Significance
RNA helicases play critical roles in multiple biological processes. However, little in vivo data are available because of the lethality of mice completely deficient in RNA helicases. Here, we generate mice with conditional knockout of DEAD-box Helicase 15 (Dhx15) in the intestine, in which we found a specific defect in antimicrobial peptide (AMP) α-defensins in Paneth cells. Additionally, we found that Dhx15-specific depletion in the intestine leads to susceptibility to enteric bacterial infection as well as dextran sulfate sodium-induced colitis in mice. In humans, we also found reduced protein levels of Dhx15 in ulcerative colitis patients.
Keywords: DEAD-box helicase 15 (Dhx15), alpha-defensins, Paneth cells, intestinal inflammation, inflammatory bowel diseases (IBD)
Abstract
RNA helicases play roles in various essential biological processes such as RNA splicing and editing. Recent in vitro studies show that RNA helicases are involved in immune responses toward viruses, serving as viral RNA sensors or immune signaling adaptors. However, there is still a lack of in vivo data to support the tissue- or cell-specific function of RNA helicases owing to the lethality of mice with complete knockout of RNA helicases; further, there is a lack of evidence about the antibacterial role of helicases. Here, we investigated the in vivo role of Dhx15 in intestinal antibacterial responses by generating mice that were intestinal epithelial cell (IEC)-specific deficient for Dhx15 (Dhx15 f/f Villin1-cre, Dhx15ΔIEC). These mice are susceptible to infection with enteric bacteria Citrobacter rodentium (C. rod), owing to impaired α-defensin production by Paneth cells. Moreover, mice with Paneth cell-specific depletion of Dhx15 (Dhx15 f/f Defensinα6-cre, Dhx15ΔPaneth) are more susceptible to DSS (dextran sodium sulfate)-induced colitis, which phenocopy Dhx15ΔIEC mice, due to the dysbiosis of the intestinal microbiota. In humans, reduced protein levels of Dhx15 are found in ulcerative colitis (UC) patients. Taken together, our findings identify a key regulator of Wnt-induced α-defensins in Paneth cells and offer insights into its role in the antimicrobial response as well as intestinal inflammation.
RNA helicases comprise over 100 members, including DEAH, DEAD, and DExH box helicases, and are essential for all kinds of cellular processes involving RNA. These proteins catalyze the unwinding of RNA duplexes and the structural rearrangement of RNAs and ribonucleoprotein (RNPs) in an ATP-dependent manner (1). They play important roles in viral infections, in which they act as sensors toward RNA viruses or are hijacked by retroviruses that lack their own RNA helicases (2). For example, retinoic-acid-inducible protein I (RIG-I, also known as DDX58) and melanoma differentiation-associated gene-5 (Mda5), are well-known pattern recognition receptors (PRRs) that recognize cytoplasmic viral RNA in the antiviral response (3). The helicase DDX24 interacts with HIV-1 Gag and Rev to facilitate viral RNA trafficking and genome packaging (4). Furthermore, another helicase, DDX3X, that is the target of multiple viruses, including HCV and HIV, interacts with NLRP3 to drive inflammasome activation in stressed cells (5, 6). However, little is known about the role of helicases in the antibacterial immune response.
Paneth cells, which play a central role in the intestinal antibacterial immune response, are specialized secretory epithelial cells of the small intestine that occupy the base of intestinal crypts (7, 8). They release antimicrobial peptides (AMPs) at mucosal surfaces to form the first line of defense against bacterial antigens in the lumen of the small intestine. AMPs include lysozyme, Reg3s, lectins, and α/β-defensins (9), whose induction is controlled by the TLR/Wnt pathways (10–12).
Dhx15 is an important member of the DEAH-box RNA helicases. Recent studies suggest that Dhx15 is involved in the antiviral immune response, tumor progression, cell proliferation, and apoptosis (13–16). Dhx15 is reported to sense double-stranded RNA to activate type I IFN, together with another helicase, namely Dhx9 (17). It has also been shown to activate NF-κB and MAPK downstream of MAVS (16). In addition, previous studies by us and others show that Dhx15 act as a coreceptor for Nlrp6 or Rig-I to promote antiviral defense against RNA viruses (18, 19). However, whether Dhx15 can regulate intestinal antibacterial response remains unexplored.
To investigate this, we generated Dhx15-floxed mice and crossed them with Villin1-cre mice or Defa6-cre mice to specifically deplete Dhx15 in intestinal epithelial cells (IECs) (Dhx15ΔIEC) or Paneth cells (Dhx15ΔPaneth), respectively. We infected these mice with the enteric bacterium Citrobacter rodentium (C. rod) and elucidated the antibacterial immune response by RNA sequencing. After identifying the defects in α-defensin production in Dhx15ΔIEC and Dhx15ΔPaneth mice, we investigated the potential involvement of Dhx15 in Wnt signaling, which drives α-defensin production. Because AMPs play a crucial role in the development of inflammatory bowel diseases (IBD), we further evaluated the role of Dhx15 in the dextran sodium sulfate (DSS)-induced colitis mouse model and analyzed the expression of DHX15 in IBD specimens.
Results
Dhx15ΔIEC Mice Are Susceptible to C. rod Infection.
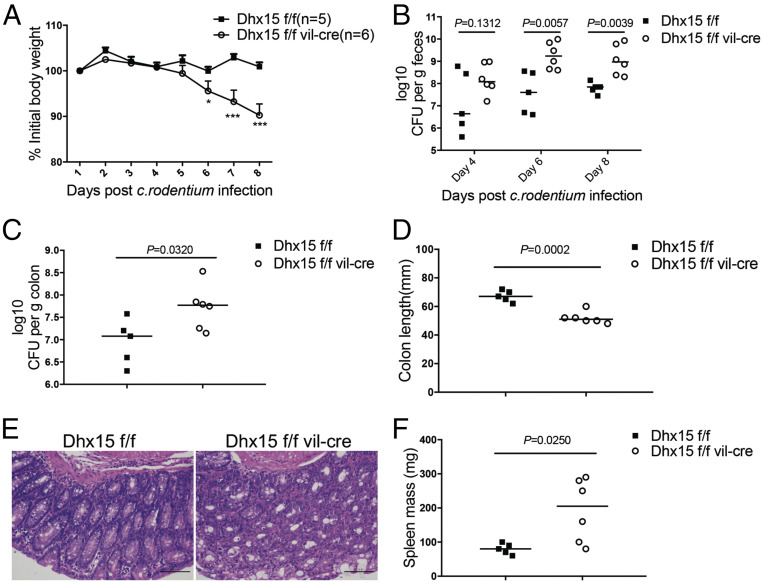
To explore whether Dhx15 plays an important role in mucosal immunity against enteric bacteria, we generated Dhx15ΔIEC mice (SI Appendix, Fig. S1) and infected the mice with C. rod. C. rod is a predominant mucosal enteric murine pathogen that is similar to two human pathogens, namely enteropathogenic Escherichia coli (EPEC) and enterohemorrhagic E. coli (EHEC) (16). After infection, the body weights of mice were monitored for 8 d and then were subjected to immunological and histological analysis. Dhx15ΔIEC mice exhibited progressive weight loss, especially after 7 d postinfection, compared with their littermate WT mice (Fig. 1A). Further, we found that Dhx15ΔIEC mice were incapable of clearing the pathogen, as reflected by a significantly increased burden of C. rod in the stool collected at days 6 and 8 as well as in the colon tissue (Fig. 1 B and C). Accordingly, Dhx15ΔIEC mice also showed increased colonic shortening (Fig. 1D), severe colonic inflammation (Fig. 1E), and splenic enlargement (Fig. 1F) in comparison with their littermate controls. Taken together, these data show that Dhx15 expression in IECs is essential to control enteric bacteria.
