Abstract
Aberrant expression of microRNA-152 (miR-152) is frequently observed in human cancers including ovarian cancer, breast cancer, prostate cancer, and gastric cancer. However, its expression and functional role in cervical cancer (CC) are poorly understood. Also, the association between miR-152 and Krüppel-like factor 5 (KLF5) expression in CC remains unclear. In this study, analyzing the expression of miR-152 by quantitative real-time PCR (qRT-PCR) revealed it was sharply reduced in CC tissues and cell lines. In addition, the negative correlation of miR-152 expression and KLF5 expression was observed. The dual-luciferase reporter assay validated that KLF5 was a target of miR-152. In vitro functional assays revealed that miR-152 could inhibit cell proliferation and cell cycle progression through regulating the expression of KLF5. Taken together, our study suggested that miR-152 functions as a tumor suppressor in CC, and the miR-152/KLF5 axis may provide novel therapeutic targets for CC treatment.
Key words: miR-152, Cervical cancer (CC), Krüppel-like factor 5 (KLF5), Proliferation, Cell cycle
INTRODUCTION
Cervical cancer (CC) is a female-specific cancer type but with a high incidence and mortality1. It was estimated that there are 529,800 new cases and 275,100 deaths annually2. The infection of human papillomavirus (HPV) was regarded to be related with the development of CC3,4. However, evidence showed that infection of HPV is not sufficient to cause CC, and therefore additional genetic changes are required4,5. A better understanding of the aberrant gene expression in CC will definitely help to improve the prognosis of CC patients.
MicroRNAs (miRNAs) are noncoding RNAs with the length of about 20–23 nucleotides6. It was reported that miRNAs were involved in diverse cellular processes by posttranscriptional regulation of gene expression7. The development of cancer is accomplished with abnormal status of cell proliferation, migration, invasion, or metastasis8,9. The published literature has already investigated the value of miRNAs as cancer diagnostics or target therapy biomarkers9–11. miRNAs were reported to function as either oncogenic miRNAs (oncomiRs) or tumor suppressor miRNAs (tumor suppressor miRs)12,13. A previous study demonstrated that miR-152 inhibits the proliferation of human endometrial cancer cells via inducing G2/M phase arrest through suppressing CDC25B expression14. It was also reported that miR-152 could regulate the immune response in gastric cancer through regulating B7-H115. In gynecology, it was recently reported to be involved in the proliferation and metastasis of ovarian cancer through suppression of ERBB316. A study showed that miR-152 expression could be induced by hypoxia, and its expression can suppress the expression of WNT1 and ERBB3 and therefore inhibit the proliferation of CC cells17. However, the clinical significance of miR-152 expression and its underlying mechanism in the progression of CC remain to be investigated.
In this present study, we found that miR-152 expression in CC tissues was lower than in matched noncancerous tissues. Furthermore, miR-152 expression inhibited cell proliferation and suppressed the cell cycle progression. Moreover, we identified that Krüppel-like factor 5 (KLF5) was a direct target of miR-152 in CC and a mediator for the tumor suppressor role of miR-152. Collectively, our results showed that artificially upregulating the expression of miR-152 might be a treatment strategy for CC patients.
MATERIALS AND METHODS
Patients and Tissue Specimens
The human CC tissues and matched noncancerous tissues were collected from 57 patients, with informed consent obtained from all the patients. This study was approved by the review board of Affiliated Qilu Hospital of Shandong University. None of the patients had ever received immunotherapy, chemotherapy, or other types of anticancer treatment methods. Tissue samples were snap frozen in liquid nitrogen and stored in −80°C until RNA and protein extraction.
Cell Culture and Transfection
HeLa and C33A CC cells obtained from the ATCC (Manassas, VA, USA) were maintained in Dulbecco’s modified Eagle’s medium (DMEM; Invitrogen, Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10% fetal bovine serum. The human cervical epithelial cell line End1 was also obtained from the ATCC and cultured in keratinocyte serum-free medium (K-SFM; Invitrogen, Thermo Fisher Scientific, Inc.). These cells were maintained in a humidified incubator at 37°C containing 5% of CO2.
Presynthesized miR-152 mimic, inhibitor, and control were purchased from RiboBio Inc. (Guangzhou, P.R. China). The siRNA against KLF5 (si-KLF5) and control were also obtained from RiboBio Inc. The cell transfection was conducted using Lipofectamine 2000 (Invitrogen, Thermo Fisher Scientific, Inc.) according to the manufacture’s protocols.
