Abstract
Recently, novel therapeutic strategies have been designed with the aim of killing cancer stem-like cells (CSCs), and considerable interest has been generated in the development of specific therapies that target stemness-related marker of CSCs. In this study, nonsteroidal anti-inflammatory drugs (NSAIDs) significantly potentiated Hsp90 inhibitor 17-allylamino-17-demethoxygeldanamycin (17-AAG)-mediated cytotoxicity through apoptotic and autophagic cell death induction, but COX-2-inhibitory function was not required for NSAID-induced autophagy in CD44-overexpressing human chronic myeloid leukemia K562 (CD44highK562) cells. Importantly, we found that treatment with NSAIDs resulted in a dose-dependent increase in LC3-II level and decrease in p62 level and simultaneous reduction in multiple stemness-related markers including CD44, Oct4, c-Myc, and mutant p53 (mutp53) in CD44highK562 cells, suggesting that NSAIDs could induce autophagy, which might mediate degradation of stemness-related marker proteins. Activation of AMPK and inhibition of Akt/mTOR/p70S6K/4EBP1 participated in NSAID-induced autophagy in CD44highK562 cells. In addition, treatment of CD44highK562 cells with NSAIDs inhibited expression of HSF1/Hsps, which resulted in suppression of 17-AAG-induced activation of Hsp70, leading to reversal of 17-AAG resistance and sensitization of CD44highK562 cells to 17-AAG by NSAIDs. In conclusion, combining NSAIDs with Hsp90 inhibitor may offer one of the most promising strategies for eradication of CD44-overexpressing CSCs.
Key words: CD44, Hsp90 inhibitor, Nonsteroidal anti-inflammatory drugs (NSAIDs), Stemness-related markers, Autophagy
INTRODUCTION
Cancer stem (-like) cells (CSCs) are characterized by an ability to self-renew and have a profound effect on tumorigenesis and progression1. In addition, CSCs resist chemo- and radiotherapy via their powerful self-renewal capacity, drug effluxion, and antiapoptotic ability. Conventional anticancer drugs kill rapidly proliferating non-CSCs, but have less effect on CSCs. Therefore, therapeutic strategies targeting CSCs may bring new and effective cancer therapy2.
Heat shock protein 90 (Hsp90) is an ATP-dependent molecular chaperone that is exploited by cancer cells to support activated oncoproteins, including many cancer-associated kinases and transcription factors, and therefore Hsp90 may serve as a therapeutic target for the treatment of cancer3. Hsp90 inhibitor treatment leads to activation of the heat shock factor (HSF1). HSF1 is the master regulator of genes encoding molecular chaperones, and it upregulates heat shock proteins such as Hsp70, which causes a reduced Hsp90-targeted drug efficacy and resistance to Hsp90 inhibitor4. HSF1 also controls the stability of mutant p53 (mutp53) protein, a client of Hsp90, in human cancer cells through activation of Hsp905, and inhibition of Hsp90 has been shown to promote the degradation of mutp53 protein6. Therefore, the oncogenicity of mutp53 critically depends on HSF1 and/or a HSF1-mediated transcriptional program7. It has been reported that mutp53 is frequently expressed in a variety of human tumors and has a major role in formation of CSCs, and also HSF1 is necessary to regulate and maintain CSC phenotype in breast cancer cells, which makes mutp53 and HSF1 potential targets to develop CSC-specific therapies4,8. In addition, mutp53 contributes to activation of HSF1 that increases expression level of multidrug resistance 1 (MDR1)/P-glycoprotein (P-gp)9, indicating the role of mutp53 expression in drug resistance of CSCs. Since it has been reported that P-gp-mediated efflux of Hsp90 inhibitors is a limiting factor affecting the sensitivity of Hsp90 inhibitors against cancer cells10, downregulation of mutp53 and HSF1, which could reduce P-gp, might be involved in reversal of resistance against Hsp90 inhibitors. In addition, positive expression of mutp53 and CD44 is directly associated with clinicopathological features and poor prognosis of oral squamous cell carcinoma11. CD44 is the most common CSC surface marker and has a pivotal role in CSC communication with the microenvironment and in regulating CSC stemness properties, indicating that targeting CD44 is a promising approach with the potential to eliminate CSCs.
The effectiveness of long-term and regular use of nonsteroidal anti-inflammatory drugs (NSAIDs) in the prevention and treatment of certain cancers, including prostate, colon, breast, lung, and gastric cancers, has been suggested12. Celecoxib (CCB), a cyclooxygenase-2 (COX-2)-selective NSAID, has been reported to increase the sensitivity of tumor cells to chemotherapy and radiotherapy in preclinical investigations, and therefore CCB is an attractive drug for anticancer treatment13,14. Ibuprofen (IBU), a nonselective NSAID, also reduced the cancerous characteristics of the adenocarcinoma gastric cells by inducing apoptosis, inhibition of cell proliferation, angiogenesis, and stemness of the cells15. Moreover, NSAIDs can sensitize cancer cells to the antiproliferative effects of cytotoxic drugs via P-gp modulator activity16–19.
Eradicating CSCs by efficient targeting agents may have the potential to cure cancer. More complete elucidation of the mechanisms underlying resistance of CSCs to treatment is necessary and may provide a more effective therapy to overcome such resistance. Herein we show that NSAIDs considerably potentiate sensitivity of CD44-overexpressing CD44highK562 cells to Hsp90 inhibitor 17-AAG by downregulation of multiple stemness-related markers and suppression of 17-AAG-induced Hsp70 activation in CD44highK562 cells, suggesting one of new therapeutic approaches to eradicate CSCs.
MATERIALS AND METHODS
Cell Culture and Reagents
CD44highK562 cells isolated from human K562 chronic myeloid leukemia cell line showed higher protein levels of stemness-related markers and ABC transporters compared with those of the parental K562 cells20. CD44highK562 cells maintained characteristics of cell morphology, cell proliferation ability, and high expression of stemness-related markers through serial culture until passage 15 after recovery from stocks. Human lymphoblastic leukemia multidrug-resistant (MDR) CEM/VLB100 and MCF7-MDR (originally named MCF-7/Adr) cells were kindly provided by Dr. Fiedler (MD Anderson, Houston, TX, USA)21. Cells were maintained in RPMI or DMEM medium (Invitrogen, Carlsbad, CA, USA) supplemented with 10% FBS and maintained at 37°C in humidified 5% CO2 atmosphere. 17-Allylamino-17-demethoxygeldanamycin (17-AAG) was purchased from Enzo Life Sciences (Farmingdale, NY, USA). Celecoxib (CCB), ibuprofen (IBU), cycloheximide (CHX), 3-methyladenine (3-MA), chloroquine (CQ), 2,5-dimethyl-celecoxib (DMC), and rapamycin were purchased from Sigma-Aldrich (St. Louis, MO, USA). Z-DEVD-FMK was purchased from R&D Systems (Minneapolis, MN, USA). OSU-03012 was purchased from Selleckchem (Houston, TX, USA).
