-18a-5p (Fig. 2C). Moreover, the luciferase reporter assay confirmed that the luciferase activity in cells with CASC2-wt was significantly inhibited by miR-18a-5p mimic (Fig. 2D). To evaluate the effect of miR-18a-5p on MM, the miR-18a-5p level was altered in A375 cells. As shown in Figure 2E, the miR-18a-5p level was significantly inhibited in cells with miR-18a-5p inhibitor compared with cells with inhibitor control (p < 0.001). Then colony formation assay found that the number of colonies was obviously lower in cells with miR-18a-5p inhibitor than that in cells with inhibitor control (p < 0.05) (Fig. 2F). Moreover, the Transwell assay revealed that, compared with cells with inhibitor control, both migratory and invasive cell numbers were remarkably reduced in cells with miR-18a-5p inhibitor (p < 0.05) (Fig. 2G and H). The results indicated that CASC2 acts as a molecular sponge for miR-18a-5p and miR-18a-5p promoted MM cell proliferation, migration, and invasion.
Figure 2.
CASC2 inhibits miR-18a-5p level, and downregulating miR-18a-5p inhibits MM cell proliferation, migration, and invasion. (A) The miR-18a-5p level in MM tissues and normal skin tissues using qRT-PCR. (B) The miR-18a-5p level in A375 cells with pc-CASC2 or pcDNA3.1 (blank) using qRT-PCR. (C) The binding site between CASC2 and miR-18a-5p. (D) The luciferase reporter assay in cells with CASC2-wild type (wt) or CASC2-mutant (mut). (E) The miR-18a-5p level in A375 cells with miR-18a-5p inhibitor or inhibitor control using qRT-PCR. (F) The colony number of A375 cells with miR-18a-5p inhibitor and inhibitor control using the colony formation assay. (G) The migratory cell number of A375 cells with miR-18a-5p inhibitor or inhibitor control using Transwell analysis. (H) The invasive cell number of A375 cells with miR-18a-5p inhibitor or inhibitor control using Transwell analysis. *p < 0.05 and ***p < 0.001 compared with normal, blank, or inhibitor control.
Confirmation of the Targeting Effects of miR-18a-5p
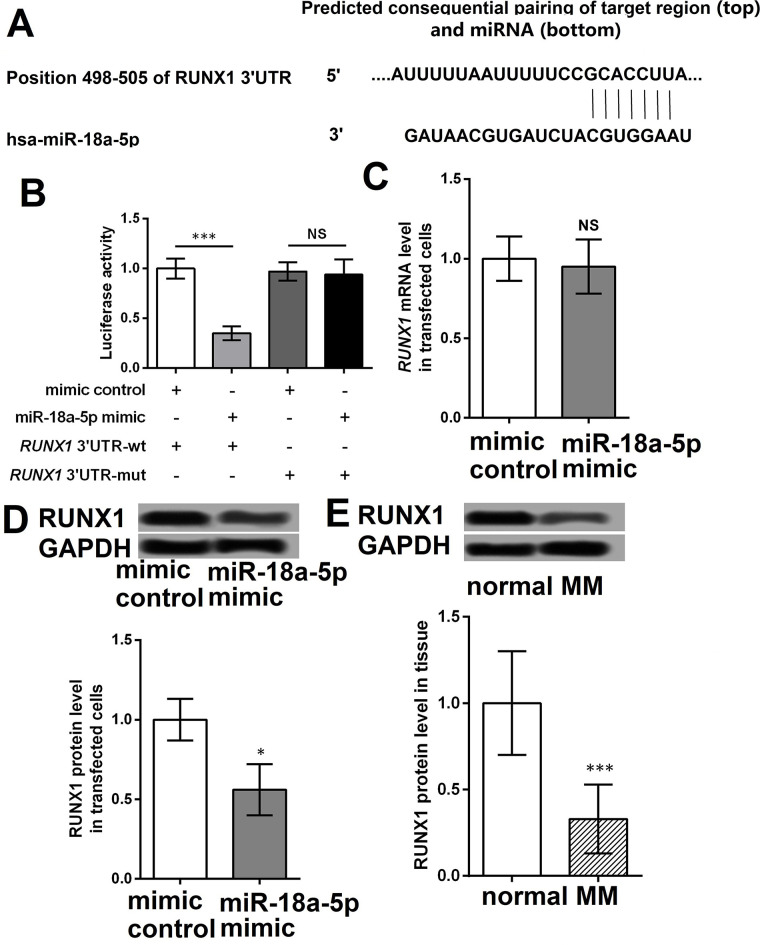
Sequence analysis revealed that RUNX1 was a potential target gene of miR-18a-5p (Fig. 3A). The luciferase reporter assay showed that miR-18a-5p mimic significantly inhibited the luciferase activity in cells with wt (p < 0.001) but not mutant RUNX1 compared with untreated cells (Fig. 3B). Compared with cells with mimic control, miR-139-5p mimic obviously inhibited the protein expression of RUNX1 but not the mRNA level (p < 0.05) (Fig. 3C and D). Furthermore, we found that the protein level of RUNX1 was significantly decreased in MM tissues compared with that in normal skin tissues (p < 0.001) (Fig. 3E).
