Abstract
LACTB, a mitochondrial protein, was ubiquitously expressed in different mammalian tissues, such as liver, heart, and skeletal muscle. It has been shown that LACTB is downexpressed in breast cancers, and it suppresses the proliferation and promotes the apoptosis of breast cancers. However, its role in the progression and prognosis of glioma remains unknown. In this study, we analyzed the clinicopathological features and outcomes of LACTB expression in 98 glioma patients and investigated the effects of LACTB overexpression on the proliferation, invasion, and angiogenesis of glioma cells in vitro. We observed a significant decrease in LACTB expression in glioma, and downexpression of LACTB is correlated with a poor prognosis of glioma patients. Moreover, Cox regression analysis reveals that the LACTB is an independent prognostic indicator for glioma patients. Overexpression of LACTB could suppress the proliferation, invasion, and angiogenesis of glioma cells. In addition, overexpression of LACTB could inhibit the expression of PCNA, MMP2, MMP9, and VEGF. Taken together, these data indicate that LACTB may serve as a promising therapeutic target for gliomas.
Key words: LACTB, Glioma, Proliferation, Invasion, Angiogenesis, Prognosis
INTRODUCTION
Gliomas, accounting for 80% of all malignant brain tumors and resulting in poor prognosis, are the most common brain tumor1–3. Despite the great advances in neurosurgery, radiotherapy, and chemotherapies that have been made during the past decades, the median survival duration for patients with glioma is less than 12–15 months4–6. Considering that glioma is a heterogeneous disease, with the abnormalities of oncogenes and tumor suppressor genes7, there is an urgent need to identify novel diagnostic and prognostic markers during the development of gliomas to help in providing an effective treatment method and enhancing the prognosis of glioma patients.
LACTB (lactamase β) is a mitochondrial protein that is widely expressed in various mammalian tissues8–12. Previous research reported that LACTB could promote intramitochondrial membrane organization and regulate complex I of the mitochondrial electron transport chain and cellular metabolic processes13–15. Overexpression of LACTB in mouse resulted in an obese phenotype14. More recently, it was reported that LACTB positively contributed to the apoptosis and suppression of the proliferation of breast cancer cells16. However, up to now, the expression of LACTB in gliomas and its clinicopathological significance have not been elucidated. Therefore, to explore the role of LACTB in gliomas is greatly significant.
In this study, we demonstrated the downregulation of LACTB in gliomas and analyzed the relationship between clinicopathological characteristics and glioma patients’ survival. Next, we then investigated the effects of overexpressing LACTB on the proliferation, invasion, and angiogenesis of glioma cells in vitro, which suggested that LACTB might play a critical role in glioma progression and might function as a novel prognostic factor and a promising treatment target for glioma.
MATERIALS AND METHODS
Cell Culture and Reagents
Human glioma cell lines were purchased from Type Culture Collection of the Chinese Academy of Sciences. All cell lines were maintained in Dulbecco’s modified Eagle’s medium (DMEM) with 10% fetal bovine serum (FBS), 100 mg/ml streptomycin, and 100 U/ml penicillin. The cells were cultured at 37°C and 5% CO2 in a humidified atmosphere. Every 2–3 days, the cells were subcultured in 1 mM EDTA and 0.25% trypsin. The cells were routinely screened and were found to be free of mycoplasma. Antibodies against LACTB were obtained from Abcam (Cambridge, MA, USA); PCNA, MMP2, MMP9, VEGF, p-AKT, t-AKT, p-STAT3, and t-STAT3 were obtained from Cell Signaling Technology (Danvers, MA, USA). GAPDH was purchased from Beyotime (Shanghai, P.R. China).
Patients and Tissue Preparation
Specimens of 98 gliomas and 10 nontumor brains were obtained from The First People’s Hospital of Chongqing Liang Jiang New Area between 2009 and 2015. Dissected samples were preserved in formalin immediately after surgery and stored at room temperature. Approval from The First People’s Hospital of Chongqing Liang Jiang New Area Ethics Committee was obtained before the research. All experimental protocols were carried out in accordance with the guidelines approved by The First People’s Hospital of Chongqing Liang Jiang New Area Ethics Committee. Informed consent was obtained from all of the patients in this study.
