Abstract
miR-223-5p has been demonstrated to regulate the development and progression of various cancers, such as hepatocellular carcinoma, breast cancer, and gastric carcinoma. However, the role of miR-223-5p in non-small cell lung cancer (NSCLC) requires further investigation. In this study, we found that the expression of miR-223-5p was significantly downregulated in NSCLC tissues and cell lines. Moreover, the expression level of miR-223-5p is negatively correlated with the malignance of NSCLC. We found that overexpression of miR-223-5p remarkably suppressed the proliferation of NSCLC cells in vitro and in vivo. miR-223-5p overexpression also led to reduced migration and invasion in NSCLC cells. Mechanistically, we found that E2F8, a key transcription factor involved in many kinds of biological processes, was a direct target gene of miR-223-5p. Overexpression of miR-223-5p significantly decreased the mRNA and protein levels of E2F8 in NSCLC cells. We also showed that restoration of E2F8 rescued the proliferation, migration, and invasion of miR-223-5p-overexpressing NSCLC cells. Taken together, our findings demonstrated that miR-223-5p suppressed NSCLC progression through targeting E2F8.
Key words: miR-223-5p, Non-small cell lung cancer (NSCLC), Growth, Metastasis, E2F8
INTRODUCTION
Lung cancer is one of the most common and aggressive cancers among both men and women1. Non-small cell lung cancer (NSCLC) accounts for almost 80% of all kinds of lung cancers and gives rise to large amounts of cancer-related deaths worldwide every year2. Surgical resection is the main treatment method for NSCLC. Despite some advances in the experimental oncology and treatment approach, the 5-year survival rate of NSCLC patients remains unsatisfactory3. The number of NSCLC patients is quickly increasing year by year. NSCLC is still a big challenging and major problem for public health4. Therefore, a detailed and improved understanding about the underlying pathogenesis of NSCLC development and progression is urgently required.
MicoRNAs (miRNAs) belongs to a class of small noncoding RNAs and possess a length of about 22 nucleotides5. By associating with the complementary sequence of the target 3′-UTR of mRNAs, miRNAs are demonstrated to guide these mRNA for degradation and consequently regulate gene expression in various tissues or cell types6. miRNAs have been shown to exert a wide range of functions, including regulating cell survival, death, and mobility7–10. Because of their extensive expression and vital importance, miRNAs are involved in the development and progression of various cancers, including NSCLC11. Many miRNAs are dysregulated in cancers and are identified as promising biomarkers for tumor diagnosis or prognosis12,13. For example, Chen et al. reported that miR-145 was downregulated in NSCLC and inhibits cancer cell proliferation by targeting c-Myc14. Therefore, determining the relationship between miRNA expression and cancer occurrence will be of particular significance.
Evidence indicates that miR-223 is an important regulator in various cancers, including esophageal carcinoma15, hepatocellular carcinoma16, gastric cancer17, pancreatic cancer18, and chronic lymphocytic leukemia19. The role of miR-223 in NSCLC requires further investigation. In this study, we found that miR-223-5p was significantly downregulated in NSCLC tissues and correlated with clinical characteristics. Moreover, overexpression of miR-223-5p suppressed the proliferation, migration, and invasion of NSCLC cells. Mechanistically, we showed that E2F8 was a target of miR-223-5p and could reverse the effects of miR-223-5p on NSCLC cells. Taken together, our study demonstrated the key role and mechanism of miR-223-5p in NSCLC progression.
MATERIALS AND METHODS
Clinical Specimens and Cell Lines
Thirty-one NSCLC tumors and adjacent normal tissues were obtained from Harbin Medical University. Tissue samples were immediately frozen in liquid nitrogen after resection and stored at −80°C until use. Both tumor and nontumor samples were confirmed by pathological examinations. Patients were excluded if they had recurrent NSCLC or had primary NSCLC but received chemoradiotherapy before surgical operation. This study was approved by the Human Research Ethics Committee of Harbin Medical University. All patients provided written informed consent. The clinical stage was defined according to the revised International Staging System. The clinicopathological characteristics of the NSCLC samples are shown in Table 1.
Table 1.
