Significance
The small size of short ORF-encoded peptides (SEPs) has made it difficult to identify their cellular binding partners using standard methods. Here, we show that the photo-crosslinking amino acid AbK can be incorporated into overexpressed, epitope-tagged SEP transgenes, allowing covalent bond formation between SEPs and their interactors in live cells. From an AP-MS–based screen of conserved mammalian SEPs, we identified short ORF-encoded histone binding protein (SEBHP), a micropeptide which acts as a transcriptional regulator, capable of modulating more than 15% of the active transcriptome. The methodology described herein will likely be of broad utility in annotating the essential physiological roles of SEPs.
Keywords: short open reading frame-encoded peptide, expanded genetic code, photo-crosslinking, transcriptional regulation
Abstract
Recent technological advances have expanded the annotated protein coding content of mammalian genomes, as hundreds of previously unidentified, short open reading frame (ORF)-encoded peptides (SEPs) have now been found to be translated. Although several studies have identified important physiological roles for this emerging protein class, a general method to define their interactomes is lacking. Here, we demonstrate that genetic incorporation of the photo-crosslinking noncanonical amino acid AbK into SEP transgenes allows for the facile identification of SEP cellular interaction partners using affinity-based methods. From a survey of seven SEPs, we report the discovery of short ORF-encoded histone binding protein (SEHBP), a conserved microprotein that interacts with chromatin-associated proteins, localizes to discrete genomic loci, and induces a robust transcriptional program when overexpressed in human cells. This work affords a straightforward method to help define the physiological roles of SEPs and demonstrates its utility by identifying SEHBP as a short ORF-encoded transcription factor.
Advances in proteomics and next generation sequencing have indicated that in addition to the ∼19,000 nuclear-encoded human genes hundreds to thousands of short open reading frames (sORFs, also called smORFs) likely contribute to the functional proteome (1–3). With a high level of ribosomal occupancy, sORFs give rise to an abundant protein class, sORF-encoded peptides (SEPs, often called microproteins), which are present at tens to thousands of molecules per cell (2). Historically, SEPs have evaded detection by virtue of their small size (<150 codons) and due to the stringent criteria employed by gene prediction algorithms (1, 4). Translation of SEPs is often initiated at non-AUG start sites (5), and sORFs can be located within annotated reference open reading frames (ORFs) as well as within 5′ UTRs and in noncoding RNAs (6). SEPs have been identified in most organisms studied (1) and frequently display a high degree of conservation among higher eukaryotes (6–8). A handful of studies have recently revealed essential cellular functions for SEPs, including roles in metabolism (9), muscle performance (10), organismal development (11, 12), apoptosis (13), and DNA repair (14, 15). Despite the growing importance of SEPs, a general, robust strategy to identify their cellular functions is lacking.
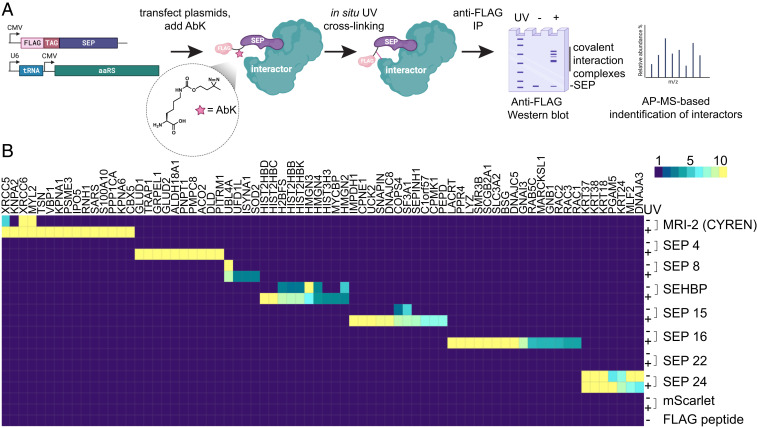
With a median length of less than 50 amino acids, SEPs themselves likely do not possess enzymatic activity, but instead act by modulating the functions of their client proteins. As such, a key step in annotating the roles of SEPs is to first define their cellular interactomes. Although a number of methods have been used to elucidate the targets of SEPs, including appending an engineered peroxidase to biotinylate potential interactors (APEX-tagging) and using CRISPR-Cas to edit an ORF’s native locus (10, 16, 17), identification of SEP interactors remains a challenge. Here, we describe an enhanced affinity purification mass spectrometry (AP-MS)–based mapping strategy to covalently fix SEP interactors in living cells, which was inspired by our previous work in deconvoluting the cellular targets of biologically active small molecules identified from phenotypic screens (18, 19). These approaches typically use a bifunctional photo-activatable affinity probe molecule bearing a small photo-crosslinking group (e.g., diazirine, aryl azide) to covalently link target to probe in live cells and an affinity or reactivity handle (e.g., biotin, alkyne) for isolation and detection. Analogously, genetic encoding of a photo-crosslinking noncanonical amino acid (ncAA) into an epitope-tagged SEP transgene would allow for covalent bond formation between a SEP and its interactor in situ and would provide a means to detect and enrich SEP complexes (Fig. 1A). In contrast to methods involving the appendage of a much larger fusion protein (e.g., APEX, eGFP), such a strategy would, in principle, allow for enhanced enrichment of low affinity or transient interaction complexes while minimally interfering with the small binding surface of the SEP.