Immunohistochemistry
Immunohistochemistry (IHC) was performed as previously described17. The IHC results were independently evaluated in a blinded manner by two pathologists. The signal was assessed by a semiquantitative scoring system, which represented the percentage of positive tumor cells and the intensity of staining. The intensity of staining of cells was scored and graded as follows: 0 (negative), 1 (faint yellow), 2 (yellow or deep yellow), and 3 (tan or brown). The proportion score according to the percentage of positively stained cells was as follows: 0 (0%–25%), 1 (26%–50%), 2 (51%–75%), and 3 (76%–100%). The expression of LACTB was measured by multiplying the staining intensity by the percentage of positively stained cells. The samples with a final score ≥5 were defined as high expression, and those with a final score <5 were defined as low expression.
Cell Transfections
LACTB lentivirus used for LACTB overexpression, corresponding negative control lentivirus, lentiviral shRNA, and matched empty lentivirus were purchased from GeneChem Co. (Shanghai, P.R. China). For cell infection, viral supernatants and 8 μg/ml polybrene were added to the culture medium, and the medium was incubated for 24 h. The target sequence of sh-LACTB was 5′-GATTACTGATTTCCCATTTAA-3′. Green fluorescence was detected 96 h later, and the expression of LACTB was verified by Western blot and real-time reverse transcription quantitative PCR (RT-qPCR).
Real-Time PCR
Total RNA was extracted from tissues and cells using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) and reverse transcribed to cDNA using a Revert-Aid™ First-Strand cDNA synthesis kit (Fermentas, Waltham, MA, USA) following the supplier’s instructions. The primer sequence and product size for LACTB were 5′-CACGGTTCTCAACATCAGCA-3′ (forward) and 5′-GCACAGGATCAAGGATGAGG-3′ (reverse), 179 base pairs (bp), and for GAPDH, which was used as a standard, was 5′-ACATCAAGAAGGTGGTGA AG-3′ (forward) and 5′-ATACCAGGAAATGAGCTTGA-3′ (reverse), 173 bp. All primers were synthesized by Takara (Dalian, P.R. China).
Western Blot
Total proteins were extracted from tissues and cells. From each sample, 30 μg of total protein lysates was loaded onto a 10% acrylamide gel, and SDS-PAGE was performed, before being transferred to polyvinylidine difluoride membranes (Millipore, Billerica, MA, USA). The membrane was blocked in 5% nonfat milk and incubated with primary antibody overnight at 4°C. The membranes were washed three times with TBST buffer, followed by incubation with secondary antibody. Then the membranes were washed three times with TBST buffer and quantified using the Quantity One 4.6 computer software.
Cell Proliferation, Invasion, and Tube Formation Assay
The proliferation, invasion, and tube formation were determined as previously described17,18.
ELISA Assay
Concentration of VEGF in supernatants of glioma cells was tested by VEGF ELISA kit according to the manufacturer’s instructions (R&D, Minneapolis, MN, USA).
The Gene Expression Omnibus and The Cancer Genome Atlas Analysis
Data of patients with glioma were downloaded from the Gene Expression Omnibus (GEO) and The Cancer Genome Atlas (TCGA) data portal website (http://cancergenome.nih.gov). Clinical data, including survival time, age, gender, grade, and Karnofsky performance score (KPS), were also obtained from TCGA. Patients with incomplete clinical data were excluded from this analysis.
Statistical Analysis
All of the statistical analyses were performed using the SPSS 17.0 software package. Statistical differences among groups were analyzed by ANOVA, Kruskal–Wallis, t-test, or chi-square test. The prognostic significance analysis was performed using the Kaplan–Meier method and log rank tests. Cox’s proportional hazards regression model was used for multivariate survival analysis. A value of p < 0.05 was considered significant. All data are presented as mean ± standard deviation (SD).
