Significance
T-lineage specification requires waves of gene network changes to terminate multipotency and establish T identity. Circumstantial evidence has suggested roles for Runt domain-related (Runx) factors in these regulatory events; Runx1 is strongly expressed, but Runx1 knockouts have previously shown little impact during stages around T-lineage commitment. We show that Runx1 and Runx3 function together to drive T-lineage commitment, binding the same sites. Double knockouts block both activation of T-lineage genes and repression of progenitor genes. Contrasting with the stable activity of Runx proteins throughout these stages, Runx factors preferentially regulate target genes that sharply change expression during T commitment. We show that this reflects pronounced, global redistributions of Runx binding choices across the genome before and after T-lineage commitment.
Keywords: Runx transcription factors, early T lymphocyte development, transcriptional regulation, DNA binding site choice, functional genomics
Abstract
Runt domain-related (Runx) transcription factors are essential for early T cell development in mice from uncommitted to committed stages. Single and double Runx knockouts via Cas9 show that target genes responding to Runx activity are not solely controlled by the dominant factor, Runx1. Instead, Runx1 and Runx3 are coexpressed in single cells; bind to highly overlapping genomic sites; and have redundant, collaborative functions regulating genes pivotal for T cell development. Despite stable combined expression levels across pro-T cell development, Runx1 and Runx3 preferentially activate and repress genes that change expression dynamically during lineage commitment, mostly activating T-lineage genes and repressing multipotent progenitor genes. Furthermore, most Runx target genes are sensitive to Runx perturbation only at one stage and often respond to Runx more for expression transitions than for maintenance. Contributing to this highly stage-dependent gene regulation function, Runx1 and Runx3 extensively shift their binding sites during commitment. Functionally distinct Runx occupancy sites associated with stage-specific activation or repression are also distinguished by different patterns of partner factor cobinding. Finally, Runx occupancies change coordinately at numerous clustered sites around positively or negatively regulated targets during commitment. This multisite binding behavior may contribute to a developmental “ratchet” mechanism making commitment irreversible.
Runt domain-related (Runx) family transcription factors regulate transcriptional and epigenetic programs essential for multiple developmental processes (1–6). T cell development is particularly dependent on Runx family factor activity from its earliest pro-T cell stages (7, 8). Both Runx-dependent activation and Runx-dependent repression are important (9–12). The recognition motif for Runx factors is also frequently enriched in cis-regulatory regions of genes used in early T cell development (11, 13–17), although the targets most sensitive to Runx activity are still not fully defined.
As Runx1 is the predominant Runx factor expressed in early T-lineage development (17, 18), it could be assumed to mediate these activities. Runx1 is critical for hematopoietic development from the earliest embryonic stem cell stage onward (19, 20), and among hematopoietic cells tested, intrathymic T cell precursors express the highest levels of Runx1 (17, 18). Runx1 plays a long-established role in opening the T cell receptor (TCR) coding loci for recombination (21–24). Runx1 is also a functional regulatory collaborator both of transcription factors that are active after pro-T cell lineage commitment (11) and of those acting before (refs. 14 and 15; reviewed in ref. 25). However, although pro-T cell stages are all nearly eliminated when core binding factor β (CBFβ), the common partner of all Runx factors, is down-regulated (8), the acute disruption of Runx1 caused only mild transcriptome effects in pro-T cells (14). Furthermore, impacts of Runx1 deletion in vivo are mainly seen in postcommitment pro-T cell stages (10, 26), later than the effect of deleting Cbfb. Therefore, we have tested whether other Runx family members compete, complement, or collaborate with Runx1 in shaping the gene regulatory programs involved in pro-T cell specification.
Three Runx paralogs Runx1, Runx2, and Runx3 (4, 27) have diverged to distinct, often reciprocal tissue expression patterns, which restrict their functional redundancy (28–31). Different Runx paralogs can mediate nonredundant functions (in Natural Killer cell responses), maintain mutual exclusion (in B cells), or compensate for one another (in leukemia) (32–34). The relationship and division of labor between different Runx proteins thus need to be defined in a context-dependent manner.
Early T cell lineage development actually provides more than one regulatory context in which Runx factors can work (25). Driven by Notch signaling, the early precursor cells in the thymus, CD4−CD8− double-negative (DN) pro-T cells, progress in stages from DN1 or “early thymic progenitor” (ETP) to DN2a, DN2b, and DN3 (Fig. 1A) and after acquiring a form of TCR, to DN4 and beyond (35–38). In ETP and DN2a stages, pro-T cells are still developmentally multipotent, express the transcription factor PU.1, and epigenetically resemble prethymic multipotent progenitors (15, 17, 39–42). These uncommitted stages are referred to as “Phase1.” Commitment to the T cell fate occurs in transition from DN2a to DN2b stage and coincides with epigenetic changes defining entry into “Phase2” (25). This depends, at least in part, on up-regulation of the transcription factor Bcl11b (41, 43, 44), which mediates changes in chromatin states and transcriptional signatures (11, 42). In Phase2, committed DN2b and DN3 cells establish expression of T cell identity genes and begin TCR assembly (Fig. 1A). Importantly, signature transcription factors for Phase1 and Phase2, PU.1 and Bcl11b, respectively, both use Runx1 as a cofactor (11, 14).
Fig. 1.
Enrichment of Runx motif sequences at accessible chromatin (ATAC) peaks from pro-T cells and Runx1 and Runx3 coexpression in individual thymic progenitor cells. (A) Schematic of T cell development and ATAC peak groups. P1: Phase 1; P2: Phase 2. (B) Motif density histograms show frequency of motif occurrences around ATAC peaks of indicated sites. (C) Gene expression kinetics of Spi1, Bcl11b, Tcf7, Runx1, and Runx3 during T cell development (http://www.immgen.org/) are shown. (D and E) ETP-DN2 (cKithigh CD44+) and DN3 (cKitlow CD44− CD25+) cells were isolated from 4-wk-old animals. Expression of Runx1 and Runx3 transcripts was measured by seqFISH in ETP (Kit copies ≥ 5, Il2ra copies ≤ 3, n = 890), DN2 (Kit copies ≥ 5, Il2ra copies >3, n = 1,984), and separate DN3 (n = 1,587) cells. Transcript counts of Runx1 and Runx3 are enumerated and shown with median (D). Gene–gene Pearson distance heat maps of Runx1 and Runx3 coexpression in thymic progenitor cells are displayed (E). LT-, ST-HSC: long-term, short-term hematopoietic stem cell; DP: double positive; SP: single positive.
This study focused on three central questions. First, is Runx1 the sole member of the Runx family that contributes to early T cell development, or is its activity reinforced or modulated by actions of other Runx family members? Second, does the Runx family regulate a constant core of target genes throughout early T cell development, or does its mode of activity shift from Phase1 to Phase2? As the three Runx paralogs are expressed in different developmental patterns in early pro-T cells, the answers to these two questions may be linked. Finally, how does genomic binding of Runx factors explain their actions?
Our results show that at each pro-T cell stage, Runx1 and Runx3 are largely concordant in their binding and effects on target genes, but they bind to substantially different sites before and after commitment. Their target genes are highly stage specific in their functional responses and are highly developmentally dynamic in normal expression. Thus, despite apparently constant activity levels, Runx factors work via developmentally changing genomic sites and preferentially drive developmental state transitions.
Results
Runx Motifs Are Highly Enriched in the Open Chromatin Regions of Thymic Precursor Cells before and after Commitment.
The sequence motif recognized by Runx proteins has been found enriched at DNA sites occupied by E2A, PU.1, GATA3, or Bcl11b in T-lineage precursor cells (11, 13–16, 45). To measure the prevalence of Runx motifs in an unbiased way across all genomic regions likely to be active in early pro-T cells, we screened for the most enriched transcription factor binding motifs in the open chromatin sites reported for thymic ETP, DN2a, DN2b, and DN3 cells (17) (SI Appendix, Fig. S1A and Dataset S1). Globally, there is a sharp transition in genomic activity and conformation from Phase1 (ETP and DN2a) to Phase2 (DN2b and DN3), although many sites maintain accessibility through both stages (Phase1&2 in common) (Fig. 1A) (17, 42). Motifs for Spi1 (PU.1) and Tcf7 (high mobility group [HMG] box) family factors showed strong reciprocal changes in enrichment between Phase1-specific and Phase2-specific sites (Fig. 1B). However, consensus binding motifs for Runx factors were consistently among the top three at open sites at all these stages (Fig. 1B and SI Appendix, Fig. S1A), supporting roles for Runx motif binding factors in pro-T cells before and after commitment.
Runx1 and Runx3 Are Coexpressed in Individual Thymic T Progenitor Cells.
All Runx paralogs use a Runt homology domain to bind to the same motif (1, 4). Whereas in most cell types, distinct Runx paralogs are expressed in different patterns, all three Runx paralogs are expressed in bone marrow hematopoietic precursors before they enter the thymus, in data from http://www.immgen.org/ (Fig. 1C and SI Appendix, Fig. S1B) (17). Similar levels of Runx1, Runx2, and Runx3 RNAs all appear to be expressed in the ETP population, which also expresses Tcf7 and high Spi1 (encoding PU.1) but little if any Bcl11b (Fig. 1C). Phase1 pro-T cells also express Runx3 transcripts from the distal promoter (SI Appendix, Fig. S1C) (17), which may be preferentially translated into protein (46). After T-lineage commitment, as Spi1 falls and Bcl11b rises, levels of Runx1 increase, whereas Runx3 expression declines and Runx2 expression disappears completely (17, 18, 47) (Fig. 1C and SI Appendix, Fig. S1B) (http://www.immgen.org/).