Fig. 1.
A genetically encoded photo-crosslinker identifies cellular interactors of unexplored SEPs. (A) Schematic depicting the strategy used in this work to introduce the photo-crosslinking amino acid AbK (structure within inset circle) into SEP transgenes and identify their protein targets in cells. (B) Heatmap of the relative spectral count enrichment corresponding to protein preys identified from AP-MS experiments with the indicated SEP baits in the presence or absence of UV (mean of three biological replicates).
Here, we demonstrate that a photo-crosslinking amino acid can be incorporated into SEPs at high levels and subsequently used to enrich putative binding partners in pull down experiments. From an AP-MS–based screen of uncharacterized SEPs, we demonstrate the utility of this approach with the discovery of a sORF-encoded peptide that binds chromatin-associated proteins and regulates transcription in human cells.
Results
A Genetically Encoded Photo-Crosslinking Strategy for Identifying the Cellular Targets of SEPs.
We first sought to determine the feasibility of this mapping methodology in the context of a SEP with defined binding partners. The nuclear-localized SEP MRI-2 (also called CYREN) determines at which phases of the cell cycle nonhomologous end joining repairs double-stranded DNA breaks by binding the XRCC6 and XRCC5 heterodimer (X-ray Repair Cross Complementing 6/5; often called Ku70/Ku80, respectively) (14). To introduce a photo-crosslinker into MRI-2 in mammalian cells, we used an evolved mutant of the Methanosarcina barkeri pyrrolysyl aminoacyl transfer RNA (tRNA) synthetase/tRNA pair (expressed from the vector pCMV-AbK) to genetically encode the diazirine functionalized ncAA N6-[[2-(3-Methyl-3H-diazirin-3-yl)ethoxy]carbonyl]-l-lysine (AbK) (20). Photoactivation leads to a highly reactive carbene species, which can insert into a wide number of C-H and unsaturated C-C bonds. We next generated a transient mammalian expression vector encoding a FLAG-tagged MRI-2 transgene harboring an amber codon for ncAA suppression (Fig. 1A). Transient transfection of the two vectors in HEK293T cells in the presence of AbK (1 mM) allowed for robust expression of a WT FLAG-tagged MRI-2 (MRI-2-FLAG) and an AbK-containing transgene (MRI-2-AbK-FLAG), which were detected after anti-FLAG immunoprecipitation (SI Appendix, Fig. S1A).
As expected, exposure of MRI-2-AbK-FLAG–expressing cells to long-wave ultraviolet (UV) light (365 nm) induced the appearance of high molecular weight adducts in denaturing anti-FLAG Western blots after immunoprecipitation, suggesting successful formation of covalent adducts between MRI-2 and its interactors (SI Appendix, Fig. S1A). Analysis of the immunoprecipitated material by shotgun proteomics revealed that MRI-2-Abk-FLAG formed crosslinks to XRCC6 and XRCC5 (∼20-fold and ∼5-fold over background, respectively; SI Appendix, Fig. S1B). In samples exposed to UV, the enrichment of XRCC6 and XRCC5 was significantly increased relative to nonirradiated samples (>10-fold and 2-fold, respectively; SI Appendix, Fig. S1B). Further, only under UV irradiation were a number of other potential interactors identified, such as members of the importin complex (Importin 5 [IPO5], Karyopherin alpha 1 and 2 [KPNA1 and KNPA2]) and the DNA-binding protein Translin (TSN; SI Appendix, Fig. S1B) (21). Together, these results suggested our mapping strategy provides a useful method for detecting cellular binding partners of SEPs.
A Proteomics-Based Screen to Identify the Targets of Conserved Mammalian SEPs.
We next applied this methodology in an unbiased fashion to deconvolute the targets of previously uncharacterized SEPs. We used a recently described an approach for integrating de novo transcriptome assembly and Ribo-sequencing (seq), which resulted in the annotation of more than 1,000 sORFs with high translational potential in mammalian cells (3). From this set, we selected a subset of 24 high confidence SEPs which had no annotated cellular functions, that displayed high coding sequence similarity across mammals, and whose presence was cellularly validated by Ribo-seq (SI Appendix, Table S1). We next created a library of overexpression vectors encoding transgenes from this subset containing an N-terminal 3xFLAG sequence, a GGSG linker, and an amber codon to encode AbK before readthrough of a C-terminal SEP (SI Appendix, Table S2). A Western blotting-based screen for overexpression in HEK293T cells demonstrated that seven members (SEP 4, 8, 10, 15, 16, 22, and 24) expressed at detectable levels and 4 SEPs of this subset (SEP 4, 16, 22, and 24) exhibited high molecular weight adducts when exposed to UV (SI Appendix, Fig. S2).
