Abstract
miR-1284 has been reported to inhibit tumor growth in some human cancers, including lung cancer, ovarian cancer, and gastric cancer. Whether it regulates breast cancer progression remains elusive. In this study, we found that miR-1284 was downregulated in breast cancer tissues and cell lines compared to normal control cells. Moreover, we showed that overexpression of miR-1284 significantly inhibited the proliferation, migration, and invasion of breast cancer cells while promoting apoptosis. In terms of mechanism, we found that transcription factor ZIC2 was a target of miR-1284 in breast cancer cells. Through the luciferase reporter assay, we demonstrated their direct interaction. RT-qPCR and Western blot also indicated that miR-1284 overexpression inhibited the protein levels of ZIC2 in breast cancer cells. Moreover, we found that ZIC2 knockdown inhibited the proliferation, migration, and invasion of breast cancer cells, whereas restoration of ZIC2 reversed the effects of miR-1284 on breast cancer cells. Taken together, our findings demonstrated that miR-1284 suppressed the proliferation, migration, and invasion of breast cancer cells via targeting ZIC2, which provided a new insight on the development of therapeutic targets for breast cancer treatment.
Key words: Breast cancer, miR-1284, Proliferation, Migration, ZIC2
INTRODUCTION
Breast cancer is the most prevalent and life-threatening cancer diagnosed in women, and remains a leading cause of cancer-related death among women worldwide1. The incidence of breast cancer is increasing year by year. Tumor metastasis is often developed after initial diagnosis, which greatly contributes to the poor outcomes of patients with breast cancer2. The factors that induce breast cancer development still need to be investigated, and the pathological mechanisms regulating breast cancer progression remain largely unknown. Many proteins, such as HER23, have been shown to be linked with breast cancer. In order to look for novel diagnostic and prognostic biomarkers and therapeutic targets, it is crucial to identify the key regulators in breast cancer.
MicroRNAs (miRNAs) represent a class of endogenous noncoding RNAs and repress gene expression posttranscriptionally by interacting with complementary sequence of the 3′-UTR of target mRNAs4,5. Accumulating evidence indicates that numerous miRNAs function as oncogenes6 or tumor suppressors7 via regulating cell cycle, proliferation, apoptosis, invasion, and migration8,9. miR-1284 is located at chromosome region 3p13 and has been reported to suppress tumor progression in various human cancers. For instance, Li et al. reported that miRNA-1284 inhibits cell growth and induces apoptosis of lung cancer cells10. Pan et al. indicated that miRNA-1284 inhibits cell viability and induces apoptosis of ovarian cancer cells11. Huang et al. showed that miRNA-1284 inhibits proliferation and induces apoptosis in SGC-7901 human gastric cancer cells12. However, whether miR-1284 regulates breast cancer progression remains to be investigated.
In this study, we found that miR-1284 was significantly downregulated in breast cancer cell lines and human samples. Moreover, we found that overexpression of miR-1284 remarkably inhibited the proliferation, migration, and invasion of breast cancer cells while promoting cellular apoptosis. Mechanistically, we identified ZIC2 as a target gene of miR-1284. We found that overexpression of miR-1284 significantly inhibited the protein levels of ZIC2 in breast cancer cells, and there was an inverse correlation between miR-1284 expression and ZIC2 mRNA levels in breast cancer tissues. Furthermore, we demonstrated that ZIC2 knockdown inhibited the proliferation, migration, and invasion of breast cancer cells, whereas restoration of ZIC2 partially rescued the effects of miR-1284. Taken together, our study indicated that miR-1284 serves as a tumor suppressor by targeting ZIC2 in breast cancer.
MATERIALS AND METHODS
Patient Samples
We obtained breast tumor tissues and the paired normal adjacent tissues from patients without preoperative chemotherapy, hormone therapy, or radiotherapy who had undergone tumor resection at Renmin Hospital of Wuhan University between 2012 and 2016. All patients provided written informed consent. Renmin Hospital of Wuhan University clinical research ethics committee approved the study protocol.
Cell Lines and Cell Culture
The MCF-10A, MDA-MB-231, Hs 578T, and BT549 breast cancer cell lines were purchased from Shanghai Cell Bank (Shanghai, P.R. China) and were cultured in RPMI-1640 medium (Gibco, Grand Island, NY, USA) supplemented with 10% fetal bovine serum (FBS; Sigma-Aldrich, St. Louis, MO, USA) and 100 U/ml penicillin and streptomycin in a humidified atmosphere of 5% CO2 at 37°C.