Cell Proliferation Assay
Cell proliferation was measured using the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay. Exponentially growing cells were plated in a 96-well plate and incubated in growth medium containing the indicated concentrations of 17-AAG and/or CCB/IBU at 37°C for 96 h. Inhibition of cell proliferation was expressed as a percentage of the untreated control cell growth. Interactions between 17-AAG and CCB/IBU were assessed using the CompuSyn Software program (ComboSyn, Paramus, NJ, USA), which utilizes the Chou–Talalay equation. Combination index (CI) was used to determine synergy (CI < 0.9), additivity (0.9 < CI < 1.1), and antagonism (CI > 1.1) of the drug combinations tested. All experiments were carried out in triplicate.
Western Blot and Coimmunoprecipitation Analysis
Protein samples were separated by SDS-PAGE and blotted to a nitrocellulose membrane (Hybond-ECL, GE Healthcare, USA). The membrane was incubated with antibody as specified, followed by secondary antibody conjugated with horseradish peroxidase. Specific antigen–antibody complexes were detected by enhanced chemiluminescence (PerkinElmer, Life Science, Waltham, MA, USA). Western blot analysis was performed with the following antibodies: LC3B (LC3), p62 (Novus Biologicals, Littleton CO, USA), CD44, Oct4, Atg7, Beclin1, Mcl-1, STAT3, phospho-STAT3 (Tyr705), AMPK, phospho-AMPK (Thr172), Akt, phospho-Akt (Thr308), mTOR, phospho-mTOR (Ser2448), 4E-BP1, phospho-4E-BP1 (Thr37/46), p70-S6 kinase (p70S6K), phospho-p70S6K (Thr389), caspase 3 (Cell Signaling, Danvers, MA, USA), c-Myc (Epitomics, CA, USA), β-actin (Sigma-Aldrich), p53, HSF1, P-gp (Santa Cruz Biotechnology, Santa Cruz, CA, USA), Hsp27, Hsp70, and Hsp90 (Enzo Life Sciences). The p53 antibody (DO-1) is a mouse monoclonal antibody raised against amino acids 11–25 of p53 of human origin, as recommended for detection of wild and mutp53 of human origin. For coimmunoprecipitation, whole cell extracts from CD44highK562 cells treated with or without CCB were incubated with indicated antibody overnight at 4°C. Protein G-Sepharose beads with immunocomplexes were boiled, electrophoresed on 8% SDS-polyacrylamide gels, and analyzed by Western blotting using the indicated antibody. Data shown are representative results from at least two independent experiments.
Apoptosis Assessment by Annexin V Staining
CD44highK562 cells were treated with 17-AAG in the presence/absence of CCB/IBU or autophagy/apoptosis inhibitor under indicated conditions for 24 h. Subsequently, cells were centrifuged and resuspended in 100 μl of the kit’s staining solution containing annexin V–fluorescein (FITC Apoptosis detection kit; BD PharMingen San Diego, CA, USA) and propidium iodide (PI; Sigma-Aldrich) in HEPES buffer. After incubation at room temperature for 20 min, the percentages of early and late apoptotic cells were quantified by performing FACS for annexin V and PI staining.
Flow Cytometric Analysis for CD44 Surface Expression
Surface expression of CD44 on cells treated with or without NSAIDs was determined by flow cytometry. Briefly, cells were washed once at the time of harvesting with PBS/0.1% sodium azide and aliquoted into polystyrene tubes. Cells were stained with FITC-labeled anti-CD44 mAbs (BD PharMingen). Autofluorescence and isotype (IgG2b)-matched control Abs (BD Biosciences, San Jose, CA, USA) were included. Data were acquired on a CANTO II and Calibur (BD Biosciences) and analyzed using FlowJo (ver. 10; Tree Star, Ashland, OR, USA).
Statistical Analysis
A Student’s t-test was used to calculate the statistical significance of the experimental data, and the level of significance was set as p < 0.05, p < 0.01, and p < 0.001.
RESULTS
Potentiation of Hsp90 Inhibitor-Mediated Cytotoxicity and Apoptosis by NSAIDs
CD44highK562 cells exhibited highly enriched in CD44+ population (92.1%) compared with parental K562 cells (0.3%) (Fig. 1A). Indeed, we reported that CD44highK562 cells exhibited high basal expression of stemness-related markers such as Oct4, CD34, mutp53, c-Myc, and ABC transporters as well as CD44, and resistance to multiple anticancer drugs, and inhibition of P-gp by SIRT1 inhibitor promoted reversal of drug resistance20. Since some NSAIDs displayed inhibitory activity of P-gp18,19,22, we determined whether clinically useful NSAIDs could act as a new class of chemosensitizer for Hsp90 inhibitor-resistant CD44highK562 cells. CD44highK562 cells treated with 17-AAG, an ansamycin Hsp90 inhibitor, in the presence or absence of CCB showed that combined treatment of 17-AAG and CCB significantly increased cytotoxicity compared to either of the drugs alone, and average CI values for combination treatments with 17-AAG and CCB were lower than 1 at all concentrations, indicating a synergistic effect in CD44highK562 cells (Fig. 1B), even though CCB sensitivity of CD44highK562 cells (IC50; 15.6 μM) was less sensitive than that of parental K562 cells (IC50; 8.5 μM; data not shown). This result indicates that CCB synergistically enhances celecoxib-induced growth and could sensitize CD44highK562 cells to 17-AAG. Next, the combined effect of Hsp90 inhibitor and NSAIDs on apoptosis was further confirmed. Induction of apoptosis was monitored with flow cytometry using PI and annexin V. When CD44highK562 cells were treated with 17-AAG (1 or 10 μM) in the presence or absence of CCB (10 μM) or IBU (400 μM), both CCB and IBU significantly enhanced 17-AAG-induced apoptosis in CD44highK562 cells, as determined by summing percentages in the second and fourth quadrants that are considered to be the percentage of early and late apoptotic cells (Fig. 2A and B). These results suggest that NSAIDs could potentiate Hsp90 inhibitor-mediated cytotoxicity and apoptosis and thus sensitize CD44highK562 cells to Hsp90 inhibitor.
Figure 1.
Potentiation of 17-allylamino-17-demethoxygeldanamycin (17-AAG) cytotoxicity by nonsteroidal anti-inflammatory drugs (NSAIDs). (A) Cell surface expression of CD44 in CD44high and parental K562 cells was quantified by flow cytometry after labeling both cells with anti-CD44 antibody. (B) CD44highK562 cells were treated with serial doses of 17-AAG in the presence or absence of celecoxib (CCB). The percentage of cell survival was determined after 96 h of incubation using MTT assay. Each bar represents the mean ± SD of triplicate experiments. *p < 0.05, **p < 0.01, and ***p < 0.001.
Figure 2.