The Correlation Between miR-233-5p Expression and Clinicopathological Characteristics of NSCLC Patients
| Characteristics | Patients (n = 31) | Low (n = 15) | High (n = 16) | p Value |
|---|---|---|---|---|
| Gender | 1.00 | |||
| Male | 21 | 10 | 11 | |
| Female | 10 | 8 | 8 | |
| Age (years) | 0.458 | |||
| ≤60 | 21 | 9 | 12 | |
| >60 | 10 | 6 | 4 | |
| Differentiation | 0.032 | |||
| Moderate and well | 13 | 3 | 10 | |
| Poor | 16 | 11 | 5 | |
| Metastasis | 0.029 | |||
| Negative | 13 | 3 | 10 | |
| Positive | 18 | 12 | 6 | |
| TNM stage | 0.011 | |||
| I–II | 14 | 3 | 11 | |
| III–IV | 17 | 12 | 5 |
The human NSCLC cell lines A549, H2170 and H1299 and primary human bronchial epithelial cells (NHBE; ScienCell, Research Laboratories, Carlsbad, CA, USA) were grown in RPMI-1640 (Invitrogen, Carlsbad, CA, USA) in the presence of 10% heat-inactivated fetal bovine serum, 100 IU/ml penicillin, and 100 μg/ml streptomycin in a humidified 5% (v/v) atmosphere of CO2 at 37°C. Transfection was performed using Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer’s protocol.
Cell Proliferation Assay
We used Cell Counting Kit-8 (CCK-8; Dojindo Laboratories, Tokyo, Japan) to determine the viability of cells. Cells were plated in 96-well plates at 5,000 cells per well and incubated for 48 h after transfection of 50 nmol/L miR-223-5p mimic or negative control. At the end of incubation, the cell proliferation reagent WST-8 (10 μl) was added to each well and incubated for 3 h at 37°C. Viable cell numbers were estimated by measurement of optical density (OD) at 450 nm.
Transwell Invasion Assay
Cell invasion was evaluated using Chemicon QCM™ 24-Well Collagen-Based Cell Invasion Assay (Millipore, Billerica, MA, USA) according to the manufacturer’s instructions. In brief, 0.3 ml of serum-free medium was added into the interior of each insert to rehydrate the collagen layer for 30 min at room temperature, and then it was replaced with 0.3 ml of prepared serum-free cell suspension containing 3.0 × 105 cells transfected with miR-223-5p mimics or negative control. Medium (0.5 ml) containing 10% FBS was added to the lower chamber. Cells were incubated for 24 h at 37°C. After 24 h, all noninvaded cells were removed from the interior of the insert, and the invaded cells were stained with cell stain. The stained cells were dissolved in extraction buffer, and solutions were transferred to a 96-well culture plate for colorimetric reading of OD at 560 nm. The OD value represents the invasive ability.
Reverse Transcription and Real-Time PCR
Total RNA was extracted from cultured cells using TRIzol reagent (Invitrogen) according to the manufacturer’s protocol, and cDNA was synthesized from total RNA by a PrimerScript RT Reagent Kit (TaKaRa, Shiga, Japan). miRNA from total RNA was reverse transcribed using the PrimeScript miRNA cDNA Synthesis Kit (TaKaRa). Real-Time (RT)-PCR was performed with the SYBR Green Premix Ex Taq II (TaKaRa) on Applied Biosystems Step One Plus Real-Time PCR System (Applied Biosystems, Carlsbad, CA, USA). GAPDH was used as the endogenous control for the detection of mRNA expression level, and U6 was used as endogenous control for miRNA expression analysis.
Luciferase Reporter Assay
The wild-type or mutant 3′-UTRs of E2F8 were designed and prepared by GenePharma Co., Ltd. (Guangzhou, P.R. China) and then cotransfected into the cells with miR-150. Relative luciferase activity was calculated 48 h posttransfection by the dual-luciferase reporter assay (Promega, Madison, WI, USA).
In Vivo Studies
Animal studies were performed according to institutional guidelines. H1299 cells (5.0 × 106) were suspended in 100 μl of phosphate-buffered saline and then injected subcutaneously into either side of the posterior flank of the same female BALB/c athymic nude mouse (Charles River Breeding Laboratories, Wilmington, MA, USA) at 5–6 weeks of age. Tumor growth was examined. After 30 days, the mice were killed, and the tumors were weighed.
Statistical Analysis
Statistical analysis was performed using the SPSS 17.0 (SPSS Inc., Chicago, IL, USA) and done by analysis of variance (ANOVA) or Student’s t-test. Statistical significance was defined as a value of p < 0.05.