Runx1;Runx3 double knockouts (dKOs) lose all T cell development in vivo (48), but Runx1 and Runx3 have contrasting roles and expression in later thymocytes (46). Although both appeared highly expressed in ETP and DN2a populations, we asked whether Runx1 and Runx3 expression might be mutually exclusive in single cells. For maximally sensitive detection of Runx transcripts, we used sequential fluorescent in situ hybridization (seqFISH) datasets (39) for efficient detection of low-copy number RNA transcripts, important for genes encoding transcription factors (49–51). In fact, single-molecule transcript counts in single cells (Dataset S2) agreed with bulk RNA sequencing (RNA-seq) for Runx1 and Runx3 expression profiles overall (Fig. 1D) and confirmed that both Runx1 and Runx3 transcripts are expressed simultaneously in the same individual pro-T cells (Fig. 1E). As expected, the ratio of Runx3 to Runx1 transcript counts among individual cells in the population decreased with developmental stage (Fig. 1E), with Runx3 transcript counts slightly higher in most ETP and Runx1 transcript counts higher in most DN3 cells. Even so, >85% of ETPs and >93% of DN2 cells expressed three or more copies of both (57% of ETPs and >74% of DN2 cells had five or more copies of both) (Dataset S2). Runx1 and Runx3 proteins were also clearly coexpressed in thymic pro-T cells, especially in DN2 stages (SI Appendix, Fig. S1D). Thus, despite changing developmental patterns, Runx1 and Runx3 are substantially coexpressed in nearly all individual early intrathymic T cell precursors.
Runx1 and Runx3 Bind Shared Genomic Loci in Pro-T Cells but Have Stage-Specific Patterns of Occupancy.
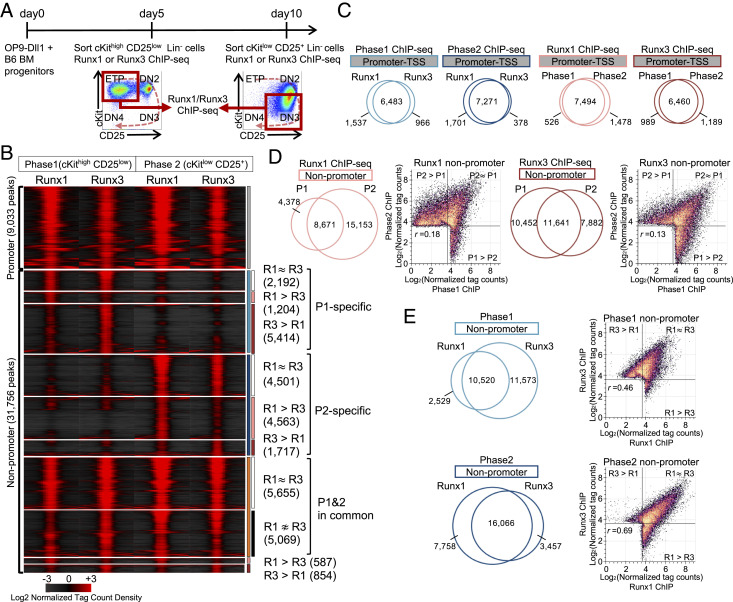
Despite coexpression, Runx factors in pro-T cells might have paralog-specific functions if they interacted with distinct genomic regions. We compared Runx1 and Runx3 occupancies directly by chromatin immune precipitation and deep sequencing (ChIP-seq) for Runx1 and Runx3 in primary Phase1 (cKithigh CD25low) and Phase2 (cKitlow CD25+) pro-T cells. Populations were generated using the OP9-Delta-like 1 (Dll1; a Notch ligand) coculture in vitro differentiation system (52) (Fig. 2A). This mimics closely the early pro-T cell stages in vivo before TCR gene assembly; a direct transcriptomic comparison is also shown (SI Appendix, Fig. S4D). Total Runx peak counts in Phase1 and Phase2 (reproducible peaks, as defined in Materials and Methods) were broadly consistent with the Runx gene expression patterns (Phase1: 21,069 Runx1; 29,541 Runx3; Phase2: 32,796 Runx1; 27,172 Runx3).
Fig. 2.
Runx1 and Runx3 interact with shared genomic sites that shift at different developmental stages. (A) Progenitor cells from normal bone marrow (BM) were cultured with OP9-Dll1 stromal cells for 5 d (Phase1) or for 10 d (Phase2); then, pro-T cells in Phase1 (cKithigh CD25low) or Phase2 (cKitlow CD25+) were fluorescence-activated cell-sorted for chromatin immunoprecipitation with sequencing (ChIP-seq) as described in Materials and Methods. Experiment schematic and gating strategy are shown. (B) Tag count distributions for Runx1 and Runx3 in Phase1 and Phase2 stages are represented by peak-centered heat map. P1: Phase1; P2: Phase2; R1: Runx1; R3, Runx3. (C–E) Area-proportional Venn diagrams illustrate the peak overlaps of Runx1 and Runx3 ChIP-seq peaks in Phase1 and Phase2, with numbers of peaks in each category. Scatterplots compare log2 normalized tag counts per 10 million tags for peaks in indicated categories with Pearson correlation r. (C) Promoter region peaks (TSS: transcription start site); (D) nonpromoter region peak comparison for Runx1 and Runx3; and (E) nonpromoter region comparison for Phase1 and Phase2. Data are based on ChIP-seq peaks scored as reproducible in two replicate samples (B–E).
In each phase, Runx1 and Runx3 occupied most of the same sites (Fig. 2 B–E) and usually bound to those sites with similar intensities (Fig. 2B, subgroups denoted as “R1∼R3”). However, despite similar overall levels of Runx binding at both phases, the patterns of occupancy for both factors shifted substantially between Phase1 and Phase2 (Fig. 2B). The statistics were different for promoter and nonpromoter regions, in accord with the globally stronger correlation reported between nonpromoter (enhancer) activity and developmentally dynamic gene expression (17). Most of the promoter regions (22% of Runx-interacting sites) were occupied similarly in both Phase1 and Phase2, in common by Runx1 and Runx3 (Fig. 2C and SI Appendix, Fig. S2 A and B). In contrast, nonpromoter sites comprised multiple groups with distinctive stage-specific Runx binding patterns, and we focused on these (Fig. 2 B, D, and E). Three major groups were distinguished: a Phase1-specific group (8,810 peaks total, 27% of nonpromoter sites), a Phase2-specific group (10,781 peaks total, 34% of nonpromoter sites), and a separate Stable group common to both Phase1 and Phase2 (10,724 peaks total, 34% of nonpromoter sites). The Phase1-specific peaks were also highly enriched for PU.1 (Spi1) motifs as well as Runx motifs, whereas at the Phase2-specific peaks, Runx, E26 transformation-specific (ETS), Tcf7 (HMG), and basic helix-loop-helix (bHLH) motifs were all overrepresented (SI Appendix, Fig. S2 C and D). Thus, Runx1 and Runx3 behaved similarly in DNA binding activity, and they collectively occupied similar numbers of sites before and after T cell lineage commitment but markedly shifted their binding site choices across commitment in a way that was not explained by availability of Runx factors alone.
Functional Differences between Subsets of Runx1 and Runx3 Sites.
Despite sharing occupancy of 10,000 to 16,000 peaks, Runx1 or Runx3 showed preferential occupancy at some genomic regions (Fig. 2 B and E, “R1 > R3” and “R1 < R3”). In Phase1 when Runx3 expression is higher, ∼47% of all Phase1 peaks (11,573 peaks of 24,622) showed stronger binding of Runx3 than of Runx1, and very few bound Runx1 preferentially. In Phase2, ∼28% of the Phase2 peaks (7,758 of 27,281) bound more Runx1 than Runx3. Interestingly, these select, paralog binding-biased genomic regions displayed slightly different motif enrichments (Dataset S1). Runx3-preferring sites always gave the Runx motif as the most frequently discovered consensus sequence, both in Phase1 and Phase2, whereas Runx1-preferring sites showed similar or higher enrichments for motifs of other partners such as bHLH or ETS factors (SI Appendix, Fig. S2 E and F) (cf ref. 14). Hence, despite the preponderance of common targets for both Runx factors in both stages, Runx1-specific DNA interactions may be more influenced by its binding partners than those of Runx3.
Runx1 and Runx3 Are Necessary and Functionally Redundant for Pro-T Cell Development in Phase1.