Figure 3.
miR-18a-5p targets and inhibits RUNX1. (A) Prediction of the target gene of miR-18a-5p by sequence analysis. (B) The luciferase activity in cells with wt or mut RUNX1 when cotreated with miR-18a-5p mimic or mimic control using luciferase reporter assay. (C) The mRNA level of RUNX1 in A375 cells with miR-18a-5p mimic or mimic control using qRT-PCR. (D) The protein level of RUNX1 in A375 cells with miR-18a-5p mimic and mimic control using Western blotting. (E) The protein level of RUNX1 in MM tissues and normal skin tissues using Western blotting. *p < 0.05 and ***p < 0.001 compared with normal or mimic control.
Effect of RUNX1 on MM Cell Proliferation, Migration, and Invasion
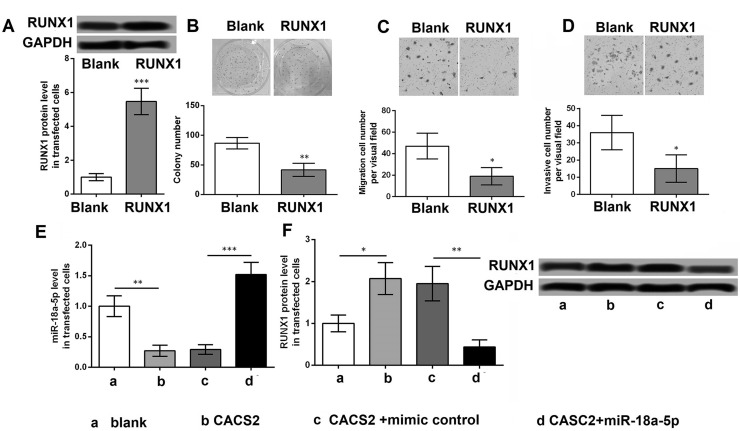
As shown in Figure 4A, the protein level of RUNX1 was significantly increased in cells with pc-RUNX1 compared with that in cells with pcDNA3.1 (p < 0.001). The results revealed that the number of colonies, as well as the migratory and invasive cell numbers, was obviously lower in cells with pc-RUNX1 compared with that in cells with pcDNA3.1 (p < 0.05) (Fig. 4B–D). Moreover, upregulated CASC2 significantly inhibited the miR-18a-5p level compared with cells with pcDNA3.1 (p < 0.01), while the cotreatment of pc-CASC2 and miR-18a-5p mimic remarkably increased the miR-18a-5p level compared with cells with cotreatment of pc-CASC2 and mimic control (p < 0.001) (Fig. 4E). Upregulated CASC2 significantly elevated the protein level of RUNX1 compared with cells with pcDNA3.1 (p < 0.05), while the cotreatment of pc-CASC2 and miR-18a-5p mimic remarkably decreased the protein level of RUNX1 compared with cells with cotreatment of pc-CASC2 and mimic control (p < 0.01) (Fig. 4F).
Figure 4.
RUNX1 inhibits MM cell proliferation, migration, and invasion. (A) The protein level of RUNX1 in A375 cells with pc-RUNX1 and pcDNA3.1 (blank) using Western blotting. (B) The colony number of A375 cells with pc-RUNX1 or pcDNA3.1 (blank) using colony formation assay. (C) The migratory cell number of A375 cells with pc-RUNX1 or pcDNA3.1 (blank) using Transwell analysis. (D) The invasive cell number of A375 cells with pc-RUNX1 or pcDNA3.1 using Transwell analysis. (E) The miR-18a-5p level in A375 cells with pc-CASC2, pcDNA3.1 (blank), pc-CASC2 + miR-18a-5p mimic, or pc-CASC2 + mimic control using qRT-PCR. (F) The protein level of RUNX1 in A375 cells with pc-CASC2, pcDNA3.1 (blank), pc-CASC2 + miR-18a-5p mimic, or pc-CASC2 + mimic control using Western blotting. *p < 0.05, **p < 0.01, and ***p < 0.001 compared with blank.