RESULTS
Expression Patterns of miR-223-5p in NSCLC Tissues and its Clinical Significance
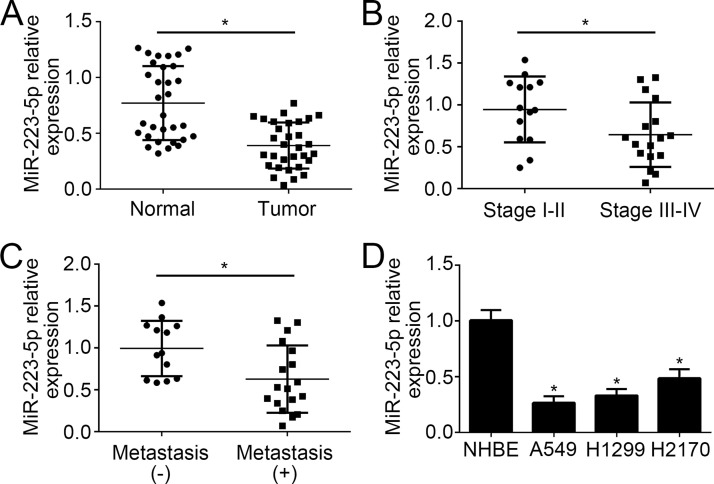
The expression level of miR-223-5p was measured in 31 pairs of NSCLC and adjacent normal tissues through qRT-PCR. The results indicated that miR-223-5p was significantly downregulated in 31 tumor tissues compared to normal tissues (Fig. 1A). According to the TNM stage, we divided these tumor tissues into two groups, namely, stages I–II and stages III–IV. qRT-PCR analysis showed that miR-223-5p was markedly downregulated in samples of stages III–IV compared to that of I–II (Fig. 1B). Moreover, the lower expression of miR-223-5p was observed in metastatic NSCLC tissues than nonmetastatic tissues (Fig. 1C). Consistently, we found that miR-223-5p was downregulated in NSCLC cell lines (H1299, H1299, and H2170 cells) compared to primary human bronchial epithelial cells (NHBE) (Fig. 1D).
Figure 1.
Expression patterns of miR-223-5p in non-small cell lung cancer (NSCLC) tissues and its clinical significance. (A) Relative expression of miR-223-5p in 31 NSCLC tissues and matched adjacent normal tissues. (B) The miR-223-5p expression was significantly downregulated in patients with stages III–IV compared to those with stages I–II. (C) The miR-223-5p expression was significantly lower in patients with distal metastasis than in those without. (D) Relative expression of miR-223-5p in NSCLC cell lines was measured by quantitative reverse transcription and real-time (qRT)-PCR. *p < 0.05.
miR-223-5p Impaired the Proliferation, Migration, and Invasion of NSCLC Cells
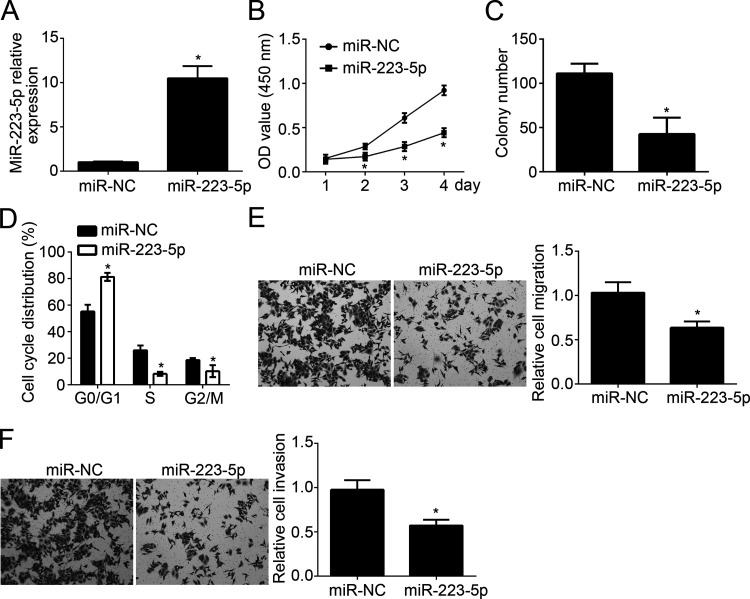
To explore the function of miR-223-5p in NSCLC, we overexpressed miR-223-5p in H1299 cells. qRT-PCR showed that miR-223-5p was significantly overexpressed in H1299 cells transfected with miR-223-5p mimics (Fig. 2A). Then CCK-8 and colony formation assays were conducted to check cellular proliferation. We found that overexpression of miR-223-5p significantly reduced the proliferation of H1299 cells and the colony number (Fig. 2B and C). Cell cycle is directly linked to proliferation. Thus, we checked the cell cycle distribution and found that overexpression of miR-223-5p significantly inhibited the cells in the GS and G2/M phases (Fig. 2D). Lung cancer metastasis leads to the poor outcomes of patients. To further analyze the effect of miR-223-5p on tumor metastasis, we performed Transwell assay. Results indicated that overexpression of miR-223-5p dramatically repressed the numbers of migrated and invaded cells (Fig. 2E and F). These data suggested that miR-223-5p served as a tumor suppressor in NSCLC.