(A) Enhancement of 17-AAG-induced apoptosis by NSAIDs. CD44highK562 cells were treated with 17-AAG in the absence (w/o) or presence of CCB or ibuprofen (IBU) for 24 h, and the percentage of apoptotic cells was quantified using FACS. The upper right quadrants contain late apoptotic cells (positive for both PI and annexin V), and the lower right quadrants represent early apoptotic cells (annexin V+ and PI). Images shown are representative of three independent experiments. (B) Bar graph shows mean percentage of apoptotic cells, and values represent the means ± SD. *p < 0.05, **p < 0.01.
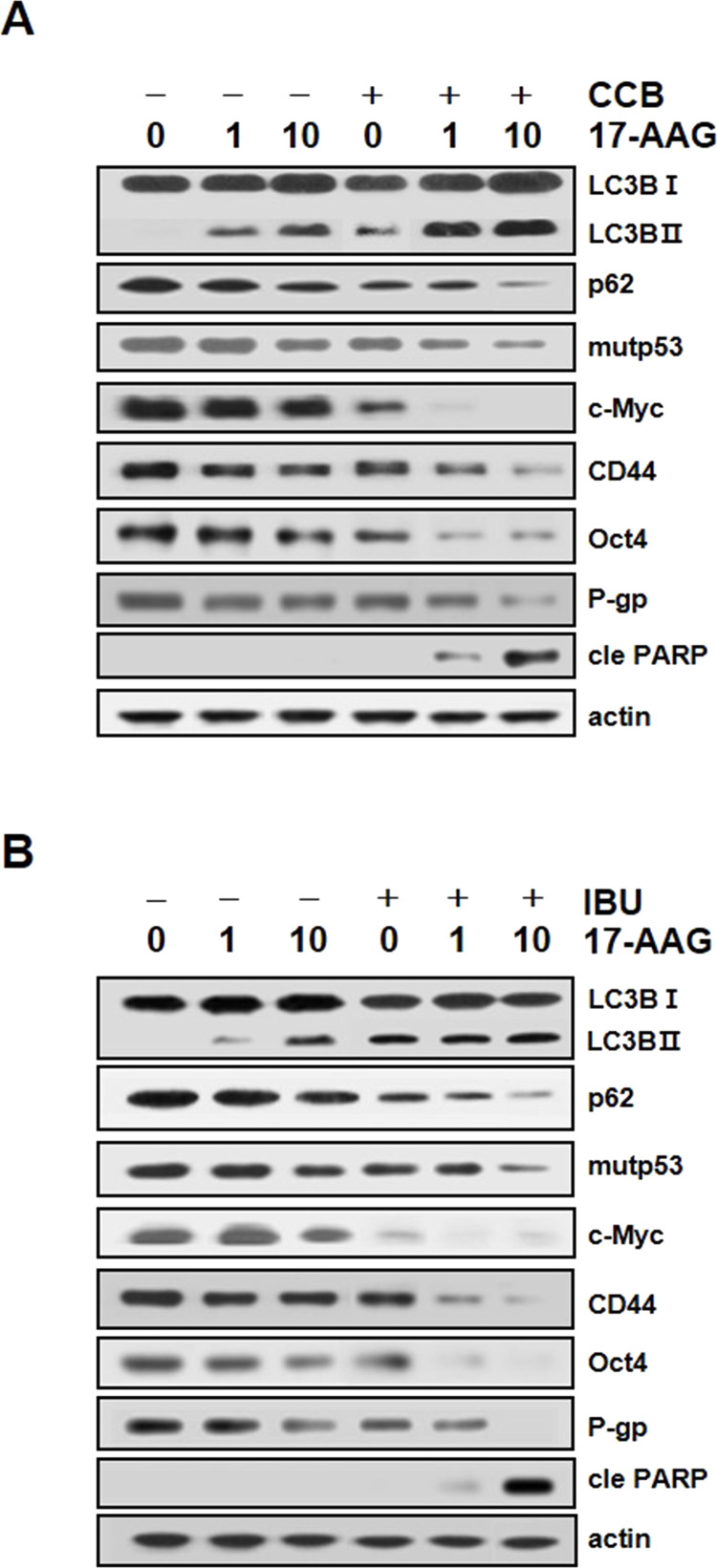
Induction of Autophagy and Subsequent Downregulation of Stemness-Related Markers and Cell Surface Expression of CD44 by NSAIDs
Since downregulation of mutp53 is involved in the reversal of Hsp90 inhibitor resistance in MDR cells7,23, and autophagy contributed to mutp53 degradation in cancer cells24,25, we examined whether NSAIDs could induce downregulation of stemness-related markers involving mutp53 and other in CD44highK562 cells via autophagy-dependent pathway (Fig. 3). To investigate whether NSAIDs could induce autophagy in CD44highK562 cells, we assessed autophagy activation by tracking the conversion of microtubule-associated protein 1 light chain 3 (LC3)-I to LC3-II26 and the level of p62 degradation27. When the levels of LC3-II and p62 in CD44highK562 cells were evaluated in the presence or absence of CCB, dose-dependent accumulation of LC3-II and reduction in p62 were observed in CCB-treated CD44highK562 cells, which was closely associated with CCB-induced downregulation of multiple stemness-related markers (Fig. 3A). Mutp53 can regulate the expression of the endogenous c-Myc gene and is a potent activator of the c-Myc promoter28,29. Moreover, degradation of c-Myc might be regulated by autophagy30,31. Our results showed that CCB-induced autophagy was associated with downregulation of c-Myc as well as mutp53. Interestingly, CCB also induce dose-dependent reduction in CD44 and Oct4 levels in CD44highK562 cells. Similarly, IBU-treated CD44highK562 cells showed that a dose-dependent increase in LC3-II level and decrease in p62, and simultaneous reduction in mutp53, c-Myc, CD44, and Oct4 (Fig. 3B), indicate that NSAID could induce autophagy, which might promote downregulation of multiple stemness-related markers. To further confirm CCB-induced downregulation in CD44 in other cancer cells with high level of CD44, cell surface expression of CD44 in CD44-high-expressing CEM/VLB100 and MCF7-MDR cells as well as CD44highK562 cells was determined using flow cytometry (Fig. 3C). Dose-dependent reduction in CD44 cell surface expression by CCB was observed in CD44highK562, CEM/VLB100, and MCF7-MDR cells. These results raised the possibility that NSAID might be effective at targeting both CD44-high-expressing cancer cells and CSCs by downregulation of stemness-related markers.
Figure 3.
Induction of autophagy and downregulation of stemness-related markers by NSAIDs, and CCB-mediated reduction in CD44 cell surface expression. CD44highK562 cells were treated with serial doses of CCB (A), IBU (B) for 24 h, and the changed levels of autophagy (LC3B-1/II and p62) and expression of stemness-related markers (mutp53, c-Myc, CD44, and Oct4) were determined by Western blot analysis. Actin was used as a loading control. (C) CD44highK562, CEM/VLB100, and MCF7-MDR cells were treated with serial doses of CCB for 24 h, and the change in CD44 cell surface expression of each cell was determined by FACS.