We then performed shotgun proteomics to analyze the potential interactomes of these seven SEPs in the presence or absence of UV irradiation in HEK293T cells. We included an overexpressed 3xFLAG and AbK-modified mScarlet fluorescent protein as well as a 3xFLAG peptide control to ensure that identified interactors were not artifacts of overexpression or nonspecific binders to the anti-FLAG resin (22). From 48 individual proteomic profiling runs, evaluation of biological triplicates allowed for the semiquantitative comparative enrichment of spectral counts across samples to identify putative interactors (Fig. 1B and Dataset S1). Overall, this dataset suggested that all of the tagged SEPs enriched specific interactomes with the exception of SEP22 (Fig. 1B). Exposure of samples to UV irradiation generally increased the magnitude of enrichment and substantially expanded the breadth of interactors identified (e.g., SEP4, Fig. 1B). Key highlights from this profiling screen included the discovery that SEP4 exclusively enriched mitochondrial localized enzymes, including glutamate dehydrogenase 1 and 2 (GLUD1 and GLUD2), likely suggesting mitochondrial localization (Fig. 1B). Additionally, SEP16 was found to bind the central signaling GTPases, RAC1, RAC2, and RAC3 (Ras-related C3 botulinum toxin substrate 1, 2, and 3) as well as multiple transmembrane and signaling related components, perhaps suggesting a role in cell signaling (Fig. 1B). Notably, the interactomes from SEP4 and SEP16 were uniquely identified in conditions exposed to UV-based crosslinking (Fig. 1B and Dataset S2).
Characterization of SEHBP, a Chromatin-Associated SEP.
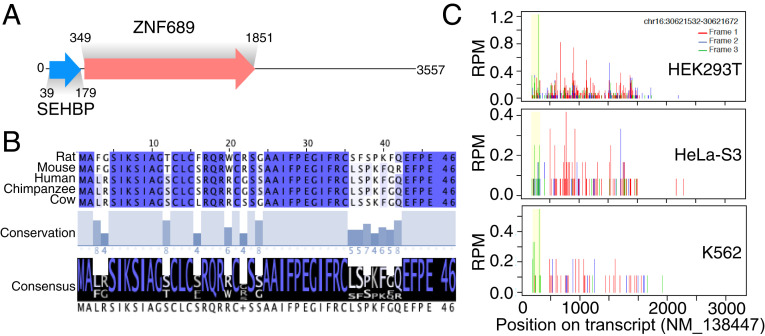
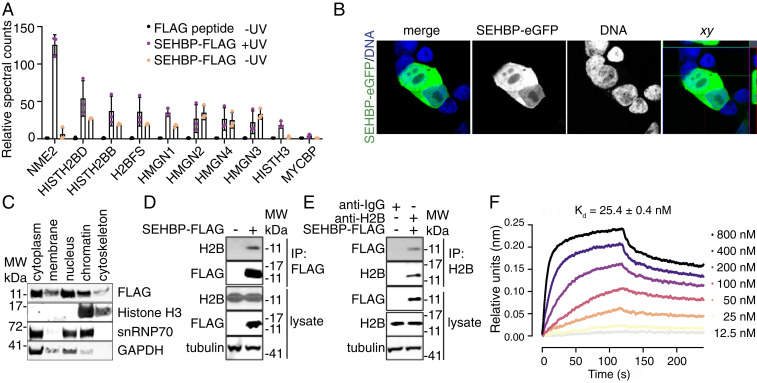
Given the large number of potential interactors identified in this single experiment, we thought it best to initially focus on characterizing SEP10, which immunoprecipitated a number of chromatin-associated proteins. SEP10 is found within the 5′UTR of ZNF689 (Zinc Finger Protein 689) and encodes a 46 amino acid gene product that displays high amino acid sequence homology across mammalian species (Fig. 2 A and B). Using data derived from HEK293T, HeLa-S3, and K562 cells, Ribo-seq–based profiling suggested that the SEP10 sORF has high ribosomal occupancy in these human cell lines (Fig. 2C) (2, 3). In our AP-MS profiling experiments, SEP10 was found to precipitate histone H2B proteins isoforms (called H2B throughout), multiple members of the high mobility group nucleosome binding domain family (HMGN1, HMGN2, HMGN3, HMGN4) as well as other nuclear proteins known to be involved with transcriptional regulation such as NME2 (nucleoside diphosphate kinase 2) and MYCBP (c-Myc binding protein; Fig. 3A) (23–25). Together these data hinted at the intriguing hypothesis that SEP10 might be a chromatin-associated protein with a potential role in epigenetic regulation—we therefore termed SEP10 as short ORF-encoded histone binding protein (SEHBP).
Fig. 2.