RESULTS
Downexpression of LACTB in Gliomas
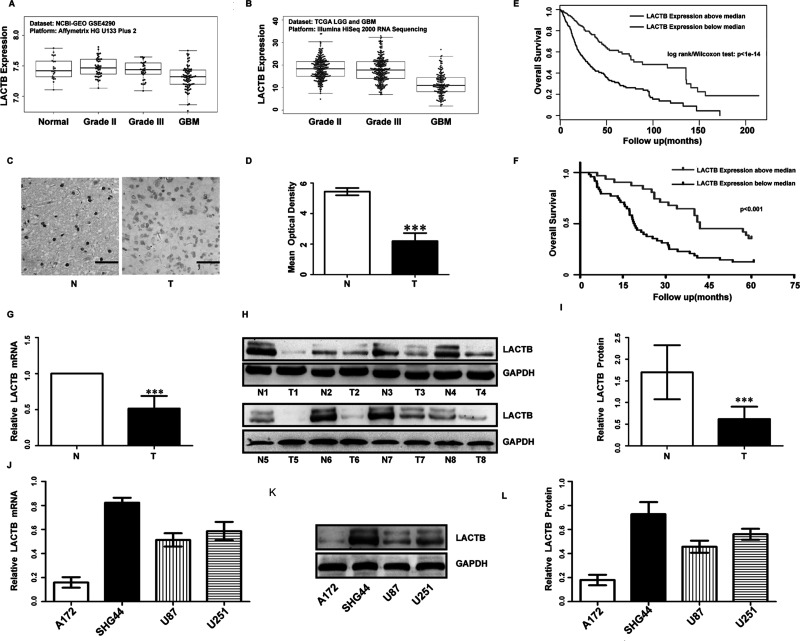
We analyzed the data from the GEO and the glioma cohort of TCGA, and we found that expression of LACTB was markedly decreased in glioma tissues compared with normal tissues, and it was lower with the increase in grade of glioma (Fig. 1A and B). Then we also tested LACTB expression in 98 glioma specimens and 10 normal brain tissues by IHC. The expression level of LACTB in the gliomas was significantly decreased compared with that in the normal brain tissues (Fig. 1C and D), which is in accordance with the result of RT-PCR (Fig. 1G) and Western blot (Fig. 1H and I).
Figure 1.
Low lactamase β (LACTB) expression predicted a worse prognosis for patients with glioma. LACTB expression in glioma tissues and normal brain tissues from the Gene Expression Omnibus (GEO) (A) and The Cancer Genome Atlas (TCGA) (B) data sets. LACTB expression in 98 glioma tissues and 10 normal brain tissues was tested by immunohistochemistry (IHC; scale bars: 50 μm, 400×) (C) and statistical quantification of the mean optical density (D) in normal brain tissues and glioma tissues (*p < 0.001). Survival analysis of patients with glioma from the TCGA data sets (E) and our data (F). Real-time reverse transcription quantitative PCR (RT-qPCR) (G) and Western blot analysis (H, I) of LACTB in eight glioma tissues and paired adjacent nontumor tissues. ***p < 0.001. RT-qPCR (J) and Western blot analysis (K, L) of LACTB in four glioma cell lines.
LACTB Expression Related to Glioma Patients’ Survival
The relationship between LACTB expression levels and the clinicopathologic features in 98 glioma patients is shown in Table 1. Chi-square test revealed that the decreased expression of LACTB was significantly correlated with KPS and World Health Organization (WHO) grade. There were no statistically significant differences in gender or age according to the staining results (data not shown). Kaplan–Meier survival curves demonstrated that downexpression of LACTB was closely related to poor overall survival (Fig. 1E and F). Furthermore, univariate and multivariate analyses indicated that KPS and LACTB expression were two prognostic factors of gliomas (data not shown).
Table 1.
Association of LACTB Expression With the Clinical Parameters
| Groups | Total | LACTB Expression | p | |
|---|---|---|---|---|
| Low | High | |||
| Gender | 0.18 | |||
| Male | 56 | 15 | 39 | |
| Female | 42 | 16 | 26 | |
| Age (years) | 0.58 | |||
| ≤60 | 49 | 14 | 35 | |
| >60 | 49 | 17 | 32 | |
| WHO grade | <0.001 | |||
| I–II | 40 | 18 | 22 | |
| III–IV | 58 | 13 | 45 | |
| KPS | 0.005 | |||
| ≤70 | 49 | 27 | 22 | |
| >70 | 49 | 4 | 45 | |
WHO, World Health Organization; KPS, Karnofsky performance score.
