Significance
The radial spoke (RS) transmits mechanochemical signals from the central pair (CP) to axonemal dynein arms to coordinate ciliary motility. The RS head, the key CP interaction domain, differs dramatically in morphology between protozoa and metazoa despite the conservation of components in evolution. In this work, we show that the murine RS head is compositionally distinct from that of Chlamydomonas and describe the cryo-EM structure of its core complex at 3.2-Å resolution. Our results provide a blueprint for understanding not only the allosteric coordination within the complex and the mechanism of the RS–CP interaction but also the etiology of numerous RS head gene mutations that have been linked to primary ciliary dyskinesia.
Keywords: radial spoke, cilia, flagella, cryo-EM, primary ciliary dyskinesia
Abstract
The radial spoke (RS) heads of motile cilia and flagella contact projections of the central pair (CP) apparatus to coordinate motility, but the morphology is distinct for protozoa and metazoa. Here we show the murine RS head is compositionally distinct from that of Chlamydomonas. Our reconstituted murine RS head core complex consists of Rsph1, Rsph3b, Rsph4a, and Rsph9, lacking Rsph6a and Rsph10b, whose orthologs exist in the protozoan RS head. We resolve its cryo-electron microscopy (cryo-EM) structure at 3.2-Å resolution. Our atomic model further reveals a twofold symmetric brake pad-shaped structure, in which Rsph4a and Rsph9 form a compact body extended laterally with two long arms of twisted Rsph1 β-sheets and potentially connected dorsally via Rsph3b to the RS stalk. Furthermore, our modeling suggests that the core complex contacts the periodic CP projections either rigidly through its tooth-shaped Rsph4a regions or elastically through both arms for optimized RS–CP interactions and mechanosignal transduction.
The majority of motile cilia and flagella are composed of nine dynein arm-containing peripheral doublet microtubules (DMTs) surrounding a central pair (CP) of MTs (the “9+2” axoneme). The radial spoke (RS) is a T-shaped protein complex with an orthogonal head pointing toward the CP and a stalk anchored on each A-tubule of the DMTs (1–5). It acts as the mechanochemical transducer between the CP and axonemal dynein arms to regulate flagellar/ciliary motility (6–11). The flagella of Chlamydomonas reinhardtii, a widely used model organism, contain two full-size RSs (RS1 and RS2) in each 96-nm repeat unit of the axoneme. In contrast, motile cilia/flagella of Tetrahymena thermophila and metazoa possess triplet RSs (RS1 to RS3) (2–4, 11). The Chlamydomonas RS is composed of at least 23 subunit proteins (RSP1 to RSP23) (2, 12, 13). Seventeen of them have mammalian homologs (14). Mutations leading to the loss of the entire RS or RS head result in immotile flagella in Chlamydomonas (6–8) but in rotatory ciliary beat in mammals, causing primary ciliary dyskinesia (PCD), a genetic syndrome characterized by recurrent respiratory infections, situs inversus, infertility, and hydrocephalus (4, 15–21).
The most striking morphological differences in the RS lie in the RS head, the key structural domain that mediates the mechanosignaling by directly contacting projections of the CP (9–11). The heads of RS1 and RS2 consist of two structurally identical, rotationally symmetric halves that differ largely from that of RS3 (3, 4). Furthermore, their morphologies differ dramatically between protozoa and metazoa. In Chlamydomonas and Tetrahymena, for instance, the heads of RS1 and RS2 are rich in lateral branches that also form a connection between the two heads (2, 4). In contrast, in sea urchin (Strongylocentrotus purpuratus) and human, the heads of RS1 and RS2 resemble a pair of ice skate blades with many fewer interfaces toward the CP (3, 4). Despite the importance of the RS and RS head in cilia/flagella motility, the structural details of the RS and the RS–CP interactions remain poorly understood, especially in mammals.
The RS heads have probably been remodeled to comply with both structural and functional alterations of the axoneme during evolution. How the morphological changes occurred, however, remains unclear. The Chlamydomonas RS head is composed of RSP1, -4, -6, -9, and -10 and part (the C terminus) of the stalk component, RSP3. Each of the symmetrical halves of the head contains one copy of these components (2, 10, 22). All the head components have mammalian orthologs (Rsph1, -4a, -6a, -9, -10b, and -3b) (11, 14). In sharp contrast to the markedly reduced surface area of metazoan RS heads, the peptides of human RSPH4A, -6A, and -10B are longer than their Chlamydomonas orthologs by 1.5-, 1.3-, and 4-fold, respectively (11). Only RSPH1 (309 amino acids [aa]) is shorter than RSP1 (814 aa) (11). The lengths of mouse RS head proteins are also similarly changed as their human counterparts (SI Appendix, Fig. S1A). Furthermore, while murine Rsph4a is essential for the head formation of RS1 to RS3 in motile multicilia of the trachea, ependyma, and oviduct (15), Rsph6a is specifically expressed in sperm for their normal flagellar formation (23). RSP4/Rsph4a and RSP6/Rsph6a are paralogs: RSP4 and RSP6 share 48% sequence identity (24), whereas murine Rsph4a is 63% identical to Rsph6a (SI Appendix, Fig. S1B). Sea urchin and Ciona, however, have only one ortholog (11, 25). These results suggest that, unlike the protozoan RS heads, the metazoan ones may not simultaneously contain Rsph4a and Rsph6a. The general shapes of the RS structure in axonemes have been determined by conventional electron microscopy (EM) (26–28) and cryo-electron tomography (cryo-ET) (2–5). Recently, a 15-Å-resolution RS structure of Chlamydomonas was resolved by cryo-EM single-particle analysis (29). The resolutions, however, do not suffice for the delineation of the locations of individual RS subunits.
In the present study, by biochemical and structural analyses, we show the murine RS head is both compositionally and morphologically distinct from that of Chlamydomonas. Our study suggests that the RS head has experienced profound remodeling to probably comply with both structural and functional alterations of the axoneme during evolution for coordinated ciliary or flagellar motility.
Results
Mouse Rsph1–Rsph3b–Rsph4a–Rsph9 Can Form a Stable RS Head Core Complex.
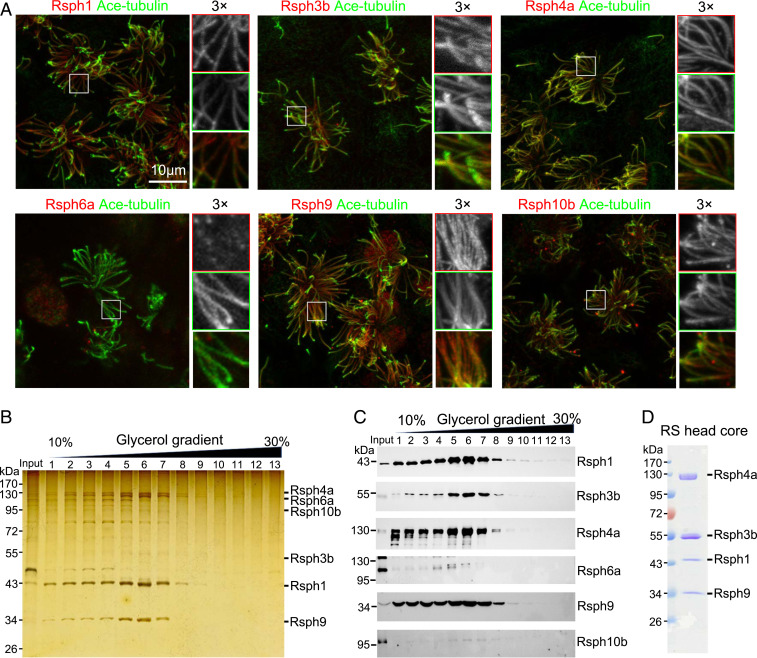
We first examined whether all the mouse homologous RS head proteins (Rsph1, -4a, -6a, -9, and -10b) and RSP3 (Rsph3b) localize in the axoneme of motile cilia in multiciliated mouse ependymal cells (mEPCs) (30, 31). All these subunits except Rsph6a, which is homologous to Rsph4a (SI Appendix, Fig. S1B), were detected in the cilia of mEPCs (Fig. 1A). Rsph6a was also not detected in the mEPC lysate, though; consistent with a previous report (23), it was readily detected in mouse sperm lysate through immunoblotting and sperm flagella through immunostaining (SI Appendix, Fig. S1 C and D). Surprisingly, Rsph4a was undetectable in mouse sperm in both immunoblotting and immunostaining (SI Appendix, Fig. S1 C and D). These results suggest that, unlike in protozoa, mammalian Rsph4a and Rsph6a are, respectively, expressed in multiciliated cells and sperm and do not coexist in the same axoneme.