Figure 2.
miR-223-5p impaired the proliferation, migration, and invasion of NSCLC cells. (A) Relative expression of miR-223-5p in H1299 cells transfected with miR-223-5p mimics or control. (B, C) Cellular proliferation of H1299 cells was determined by Cell Counting Kit-8 (CCK-8) and colony formation assay. (D) Cell cycle distribution was measured by FACS. (E, F) Cell migration and invasion of H1299 cells were determined by Transwell assay. *p < 0.05.
E2F8 Was a Target of miR-223-5p
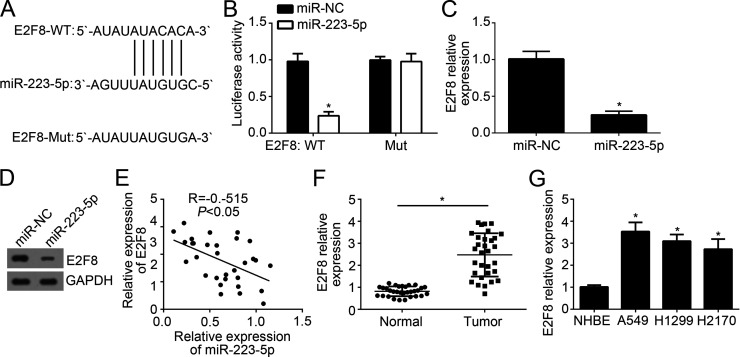
Further analysis by informatics, we found that E2F8 may be a target of miR-223-5p. We identified a potential binding site with miR-223-5p in the 3′-UTR region of E2F8 mRNA (Fig. 3A). To validate it, we constructed wild-type (WT) and mutant (Mut) luciferase reporter plasmid. Through luciferase reporter assay, we showed that overexpression of miR-223-5p significantly inhibited that luciferase activity in H1299 cells transduced with WT luciferase reporter plasmid (Fig. 3B). We found that ectopic expression of miR-223-5p effectively downregulated the mRNA and protein levels of E2F8 in H1299 cells (Fig. 3C and D). Furthermore, qRT-PCR analysis indicated that there was a negative correlation between the expression of miR-223-5p and E2F8 in NSCLC tissues (Fig. 3E). Moreover, to determine the role of E2F8, we analyzed the expression of E2F8 in NSCLC tissues and cell lines and found that E2F8 was upregulated in tumor tissues and cell lines (Fig. 3F and G).
Figure 3.
E2F8 was a target of miR-223-5p. (A) Diagram of predicted binding site of miR-223-5p in the 3′-UTR of E2F8 mRNA. (B) Luciferase reporter assay indicated that overexpression of miR-223-5p repressed the luciferase activity in H1299 cells transfected with wild-type (WT)-E2F8-3′-UTR. (C, D) qRT-PCR and Western blot analysis indicated that overexpression of miR-223-5p inhibited the mRNA and protein levels of E2F8 in H1299 cells. (E) Correlation of miR-223-5p expression with E2F8 level in NSCLC tissues. (F) Relative expression of E2F8 in NSCLC tissues and adjacent normal tissues. (G) Relative expression of E2F8 in NSCLC cell lines by qRT-PCR. *p < 0.05.