DISCUSSION
In the current study, our results showed that levels of CASC2 and RUNX1 were downregulated, while miR-18a-5p was overexpressed in MM tissues compared with that in normal skin tissues. Upregulated CASC2 could inhibit cell proliferation, migration, and invasion, which was similar to the role of downregulated miR-18a-5p in MM cells. In addition, RUNX1 was identified as a target gene of miR-18a-5p. CASC2 overexpression promoted the expression of RUNX1, while upregulated miR-18a-5p significantly reversed the effect of CASC2 on RUNX1 levels.
Previous studies have demonstrated the downregulation of CASC2 in several cancers22–24. The prognosis significance of CASC2 has been shown in non-small cell lung cancer (NSCLC)25. Consistently, this study also found low expression of CASC2 in MM patients. The study of Feng et al.23 reveals that CASC2 overexpression can inhibit cell proliferation by regulating miR-21 and its target gene PTEN in cervical cancer. Similarly, Li et al.24 and He et al.25 also suggest that cell proliferation is inhibited when CASC2 is overexpressed both in vitro and in vivo in NSCLC and gastric cancer. In addition, upregulated CASC2 inhibits cell proliferation, migration, and invasion and promotes cell apoptosis by inhibiting miR-21 levels in glioma26. Cao et al.9 also showed that downregulated CASC2 promoted cell proliferation and migration by regulating miR-21 in renal cell carcinoma. Consistent with these studies, the present study demonstrated similar inhibitor effects of CASC2 on MM cell proliferation, migration, and invasion. These results indicate that CASC2 has an antitumor effect in MM.
To further evaluate the mechanism of CASC2 in MM, miR-18a-5p levels were detected in MM. The regulatory relationship of CASC2 and miR-18a-5p has been demonstrated in colorectal cancer21. This study found that miR-18a-5p was overexpressed in MM tissues and CASC2 could inhibit the miR-18a-5p level, which are consistent with previous studies20,21. A recent meta-analysis reveals that miR-18a-5p may be considered as a promising biomarker for cancer screening. It has been reported that miR-18a expression is increased in patients with pancreatic cancer, indicating miR-18a as a tumor promoter27. However, Tsang et al. showed that miR-18a can inhibit tumor growth by suppressing K-Ras28. Thus, the effect of miR-18a is variable in different cancers, and the mechanism is also different. Song et al.29 reported that downregulated miR-18a in human glioblastoma cells can suppress cell proliferation, migration, and invasion by upregulating neogenin. In addition, Chen et al.30 suggested that miR-18a overexpression can promote cell proliferation and metastasis by targeting DICER1 in nasopharyngeal carcinoma cells. Consistently, this study showed that downregulated miR-18a-5p significantly inhibited cell proliferation, migration, and invasion in MM cells. Collectively, these results suggest that the antitumor effect of CASC2 may function by inhibiting miR-18a-5p in MM.
Furthermore, this study confirmed that RUNX1 was a target gene of miR-18a-5p, which is consistent with the predictions of a previous study20. RUNX1, also known as acute myeloid leukemia 1, plays a crucial role in the establishment of definitive hematopoiesis31. The role of RUNX1 in different cancers is controversial. Several studies have shown that RUNX1 is a tumor inhibitor in gastric cancer32 and esophageal cancer, while RUNX1 has a tumorigenic role in head and neck squamous cell cancer and oral squamous cell cancer33. It has been reported that miR-215 promotes malignant progression of gastric cancer by targeting RUNX132. However, knockdown of RUNX1 inhibits cell proliferation, migration, and invasion in epithelial ovarian cancer34. In this study, RUNX1 was downregulated in MM tissues and CASC2 could promote the level of RUNX1. These results indicate that RUNX1 may be a tumor inhibitor in MM.
The present study reveals that lncRNA CASC2 can inhibit cell proliferation, migration, and invasion by inhibiting miR-18a-5p and upregulating its target gene RUNX1, suggesting that lncRNA CASC2 may be a potential therapeutic target for MM. However, the mechanisms of CASC2 should be further investigated.
ACKNOWLEDGMENT
The authors declare no conflicts of interest.
Footnotes
The authors declare no conflicts of interest.