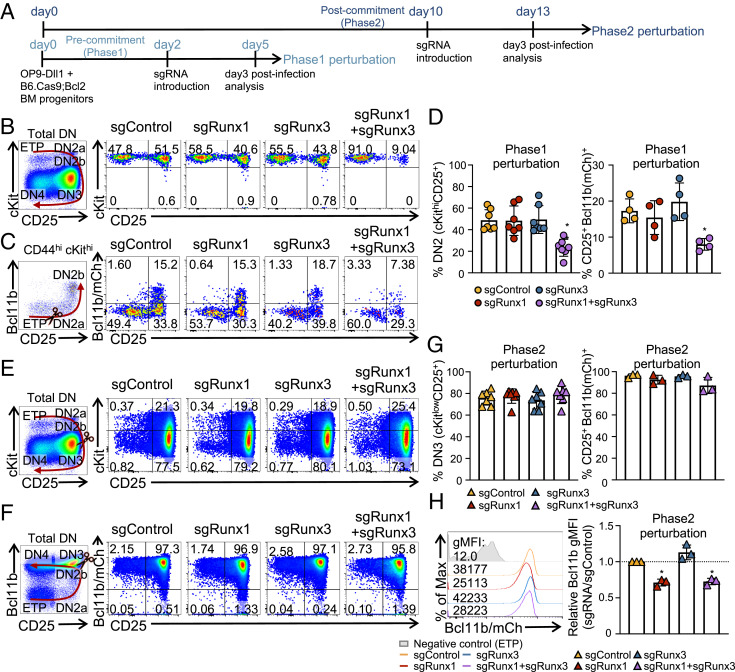
Even when binding to the same sites, Runx1 and Runx3 might exert complementary, antagonistic, or mutually reinforcing impacts. These could be distinguished by effects of acute, stage-specific Cas9-mediated single- or double-gene deletion. To test this, therefore, bone marrow progenitor cells from B6.ROSA26-Cas9 knock-in mice with a Bcl2 transgene (B6.Cas9;Bcl2) were cocultured with OP9-Dll1 cells to initiate T cell development, and guide RNAs (sgRNAs) against Runx1 and/or Runx3 or control sgRNAs were retrovirally delivered to induce deletion at specific times of development (Fig. 3). The Bcl2 transgene was included to enhance viable recovery of cells with regulatory perturbations without altering development (53), and a Bcl11bmCh allele (54), also included, provided an mCherry fluorescent marker for Bcl11b up-regulation, which coincides with T commitment in late DN2a stage (41). We induced acute deletion before commitment (Phase1 deletion) or after commitment (Phase2 deletion) and then analyzed the cells 3 d later (Fig. 3A). The introduction of sgRunx1 and/or sgRunx3 caused specific deletion at targeted sites within 3 d, as shown by point-focused deletions in RNA transcripts and loss of protein detected by intracellular immunostaining in the pro-T cells and supported by western blotting in acute Runx knockout (KO) cell lines (SI Appendix, Fig. S3 A–C). The developmental status of Runx-disrupted pro-T cells was scored with markers cKit and CD25, which distinguish pro-T stages as shown in Fig. 1A, and using Bcl11b-mCherry expression to mark commitment.
Fig. 3.
Runx factors are essential for thymic T cell development at Phase1 and Phase2. (A) Experiment schematic for Phase1 and Phase2 perturbation is displayed. (B–D) For Phase1 perturbation, bone marrow (BM) progenitor cells from Cas9;Bcl2 mice or Cas9;Bcl2 mice with Bcl11b-mCherry reporter were cultured with OP9-Dll1 cells for 2 d before transduction with sgRNAs (Phase1). (B and C) Expression levels of cKit, CD25, and Bcl11b of pro-T cells as analyzed by flow cytometry 3 d after Phase1 perturbation, gated on 7AAD− Lin− CD45+ infection marker (CFP&NGFR)+ population. Frequencies of DN2 (cKithigh CD25+) population and CD25+ Bcl11b+ population are shown (D). (E–H) Expression levels of cKit, CD25, and Bcl11b are shown 3 d after Phase2 perturbation, gated on 7AAD− Lin− CD45+ infection marker (CFP&NGFR)+ population. (E and F) Representative flow cytometry plots. (G) Frequencies of DN3 (cKitlow CD25+) cells and CD25+ Bcl11b+ populations. (H) Histogram of geometric mean fluorescence intensity (gMFI) of Bcl11b expression following Phase2 perturbation, comparing relative gMFI with control group. n = 7 for cKit, CD25 analysis (B and E), and n = 3 to 4 for Bcl11b analysis (C and F). Graphs show the average ± SD. (D, G, and H) One-way ANOVA. *P value < 0.05; **P value < 0.01.
Phase1 pro-T cells with single disruptions (single knockout [sKO]) of Runx1 or Runx3 were minimally affected. They still progressed to DN2 stage and activated levels of Bcl11b expression comparable with those of sgControl transduced cells (Fig. 3 B–D), despite slightly lower overall cell recoveries (SI Appendix, Fig. S3D). In contrast, deletion of both Runx1 and Runx3 simultaneously (dKO) severely impaired progression of Phase1 cells, yielding a significantly lower percentage of DN2 cells (Fig. 3 B and D, Left) and failure to induce Bcl11b expression (Fig. 3 C and D, Right). The recovered cell numbers were also more profoundly reduced in dKO than in control or single deletion groups, indicating that both Runx1 and Runx3 normally contribute to proliferation or survival (SI Appendix, Fig. S3D). Thus, Runx1 does not act alone in Phase1 pro-T cell development; it shows substantial redundancy and compensation with Runx3.
Runx1 Supports Optimal Expression of Bcl11b in Phase2.
The knockout effect was not superficially as obvious in Phase2 as in Phase1. When we delivered sgRNAs to B6.Cas9;Bcl2 cells after progenitor cells had become committed to the T cell fate (Fig. 3A), most cells appeared to reach DN3 stage by the time of analysis (Fig. 3E). Runx gene knockouts now caused no significant impacts on the CD25 and cKit expression phenotypes at day 3 postinfection, and the frequency of Bcl11b+ CD25+ (presumptively committed) cells was not altered either (Fig. 3 E–G). Also, unlike Phase1, the recovery of these Phase2 cells was almost insensitive to Runx disruption. Only Runx1, Runx3 double deletion yielded slightly lower numbers of cells than the other groups, and this was not statistically significant (SI Appendix, Fig. S3E). In agreement with the high percentage of Bcl11b-mCherry+ cells, the Phase2-deleted cells remained functionally committed to the T cell lineage, as shown by their response on transfer to conditions lacking Notch ligand (SI Appendix, Fig. S3F). However, the expression level of Bcl11b (detected by mCherry reporter) per cell was modestly but consistently reduced in both the Runx1 sKO and the Runx1, Runx3 dKO samples (Fig. 3H) (P value < 0.05), in agreement with a previous report (41). More substantial effects of Runx disruption in Phase2 became evident from transcriptome analysis, which is discussed next.
Runx1 and Runx3 Exert Parallel Functions in Gene Regulation in Phase1 and Phase2.
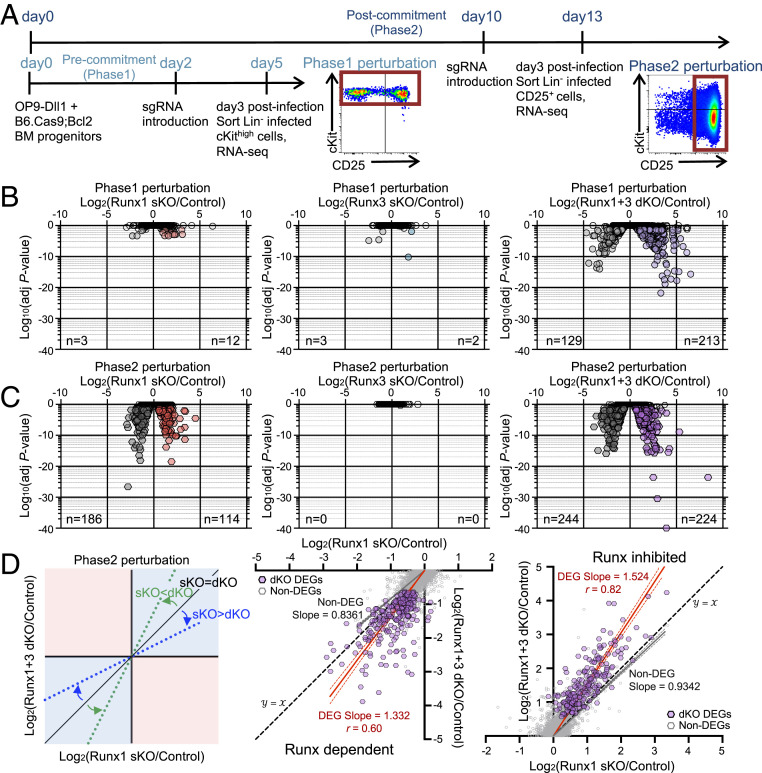
RNA-seq analysis at 3 d post-sgRNA introduction (Dataset S3) showed that sKO of either Runx factor alone caused little change in transcriptomes of Phase1 pro-T cells (Fig. 4 A and B). In contrast, when Runx1 and Runx3 were disrupted together, 342 genes were differentially expressed (differentially expressed genes [DEGs]; adjusted P value < 0.05, log2 fold change either direction >0.5) (Fig. 4B and Dataset S3). More genes in the dKO were derepressed (213 genes up-regulated) than down-regulated (129), implying that Runx factors normally have substantial direct or indirect roles in these cells in repression (“inhibition”) as well as activation (“dependency”). Similar results were seen using a more stringent threshold, |log2 fold change| > 1 (210 inferred to be Runx inhibited, 118 dependent). The synergy observed implies that previously observed Runx1 effects in Phase1 cells only appeared weak due to paralog redundancy, primarily with Runx3, in these stages.
Fig. 4.
Runx1 and Runx3 function in parallel in gene regulation. (A) Experimental schematic for RNA-seq is displayed. RNA-seq measurements were performed on fluorescence-activated cell-sorted sgControl-, sgRunx1-, sgRunx3-, and sgRunx1 + sgRunx3-transduced pro-T cells on day 3 posttransduction (7AAD− Lin− CD45+ CFP+ NGFR+ cells) for Phase1 (cKithigh population) and Phase2 (CD25+ population) perturbations. (B and C) Volcano plots show statistical significance (log10 P value) vs. log2 fold change (FC) of DEGs (average fragments per kilobase-million reads ≥ 1, adjusted P value < 0.05, log2 FC > 0.5 either way) in Phase1 perturbation (B) and Phase2 perturbation (C) groups. n = number of genes scoring as DEGs in each condition. (D) Dot plots show log2 FC of Runx1 sKO vs. Runx1 + Runx3 dKO upon Phase2 perturbation. Trend lines show linear regression results with 95% CI for Phase2 perturbation DEGs (red) and Non-DEGs (gray). Best-fit values for slopes and Pearson correlation r are noted. Data are based on two to three replicate samples of RNA-seq results.