Fig. 1.
Dhx15ΔIEC mice are susceptible to C. rod infection. (A) WT and Dhx15ΔIEC mice were orally infected with 1 × 109 cfu of C. rod, and their body weights were monitored daily. (B and C) Measurement of bacterial loads in fecal samples at days 4, 6, and 8 (B) or in colon tissue at day 8 postinfection (C). (D) Measurement of colon length at day 8 after infection. (E) Representative images of H&E-stained colon sections at day 8 after infection. (Scale bars, 100 μm.) (F) Spleen mass of infected mice. Data are representative of three independent experiments. The results are shown as mean ± SEM; *P < 0.05, ***P < 0.001, or as otherwise indicated (Student’s t test).
Dhx15 Specifically Regulates α-Defensins in the Intestine.
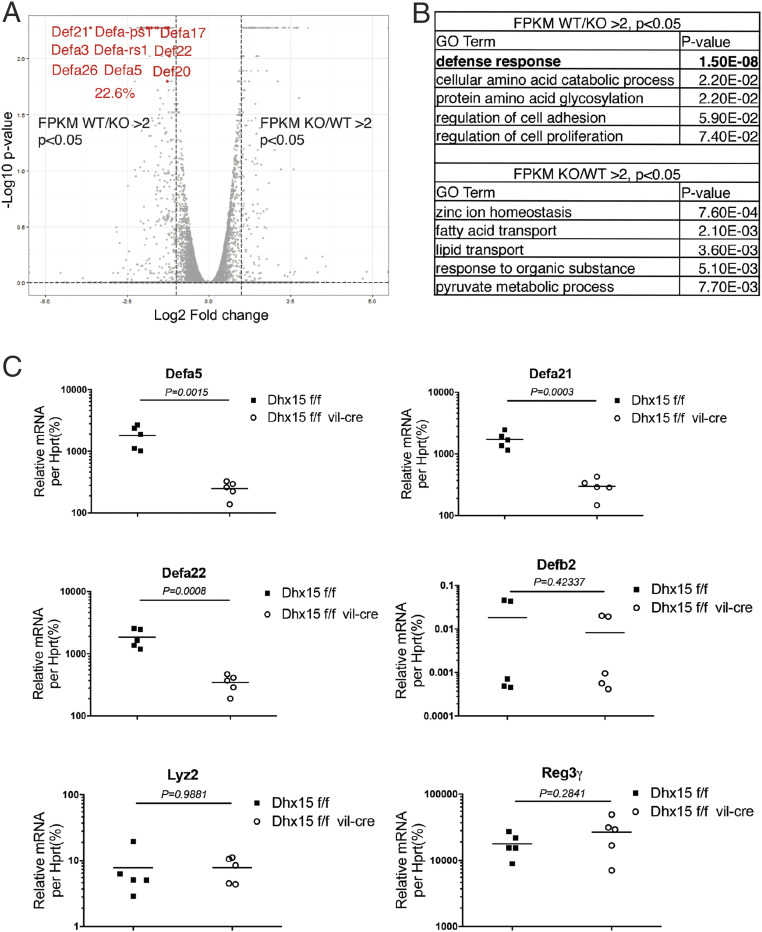
To determine the mechanism by which Dhx15 participates in limiting bacterial pathogens in the small intestine, we isolated IECs (Epcam+CD45−) from Dhx15ΔIEC and littermate WT mice for RNA sequencing (SI Appendix, Fig. S2). Gene ontology (GO) analysis identified several enriched pathways (threshold P value <0.05), with “Defense Response” most significantly (P = 1.5 × 10−8) enriched in down-regulated gene sets (Fig. 2B). Notably, a large proportion (22.6%) of the down-regulated genes in IECs from Dhx15ΔIEC mice versus their littermate WT mice belonged to the α-defensins family, which is labeled red in the volcano plot shown in Fig. 2A. α-Defensins are a group of AMPs, secreted by Paneth cells in the small intestine. Paneth cells secrete α-defensins and other AMPs such as lysozyme, Reg3g, and phospholipase A2 to orchestrate the mucosal immune response against pathogens (7, 10, 20). We then confirmed the expression levels of α-defensins and other AMPs in the small intestine epithelial cells via qPCR. Dhx15 only promoted the expression of α-defensins in a Nlrp6-independent manner, whereas that of other AMPs, such as Reg3g, lysozyme 2, or β-defensins, remained unaffected (Fig. 2C and SI Appendix, Fig. S3). Collectively, these data suggest that Dhx15 specifically regulates the expression of α-defensins in the immune response against intestinal bacterial pathogens.
Fig. 2.
Dhx15 specifically regulates α-defensins in the intestine. (A and B) RNA-seq of IECs from two Dhx15ΔIEC mice compared with their two WT littermates. Volcano plot of differentially regulated genes, with α-defensin genes labeled in red (A); GO analysis of differentially regulated gene sets (B). (C) qPCR validation of the expression levels of α-defensins and other AMPs in IECs from Dhx15ΔIEC mice compared with their WT littermates. Data are representative of three independent experiments. The results are shown as mean ± SEM (Student’s t test).
Dhx15 Induces α-Defensins in a Wnt-Dependent Manner.