Since CCB, a cyclooxygenase-2 (COX-2)-specific inhibitor, has several COX-2-independent activities32, the question arose as to whether or not its COX-2-inhibitory function is required for its autophagy-inducing property. We therefore employed 2,5-dimethyl-celecoxib (DMC), which is a close structural analog of celecoxib that lacks a COX-2 inhibitory function33. We found that DMC potently mimicked the autophagy-inducing effect of CCB (Fig. 4A). Treatment of CD44highK562 cells with DMC as well as CCB resulted in elevation of the LC3-II/LC3-I and reduction in p62 level in parallel with reduction in multiple stemness-related marker levels, even though the autophagic ability of DMC was less than that of CCB, indicating that these changes occur regardless of COX-2 inhibitory activity for autophagic degradation of stemness-related marker proteins. Since it has been reported that OSU-03012, a non-COX-inhibiting celecoxib derivative, is capable of inducing apoptosis in various cancer cell types34, we next determined whether OSU-03012 also could induce autophagy and downregulate multiple stemness-related markers. Treatment of CD44highK562 cells with OSU-03012 resulted in elevation of the LC3-II/LC3-I level in parallel with reduction in multiple stemness-related marker levels. These results strongly suggest that COX-2-inhibitory function is not required for NSAID-induced autophagic degradation of stemness-related marker proteins in CD44highK562 cells. We also found that rapamycin, a well-known inducer of autophagy35, induced downregulation of multiple stemness-related markers, coincident with autophagy-inducing NSAIDs (Fig. 4B). It seems likely that autophagic pathway may be involved in degradation of stemness-related marker proteins. Moreover, in the presence of CCB, the half-life of stemness-related marker protein in CD44highK562 cells was further reduced as observed via CHX chase assay (Fig. 4C). The levels of multiple stemness-related marker proteins in CD44highK562 cells were determined in the presence of protein synthesis inhibitor CHX after treatment with CCB. Degradation of stemness-related marker proteins in CCB-treated cells was accelerated in the presence of CHX compared with CCB-untreated cells, indicating that CCB reduced half-lives of stemness-related marker proteins possibly through autophagic degradation. To further confirm the effect of CCB on autophagic degradation of stemness-related markers, CD44highK562 cells were treated with CCB in the presence or absence of autophagy inhibitor, and the modulation of CCB-mediated p62 and stemness-related markers was determined (Fig. 5A, left). Inhibition of autophagy at an early stage using 3-MA prevented CCB-induced downregulation of p62 and subsequent reduction in CD44, mutp53, c-Myc, and Oct4 in CD44highK562 cells. Similarly, suppression of autophagy at a late stage by CQ also prevented CCB-induced downregulation of p62 and these stemness-related markers (Fig. 5A, right), indicating that autophagy might be associated with degradation of stemness-related marker proteins.
Figure 4.
Comparison of autophagic-inducing effect of CCB derivatives and rapamycin, and autophagic degradation of stemness-related markers by CCB. CD44highK562 cells were treated with CCB or 2,5-dimethyl-celecoxib (DMC) (A) or OSU-03012 or rapamycin (B) for 24 h. (C) The cells were treated with or without 25 μM CCB for 24 h and were collected at 0, 3, and 6 h after, following treatment with 20 μg/ml cycloheximide (CHX), and the levels of stemness-related marker proteins were determined by Western blot analysis.
Figure 5.
Crosstalk between CCB-induced apoptotic and autophagic cell death. (A) CD44highK562 cells were treated with 25 μM CCB in the absence or presence of 10 mM 3-methyladenine (3-MA, left) or 5 mM chloroquine (CQ, right) for 24 h, and levels of p62 and stemness-related markers were determined by Western blot analysis. (B) CD44highK562 cells were treated with CCB in the absence (w/o) or presence of Z-DEVD-FMK, and the percentages of apoptotic cells were quantified by FACS for annexin V and PI staining. Images shown are representative of three independent experiments (left). Bar graph shows mean percentage of early and late apoptotic cells, and values represent the means ± SD. ***p < 0.001.
The molecular connections between autophagy and cell death are complicated, and autophagic cell death induces independently of caspase activity when the apoptotic pathway is blocked36. To further evaluate involvement of autophagic cell death in CCB-induced cell death, CD44highK562 cells were treated with CCB in the presence of Z-DEVD-FMK, a caspase 3 inhibitor, and the degree of cell death was determined by FACS. We found that Z-DEVD-FMK did not completely block CCB-induced cell death, indicating the existence of a caspase-independent type of cell death (Fig. 5B). These results suggest that CCB can induce caspase-independent autophagic cell death, which could enhance apoptotic cell death that can occur in response to treatment with CCB.
Regulation of AMPK, Akt/mTOR/p70S6K/4EBP1, and STAT3 Pathways by NSAIDs
The autophagy-related (Atg) protein Atg7 is necessary for processing LC3-I to activate LC3-II and autophagosome formation37, and phospho-AMP-activated protein kinase (p-AMPK), which serves as an indicator of AMPK activity, is a positive regulator of autophagy38. We examined whether the expressions of Atg7 and p-AMPK could be modulated by treatment of CCB. CD44highK562 cells showed that CCB treatment induced Atg7 expression and phosphorylation of AMPK in a dose-dependent manner, which is involved in the autophagy-inducing ability of CCB (Fig. 6A, top). To elucidate further the potential pathways of CCB involved in the regulation of autophagic responses in CD44highK562 cells, we next examined the effect of CCB on the signaling pathway of Akt/mammalian target of rapamycin (mTOR) and, p70S6K and eukaryotic initiation factor 4E-binding protein 1 (4EBP1), downstream targets of mTOR, in autophagy regulation since inhibition of the Akt/mTOR pathway has been associated with triggering autophagy in cancer cells39,40. Our results showed that the phosphorylation/total protein levels of p-Akt/Akt, p-mTOR/mTOR, p-p70S6K/p70S6K, and p-4EBP/4EBP were decreased in CCB-treated CD44highK562 cells (Fig. 6A, bottom). CCB treatment markedly reduced levels of p-Akt and p-mTOR and subsequently p-P70S6K and p-4EBP in CD44highK562 cells, in a dose-dependent manner. The signal transducer and activator of transcription 3 (STAT3), another downstream signal of mTOR, and myeloid leukemia-1 (Mcl-1) have been reported to play a role in governing regulation of autophagy41. We therefore examined the changed levels of STAT3, phospho-STAT3 (p-STAT3), and Mcl-1 in CD44highK562 cells by NSAIDs. CCB significantly reduced p-STAT3, and Mcl-1, a downstream molecule of STAT3, in a dose-dependent manner. IBU also reduced p-STAT3 and Mcl-1 levels in CD44highK562 cells (Fig. 6B). Next, we investigated the interaction of Mcl-1 with Beclin-1 in CCB-treated CD44highK562 cells by coimmunoprecipitation assay (Fig. 6C). CCB reduced the interaction between Mcl-1 and Beclin-1. These results imply that CCB inhibits the expression of Mcl-1 through STAT3 inactivation, thereby relieving inhibition of Beclin-1 and promoting further formation of autophagosomes to activate autophagy in CD44highK562 cells, indicating that CCB disrupts the Mcl-1/Beclin-1 complex via inhibition of STAT3 signaling pathway. Based on the above results, we suggest that NSAIDs exert their autophagy-inducing effect through activation of AMPK and inhibition of Akt–mTOR–p70S6K–4EBP1 signaling axis and also STAT3/Mcl-1 pathways.