SEHBP is a translated 5′ ORF that is conserved in mammals. (A) Schematic depicting the position of the SEHBP and ZNF689 ORFs within transcript NM_138447. (B) Multiple sequence alignment depicting the sequence (Top), conservation (Middle), and consensus (Bottom) of the SEHBP amino acid sequence across the indicated mammalian species. (C) Representative A-site plots (Ribo-seq) from the indicated cell lines with the ORF of SEHBP shown in yellow.
Fig. 3.
SEHBP interacts with Histone H2B in cells. (A) Relative spectral counts of the indicated anti-FLAG immunoprecipitated proteins after 48-h expression of SEHBP-AbK-FLAG in HEK293T cells followed by exposure to UV (n = 3; mean ± SD). (B) Representative confocal microscopy-derived images of SEHBP-eGFP localization (green) from HEK293T cells after 48 h of SEHBP-eGFP expression followed by fixation and exposure to Hoechst 33342. (C) Representative Western blotting analysis for the indicated proteins after 48-h expression of SEHBP-FLAG followed by subcellular fractionization. (D) Western blotting for endogenous H2B content after anti-FLAG immunoprecipitation from HEK293T cells expressing SEHBP-FLAG. (E) Western blotting analysis for SEBHP-FLAG content after anti-H2B immunoprecipitation from HEK293T cells expressing SEHBP-FLAG. (F) Sensorgram plot from biolayer interferometry experiments demonstrating the interaction between recombinant H2B and immobilized SEHBP.
To explore this hypothesis, we first investigated the subcellular localization and interactors of SEHBP using independent methods. Immunofluorescent analysis of a C-terminally tagged, enhanced green fluorescent protein (eGFP) SEHBP suggested that SEHBP localizes in nuclear and cytoplasmic compartments in HEK293T cells (Fig. 3B). Further, we found that an unmodified SEHBP-FLAG transgene was also detected in the cytoplasm and nuclear compartments when subcellular fractions were evaluated by anti-FLAG Western blotting (Fig. 3C). Because SEHBP is found in the nuclear compartment, we next evaluated if its nuclear interactors could be validated by noncovalent methods. From a screen of HA-tagged preys (H2B, Histone H3, MYCBP, HMGN1, and HMGN4), H2B alone was efficiently immunoprecipitated from HEK293T cells expressing SEHBP-FLAG without the need for crosslinking (SI Appendix, Fig. S3A). Endogenous H2B protein could additionally be immunoprecipitated from HEK293T cells expressing SEHBP-FLAG, and SEHBP itself could be pulled down when an anti-H2B antibody was used for immunoprecipitation (Fig. 3 D and E). Lastly, we confirmed that recombinant SEHBP and H2B strongly associate in vitro, as biolayer interferometry experiments determined their dissociation constant to be 25 ± 0.4 nM (Fig. 3F). We also used the chemical crosslinkers disuccinimidyl suberate and N-ε-malemidocaproyl-oxysuccinimide ester as an independent method to trap other potential interactors. We found that a number of higher molecular weight adducts could be immunoprecipitated from HEK293T cells expressing SEHBP-FLAG (SI Appendix, Fig. S3B). This data suggested that SEHBP likely binds other cellular targets including those identified from our proteomics experiment but with lower affinity than H2B. Evaluating the presence of endogenous interactors from pulldowns using chemical crosslinkers, we confirmed that SEHBP additionally interacts with endogenous HMGN1 and HMGN3, as higher molecular weight adducts of these proteins were identified by Western blotting (SI Appendix, Fig. S3C).
Regulation of Transcription by SEHBP.
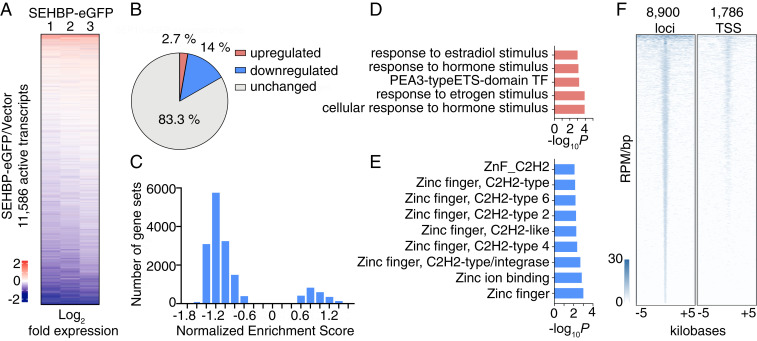
Given its association with chromatin, we next sought to understand if SEHBP might play a role in regulating transcription. Transient overexpression of SEHBP-eGFP in HEK293T cells resulted in a marked effect on gene expression, as 16.7% of the active transcriptome (1,985 of 11,586 transcripts) was significantly modulated in RNA-seq experiments when compared to a vector control (Fig. 4 A and B). Interestingly, the majority of transcripts altered by SEHBP overexpression were down-regulated (84%), suggesting SEHBP may play a largely suppressive role in regulating transcription. Gene set enrichment analysis corroborated this hypothesis, as analysis of all gene sets within the MSigDB revealed the majority of them gave negative enrichment values (Fig. 4C) (26). Evaluating the top 200 up-regulated and down-regulated transcripts by Gene Ontology term analysis suggested that SEHBP likely regulates a specific transcriptional program, as up-regulated transcripts were associated with responses to steroid hormones and down-regulated transcripts were strongly associated with the suppression of zinc finger protein encoding transcripts (Fig. 4 D and E) (27).