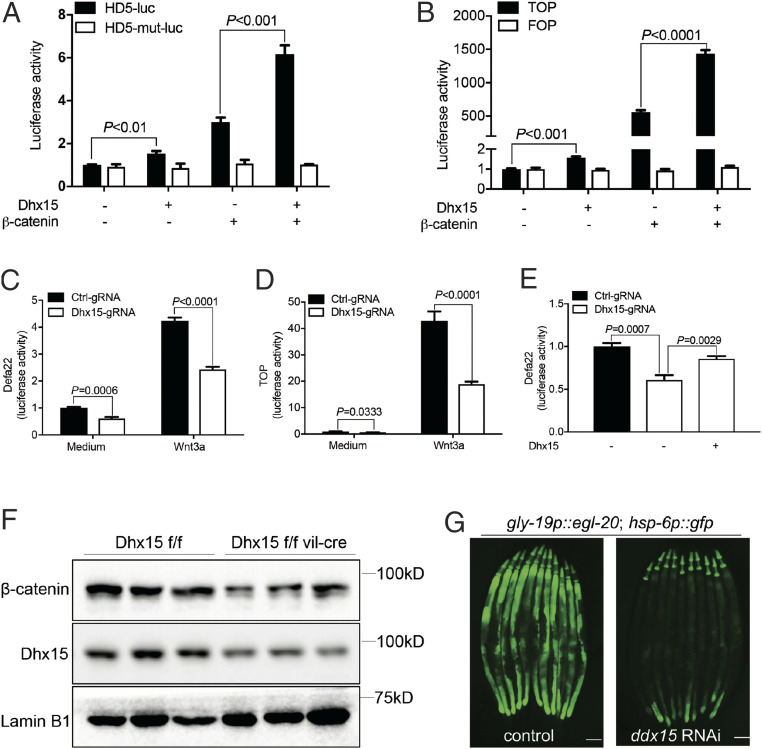
In the small intestine, the Wnt signaling pathway plays a principal role in orchestrating stem cell renewal and differentiation, as well as Paneth cell maturation and differentiation. Wnt can also transcriptionally regulate the expression of α-defensins through β-catenin (11, 21–24) (SI Appendix, Fig. S4). Thus, we attempted to determine whether, and how, Dhx15 might regulate Wnt-induced α-defensins in the intestine. We first constructed luciferase reporter plasmids for human α-defensin HD5 as well as murine α-defensin Defa22, with or without the Wnt/β-catenin–responsive elements in the promoter regions (HD5-luc and HD5-mut-luc), to investigate the role of Dhx15 in Wnt-induced α-defensin. Ectopic expression of human Dhx15 in 293T cells activated HD5 promoter-driven luciferase expression, and Dhx15 induced HD5 synergistically with β-catenin—a key transcriptional regulator downstream of Wnt (25) (Fig. 3A). Moreover, Dhx15 did not induce HD5 in the absence of the Wnt response element in the promoter (Fig. 3A), suggesting that Dhx15 promotes α-defensin expression in a Wnt-dependent manner. TOPFlash (TOP) is a luciferase reporter that contains a minimal fos promoter coupled to TCF-binding sites upstream of a modified firefly luciferase gene. FOPFlash (FOP) is similar, except that the TCF-binding sites are mutated and nonfunctional. Thus, the TOP/FOP assay provides a precise measurement of canonical Wnt/β-catenin transcriptional activity. Consistent with this finding, the same results were observed in TOP/FOP luciferase reporters (Fig. 3B). We also knocked out endogenous Dhx15 in 293T cells using CRISPR/Cas9 technology (SI Appendix, Fig. S5). The depletion of Dhx15 resulted in reduced Wnt-induced Defa22 and TOP luciferase activities (Fig. 3 C and D). The restoration of Dhx15 expression also rescued the impaired Defa22 luciferase activity due to Dhx15 depletion (Fig. 3E). Interestingly, we found reduced levels of β-catenin in the ileum of Dhx15ΔIEC mice (Fig. 3F), indicative of the potential involvement of Dhx15 in the upstream of β-catenin to regulate wnt-induced α-defensins. Moreover, it has been reported that overexpression of Wnt/EGL-20 ligand induces the mitochondrial unfolded protein response (UPRmt) in Caenorhabditis elegans (26). RNA interference against ddx-15, the C. elegans homolog of Dhx15, resulted in strong suppression of the UPRmt in C. elegans expressing Wnt/EGL-20 (Fig. 3G). These results support the conserved role of Dhx15 in regulating Wnt/β-catenin signaling.
Fig. 3.
Dhx15 induces α-defensins in a Wnt-dependent manner. (A and B) Luciferase activity analysis of Wnt reporters in HEK293T cells. Cells are transfected with HD5-luc or HD5-mut-luc (A) TOP or FOP reporters (B) along with indicated plasmids. (C–E) Luciferase activity analysis of Defa22-luc (C and E) and TOP-luc (D) by knocking out endogenous Dhx15, upon Wnt3a stimulation (C–E), and restoration of Dhx15 (E) in HEK293T cells. (F) Nucleic β-catenin expression in the ileum tissue from Dhx15ΔIEC mice and littermate WT mice. LaminB1 was used as the nucleic loading control. (G) Representative images of a Wnt homolog (egl-20)-induced mitoUPR (hsp-6p-gfp) reporter in C. elegans by silencing Dhx15. (Scale bars, 100 μm.) Data are representative of at least three independent experiments. The results are shown as mean ± SEM, ***P < 0.001, or as otherwise indicated (Student’s t test).
Severe DSS-Induced Colitis in Dhx15ΔIEC Mice.