Figure 6.
NSAID-induced activation of AMPK and inhibition of Akt/mTOR signaling and disruption of Beclin-1/Mcl-1 complex via inhibition of STAT3 signaling pathway. (A) CD44highK562 cells were treated with increasing doses of CCB for 36 h, and then levels of indicated molecules were determined by Western blot analysis. (B) CD44highK562 cells were treated with increasing doses of CCB or IBU for 24 h, and the levels of STAT3 and Mcl-1 were determined by Western blot analysis. (C) Mcl-1 or Beclin-1 was immunoprecipitated from CD44highK562 cells treated with 25 μM CCB for 24 h and analyzed for the presence of Mcl-1 or Beclin-1.
Acceleration of 17-AAG-Mediated Autophagy and Downregulation of Stemness-Related Markers by NSAIDs
Since geldanamycin, one of Hsp90 inhibitors, could induce autophagy by modulating LC3-II and p62 levels36,42, we determined whether NSAIDs could modulate Hsp90 inhibitor-mediated autophagy and expression of stemness-related markers, and consequent PARP activity when CD44highK562 cells were cotreated with CCB and 17-AAG (Fig. 7A). CCB significantly accelerated 17-AAG-mediated conversion of LC3-I to LC3-II and reduction in p62 level, indicating that CCB significantly augmented 17-AAG-mediated autophagic ability. Concomitantly, CCB accelerated 17-AAG-mediated reduction in mutp53, c-Myc, CD44, and Oct4. In addition, the expression of P-gp was significantly reduced by CCB alone, and cotreatment of 17-AAG and CCB accelerated P-gp downregulation in CD44highK562 cells. The ability of CCB to downregulate multiple stemness-related markers and P-gp consequently increased PARP activity in 17-AAG-treated cells. Similarly, IBU also accelerated 17-AAG-induced autophagy and a concomitant downregulation of multiple stemness-related markers and reduction in P-gp level, leading to PARP activation in CD44highK562 cells (Fig. 7B). These results suggest that 17-AAG-mediated elimination of CSCs could be accelerated by NSAIDs through autophagic degradation of stemness-related markers and reduction in P-gp level.
Figure 7.
Enhancement of 17-AAG-induced autophagy, acceleration of autophagic degradation of CSC marker, and PARP activation by NSAID. CD44highK562 cells were treated with 17-AAG in the presence or absence of 25 μM CCB (A) or 400 μM IBU (B) for 24 h. The changes in autophagy induction and levels of stemness-related markers, and cleaved PARP (cle PARP) were determined by Western blot analysis.
Suppression of 17-AAG-Mediated Activation of HSF1/Hsp70 by NSAIDs
Acquired resistance to Hsp90 inhibitors has been associated with activation of HSF1 and induction of Hsp7021, and IBU can regulate the expression of Hsp70 in cancer cells23. Therefore, we determined whether the expression level of HSF1/Hsps in CD44highK562 cells could be changed by NSAIDs (Fig. 8A). Treatment of CD44highK562 cells with CCB resulted in a reduction in HSF1 and subsequently downregulated Hsps such as Hsp90, Hsp70, and Hsp27. Similar results were obtained in IBU-treated CD44highK562 cells. Downregulation of HSF1 and subsequent Hsps occurred by IBU treatment. These results suggest that NSAID could block HSF1/Hsps, indicating a possibility for suppression of Hsp90 inhibitor-induced upregulation of Hsp70 by NSAIDs.
Figure 8.
Downregulation of HSF1/Hsps and suppression of 17-AAG-mediated Hsp70 by NSAID. (A) CD44highK562 cells were treated with serial doses of CCB or IBU. (B) CD44highK562 cells were treated with 17-AAG in the presence or absence of 25 μM CCB or 400 μM IBU for 24 h. The changed levels of HSF1/Hsps were determined by Western blot analysis.
Since direct inhibition of Hsp90 leads to disruption of complexes of Hsp90 with HSF1, leading to activation of HSF1/Hsp70, suppression of Hsp90 inhibitor-mediated rebound induction of Hsp70 is required to reverse Hsp90 inhibitor resistance. We therefore examined whether NSAIDs could attenuate Hsp90 inhibitor-mediated HSF1/Hsp70 activation (Fig. 8B). When CD44highK562 cells were treated with 17-AAG alone, the activation of HSF1 was evidenced by an electrophoretic mobility shift, and upregulation of Hsp70, which was attenuated by treatment of CCB or IBU, indicating that NSAIDs have the ability to suppress 17-AAG-mediated Hsp70. Therefore, our results showed that NSAIDs induced autophagic degradation of multiple stemness-related marker proteins, and further NSAIDs in combination with Hsp90 inhibitor attenuated Hsp90 inhibitor-induced Hsp70 induction, which contributes to enhance Hsp90 inhibitor activity of CD44highK562 cells.
In summary, we propose that NSAIDs induce autophagic cell death through inhibition of Akt/mTOR/p70S6K pathway and downregulation of Bcl-2/Mcl-1, which enhances NSAID-induced apoptotic cell death. NSAIDs also trigger autophagy-mediated degradation of multiple stemness-related marker proteins and suppress upregulation of Hsp70 and P-gp, consequently leading to sensitization of CD44highK562 cells to Hsp90 inhibitor (Fig. 9).
Figure 9.
A proposed model of molecular targets of NSAIDs in enhancing Hsp90 inhibitor cytotoxicity of CD44highK562 cells. Inhibition of Hsp90 leads to disruption of regulatory complexes of Hsp90 with HSF1 and client proteins involving mutp53, thereby causing HSF1-mediated induction of Hsp70, which is responsible for the resistance of CD44highK562 cells to Hsp90 inhibitor. NSAIDs inhibit phosphorylation of STAT3 that can upregulate HSF-1, resulting in reduced resistance against Hsp90 inhibitor in the cells. NSAIDs can promote autophagic cell death through inhibition of Akt/mTOR/p70S6K pathway and downregulation of Bcl-2/Mcl-1, which trigger autophagy-mediated degradation of multiple stemness-related marker proteins.