Fig. 1.
Rsph1–Rsph3b–Rsph4a–Rsph9 can form a stable RS head core complex and be purified. (A) Immunofluorescence of mEPCs showed that Rsph1, Rsph3b, Rsph4a, Rsph9, and Rsph10b, but not Rsph6a, localize in ciliary axonemes. Acetylated (Ace)-tubulin served as a ciliary axoneme marker. (B and C) Flag-tagged Rsph3b pulled down a complex abundant in Rsph1, -4a, and -9, but not -6a and -10b. The indicated proteins were coexpressed in HEK293T cells and coimmunoprecipitated with anti-Flag resins. The immunoprecipitates were subjected to a 10 to 30% glycerol gradient ultracentrifugation. Fractions collected from top to bottom were separated by SDS-PAGE, and followed by silver staining (B) and immunoblotting (C). Note that Rsph6a and Rsph10b were highly expressed in the input but poorly detected in the complex (C, fractions 5 to 7). (D) Purification of the Rsph1–Rsph3b–Rsph4a–Rsph9 complex by immunoprecipitation. The indicated proteins, in which Rsph3b and Rsph1, respectively, carried a Flag tag and a 6×His tag, were coexpressed in HEK293T cells and immunoprecipitated by using anti-Flag beads. The complex was detected by Coomassie blue staining after SDS-PAGE. This purification was repeated more than three times with similar results.
To elucidate the interaction network of the RS head subunits, we performed coimmunoprecipitation (co-IP) assays. We expressed Flag-tagged Rsph3b together with Rsph1, -4a, -6a, -9, and -10b in HEK293T cells and used anti-Flag beads to pull down the complex. Glycerol gradient ultracentrifugation indicated a complex formation among Rsph1, -3b, -4a, and -9 (Fig. 1 B and C). In contrast, although Rsph6a and Rsph10b were highly expressed in the cells, they were only weakly detected in the complex (Fig. 1 B and C). Co-IP further revealed that Rsph4a was able to form a stable binary complex with Rsph3b, -1, or -9 and a putative ternary complex with -1 and -9 (SI Appendix, Fig. S1E), in addition to the quaternary complex with -1, -3, and -9 (Fig. 1 C and D and SI Appendix, Fig. S1E). When Flag-Rsph6a was expressed for co-IP, it immunoprecipitated Rsph1 and -9 or Rsph1, -3, and -9 (SI Appendix, Fig. S1E), implying formation of ternary and quaternary complexes as well. Altogether, our data suggest that mammalian Rsph4a and Rsph6a form dually exclusive complexes with Rsph1, -3b, and -9 to function in multicilia and sperm flagella, respectively. As Rsph4a is known to be an essential RS head subunit in multiple types of multicilia (15), we chose to focus on the Rsph1–Rsph3b–Rsph4a–Rsph9 complex in the following experiments.
Architecture of the Mouse RS Head Core Complex Revealed by Cryo-EM.
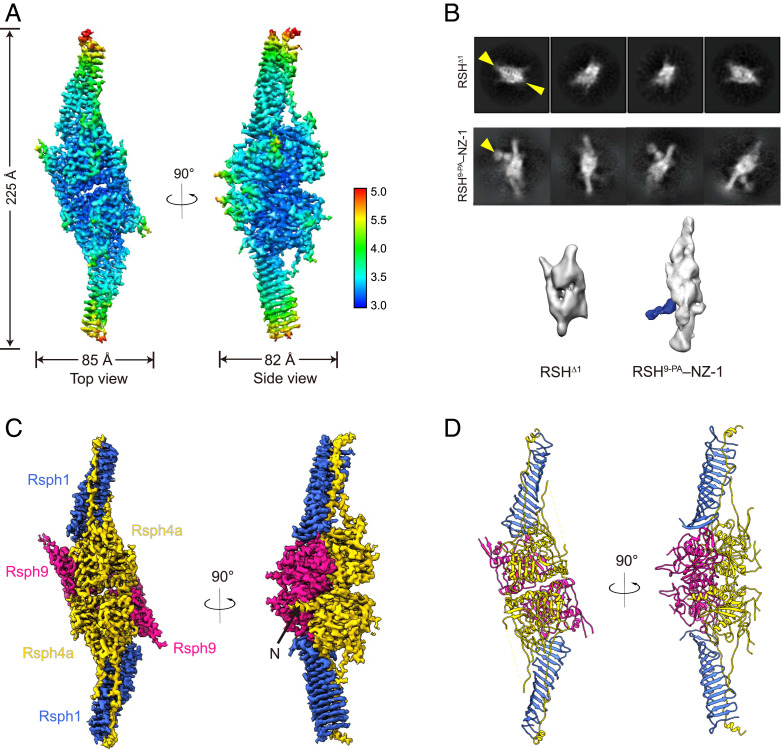
We purified the Rsph1–Rsph3b–Rsph4a–Rsph9 complex and validated the existence of all the subunits by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and mass spectrometry analysis (Fig. 1D and SI Appendix, Fig. S1F). We then carried out a cryo-EM study on the RS head core complex (SI Appendix, Figs. S2 and S3). Our initial reconstruction suggested a preferred-orientation problem associated with the RS head core complex (highly preferred “top” orientation but lacking side views; SI Appendix, Fig. S2C). To overcome this problem, we adopted the recently developed tilt stage strategy in data collection with additional data collected at 30 and 40° tilt angles (32). This allowed us to obtain a cryo-EM structure of the RS head core complex at 3.2-Å resolution without imposing any symmetry, revealing the uncharacterized architecture of the mammalian RS head core complex (Fig. 2A and SI Appendix, Figs. S2 and S3 and Table S1). After overcoming the preferred-orientation problem, the structural features of the map were very well resolved without distortions, especially for that of the side view (SI Appendix, Fig. S2D). The map appears to be ∼225 Å in height, ∼85 Å in length, and 82 Å in width (Fig. 2A). This complex has a compact body with two arms stretching out. The two arms are both ∼80 Å in height and resemble each other very well (Fig. 2A). Moreover, when visualized from the top, the map appears to have a central canyon dividing the map into two portions highly similar to each other (Fig. 2A), even though we did not impose any symmetry in the reconstruction process. Interestingly, the front side of the map appears to have a serration shape with a groove in the middle (side view in Fig. 2A).
Fig. 2.