LACTB Inhibits the Proliferation of Glioma Cells
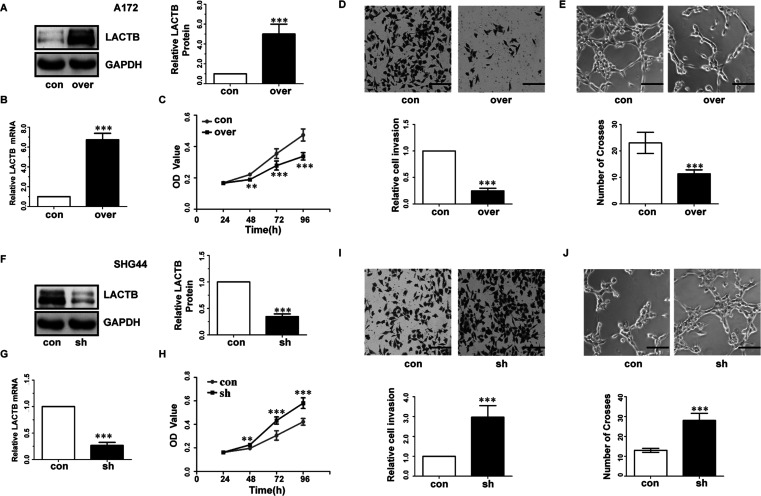
Next, Western blot and RT-qPCR were performed to examine LACTB expression levels in glioma cell lines (Fig. 1J–L). To further investigate the functions of the LACTB in glioma cells, the SHG44 and A172 cell lines were selected for further experimentation. We introduced the LACTB gene into the A172 cells using LACTB lentivirus expression vector, and sh-LACTB was transfected into SHG44 cells, as described in the Materials and Methods section. RT-PCR and Western blot results revealed that LACTB expression was increased in A172 cells and effectively silenced in SHG44 cells, compared with control cells (Fig. 2A, B, F, and G). CCK-8 cell proliferation assay was utilized to determine cell proliferation. The growth curves revealed that LACTB overexpressing significantly inhibited cell proliferation in A172 cells compared with the control group (Fig. 2C). Meanwhile, depletion of LACTB promoted cell proliferation in SHG44 lines (Fig. 2H).
Figure 2.
LACTB inhibited proliferation invasion and angiogenesis of glioma cells. (A, B, F, G) RT-qPCR and Western blot were conducted to determine the mRNA levels and protein of LACTB in A172 and SHG44 cells. (C, H) CCK-8 assay tested cell proliferation in A172 and SHG44 cells. (D, I) Representative fields of A172 and SHG44 invasive cells (scale bar: 100 μm, 200×) (up), average number of invasive cells per field from three independent experiments (down). con, transfected with empty vectors or control shRNA; over, transfected with LACTB vectors; sh, transfected with LACTB shRNA. (E, J) Representative fields of tube formation of HUVECs (scale bars: 100 μm, 200×) (up) and number of crosses of tube formation by HUVECs. Data are based on at least three independent experiments and shown as the means ± standard deviation (SD). **p < 0.01, ***p < 0.001, compared with con.
LACTB Suppresses Invasion and Angiogenesis of Glioma Cells
Transwell assay was performed to determine the effect of LACTB on the invasiveness of glioma cells. As a result, LACTB-overexpressing A172 cells showed a significantly decreased invasiveness compared with the control A172 cells (Fig. 2D). Conversely, the invasiveness of LACTB-overexpressing SHG44 cells was significantly weaker than that of the control cells (Fig. 2I).
To determine whether LACTB could affect the angiogenesis ability of glioma cells, we first performed the tube formation assay. LACTB overexpression in A172 cells elicited a significant reduction in the number of crosses (Fig. 2E) compared with control groups. Conversely, the number of crosses was significantly enhanced upon depletion of LACTB in SHG44 cells (Fig. 2J).