In Phase2 after T commitment, delivery of sgRunx1 alone was sufficient to change gene expression substantially (Fig. 4C and Dataset S3). This transcriptome impact was much stronger than was obvious from the CD25, cKit, and Bcl11b marker analysis (cf Fig. 3 E–G). Because the knockouts had little impact on cell recovery at this stage, the effects seen could not be artifacts of selective survival. The Runx1 sKO gave >300 differentially regulated genes (DEGs), of which 186 were down-regulated (Runx dependent) and 114 were up-regulated (Runx inhibited) in the sKO. In a sharp contrast, Runx3 deletion alone at this stage did not cause any changes in messenger RNA (mRNA) levels. However, Runx1, Runx3 dKO cells again showed the greatest number of DEGs. Runx-inhibited genes increased to 224 significant DEGs, while Runx-dependent DEGs increased to 244 (Fig. 4C) (with more stringent threshold, 168 inhibited and 167 dependent).
Runx1 and Runx3 worked in the same direction on individual genes in Phase2 cells, as shown by comparing the log2 fold change values of individual DEGs obtained from Runx1 sKO and from Runx1, Runx3 dKO cells. Effects in Runx1 sKO and Runx1, Runx3 dKO Phase2 cells were positively correlated for both Runx-dependent and -inhibited targets (Pearson r = 0.60 for dependent genes; r = 0.82 for inhibited genes). Comparing dKO with sKO effects on the same genes, dKO effects were more intense for both Runx-dependent (slope = 1.332) and Runx-inhibited DEGs (slope = 1.524), whereas best-fit line slopes for non-DEGs were <1, ruling out nonspecific effects (Fig. 4D). Thus, even in Phase2, the dKO augmented changes in Runx-sensitive (dependent or inhibited) gene expression, in the same directions, relative to effects of the sKO.
These data, with Fig. 3, demonstrate that Runx1 and Runx3 function similarly when they are coexpressed in early T cell development. They show strong functional redundancy in Phase1, and in Phase2, where Runx1 becomes the major contributor, even lowly expressed Runx3 plays a milder but still parallel role in transcriptional regulation. Thus, the sum of Runx1 and Runx3 activity appears most important.
Runx Factors Drive Activation of the T-Lineage Program and Suppress Alternative Lineage Signatures.
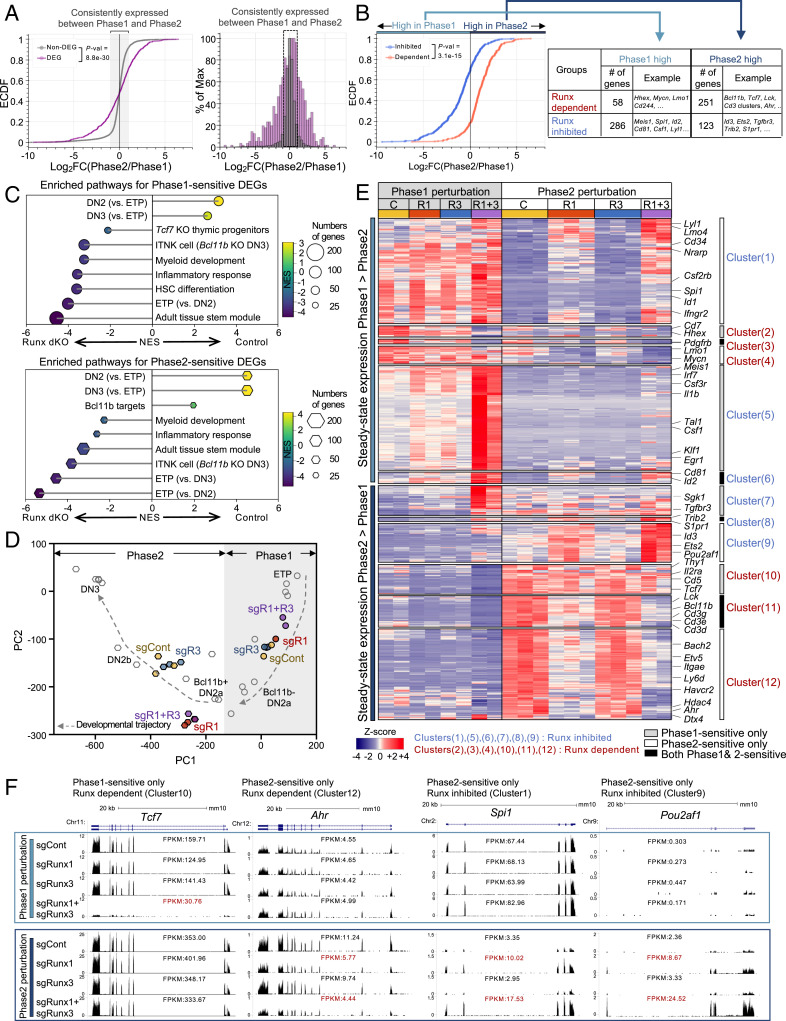
If Runx1 and Runx3 act equivalently, it was notable that the genes affected by Runx1, Runx3 dKO were a select minority of the T cell precursor transcriptome that was most developmentally dynamic. Empirical cumulative distribution frequency (ECDF) plots and histograms showed that about 60% of total Runx-responsive DEGs normally underwent more than twofold changes in expression levels between Phase1 and Phase2, much higher than the fraction of expressed Runx non-DEGs undergoing such changes (about 15%; P value < 9e-30) (Fig. 5A). Furthermore, Runx-dependent and Runx-inhibited genes had reciprocally biased developmental profiles: 80% of Runx-dependent genes (defined by dKO at either phase) were more highly expressed normally in Phase2, whereas 70% of Runx-inhibited genes (defined by dKO at either phase) had higher gene expression normally in Phase1 (P value = 3.1e-15) (Fig. 5B). Thus, despite near-constant total Runx factor DNA binding (Fig. 2 C and E), Runx activities were most important to cause gene expression changes during the Phase1–Phase2 transition.
Fig. 5.
Runx1 and Runx3 function additively to direct pro-T cell program and suppress alternative lineage potentials. (A) RNA expression changes (log2 fold change [FC]) between Phase1 and Phase2 in controls are shown, comparing Runx DEGs with non-DEGs in cumulative frequency (ECDF; Left) and histogram (Right) plots. Consistently expressed genes are genes with |log2 FC| < 1 in either direction and average expression (as fragments per kilobase-million reads [FPKM]) ≥ 1. (B) ECDF plot comparing Phase2/Phase1 mRNA expression ratios (log2 FC) in Runx-dependent genes vs. Runx-inhibited genes. “Phase1 high” indicates genes with log2 FC of Phase2/Phase1 < 0. “Phase2 high” indicates genes with log2 FCs of Phase2/Phase1 > 0. (C) Top pathways identified by GSEA and normalized enrichment score (NES) are shown for Phase1-sensitive Runx-regulated genes (Left) and Phase2-sensitive genes (Right). Size of the dots indicates the number of genes in enriched gene set. Color of dot is NES. HSC: hematopoietic stem cell. (D) Effects of indicated perturbations on developmental progression, as shown by supervised PCA plot from RNA-seq data. Fixed PC loadings were determined based on single-cell RNA-seq data as described in Materials and Methods. (E) Heat map of RNA-seq displays expression levels of the DEGs either in Phase1 perturbation and/or in Phase2 perturbation groups. Each cluster was determined by phase sensitivity (white bar, Phase1 sensitive only; gray bar, Phase2 sensitive only; black bars, Phase1 and Phase2 sensitive), steady-state expression pattern, and mode of Runx factor effects (blue, Runx inhibited; red, Runx activated). (F) Representative genome browser tracks (http://genome.ucsc.edu) for DEGs sensitive in either Phase1 or Phase2 only. FPKM values are shown. Data are based on two to three replicate samples of RNA-seq results (A–E) or are representative of two or three replicate samples (F).
Runx-dependent DEGs were enriched for T cell “identity” genes associated with DN2 and DN3 Phase2 cells in Gene Set Enrichment Analysis (GSEA). For example, Bcl11b, Cd3d, Cd3e, Cd3g, and Lck, key T cell program genes highly activated during T-lineage commitment (Fig. 5C and SI Appendix, Fig. S4A), were down-regulated by loss of Runx whether deleted in Phase1 (Phase1 sensitive) or in Phase2 (Phase2 sensitive). In contrast, Runx-inhibited DEGs were enriched for stemness signature genes (ETPs and hematopoietic stem cells; e.g., Meis1, Tal1, and Klf1 [Phase1 sensitive]) and for genes associated with alternative lineages, including Csf1, Ly9, Cd86, and Id2 (Phase1 sensitive), and Spi1, Id1, Id2, Id3, and Cd81 (Phase2 sensitive) (Fig. 5C and SI Appendix, Fig. S4B). In agreement with GSEA, the ImmGen database (17) showed that the Runx-dependent genes were expressed more highly in early T-lineage populations (SI Appendix, Fig. S4C, red labels) than in myeloid cells, whereas the Runx-inhibited genes had the reverse pattern (SI Appendix, Fig. S4C). Therefore, Runx1 and Runx3 are essential to establish the T cell program and inhibit myeloid and stem cell gene expression in this process.