Architecture of the mouse RS head core complex. (A) Local resolution estimation of the cryo-EM map of the RS head core complex, with the resolution color bar shown (Right). (B) Determination of subunit location by cryo-EM analysis on subunit-deleted or PA–NZ-1 epitope-labeled samples. Reference-free 2D analysis and 3D reconstruction showed that RSH∆1 (lacking Rsph1) retains a compact body but misses the two arms (indicated by two yellow arrowheads). RSH9-PA–NZ-1 shows an obvious extra density exposed adjacent to an arm (indicated by an arrowhead in 2D analysis and a density in blue in 3D reconstruction), corresponding to the NZ-1 Fab attached to the PA tag inserted into Rsph9. (C) Subunit arrangement of the RS head core complex. Rsph1 is in royal blue, Rsph4a is in gold, and Rsph9 is in deep pink. The color scheme is followed throughout. The remaining N-terminal region of Rsph4a is indicated by a black arrow (in the side view). (D) Atomic model of the RS head core complex.
Subunit Identification by Subunit Deletion and PA–NZ-1 Epitope Labeling Strategy.
To unambiguously locate individual subunits within the complex, we performed cryo-EM analysis on subunit-deleted or epitope-labeled RS head core complexes based on our previously developed subunit PA–NZ-1 tag–Fab labeling strategy (33–35). To determine the location of Rsph1, we first expressed the complex without Rsph1 (termed RSHΔ1). Our reference-free two-dimensional (2D) analysis and 3D reconstruction on RSHΔ1 both showed that the compact body remained while the two stretching arms disappeared in comparison with the wild-type core complex (Fig. 2B and SI Appendix, Fig. S2B). This result suggests that there are two copies of Rsph1 in the RS head core complex, and each locates in one of the arm positions. We then sought to locate Rsph9 in the map by adopting our PA–NZ-1 epitope labeling strategy (33–35). We inserted a dodecapeptide PA tag into the C terminus of Rsph9 and expressed the PA-labeled core complex (termed RSH9-PA). RSH9-PA then formed a complex with the Fab fragment of the NZ-1 antibody, termed RSH9-PA–NZ-1. Subsequent reference-free 2D analysis and 3D reconstruction on RSH9-PA–NZ-1 both showed an obvious extra density exposed outside the lower portion of the body and adjacent to the arm, most likely corresponding to the NZ-1 Fab, indicating that Rsph9 may locate in the body close to an arm (Fig. 2B). Taken together, these structural analyses suggest that there are two copies of Rsph1 residing in the two arms, with Rsph9 in the body close to an arm (Fig. 2B).
Ab Initio Model Building and the Twofold Symmetric Nature of the RS Head Core.
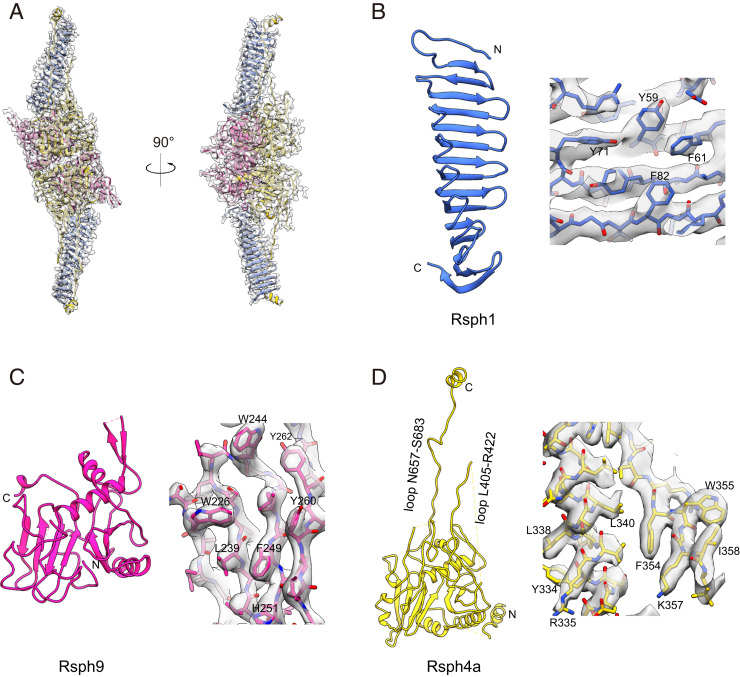
So far, template information of mammalian RS head proteins for homology model building is extremely limited. Only Rsph1 has a homologous template structure (Protein Data Bank [PDB] ID code 6JLE) (36), with ∼41% sequence coverage of the subunit. We then performed ab initio model building using the Coot software package (37) based on our 3.2-Å-resolution cryo-EM map combined with secondary structure element (SSE) prediction by Phyre2 (SI Appendix, Fig. S4) (38). In our density map, the two arms, predicted to be Rsph1 by subunit deletion analysis (Fig. 2B), appear to consist of a cluster of β-stranded structures (Fig. 2 A and C). Also, for Rsph1, the homology model shows an overall β-structure feature (SI Appendix, Fig. S4A) that matches the central portion of the arm well. Based on this initial fitting, we conducted ab initio model building for the remaining portion of Rsph1 using Coot software, and this Rsph1 model fits into the density map of the two arms very well (Fig. 3 A and B). Subsequently, based on the approximate Rsph9 C-terminal location determined through our epitope labeling strategy, and especially the side-chain information revealed in our map combined with the SSE prediction, we carried out ab initio model building for Rsph9 (Fig. 3C). For the remaining density, based on the side-chain information and SSE prediction, we allocated Rsph4a in this region and performed ab initio model building for Rsph4a (Fig. 3D). After combining the subunit models together and further refinement, we achieved an atomic model of the RS head core complex that fits in the map very well (Fig. 3 and Movie S1). Still, several long loops (i.e., Q292 to S309, E378 to E404, G505 to R518, and K563 to I584 for Rsph4a, and L116 to E129 and I194 to F213 for Rsph9), the N-terminal region of Rsph4a (before L277), and the C-terminal region of Rsph1 (L203 to D301) appear to be disordered in our density map, possibly due to the intrinsic dynamic nature of those regions.
Fig. 3.
Atomic models of individual subunits and representative high-resolution structural features of the RS head core complex. (A) Model and map fitting showing that the atomic model (ribbon in color) fits in the cryo-EM map (transparent gray) very well. (B–D) Atomic models of Rsph1 (B), Rsph9 (C), and Rsph4a (D), and representative high-resolution structural features.
In this configuration, Rsph4a and Rsph9 form a compact body, whereas Rsph1 is an elongated and twisted β-sheet constituting a stretching arm of about 80 Å (Fig. 2 C and D). Rsph1 contains a motif known as a membrane occupation and recognition nexus (MORN) (39). In other proteins, MORN repeats have been suggested to be responsible for membrane association or stabilization of protein complexes (40, 41). It is noteworthy that there are two copies for each of these three subunits in the core complex, forming a dimer of trimers. Therefore, the quaternary structure assembly appears to have a twofold symmetric nature (Fig. 2 C and D).
Subunit Interaction Network of the RS Head Core Complex.