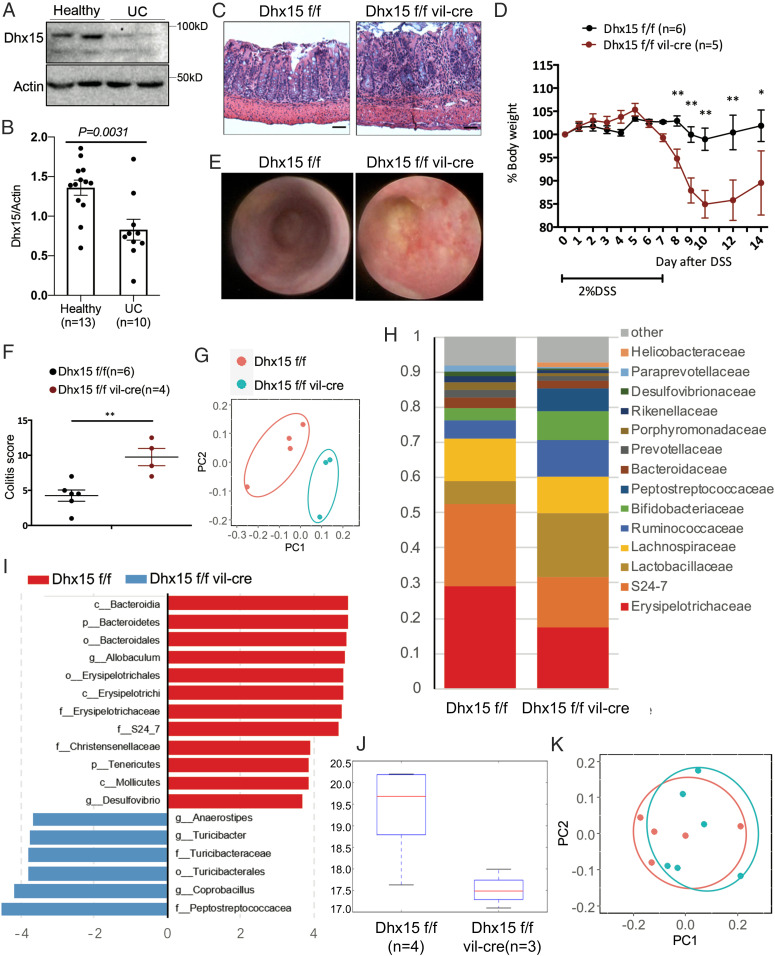
Wnt/β-catenin signaling, which acts as the central organizer of epithelial stem cell identity and maintenance, also serves as a regulator of inflammation. The dysregulated Wnt/β-catenin signaling in IBD (inflammatory bowel disease) is now well recognized (27, 28). We therefore investigated whether there was a correlation between Wnt-induced α-defensins and human intestinal diseases. Intriguingly, in Crohn’s disease (CD), the reduced expression of Wnt-induced α-defensins HD5 and HD6 is tightly correlated with small intestine symptoms (29–31). Mechanistically, the compromised expression of α-defensins HD5 and HD6 leads to an imbalanced microbial community and a defective intestinal barrier function (32–34). Notably, several key elements in Wnt/β-catenin signaling exhibited significantly aberrant expression in ulcerative colitis (UC) compared to non-IBD patients. In support of this, we found decreased expression of Dhx15 in UC patients (Fig. 4 A and B), indicative of potential involvement of Dhx15 in IBD development. To determine whether Dhx15 regulates Wnt induced α-defensins to contribute to IBD pathogenesis, we first performed experiments using a DSS-induced colitis model of Dhx15ΔIEC mice. Indeed, compared with their WT littermates, Dhx15ΔIEC mice showed spontaneous ileitis as evidenced by hematoxylin and eosin (H&E) staining of ileum tissue (Fig. 4C), as well as severe colitis following DSS treatment, which was characterized by higher body weight loss (Fig. 4D), and more rectal bleeding and colorectal polyps as evidenced by endoscopic analysis (Fig. 4E), resulting in higher colitis scores (Fig. 4F). As Dhx15 controlled α-defensin expression in the intestine, we next asked whether Dhx15 regulate microbiota homeostasis in the intestine. Indeed, by separating the Dhx15ΔIEC mice and their littermate WT mice for 9 wk after weaning, Dhx15ΔIEC mice shaped diverse microbiota in comparison to their littermate WT mice, as evidenced by principal coordinates analysis (PCoA) of the 16s sequencing data (Fig. 4G), bacterial composition (Fig. 4H), and Linear discriminant analysis Effect Size (LEfSe) analysis (Fig. 4I). Also, the diversity is much lower in Dhx15ΔIEC mice (Fig. 4J). However, by cohousing the Dhx15ΔIEC mice and their littermate WT mice for 9 wk after weaning, Dhx15ΔIEC mice and their littermate WT mice shaped similar microbiota (Fig. 4K). Together, these results indicated that Dhx15 deficiency may influence disease progression in colitis through dysregulation of Wnt-induced α-defensins as well as by inducing dysbiosis of microbiota.
Fig. 4.
Severe DSS-induced colitis in Dhx15ΔIEC mice. (A and B) Representative bands (A) and statistical analysis (B) of Western blot analysis for DHX15 expression in the gut biopsies from IBD patients and healthy individuals in whom endoscopic screening was performed. (C) Representative images of H&E-stained ileum sections from the indicated mice strain. (Scale bars, 50 μm.) (D–F) Dhx15ΔIEC and WT mice were treated with 2.5% DSS in drinking water for 7 d, followed by administration of normal drinking water for 7 d. (D) Body weight changes. (E) Representative endoscopic images of mice at day 10. (Scale bars, 50 μm.) (F) Summary of endoscopic index of each strain. (G–K) 16S sequencing analysis of stools from Dhx15ΔIEC and WT mice. (G and K) PCoA of 16s sequencing data. (H) Bacterial composition in percentage. (I) LEfSe analysis. (J) Alpha diversity. (K) PCoA of 16s sequencing data from 9-wk cohousing Dhx15ΔIEC and WT mice. Data are representative of two (A, G–K) or three independent experiments (C–F). The results are shown as mean ± SEM; *P < 0.05, **P < 0.01 (Student’s t test).
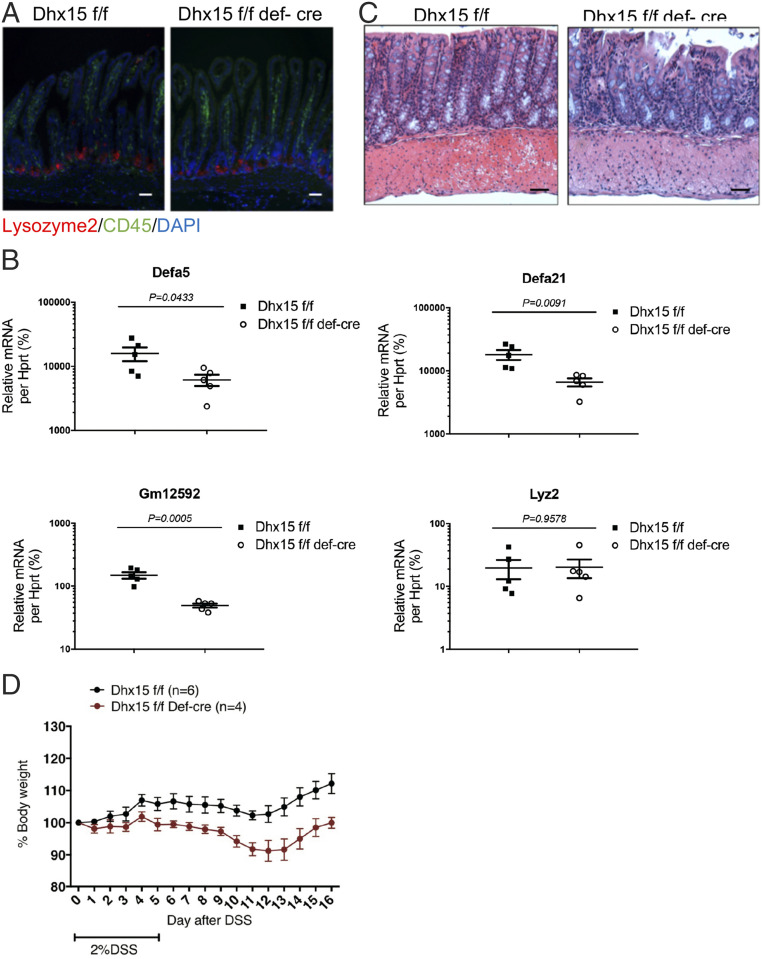
Reduced Expression of α-Defensins and Severe DSS-Induced Colitis in Dhx15ΔPaneth Mice.