RNA Extraction and Quantitative RT-PCR (qRT-PCR)
Total RNA from fresh tissues and cultured cells was harvested using TRIzol reagent (Thermo Fisher Scientific, Inc.). To analyze the relative expression level of KLF5, the first-strand cDNA was synthesized using Promega Reverse Transcription System (Promega, Madison, WI, USA). To analyze the level of miR-152, miRNA was reverse transcribed from the total RNA using a specific miRNA primer and miScript Reverse Transcription Kit (Qiagen, Düsseldorf, Germany). ABI 7500 Real-Time PCR System (Applied Biosystems, Foster City, CA, USA) was used to perform qRT-PCR with miScript SYBR Green PCR Kit (Qiagen). U6 snRNA and GAPDH were used as endogenous control for miR-152 and KLF5, respectively. The primers were synthesized by RiboBio Inc. and are summarized in Table 1.
Table 1.
Primers Used in This Study
| Gene Name | Sequence From 5′ to 3′ |
|---|---|
| miR-152 | F: ACACTCCAGCTGGGAGGTTCTGTGATACACT |
| R: AACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGAGTCGG | |
| KLF5 | F: CAGAGGACCTGGTCCAGACAAG |
| R: GAGGCCAGTTCTC-AGGTGAGTG | |
| U6 snRNA | F: GCGCGTCGTGAAGCGTTC |
| R: GTGCAGGGTCCGAGGT | |
| GAPDH | F: ATGGGGAAGGTGAAGGTCGG |
| R: GACGGTGCCATGGAATTTGC |
miR-152, microRNA-152; KLF5, Krüppel-like factor 5; GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
Western Blot
Total RNA from fresh tissues and cultured cells was harvested using RIPA lysis buffer (Thermo Fisher Scientific, Inc.) following the provided instructions. Samples were separated using SDS-PAGE and then transferred onto a PVDF membrane. The membrane was blocked with skimmed milk at room temperature. The membrane was incubated with antibodies against KLF5 (1:2,000; Abcam, Cambridge, MA, USA) and GAPDH (1:1,000; Abcam) at 4°C overnight. After that, the membrane was washed and incubated with secondary antibody (1:1,500; Santa Cruz Biotechnology, Inc., Dallas, TX, USA) for 3 h. Finally, the band signals were developed using ECL Plus Kit (Beyotime, Jiangsu, P.R. China) and analyzed using ImageJ software (NIH, Bethesda, MD, USA). GAPDH was selected as control for KLF5.
Luciferase Activity Reporter Assay
To construct the pGL3-KLF5-3′-UTR-wild type (WT) and pGL3-KLF5-3′-UTR-mutant (Mut) constructs, the 3′-UTR sequence of KLF5 was amplified and mutated by PCR and cloned into pGL3-control vector (Promega). For luciferase activity analysis, cells were cotransfected with GL3-KLF5-3′-UTR-WT and pGL3-KLF5-3′-UTR-Mut and miR-152 mimic or control using Lipofectamine 2000 (Invitrogen, Thermo Fisher Scientific, Inc.). After transfection for 48 h, the luciferase activity was measured using Dual-Glo Luciferase assay system (Promega).
Cell Proliferation Assay
CCK-8 purchased from Beyotime was used to measure the cell proliferation rate. In brief, cells were seeded in a 96-well plate and cultured in the aforementioned condition. At indicated time points (0, 24, 48, and 72 h), 10 μl of CCK-8 reagent was added to each well and incubated for an additional 4 h. The absorbance was measured at 450 nm using a microplate reader (Bio-Rad Laboratories, Gaithersburg, MD, USA).
Cell Cycle Assay
The cultured cells were harvested and fixed in 70% precold ethanol. The cells were then incubated with RNase A (1 mg/ml) in PBS at 37°C for 30 min. Propidium iodide (Beyotime) was added to the cell suspension and incubated at room temperature for 30 min avoiding the light. Finally, the samples were analyzed by a FACSCalibur Cytometer (Becton Dickinson, Franklin Lakes, NJ, USA). Cell cycle distribution was analyzed using ModiFit software (Verity Software House Company, Topsham, ME, USA).
Statistical Analysis
GraphPad Prism version 5.0 software (GraphPad, San Diego, CA, USA) was used for data analysis. The survival rates in relation to miR-152 expression were estimated by the Kaplan–Meier method, and the difference in survival curves was tested by the log-rank test. To determine the difference of two groups and multiple groups, Student’s t-test and one-way ANOVA were conducted, respectively. The corrections between miR-152 or KLF5 expression were determined by Spearman’s correlation analysis. A value of p < 0.05 was considered to indicate a statistically significant difference.