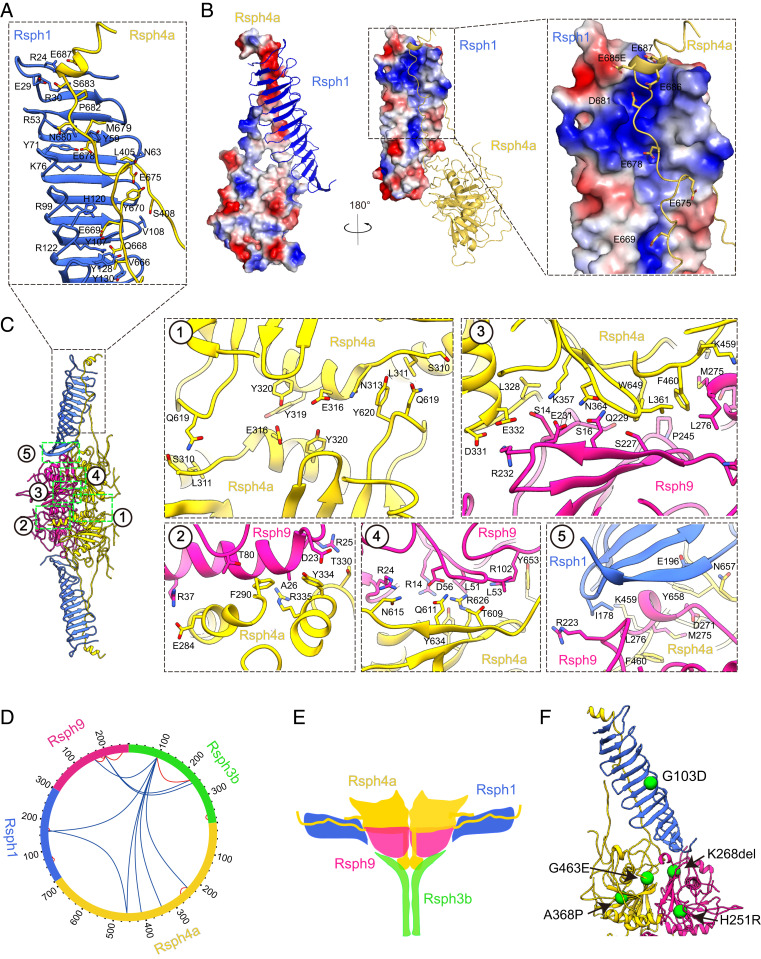
To better understand the molecular basis of the complex assembly and the allosteric network of the core complex, we inspected the subunit interaction interfaces. Surprisingly, in the twisted β-sheet of Rsph1, there appears to be a central positively charged pocket, which has been perfectly matched by the extended loop (N657 to S683) of Rsph4a with a negative electrostatic potential covering throughout the Rsph1 central pocket (Fig. 4 A and B). This interaction could greatly stabilize the binding between Rsph4a and Rsph1, substantiating the notion that the MORN motif could contribute to complex stabilization and assembly (40, 41). In addition, Rsph4a has another loop (L405 to R422) extending from the compact body with its end adjacent to the G78-to-W94 β-hairpin of Rsph1 and appearing like an adjustable string (Figs. 2 and 3D). This, together with the extremely long loop (N657 to S683) of Rsph4a, forms a triangle between the body and the arm (Figs. 3D and 4 A–C). This sophisticated allosteric network designed by nature could potentially adjust the movement of the arms relative to the body, leading to inherent plasticity of the RS head. Moreover, in the canyon between the two protomers, there appears to form a strong interaction network between Rsph4a/Rsph9 of one protomer and Rsph4a from the other protomer (Fig. 4 C, 1 and 2 and SI Appendix, Table S3), indicating efficient coordination between the two symmetric protomers of the complex. Within the protomer, we also observed rich H-bond/salt-bridge interactions between Rsph9 and Rsph4a, suggesting a strong binding between the neighboring Rsph4a and Rsph9 subunits (Fig. 4 C, 3 and 4). Although relatively rare interactions between Rsph1 and Rsph9 were identified in our structure (Fig. 4 C, 5), the disordered C-terminal portion of Rsph1 (L203 to D301) may also locate in this position and contribute to the interaction.
Fig. 4.
Subunit interaction network of the RS head core complex and locations of PCD-related mutations. (A) Interactions between the Rsph1 MORN repeats and the long extended loop of Rsph4a. (B) Electrostatic interactions between Rsph4a and Rsph1. The Rsph1 twisted β-sheet appears to have a central positively charged pocket, which can be perfectly matched by the extended N657-to-S683 loop of Rsph4a with negative electrostatic potential covering throughout the Rsph1 central pocket. (C) Interaction interfaces between the two protomers (1 and 2), of Rsph4a–Rsph9 (3 and 4), and Rsph1–Rsph4a–Rsph9 (5). (D) XL-MS analysis of the RS head core complex. Identified cross-linked intersubunit contacts are shown as blue lines, and intrasubunit contacts as red lines. We used best e value (1.00E-02) as the threshold to remove extra XL-MS data with lower confidence. (E) Proposed model showing the potential location of Rsph3b (in green) in the back of the compact body of the RS head core complex. Refer to SI Appendix, Fig. S5 for details. (F) Known PCD disease-causing mutations mapped onto the model of the complex. Each green sphere represents a mutation associated with PCD disease.
The two Rsph1 arms take up ∼70% of the height of the complex, resulting in a stretched-out configuration of the RS head complex, which may serve as a spacer between consecutive RSs and also enlarge the interaction of the RS with the apparatus of the CP (discussed below). Moreover, Rsph4a is the largest subunit localized on the top surface of the complex interacting with all the other subunits (Fig. 2 C and D). For instance, its remaining N-terminal region embraces and wraps around Rsph9 and its two extended loops attach to Rsph1 (Figs. 2 C and D and 4 A and B); it is also heavily involved in the interactions between the two symmetric protomers (Fig. 4 C, 1 and 2). This is substantiated by our co-IP assay results showing that Rsph4a interacts with all the other three subunits (SI Appendix, Fig. S1E). Thus, Rsph4a may serve as a scaffold for the proper assembly of the complex. It also forms two tooth-shaped structures hooking to the two stretched-out arms of Rsph1 (Figs. 2, 3, and 4), possibly for optimized RS–CP interaction.
We should mention that we used Flag-tagged Rsph3b for affinity purification of the head core complex and that the existence of Rsph3b has also been detected by SDS-PAGE, co-IP, and mass spectrometry analyses (Fig. 1D and SI Appendix, Fig. S1 E and F). Also, as suggested by our cross-linking mass spectrometry (XL-MS) analysis, Rsph3b was cross-linked with other subunits (Fig. 4D and SI Appendix, Table S2). Furthermore, although Rsph3b is undetectable in our RS head core complex map rendered at higher threshold, our reference-free 2D analysis implies that there is extra weak density in the back of the compact body (SI Appendix, Fig. S5A), and this extra density can be visualized in our 3D map rendered at lower threshold (SI Appendix, Fig. S5B). Indeed, this extra density appears more obvious in a 3D reconstruction from a smaller dataset after multiple rounds of 3D classification (SI Appendix, Figs. S3 and S5C). Collectively, these data suggest that very likely Rsph3b resides in the back of the compact body and is adjacent to the flexible N-terminal region of Rsph4a (Fig. 4E). Considering the SSE prediction that Rsph3b contains a long α-helix bundle (∼D154 to F278) (SI Appendix, Fig. S5D), it is possible that connected to the flexible N-terminal region of Rsph4a, this Rsph3b helix swings in the back of the compact body and is thus difficult to be captured (Fig. 4E). We propose that the core complex also contains two copies of Rsph3a (Fig. 4E) but the actual stoichiometry remains to be elucidated.
Mapping of PCD-Related Mutations on Our RS Head Core Complex.