DISCUSSION
CSCs have been isolated from many organs and confirmed to have stem cell-like abilities such as self-renewal, multilineage differentiation potential, and expression of stemness-related markers. Overexpression of stemness-related markers plays an important role in promoting CSC proliferation and the maintenance of the stem cell phenotype, and therefore targeting the suppression of stemness-related markers could be a potential therapeutic approach to eradicating CSCs. In this study, we showed effectiveness of NSAIDs to eliminate chemoresistant CD44highK562 cells expressing high levels of CD44 and other stemness-related markers. Importantly, NSAIDs involving CCB and IBU induced autophagic degradation of stemness-related markers, leading to potentiate 17-AAG-induced cytotoxicity and apoptosis of CD44highK562 cells through activation of autophagic cell death. Indeed, CD44highK562 cells were highly resistant to imatinib as well as 17-AAG20, and we confirmed that combined treatment of imatinib with CCB exerted synergistic cytotoxic effects against CD44highK562 cells (data not shown). These results suggest that NSAIDs could be an effective chemosensitizer. Moreover, cell surface expression of CD44 in CEM/VLB100 and MCF7/MDR cells as well as CD44highK562 cells was significantly decreased by CCB, dose dependently. Similar results were obtained in other CD44-overexpressing variants isolated from human hepatocellular carcinoma cells treated with CCB (unpublished data).
Autophagy is the process by which intracellular contents such as proteins or organelles are degraded through lysosomes. The role of autophagy in cancer is complex, and it can both promote and inhibit tumor development43. Autophagy has a potential role in cell death as a tumor suppressor, and its induction, especially in combination with apoptosis, could be beneficial44. We found that NSAID-induced autophagy occurred, regardless of inhibitory activity of COX-2 since the CCB derivatives such as DMC and OSU-03012 without COX-2 inhibitory activity as well as COX-2-selective CCB induced autophagy. Treatment of CD44highK562 cells with NSAIDs resulted in LC3-II/LC3-I elevation and p62 reduction, which is followed by loss of multiple stemness-related markers involving CD44, Oct4, mutp53, and c-Myc. Similarly, we found that treatment of CD44highK562 cells with autophagy activator rapamycin downregulated stemness-related markers. We further demonstrated that CCB-induced downregulation of p62 and stemness-related markers was prevented by treatment of autophagy inhibitor 3-MA or CQ, and also CCB reduced half-lives of stemness-related marker proteins, indicating that NSAIDs may induce the degradation of stemness-related marker proteins via autophagic process. Indeed, it has been reported that autophagy provides a route for mutp53 degradation24, and lysosomal degradation of CD44 in CD44-expressing cancer stem cells was prevented by autophagy inhibitors45, indicating autophagic degradation of mutp53 and CD44. It was also reported that degradation of c-Myc31,46 and Oct447 was controlled by autophagy. Our data suggest that NSAIDs can promote autophagic degradation of multiple stemness-related markers, which might facilitate elimination of CSCs.
AMP-activated protein kinase (AMPK) serves as a positive regulator of autophagy48. Indeed, NSAIDs can induce autophagy in various pathways, and the most important signaling pathway that controls autophagy is mTOR signaling pathway including PI3K/AKT and AMPK/mTOR signaling axis in various cancers49. Recently, Mcl-1, a downstream target of STAT3, has been reported to have an important role in the regulation of autophagy, and the degradation of Mcl-1 relieves Beclin-150,51, indicating that STAT3/Mcl-1 pathway is also involved in regulation of autophagy. Previously, we also reported that autophagy inducing the ability of NSAIDs through inhibition of Akt/mTOR and STAT3 pathways was effective for the sensitization of MDR cells to 17-AAG52. Similarly, the current study showed that activation of AMPK and inhibition of Akt/mTOR/p70S6K/4EBP1 signaling axis and STAT3/Mcl-1 pathways participate in NSAID-induced autophagy in CD44highK562 cells. CCB and IBU reduced pSTAT3 level and subsequent Mcl-1 expression, which was followed by disruption of the Beclin-1/Mcl-1 complex and promotion of the formation of autophagosomes, indicating that the inactivation of STAT3/Mcl-1 by NSAIDs might be key events in the activation of apoptotic and autophagic cell death. We also showed that CCB induced caspase 3-independent autophagic cell death, and autophagy inhibition significantly blocked CCB-induced apoptosis in CD44highK562 cells. In addition, downregulation of HSF1/Hsp70 expression by STAT3 inhibitor is associated with reduction in Mcl-1 via STAT3 signaling51. Despite their efficacy, the ineffectiveness of Hsp90 inhibitors was involved in activation of HSF1, leading to an increase in the expression of Hsp70, and P-gp-mediated efflux that contribute to resistance to Hsp90 inhibitors20. HSF1 stabilizes mutp53 protein in human cancer cells via activation of Hsp905. Accumulated mutp53 protein is actively involved in resistance to multiple anticancer drugs53. Our data indicated that downregulation of STAT3/Mcl-1 and mutp53 by NSAIDs might be associated with downregulation of HSF1/Hsp70 and P-gp.
NSAIDs are commonly used to manage pain and inflammation, and they have potent antitumor activity in a wide variety of human cancers54. We showed that NSAID-induced suppression of HSF1/Hsp70 blocked 17-AAG-mediated Hsp70 activation, contributing to reverse 17-AAG resistance. Moreover, NSAIDs significantly potentiated 17-AAG-mediated autophagic ability and subsequent downregulation of multiple stem cell markers, and consequently enhanced cell death, resulting in sensitization of CD44highK562 cells to 17-AAG, indicating autophagic effectiveness of CCB for downregulation of multiple stem cell markers might enhance Hsp90 inhibitor-mediated cytotoxicity and apoptosis for eradiating CSCs. Therefore, combined use of NSAIDs and Hsp90 inhibitor may offer one of the most promising strategies for targeting CD44-overexpressing cancer cells involving CSCs.
ACKNOWLEDGMENT
This work was supported by the Basic Science Research Program through the Korean National Research Foundation (NRF) funded by the Ministry of Education (No. 2017R1D1A1A09000515).
Footnotes
The authors declare no conflicts of interest.