Overexpression of E2F8 Alleviated the Effects of miR-223-5p
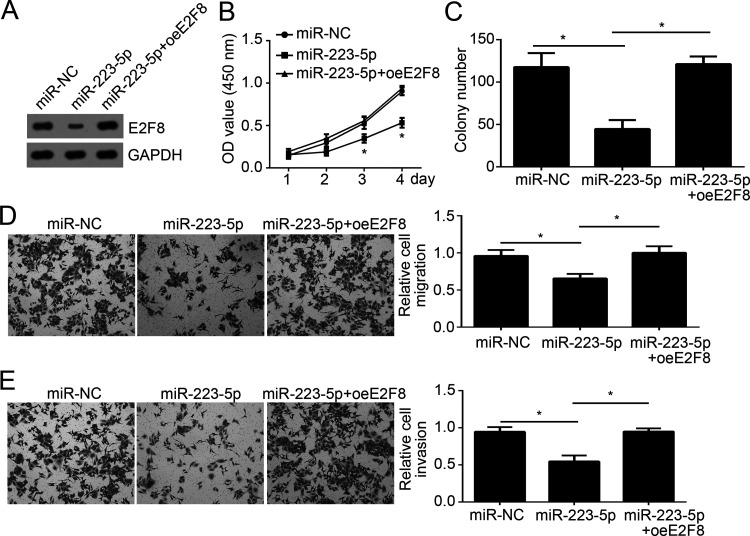
Then in order to determine whether E2F8 expression is responsible for miR-223-5p-mediated effects on NSCLC cells, we rescued the protein levels of E2F8 in H1299 cells transfected with miR-223-5p mimics. The Western blot indicated that E2F8 was significantly upregulated in H1299 cells (Fig. 4A). CCK-8 and colony formation assays indicated that overexpression of E2F8 promoted the proliferation and colony number (Fig. 4B and C). Moreover, the Transwell assay indicated that restoration of E2F8 enhanced the migration and invasion of H1299 cells transfected with miR-223-5p (Fig. 4D and E). Taken together, these data indicated that E2F8 downregulation was indispensable for miR-223-5p-mediated inhibition on NSCLC cell proliferation, migration, and invasion.
Figure 4.
Overexpression of E2F8 alleviated the effects of miR-223-5p. (A) Western blot indicated that the expression of E2F8 was restored in H1299 cells. (B, C) CCK-8 and colony formation assays showed that overexpression of E2F8 restored the proliferation ability of H1299 cells. (D, E) Transwell assay was used to evaluate the migration and invasion of H1299 cells. *p < 0.05.
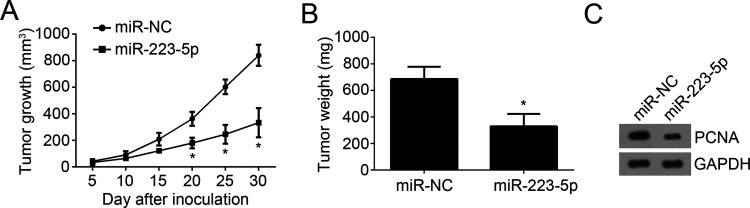
Effect of miR-223-5p on NSCLC Cell Proliferation In Vivo
We then assessed the effects of miR-223-5p on NSCLC cell proliferation in vivo by a xenograft model. We injected H1299 cells transfected with miR-223-5p mimics or control into nude recipient mice. Every other 5 days, we measured the tumor volumes and found that overexpression of H1299 significantly inhibited the tumor growth (Fig. 5A). Moreover, the tumor weight was determined after 30 days. We found that the weights of tumors derived from miR-223-5p-overexpressed H1299 cells were lighter than those of the control group (Fig. 5B). Furthermore, the proliferation of tumor tissues was measured by analysis of PCNA expression. As shown in Figure 5C, overexpression of miR-223-5p significantly decreased the expression of PCNA in tumor tissues. Summarily, our data demonstrated that miR-223-5p suppressed NSCLC progression by targeting E2F8 in vitro and in vivo.
Figure 5.
Effect of miR-223-5p on NSCLC cell proliferation in vivo. (A) Tumor volume was measured every 5 days. (B) Tumor weight was determined at the end of experiment. (C) The protein level of PCNA was measured in the tumor tissues by Western blot. *p < 0.05.
DISCUSSION
In the past decades, much effort has been made to investigate the biological functions of miRNAs. Increasing evidence demonstrates the essential role of miRNAs in various biological processes and implies the close correlation between miRNA expression and human cancers20. Until now, the functions of only a small fraction of miRNAs have been elucidated. The mechanisms of most miRNAs in the regulation of tumorigenesis, cell proliferation, angiogenesis, tumor metastasis, and chemoresistance remain largely unknown. Reports suggest that miRNAs are effective biomarkers for tumor diagnosis and prognosis21,22. Also, some studies indicate that miRNAs are promising therapeutic targets for cancer intervention23. Therefore, the research on miRNA functions is of clinical importance. In this study, we investigated the function and mechanism of miR-223-5p on NSCLC progression.