As multiple pathogenic patient mutations of the RS head complex have been documented (16, 18, 42), resolving the atomic model of the complex has enabled us to map the mutations on the complex structure (Fig. 4F). As discussed before, the long C-terminal extended loop of Rsph4a is embraced by the MORN repeat β-sheet of Rsph1, forming strong interactions through complementary electrostatic interactions (Fig. 4 A and B). The pathogenic G103D mutation of Rsph1 (18), in which the hydrophobic G103 without a side chain is replaced with the charged hydrophilic acidic D (Fig. 4F), might interfere with normal RS head formation by disturbing the original interaction network. The K268del mutation in Rsph9 (16) is located in the interface between Rsph9, Rsph1, and Rsph4a and may disturb the interaction among these subunits (Fig. 4F). The loss-of-function H251R mutation in Rsph9, which abolishes the ciliary localization of Rsph9 (42), is located in a β-sheet of Rsph9 near Rsph4a and the protomer interface and may thus disturb the interaction with Rsph4a and the protomer association (Fig. 4F). The pathogenic A368P mutation in Rsph4a is located in the core position near a β-sheet. Furthermore, the G463E mutation in Rsph4a, which abolishes the RS complex assembly (42), resides near the Rsph4a L465-to-G477 loop and may disturb the loop interaction with the G148-to-F163 β-hairpin of Rsph1 (Fig. 4F).
Conformational Dynamics of the RS Core Complex Determined by Multibody Refinement.
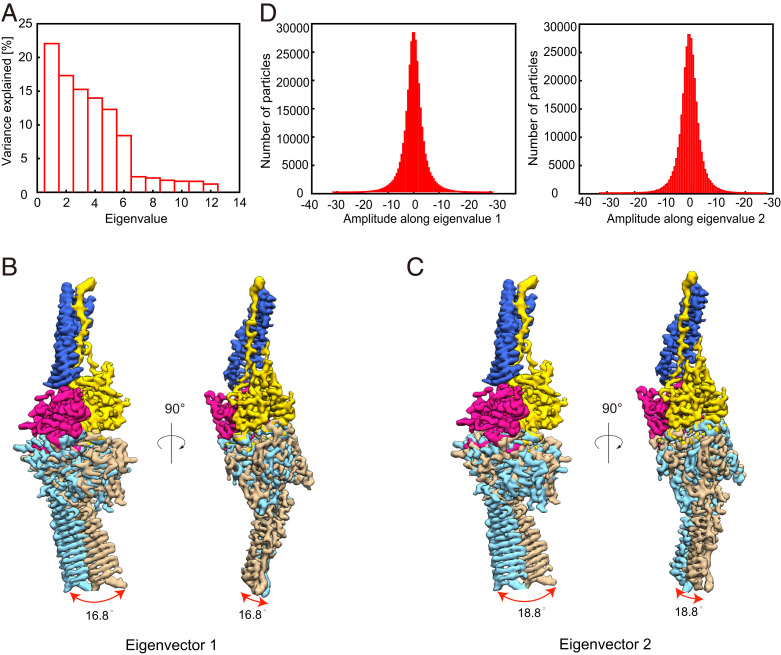
The local resolution analysis suggested that overall the two Rsph1 arms show slightly lower local resolution especially in the distal ends, implying relative dynamics between the two protomers of the complex (Fig. 2A). To better delineate the conformational space of the RS head core complex, we performed multibody refinement in Relion 3.1 (Fig. 5) (43). Principal-component analysis of the movement revealed that ∼50% of the movement of the core complex can be described by the first two eigenvectors representing relative motions in distinct directions between the two protomers (Fig. 5A). Eigenvector 1 describes an open–close motion between the two protomers with an angular range of 16.8° (Fig. 5B and Movie S2). Eigenvector 2 corresponds more to the swing combined with open–close motions between the two protomers with an angular range of 18.8° (Fig. 5C and Movie S2). Moreover, histograms of the amplitudes along the two eigenvectors are unimodal, indicating the two motions are continuous (Fig. 5D). Collectively, our multibody analysis suggests a combined continuous open–close with slight swing motions between the two protomers of the complex.
Fig. 5.
Conformational dynamics of the RS head core complex determined by multibody refinement. (A) Contributions of all eigenvectors to the motions between the two protomers of the RS head core complex. Approximately 50% of the movement of the core complex can be described by the first two eigenvectors representing relative motions in distinct directions between the two protomers. (B and C) Side and top views of the map showing the motions of the first two eigenvectors, with the upper protomer following the color schema as in Fig. 2, and the two extreme locations of the lower protomer illustrated in sky blue and tan densities. The swing angular range and direction are indicated by red arrows. Eigenvector 1 describes an open–close motion between the two protomers with an angular range of 16.8° (B). Eigenvector 2 more corresponds to the swing combined with open–close motions between the two protomers with an angular range of 18.8° (C). Also see Movie S2. (D) Histograms of the amplitudes along the first two eigenvectors.
The Cryo-EM Map of the RS Head Core Complex Fits Well with the RS1 and RS2 Head Envelope Density from a Previous Cryo-ET Human Cilia Structure.
We then fitted our RS head core cryo-EM map into the head region of the T-shaped RS density from the previous cryo-ET map of human airway cilia (Electron Microscopy Data Bank [EMDB] ID code EMD-5950) (4). The cryo-ET density of each head of RS1 and RS2 is able to hold two copies of our RS head core cryo-EM map very well (Fig. 6A). In our fitting, the RS head core complexes are oriented with the serration-shaped Rsph4a side facing the CP and the Rsph9 side connected to the RS stalk. In contrast, our RS head core structure does not match the head density of RS3 (SI Appendix, Fig. S6), suggesting that RS3 may not contain exactly the same head components as RS1 and RS2, although all three of them share Rsph4a (15). Chlamydomonas could accordingly lack a certain RS3-specific component(s) and is therefore only able to assemble a stalk-like structure at the supposed position of RS3 in the flagella (2–4).
Fig. 6.
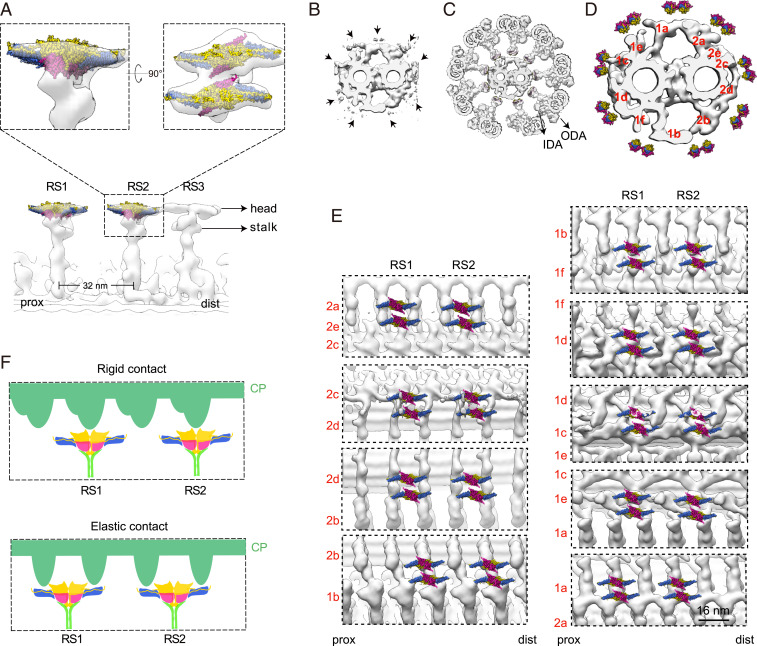
Proposed model of interactions between RS heads and CP projections. (A) Longitudinal view of the cryo-ET map of an RS triplet from the wild-type human DMT (transparent gray; ID code EMD-5950) fitted with our cryo-EM map of the RS head core complex (in color, for RS1 and RS2), and magnified views of RS2 fitted with our map in different orientations. The ice skate blade-shaped RS head density (please see Top) appears to hold two copies of our RS head core complex very well. In this fitting, the core complex is oriented with the Rsph4a side up to face the CP and the Rsph9 side down to be connected to the RS stalk. (B–D) Coordination of the RS head core complex-fitted human DMT (A) and the previous cryo-ET map of sea urchin CP (ID code EMD-9385) (B) into the framework of a 9+2 axoneme (C and D). The locations of the outer dynein arm (ODA) and inner dynein arm (IDA) are indicated (C and D). Positioning of the RS (C) and the RS head core complex (D) relative to the CP projections was mostly based on the remaining RS densities of the CP (B, arrows). Please also refer to Movie S3. (E) Longitudinal views of locations of the dual-core complexes of the RS head relative to the indicated periodic CP projections. Note that portions of the RS heads are visible at the 2c–2d and especially the 1c–1d junctions in the cryo-ET maps of the CP projections. (F) Based on this fitting, we proposed two modes of RS–CP interactions (illustrated in the cartoon diagram), including a rigid contact mode with the periodic projections of the CP (in green) appearing to be able to firmly hold the Rsph4a teeth, and an elastic contact mode with the periodic projections of the CP contacting the two arms of the RS head.