LACTB Inhibits Expression of PCNA, MMP9, MMP2, and VEGF in Glioma Cells
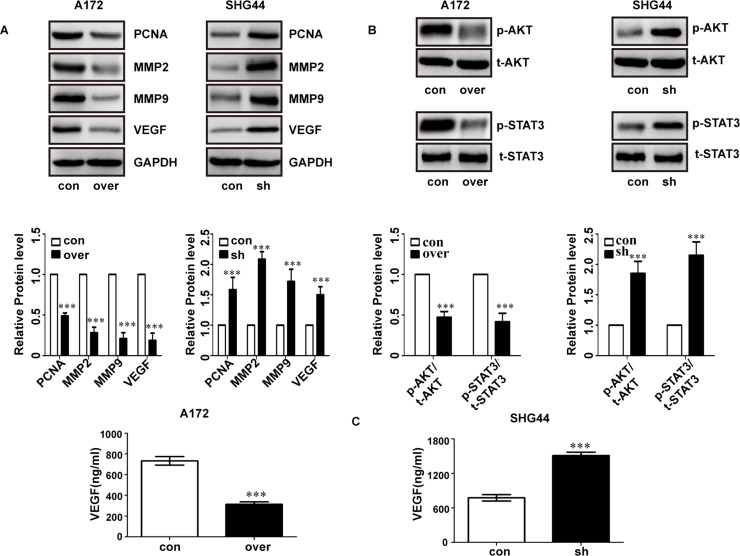
Next, we tested the expression of PCNA, MMP9, MMP2, and VEGF and found they were suppressed in LACTB-overexpressing A172 cells (Fig. 3A, left). Conversely, depletion of LACTB in SHG44 cells promoted expression of these molecules (Fig. 3A, right). We also tested the expression of p-AKT and p-STAT3, which was closely related to the invasion and proliferation. The results showed that p-AKT and p-STAT3 expression was significantly lower in LACTB-overexpressing A172 cells compared with the control cells (Fig. 3B, left). Conversely, depletion of LACTB in SHG44 cells significantly promoted the expression of p-AKT and p-STAT3 (Fig. 3B, right).
Figure 3.
LACTB suppressed PCNA, MMP9, MMP2, and VEGF in glioma cells. (A) LACTB overexpression inhibits the expression of PCNA, MMP9, MMP2, and VEGF in A172 cells. (B) LACTB downexpression promotes the expression of PCNA, MMP9, MMP2, and VEGF in SHG44 cells. (C) The level of secreted VEGF in A172 and SHG44 cells was detected by ELISA. ***p < 0.001.
In addition, an ELISA assay was used to detect expression of VEGF in the culture supernatants of the glioma cells. Our results showed that VEGF expression was markedly lower in LACTB-overexpressing A172 cells compared with the control cells (Fig. 3C, left). In contrast, depletion of LACTB in SHG44 cells significantly promoted VEGF expression (Fig. 3C, right).
DISCUSSION
LACTB is a mitochondrial protein that is related with cellular metabolism18. It is a newly found tumor suppressor gene. However, it remains unknown whether LACTB has important biological functions in gliomas. In this study, we investigated the expression level and biological function of LACTB in glioma tissues. We found that LACTB was frequently downexpressed in the glioma tissues, which is in accordance with the data from TCGA and GEO. We hypothesized that LACTB may function as a tumor suppressor gene, and downexpression of LACTB may promote the development of glioma. In order to test our hypothesis, we analyzed the relationship between the expression level of LACTB and the clinicopathologic features of gliomas, and investigated the biological role of its expression in glioma cells. The results demonstrated that LACTB expression levels were inversely associated with KPS and WHO grade. In addition, the Kaplan–Meier survival curve data revealed that LACTB downexpression predicted poor survival of glioma, and univariate and multivariate analyses demonstrated that LACTB was an independent prognostic factor for the overall survival of patients with glioma.
Recent studies suggested that downexpression of LACTB promoted cell proliferation and inhibited apoptosis in breast cancer cells16. However, to the best of our knowledge, there is no research to study the biological role of LACTB in glioma cells. Thus, we determined the effects of LACTB on the proliferation, invasion, and angiogenesis of glioma cells. Consequently, we found that suppression of LACTB resulted in a significantly promoted proliferation, invasion, and angiogenesis of glioma cells. Furthermore, we observed that overexpression of LACTB significantly decreased the protein levels of PCNA, MMP2, MMP9, and VEGF, which are considered to be important in glioma cell proliferation, invasion, and angiogenesis19–25. The results proved that LACTB plays an important function in the progression of the glioma cells.
