Significance
Glycoconjugates and glycopolymers are involved in critical and varied biological functions across domains of life. Many of these complex molecules are biosynthesized via multistep membrane-associated pathways initiated through the action of phosphoglycosyl transferases (PGTs) and propagated by the collective action of glycosyl transferases. PGTs comprise two distinct superfamilies of enzymes, and, although the ubiquitous polytopic PGTs exhibit a familiar multi-transmembrane helical structure, the strictly prokaryotic monotopic PGTs feature a core signature structure with a reentrant membrane helix and a highly conserved soluble domain. In the presented analysis these motifs provide a “lynch pin” for positively identifying over 38,000 nonredundant superfamily members and their genome neighborhoods. From this foundation, fusions of the monotopic PGT reveal insight into pathway function and regulation.
Keywords: glycan biosynthetic pathway, sequence similarity network, phylogenetic reconstruction, enzyme evolution, membrane-associated pathway
Abstract
The monotopic phosphoglycosyl transferase (monoPGT) superfamily comprises over 38,000 nonredundant sequences represented in bacterial and archaeal domains of life. Members of the superfamily catalyze the first membrane-committed step in en bloc oligosaccharide biosynthetic pathways, transferring a phosphosugar from a soluble nucleoside diphosphosugar to a membrane-resident polyprenol phosphate. The singularity of the monoPGT fold and its employment in the pivotal first membrane-committed step allows confident assignment of both protein and corresponding pathway. The diversity of the family is revealed by the generation and analysis of a sequence similarity network for the superfamily, with fusion of monoPGTs with other pathway members being the most frequent and extensive elaboration. Three common fusions were identified: sugar-modifying enzymes, glycosyl transferases, and regulatory domains. Additionally, unexpected fusions of the monoPGT with members of the polytopic PGT superfamily were discovered, implying a possible evolutionary link through the shared polyprenol phosphate substrate. Notably, a phylogenetic reconstruction of the monoPGT superfamily shows a radial burst of functionalization, with a minority of members comprising only the minimal PGT catalytic domain. The commonality and identity of the fusion partners in the monoPGT superfamily is consistent with advantageous colocalization of pathway members at membrane interfaces.
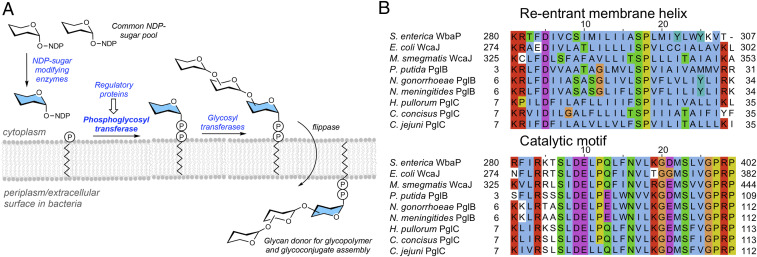
Bacterial glycoconjugate biosynthetic pathways produce complex biopolymers that are essential for cell wall integrity as well as for facilitating interactions among bacterial symbionts and pathogens and their respective hosts (1). A prevalent mechanism of glycoconjugate biosynthesis involves an en bloc strategy in which a glycan is built sequentially on an amphiphilic polyprenol phosphate (PrenP) anchored in the membrane (Fig. 1A) (2, 3). Bacterial enzymes that catalyze reactions in a distinct glycoconjugate pathway are frequently encoded in the same operon (1), advantageously providing clues to associated pathway members. A variety of pathways afford diverse glycoconjugate products including peptidoglycan (PG) (4), wall teichoic acid (WTA) (5), N-/O-linked glycoproteins (6), O-antigen of lipopolysaccharide (LPS) (7), and capsular polysaccharide (CPS) (8, 9).
Fig. 1.
Phosphoglycosyl transferases initiate glycan biosynthesis at the membrane interface by catalyzing the first membrane-committed step. (A) Bacterial en bloc biosynthesis of glycoconjugates requires GTs downstream of the PGT to build the glycan and often sugar-modifying enzymes to transform the common sugar pool to unique sugars. (B) Multiple sequence alignment of the reentrant membrane helix and catalytic motifs of representative enzymes from all three families of the monoPGT superfamily. UniProt IDs for the large family are as follows: S. enterica WbaP (P26406), E. coli WcaJ (P71241), and Mycobacterium smegmatis WcaJ (A0A0D6J209); bifunctional (acetyltransferase) family: Pseudomonas putida sugar transferase (A0A0P7CW64), N. gonorrhoeae PglB (D6HAT8), and N. meningitides PglB (Q7DD76); and small family: Helicobacter pullorum PglC (E1B268), C. concisus PglC (A7ZET4), and C. jejuni PglC (Q0P9D0). The sequences are colored as follows: basic (red), polar (green), acidic (magenta), hydrophobic (blue), tyrosine (cyan), glycine (orange), and proline (yellow) using Jalview (63) (Clustal coloring scheme). White residues do not follow the consensus sequence at that position based on Clustal coloring-scheme thresholds.