miR-223 has been shown to be dysregulated in a diversity of cancers, including esophageal carcinoma15, hepatocellular carcinoma16, gastric cancer17, pancreatic cancer18, chronic lymphocytic leukemia19, prostate cancer24,25, breast cancer26, osteosarcoma27, nasopharyngeal carcinoma28, and colorectal cancer29. Additionally, some reports indicated that miR-223 overexpression leads to poor prognosis in specific cancers30. A previous study also indicated that miR-223-3p inhibited NSCLC cell proliferation and induced apoptosis31. In our study, we found that miR-223-5p was downregulated in NSCLC tissues compared with adjacent normal tissues. Moreover, we indicated that miR-223-5p expression was correlated with tumor progression and metastasis. As we have shown, miR-223-5p was expressed more lowly in NSCLC tissues of stages III–IV or metastasis. By CCK-8 and colony formation assay, we demonstrated that overexpression of miR-223-5p suppressed cellular proliferation. We validated the inhibition of miR-223-5p on proliferation by in vivo assay. Moreover, we found that overexpression of miR-223-5p significantly inhibited cell migration and invasion, which has not been reported previously. Therefore, our study, for the first time, indicated the correlation between miR-223-5p expression and NSCLC progression.
E2F8 belongs to the E2F transcription factor family that is essential for the regulation of cell cycle progression32. Increasing evidence indicates that E2F8 acts as an oncogene in various tumors, such as hepatocellular carcinoma33, lung cancer34, breast cancer35, prostate cancer36, and ovarian cancer37. Furthermore, Sun et al. reported that E2F8 targeted by miR-144 promotes papillary thyroid cancer progression via regulating cell cycle32. E2F8 has been acknowledged to regulate cell cycle and tumor progression32. In our study, we found that E2F8 was a direct target of miR-223-5p in NSCLC cells. We showed that overexpression of miR-223-5p significantly inhibited the mRNA and protein levels of E2F8 in NSCLC cells. Moreover, we found that there was an inverse relationship between the expression of miR-223-5p and E2F8 in NSCLC tissues. We also found that restoration of E2F8 in miR-223-5p-overexpressing NSCLC cells significantly promoted the proliferation, migration, and invasion of cancer cells. Our data, for the first time, demonstrated the correlation of miR-223-5p with E2F8 in NSCLC cells.
In summary, our study demonstrated that miR-223-5p suppressed the proliferation, migration, and invasion of NSCLC cells through direct inhibition of E2F8 expression. Therefore, our findings will contribute to the knowledge about the molecular mechanism of NSCLC progression.
ACKNOWLEDGMENT
The authors declare no conflicts of interest.
Footnotes
The authors declare no conflicts of interest.
REFERENCES
- 1. Jemal A, Siegel R, Ward E, Hao YP, Xu JQ, Thun MJ. Cancer statistics, 2009. CA Cancer J Clin. 2009;59(4):225–49. [DOI] [PubMed] [Google Scholar]
- 2. Jemal A, Siegel R, Xu JQ, Ward E. Cancer statistics, 2010. CA Cancer J Clin. 2010;60(5):277–300. [DOI] [PubMed] [Google Scholar]
- 3. Verdecchia A, Francisci S, Brenner H, Gatta G, Micheli A, Mangone L, Kunkler I. Recent cancer survival in Europe: A 2000-02 period analysis of EUROCARE-4 data. Lancet Oncol. 2007;8(9):784–96. [DOI] [PubMed] [Google Scholar]
- 4. Mercier O, Fadel E, de Perrot M, Mussot S, Stella F, Chapelier A, Dartevelle P. Surgical treatment of solitary adrenal metastasis from non-small cell lung cancer. J Thorac Cardiovasc Surg. 2005;130(1):136–40. [DOI] [PubMed] [Google Scholar]
- 5. Valencia-Sanchez MA, Liu JD, Hannon GJ, Parker R. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev. 2006;20(5):515–24. [DOI] [PubMed] [Google Scholar]
- 6. Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004;116(2):281–97. [DOI] [PubMed] [Google Scholar]