Runx-gene dKO at different phases distorted the normal developmental trajectory, as shown by a supervised principal component analysis (PCA) of control and Runx KO pro-T cell transcriptomes (Fig. 5D). We used 65 highly informative developmental regulatory genes from a reference single-cell RNA-seq analysis of normal in vivo thymocytes (39) to define a fixed frame of principal component (PC) loadings (Fig. 5D, open symbols; also Materials and Methods and Dataset S4). As expected, RNA-seq results from control in vitro-generated pro-T cell populations (no sgRNA) mapped into this PC space close to their in vivo counterparts (SI Appendix, Fig. S4D), validating the similarity of in vitro- and in vivo-generated pro-T cells. In this framework (Fig. 5D), transcriptomes of populations transduced in Phase1 with sgControl, sgRunx1 alone, or sgRunx3 alone all clustered together, between those of normal ETP and Bcl11b-negative DN2a thymocytes. In contrast, transcriptomes from Phase1 dKO populations (sgR1 + R3) were clearly shifted backward toward ETP, indicating a defect in progression. In Phase2 perturbations, samples transduced with sgControl or sgRunx3 mapped close to DN2b references as expected, but here, both Runx1 sKO and Runx1, Runx3 dKO samples were retarded (Fig. 5D). Taken together (Dataset S4), these data confirm that Runx regulates targets that promote T-lineage identity and developmental progression before and after commitment.
Specific Phase-Dependent Regulation of Runx Factor Functional Targets.
In principle, transcription factors stably expressed across a developmental stage transition like Runx1 + 3 might be expected to regulate their target genes similarly in both stages. However, among 312 Runx-dependent genes, only 61 (19%) were sensitive to Runx disruption at both stages in common. The targets of Runx inhibition showed even less overlap, as only 27 of 410 (6.5%) Runx-inhibited genes were sensitive in both (Fig. 5E and SI Appendix, Fig. S4E). The rest responded in stage-specific ways.
Runx-dependent and Runx-inhibited targets showed stage-specific responses to Runx deletion, whether they were developmentally increasing or decreasing in expression (Fig. 5E). The varied patterns argue against systematic detection biases, and RNA-seq tracks show the robustness of these effects (Fig. 5F and SI Appendix, Fig. S5). Many Runx-dependent targets showed stage-specific responses, including genes with key ongoing roles in T cell development (e.g., Tcf7, Thy1, Il2ra, and Cd5 [Cluster10], Phase1 sensitive only), while Ahr, Hdac4, and Hmgcs2 (Cluster12) were sensitive in Phase2 only (Fig. 5F and SI Appendix, Fig. S5 C and E; note scale changes between phases). Differentially stage-sensitive groups were especially obvious among the repression targets, as many Runx-inhibited genes were more up-regulated when Runx was removed in Phase1 (Fig. 5F and SI Appendix, Fig. S5D) (Cluster5 and Cluster7; e.g., Tal1, Sgk1, Meis1, Irf7, and Csf1), while others responded more in Phase2 (Cluster1 and Cluster9; Spi1, Pou2af1, Lyl1, CD34, Id1, and Id3) (Fig. 5F and SI Appendix, Fig. S5F). The stage-specific targets Tcf7 (Runx dependent only in Phase1 [Cluster10]) and Spi1 (PU.1; Runx inhibited only in Phase2 [Cluster1]) (Fig. 5F) have particularly important roles in T cell development. Thus, unexpectedly, a majority of both Runx-dependent and Runx-inhibited targets responded to Runx input selectively during specific developmental phases, often during expression changes, rather than for maintenance of a given expression level.
Runx Binding Is Enriched at DEGs and Enriched at Phase of Highest DEG Expression.
Given the near-constant levels of Runx1 + Runx3 activity across commitment, these results raised questions. First, were most effects of Runx activity mediated directly, and were stage-specific changes in binding sites (Fig. 2) responsible for developmental changes in action on different sets of target genes? Second, could the stage-specific positive and negative impacts of Runx on particular target genes (Fig. 5E) be linked to Runx binding to different function-specific sites or to altering Runx function at the same sites? For insight, we tested whether particular clusters of genes showing different patterns of developmental expression and Runx responsiveness (Fig. 5 B and E) might be correlated with particular patterns of Runx factor occupancies at sites linked to them, from Phase1 to Phase2 (Dataset S5).
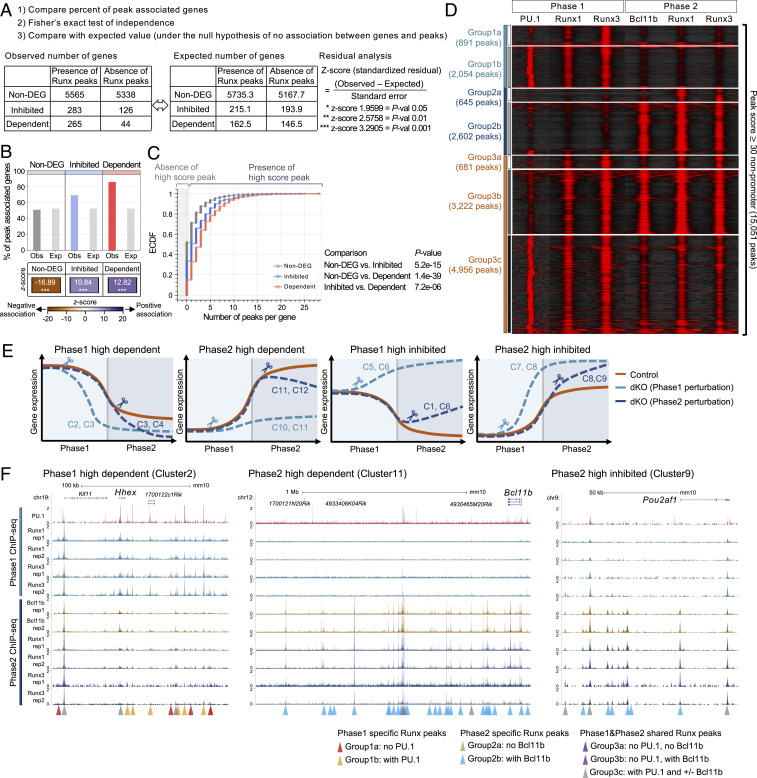
We first determined whether each transcript in DEG and non-DEG groups had any linked high-scoring promoter and nonpromoter Runx peak(s) in its surrounding genomic regions (Materials and Methods). Then, we compared 1) the percentage of genes in each category linked to such Runx binding and 2) the z score showing positive or negative association, by comparing the difference between random frequencies of Runx peaks among all expressed genes and the observed frequencies in each group (Fig. 6A). Non-DEGs were more likely than DEGs to possess Runx binding sites at promoters, indicating that Runx occupancy near the promoter was a poor indicator of functional targets (SI Appendix, Fig. S6). In contrast, a higher percentage of DEGs (70 to 90%) than of non-DEGs (∼50%) was linked to at least one nonpromoter Runx peak (Fig. 6B). Both Runx-inhibited and Runx-dependent genes had more local Runx binding at nonpromoter regions than random expressed genes (z scores +10.8 and +12.8), whereas expressed but Runx-insensitive non-DEGs showed less binding (z score −16.9) (Fig. 6B and Dataset S5). This was also evident in ECDF of Runx occupancies per gene (Fig. 6C). Runx-dependent genes showed higher numbers of linked Runx binding sites/gene than the Runx-inhibited genes (Fig. 6 B and C). This suggests that both activation and repression may often be direct and that higher numbers of high-scoring nonpromoter peaks were more associated with positive transcriptional regulation.
Fig. 6.
Phase-specific collaboration with PU.1 and Bcl11b is associated with Runx functional target genes. (A) Bar graph displays the percentage of the genes associated with peaks, comparing observed values (Obs) and expected values (Exp). z scores (standardized residuals) across categories are shown by the color map. (B) Cumulative frequency of the number of high score peaks found per gene in indicated group. Gray area indicates zero high-score peaks per gene. P values: Kolmogorov–Smirnov test. (C) Peak-centered heat map illustrates ChIP-seq tag count distributions for indicated transcription factors across 15,051 high-quality genomic sites of Runx factor binding in Phase1 and Phase2 stages. High-score peaks indicate nonpromoter Runx binding sites with peak scores ≥ 30. (D) ChIP-seq and RNA-seq integrative analysis strategy is shown. As an example, observed numbers of genes in non-DEG, Runx-inhibited, and Runx-dependent categories with or without any Runx binding peaks are given, compared with a table of expected values if sites were distributed randomly. (E) Diagrams illustrate expression patterns of genes in different clusters from Fig. 5E and SI Appendix, Fig. S9B. C, cluster. (F) Representative genome browser profiles (http://genome.ucsc.edu) are shown with highlighted stage-specific peaks. (Left) Phase1-high Runx-dependent gene (Hhex locus), (Center) Phase2-high Runx-dependent gene (Bcl11b locus), and (Right) Phase2-high Runx-inhibited gene (Pou2af1 locus). Data are based on ChIP-seq peaks scored as reproducible in two biological replicate ChIP-seq samples and two or three replicate samples of RNA-seq results.
To test how Runx occupancy shifts (Fig. 2B) might be related to different functions, we classified nonpromoter binding sites by their occupancy with Runx factors in Phase1 only (Group 1), Phase2 only (Group 2), or both (Group 3) (cf Fig. 2B). We then asked which site groups were preferentially enriched near Runx-activated and Runx-inhibited DEGs of distinct expression and sensitivity features (details and caveats are in SI Appendix, Fig. S7). In general, the Runx sites with stage-specific binding had higher enrichment near Runx-responsive target genes that were expressed most strongly in that stage. Phase2-specific (Group 2) peaks especially were highly enriched near targets expressed highly in Phase2 (SI Appendix, Fig. S7 A and B). Peaks shared between Phase1 and Phase2 (Group 3) were also more enriched at Phase2-high DEGs. Runx binding was also slightly higher near the Runx-dependent targets with high expression in the matching stage than near Runx-inhibited ones (SI Appendix, Fig. S7B), both for Phase2-specific Group 2 Runx peaks and for Phase1-specific Group 1 peaks.