REFERENCES
- 1. Gambichler T, Scholl L, Stücker M, Bechara FG, Hoffmann K, Altmeyer P, Othlinghaus N. Clinical characteristics and survival data of melanoma patients with nevus cell aggregates within sentinel lymph nodes. Am J Clin Pathol. 2013;139(5):566–73. [DOI] [PubMed] [Google Scholar]
- 2. Linos E, Swetter SM, Cockburn MG, Colditz GA, Clarke CA. Increasing burden of melanoma in the United States. J Invest Dermatol. 2009;129(7):1666–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Shi X, Sun M, Liu H, Yao Y, Song Y. Long non-coding RNAs: A new frontier in the study of human diseases. Cancer Lett. 2013;339(2):159–66. [DOI] [PubMed] [Google Scholar]
- 4. Heward JA, Lindsay MA. Long non-coding RNAs in the regulation of the immune response. Trends Immunol. 2014;35(9):408–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wu GC, Pan HF, Leng RX, Wang DG, Li XP, Li XM, Ye DQ. Emerging role of long noncoding RNAs in autoimmune diseases. Autoimmun Rev. 2015;14(9):798–805. [DOI] [PubMed] [Google Scholar]
- 6. Congrains A, Kamide K, Oguro R, Yasuda O, Miyata K, Yamamoto E, Kawai T, Kusunoki H, Yamamoto H, Takeya Y. Genetic variants at the 9p21 locus contribute to atherosclerosis through modulation of ANRIL and CDKN2A/B. Atherosclerosis 2012;220(2):449–55. [DOI] [PubMed] [Google Scholar]
- 7. Johnson R. Long non-coding RNAs in Huntington’s disease neurodegeneration. Neurobiol Dis. 2012;46(2):245–54. [DOI] [PubMed] [Google Scholar]
- 8. Lawson J, Singh R, Hultner M, Ariyur K, Martin L. Long non-coding RNAs and cancer: A new frontier of translational research? Oncogene 2012;31(43):4577–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cao Y, Xu R, Xu X, Zhou Y, Cui L, He X. Downregulation of lncRNA CASC2 by microRNA-21 increases the proliferation and migration of renal cell carcinoma cells. Mol Med Rep. 2016;14(1):1019–25. [DOI] [PubMed] [Google Scholar]
- 10. Yiwei L, Liangfang S, Haiting Z, Qing L, Jun F, Yong G, Renjun P, Lei C. LncRNA CASC2 interacts with miR-181a to modulate glioma growth and resistance to TMZ through PTEN pathway. J Cell Biochem. 2017;118(7):1889–99. [DOI] [PubMed] [Google Scholar]
- 11. Pei Z, Xian D, Song Y, Lin F, Li F, Yan G, Wu R, Chen Y, Wei L, Hong Z. Down-regulation of lncRNA CASC2 promotes cell proliferation and metastasis of bladder cancer by activation of the Wnt/β-catenin signaling pathway. Oncotarget 2017;8(11):18145–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004;116(2):281–97. [DOI] [PubMed] [Google Scholar]
- 13. Kloosterman WP, Plasterk RH. The diverse functions of microRNAs in animal development and disease. Dev Cell 2006;11(4):441–50. [DOI] [PubMed] [Google Scholar]
- 14. Jovanovic M, Hengartner M. miRNAs and apoptosis: RNAs to die for. Oncogene 2006;25(46):6176–87. [DOI] [PubMed] [Google Scholar]
- 15. Michael MZ, O’Connor SM, van Holst Pellekaan NG, Young GP, James RJ. Reduced accumulation of specific microRNAs in colorectal neoplasia. Mol Cancer Res. 2003;1(12):882–91. [PubMed] [Google Scholar]
- 16. Murakami Y, Yasuda T, Saigo K, Urashima T, Toyoda H, Okanoue T, Shimotohno K. Comprehensive analysis of microRNA expression patterns in hepatocellular carcinoma and non-tumorous tissues. Oncogene 2006;25(17):2537–45. [DOI] [PubMed] [Google Scholar]
- 17. Calin GA, Liu C-G, Sevignani C, Ferracin M, Felli N, Dumitru CD, Shimizu M, Cimmino A, Zupo S, Dono M. MicroRNA profiling reveals distinct signatures in B cell chronic lymphocytic leukemias. Proc Natl Acad Sci USA 2004;101(32):11755–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Iorio MV, Ferracin M, Liu C-G, Veronese A, Spizzo R, Sabbioni S, Magri E, Pedriali M, Fabbri M, Campiglio M. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65(16):7065–70. [DOI] [PubMed] [Google Scholar]