To more specifically define the intrinsic role of dhx15 in regulation of α-defensin in intestinal Paneth cells, we next crossed Dhx15-floxed mice with Paneth cell-specific cre mice-defensinα6-cre mice (Dhx15ΔPaneth). The loss or dysfunction of Paneth cells is known to occur during IBD pathogenesis (35, 36); therefore, we first examined whether there is a defect in Paneth cell number in Dhx15ΔPaneth mice. The immunological staining of ileal tissue from Dhx15ΔPaneth mice and WT littermates with anti-lysozyme antibody showed that the Paneth cell number remained unchanged in Dhx15ΔPaneth mice (Fig. 5A). However, similar to Dhx15ΔIEC mice, Dhx15ΔPaneth mice also exhibited defects in the messenger RNA (mRNA) levels of α-defensins in the small intestine (Fig. 5B). Accordingly, Dhx15ΔPaneth mice also developed spontaneous ileitis, as well as severe colitis upon DSS treatment in comparison to WT littermates, which exactly phenocopied Dhx15ΔIEC mice (Fig. 5 C and D). Overall, our data suggest that the loss of Dhx15 in Paneth cells results in reduced α-defensin production and enteritis.
Fig. 5.
Deficiency of Wnt-induced α-defensins and impaired DSS-induced colitis in Dhx15ΔPaneth mice. (A) Immunostaining of Paneth cells by anti-lysozyme antibody. (Scale bars, 50 μm.) (B) qPCR analysis of the expression levels of α-defensins and other AMPs in IECs from Dhx15ΔPaneth mice compared with that of their WT littermates, using qPCR. (C) Representative images of H&E-stained ileum sections from the indicated mice strain. (Scale bars, 50 μm.) (D) Changes in body weight in Dhx15ΔPaneth mice and their littermate controls upon DSS treatment. Data are representative of two (A, C, and D) or three independent experiments (B). The results are shown as mean ± SEM; *P < 0.05, **P < 0.01, ***P < 0.001 (Student’s t test).
Discussion
RNA helicases are essential for RNA-related biological processes. They act as sensors of viral RNA, regulate the microbial life cycle, initiate and modulate immune responses, and assist many other important cellular processes (1, 3). Some of them have been well studied in the context of the immune system, such as RIG-I like proteins, MDA-5, and HEL-1. Although most RNA helicases are so far reported to be involved in the antiviral response, our work shows that a helicase can regulate AMPs in Paneth cells in the ileum, by promoting Wnt-induced α-defensin secretion.
RNA helicases are also prognostic and diagnostic markers and potential drug targets, predominantly for clinical therapies (37, 38). Thus, it is of interest to elucidate the underlying mechanism by which RNA helicases participate in biological processes. Here, we found that the disruption of antibacterial response caused by Dhx15 depletion in IECs is attributable to a previously unanticipated role of Dhx15 in modulating Wnt/β-catenin signaling. The importance of Wnt/β-catenin signaling in a variety of human diseases is well known, and previous studies have also shown other RNA helicases (DDX3 and DDX5) can regulate Wnt/β-catenin signaling in cell-based experiments (39–42). Our work confirms that another RNA helicase, Dhx15, affects Wnt/β-catenin signaling in vivo. Together, these findings indicate promise for potential therapeutic applications involving targeting these RNA helicases to regulate Wnt/β-catenin signaling in order to treat diseases such as CRC (43, 44).
Our previous work revealed that, in response to enteric viruses, Dhx15 associated with Nlrp6 senses double-stranded RNA to activate IFN and ISG production (18, 45). However, in this study, we found that α-defensin production controlled by Dhx15 is Nlrp6-independent, and also inflammasome-independent (SI Appendix, Fig. S6), indicating that Dhx15 might be involved in multiple pathways involving diverse mechanisms. Dhx15 and Dhx9 are shown to form a complex to sense viral RNA (17), and, interestingly, Dhx9 was shown to associate with another intestinal-specific NLR-Nlrp9b to activate pyroptosis in respond to rotaviral infection (46). It would be of interest to analyze AMP production in Dhx9-deficient and Nlrp9b-deficient mice.
In this study, we generated conditional KO mice for Dhx15 to study its in vivo function, and, in addition to its role in antiviral signaling pathways, we found that Dhx15 plays a critical role in the antibacterial response in the intestine by regulating Wnt-induced α-defensin production in Paneth cells. Furthermore, we found reduction of expression of Dhx15 in UC patients, and that Dhx15 deficiency in IECs impairs the intestinal barrier and causes severe DSS-induced colitis. Our work elucidates the in vivo function of Dhx15 in the intestine and the manner in which it regulates Wnt signaling. Further research is warranted to elucidate the mechanisms by which it coordinates these antiviral and antibacterial responses.
Materials and Methods
Mice.
Dhx15-floxed mice were generated by CRISPR technology, the guide (g)RNA sequences for each gene with low off-target score were selected using https://crispr.cos.uni-heidelberg.de/. Two gRNAs chosen were: TGAACTGCAGCAGCGTTCCT and AGATATTATTAGAGTAGCCG. The corresponding donor sequences were:
catctaacagaccttttgtatcatcatcattcagagtagccagtgtgaactgcagcagcgttataactt‐cgtataatgtatgctatacgaagttatcctaggcattgcatcagttttctgcatatccctaaatgtttc‐ttgatagttgatgttttttc
and cttttcagttgcttgaatcccccagcacaaaacaatatactacagatattattagagtagata‐acttcgtataatgtatgctatacgaagttatccgaggaagaccaacctagttctacattttttttaaag‐atttatttatctatatgaatatac.