Reverse Transcription Quantitative Polymerase Chain Reaction (RT-qPCR)
Total RNA was extracted from the breast cancer tissues and cell lines using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) following the manufacturer’s protocol. A total of 2 μg of RNA was used for RT-qPCR with the Qiagen OneStep RT-PCR Kit (Qiagen Benelux BV, Venlo, The Netherlands) in the following conditions: 37°C for 30 min, 85°C for 5 s, and kept at 4°C until use. TaqMan miRNA assays (Applied Biosystems; Thermo Fisher Scientific, Inc.) were performed to quantify the relative expression of miR-1284. U6 was used as the normalization control.
shRNA-Mediated Interference
MDA-MB-231 or BT549 cells were seeded in six-well plates at a density of 5 × 106 cells/well. The cells in each well were transfected with a solution of 3 μg of pcDNA3.1-ZIC2 (GenePharma Co., Ltd., Shanghai, P.R. China) or 20 nM RNA (miR-1284 mimics or negative control) using Lipofectamine 3000 (Invitrogen; Thermo Fisher Scientific) according to the manufacturer’s protocol. Subsequent experiments were conducted 24 h after transfection.
Cellular Proliferation Assays
Cells were seeded into 96-well plates at a density of 1 × 103 cells/well. After 24, 48, and 72 h of incubation at 37°C, cellular viability was evaluated using a Cell Counting Kit-8 (CCK-8) assay (Dojindo Molecular Technologies, Inc., Kumamoto, Japan), according to the manufacturer’s protocol. The absorbance was measured at a wavelength of 450 nm using a Multiskan™ GO Microplate Spectrophotometer (Thermo Fisher Scientific, Inc.).
A colony formation assay was also performed. Cells were seeded into six-well plates and cultured at 37°C with 5% CO2 for 14 days. Colonies were fixed with methanol at room temperature for 20 min and stained with 0.1% crystal violet (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany). The total number of visible colonies was determined under an optical microscope (Olympus Corporation, Tokyo, Japan). All experiments were repeated three times.
Cellular Migration and Invasion Assays
The cell migration assay was performed in vitro using 24-well Transwell chambers. Cells (2 × 104) were seeded in the top chambers, and the bottom chambers were filled with RPMI-1640 medium (600 μl) containing 10% FBS to stimulate migration. After 18 h of incubation, the cells were stained with 0.1% crystal violet. The cells that had migrated through the ostioles to the reverse side were counted under a microscope in five predetermined fields at 200× magnification. Each assay was performed in triplicate.
The cell invasion assay was performed using 24-well Transwell chambers. Cells (1 × 105) were seeded in the Matrigel-coated (BD Biosciences, San Jose, CA, USA) top chambers. The bottom chambers were filled with RPMI-1640 medium (600 μl) containing 10% FBS to stimulate invasion. After 16 h of incubation, the cells were stained with 0.1% crystal violet. The cells that had invaded through the Matrigel to the reverse side were counted under a microscope in five predetermined fields at 200× magnifications. Each assay was performed in triplicate.
Luciferase Assay
Fragments of the ZIC2 mRNA 3′-UTRs containing the putative or mutated miRNA binding sites for miR-1284 were cloned into the GV306 luciferase reporter vector (GeneChem, Shanghai, P.R. China). The constructs were then cotransfected with miR-1284 mimic or negative control oligos into cells according to the manufacturer’s instructions. Luciferase activity was measured 48 h after transfection using the Dual-Luciferase® Reporter Assay System (Promega, Madison, WI, USA) according to the manufacturer’s protocol.
Statistical Analysis
Statistical Package for Social Science (SPSS; v. 22) was used to investigate the correlation between miR-1284 relative expression levels and ZIC2 relative expression levels in clinical breast cancer tissues. All data are presented as the mean ± SD; groups were compared using two-tailed Student’s t-test. Values of p < 0.05 were considered significant.