REFERENCES
- 1. Chen K, Huang YH, Chen JL. Understanding and targeting cancer stem cells: Therapeutic implications and challenges. Acta Pharmacol Sin. 2013;34:732–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ning XY, Shu JC, Du YQ, Ben QW, Li ZS. Therapeutic strategies targeting cancer stem cells. Cancer Biol Ther. 2013;14:295–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Neckers L, Workman P. Hsp90 molecular chaperone inhibitors: Are we there yet? Clin Cancer Res. 2012;18:64–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wang B, Lee CW, Witt A, Thakkar A, Ince TA. Heat shock factor 1 induces cancer stem cell phenotype in breast cancer cell lines. Breast Cancer Res Treat. 2015;153:57–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Li D, Yallowitz A, Ozog L, Marchenko N. A gain-of-function mutant p53-HSF1 feed forward circuit governs adaptation of cancer cells to proteotoxic stress. Cell Death Dis. 2014;5:e1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Muller P, Hrstka R, Coomber D, Lane DP, Vojtesek B. Chaperone-dependent stabilization and degradation of p53 mutants. Oncogene 2008;27:3371–83. [DOI] [PubMed] [Google Scholar]
- 7. Alexandrova EM, Marchenko ND. Mutant p53-heat shock response oncogenic cooperation: A new mechanism of cancer cell survival. Front Endocrinol. (Lausanne) 2015;6:53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Shetzer Y, Solomon H, Koifman G, Molchadsky A, Horesh S, Rotter V. The paradigm of mutant p53-expressing cancer stem cells and drug resistance. Carcinogenesis 2014;35:1196–208. [DOI] [PubMed] [Google Scholar]
- 9. Vilaboa NE, Galan A, Troyano A, de Blas E, Aller P. Regulation of multidrug resistance 1 (MDR1)/P-glycoprotein gene expression and activity by heat-shock transcription factor 1 (HSF1). J Biol Chem. 2000;275:24970–6. [DOI] [PubMed] [Google Scholar]
- 10. Piper PW, Millson SH. Mechanisms of resistance to Hsp90 inhibitor drugs: A complex mosaic emerges. Pharmaceuticals (Basel) 2011;4:1400–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lee HJ, Kang YH, Lee JS, Byun JH, Kim UK, Jang SJ, Rho GJ, Park BW. Positive expression of NANOG, mutant p53, and CD44 is directly associated with clinicopathological features and poor prognosis of oral squamous cell carcinoma. BMC Oral Health 2015;15(1):153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Signore M, Ricci-Vitiani L, De Maria R. Targeting apoptosis pathways in cancer stem cells. Cancer Lett. 2013;332:374–82. [DOI] [PubMed] [Google Scholar]
- 13. Jendrossek V. Targeting apoptosis pathways by celecoxib in cancer. Cancer Lett. 2013;332:313–24. [DOI] [PubMed] [Google Scholar]
- 14. Waskewich C, Blumenthal RD, Li HL, Stein R, Goldenberg DM, Burton J. Celecoxib exhibits the greatest potency amongst cyclooxygenase (COX) inhibitors for growth inhibition of COX-2-negative hematopoietic and epithelial cell lines. Cancer Res. 2002;62:2029–33. [PubMed] [Google Scholar]
- 15. Akrami H, Aminzadeh S, Fallahi H. Inhibitory effect of ibuprofen on tumor survival and angiogenesis in gastric cancer cell. Tumor Biol. 2015;36:3237–43. [DOI] [PubMed] [Google Scholar]
- 16. Dharmapuri G, Doneti R, Philip GH, Kalle AM. Celecoxib sensitizes imatinib-resistant K562 cells to imatinib by inhibiting MRP1-5, ABCA2 and ABCG2 transporters via Wnt and Ras signaling pathways. Leuk Res. 2015;39:696–701. [DOI] [PubMed] [Google Scholar]
- 17. Takara K, Hayashi R, Kokufu M, Yamamoto K, Kitada N, Ohnishi N, Yokoyama T. Effects of nonsteroidal anti-inflammatory drugs on the expression and function of P-glycoprotein/MDR1 in Caco-2 cells. Drug Chem Toxicol. 2009;32:332–7. [DOI] [PubMed] [Google Scholar]
- 18. Xu HB, Shen FM, Lv QZ. Celecoxib enhanced the cytotoxic effect of cisplatin in drug-resistant human gastric cancer cells by inhibition of cyclooxygenase-2. Eur J Pharmacol. 2015;769:1–7. [DOI] [PubMed] [Google Scholar]
- 19. Zrieki A, Farinotti R, Buyse M. Cyclooxygenase-2 inhibitors prevent trinitrobenzene sulfonic acid-induced P-glycoprotein up-regulation in vitro and in vivo. Eur J Pharmacol. 2010;636:189–97. [DOI] [PubMed] [Google Scholar]
- 20. Kim HB, Lee SH, Um JH, Kim MJ, Hyun SK, Gong EJ, Oh WK, Kang CD, Kim SH. Sensitization of chemo-resistant human chronic myeloid leukemia stem-like cells to Hsp90 inhibitor by SIRT1 inhibition. Int J Biol Sci. 2015;11:923–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kim DY, Kim MJ, Kim HB, Lee JW, Bae JH, Kim DW, Kang CD, Kim SH. Suppression of multidrug resistance by treatment with TRAIL in human ovarian and breast cancer cells with high level of c-Myc. Biochim Biophys Acta 2011;1812:796–805. [DOI] [PubMed] [Google Scholar]
- 22. Arisawa M, Kasaya Y, Obata T, Sasaki T, Nakamura T, Araki T, Yamamoto K, Sasaki A, Yamano A, Ito M, Abe H, Ito Y, Shuto S. Design and synthesis of indomethacin analogues that inhibit P-glycoprotein and/or multidrug resistant protein without Cox inhibitory activity. J Med Chem. 2012;55:8152–63. [DOI] [PubMed] [Google Scholar]
- 23. Kim HB, Lee SH, Um JH, Oh WK, Kim DW, Kang CD, Kim SH. Sensitization of multidrug-resistant human cancer cells to Hsp90 inhibitors by down-regulation of SIRT1. Oncotarget 2015;6:36202–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Choudhury S, Kolukula VK, Preet A, Albanese C, Avantaggiati ML. Dissecting the pathways that destabilize mutant p53 The proteasome or autophagy? Cell Cycle 2013;12:1022–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Foggetti G, Ottaggio L, Russo D, Monti P, Degan P, Fronza G, Menichini P. Gambogic acid counteracts mutant p53 stability by inducing autophagy. Biochim Biophys Acta Mol Cell Res. 2017;1864:382–92. [DOI] [PubMed] [Google Scholar]
- 26. Mizushima N, Yoshimori T. How to interpret LC3 immunoblotting. Autophagy 2007;3:542–5. [DOI] [PubMed] [Google Scholar]
- 27. Bjorkoy G, Lamark T, Pankiv S, Overvatn A, Brech A, Johansen T. Monitoring autophagic degradation of P62/Sqstm1. Methods Enzymol. 2009;452:181–97. [DOI] [PubMed] [Google Scholar]
- 28. Frazier MW, He X, Wang J, Gu Z, Cleveland JL, Zambetti GP. Activation of c-myc gene expression by tumor-derived p53 mutants requires a discrete C-terminal domain. Mol Cell Biol. 1998;18:3735–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Shen W, Chen D, Liu S, Chen L, Yu A, Fu H, Sun X. S100A4 interacts with mutant p53 and affects gastric cancer MKN1 cell autophagy and differentiation. Int J Oncol. 2015;47:2123–30. [DOI] [PubMed] [Google Scholar]