Vil-cre (villin-cre) mice were purchased from JAX, and def-cre (defensinα6-cre) mice were kindly provided by Richard Blumberg, Harvard Medical School, Boston, MA. The cohoused littermate mice were used as the control in the present study, and mice were maintained under a strict 12-h light cycle under specific pathogen-free conditions. All animal experiments were approved by the Ethics Committee of University of Science and Technology of China.
Human Subjects and Western Blot Analysis.
Informed consent was obtained from all human subjects enrolled in the study for biopsy sampling. Biopsies were obtained from the ileum of individuals undergoing screening colonoscopy. These samples were deidentified prior to use in the study. This study was approved by the institutional review board at the University of Science and Technology of China.
Biopsy tissue was harvested from human subjects and resuspended in 500 μL of ice-cold Pierce IP buffer (87787) containing protease inhibitors (Complete Mini EDTA-free, Roche and PMSF) and phosphatase inhibitor (NaF and NaVO3). Cells were lysed for 30 min at 4 °C, and lysates were spun for 30 min at maximum speed at 4 °C. Samples were separated on 8% Tris-Bis gels and transferred onto polyvinylidene fluoride (PVDF) membranes (Millipore). Western blot analysis was performed using antibodies against Dhx15 (Santa Cruz, sc-271686), β-actin (Proteintech, 60008-1-Ig).
Targeting of Dhx15 Using CRISPR–Cas9-Mediated Genome Editing in HEK-293T Cells.
Lentiviral CRISPR–Cas9 targeting gRNA expressing the vector lentiCRISPRv2 was obtained from Addgene (52961). The gRNA sequence for Dhx15 was “caccgGATGTCCAAGCGGCACCGGT.” Dhx15 gRNAs and Cas9 were transduced into HEK-293T cell lines by lentiviral delivery with lentiCRISPRv2, followed by selection of gRNA-expressing cells with puromycin (Life Technologies).
C. rod Infection.
C. rod was cultured in prewarmed Luria-Bertani (LB) broth supplemented with nalidixic acid (100 μg/mL), with shaking for 9 h at 37 °C. Mice were fasted overnight prior to infection with 1 × 109 colony forming unit (cfu) per mouse by oral gavage. Body weight was recorded daily for 8 d. Mice were killed at day 8. Spleen mass and colon length were assessed. Furthermore, spleen, liver, colon, mesenteric lymph node, and fecal pellets were harvested to determine bacterial counts. Serial dilutions of homogenized tissues and fecal pellets were plated on MacConkey agar plates and incubated for 24 h at 37 °C.
DSS.
Acute colitis was induced via the administration of DSS (molecular mass 36–50 kDa; MP Biomedicals) in drinking water (2.5%, wt/vol). Mice were weighed daily.
Endoscopic Procedures.
For monitoring colitis in vivo, mice were anesthetized using an intraperitoneal injection of ketamine (Ratiopharm)/Rompun (Bayer) and a high-resolution coloview miniendoscope (Karl-Storz) was used as previously described. The mean endoscopic index of colitis severity (MEICS) was determined in a blinded fashion by evaluating stool consistency, fibrin deposition, vasculature, translucency, and granularity of the colon wall. Each of these five different parameters of inflammation was given a score from 0 to 3, resulting in a total MEICS ranging from 0 to 15.
Plasmids.
HD5 and Defa22 luciferase constructs were synthesized from human or mouse genomic DNA by PCR and subcloned into the pGL4.27 luciferase reporter vector (Promega). HD5-mut-luc mutant plasmid was constructed using Agilent’s QuikChange method, for the mutagenesis of TCF consensus elements. PCR primers are as follows:
HD5 forward primer: TGGGTACCTCCAGGGACTAGGAGAAAGTGT
HD5 reverse primer: CCAAGCTTCAGGAGAATGGCAGCAAGGA
Defa22 forward primer: TTGGTACCCTCCCATGTGCCTAAGTGGG
Defa22 Reverse primer: CCGATATCGGAGCCCAATGTGTGGAGAA
HD5-mut forward primer: GCTGGGAGAAAGAGGAGCGCCAAAGGGATC‐TTGAGAAC
HD5-mut reverse primer: GTTCTCAAGATCCCTTTGGCGCTCCTCTTTCTCCCAGC
Cell Culture.
HEK293T (ATCC CRL-3216) cells were cultured in Dulbecco's Modified Eagle medium (DMEM) with 10% fetal bovine serum (FBS). These cell lines are not listed in the database of commonly misidentified cell lines maintained by International Cell Line Authentication Committee and not authenticated or tested for mycoplasma contamination.
Cell Sorting.
Small intestines were excised and flushed thoroughly three times with phosphate-buffered saline (PBS). They were turned inside out and cut into ∼1-cm sections; then, sections were transferred into RPMI with 2 mM EDTA and shaken for 20 min at 37 °C. Supernatants were collected through a 100-mm cell strainer to obtain single-cell suspensions. Cells were collected as the IEC fraction, which contained both epithelial cells (∼90%) and lymphocytes (IEL, ∼10%). Single-cell suspensions were collected and stained with fluorescence-activated cell sorting (FACS) antibodies in FACS buffer (PBS with 2% bovine serum albumin (BSA) and 5 mM EDTA) for 20 min at 4 °C for surface staining, and then sorted on a BD FACSAria flow cytometer. CD45.2- EpCAM+ cells were sorted as IECs. The FACS antibodies used are as follows: CD45.2-FITC (104), EpCAM-APC, and EpCAM-PE/cy7(G8.8),
Quantitative PCR with Reverse Transcription.
Total RNA was extracted from IECs, lamina propria leukocytes (LPLs), or FACS-sorted cells, using TRIzol reagent (TaKaRa). RNA was purified according to the manufacturer’s instructions. Reverse transcription (RT) was performed according to the manufacturer’s instructions for the TaKaRa Advantage RT-for-PCR kit. After RT, qPCR (SYBR premix EX Taq, TaKaRa) was performed using the CFX384 Real-Time System (Bio-Rad). The sequences of primers used for the detection of mRNA transcripts is listed below:
Defa5-F:TTGGGCTCCTGCTCAACAAT
Defa5-R:GACACAGCCTGGTCCTCTTC
Defa21-F:CCAGGGGAAGATGACCAGGCTG
Defa21-R:TGCAGCGACGATTTCTACAAAGGC
Defa22 -F:ACGTCGCTGCAATAGAGGAG
Defa22 -R:GCCTCAGAGCTGATGGTTGT
Reg3γ -F:CAGACAAGATGCTTCCCCGT
Reg3γ -R:GCAACTTCACCTTGCACCTG
Lyz2 -F:GAATGGAATGGCTGGCTACT
Lyz2 -R:CGTGCTGAGCTAAACACACC
Defb2 -F: TGGAGTCTGAGTGCCCTTTC
Defb2 -R: AGTGGTCAAGTTCTGCTTCGT
Gm12592 -F: AGAGGAAGAGGAGCAGGCTA
Gm12592 -R: CCCTTTCTGCAGGTCCCATT
Vil1 -F: ATCTCCCTGAGGGTGTGGA
Vil1 -R: AGTGAAGTCTTCGGTGGACAG
CD45 -F: ATGGTCCTCTGAATAAAGCCCA
CD45 -R: TCAGCACTATTGGTAGGCTCC
Gapdh -F: TGCACCACCAACTGCTTAGC
Gapdh -R: GGAAGGCCATGCCAGTGA
Data were analyzed using the Sequence Detection Software according the ΔCt method. All results were normalized to the corresponding values for Hprt or Gapdh quantified in parallel amplification reactions.