Our long-standing interests in bacterial glycoconjugate biosynthesis, and membrane-associated enzymes in general, led us to target the phosphoglycosyl transferase (PGT) from the N-linked protein glycosylation (pgl) pathway of Campylobacter jejuni for global sequence network analysis. This enzyme catalyzes the first membrane-committed step in a biochemically well-defined pathway, which is prototypical of many glycan-assembly processes. PGTs transfer a phosphosugar from a nucleoside diphosphosugar (NDP-sugar) to a membrane-resident PrenP. The product of this step is then elaborated by a series of glycosyl transferases (GTs) (10) followed by translocation from the cytoplasmic to the periplasmic face by a flippase (11). Finally, the completed glycan is transferred to its protein target by an oligosaccharyl transferase (Fig. 1A) (12, 13)
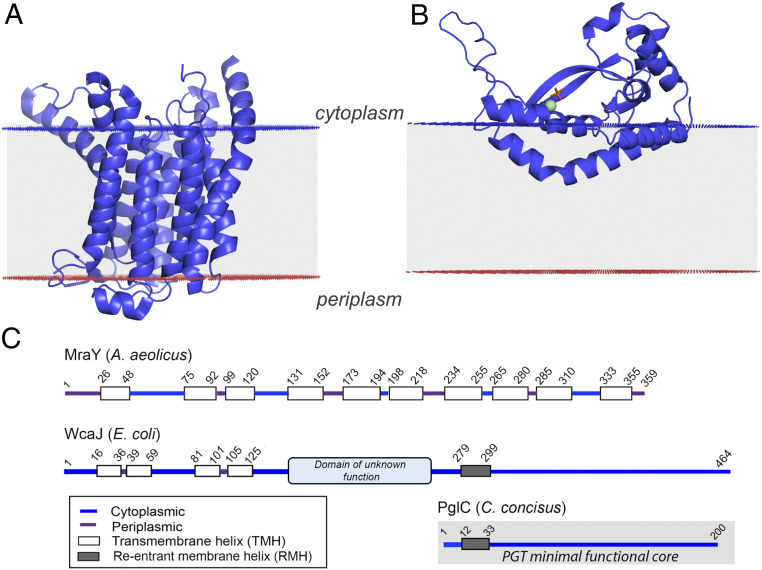
There are two structurally and mechanistically distinct superfamilies of PGTs, the monotopic PGTs (monoPGTs) and the polytopic PGTs (polyPGTs), which catalyze the same transformation. Named for their topology with respect to the membrane, the monoPGTs penetrate a single leaflet of the bilayer whereas the polyPGTs include multiple membrane-spanning helices. These enzyme superfamilies often co-occur in a single organism, allowing for biosynthesis of different glycoconjugate products through distinct pathways (1, 2). Whereas, as defined herein, the monoPGTs are strictly prokaryotic, the polyPGTs are found in both prokaryotes and eukaryotes (2). PolyPGT superfamily members most commonly include 10 to 11 transmembrane helices (TMHs) and extended cytoplasmic loops, with the active site proximal to the plane of the membrane (Fig. 2A). Prokaryotic members of the polyPGT superfamily are exemplified by MraY, WecA, and TagO subclasses—enzymes involved in PG, LPS, and WTA biosynthesis, respectively (14–16). MraY, which catalyzes the transfer of phospho-N-acetylmuramoyl (MurNAc)-pentapeptide onto undecaprenol phosphate (UndP), producing Und-PP-MurNAc-pentapeptide, is the only prokaryotic member of the polyPGTs to be structurally characterized (17, 18). The reactions catalyzed by WecA, MraY, and the eukaryotic N-acetylglucosamine (GlcNAc) phosphotransferase (DPAGT1) have been shown to proceed through a ternary complex intermediate (19, 20). The monoPGTs, in contrast, utilize a Bi-Bi ping-pong mechanism involving a covalent phosphoglycosyl intermediate (21).
Fig. 2.