In conclusion, we found for the first time that LACTB was downexpressed in gliomas. Downregulation of LACTB predicted poor survival of glioma and promoted cell proliferation, invasion, and angiogenesis of gliomas. Our data demonstrated that LACTB may serve as a promising therapeutic target of glioma treatment.
ACKNOWLEDGMENTS
The authors feel deeply grateful that patients and their families participated in this study. Haitao Li, Daoyong Dong, Qiang Liu, Yiqin Xu, and Langbo Chen conceived and designed the study; Haitao Li performed the majority of the experiments and wrote the manuscript.
Footnotes
The authors declare no conflicts of interest.
REFERENCES
- 1. Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, Burger PC, Jouvet A, Scheithauer BW, Kleihues P. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007;114:97–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ostrom QT, Bauchet L, Davis FG, Deltour I, Fisher JL, Langer CE, Pekmezci M, Schwartzbaum JA, Turner MC, Walsh KM. Response to “The epidemiology of glioma in adults a ‘state of the science’ review”. Neuro-Oncology 2015;17:896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Goodenberger ML, Jenkins RB. Genetics of adult glioma. Cancer Genetics 2012;205:613–21. [DOI] [PubMed] [Google Scholar]
- 4. Thorne AH, Meisen WH, Russell L, Ji YY, Bolyard CM, Lathia JD, Rich J, Puduvalli VK, Mao H, Yu J. Role of cysteine-rich 61 protein (CCN1) in macrophage-mediated oncolytic Herpes simplex virus clearance. Mol Ther. 2014;22:1678–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Liu Q, Li G, Li R, Shen J, He Q, Deng L, Zhang C, Zhang J. IL-6 promotion of glioblastoma cell invasion and angiogenesis in U251 and T98G cell lines. J Neuro-oncol. 2010;100:165–76. [DOI] [PubMed] [Google Scholar]
- 6. Starkweather AR, Sherwood P, Lyon DE, Mccain NL, Bovbjerg DH, Broaddus WC. A biobehavioral perspective on depressive symptoms in patients with cerebral astrocytoma. J Neurosurg Nurs. 2011;43:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kraus JA, Lamszus K, Glesmann N, Beck M, Wolter M, Sabel M, Krex D, Klockgether T, Reifenberger G, Schlegel U. Molecular genetic alterations in glioblastomas with oligodendroglial component. Acta Neuropathol. 2001;101:311–20. [DOI] [PubMed] [Google Scholar]
- 8. Smith TS, Southan C, Ellington K, Campbell D, Tew DG, Debouck C. Identification, genomic organization, and mRNA expression of LACTB, encoding a serine β-lactamase-like protein with an amino-terminal transmembrane domain. Genomics 2001;78:12–4. [DOI] [PubMed] [Google Scholar]
- 9. Mootha VK, Bunkenborg J, Olsen JV, Hjerrild M, Wisniewski JR, Stahl E, Bolouri MS, Ray HN, Sihag S, Kamal M, Patterson N, Lander ES, Mann M. Integrated analysis of protein composition, tissue diversity, and gene regulation in mouse mitochondria. Cell 2003;115:629–40. [DOI] [PubMed] [Google Scholar]
- 10. Rome S, Clément K, Rabasa-Lhoret R, Loizon E, Poitou C, Barsh GS, Riou JP, Laville M, Vidal H. Microarray profiling of human skeletal muscle reveals that insulin regulates 800 genes during a hyperinsulinemic clamp. J Biol Chem. 2003;278:18063–8. [DOI] [PubMed] [Google Scholar]
- 11. Taylor SW, Fahy E, Zhang B, Glenn GM, Warnock DE, Wiley S, Murphy AN, Gaucher SP, Capaldi RA, Gibson BW, Ghosh SS. Characterization of the human heart mitochondrial proteome. Nat Biotechnol. 2003;21:281–6. [DOI] [PubMed] [Google Scholar]