- 7. Pogue AI, Cui JG, Li YY, Zhao Y, Culicchia F, Lukiw WJ. Micro RNA-125b (miRNA-125b) function in astrogliosis and glial cell proliferation. Neurosci Lett. 2010;476(1):18–22. [DOI] [PubMed] [Google Scholar]
- 8. Verduci L, Simili M, Rizzo M, Mercatanti A, Evangelista M, Mariani L, Rainaldi G, Pitto L. MicroRNA (miRNA)-mediated interaction between leukemia/lymphoma-related factor (LRF) and alternative splicing factor/splicing factor 2 (ASF/SF2) affects mouse embryonic fibroblast senescence and apoptosis. J Biol Chem. 2010;285(50):39551–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Baranwal S, Alahari SK. miRNA control of tumor cell invasion and metastasis. Int J Cancer 2010;126(6):1283–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zavala-Yoe R, Ramirez-Mendoza RA, Cordero LM. Entropy measures to study and model long term simultaneous evolution of children in Doose and Lennox-Gastaut syndromes. J Integra Neurosci. 2016;15(2):205–21. [DOI] [PubMed] [Google Scholar]
- 11. Gallach S, Jantus-Lewintre E, Montaner D, Uso M, Sanmartin E, Sirera R, Blasco A, Guijarro R, Martorell M, Camps C. miRNA profiling in respectable NSCLC by multiplex next-generation sequencing. J Clin Oncol. 2012;30(15):7060. [Google Scholar]
- 12. Rubio-Briones J, Fernandez-Serra A, Casanova-Salas I, Casanova J, Rubio-Martinez L, Soriano P, Garcia-Casado Z, Iborra I, Solsona E, Lopez-Guerrero JA. MiRNA profiling in the screening of potential biomarkers for prostate cancer diagnosis. Eur Urol Suppl. 2012;11(1):E541–U167. [Google Scholar]
- 13. Pichler M, Winter E, Stotz M, Eberhard K, Samonigg H, Lax S, Hoefler G. Down-regulation of KRAS-interacting miRNA-143 to predict prognosis and response to EGFR-targeted agents in colorectal cancer. Br J Cancer 2012;106(11):1826–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chen Z, Zeng HZ, Guo Y, Liu P, Pan H, Deng AM, Hu JA. miRNA-145 inhibits non-small cell lung cancer cell proliferation by targeting c-Myc. J Exp Clin Cancer Res. 2010;29:151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Li SJ, Li ZG, Guo FJ, Qin XB, Liu B, Lei Z, Song ZQ, Sun LY, Zhang HT, You JC, Zhou QH. miR-223 regulates migration and invasion by targeting Artemin in human esophageal carcinoma. J Biomed Sci. 2011;18:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wong QWL, Lung RWM, Law PTY, Lai PBS, Chan KYY, To KF, Wong N. MicroRNA-223 is commonly repressed in hepatocellular carcinoma and potentiates expression of Stathmin1. Gastroenterology 2008;135(1):257–69. [DOI] [PubMed] [Google Scholar]
- 17. Li XH, Zhang Y, Zhang HW, Liu XN, Gong TQ, Li MB, Sun L, Ji G, Shi YQ, Han ZY, Han S, Nie YZ, Chen X, Zhao QC, Ding J, Wu KC, Daiming F. miRNA-223 promotes gastric cancer invasion and metastasis by targeting tumor suppressor EPB41L3. Mol Cancer Res. 2011;9(7):824–33. [DOI] [PubMed] [Google Scholar]
- 18. Ma J, Fang BB, Zeng FP, Ma C, Pang HJ, Cheng L, Shi Y, Wang H, Yin B, Xia J, Wang ZW. Down-regulation of miR-223 reverses epithelial-mesenchymal transition in gemcitabine-resistant pancreatic cancer cells. Oncotarget 2015;6(3):1740–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Stamatopoulos B, Meuleman N, Haibe-Kains B, Saussoy P, Van Den Neste E, Michaux L, Heimann P, Martiat P, Bron D, Lagneaux L. microRNA-29c and microRNA-223 down-regulation has in vivo significance in chronic lymphocytic leukemia and improves disease risk stratification. Blood 2009;113(21):5237–45. [DOI] [PubMed] [Google Scholar]
- 20. Ju JF, Jiang JT, Fesler A. miRNA: The new frontier in cancer medicine. Future Med Chem. 2013;5(9):983–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Izquierdo L, Ingelmo M, Mallofre C, Lozano JJ, Verhasselt-Crinquette M, Leroy X, Colin P, Comperat E, Roupret M, Alcaraz A, Mengual L. Development of miRNA signature for the prognosis of upper urinary tract tumors. J Urol. 2013;189(4):E371–2. [Google Scholar]