Fig. 4.
SEHBP modulates transcription at distinct loci. (A) Rank-ordered heat map depicting the log2-fold change of mRNA levels in response to SEHBP overexpression relative to a vector control in RNA-seq experiments using HEK293T cells (three biological replicates). (B) Pie chart depicting the percentage of active transcripts significantly changed by SEHBP overexpression (P < 0.05). (C) Histogram comparing the normalized enrichment scores of all MSigDB gene sets using GSEA of RNA-seq data in response to SEHBP overexpression. Plots depicting the P values of representative up-regulated (D) and down-regulated (E) gene sets from RNA-seq analysis of HEK293T cells expressing SEHBP. (F) Rank-ordered heat map of ChIP-seq derived reads of SEHBP-based enrichment at 8,900 distinct loci (Left) and 1,786 TSSs (Right).
Lastly, we sought to understand if SEHBP might itself occupy distinct loci in the genome. In chromatin immunoprecipitation (ChIP)-seq experiments from HEK293T cells, SEBHP-eGFP was found to localize throughout the genome: SEHBP was associated with 8,900 loci (Fig. 4 F, Left). Consistent with its role as a transcriptional regulator, SEHBP also occupied 1,786 transcriptional start sites (TSSs, Fig. 4 F, Right). Importantly, from a survey of some of the most dynamically regulated transcripts by RNA-seq analysis, SEHBP was found to occupy these genomic loci with high coverage at the TSSs and throughout CDS (SI Appendix, Fig. S4).
Discussion
The ability to genetically encode ncAAs in mammalian cells has provided a large repertoire of useful tools to probe the functions of proteins in their native environment (28). Here, we have exploited our ability to site-specifically incorporate the photoactivated amino acid AbK into epitope-tagged SEP transgenes to covalently crosslink interacting proteins in situ. Performing coimmunoprecipitation after UV exposure enables efficient enrichment of covalent SEP complexes and Western blotting allows for facile detection of higher molecular weight species. In contrast to standard coimmunoprecipitation, forming a covalent bond between interacting proteins allows for the enhanced capture of transient or weak cellular interactions, negating typical concerns about buffer choice and interaction affinity. While chemical crosslinkers have also been used in coimmunoprecipitation experiments, high concentrations of compound are typically required (1–10 mM) that are toxic to living cells and only nucleophilic amino acids are amenable to labeling. The genetic encoding of the diazirine-containing ncAA AbK overcomes these limitations.
Although we were able to overexpress a number of SEPs at high levels, some SEPs did not express or did not display visible formation of high molecular weight adducts in response to UV treatment (SI Appendix, Fig. S2). While low expression may derive from intrinsic cellular regulation, changing the placement of the epitope tag (C- vs. N-terminal) or encoding AbK at an alternative site within the SEP may allow for increased expression and target engagement. A potential limitation of the current study was the reliance upon the 3xFLAG epitope tag for immunoprecipitation, which may alter the physical properties of a SEP and sterically block certain interactions in the cell. We have recently described methodology to genetically encode multiple ncAAs at distinct sites within a single protein in mammalian cells (29). As such, future efforts to simultaneously incorporate a photo-crosslinker (e.g., AbK, p-benzoyl-phenylalanine) and an orthogonal reactivity handle (e.g., homopropargylglycine, p-acetylphenylalanine) would likely allow for enhanced identification of binding partners without the need for epitope tagging.
We have shown that coupling our photo-crosslinking strategy to AP-MS–based detection increases the ability to identify known interactors of established SEPs and considerably expands the potential to identify novel interactors of this protein class. We initially evaluated the nuclear-localized SEP MRI-2 and identified its reported interactors XRCC6/XRCC5 (Ku70/80) as well as a series of unreported targets including members of the importin complex and the protein Translin (TSN). As part of a heterodimeric complex with Translin-associated factor X (TRAX), TSN-TRAX mediates signaling in response to sensing DNA double-strand breaks (21, 30). Given that MRI-2 has recently been reported to be a cell-cycle–dependent inhibitor of the NHEJ machinery XRCC6/XRCC5, our study raises the interesting possibility that MRI-2 may additionally engage other proteins to modulate DNA repair (14). In addition, we have used an AP-MS–based screen to annotate the interactomes of seven previously uncharacterized SEPs, resulting in a dataset of hundreds of interactions (Dataset S2). On the basis of this data, we hypothesize that SEP4 likely localizes to the mitochondria, and that SEP16 may play a role in cell signaling. Importantly, the vast majority of interactors identified from this screen were from UV-exposed samples, suggesting that photo-crosslinking markedly augments the potential to identify novel interactions by immunoprecipitation. We anticipate this dataset will be a rich source of hypothesis-generating material for future lines of inquiry and that this methodology will serve as a generalizable tool to characterize the interactomes of SEPs in future studies.