RESULTS
Expression of miR-152 and its Correlation With Poor Prognosis of CC Patients
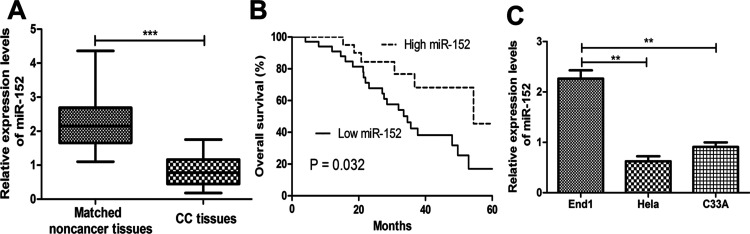
To investigate the clinical significance of miR-152 in CC, we first examined the expression of miR-152 by qRT-CPR in 57 pairs of CC tissues and matched noncancerous tissues. We found that miR-152 levels were significantly reduced in CC tissues compared with their corresponding noncancerous tissues (Fig. 1A). To further investigate the significance of miR-152 in CC, we divided the CC samples into two groups according to the levels of miR-152. As expected, the low miR-152 predicts a poorer overall survival than those with high miR-152 (Fig. 1B). To further investigate the role of miR-152 in CC, we examined its expression in CC cell lines and human cervical epithelial cell line. Not surprisingly, expression of miR-152 was lower in CC cells compared with that of human cervical epithelial cell line (Fig. 1C). Taken together, these findings indicate that miR-152 correlates with CC prognosis.
Figure 1.
Reduced microRNA-152 (miR-152) expression in cervical cancer (CC). (A) Relative miR-152 expression in CC tissues and matched noncancer tissues. (B) Reduced expression of miR-152 means poorer overall survival of CC patients. (C) Relative miR-152 expression in CC cell lines and human cervical epithelial cell line. **p < 0.01, ***p < 0.001.
miR-152 Inhibits KLF5 Expression by Binding its 3′-UTR
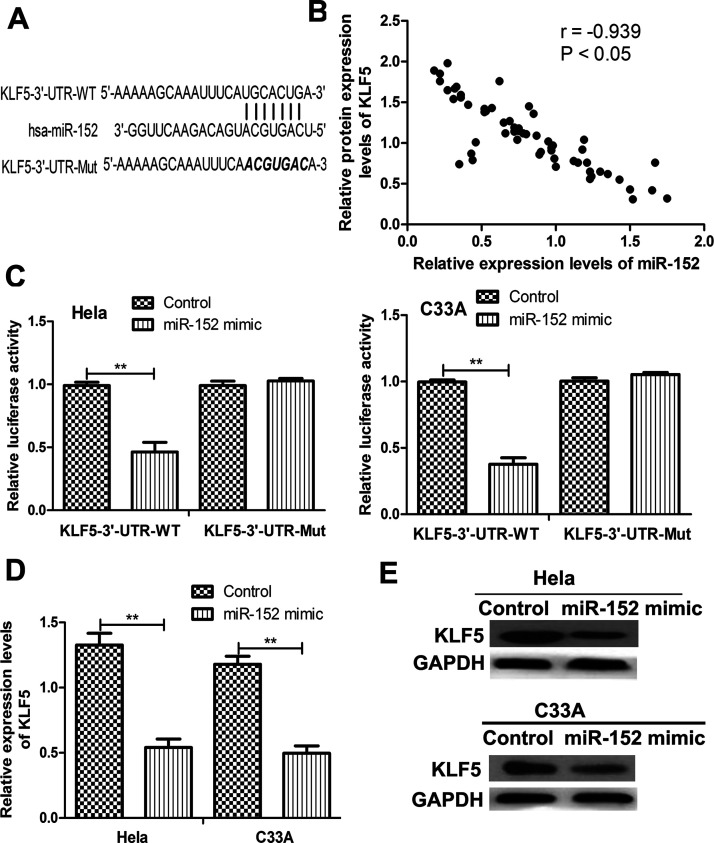
In order to investigate the possible mechanism of miR-152 in CC, we searched the candidate target genes of miR-152 using TargetScan. We found that the 3′-UTR of KLF5 contains a sequence that can complementarily bind with miR-152 (Fig. 2A). Therefore, we examined the correlation of miR-152 and KLF5 in CC tissues. As shown in Figure 2B, the protein expression of KLF5 was found inversely correlated with miR-152 in CC tissues. To validate that KLF5 was a direct target of miR-152, the luciferase activity assay was performed. The results presented in Figure 2C demonstrated that miR-152 decreased the luciferase activity of the WT KLF5 3′-UTR group when compared with the control-transfected cells. No significant change in luciferase activity in the mutant groups (Fig. 2C) was observed. Furthermore, qRT-PCR was conducted to measure the expression of KLF5 in the miR-152 mimic- and control-transfected cells. As expected, both the mRNA and protein levels of KLF5 in miR-152 mimic-transfected cells were lower than those in the control-transfected cells (Fig. 2D and E). These results implicate that KLF5 was a direct target of miR-152.