The Core Complex-Fitted RS1 and RS2 Heads Match Geometries of Periodic CP Projections.
The CP projections that interact with the RS heads are evolutionarily conserved proteinous structures (44, 45). They are named 1a to 1f and 2a to 2e according to their locations on the two central (C1 and C2) MTs and arranged along the CP in 16- or 32-nm periodicity (44). To gain insights into the CP contact sites of the RS head and their contact patterns, we incorporated the RS head core complex-fitted DMT (Fig. 6A) and the cryo-ET maps of sea urchin CP (ID code EMD-9385) (44) into a 9+2 axoneme by matching the densities of their RS in a cross-sectional view (Fig. 6 B–D and Movie S3). We found that, except the one facing projection 1d, the remaining eight RS heads are located at or close to the junctions of neighboring projections (Fig. 6D). The two heads at the 1c–1d and the 2c–2d junctions appear to be the closest ones to the projections (Fig. 6E and Movie S3).
When we slightly adjusted the position of each DMT along the CP to properly match the cryo-ET densities of the RS in both the DMT and CP, we generated longitudinal views for the possible CP contact sites of each RS head at a static state. Five of the positions, namely those at the projections of 2a–2e, 2c–2d, 1d, 1e–1a, and 1a–2a (Fig. 6E), are likely reliable due to the existence of clearly visible RS head densities in the cryo-ET CP maps (Fig. 6B, arrows), while the remaining four positions could be relatively ambiguous (Fig. 6E). We noticed striking complementary geometries of the CP projections to the heads: the periodic projections formed structures that appear to be able to firmly hold either the Rsph4a teeth or both arms of each RS head core complex when they contact during the ciliary beat (Fig. 6E). Such CP–RS relationships would lead to “rigid” or “elastic” contact (Fig. 6F), respectively, because mechanical pressures on the teeth are expected to strengthen the contact, whereas pressures on both arms are expected to induce motions between the two protomers of the RS head core complex (Fig. 5 and Movie S2). Furthermore, the dual-core complexes in each head assumed contact modes that appear to be both elastic (e.g., those at the 2d–2b junction), both rigid (e.g., those at the 1a–2a junction), or combinatory (e.g., those at the 2c–2d junction). The complicated geometries of projections at and around each presumed contact site (Fig. 6E) might accordingly facilitate the formation of asymmetric planar beat patterns of cilia (9).
Discussion
We demonstrate that the head of mouse RS1/RS2 is compositionally distinct from the Chlamydomonas one. The mouse RS head core complex in multicilia consists of Rsph1, -3b, -4a, and -9 (Fig. 2), lacking -6a and -10b, whose orthologs exist in the Chlamydomonas RS head (2, 10, 22). This is supported by the fact that sea urchin has RS heads highly similar in morphology to mammalian ones but contains only one ortholog of Rsph4/6 (4, 11). In addition, our results suggest that Rsph1, -3b, -6a, and -9 may also form a head complex specific for sperm flagella (Fig. 1A and SI Appendix, Fig. S1), though the structural details are still unclear. It is possible that the Rsph4a-based RS head complex in multicilia controls the generation of a planar beat (15, 21), whereas the Rsph6a-based one functions in male germ cells for flagellar elongation and motility (23). Why the head complex lacks Rsph10b is unknown. In contrast to the small size of RSP10 (216 aa in Chlamydomonas and 227 aa in Tetrahymena), metazoan Rsph10b somehow becomes quite large (870 aa in human and 876 aa in mouse) (11). Although Rsph10b was readily observed in ependymal multicilia (Fig. 1A), it was only faintly detected in the core complex according to the ultracentrifugation results (Fig. 1 B and C). It might only weakly associate with the core complex in vitro and thus be prone to dissociation. Alternatively, it might be specifically involved in the head formation of RS3. The head of RS3 is morphologically distinct from that of RS1/RS2 throughout evolution but its components are still poorly understood (3, 4, 11). While Rsph4a is a common component of the RS triplet (15), Rsph1 is only essential for the head of RS1/RS2 but dispensable for that of RS3 (4). Rsph10b might thus replace Rsph1 in RS3 because both proteins contain the MORN motif (12). Furthermore, as studies in Ciona intestinalis suggested the existence of metazoan-specific RS components (25, 46), future investigations are required to clarify whether metazoan RS heads contain extra subunits.
We resolved the cryo-EM structure of the mammalian RS head core complex (Rsph1–Rsph3b–Rsph4a–Rsph9) at 3.2-Å resolution (Figs. 2, 3, and 4). Through subunit deletion and PA–NZ-1 epitope labeling strategies, especially based on the side-chain densities resolved in our cryo-EM map, we located the subunits and built the atomic model of the complex (Figs. 2 and 3). We further delineated the subunit interaction network by combined information from the structural model, XL-MS, and co-IP analyses (Fig. 4 and SI Appendix, Fig. S1). The structure is characterized by multiple surprising architectural features: 1) there are two copies of Rsph1, -4a, and -9 in the complex, forming a dimer of trimers (Fig. 2) capable of open–close motions between the two protomers (Fig. 5 and Movie S2); 2) Rsph4a and Rsph9 form a compact body, with Rsph4a interacting with all the other subunits and forming two tooth-shaped structures facing the CP (Figs. 3 and 4); 3) the twisted β-sheets of Rsph1 stretch out to form the two arms of the RS head (Figs. 2, 3, and 4); 4) two loops of Rsph4a protrude throughout the sheets of Rsph1 to probably function as pulling strings to strengthen the arms and adjust the movement of the two protomers (Figs. 2 and 5); and 5) Rsph3b may locate at the back (Rsph9 side) of the core region to bridge the RS head with the stalk (Fig. 4E and SI Appendix, Fig. S5), similar to RSP3 (10). Although we used Rsph3b to pull down the core complex, we were unable to clearly extract its structural information, suggesting the requirement of other stalk components to stabilize its conformation. This may also be related to the evolutionary changes of the Rsph3b protein. RSP3 is rod-like, with its C terminus locating in the RS head and its N terminus protruding toward the base of the RS stalk (10), while Rsph3b is shorter by more than 200 aa at the C terminus as compared with RSP3 (11), and potentially forms a contact with the RS head instead of being able to protrude into the RS head. Such a compact, intricate RS head structure also explains why the entire head is disassembled when either protein is depleted or inactivated (4, 15, 17, 31). In addition, we found that multiple pathogenic mutations occur at sites potentially critical for the proper formation or stabilization of the complex (Fig. 4F).