RESULTS
miR-1284 Was Downregulated in Breast Cancer Tissues
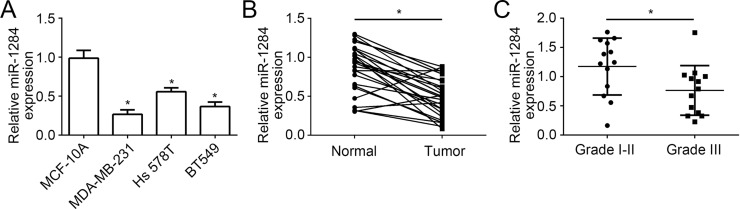
To analyze the function of miR-1284 in breast cancer, we first measured miR-1284 levels in breast cancer cell lines and normal breast epithelial cells by RT-qPCR. The results indicated that miR-1284 expression was significantly downregulated in breast cancer cell lines, including MDA-MB-231, Hs 578T, and BT549, compared to the normal breast epithelial cell line MCF-10A (Fig. 1A). In addition, we analyzed its expression in 26 pairs of breast cancer tissues and adjacent normal tissues by RT-qPCR. We found that miR-1284 was also downregulated in breast cancer tissues compared with adjacent normal tissues (Fig. 1B). Moreover, miR-1284 expression was significantly lower in advanced breast cancer tissues (n = 13) than that in early breast cancer tissues (n = 13) (Fig. 1C). According to these data, we demonstrated that miR-1284 was downregulated in breast cancer cells, which suggested that miR-1284 may be involved in breast cancer progression.
Figure 1.
MicroRNA-1284 (miR-1284) was downregulated in breast cancer tissues. (A) Reverse transcription quantitative polymerase chain reaction (RT-qPCR) was used to analyze the expression of miR-1284 in breast cancer cell lines (MDA-MB-231, Hs 578T, and BT549 cells) and MCF-10A. (B) Relative expression levels of miR-1284 in 25 pairs of breast cancer tissues and adjacent normal tissues were measured by RT-qPCR. (C) miR-1284 levels were determined in advanced breast cancer (stage III) tissues and early breast cancer (stages I–II) tissues by RT-qPCR. *p < 0.05.
miR-1284 Overexpression Inhibited Cell Proliferation, Migration, and Invasion
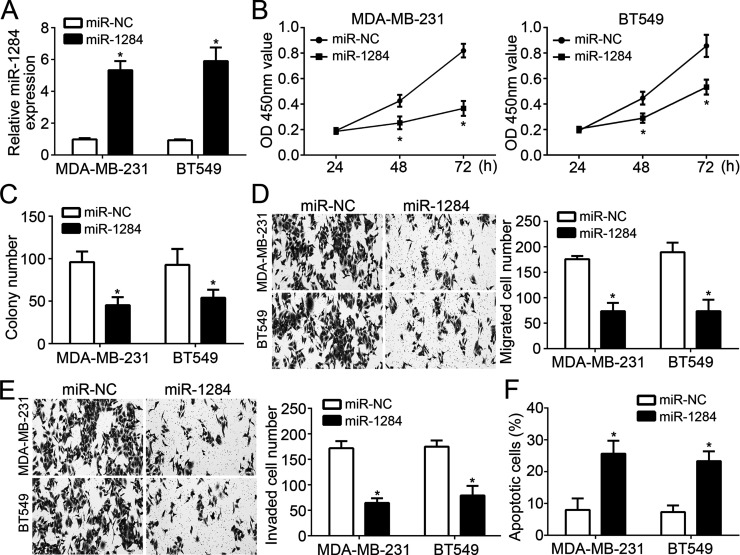
Then we chose MDA-MB-231 and BT549 cells to explore the functions of miR-1284 in breast cancer. We overexpressed miR-1284 in MDA-MB-231 and BT549 cells by transfection with miR-1284 mimics or negative controls (miR-NC). RT-qPCR analysis indicated that miR-1284 was effectively upregulated after transfection (Fig. 2A). Then we performed CCK-8 and colony formation assays to evaluate cell proliferation. The results indicated that overexpression of miR-1284 significantly inhibited the proliferation and colony formation of MDA-MB-231 and BT549 cells (Fig. 2B and C). Furthermore, Transwell assay was used to determine the effects of miR-1284 on cell migration and invasion. We found that overexpression of miR-1284 significantly inhibited the numbers of migrated and invaded cells (Fig. 2D and E). Notably, we found that miR-1284 overexpression remarkably increased the cellular apoptosis in MDA-MB-231 and BT549 cells (Fig. 2F).
Figure 2.
miR-1284 overexpression inhibited cell proliferation, migration, and invasion. (A) RT-qPCR was used to check the expression of miR-1284 in MDA-MB-231 and BT549 cells transduced with miR-1284 mimics or controls. (B) Cell counting kit-8 (CCK-8) assays for cellular proliferation analysis. (C) Colony formation assays indicated that overexpression of miR-1284 significantly inhibited the colony number. (D, E) Transwell assays were utilized to evaluate the migration and invasion of MDA-MB-231 and BT549 cells transfected with miR-1284 mimics or controls. (F) The effect of miR-1284 on MDA-MB-231 and BT549 cell apoptosis was determined by annexin V/PI staining. *p < 0.05.