- 19. Liz J, Esteller M. lncRNAs and microRNAs with a role in cancer development. Biochim Biophys Acta 2016;1859(1):169–76. [DOI] [PubMed] [Google Scholar]
- 20. Mishra RR, Kneitz S, Schartl M. Comparative analysis of melanoma deregulated miRNAs in the medaka and Xiphophorus pigment cell cancer models. Comp Biochem Physiol C Toxicol Pharmacol. 2014;163(18):64–76. [DOI] [PubMed] [Google Scholar]
- 21. Huang G, Wu X, Shi L, Xu X, Hua Z, Chen X. The long noncoding RNA CASC2 functions as a competing endogenous RNA by sponging miR-18a in colorectal cancer. Sci Rep. 2016;6:26524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Huang G, Ke ZP, Hu HB, Gu B. Co-expression network analysis of long noncoding RNAs (IncRNAs) and cancer genes reveals SFTA1P and CASC2 abnormalities in lung squamous cell carcinoma. Cancer Biol Ther. 2017;18(2):115–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Feng Y, Zou W, Hu C, Li G, Zhou S, He Y, Ma F, Deng C, Sun L. Modulation of CASC2/miR-21/PTEN pathway sensitizes cervical cancer to cisplatin. Arch Biochem Biophys. 2017;623/624:20–30. [DOI] [PubMed] [Google Scholar]
- 24. Li P, Xue WJ, Feng Y, Mao QS. Long non-coding RNA CASC2 suppresses the proliferation of gastric cancer cells by regulating the MAPK signaling pathway. Am J Transl Res. 2016;8(8):3522–9. [PMC free article] [PubMed] [Google Scholar]
- 25. He X, Liu Z, Su J, Yang J, Yin D, Han L, De W, Guo R. Low expression of long noncoding RNA CASC2 indicates a poor prognosis and regulates cell proliferation in non-small cell lung cancer. Tumor Biol. 2016;37(7):9503–10. [DOI] [PubMed] [Google Scholar]
- 26. Wang P, Liu YH, Yao YL, Li Z, Li ZQ, Ma J, Xue YX. Long non-coding RNA CASC2 suppresses malignancy in human gliomas by miR-21. Cell Signal. 2015;27(2):275–82. [DOI] [PubMed] [Google Scholar]
- 27. Liu JQ, Gao J, Ren Y, Wang XW, Wang WW, Lu H. Diagnostic value of plasma miR-21 in pancreatic cancer. World Chinese J Digestol. 2011;19(8):860–3. [Google Scholar]
- 28. Tsang WP, Kwok TT. The miR-18a* microRNA functions as a potential tumor suppressor by targeting on K-Ras. Carcinogenesis 2009;30(6):953–9. [DOI] [PubMed] [Google Scholar]
- 29. Song Y, Wang P, Zhao W, Yao Y, Liu X, Ma J, Xue Y, Liu Y. MiR-18a regulates the proliferation, migration and invasion of human glioblastoma cell by targeting neogenin. Exp Cell Res. 2014;324(1):54–64. [DOI] [PubMed] [Google Scholar]
- 30. Chen X, Wang J, Cheng L, Lu MP. miR-18a downregulates DICER1 and promotes proliferation and metastasis of nasopharyngeal carcinoma. Int J Clin Exp Med. 2014;7(4):847–55. [PMC free article] [PubMed] [Google Scholar]
- 31. De Braekeleer E, Douet-Guilbert N, Morel F, Le Bris MJ, Férec C, De Braekeleer M. RUNX1 translocations and fusion genes in malignant hemopathies. Future Oncol. 2011;7(1):77–91. [DOI] [PubMed] [Google Scholar]
- 32. Na L, Zhang QY, Zou JL, Li ZW, Tian TT, Dong B, Liu XJ, Ge S, Yan Z, Jing G. miR-215 promotes malignant progression of gastric cancer by targeting RUNX1. Oncotarget 2016;7(4):4817–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Schuijers J, Clevers H. Adult mammalian stem cells: The role of Wnt, Lgr5 and R-spondins. EMBO J. 2012;31(12):2685–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Keita M, Bachvarova M, Morin C, Plante M, Gregoire J, Renaud MC, Sebastianelli A, Xuan BT, Bachvarov D. The RUNX1 transcription factor is expressed in serous epithelial ovarian carcinoma and contributes to cell proliferation, migration and invasion. Cell Cycle 2013;12(6):972–86. [DOI] [PMC free article] [PubMed] [Google Scholar]