H&E Staining.
For histology, colon tissue was flushed thoroughly with ice cold PBS for 3 times and fixed in 10% neutral-buffered formalin and embedded in paraffin. Intestine sections were affixed into 5-µm slides, deparaffinized, and stained with H&E. Stained sections were examined under a light microscope (BX53, Olympus) for morphology.
Luciferase Reporter Assays for Wnt/β-Catenin Signaling.
TOP is a luciferase reporter that contains a minimal fos promoter coupled to TCF-binding sites upstream of a modified firefly luciferase gene. FOP is similar, except that the TCF-binding sites are mutated and nonfunctional. Thus, this assay provides a precise measurement of canonical Wnt-specific transcriptional activity.
The plasmids were each cotransfected with pRLTK, the Renilla luciferase plasmid that controls for transfection efficiency. Transfections were carried out with Lipofectamine 2000 in 24-well plates. 293T cells were harvested after 24 h in culture, and both firefly (TOP) and Renilla luciferase activity was measured in duplicate or triplicate according to the manufacturer’s instructions. The firefly luciferase activity was normalized against the Renilla luciferase (pRLTK) activity.
RNA-Sequencing and Data Analysis.
RNA-Sequencing (RNA-Seq) library preparation and sequencing for IEC samples were performed by Berry Genomics on an Illumina 2500 machine. RNA-Seq fastq files were first processed by fastp to remove adaptor and low-quality reads, and then aligned to the mm10 mouse genome assembly by STAR (v2.5.3a), followed by quantification with HT-seq (v0.9.0). The resultant gene count data were first quantile-adjusted using conditional maximum likelihood model (estimateCommonDisp function of EdgeR [v3.29.2]), then differentially expressed genes between the experimental groups were identified by edgeR (v3.29.2), and further filtered for at least 1.5-fold changes. GO analysis was performed using Metascape (http://metascape.org/gp/index.html#/main/step1).
Statistical Analysis.
The sample size chosen for the animal experiments in this study was estimated based on our prior experience of performing similar sets of experiments. All animal results were included, and no method of randomization was applied. For all the bar graphs, data were expressed as mean ± SEM. Statistical analyses were performed with a standard two-tailed unpaired Student’s t test or paired Student’s t test using GraphPad Prism 5. To compare two nonparametric datasets, a Mann–Whitney u test was used. P values ≤0.05 were considered to indicate significant results. The sample sizes (biological replicates), specific statistical tests used, and the main effects of our statistical analyses for each experiment are shown in each figure legend.
Supplementary Material
Acknowledgments
We thank Xingxing Ren and Runzhi Li for maintaining some of the mouse colonies and Min Huang for plasmid construction. This work was supported by National Key R&D Program of China Grant 2018YFA0508000 (to S.Z.), Strategic Priority Research Program of the Chinese Academy of Sciences Grant XDB29030101 (to S.Z.), National Natural Science Foundation of China Grants 81822021, 91842105, 31770990, 81821001 (to S.Z.), and 91753141/82030042/32070917 (to H.-B.L.), and Fundamental Research Funds for the Central Universities Grants WK2070000159 (to S.Z.). R.A.F. is an Investigator at the Howard Hughes Medical Institute.
Footnotes
Competing interest statement: R.F. is a cofounder of Ventus, which studies inflammatory pathways.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2017432118/-/DCSupplemental.
Data Availability Statement.
The data that support the findings of this study are available from the corresponding author upon reasonable request. RNA-seq datasets have been deposited in Gene Expression Omnibus (Bioproject no. PRJNA684445).
References
- 1.Steimer L., Klostermeier D., RNA helicases in infection and disease. RNA Biol. 9, 751–771 (2012). [DOI] [PubMed] [Google Scholar]
- 2.Krishnan V., Zeichner S. L., Alterations in the expression of DEAD-box and other RNA binding proteins during HIV-1 replication. Retrovirology 1, 42 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yoneyama M., et al. , The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat. Immunol. 5, 730–737 (2004). [DOI] [PubMed] [Google Scholar]
- 4.Ma J., et al. , The requirement of the DEAD-box protein DDX24 for the packaging of human immunodeficiency virus type 1 RNA. Virology 375, 253–264 (2008). [DOI] [PubMed] [Google Scholar]
- 5.Samir P., et al. , DDX3X acts as a live-or-die checkpoint in stressed cells by regulating NLRP3 inflammasome. Nature 573, 590–594 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Szappanos D., et al. , The RNA helicase DDX3X is an essential mediator of innate antimicrobial immunity. PLoS Pathog. 14, e1007397 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clevers H. C., Bevins C. L., Paneth cells: Maestros of the small intestinal crypts. Annu. Rev. Physiol. 75, 289–311 (2013). [DOI] [PubMed] [Google Scholar]
- 8.Ghosh D., et al. , Paneth cell trypsin is the processing enzyme for human defensin-5. Nat. Immunol. 3, 583–590 (2002). [DOI] [PubMed] [Google Scholar]
- 9.Sankaran-Walters S., Hart R., Dills C., Guardians of the gut: Enteric defensins. Front. Microbiol. 8, 647 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Muniz L. R., Knosp C., Yeretssian G., Intestinal antimicrobial peptides during homeostasis, infection, and disease. Front. Immunol. 3, 310 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van Es J. H., et al. , Wnt signalling induces maturation of Paneth cells in intestinal crypts. Nat. Cell Biol. 7, 381–386 (2005). [DOI] [PubMed] [Google Scholar]
- 12.Menendez A., et al. , Bacterial stimulation of the TLR-MyD88 pathway modulates the homeostatic expression of ileal Paneth cell α-defensins. J. Innate Immun. 5, 39–49 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen X. L., et al. , ETS1 and SP1 drive DHX15 expression in acute lymphoblastic leukaemia. J. Cell. Mol. Med. 22, 2612–2621 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ito S., Koso H., Sakamoto K., Watanabe S., RNA helicase DHX15 acts as a tumour suppressor in glioma. Br. J. Cancer 117, 1349–1359 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jing Y., et al. , DHX15 promotes prostate cancer progression by stimulating Siah2-mediated ubiquitination of androgen receptor. Oncogene 37, 638–650 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mosallanejad K., et al. , The DEAH-box RNA helicase DHX15 activates NF-κB and MAPK signaling downstream of MAVS during antiviral responses. Sci. Signal. 7, ra40 (2014). [DOI] [PubMed] [Google Scholar]