ZIC2 Was a Target of miR-1284
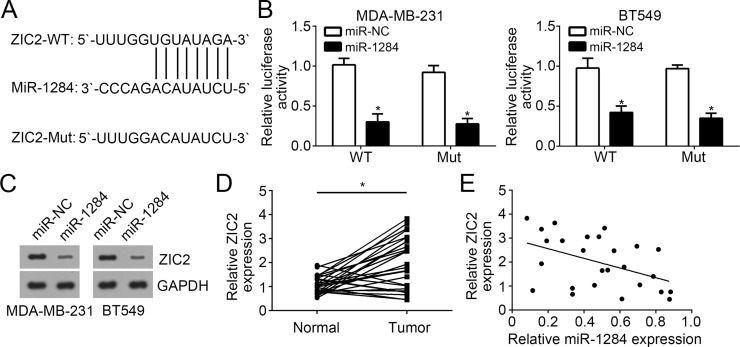
We utilized miRanda (http://www.microrna.org/microrna) to analyze potential target genes of miR-1284. Among all candidates, transcription factor ZIC2 was selected for further investigation because it has been reported to serve as an oncogene in other human cancers13. The ZIC2 3′-UTR region containing a putative wild-type (WT) or mutant miR-1284 binding site was cloned into luciferase reporter vector (Fig. 3A). MDA-MB-231 and BT549 cells were transfected with miR-1284 mimics as well as WT or mutant luciferase reporter plasmid. Results from luciferase reporter assays indicated that overexpression of miR-1284 significantly inhibited the luciferase activity of WT ZIC2 3′-UTR (Fig. 3B). Moreover, we found that overexpression of miR-1284 repressed the protein levels of ZIC2 in MDA-MB-231 and BT549 cells (Fig. 3C). We also detected the expression of ZIC2 in breast cancer and found that ZIC2 expression was upregulated in breast cancer tissues compared with matched normal tissues (Fig. 3D). Additionally, RT-qPCR analysis showed that miR-1284 expression was negatively correlated with that of ZIC2 in breast cancer tissues (Fig. 3E).
Figure 3.
ZIC2 was a target of miR-1284. (A) Base complementarity between miR-1284 and the 3′-UTR of ZIC2 mRNA. (B) Luciferase reporter assays showed that miR-1284 overexpression repressed the luciferase activity in MDA-MB-231 and BT549 cells transduced with wild-type (WT) 3′-UTR of ZIC2 mRNA. (C) Western blot was used to analyze the protein level of ZIC2 in MDA-MB-231 and BT549 cells transduced with miR-1284 mimics or controls. (D) Relative expression of ZIC2 in 26 pairs of breast cancer tissues and adjacent normal tissues. (E) ZIC2 was negatively correlated with miR-1284 at mRNA level in breast cancer tissues. *p < 0.05.
ZIC2 Knockdown Inhibited the Proliferation, Migration, and Invasion of Breast Cancer Cells
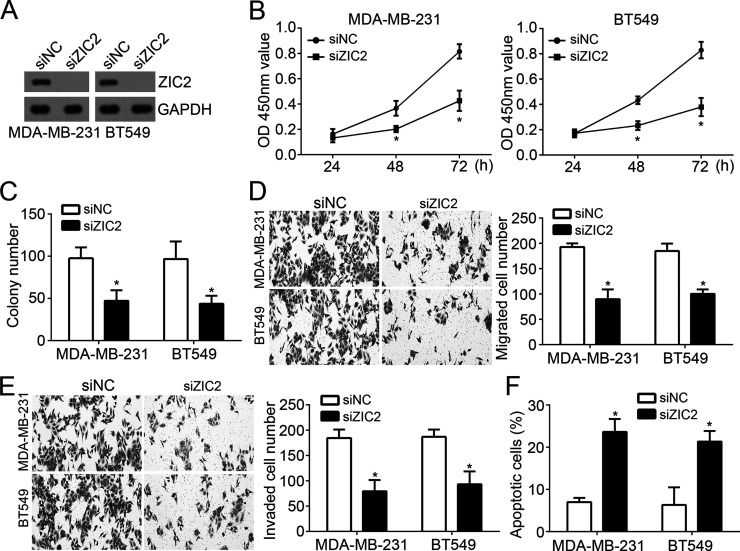
To identify whether ZIC2 is involved in miR-1284-mediated inhibition on breast cancer progression, we knocked down ZIC2 by specific siRNA in MDA-MB-231 and BT549 cells. Western blot analysis indicated that ZIC2 was significantly downregulated in MDA-MB-231 and BT549 cells transfected with siZIC2 (Fig. 4A). As CCK-8 and colony formation assays showed, ZIC2 depletion significantly suppressed the proliferation of breast cancer cells (Fig. 4B and C). Furthermore, knockdown of ZIC2 also inhibited the migration and invasion of MDA-MB-231 and BT549 cells (Fig. 4D and E). In addition, we found that ZIC2 knockdown remarkably increased the cellular apoptosis in MDA-MB-231 and BT549 cells (Fig. 2F). In summary, ZIC2 knockdown suppressed the breast cancer progression by regulating cell proliferation, migration, invasion, and apoptosis.