- 22. Ratert N, Meyer HA, Jung M, Lioudmer P, Mollenkopf HJ, Wagner I, Miller K, Kilic E, Erbersdobler A, Weikert S, Jung K. miRNA profiling identifies candidate miRNAs for bladder cancer diagnosis and clinical outcome. J Mol Diagn. 2013;15(5):695–705. [DOI] [PubMed] [Google Scholar]
- 23. Weidle UH, Birzele F, Kollmorgen G, Nopora A. Potential microRNA-related targets for therapeutic intervention with ovarian cancer metastasis. Cancer Genomics Proteomics 2018;15(1):1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wei YB, Yang JR, Yi L, Wang YH, Dong ZT, Liu ZT, Ou-Yang SF, Wu HT, Zhong ZH, Yin Z, Zhou KQ, Gao YL, Yan B, Wang Z. MiR-223-3p targeting SEPT6 promotes the biological behavior of prostate cancer. Sci Rep. 2014;4:7546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kurozumi A, Goto Y, Matsushita R, Fukumoto I, Kato M, Nishikawa R, Sakamoto S, Enokida H, Nakagawa M, Ichikawa T, Seki N. Tumor-suppressive microRNA-223 inhibits cancer cell migration and invasion by targeting ITGA3/ITGB1 signaling in prostate cancer. Cancer Sci. 2016;107(1):84–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pinatel EM, Orso F, Penna E, Cimino D, Elia AR, Circosta P, Dentelli P, Brizzi MF, Provero P, Taverna D. miR-223 is a coordinator of breast cancer progression as revealed by bioinformatics predictions. PLoS One 2014;9(1):e84859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Xu JL, Yao Q, Hou Y, Xu M, Liu S, Yang LQ, Zhang L, Xu H. MiR-223/Ect2/p21 signaling regulates osteosarcoma cell cycle progression and proliferation. Biomed Pharmacother. 2013;67(5):381–6. [DOI] [PubMed] [Google Scholar]
- 28. Yang WY, Lan X, Li DM, Li T, Lu SM. MiR-223 targeting MAFB suppresses proliferation and migration of nasopharyngeal carcinoma cells. BMC Cancer 2015;15:461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Li ZW, Yang YM, Du LT, Dong ZG, Wang LL, Zhang X, Zhou XJ, Zheng GX, Qu AL, Wang CX. Overexpression of miR-223 correlates with tumor metastasis and poor prognosis in patients with colorectal cancer. Med Oncol. 2014;31(11):256. [DOI] [PubMed] [Google Scholar]
- 30. Wang H, Wang L, Wu Z, Sun R, Jin HF, Ma JF, Liu LL, Ling R, Yi J, Wang L, Bian JF, Chen JH, Li NL, Yuan SF, Yun J. Three dysregulated microRNAs in serum as novel biomarkers for gastric cancer screening. Med Oncol. 2014;31(12):298. [DOI] [PubMed] [Google Scholar]
- 31. Zhou Z, Meng ZL, Hong YQ, Xiong YQ. miR-223 inhibits tumor development of non-small cell lung cancer and sensitizes cancer cells to gefitinib via targeting E2F1. Int J Clin Exp Pathol. 2017;10(3):2723–33. [Google Scholar]
- 32. Sun J, Shi R, Zhao S, Li XN, Lu S, Bu HM, Ma XH, Su C., E2F8, a direct target of miR-144, promotes papillary thyroid cancer progression via regulating cell cycle. J Exp Clin Cancer Res. 2017;36(1):40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Deng Q, Wang Q, Zong WY, Zheng DL, Wen YX, Wang KS, Teng XM, Zhang X, Huang JA, Han ZG. E2F8 contributes to human hepatocellular carcinoma via regulating cell proliferation. Cancer Res. 2010;70(2):782–91. [DOI] [PubMed] [Google Scholar]
- 34. Park SA, Platt J, Lee JW, Lopez-Giraldez F, Herbst RS, Koo JS. E2F8 as a novel therapeutic target for lung cancer. J Natl Cancer Inst. 2015;107(9):pii:djv151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ye LP, Guo L, He ZY, Wang X, Lin CY, Zhang X, Wu S, Bao Y, Yang Q, Song LB, Lin HX. Upregulation of E2F8 promotes cell proliferation and tumorigenicity in breast cancer by modulating G1/S phase transition. Oncotarget 2016;7(17):23757–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lee S, Park YR, Kim SH, Park EJ, Kang MJ, So I, Chun JN, Jeon JH. Geraniol suppresses prostate cancer growth through down-regulation of E2F8. Cancer Med. 2016;5(10):2899–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Petru E, Moinfar F, Graf S. Regulation of the E2F family—A further step in understanding ovarian cancer biology. Cancer Biol Ther. 2006;5(7):777–8. [DOI] [PubMed] [Google Scholar]