To demonstrate the utility of our approach in identifying biologically relevant interactions, we have reported the initial characterization of SEHBP, a tight binder of histone H2B that interacts with chromatin-associated proteins including HMGN1 and HMGN3. HMGN proteins are a family of small (<20 kDa for HMGN1-4), ubiquitously expressed scaffolding proteins which bind and dynamically space nucleosomes to modulate chromatin accessibility (23). HMGNs do not sequence specifically bind DNA but localize to enhancers and promoters through interactions with nucleosomes and other chromatin-associated proteins, resulting in transcriptional modulation essential for development and stemness (31–33). Because SEHBP localizes to distinct genomic loci and binds H2B and HMGNs, we hypothesize that SEHBP may act analogously to or assist in the functions of HMGN family proteins. Further, we found that SEHBP overexpression induces a robust transcriptional program in HEK293T cells, which included up-regulating multiple genetic targets also induced by hormonal stimuli, like estradiol. Curiously, these analyses also indicated that SEHBP down-regulates a large number of largely unclassified zinc finger-containing proteins (e.g., ZNF76, ZNF385C, ZNF783, SI Appendix, Fig. S4) and it is itself transcribed from a messenger RNA (mRNA) encoding a zinc finger protein (ZNF689). Determining whether this transcriptional program is a result of modulating active chromatin or derives from sequence specific targeting through yet-unknown interactions will necessarily be the work of future studies.
In addition to its activities in the nucleus, SEHBP localizes to the cytoplasm, suggesting this microprotein could play other cellular roles or may be regulated by cytoplasmic factors. Interestingly, our AP-MS screen indicated that SEHBP interacts with MYCBP, a protein that cell-cycle dependently shuttles from the cytoplasm to the nucleus to augment the activity of the transcriptional amplifier c-Myc during S phase (34). These observations raise the intriguing hypothesis that SEHBP may act similarly to or aid in the activity of MYCBP by modulating transcription in a cell cycle-dependent manner. Subsequent experiments aimed at understanding the structure and context specific activity of SEHBP will undoubtedly shed more light on its functions in regulating transcription and cellular physiology.
Materials and Methods
Cell Culture.
HEK293T and HeLa-S3 cells were purchased from American Type Culture Collection and propagated in Dulbecco's Modified Eagle Medium (DMEM) (Corning) supplemented with 10% fetal bovine serum (FBS, Corning) and 1% penicillin/streptomycin (Gibco). K562 cells were purchased from Millipore Sigma and propagated in RPMI 1640 (Corning) supplemented with 10% FBS (Corning) and 1% penicillin/streptomycin (Gibco). All cells were maintained at 37 °C with 5% CO2.
Incoporation of AbK and Photo-Crosslinking.
HEK293T cells were plated at 2 × 105 cells per well in six-well plates in 2 mL of growth medium. After 24 h, each well was transfected with 1,000 ng of pCMV6-FLAG-MRI-2-Abk and 1,000 ng of pCMV6-Abk in 100 μL of OptiMEM medium (Gibco) along with 6 μL of FuGENE HD (Promega). AbK (0.5 mM, Tocris) was added 1 h after transfection, and after an additional 48-h incubation, cells were washed with phosphate-buffered saline (PBS). Cells were then irradiated with UV (365 nm) for 30 min at 4 °C using a Stratalinker 1800 (Stratagene). Samples were then disrupted by the addition of cold RIPA buffer and subsequent sonication (Branson, SFX150) for 5 s followed by a 10-min centrifugation at 4 °C at 16,000 × g. The supernatant was incubated overnight at 4 °C with anti-FLAG M2 magnetic beads (Sigma) in radioimmunoprecipitation assay (RIPA) buffer in the presence of protease inhibitors (Halt Protease Inhibitor Mixture, Thermo Fisher Scientific). Beads were then washed with RIPA buffer and eluted with 0.1 M glycine (pH 3.5). The eluates were then used for proteomics analysis.
Affinity Purification Mass Spectrometry.
Enriched proteins were digested with trypsin prior to analysis on a Q Exactive Hybrid Quadrupole-Orbitrap mass spectrometer using previously established conditions (15). Peptides were queried against the human UniProt database supplemented with the sequences of the baits themselves. To probe this dataset for interactors uniquely enriched for each bait, we normalized spectral counts of all enriched interactors against the FLAG-linker background control. The filtering criteria for identifying interactors for each bait was 1) detection in all three replicates and 2) ≥fourfold change in spectral counts with respect to every other bait.
Coimmunoprecipitation Experiments.