In contrast, the timing of DEG sensitivity to Runx was not necessarily correlated with the time of highest Runx binding, and this showed a much more complex pattern (SI Appendix, Fig. S7C), dissected further below. Of note, although sites gaining or maintaining maximal Runx binding at Phase2 (Groups 2 and 3) showed the most biased linkage with genes that were Runx dependent, not Runx inhibited, this bias was surprisingly strongest among genes that showed Runx dependence only in Phase1 (Phase1-sensitive Dep vs. Inhib) (SI Appendix, Fig. S7 C, Middle and Bottom). These patterns suggest that a target gene’s maximum sensitivity to Runx perturbation could occur during an Runx binding transition (e.g., during increase of local binding) rather than at the time of maximum Runx binding.
Association of Chromatin Accessibility with Stage-Specific Runx Binding.
To explore how the developmental shifts in Runx binding (Fig. 2B) could be associated with function, we asked whether Runx factors require or induce chromatin opening, testing the relationship between chromatin accessibility (assay of transposase-accessible chromatin by sequencing [ATAC-seq]) and Runx occupancy (cf Fig. 2B) at nonpromoter Runx binding sites (SI Appendix, Fig. S8). Not surprisingly, about 85% of the Group 3 sites, which are commonly occupied by Runx factors both in Phase1 and Phase2, were constantly open from ETP through DN3. Group 1 sites also showed binding primarily associated with open chromatin, with about 46% of Group 1 sites losing accessibility as Runx binding declined. In sharp contrast, the majority of Group 2 sites were either open (55% of Group 2 peaks) or closed (25% of Group 2 peaks) at all times; less than 12% opened specifically at the time of Runx binding. Thus, Phase2-specific Runx binding neither awaits nor causes chromatin opening (SI Appendix, Fig. S8).
Distinct Stage-Specific Runx-Partner Factors Preferentially Associate with Different Regulatory Effects.
To distinguish between Runx-bound sites that mediate positive and negative gene regulation, we asked whether cobinding with prominent partner factors could be important. As previously noted (11, 14, 15), most Runx1 occupancy sites were co-occupied with PU.1 (53%) and/or Bcl11b (66%), and Runx3 sites overlapped similarly with PU.1 (43%) or Bcl11b (67%) (SI Appendix, Fig. S9A). We subdivided the site groups based on ChIP-seq evidence for Runx cobinding with PU.1 in Phase1 and/or with Bcl11b in Phase2 (Fig. 6D, SI Appendix, Fig. S9A, and Dataset S5), thus resolving seven site groups (Fig. 6D: 1a, 1b, 2a, 2b, 3a, 3b, and 3c). We then tested each of these finer groups for preferential enrichment near DEG types defined in Fig. 5E (Fig. 6E and SI Appendix, Fig. S9B), first using the coarse DEG clusters defined in SI Appendix, Fig. S7B (SI Appendix, Fig. S9 C–E) and then using the finer DEG clusters (SI Appendix, Fig. S9 F–J). As simplified in Fig. 6E, genes in these DEG clusters had separable responses (Fig. 6E, broken lines) to Runx dKO at different stages (scissors), which were superimposed upon their different normal developmental patterns of expression (solid brown lines). Nearly all these Runx site groups were enriched in most DEG clusters relative to the non-DEG background (SI Appendix, Fig. S9 C–H, horizontal broken lines). Some special functional relationships stood out with particular DEG response types. This was shown when each site group’s enrichment in linkage to genes of a particular DEG response type was compared with its enrichment among DEGs overall (i.e., values expected if the frequency of that site group was randomized among all DEGs) (SI Appendix, Fig. S9 C–H, Rdm DEG bar; z scores for comparisons between DEGs are shown in color scale). These results showed that co-occupancy of an Runx site with PU.1 or Bcl11b was associated with markedly different linkages to specific DEG types (SI Appendix, Fig. S9 C and D, Group 1a vs. 1b and Group 2a vs. 2b, respectively).
Both cobinding partners, PU.1 and Bcl11b, were associated with more frequent binding in DEGs as compared with non-DEGs (SI Appendix, Fig. S9 C–E; compare 1a vs. 1b, 2a vs. 2b, and 3a vs. 3b and 3c). However, the rare Phase1-high genes positively regulated by Runx were markedly enriched for Phase1-specific Runx sites without PU.1 cobinding (SI Appendix, Fig. S9C, Group 1a). Cluster C2 genes, Runx dependent in Phase1, were especially enriched for Group 1a (no PU.1) and not Group 1b sites (with PU.1). In contrast, Phase1 Runx cobinding with PU.1 (Group 1b) was found more broadly with weaker, varied functional associations (SI Appendix, Fig. S9F).
Cobinding with the Phase2 partner, Bcl11b, had markedly greater predictive value. Sites of Runx cobinding with Bcl11b (Group 2b, 3b, 3c) were strongly enriched for both overall DEG binding and overall association with Phase2-specific, Runx-dependent DEGs as compared with those without Bcl11b (SI Appendix, Fig. S9 D and E, cf Groups 2a and 3a). Fine-grained cluster comparison showed that sites with Bcl11b cobinding (SI Appendix, Fig. S9G, Group 2b) could be associated with Phase2 timing of repressive as well as activating functions (i.e., both for Phase2-high Runx-dependent DEGs [C10, 12] and for the few Phase2-high genes that were constrained by Runx inhibition in Phase2 [C9]). Of note, Bcl11b-linked Phase2 Runx binding did not seem as important for silencing Phase1-high Runx repression targets, as these sites were systematically underrepresented near Phase1-high Runx-inhibited loci (SI Appendix, Fig. S9 D and E, Groups 2b and 3b; negative z scores). Their association fell to background levels near C5 DEGs, which were only Runx sensitive in Phase1 (SI Appendix, Fig. S9 G, Group 2b and H, Groups 3b and 3c). Thus, “new” Runx binding sites in Phase2 are probably not needed to maintain silence of genes originally repressed in Phase1. SI Appendix, Fig. S9 I and J summarizes all these patterns, showing the representation of each peak group with each DEG cluster as its percentage of all peaks linked to genes in that cluster.
We asked whether sites with preferences for Runx3 or Runx1 are linked to different functions (SI Appendix, Fig. S10 A–D). Most Runx-sensitive loci were linked to sites with both Runx1 and Runx3 occupancy (Runx1∼Runx3), and overall results with these and the Phase2-specific Runx1-preferring sites were as described above (SI Appendix, Fig. S10 B and D). However, Phase1 peaks that preferred Runx3 over Runx1 distinctively were enriched near Runx-dependent genes with highest expression in Phase1, not Runx-inhibited genes (SI Appendix, Fig. S10C). They thus resembled Phase1 occupancy sites without PU.1 (Group 1a: cf SI Appendix, Fig. S9 C and F). Thus, Runx3 may have specific, limited positive roles in Phase1.
Thus, different classes of Runx sites are associated with different target gene response types. Bcl11b cobinding potentiates Phase2-specific Runx effects and initiation of Phase2-high expression, whereas PU.1 cobinding may mask Phase1-specific Runx effects with its own generalized pioneering and regulatory roles (15). In addition, the need for Runx to inhibit Spi1 itself in Phase2 may create indirect pathways through which Phase1-high genes that depend on PU.1-bound sites (C1) can be inhibited by Runx in Phase2.
Runx-Sensitive Target Genes Recruit Stage-Specific Runx Binding in Clusters.
The pattern of stage-specific Runx binding to functional target loci showed one more contrast with the overall stability of Runx activity. Occupancy patterns across the regulatory elements of Runx DEGs (SI Appendix, Figs. S10E and S11) underwent coordinated developmental changes in Runx binding during commitment, at multiple sites around target loci spanning large genomic regions. Runx-dependent genes specifically expressed in Phase1 (e.g., Hhex [C2], Clnk [C4], and Itgax [C2]) displayed dramatic losses of Runx1 and Runx3 binding as clusters of Group1 Runx peaks disappeared after commitment (Fig. 6 F, Left and SI Appendix, Fig. S11A, orange and red arrowheads). Runx-inhibited genes with high expression in Phase1 also lost multiple Phase1 occupancy peaks during transition to Phase2 (e.g., Csf1 [C5], Meis1 [C5], and especially Plek [C6]) (SI Appendix, Fig. S11 B and C), although statistical inference for this group of DEG was weaker.
The Runx-dependent loci highly expressed in Phase2 showed the opposite pattern. The entire regulatory region of Bcl11b, spanning >1 Mb, was almost devoid of Runx binding in Phase1 cells; then, >30 Group2 sites were occupied by Runx proteins and Bcl11b, a few days later, in Phase2 (Fig. 6 F, Center, light blue arrowheads). Thus, the early sensitivity of Bcl11b expression to Runx dKO in Phase1 could reflect the need to assemble a new activation structure for Bcl11b expression for transition to Phase2. Similar concerted Runx binding was found at Thy1 (C10), the Cd3 cluster (C11), and Hdac4 (C12) (SI Appendix, Fig. S11D). Runx-inhibited genes normally expressed in Phase2 also gained multisite binding in Phase2, as represented at Pou2af1 and Id3 (Fig. 6F and SI Appendix, Fig. S11 C, Right). Such concerted binding of Group2 sites was striking, considering that most of these sites did not change in chromatin ATAC accessibility (SI Appendix, Fig. S8).
Thus, taken together, Runx1 and Runx3 show phase-specific DNA binding patterns around both positively and negatively regulated targets, which often appear as large groups of linked sites that dynamically change occupancy together during T cell development (SI Appendix, Fig. S10). These distinctive genomic interactions in Phase1 and Phase2 are closely associated with stage-specific gene regulation, which is also correlated with patterns of co-occupancy with PU.1 or Bcl11b.