Figure 2.
Krüppel-like factor 5 (KLF5) was a direct target of miR-152 in CC. (A) The wild-type (WT) and mutant (Mut) miR-152 binding sequence in the 3′-UTR of KLF5. (B) Correlation between miR-152 and KLF5 expression in CC tissues. (C) Relative luciferase activity of HeLa and C33A cells cotransfected with miR-152 mimic or control and WT or Mut KLF5 3′-UTR. Relative KLF5 (D) mRNA and (E) protein expressions of KLF5 in HeLa and C33A cells transfected with miR-152 mimic and control. **p < 0.01.
miR-152 Suppresses CC Cell Proliferation Through Inhibiting KLF5
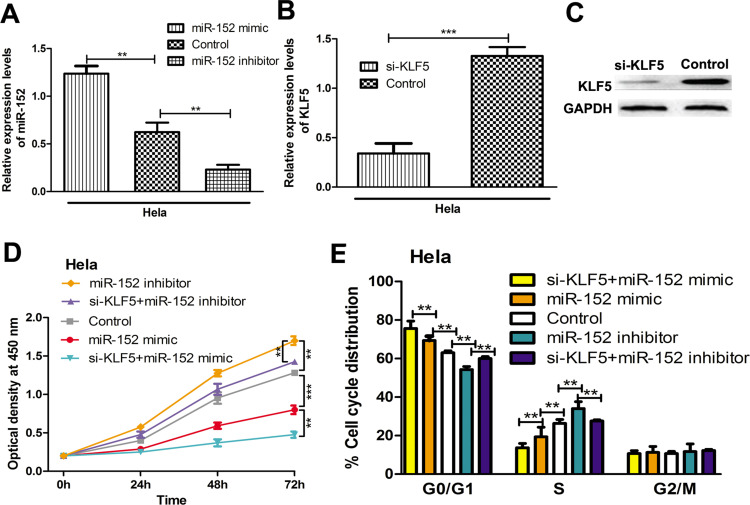
To investigate the functions of miR-152 and KLF5 in CC cells, we examined the cell proliferation of HeLa cells transfected with synthetic miRNAs and siRNAs. qRT-PCR revealed that the levels of miR-152 can be upregulated by miR-152 mimic but reduced by miR-152 inhibitor (Fig. 3A). Not surprisingly, we found that si-KLF5 could downregulate both the mRNA and protein expression of KLF5 (Fig. 3B and C). Cell proliferation assay revealed that overexpression of miR-152 reduced the proliferation rate of HeLa cells, whereas the downregulation of miR-152 has the opposite effects (Fig. 3D). Furthermore, we examined the effect of KLF5 on cell proliferation by cotransfecting the si-KLF5 with miR-152 mimic or inhibitor. We showed that the knockdown of KLF5 could partially reverse the effect of miR-152 inhibitor but strengthen the effect of miR-152 mimic on cell proliferation (Fig. 3D). These results suggest that miR-152 impairs the proliferation of CC cells through downregulation of KLF5.
Figure 3.
miR-152 inhibits cell proliferation and arrest cell cycle through regulating KLF5. (A) Relative miR-152 expression in HeLa cells transfected with miR-152 mimic, inhibitor, or control. Relative KLF5 (B) mRNA and (C) protein expressions of KLF5 in HeLa cells transfected with si-KLF5 and control. (D) Cell proliferation rate of HeLa cells transfected with miR-152 mimic, inhibitor, siRNA targeting KLF5 (si-KLF5), or control. (E) Cell cycle distribution of HeLa cells transfected with miR-152 mimic, inhibitor, si-KLF5, or control. **p < 0.01, ***p < 0.001.
miR-152 Induces Cell Cycle Arrest in CC Cells
To investigate the mechanism of the suppressive role of miR-152 in CC cell proliferation, we examined the cell cycle distribution of HeLa cells with synthetic miRNAs or siRNAs transfection. We found that overexpression of miR-152 arrested the cell cycle at the G0/G1 phase (Fig. 3E). In contrast, downregulation of miR-152 led to a reduction in the G0/G1 phase (Fig. 3E). We also found that knockdown of miR-152 reversed the effect of miR-152 inhibitor on cell cycle distribution, that is, the cells at the G0/G1 phase were reduced compared with the miR-152-transfected group (Fig. 3E). Collectively, these data suggest that miR-152 arrested cell cycle at the G0/G1 phase in CC cells.