Structural overview of phosphoglycosyl transferases. (A) The X-ray crystal structure of polyPGT Aquifex aeolicus MraY (PDB: 5CKR). (B) The X-ray crystal structure of small monoPGT C. concisus PglC (PDB: 5W7L), which interacts with the membrane via the RMH. Structures were placed relative to the membrane using the positioning of proteins in the membrane server (64). (C) Experimentally verified membrane topology of polyPGTs (MraY), large monoPGTs (WcaJ), and small monoPGTs (PglC).
The monoPGT superfamily can be classified, based on domain structure, into three distinct families of enzymes: small “PglC-like,” large “WcaJ-like,” and bifunctional enzymes (3). Recent structural characterization of Campylobacter concisus PglC by Ray et al. elucidated a distinct mode of membrane association involving a reentrant membrane helix (RMH) as part of the monoPGT core catalytic domain (22). In this unique monotopic topology, PglC interacts with only one leaflet of the membrane (Fig. 2B). Large monoPGTs share the PGT core domain, represented by C. concisus PglC, with the N-terminal addition of four predicted TMHs and a cytoplasmic domain of unknown function (as shown herein and in Lukose et al. (23) and Furlong et al. (24)) (Fig. 2C). Bifunctional members include an accessory domain, fusing a sugar-modifying enzyme that acts before the PGT reaction to the core domain. For example, in Neisseria gonorrhoeae, the bifunctional PglB replaces the activities of the Campylobacter PglD, a uridine diphosphate (UDP) sugar-modifying enzyme, and the monoPGT, PglC (25).
Initial sequence-based analysis of the monoPGT superfamily identified the conservation of a catalytic motif comprising an Asp-Glu dyad and key residues completing the active site machinery as well as the RMH sequence (Fig. 1B) (23, 26). These highly conserved motifs provide a signature for positively identifying the entirety of the family. Moreover, the singularity of the monoPGT fold allows confident assignment of its role in the first membrane-associated step in the pathway. This is in stark contrast to the GT-A and GT-B fold GT enzymes where the ubiquitous Rossmann-fold proteins are utilized for multiple and varied glycosyl transfer steps within any one pathway. The work described herein highlights the diversity of the prokaryotic monoPGT superfamily through the identification and analysis of bifunctional and fusion monoPGT enzymes. We generated a sequence similarity network (SSN), genome neighborhood network and diagram, and phylogenetic reconstruction to elucidate clustering of unique bifunctional and fusion enzymes and to predict the glycosylation pathways in which they act. Fusion enzymes containing monoPGT domains include the following: sugar-modifying enzymes, GTs, regulatory domains, and, notably, polyPGT enzymes. Large-scale sequence analyses from the SSN serve to identify nonfunctional, pseudoenzyme domains of monoPGTs present in some of the fusion enzymes. These bioinformatic analyses provide a framework for further evolutionary and biochemical exploration of the monoPGT superfamily.