- 30. Cianfanelli V, Fuoco C, Lorente M, Salazar M, Quondamatteo F, Gherardini PF, De Zio D, Nazio F, Antonioli M, D’Orazio M, Skobo T, Bordi M, Rohde M, Dalla Valle L, Helmer-Citterich M, Gretzmeier C, Dengjel J, Fimia GM, Piacentini M, Di Bartolomeo S, Velasco G, Cecconi F. AMBRA1 links autophagy to cell proliferation and tumorigenesis by promoting c-Myc dephosphorylation and degradation. Nat Cell Biol. 2015;17:20–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Puustinen P, Rytter A, Mortensen M, Kohonen P, Moreira JM, Jaattela M. CIP2A oncoprotein controls cell growth and autophagy through mTORC1 activation. J Cell Biol. 2014;204:713–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Cervello M, Bachvarov D, Cusimano A, Sardina F, Azzolina A, Lampiasi N, Giannitrapani L, McCubrey JA, Montalto G. COX-2-dependent and COX-2-independent mode of action of celecoxib in human liver cancer cells. OMICS 2011;15(6):383–92. [DOI] [PubMed] [Google Scholar]
- 33. Schonthal AH. Exploiting cyclooxygenase-(in) dependent properties of COX-2 inhibitors for malignant glioma therapy. Anticancer Agents Med Chem. 2010;10:450–61. [DOI] [PubMed] [Google Scholar]
- 34. Liu J, Qin CK, Lv W, Zhao Q, Qin CY. OSU-03012, a non-cox inhibiting celecoxib derivative, induces apoptosis of human esophageal carcinoma cells through a p53/Bax/cytochrome c/caspase-9-dependent pathway. Anticancer Drugs 2013;24:690–8. [DOI] [PubMed] [Google Scholar]
- 35. Harder LM, Bunkenborg J, Andersen JS. Inducing autophagy A comparative phosphoproteomic study of the cellular response to ammonia and rapamycin. Autophagy 2014;10:339–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yonekawa T, Thorburn A. Autophagy and cell death. Essays Biochem. 2013;55:105–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Komatsu M, Waguri S, Ueno T, Iwata J, Murata S, Tanida I, Ezaki J, Mizushima N, Ohsumi Y, Uchiyama Y, Kominami E, Tanaka K, Chiba T. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J Cell Biol. 2005;169:425–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Song X, Kim SY, Zhang L, Tang D, Bartlett DL, Kwon YT, Lee YJ. Role of AMP-activated protein kinase in cross-talk between apoptosis and autophagy in human colon cancer. Cell Death Dis. 2014;5:e1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Degtyarev M, De Maziere A, Orr C, Lin J, Lee BB, Tien JY, Prior WW, van Dijk S, Wu H, Gray DC, Davis DP, Stern HM, Murray LJ, Hoeflich KP, Klumperman J, Friedman LS, Lin K. Akt inhibition promotes autophagy and sensitizes PTEN-null tumors to lysosomotropic agents. J Cell Biol. 2008;183:101–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sarkar S. Regulation of autophagy by mTOR-dependent and mTOR-independent pathways: Autophagy dysfunction in neurodegenerative diseases and therapeutic application of autophagy enhancers. Biochem Soc Trans. 2013;41:1103–30. [DOI] [PubMed] [Google Scholar]
- 41. You LK, Wang ZG, Li HS, Shou JW, Jing Z, Xie JS, Sui XB, Pan HM, Han WD. The role of STAT3 in autophagy. Autophagy 2015;11:729–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Mori M, Hitora T, Nakamura O, Yamagami Y, Horie R, Nishimura H, Yamamoto T. Hsp90 inhibitor induces autophagy and apoptosis in osteosarcoma cells. Int J Oncol. 2015;46:47–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Muller PA, Vousden KH. Mutant p53 in cancer: New functions and therapeutic opportunities. Cancer Cell 2014;25:304–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Shimizu S, Yoshida T, Tsujioka M, Arakawa S. Autophagic cell death and cancer. Int J Mol Sci. 2014;15:3145–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Haakenson JK, Khokhlatchev AV, Choi YJ, Linton SS, Zhang P, Zaki PM, Fu CL, Cooper TK, Manni A, Zhu JJ, Fox TE, Dong C, Kester M. Lysosomal degradation of CD44 mediates ceramide nanoliposome-induced anoikis and diminished extravasation in metastatic carcinoma cells. J Biol Chem. 2015;290:8632–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Cianfanelli V, Fuoco C, Lorente M, Salazar M, Quondamatteo F, Gherardini PF, De Zio D, Nazio F, Antonioli M, D Orazio M, Skobo T, Bordi M, Rohde M, Dalla Valle L, Helmer-Citterich M, Gretzmeier C, Dengjel J, Fimia GM, Piacentini M, Di Bartolomeo S, Velasco G, Cecconi F. AMBRA1 links autophagy to cell proliferation and tumorigenesis by promoting c-Myc dephosphorylation and degradation. Nat Cell Biol. 2015;17:706. [DOI] [PubMed] [Google Scholar]
- 47. Yang Y, Yu L, Li J, Yuan YH, Wang XL, Yan SR, Li DS, Ding Y. Autophagy regulates the stemness of cervical cancer stem cells. Biologics 2017;11:71–9. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 48. Yang ZF, Klionsky DJ. Mammalian autophagy: Core molecular machinery and signaling regulation. Curr Opin Cell Biol. 2010;22:124–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Yu C, Li WB, Liu JB, Lu JW, Feng JF. Autophagy: Novel applications of nonsteroidal anti-inflammatory drugs for primary cancer. Cancer Med. 2018;7:471–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Tai WT, Shiau CW, Chen HL, Liu CY, Lin CS, Cheng AL, Chen PJ, Chen KF. Mcl-1-dependent activation of Beclin 1 mediates autophagic cell death induced by sorafenib and SC-59 in hepatocellular carcinoma cells. Cell Death Dis. 2013;4:e485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Granato M, Chiozzi B, Filardi MR, Lotti LV, Di Renzo L, Faggioni A, Cirone M. Tyrosine kinase inhibitor tyrphostin AG490 triggers both apoptosis and autophagy by reducing HSF1 and Mcl-1 in PEL cells. Cancer Lett. 2015;366:191–7. [DOI] [PubMed] [Google Scholar]
- 52. Moon HJ, Kim HB, Lee SH, Jeun SE, Kang CD, Kim SH. Sensitization of multidrug-resistant cancer cells to Hsp90 inhibitors by NSAIDs-induced apoptotic and autophagic cell death. Oncotarget 2018;9:11303–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Vakifahmetoglu-Norberg H, Kim M, Xia HG, Iwanicki MP, Ofengeim D, Coloff JL, Pan L, Ince TA, Kroemer G, Brugge JS, Yuan J. Chaperone-mediated autophagy degrades mutant p53. Genes Dev. 2013;27:1718–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Rayburn ER, Ezell SJ, Zhang R. Anti-inflammatory agents for cancer therapy. Mol Cell Pharmacol. 2009;1:29–43. [DOI] [PMC free article] [PubMed] [Google Scholar]