- 17.Lu H., et al. , DHX15 senses double-stranded RNA in myeloid dendritic cells. J. Immunol. 193, 1364–1372 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang P., et al. , Nlrp6 regulates intestinal antiviral innate immunity. Science 350, 826–830 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pattabhi S., Knoll M. L., Gale M. Jr, Loo Y. M., DHX15 is a coreceptor for RLR signaling that promotes antiviral defense against RNA virus infection. J. Interferon Cytokine Res. 39, 331–346 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ostaff M. J., Stange E. F., Wehkamp J., Antimicrobial peptides and gut microbiota in homeostasis and pathology. EMBO Mol. Med. 5, 1465–1483 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wallmen B., Schrempp M., Hecht A., Intrinsic properties of Tcf1 and Tcf4 splice variants determine cell-type-specific Wnt/β-catenin target gene expression. Nucleic Acids Res. 40, 9455–9469 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haegebarth A., Clevers H., Wnt signaling, lgr5, and stem cells in the intestine and skin. Am. J. Pathol. 174, 715–721 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gregorieff A., et al. , Expression pattern of Wnt signaling components in the adult intestine. Gastroenterology 129, 626–638 (2005). [DOI] [PubMed] [Google Scholar]
- 24.Miyoshi H., Wnt-expressing cells in the intestines: Guides for tissue remodeling. J. Biochem. 161, 19–25 (2017). [DOI] [PubMed] [Google Scholar]
- 25.Nusse R., Clevers H., Wnt/β-Catenin signaling, disease, and emerging therapeutic modalities. Cell 169, 985–999 (2017). [DOI] [PubMed] [Google Scholar]
- 26.Zhang Q., et al. , The mitochondrial unfolded protein response is mediated cell-non-autonomously by retromer-dependent Wnt signaling. Cell 174, 870–883.e17 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Serafino A., et al. , WNT-pathway components as predictive markers useful for diagnosis, prevention and therapy in inflammatory bowel disease and sporadic colorectal cancer. Oncotarget 5, 978–992 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moparthi L., Koch S., Wnt signaling in intestinal inflammation. Differentiation 108, 24–32 (2019). [DOI] [PubMed] [Google Scholar]
- 29.Courth L. F., et al. , Crohn’s disease-derived monocytes fail to induce Paneth cell defensins. Proc. Natl. Acad. Sci. U.S.A. 112, 14000–14005 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Armbruster N. S., Stange E. F., Wehkamp J., In the Wnt of Paneth cells: Immune-epithelial crosstalk in small intestinal Crohn’s disease. Front. Immunol. 8, 1204 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Beisner J., et al. , TCF-1-mediated Wnt signaling regulates Paneth cell innate immune defense effectors HD-5 and -6: Implications for Crohn’s disease. Am. J. Physiol. Gastrointest. Liver Physiol. 307, G487–G498 (2014). [DOI] [PubMed] [Google Scholar]
- 32.Wehkamp J., et al. , Reduced Paneth cell alpha-defensins in ileal Crohn’s disease. Proc. Natl. Acad. Sci. U.S.A. 102, 18129–18134 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Williams A. D., et al. , Human alpha defensin 5 is a candidate biomarker to delineate inflammatory bowel disease. PLoS One 12, e0179710 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wehkamp J., et al. , The Paneth cell alpha-defensin deficiency of ileal Crohn’s disease is linked to Wnt/Tcf-4. J. Immunology 179, 3109–3118 (2007). [DOI] [PubMed] [Google Scholar]
- 35.Adolph T. E., et al. , Paneth cells as a site of origin for intestinal inflammation. Nature 503, 272–276 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cadwell K., et al. , A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells. Nature 456, 259–263 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Frick D. N., The hepatitis C virus NS3 protein: A model RNA helicase and potential drug target. Curr. Issues Mol. Biol. 9, 1–20 (2007). [PMC free article] [PubMed] [Google Scholar]
- 38.Bol G. M., et al. , Targeting DDX3 with a small molecule inhibitor for lung cancer therapy. EMBO Mol. Med. 7, 648–669 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cruciat C. M., et al. , RNA helicase DDX3 is a regulatory subunit of casein kinase 1 in Wnt-β-catenin signaling. Science 339, 1436–1441 (2013). [DOI] [PubMed] [Google Scholar]
- 40.Yang F., et al. , Cis-acting circ-CTNNB1 promotes β-Catenin signaling and cancer progression via DDX3-mediated transactivation of YY1. Cancer Res. 79, 557–571 (2019). [DOI] [PubMed] [Google Scholar]
- 41.Zhang M., et al. , The lncRNA NEAT1 activates Wnt/β-catenin signaling and promotes colorectal cancer progression via interacting with DDX5. J. Hematol. Oncol. 11, 113 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen Y., et al. , DEAD-box helicase 5 interacts with transcription factor 12 and promotes the progression of osteosarcoma by stimulating cell cycle progression. Front. Pharmacol. 9, 1558 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 43.Bass A. J., et al. , Genomic sequencing of colorectal adenocarcinomas identifies a recurrent VTI1A-TCF7L2 fusion. Nat. Genet. 43, 964–968 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang X., et al. , Mutations in X-linked PORCN, a putative regulator of Wnt signaling, cause focal dermal hypoplasia. Nat. Genet. 39, 836–838 (2007). [DOI] [PubMed] [Google Scholar]
- 45.Li R., Zhu S., NLRP6 inflammasome. Mol. Aspects Med. 76, 100859 (2020). [DOI] [PubMed] [Google Scholar]
- 46.Zhu S., et al. , Nlrp9b inflammasome restricts rotavirus infection in intestinal epithelial cells. Nature 546, 667–670 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request. RNA-seq datasets have been deposited in Gene Expression Omnibus (Bioproject no. PRJNA684445).