Figure 4.
ZIC2 knockdown inhibited the proliferation, migration, and invasion of breast cancer cells. (A) Western blot was used to assess the protein levels of ZIC2 in MDA-MB-231 and BT549 cells transduced with siZIC2 or control. (B, C) CCK-8 and colony formation assays were used to measure the proliferation of MDA-MB-231 and BT549 cells transduced with siZIC2 or control. (D, E) The migration and invasion of MDA-MB-231 and BT549 cells were determined by Transwell assays. (F) The effect of ZIC2 on MDA-MB-231 and BT549 cell apoptosis was determined by annexin V/PI staining. *p < 0.05.
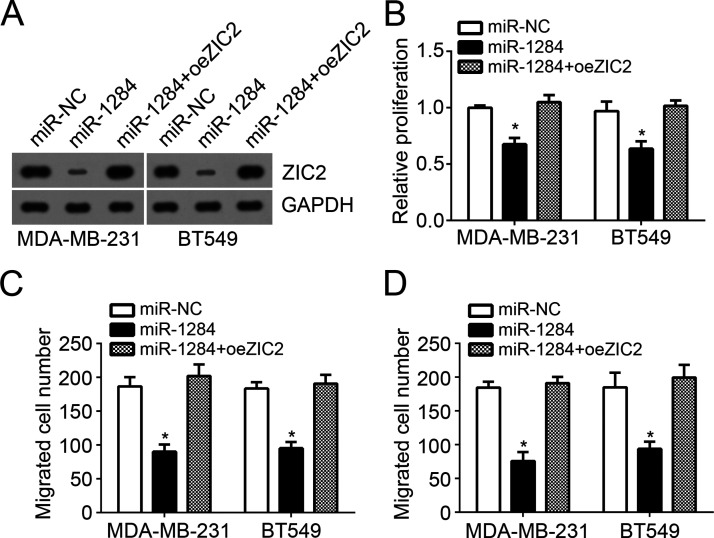
ZIC2 Reversed the Anticancer Function of miR-1284 in Breast Cancer Cells
To further demonstrate ZIC2 as a functional target gene of miR-1284, we performed rescue experiments with MDA-MB-231 and BT549 cells. We restored ZIC2 expression in miR-1284-overexpressing MDA-MB-231 and BT549 cells by transfection with ZIC2 ectopic expressing vector. Western blot assays indicated that ZIC2 was effectively overexpressed (Fig. 5A). Then CCK-8 and Transwell assays were performed. The results indicated that restoration of ZIC2 promoted the proliferation, migration, and invasion of miR-1284-overexpressing MDA-MB-231 and BT549 cells (Fig. 5B–D). Taken together, our results reinforced the idea that miR-1284 suppressed breast cancer progression by regulating ZIC2 expression.
Figure 5.
ZIC2 reversed the anticancer function of miR-1284 in breast cancer cells. (A) Western blot assay was used to determine the protein levels of ZIC2 in MDA-MB-231 and BT549 cells. (B) CCK-8 assays were utilized to examine cell proliferation. (C, D) Transwell assays were used to determine the migration and invasion of MDA-MB-231 and BT549 cells. *p < 0.05.