Although RS–CP interactions are known to be critical for ciliary/flagellar motilities (9–11), how they contact is unclear. Based on the fitting of our cryo-EM map into the framework of DMT–CP density of previous cryo-ET studies (Fig. 6 and Movie S3) (4, 44), we propose that the dual-core complexes in each RS head resemble a pair of brake pads pressed from the DMT toward the CP through the RS stalk. The capabilities of the open–close motion and lateral swing of the two protomers relative to each other in each core complex (Fig. 5 and Movie S2) probably enable optimal contacts with various geometries of the CP projections during ciliary beats (Fig. 6). The two “neck” structures that connect each core complex to the stalk (Fig. 6A) (3, 4) might allow each “brake pad” to shift individually for optimal function. Our modeling also suggests that, when contacts occur, the periodic CP projections (44) are able to firmly hold the RS heads (Fig. 6E). As different mechanical pressures exerted on the arms through elastic contact (Fig. 6 E and F) are expected to induce corresponding motions between the two protomers, the elastic contacts would allow changes in the distance according to mechanical pressures. In rigid contacts (Fig. 6 E and F), on the other hand, the Rsph4a teeth would bite into the CP projections to tighten the RS–CP interaction and also keep a persistent DMT–CP distance. These distinct modes of contacts would in turn sense and transmit the mechanical pressures differentially to DMTs to coordinate axonemal dynein activities (10, 11, 47). As metazoan cilia beat perpendicularly to the C1–C2 plane of the CP (9), the two RS heads positioned close to the 1c–1d and the 2c–2d projections (Fig. 6E and Movie S3) might function to restrict the beat plane, whereas the others would facilitate the generation of detailed beat patterns. Apparently, future research is required to achieve comprehensive knowledge on these elegant regulatory machineries of ciliary motility.
Materials and Methods
Protein Expression and Purification.
To express and purify the RS head complex, HEK293T cells were transiently transfected with plasmids of radial spoke components using polyethyleneimine (Polysciences) when reaching 80 to 90% fluency. The harvested cells were lysed by sonication on ice and centrifugation at 20,000 × g for 1 h, and the supernatants were collected, followed by the addition of anti-Flag M2 affinity resins (Sigma) and incubation for 3 h at 4 °C. The resins were rinsed three times with wash buffer (20 mM Hepes, pH 7.5, 150 mM KCl, 10% glycerol, and protease inhibitors). The bound proteins were eluted with wash buffer plus Flag peptide (1 mg/mL). The eluted protein complexes were cross-linked with 0.1% glutaraldehyde (Sigma) for 3 h at 4 °C and neutralized by adding glycine (pH 7.5) to a final concentration of 50 mM for 1 h at 4 °C. After concentration, the sample was subjected to a 10 to 30% glycerol gradient ultracentrifugation.
Cryo-EM Sample Preparation and Data Collection.
Freshly purified RS head core complex (0.4 mg/mL) was placed onto plasma-cleaned grids (Quantifoil R1.2/1.3 200-mesh Cu grids) and plunge-frozen into liquid ethane, cooled with liquid nitrogen, using a Vitrobot Mark IV (Thermo Fisher Scientific). The vitrified RS head core complex showed an obvious preferred-orientation problem. To overcome this problem, we tried several vitrification conditions, including using graphene oxide supports, grids coated with polylysine (35, 48, 49), or sample with an added trace amount of detergent n-Octyl-β-D-glucopyranoside (OG) or n-Dodecyl-β-D-Maltopyranoside (DDM), which allowed a slightly improved broader orientation distribution of the complex. For the PA tag-inserted RS head core complex, the sample was incubated with NZ-1 Fab at a molar ratio of 1:2 (RS head core vs. NZ-1 Fab) on ice for 30 min, and then prepared for vitrification as described.
Selected grids were imaged on a Titan Krios transmission electron microscope (Thermo Fisher Scientific) operated at 300 kV at liquid nitrogen temperature. A total of 6,917 nontilt movies and 2,295 tilt movies (including 1,932 movies at 30° tilt and 363 movies at 40° tilt) were collected with a nominal magnification of 22,500×. The images were recorded on a K2 Summit direct electron detector (Gatan) operated in superresolution mode, yielding a pixel size of 1.02 Å after binning two times. Each frame was exposed for 0.2 s, with an accumulation time of 7.6 s for each movie, leading to a total accumulated dose of 58 e−/Å2 on the specimen.
More detailed procedures for methods are provided in SI Appendix, Materials and Methods.
Supplementary Material
Acknowledgments
We thank Z. Zhou, L. Bao, D. Li, J. Shuai, J. Li, and S. Wang (Institute of Biochemistry and Cell Biology), J. Pan (Tsinghua University), and J. Lin and D. Nicastro (University of Texas Southwestern Medical Center) for their suggestions and support of this project. We are grateful to the staff of the National Center for Protein Science Shanghai Electron Microscopy facility, Database and Computing facility, Mass Spectrometry facility, and Protein Expression and Purification facility for instrument support or technical assistance. This work was supported by the National Key R&D Program of China (2017YFA0503503 to Y.C., X.Y., and X.Z.), Strategic Priority Research Program of the Chinese Academy of Sciences (CAS) (XDB37040103 to Y.C.; XDB19020102 to X.Z.), National Natural Science Foundation of China (31861143028, 31670754, and 31872714 to Y.C.; 31991192 to X.Z.; 31800623 to Z.D.), CAS Major Science and Technology Infrastructure Open Research Projects, and CAS-Shanghai Science Research Center (CAS-SSRC-YH-2015-01 and DSS-WXJZ-2018-0002 to Y.C.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2021180118/-/DCSupplemental.
Data Availability.
All data presented in this study are available within the figures and the supplementary information. A cryo-EM map has been deposited in the EMDB (ID code EMD-30766), and the associated model has been deposited in the PDB (ID code 7DMP).