Discussion
Functional genomics and gene network predictions often assume that the target genes of a transcription factor should be expressed in a pattern matching that of the transcription factor itself (55). Here, by dissecting the roles of Runx transcription factors in T cell development, we find a strong counterexample to this prediction. The roles of Runx proteins in early T cell development have long been known to be important but were not clearly defined before. One problem was that the dominant factor, Runx1, was expressed very broadly with little change across periods of strong transcriptome change; another was that Runx1 deletion in early T-lineage cells appeared to have only modest effects until later stages. Here, RNA, protein, and functional data show that coexpressed Runx1 and Runx3 work collaboratively and redundantly in individual intrathymic pro-T cells before T-lineage commitment, providing a crucial input that activates or represses key genes to drive pro-T cell progression. Runx2, although not tested here, might also contribute during Phase1. Importantly, despite near-constant Runx activity levels, the target genes preferentially regulated by Runx1 and Runx3 are stage specific and enriched for strong expression changes across commitment. Runx action on these targets reflects major redistributions of Runx binding across the genome during commitment. A distinctive mode of Runx action is its stage-dependent, coordinate gain or loss of binding to multiple neighboring sites in large genomic regions. Thus, Runx site binding shifts, rather than changes in Runx expression, generate momentous changes in T cell precursor gene expression.
Paralogous factors Runx1 and Runx3 direct development and function of various hematopoietic cells (3, 9, 12, 18, 46, 48, 56, 57). If these factors were antagonists as in later thymocytes (46), decreasing Runx3:Runx1 ratios from Phase1 to Phase2 might be a switch between precommitment and postcommitment regulatory patterns. Small differences between the RUNT1 and RUNT3 domains reportedly give Runx3 a stronger affinity for the consensus Runx binding motif than Runx1 (27). Indeed, some of our results would support Runx3 acting more independently than Runx1 on Phase1 target sites. Conversely, fine specificity of binding by Runx1 may be more dependent on partners. bHLH target motifs are enriched at Runx1-preferring sites, and partner Bcl11b may also interact preferentially with Runx1, even in an Innate Lymphoid Cell Type 2 context expressing more Runx3 than Runx1 (58). However, these small differences between Runx1 and Runx3 were dwarfed in pro-T cells by their coexpression, highly overlapping binding sites, and functional parallelism in activating T cell identity genes while suppressing alternative lineage genes.
Although Runx1 and Runx3 together give pro-T cells nearly constant levels of Runx activity across commitment, they preferentially regulate genes that change expression sharply between Phase1 and Phase2. Furthermore, Runx target genes responded to Runx factor perturbations with notably distinct stage sensitivities. Many genes were Runx sensitive (for positive or negative regulation) only at one phase of their expression, often during a transition in their expression. This strongly recalled previous evidence for hit-and-run Runx activity at specific hematopoietic loci, where transient binding of Runx1 to key target sites was sufficient to reorganize chromatin structure and sustained presence of Runx1 was dispensable for maintaining chromatin accessibility (2). Thus, Runx1 + Runx3 made the greatest difference for genes that were temporarily “destabilized” and poised for sharp developmental changes in expression. This suggests that Runx factors primarily regulate change rather than steady-state expression levels in pro-T cells.
A major contributor to this dynamism is the physical relocation of Runx binding from an initial set of physiological genomic sites to another set, often accompanied by different transcription factor partners. We have previously shown that factor–factor interaction (“theft”) plays a role in this redistribution (14, 25), but not all Phase1 sites are associated with the same “thieving” partner. Both Phase1 and Phase2 sites appear functional. Notably, also, some of the most highly enriched site–response associations suggest that target gene sensitivity to Runx perturbation is highest when Runx is making the site shift, more than when its configuration is stable.
These results do not rule out indirect regulation, especially for some classes of Runx-inhibited genes. Also, PU.1 or antagonism of E proteins could also cause indirect inhibition of some apparently Runx-dependent genes since loss of Runx factors in Phase2 unleashed Spi1 and ID protein expression (Id1, Id2, and Id3). However, Runx binding overall and specific Runx site classes were clearly enriched with repressed as well as with activated targets.
Finally, the shifting of Runx factors between Phase1 and Phase2 showed striking group behavior. Multiple peaks of Runx binding appeared or disappeared together across large genomic domains, such as Bcl11b distal enhancer regions, where >30 sites over >1 Mb dramatically appeared after T-lineage commitment. This suggests that Runx factors may recognize chromatin three-dimensional structures, although ATAC accessibility per se did not appear to be a major constraint. Notably, previous studies showed that Runx1 regulates hematopoiesis by globally redistributing transcription factors and remodeling chromatin structures itself (2, 59–61). Thus, as responders or drivers, Runx factors are illuminating probes for the regulatory mechanisms that control remodeling of chromatin architecture in the most transformative phases of cell-type development.
Materials and Methods
Mouse strains, reagents, Cas9-mediated deletion, in vitro T-lineage differentiation, RNA-seq, ChIP-seq, and analytical methods used in this study were essentially similar to those we have described previously (11, 14, 58, 62). Detailed methods and statistics are provided in SI Appendix. Newly generated RNA-seq and ChIP-seq data for this study are deposited in the Gene Expression Omnibus (accession no. GSE154304). Mice were purchased from the Jackson Laboratory or generated by our laboratory at Caltech as described previously (54). Both male and female mice were used. All experiments shown were performed in at least two separate biological replicates (deep sequencing samples) or at least three independent experiments (all cell biological samples). Numbers of replicates, statistical tests, and P values are given in the figures.
Supplementary Material
Acknowledgments
We thank Xun Wang (Caltech) and Joseph Lotem (Weizmann Institute) for helpful critiques of the manuscript; E. Janielle Cuala and Suin Jo for preliminary experiments; Diana Perez, Jamie Tijerina, and Patrick Cannon of the Caltech Flow Cytometry and Cell Sorting Facility for cell sorting; Igor Antoshechkin and Vijaya Kumar of the Jacobs Genetics and Genomics Center for sequencing; Henry Amrhein and Diane Trout for sequence curation and computer support; Ingrid Soto for animal care; Maria Quiloan for mouse breeding supervision; Rochelle Diamond for laboratory management and sorting supervision; and the members of the group of E.V.R. for sharing advice, reagents, and preliminary results. This work was supported by Cancer Research Institute Irvington Postdoctoral Fellowship CRI.SHIN (to B.S.), the Japan Society for the Promotion of Science KAKENHI Grant JP19H03692 (to H.H.), The Mochida Memorial Foundation for Medical and Pharmaceutical Research (H.H.), The Naito Foundation (H.H.), The Yasuda Medical Foundation (H.H.), the SENSHIN Medical Research Foundation (H.H.), the Takeda Science Foundation (H.H.), and US Public Health Service Grants R01AI135200 and R01HD076915 (to E.V.R.). V.R.T. was supported by a Students Training in Advanced Research award from the University of California, Davis. Flow cytometry, sequencing, and bioinformatics facility support were from the Beckman Institute at Caltech. Support was also from the California Institute of Regenerative Medicine Bridges to Stem Cell Research Program (Pasadena City College and Caltech; M.R.-W.), the L. A. Garfinkle Memorial Laboratory Fund, the Al Sherman Foundation, and the Albert Billings Ruddock Professorship (E.V.R.).
Footnotes
Competing interest statement: E.V.R. is a member of the Scientific Advisory Board of Century Therapeutics, LLC.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2019655118/-/DCSupplemental.
Data Availability.
ChIP-seq and RNA-seq data have been deposited in the Gene Expression Omnibus (accession no. GSE154304).