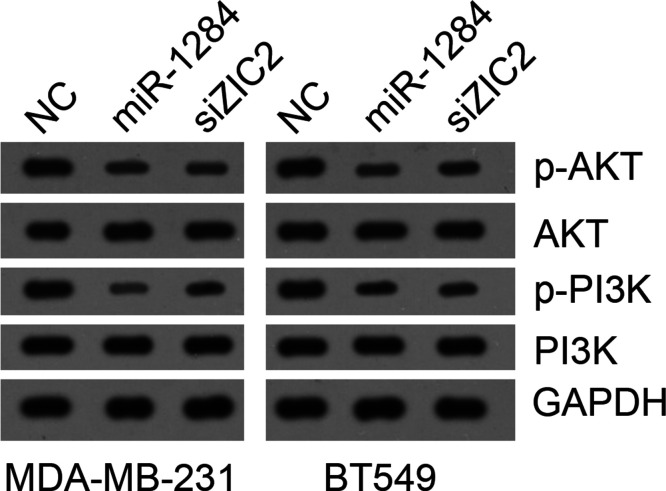
A previous study14 implied a correlation between ZIC2 and the PI3K/AKT pathway in bladder cancer. Thus, we sought to determine whether miR-1284 regulates the PI3K/AKT pathway by targeting ZIC2 in breast cancer. We found that miR-1284 overexpression or ZIC2 knockdown significantly suppressed the activation of the PI3K/AKT pathway in MDA-MB-231 and BT549 cells (Fig. 6), suggesting that miR-1284 might inhibit PI3K/AKT signaling via targeting ZIC2 to impede breast cancer progression.
Figure 6.
miR-1284 suppressed PI3K/AKT pathway by targeting ZIC2 in breast cancer cells. Western blot was used to measure the activation of the PI3K/AKT pathway.
DISCUSSION
Tumor development and progression are usually regulated by a complicated network containing expression changes of many genes. The gene expression patterns in specific types of cancers are substantially different15. Therefore, identifying essential regulators in cancer is very important for the development of therapeutic targets. In this study, we found that miR-1284 was downregulated in breast cancer tissues and cell lines. In addition, overexpression of miR-1284 significantly inhibited the proliferation, migration, and invasion of breast cancer cells. We demonstrated that ZIC2 was a direct target of miR-1284. We also showed that ZIC2 was upregulated in breast cancer tissues, and knockdown of ZIC2 suppressed breast cancer cell proliferation, migration, and invasion. Furthermore, we proved that miR-1284 inhibited breast cancer progression through targeting ZIC2 by rescue experiments.
Several studies have demonstrated that miR-1284 serves as a tumor suppressor in a diversity of human cancers, including lung cancer, ovarian cancer, and gastric cancer10–12. However, the role of miR-1284 in breast cancer is largely unclear. Our data indicated that miR-1284 was downregulated in breast cancer tissues and suppressed the proliferation of cancer cells. Tumor metastasis is a critical factor of malignance16. Inhibition of metastasis will contribute to the good outcomes of breast cancer patients. In our study, through Transwell assays, we demonstrated that overexpression of miR1284 significantly inhibited the migration and invasion of breast cancer cells, which suggested that miR-1284 might be a promising target for breast cancer intervention. However, the underlying molecular mechanism of miR-1284 needs to be investigated.
The study about the relationship between ZIC2 and breast cancer development is very limited. Sakuma et al. showed that ZIC2 expression was significantly upregulated in oral squamous cell carcinoma and may act as a prognostic marker17. Chan et al. reported that Zic2 synergistically enhances Hedgehog signaling through nuclear retention of Gli1 in cervical cancer cells18. Marchini et al. indicated that ZIC2 was overexpressed in epithelial ovarian cancer tissues and acted as an oncogene19. Another study demonstrated that ZIC2 promotes nasopharyngeal carcinoma cell proliferation and invasion20. A recent study also reported that Zic2 promotes tumor growth and metastasis via PAK4 in hepatocellular carcinoma21. In addition, Wang et al. reported that ZIC2 was involved in bladder cancer development22. These researches all indicate ZIC2 as an oncogene. However, whether ZIC2 promotes tumor occurrence in breast cancer remains elusive. Our study indicated that ZIC2 was a target gene of miR-1284 in breast cancer. Moreover, our data demonstrated that ZIC2 was upregulated in breast cancer tissues compared to adjacent normal tissues. In addition, knockdown of ZIC2 also suppressed the proliferation, migration, and invasion of breast cancer cells. Finally, through rescue experiments, we proved that restoration of ZIC2 could rescue the effects of miR-1284 on breast cancer cells, which suggested that miR-1284 repressed breast cancer progression by regulating ZIC2 expression. Aberrant activation of the PI3K/AKT signaling pathway is usually observed in many tumors, including lung carcinoma23, colon cancer24, bladder cancer25, pancreatic cancer26, and breast cancer27. Furthermore, a previous study14 revealed a correlation between ZIC2 and the PI3K/AKT pathway in bladder cancer. Therefore, we investigated the potential relationship between the miR-1284/ZIC2 axis and the PI3K/AKT pathway in breast cancer. We showed that miR-1284 suppressed PI3K/AKT activation via targeting ZIC2. Thus, our study suggested that miR-1284 repressed breast cancer progression by impeding activation of the PI3K/AKT pathway at least partially.