For coimmunoprecipitation, HEK293T cells were plated at 2 × 105 cells per well in six-well plates in 2 mL of growth medium. After 24 h, each well was transfected with 1,000 ng of pCMV6 vector encoding HA-tagged transgenes and 1,000 ng of pCMV6-SEHBP-FLAG in 100 μL of OptiMEM medium along with 6 μL of FuGENE HD. After 48-h incubation, cells were washed with PBS and then disrupted with cold RIPA buffer and sonication for 5 s followed by a 10-min centrifugation at 4 °C with 16,000 × g. The supernatant was incubated overnight at 4 °C with anti-FLAG M2 magnetic beads in RIPA buffer in the presence of protease inhibitors followed by washing with PBS and RIPA buffer, then eluted with 0.1 M glycine (pH 3.5). The eluate was subjected to sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS-PAGE) and Western blotting using anti-HA and anti-FLAG antibodies. The interaction of SEHBP-FLAG with endogenous H2B was performed as above but without the addition of HA-tagged transgene-encoding plasmids. Coimmunoprecipitation of H2B was performed by first preincubating the lysate with 5 µg of an anti-H2B antibody or with a nontargeting IgG control before the addition of protein G agarose beads (Invitrogen).
Biolayer Interferometry Experiments.
Biotinylated SEHBP (20 pM, RS Synthesis LLC) was loaded onto streptavidin-coated biosensors in kinetic buffer (DPBS, 1% bovine serum albumin, and 0.1% Tween 20) for 600 s using an Octet RED96 instrument (ForteBio). The membrane was exposed to a dilution series of H2B (800 nM, 400 nM, 200 nM, 100 nM, 50 nM, 25 nM, and 12.5 nM) and blank buffer for 120 s followed by exposing to the kinetic buffer for additional 120 s at room temperature. The traces were globally fitted on 1–1 binding model to calculate kinetic parameters.
Ribo-Seq Experiments.
Ribo-seq experiments were performed as described previously (3).
RNA-Seq Experiments.
HEK293T cells were plated at 2 × 105 cells per well in six-well plates in 2 mL of growth medium. After 24 h each well was transfected with 1,000 ng of pCMV6-SEHBP-EGFP in 100 μL of OptiMEM medium along with 3 µL of FuGENE HD. After 48-h incubation, cells were washed with PBS and suspended with cold PBS. The cells were pelleted at 300 × g, and 5 × 106 cells were used for total RNA purification using an RNeasy kit (Qiagen). Three biological replicates were submitted for library preparation and sequencing at BGI. Transcript abundance was estimated using Kallisto (https://pachterlab.github.io/kallisto).
Subcellular Protein Fractionation.
HEK293T cells transfected with pCMV6-SEHBP-FLAG were harvested and fractionated with subcellular protein fractionation kit for cultured cells (Thermo Fisher Scientific) before performing Western blotting as above.
ChIP-Seq Experiments.
ChIP-seq was conducted according to established protocols (35) with minor modifications. Briefly, 5 × 106 cells of HEK293T cells were plated on poly-d-lysine–coated 150-mm dishes in 25 mL of growth medium. After 24 h, 10 μg of pCMV6-SEHBP-EGFP in 600 μL of OptiMEM medium was transfected using 30 μL of FuGENE HD. After an additional 48-h incubation, 50 million cells were crosslinked, sonicated, and enriched with anti-GFP (Abcam, ab290): magnetic protein G beads (Dynabeads, Thermo Fisher Scientific) complex. The samples were eluted and reverse crosslinked. Then, the DNA fragments were purified by MinElute Reaction Cleanup Kit (Qiagen). Libraries for Illumina sequencing were prepared using ThruPLEX DNA-seq Kit (Rubicon) and purified using AMPure beads (Beckman Coulter).
Supplementary Material
Acknowledgments
This work was supported by NIH Grants R01 GM132071 (to P.G.S.) and R01 GM102491 (to A.S.). We thank Kristen Williams for assistance with manuscript submission.
Footnotes
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2021943118/-/DCSupplemental.
Data Availability.
RNA-sequencing data have been deposited in Gene Expression Omnibus (GEO), https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE164239.