References
- 1.Luck D., Piperno G., Ramanis Z., Huang B., Flagellar mutants of Chlamydomonas: Studies of radial spoke-defective strains by dikaryon and revertant analysis. Proc. Natl. Acad. Sci. U.S.A. 74, 3456–3460 (1977). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pigino G., et al. , Cryoelectron tomography of radial spokes in cilia and flagella. J. Cell Biol. 195, 673–687 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lin J., Heuser T., Carbajal-González B. I., Song K., Nicastro D., The structural heterogeneity of radial spokes in cilia and flagella is conserved. Cytoskeleton (Hoboken) 69, 88–100 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lin J., et al. , Cryo-electron tomography reveals ciliary defects underlying human RSPH1 primary ciliary dyskinesia. Nat. Commun. 5, 5727 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nicastro D., et al. , The molecular architecture of axonemes revealed by cryoelectron tomography. Science 313, 944–948 (2006). [DOI] [PubMed] [Google Scholar]
- 6.Witman G. B., Plummer J., Sander G., Chlamydomonas flagellar mutants lacking radial spokes and central tubules. Structure, composition, and function of specific axonemal components. J. Cell Biol. 76, 729–747 (1978). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang B., Piperno G., Ramanis Z., Luck D. J., Radial spokes of Chlamydomonas flagella: Genetic analysis of assembly and function. J. Cell Biol. 88, 80–88 (1981). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang P., Yang C., Sale W. S., Flagellar radial spoke protein 2 is a calmodulin binding protein required for motility in Chlamydomonas reinhardtii. Eukaryot. Cell 3, 72–81 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Satir P., Heuser T., Sale W. S., A structural basis for how motile cilia beat. Bioscience 64, 1073–1083 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oda T., Yanagisawa H., Yagi T., Kikkawa M., Mechanosignaling between central apparatus and radial spokes controls axonemal dynein activity. J. Cell Biol. 204, 807–819 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhu X., Liu Y., Yang P., Radial spokes—A snapshot of the motility regulation, assembly, and evolution of cilia and flagella. Cold Spring Harb. Perspect. Biol. 9, a028126 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang P., et al. , Radial spoke proteins of Chlamydomonas flagella. J. Cell Sci. 119, 1165–1174 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Piperno G., Huang B., Ramanis Z., Luck D. J., Radial spokes of Chlamydomonas flagella: Polypeptide composition and phosphorylation of stalk components. J. Cell Biol. 88, 73–79 (1981). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.O’Toole E. T., Giddings T. H. Jr, Porter M. E., Ostrowski L. E., Computer-assisted image analysis of human cilia and Chlamydomonas flagella reveals both similarities and differences in axoneme structure. Cytoskeleton (Hoboken) 69, 577–590 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yoke H., et al. , Rsph4a is essential for the triplet radial spoke head assembly of the mouse motile cilia. PLoS Genet. 16, e1008664 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Castleman V. H., et al. , Mutations in radial spoke head protein genes RSPH9 and RSPH4A cause primary ciliary dyskinesia with central-microtubular-pair abnormalities. Am. J. Hum. Genet. 84, 197–209 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jeanson L., et al. , RSPH3 mutations cause primary ciliary dyskinesia with central-complex defects and a near absence of radial spokes. Am. J. Hum. Genet. 97, 153–162 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kott E., et al. , Loss-of-function mutations in RSPH1 cause primary ciliary dyskinesia with central-complex and radial-spoke defects. Am. J. Hum. Genet. 93, 561–570 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Horani A., Ferkol T. W., Dutcher S. K., Brody S. L., Genetics and biology of primary ciliary dyskinesia. Paediatr. Respir. Rev. 18, 18–24 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Knowles M. R., et al. , Mutations in RSPH1 cause primary ciliary dyskinesia with a unique clinical and ciliary phenotype. Am. J. Respir. Crit. Care Med. 189, 707–717 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shinohara K., et al. , Absence of radial spokes in mouse node cilia is required for rotational movement but confers ultrastructural instability as a trade-off. Dev. Cell 35, 236–246 (2015). [DOI] [PubMed] [Google Scholar]
- 22.Kohno T., Wakabayashi K., Diener D. R., Rosenbaum J. L., Kamiya R., Subunit interactions within the Chlamydomonas flagellar spokehead. Cytoskeleton (Hoboken) 68, 237–246 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abbasi F., et al. , RSPH6A is required for sperm flagellum formation and male fertility in mice. J. Cell Sci. 131, jcs221648 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Curry A. M., Williams B. D., Rosenbaum J. L., Sequence analysis reveals homology between two proteins of the flagellar radial spoke. Mol. Cell. Biol. 12, 3967–3977 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Satouh Y., Inaba K., Proteomic characterization of sperm radial spokes identifies a novel spoke protein with an ubiquitin domain. FEBS Lett. 583, 2201–2207 (2009). [DOI] [PubMed] [Google Scholar]
- 26.Goodenough U. W., Heuser J. E., Substructure of inner dynein arms, radial spokes, and the central pair/projection complex of cilia and flagella. J. Cell Biol. 100, 2008–2018 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Warner F. D., New observations on flagellar fine structure. The relationship between matrix structure and the microtubule component of the axoneme. J. Cell Biol. 47, 159–182 (1970). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Warner F. D., Satir P., The structural basis of ciliary bend formation. Radial spoke positional changes accompanying microtubule sliding. J. Cell Biol. 63, 35–63 (1974). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Poghosyan E., et al. , The structure and symmetry of the radial spoke protein complex in Chlamydomonas flagella. J. Cell Sci. 133, jcs245233 (2020). [DOI] [PubMed] [Google Scholar]
- 30.Delgehyr N., et al. , Ependymal cell differentiation, from monociliated to multiciliated cells. Methods Cell Biol. 127, 19–35 (2015). [DOI] [PubMed] [Google Scholar]
- 31.Zhu L., Liu H., Chen Y., Yan X., Zhu X., Rsph9 is critical for ciliary radial spoke assembly and central pair microtubule stability. Biol. Cell 111, 29–38 (2019). [DOI] [PubMed] [Google Scholar]
- 32.Tan Y. Z., et al. , Addressing preferred specimen orientation in single-particle cryo-EM through tilting. Nat. Methods 14, 793–796 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang H., Han W., Takagi J., Cong Y., Yeast inner-subunit PA-NZ-1 labeling strategy for accurate subunit identification in a macromolecular complex through cryo-EM analysis. J. Mol. Biol. 430, 1417–1425 (2018). [DOI] [PubMed] [Google Scholar]
- 34.Wang Y., et al. , Architecture and subunit arrangement of the complete Saccharomyces cerevisiae COMPASS complex. Sci. Rep. 8, 17405 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zang Y., et al. , Staggered ATP binding mechanism of eukaryotic chaperonin TRiC (CCT) revealed through high-resolution cryo-EM. Nat. Struct. Mol. Biol. 23, 1083–1091 (2016). [DOI] [PubMed] [Google Scholar]
- 36.Li J., et al. , Structure of the MORN4/myo3a tail complex reveals MORN repeats as protein binding modules. Structure 27, 1366–1374.e3 (2019). [DOI] [PubMed] [Google Scholar]
- 37.Emsley P., Cowtan K., Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004). [DOI] [PubMed] [Google Scholar]
- 38.Kelley L. A., Mezulis S., Yates C. M., Wass M. N., Sternberg M. J., The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 10, 845–858 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kwon T., et al. , Mechanism of histone lysine methyl transfer revealed by the structure of SET7/9-AdoMet. EMBO J. 22, 292–303 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ma H., Lou Y., Lin W. H., Xue H. W., MORN motifs in plant PIPKs are involved in the regulation of subcellular localization and phospholipid binding. Cell Res. 16, 466–478 (2006). [DOI] [PubMed] [Google Scholar]
- 41.Sajko S., et al. , Structures of three MORN repeat proteins and a re-evaluation of the proposed lipid-binding properties of MORN repeats. 10.1101/826180 (23 June 2020). [DOI] [PMC free article] [PubMed]
- 42.Frommer A., et al. , Immunofluorescence analysis and diagnosis of primary ciliary dyskinesia with radial spoke defects. Am. J. Respir. Cell Mol. Biol. 53, 563–573 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fernandez-Leiro R., Scheres S. H. W., A pipeline approach to single-particle processing in RELION. Acta Crystallogr. D Struct. Biol. 73, 496–502 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Carbajal-González B. I., et al. , Conserved structural motifs in the central pair complex of eukaryotic flagella. Cytoskeleton (Hoboken) 70, 101–120 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Teves M. E., Nagarkatti-Gude D. R., Zhang Z., Strauss J. F. III, Mammalian axoneme central pair complex proteins: Broader roles revealed by gene knockout phenotypes. Cytoskeleton (Hoboken) 73, 3–22 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Satouh Y., et al. , Molecular characterization of radial spoke subcomplex containing radial spoke protein 3 and heat shock protein 40 in sperm flagella of the ascidian Ciona intestinalis. Mol. Biol. Cell 16, 626–636 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ma M., et al. , Structure of the decorated ciliary doublet microtubule. Cell 179, 909–922.e12 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jin M., et al. , An ensemble of cryo-EM structures of TRiC reveal its conformational landscape and subunit specificity. Proc. Natl. Acad. Sci. U.S.A. 116, 19513–19522 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ding Z., et al. , Structural snapshots of 26S proteasome reveal tetraubiquitin-induced conformations. Mol. Cell 73, 1150–1161.e6 (2019). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data presented in this study are available within the figures and the supplementary information. A cryo-EM map has been deposited in the EMDB (ID code EMD-30766), and the associated model has been deposited in the PDB (ID code 7DMP).