In conclusion, our findings demonstrated that miR-1284 suppressed the proliferation, migration, and invasion of breast cancer cells through a ZIC2-dependent manner, which implied that the miR-1284/ZIC2 signal might be a promising therapeutic target for breast cancer treatment.
ACKNOWLEDGMENT
The authors declare no conflicts of interest.
Footnotes
The authors declare no conflicts of interest.
REFERENCES
- 1. Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer 2015;136(5):E359–86. [DOI] [PubMed] [Google Scholar]
- 2. Weigelt B, Peterse JL, van’t Veer LJ. Breast cancer metastasis: Markers and models. Nat Rev Cancer 2005;5(8):591–602. [DOI] [PubMed] [Google Scholar]
- 3. Pegram MD, Konecny G, Slamon DJ. The molecular and cellular biology of HER2/neu gene amplification/overexpression and the clinical development of herceptin (trastuzumab) therapy for breast cancer. Cancer Treat Res. 2000;103:57–75. [DOI] [PubMed] [Google Scholar]
- 4. Plaisance-Bonstaff K, Renne R. Viral miRNAs. Methods Mol Biol. 2011;721:43–66. [DOI] [PubMed] [Google Scholar]
- 5. Shukla GC, Singh J, Barik S. MicroRNAs: Processing, maturation, target recognition and regulatory functions. Mol Cell Pharmacol. 2011;3(3):83–92. [PMC free article] [PubMed] [Google Scholar]
- 6. Guo J, Xiao Z, Yu X, Cao R. miR-20b promotes cellular proliferation and migration by directly regulating phosphatase and tensin homolog in prostate cancer. Oncol Lett. 2017;14(6):6895–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Tan H, Zhu G, She L, Wei M, Wang Y, Pi L, Chen C, Zhang D, Tan P, Chen J, Huang D, Tian Y, Liu Y, Zhang X. MiR-98 inhibits malignant progression via targeting MTDH in squamous cell carcinoma of the head and neck. Am J Cancer Res. 2017;7(12):2554–65. [PMC free article] [PubMed] [Google Scholar]
- 8. Zhang Y, Wei Y, Li X, Liang X, Wang L, Song J, Zhang X, Zhang C, Niu J, Zhang P, Ren Z, Tang B. microRNA-874 suppresses tumor proliferation and metastasis in hepatocellular carcinoma by targeting the DOR/EGFR/ERK pathway. Cell Death Dis. 2018;9(2):130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chi Y, Ding F, Zhang W, Du L. microRNA-503 suppresses the migration, proliferation and colony formation of prostate cancer cells by targeting tumor protein D52 like 2. Exp Ther Med. 2018;15(1):473–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Li J, Jin H, Yu H, Wang B, Tang J. miRNA1284 inhibits cell growth and induces apoptosis of lung cancer cells. Mol Med Rep. 2017;16(3):3049–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Pan C, Wang D, Zhang Y, Yu W. MicroRNA-1284 inhibits cell viability and induces apoptosis of ovarian cancer cell line OVCAR3. Oncol Res. 2016;24(6):429–35. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 12. Huang M, Wu L, Luo S, Qin H, Yang Y, Chen J, Li Z, Qin Y. MicroRNA-1284 inhibits proliferation and induces apoptosis in SGC-7901 human gastric cancer cells. Biotechnol Lett. 2017;39(1):33–8. [DOI] [PubMed] [Google Scholar]
- 13. Lu SX, Zhang CZ, Luo RZ, Wang CH, Liu LL, Fu J, Zhang L, Wang H, Xie D, Yun JP. Zic2 promotes tumor growth and metastasis via PAK4 in hepatocellular carcinoma. Cancer Lett. 2017;402:71–80. [DOI] [PubMed] [Google Scholar]
- 14. Wang J, Ma W, Liu Y. Long non-coding RNA HULC promotes bladder cancer cells proliferation but inhibits apoptosis via regulation of ZIC2 and PI3K/AKT signaling pathway. Cancer Biomark. 2017;20(4):425–34. [DOI] [PubMed] [Google Scholar]