DISCUSSION
It was reported that a combination of HPV vaccination and screening could almost eradicate CC and reduce the burden of other tumors and diseases related to HPV18. However, at present, CC is still an important health problem worldwide1. Although early stage CC can be curable with surgery, prognosis of patients with recurrence remains poor, with limited treatment options19. Therefore, it is important to investigate the mechanisms related to CC progression.
To date, over 60 miRNAs including oncomiRs and tumor suppressor miRs have been identified to have abnormal expression in CC by comparing miRNA expression in normal and tumor tissues20,21. The investigation on the biological functions of miRNA in CC has accelerated the development of new anticancer technology22,23. For example, miR-21 was proved to be an oncomiR in CC, and therefore antisense miRNA oligonucleotides (AMOs) targeting miR-21 were developed by Wang et al.22. They found that the transfection of AMOs decreased the expression of miR-21 and suppressed tumor growth22. In contrast, miR-143 was found as a tumor suppressor miR in CC, and its supplementation may contribute to the treatment of CC. Liu et al. found that the introduction of miR-143 inhibited CC cell growth but enhanced cell apoptosis in vitro and in vivo23. Collectively, these attempts have provided a new strategy for anticancer treatment. In light of this, we realized that it is imperative to identify miRNAs that were abnormally expressed in CC and correlated with the initiation or progression of CC.
Previous studies demonstrated that miR-152 mainly functioned as a tumor suppressor miR in cancers including gastric cancer, endometrial cancer, and ovarian cancer14–17. However, its expression and clinical significance in CC have been rarely investigated. Therefore, in this study, we first investigated the expression pattern of miR-152 in CC tissues and made a comparison with the matched noncancerous tissues. We found that miR-152 was sharply reduced in CC tissues compared with the noncancerous tissues. Also, its downregulation was correlated with poorer overall survival of CC patients. The in vitro functional assay revealed that miR-152 could inhibit cell proliferation and arrest the cell cycle in the G0/G1 phase through directly regulating the expression of KLF5. In a recent study, Xie et al. found that miR-152 inhibits proliferation of human endometrial cancer cells via inducing G2/M phase arrest by suppressing CDC25B expression14. The difference in the effect of miR-152 on cell cycle distribution might be due to the difference in cancer type or its downstream targets.
In conclusion, we demonstrated that miR-152 was reduced in CC. miR-152 could inhibit the cell proliferation and cell cycle in vitro, at least in part, by downregulating the expression of KLF5. These results together indicate that miR-152 expression could be a novel strategy for CC treatment.