- 12. Pagliarini DJ, Calvo SE, Chang B, Sheth SA, Vafai SB, Ong SE, Walford GA, Sugiana C, Boneh A, Chen WK, Hill DE, Vidal M, Evans JG, Thorburn DR, Carr SA, Mootha VK. A mitochondrial protein compendium elucidates complex I disease biology. Cell 2008;134:112–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Polianskyte Z, Peitsaro N, Dapkunas A, Liobikas J, Soliymani R, Lalowski M, Speer O, Seitsonen J, Butcher S, Cereghetti GM, Linder MD, Merckel M, Thompson J, Eriksson O. LACTB is a lament-forming protein localized in mitochondria. Proc Nat Acad Sci USA 2009;106:18960–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chen Y, Zhu J, Lum PY, Yang X, Pinto S, MacNeil DJ, Zhang C, Lamb J, Edwards S, Sieberts SK, Leonardson A, Castellini LW, Wang S, Champy MF, Zhang B, Emilsson V, Doss S, Ghazalpour A, Horvath S, Drake TA, Lusis AJ, Schadt EE. Variations in DNA elucidate molecular networks that cause disease. Nature 2008;452:429–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yang X, Deignan JL, Qi H, Zhu J, Qian S, Zhong J, Torosyan G, Majid S, Falkard B, Kleinhanz RR, Karlsson J, Castellani LW, Mumick S, Wang K, Xie T, Coon M, Zhang C, Estrada-Smith D, Farber CR, Wang SS, van Nas A, Ghazalpour A, Zhang B, Macneil DJ, Lamb JR, Dipple KM, Reitman ML, Mehrabian M, Lum PY, Schadt EE, Lusis AJ, Drake TA. Validation of candidate causal genes for obesity that affect shared metabolic pathways and networks. Nat Genet. 2009;41:415–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Keckesova Z, Donaher JL, De Cock J, Freinkman E, Lingrell S, Bachovchin DA, Bierie B, Tischler V, Noske A, Okondo MC, Reinhardt F, Thiru P, Golub TR, Vance JE, Weinberg RA. LACTB is a tumour suppressor that modulates lipid metabolism and cell state. Nature 2017;543:681–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yang D, Wang LL, Dong TT, Shen YH, Guo XS, Liu CY, Liu J, Zhang P, Li J, Sun YP. Progranulin promotes colorectal cancer proliferation and angiogenesis through TNFR2/Akt and ERK signaling pathways. Am J Cancer Res. 2015;5:3085–97. [PMC free article] [PubMed] [Google Scholar]
- 18. Li B, Wang Y, Li S, He H, Sun F, Wang C, Lu Y, Wang X, Tao B. Decreased expression of miR-378 correlates with tumor invasiveness and poor prognosis of patients with glioma. Int J Clin Exp Pathol. 2015;8:701–6. [PMC free article] [PubMed] [Google Scholar]
- 19. Sherr CJ. Mammalian G1 cyclins. Cell 1993;73:1059–65. [DOI] [PubMed] [Google Scholar]
- 20. Meyerson M, Harlow E. Identification of G1 kinase activity for cdk6, a novel cyclin D partner. Mol Cell Biol. 1994;14:2077–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Weinberg RA. The retinoblastoma protein and cell cycle control. Cell 1995;81:323–30. [DOI] [PubMed] [Google Scholar]
- 22. Harbour JW, Dean DC. Rb function in cell-cycle regulation and apoptosis. Nat Cell Biol. 2000;2:E65–7. [DOI] [PubMed] [Google Scholar]
- 23. Daniel P, Filiz G, Brown DV, Hollande F, Gonzales M, Abaco DG, Papalexis N, Phillips WA, Malaterre J, Ramsay RG, Mantamadiotis T. Selective CREB-dependent cyclin expression mediated by the PI3K and MAPK pathways supports glioma cell proliferation. Oncogenesis 2014;3:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mikulić D, Ilić I, Cepulić M, Orlić D, Giljević JS, Fattorini I, Seiwerth S. Tumor angiogenesis and outcome in osteosarcoma. Pediatr Hematol Oncol. 2004;21:611–9. [DOI] [PubMed] [Google Scholar]
- 25. Jin X, Yin J, Kim SH, Sohn YW, Beck S, Lim YC, Nam DH, Choi YJ, Kim H. EGFR-AKT-Smad signaling promotes formation of glioma stem-like cells and tumor angiogenesis by ID3-driven cytokine induction. Cancer Res. 2011;71:7125–34. [DOI] [PubMed] [Google Scholar]