References
- 1.Saghatelian A., Couso J. P., Discovery and characterization of smORF-encoded bioactive polypeptides. Nat. Chem. Biol. 11, 909–916 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Slavoff S. A., et al. , Peptidomic discovery of short open reading frame-encoded peptides in human cells. Nat. Chem. Biol. 9, 59–64 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Martinez T. F., et al. , Accurate annotation of human protein-coding small open reading frames. Nat. Chem. Biol. 16, 458–468 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chu Q., Ma J., Saghatelian A., Identification and characterization of sORF-encoded polypeptides. Crit. Rev. Biochem. Mol. Biol. 50, 134–141 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aspden J. L., et al. , Extensive translation of small open reading frames revealed by poly-ribo-seq. eLife 3, e03528 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nielsen M. M., et al. , Identification of expressed and conserved human noncoding RNAs. RNA 20, 236–251 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bazzini A. A., et al. , Identification of small ORFs in vertebrates using ribosome footprinting and evolutionary conservation. EMBO J. 33, 981–993 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mackowiak S. D., et al. , Extensive identification and analysis of conserved small ORFs in animals. Genome Biol. 16, 179 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee C., et al. , The mitochondrial-derived peptide MOTS-c promotes metabolic homeostasis and reduces obesity and insulin resistance. Cell Metab. 21, 443–454 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Anderson D. M., et al. , A micropeptide encoded by a putative long noncoding RNA regulates muscle performance. Cell 160, 595–606 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kondo T., et al. , Small peptides switch the transcriptional activity of Shavenbaby during Drosophila embryogenesis. Science 329, 336–339 (2010). [DOI] [PubMed] [Google Scholar]
- 12.Zanet J., et al. , Pri sORF peptides induce selective proteasome-mediated protein processing. Science 349, 1356–1358 (2015). [DOI] [PubMed] [Google Scholar]
- 13.Guo B., et al. , Humanin peptide suppresses apoptosis by interfering with Bax activation. Nature 423, 456–461 (2003). [DOI] [PubMed] [Google Scholar]
- 14.Arnoult N., et al. , Regulation of DNA repair pathway choice in S and G2 phases by the NHEJ inhibitor CYREN. Nature 549, 548–552 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Slavoff S. A., Heo J., Budnik B. A., Hanakahi L. A., Saghatelian A., A human short open reading frame (sORF)-encoded polypeptide that stimulates DNA end joining. J. Biol. Chem. 289, 10950–10957 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chu Q., et al. , Identification of microprotein-protein interactions via APEX tagging. Biochemistry 56, 3299–3306 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yeasmin F., Yada T., Akimitsu N., Micropeptides encoded in transcripts previously identified as long noncoding RNAs: A new chapter in transcriptomics and proteomics. Front. Genet. 9, 144 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bollong M. J., et al. , A metabolite-derived protein modification integrates glycolysis with KEAP1-NRF2 signalling. Nature 562, 600–604 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bollong M. J., et al. , A vimentin binding small molecule leads to mitotic disruption in mesenchymal cancers. Proc. Natl. Acad. Sci. U.S.A. 114, E9903–E9912 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ai H. W., Shen W., Sagi A., Chen P. R., Schultz P. G., Probing protein-protein interactions with a genetically encoded photo-crosslinking amino acid. ChemBioChem 12, 1854–1857 (2011). [DOI] [PubMed] [Google Scholar]
- 21.Gupta A., Pillai V. S., Chittela R. K., Translin: A multifunctional protein involved in nucleic acid metabolism. J. Biosci. 44, 139 (2019). [PubMed] [Google Scholar]
- 22.Bindels D. S., et al. , mScarlet: a bright monomeric red fluorescent protein for cellular imaging. Nat. Methods 14, 53–56 (2017). [DOI] [PubMed] [Google Scholar]
- 23.Nanduri R., Furusawa T., Bustin M., Biological functions of HMGN chromosomal proteins. Int. J. Mol. Sci. 21, 449 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xiong J., Du Q., Liang Z., Tumor-suppressive microRNA-22 inhibits the transcription of E-box-containing c-Myc target genes by silencing c-Myc binding protein. Oncogene 29, 4980–4988 (2010). [DOI] [PubMed] [Google Scholar]
- 25.Zhu S., et al. , A small molecule primes embryonic stem cells for differentiation. Cell Stem Cell 4, 416–426 (2009). [DOI] [PubMed] [Google Scholar]
- 26.Subramanian A., et al. , Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang W., Sherman B. T., Lempicki R. A., Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 (2009). [DOI] [PubMed] [Google Scholar]
- 28.Young D. D., Schultz P. G., Playing with the molecules of life. ACS Chem. Biol. 13, 854–870 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xiao H., et al. , Genetic incorporation of multiple unnatural amino acids into proteins in mammalian cells. Angew. Chem. Int. Ed. Engl. 52, 14080–14083 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McFarlane R. J., Wakeman J. A., Translin-Trax: Considerations for oncological therapeutic targeting. Trends Cancer 6, 450–453 (2020). [DOI] [PubMed] [Google Scholar]
- 31.He B., et al. , Binding of HMGN proteins to cell specific enhancers stabilizes cell identity. Nat. Commun. 9, 5240 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kugler J. E., Deng T., Bustin M., The HMGN family of chromatin-binding proteins: Dynamic modulators of epigenetic processes. Biochim. Biophys. Acta 1819, 652–656 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martínez de Paz A., Ausió J., HMGNs: The enhancer charmers. BioEssays 38, 226–231 (2016). [DOI] [PubMed] [Google Scholar]
- 34.Taira T., et al. , AMY-1, a novel C-MYC binding protein that stimulates transcription activity of C-MYC. Genes Cells 3, 549–565 (1998). [DOI] [PubMed] [Google Scholar]
- 35.Erb M. A., et al. , Transcription control by the ENL YEATS domain in acute leukaemia. Nature 543, 270–274 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-sequencing data have been deposited in Gene Expression Omnibus (GEO), https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE164239.