REFERENCES
- 1. International Collaboration of Epidemiological Studies of Cervical Cancer, Appleby P, Beral V, Berrington de Gonzalez A, Colin D, Franceschi S, Goodhill A, Green J, Peto J, Plummer M, Sweetland S. Cervical cancer and hormonal contraceptives: Collaborative reanalysis of individual data for 16,573 women with cervical cancer and 35,509 women without cervical cancer from 24 epidemiological studies. Lancet 2007;370(9599):1609–21. [DOI] [PubMed] [Google Scholar]
- 2. Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer 2010;127(12):2893–917. [DOI] [PubMed] [Google Scholar]
- 3. Munoz N, Bosch FX, de Sanjose S, Herrero R, Castellsague X, Shah KV, Snijders PJ, Meijer CJ. Epidemiologic classification of human papillomavirus types associated with cervical cancer. N Engl J Med. 2003;348(6):518–27. [DOI] [PubMed] [Google Scholar]
- 4. Petry KU. HPV and cervical cancer. Scand J Clin Lab Invest Suppl. 2014;244:59–62. [DOI] [PubMed] [Google Scholar]
- 5. Schiffman M, Castle PE, Jeronimo J, Rodriguez AC, Wacholder S. Human papillomavirus and cervical cancer. Lancet 2007;370(9590):890–907. [DOI] [PubMed] [Google Scholar]
- 6. Banno K, Yanokura M, Kisu I, Yamagami W, Susumu N, Aoki D. MicroRNAs in endometrial cancer. Int J Clin Oncol. 2013;18(2):186–92. [DOI] [PubMed] [Google Scholar]
- 7. Pasquinelli AE. MicroRNAs and their targets: Recognition, regulation and an emerging reciprocal relationship. Nat Rev Genet. 2012;13(4):271–82. [DOI] [PubMed] [Google Scholar]
- 8. Hanahan D, Weinberg RA. Hallmarks of cancer: The next generation. Cell 2011;144(5):646–74. [DOI] [PubMed] [Google Scholar]
- 9. Gao D, Chen Y. Organoid development in cancer genome discovery. Curr Opin Genet Dev. 2015;30:42–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yang Z, He J, Gao P, Niu Y, Zhang J, Wang L, Liu M, Wei X, Liu C, Zhang C, Wang W, Du J, Li H, Hu W, Sun G. miR-769-5p suppressed cell proliferation, migration and invasion by targeting TGFBR1 in non-small cell lung carcinoma. Oncotarget 2017;8(69):113558–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhu QN, Renaud H, Guo Y. Bioinformatics-based identification of miR-542-5p as a predictive biomarker in breast cancer therapy. Hereditas 2018;155:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Esquela-Kerscher A, Slack FJ. Oncomirs-microRNAs with a role in cancer. Nat Rev Cancer 2006;6(4):259–69. [DOI] [PubMed] [Google Scholar]
- 13. Schoniger C, Arenz C. Perspectives in targeting miRNA function. Bioorg Med Chem. 2013;21(20):6115–8. [DOI] [PubMed] [Google Scholar]
- 14. Xie D, Liang Y, Su Y, An Y, Qu P. miR-152 inhibits proliferation of human endometrial cancer cells via inducing G2/M phase arrest by suppressing CDC25B expression. Biomed Pharmacother. 2018;99:299–305. [DOI] [PubMed] [Google Scholar]
- 15. Wang Y, Wang D, Xie G, Yin Y, Zhao E, Tao K, Li R. MicroRNA-152 regulates immune response via targeting B7-H1 in gastric carcinoma. Oncotarget 2017;8(17):28125–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Li LW, Xiao HQ, Ma R, Yang M, Li W, Lou G. miR-152 is involved in the proliferation and metastasis of ovarian cancer through repression of ERBB3. Int J Mol Med. 2018;41(3):1529–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tang XL, Lin L, Song LN, Tang XH. Hypoxia-inducible miR-152 suppresses the expression of WNT1 and ERBB3, and inhibits the proliferation of cervical cancer cells. Exp Biol Med. (Maywood) 2016;241(13):1429–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Borcoman E, Le Tourneau C. Pembrolizumab in cervical cancer: Latest evidence and clinical usefulness. Ther Adv Med Oncol. 2017;9(6):431–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kamangar F, Dores GM, Anderson WF. Patterns of cancer incidence, mortality, and prevalence across five continents: Defining priorities to reduce cancer disparities in different geographic regions of the world. J Clin Oncol. 2006;24(14):2137–50. [DOI] [PubMed] [Google Scholar]
- 20. Gocze K, Gombos K, Juhasz K, Kovacs K, Kajtar B, Benczik M, Gocze P, Patczai B, Arany I, Ember I. Unique microRNA expression profiles in cervical cancer. Anticancer Res. 2013;33(6):2561–7. [PubMed] [Google Scholar]
- 21. Pereira PM, Marques JP, Soares AR, Carreto L, Santos MA. MicroRNA expression variability in human cervical tissues. PLoS One 2010;5(7):e11780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wang XM, Xu J, Cheng ZQ, Peng QZ, Hu JT, Gao LK, Zhang SF, Jin HT. Study on effects of microRNA-21 antisense oligonucleotide in vivo and in vitro on bionomics of human cervical squamous carcinoma cell lines SiHa. Chinese J Pathol. 2012;41(4):254–9. [DOI] [PubMed] [Google Scholar]
- 23. Liu L, Yu X, Guo X, Tian Z, Su M, Long Y, Huang C, Zhou F, Liu M, Wu X, Wang X. miR-143 is downregulated in cervical cancer and promotes apoptosis and inhibits tumor formation by targeting Bcl-2. Mol Med Rep. 2012;5(3):753–60. [DOI] [PubMed] [Google Scholar]