References
- 1.Mevel R., Draper J. E., Lie A. L.M., Kouskoff V., Lacaud G., RUNX transcription factors: Orchestrators of development. Development 146, dev.148296 (2019). [DOI] [PubMed] [Google Scholar]
- 2.Hoogenkamp M., et al. , Early chromatin unfolding by RUNX1: A molecular explanation for differential requirements during specification versus maintenance of the hematopoietic gene expression program. Blood 114, 299–309 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de Bruijn M., Dzierzak E., Runx transcription factors in the development and function of the definitive hematopoietic system. Blood 129, 2061–2069 (2017). [DOI] [PubMed] [Google Scholar]
- 4.Levanon D., Groner Y., Structure and regulated expression of mammalian RUNX genes. Oncogene 23, 4211–4219 (2004). [DOI] [PubMed] [Google Scholar]
- 5.Wang Q., et al. , Disruption of the Cbfa2 gene causes necrosis and hemorrhaging in the central nervous system and blocks definitive hematopoiesis. Proc. Natl. Acad. Sci. U.S.A. 93, 3444–3449 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Komori T., et al. , Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell 89, 755–764 (1997). [DOI] [PubMed] [Google Scholar]
- 7.Talebian L., et al. , T-lymphoid, megakaryocyte, and granulocyte development are sensitive to decreases in CBFbeta dosage. Blood 109, 11–21 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guo Y., Maillard I., Chakraborti S., Rothenberg E. V., Speck N. A., Core binding factors are necessary for natural killer cell development and cooperate with Notch signaling during T-cell specification. Blood 112, 480–492 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Telfer J. C., Hedblom E. E., Anderson M. K., Laurent M. N., Rothenberg E. V., Localization of the domains in Runx transcription factors required for the repression of CD4 in thymocytes. J. Immunol. 172, 4359–4370 (2004). [DOI] [PubMed] [Google Scholar]
- 10.Kawazu M., et al. , Functional domains of Runx1 are differentially required for CD4 repression, TCRbeta expression, and CD4/8 double-negative to CD4/8 double-positive transition in thymocyte development. J. Immunol. 174, 3526–3533 (2005). [DOI] [PubMed] [Google Scholar]
- 11.Hosokawa H., et al. , Bcl11b sets pro-T cell fate by site-specific cofactor recruitment and by repressing Id2 and Zbtb16. Nat. Immunol. 19, 1427–1440 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Taniuchi I., et al. , Differential requirements for Runx proteins in CD4 repression and epigenetic silencing during T lymphocyte development. Cell 111, 621–633 (2002). [DOI] [PubMed] [Google Scholar]
- 13.Miyazaki M., et al. , The E-Id protein axis specifies adaptive lymphoid cell identity and suppresses thymic innate lymphoid cell development. Immunity 46, 818–834.e4 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hosokawa H., et al. , Transcription factor PU.1 represses and activates gene expression in early T cells by redirecting partner transcription factor binding. Immunity 48, 1119–1134.e7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ungerbäck J., et al. , Pioneering, chromatin remodeling, and epigenetic constraint in early T-cell gene regulation by SPI1 (PU.1). Genome Res. 28, 1508–1519 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Van de Walle I., et al. , GATA3 induces human T-cell commitment by restraining Notch activity and repressing NK-cell fate. Nat. Commun. 7, 11171 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yoshida H. et al.; Immunological Genome Project , The cis-regulatory Atlas of the mouse Immune system. Cell 176, 897–912.e20 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mingueneau M. et al.; Immunological Genome Consortium , The transcriptional landscape of αβ T cell differentiation. Nat. Immunol. 14, 619–632 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ebihara T., Seo W., Taniuchi I., Roles of RUNX complexes in immune cell development. Adv. Exp. Med. Biol. 962, 395–413 (2017). [DOI] [PubMed] [Google Scholar]
- 20.Yzaguirre A. D., de Bruijn M. F., Speck N. A., The role of Runx1 in embryonic blood cell formation. Adv. Exp. Med. Biol. 962, 47–64 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wotton D., Ghysdael J., Wang S., Speck N. A., Owen M. J., Cooperative binding of Ets-1 and core binding factor to DNA. Mol. Cell. Biol. 14, 840–850 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Levanon D., et al. , Transcriptional repression by AML1 and LEF-1 is mediated by the TLE/Groucho corepressors. Proc. Natl. Acad. Sci. U.S.A. 95, 11590–11595 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oestreich K. J., et al. , Regulation of TCRβ gene assembly by a promoter/enhancer holocomplex. Immunity 24, 381–391 (2006). [DOI] [PubMed] [Google Scholar]
- 24.Hernández-Munain C., Krangel M. S., Distinct roles for c-Myb and core binding factor/polyoma enhancer-binding protein 2 in the assembly and function of a multiprotein complex on the TCR δ enhancer in vivo. J. Immunol. 169, 4362–4369 (2002). [DOI] [PubMed] [Google Scholar]
- 25.Hosokawa H., Rothenberg E. V., How transcription factors drive choice of the T cell fate. Nat. Rev. Immunol., 10.1038/s41577-020-00426-6 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Growney J. D., et al. , Loss of Runx1 perturbs adult hematopoiesis and is associated with a myeloproliferative phenotype. Blood 106, 494–504 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bruno L., et al. , Selective deployment of transcription factor paralogs with submaximal strength facilitates gene regulation in the immune system. Nat. Immunol. 20, 1372–1380 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Appel E., et al. , An ensemble of regulatory elements controls Runx3 spatiotemporal expression in subsets of dorsal root ganglia proprioceptive neurons. Genes Dev. 30, 2607–2622 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goyama S., et al. , The transcriptionally active form of AML1 is required for hematopoietic rescue of the AML1-deficient embryonic para-aortic splanchnopleural (P-Sp) region. Blood 104, 3558–3564 (2004). [DOI] [PubMed] [Google Scholar]
- 30.Levanon D., et al. , Spatial and temporal expression pattern of Runx3 (Aml2) and Runx1 (Aml1) indicates non-redundant functions during mouse embryogenesis. Mech. Dev. 109, 413–417 (2001). [DOI] [PubMed] [Google Scholar]
- 31.Fukushima-Nakase Y., et al. , Shared and distinct roles mediated through C-terminal subdomains of acute myeloid leukemia/Runt-related transcription factor molecules in murine development. Blood 105, 4298–4307 (2005). [DOI] [PubMed] [Google Scholar]
- 32.Spender L. C., Whiteman H. J., Karstegl C. E., Farrell P. J., Transcriptional cross-regulation of RUNX1 by RUNX3 in human B cells. Oncogene 24, 1873–1881 (2005). [DOI] [PubMed] [Google Scholar]
- 33.Rapp M., et al. , Core-binding factor β and Runx transcription factors promote adaptive natural killer cell responses. Sci. Immunol. 2, aan3796 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morita K., et al. , Genetic regulation of the RUNX transcription factor family has antitumor effects. J. Clin. Invest. 127, 2815–2828 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thompson P. K., Zúñiga-Pflücker J. C., On becoming a T cell, a convergence of factors kick it up a Notch along the way. Semin. Immunol. 23, 350–359 (2011). [DOI] [PubMed] [Google Scholar]
- 36.Seo W., Taniuchi I., Transcriptional regulation of early T-cell development in the thymus. Eur. J. Immunol. 46, 531–538 (2016). [DOI] [PubMed] [Google Scholar]
- 37.Yui M. A., Rothenberg E. V., Developmental gene networks: A triathlon on the course to T cell identity. Nat. Rev. Immunol. 14, 529–545 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang Q., Jeremiah Bell J., Bhandoola A., T-cell lineage determination. Immunol. Rev. 238, 12–22 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou W., et al. , Single-cell analysis reveals regulatory gene expression dynamics leading to lineage commitment in early T cell development. Cell Syst. 9, 321–337.e9 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Champhekar A., et al. , Regulation of early T-lineage gene expression and developmental progression by the progenitor cell transcription factor PU.1. Genes Dev. 29, 832–848 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kueh H. Y., et al. , Asynchronous combinatorial action of four regulatory factors activates Bcl11b for T cell commitment. Nat. Immunol. 17, 956–965 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hu G., et al. , Transformation of accessible chromatin and 3D nucleome underlies lineage commitment of early T cells. Immunity 48, 227–242.e8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li P., et al. , Reprogramming of T cells to natural killer-like cells upon Bcl11b deletion. Science 329, 85–89 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ikawa T., et al. , An essential developmental checkpoint for production of the T cell lineage. Science 329, 93–96 (2010). [DOI] [PubMed] [Google Scholar]
- 45.Johnson J. L., et al. , Lineage-determining transcription factor TCF-1 Initiates the epigenetic Identity of T cells. Immunity 48, 243–257.e10 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Egawa T., Tillman R. E., Naoe Y., Taniuchi I., Littman D. R., The role of the Runx transcription factors in thymocyte differentiation and in homeostasis of naive T cells. J. Exp. Med. 204, 1945–1957 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.David-Fung E. S., et al. , Transcription factor expression dynamics of early T-lymphocyte specification and commitment. Dev. Biol. 325, 444–467 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang C. Q., et al. , Disruption of Runx1 and Runx3 leads to bone marrow failure and leukemia predisposition due to transcriptional and DNA repair defects. Cell Rep. 8, 767–782 (2014). [DOI] [PubMed] [Google Scholar]
- 49.Lubeck E., Coskun A. F., Zhiyentayev T., Ahmad M., Cai L., Single-cell in situ RNA profiling by sequential hybridization. Nat. Methods 11, 360–361 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shah S., et al. , Single-molecule RNA detection at depth by hybridization chain reaction and tissue hydrogel embedding and clearing. Development 143, 2862–2867 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shah S., Lubeck E., Zhou W., Cai L., In situ transcription profiling of single cells reveals spatial organization of cells in the mouse hippocampus. Neuron 92, 342–357 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schmitt T. M., Zúñiga-Pflücker J. C., Induction of T cell development from hematopoietic progenitor cells by delta-like-1 in vitro. Immunity 17, 749–756 (2002). [DOI] [PubMed] [Google Scholar]
- 53.Yui M. A., Feng N., Rothenberg E. V., Fine-scale staging of T cell lineage commitment in adult mouse thymus. J. Immunol. 185, 284–293 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ng K. K., et al. , A stochastic epigenetic switch controls the dynamics of T-cell lineage commitment. eLife 7, e37851 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Brent M. R., Past roadblocks and new opportunities in transcription factor network mapping. Trends Genet. 32, 736–750 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Okuda T., van Deursen J., Hiebert S. W., Grosveld G., Downing J. R., AML1, the target of multiple chromosomal translocations in human leukemia, is essential for normal fetal liver hematopoiesis. Cell 84, 321–330 (1996). [DOI] [PubMed] [Google Scholar]
- 57.Woolf E., et al. , Runx3 and Runx1 are required for CD8 T cell development during thymopoiesis. Proc. Natl. Acad. Sci. U.S.A. 100, 7731–7736 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hosokawa H., et al. , Cell type-specific actions of Bcl11b in early T-lineage and group 2 innate lymphoid cells. J. Exp. Med. 217, e20190972 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lichtinger M., et al. , RUNX1 reshapes the epigenetic landscape at the onset of haematopoiesis. EMBO J. 31, 4318–4333 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gilmour J., et al. , The co-operation of RUNX1 with LDB1, CDK9 and BRD4 drives transcription factor complex relocation during haematopoietic specification. Sci. Rep. 8, 10410 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bonifer C., Levantini E., Kouskoff V., Lacaud G., Runx1 structure and function in blood cell development. Adv. Exp. Med. Biol. 962, 65–81 (2017). [DOI] [PubMed] [Google Scholar]
- 62.Romero-Wolf M., et al. , Notch2 complements Notch1 to mediate inductive signaling that initiates early T cell development. J. Cell Biol. 219, e202005093 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
ChIP-seq and RNA-seq data have been deposited in the Gene Expression Omnibus (accession no. GSE154304).