- 15. Li J, Lai Y, Ma J, Liu Y, Bi J, Zhang L, Chen L, Yao C, Lv W, Chang G, Wang S, Ouyang M, Wang W. miR-17-5p suppresses cell proliferation and invasion by targeting ETV1 in triple-negative breast cancer. BMC Cancer 2017;17(1):745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Liang YJ, Wu H, Lei R, Chong RA, Wei Y, Lu X, Tagkopoulos I, Kung SY, Yang QF, Hu GH, Kang YB. Transcriptional network analysis identifies BACH1 as a master regulator of breast cancer bone metastasis. J Biol Chem. 2012;287(40):33533–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sakuma K, Kasamatsu A, Yamatoji M, Yamano Y, Fushimi K, Iyoda M, Ogoshi K, Shinozuka K, Ogawara K, Shiiba M, Tanzawa H, Uzawa K. Expression status of Zic family member 2 as a prognostic marker for oral squamous cell carcinoma. J Cancer Res Clin Oncol. 2010;136(4):553–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chan DW, Liu VWS, Leung LY, Yao KM, Chan KKL, Cheung ANY, Ngan HYS. Zic2 synergistically enhances Hedgehog signalling through nuclear retention of Gli1 in cervical cancer cells. J Pathol. 2011;225(4):525–34. [DOI] [PubMed] [Google Scholar]
- 19. Marchini S, Poynor E, Barakat RR, Clivio L, Cinquini M, Fruscio R, Porcu L, Bussani C, D’Incalci M, Erba E, Romano M, Cattoretti G, Katsaros D, Koff A, Luzzatto L. The zinc finger gene ZIC2 has features of an oncogene and its overexpression correlates strongly with the clinical course of epithelial ovarian cancer. Clin Cancer Res. 2012;18(16):4313–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Shen ZH, Zhao KM, Du T. HOXA10 promotes nasopharyngeal carcinoma cell proliferation and invasion via inducing the expression of ZIC2. Eur Rev Med Pharmacol Sci. 2017;21(5):945–52. [PubMed] [Google Scholar]
- 21. Lu SX, Zhang CZY, Luo RZ, Wang CH, Liu LL, Fu J, Zhang LJ, Wang HM, Xie D, Yun JP. Zic2 promotes tumor growth and metastasis via PAK4 in hepatocellular carcinoma. Cancer Lett. 2017;402:71–80. [DOI] [PubMed] [Google Scholar]
- 22. Wang JT, Ma WM, Liu YD. Long non-coding RNA HULC promotes bladder cancer cells proliferation but inhibits apoptosis via regulation of ZIC2 and PI3K/AKT signaling pathway. Cancer Biomark. 2017;20(4):425–34. [DOI] [PubMed] [Google Scholar]
- 23. Song G, Lu H, Chen F, Wang Y, Fan W, Shao W, Lu H, Lin B. Tetrahydrocurcumin-induced autophagy via suppression of PI3K/Akt/mTOR in nonsmall cell lung carcinoma cells. Mol Med Rep. 2018;17(4):5964–9. [DOI] [PubMed] [Google Scholar]
- 24. Qin Y, Huo Z, Song X, Chen X, Tian X, Wang X. mir-106a regulates cell proliferation and apoptosis of colon cancer cells through targeting the PTEN/PI3K/AKT signaling pathway. Oncol Lett. 2018;15(3):3197–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Li Z, Zhang TB, Jia DH, Sun WQ, Wang CL, Gu AZ, Yang XM. Genipin inhibits the growth of human bladder cancer cells via inactivation of PI3K/Akt signaling. Oncol Lett. 2018;15(2):2619–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nath S, Mandal C, Chatterjee U, Mandal C. Association of cytosolic sialidase Neu2 with plasma membrane enhances Fas-mediated apoptosis by impairing PI3K-Akt/mTOR-mediated pathway in pancreatic cancer cells. Cell Death Dis. 2018;9(2):210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yang Y, Gao M, Lin Z, Chen L, Jin Y, Zhu G, Wang Y, Jin T. DEK promoted EMT and angiogenesis through regulating PI3K/AKT/mTOR pathway in triple-negative breast cancer. Oncotarget 2017;8(58):98708–22. [DOI] [PMC free article] [PubMed] [Google Scholar]