Results
Global Mapping of the MonoPGT Superfamily.
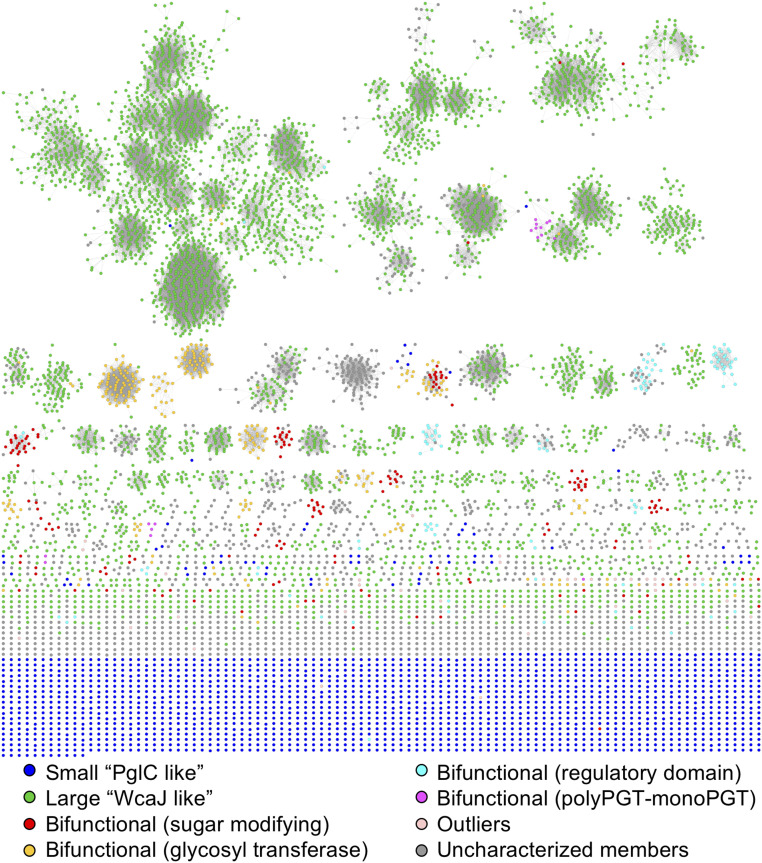
Our goal is to investigate the architectural and sequence diversity of the monoPGT superfamily, which comprises three families. Application of the SSN allows far greater granularity within the superfamily in terms of glycoconjugate pathway product, substrate specificity, organism of origin, domain architecture, and evolutionary relationships. The sequence dataset for the monoPGT superfamily was curated from the InterPro family (IPR003362) using methodology described by the Babbitt and Copp laboratories (27) followed by an all-by-all pairwise alignment at an E-value cutoff of 1 × 10−90 using the Enzyme Function Initiative-Enzyme Similarity Tool (EFI-EST) webtools (28) (Fig. 3). The 40% representative node network represents 38,878 nonredundant sequences (<95% sequence identity) comprising 8,927 nodes (18,571 sequences) and 58,663 edges. Nodes are grouped into 1,359 unique clusters comprising a total of 6,833 unique nodes. The remaining nodes (2,094 nodes) are classified as singletons, meaning that they are not connected by an edge to any other node. The ideal SSN parameters were determined empirically by iteratively changing the percent identity for representative nodes and the alignment threshold. The final 40% representative node network provides the best visualization and separation of bifunctional subclasses based on predicted functions while minimizing the total number of singleton nodes. (SI Appendix, Fig. S1). Because the bifunctional family contains the most divergent domains outside of the PGT core domain, we followed the clustering of these subclasses to select the optimal alignment score.
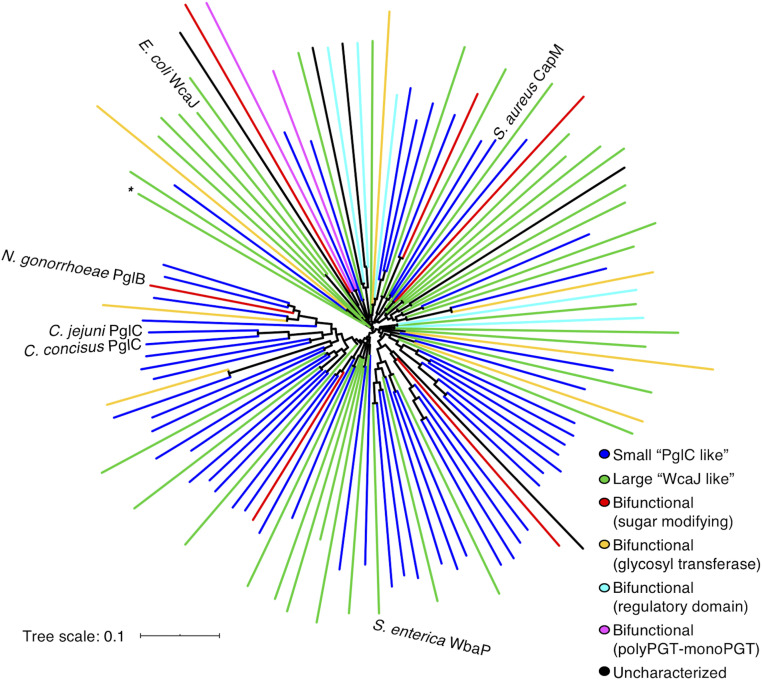
Fig. 3.
Sequence similarity network of the monoPGT superfamily colored by monoPGT family and subclasses. The network is a 40% representative node network with edges representing an E-value cutoff of 1 × 10−90.
Visualization of the network by domain of life using Cytoscape (29) shows the superfamily as predominantly bacterial with only 93 archaeal sequences in all (SI Appendix, Fig. S2), 0.5% of all sequences included in the network. Specifically, all archaeal sequences come from the phylum Euryarchaeota. The halobacterial sequences comprise a single cluster, whereas the methanobacterial sequences are in nodes contained within the largest cluster in the SSN. Most of the archaeal enzymes fall within the large monoPGT family, sharing the same predicted topology of four TMHs, a cytoplasmic domain, and the core catalytic domain. Although the UniProt database reveals 28 eukaryotic sequences corresponding to the monoPGT superfamily (IPR003362), investigation of codon usage and frequencies determined that they are misannotated as eukaryotic and in fact are bacterial in origin. Notably, the network includes three families of monoPGT enzymes: small, large, and bifunctional. Upon further analysis of domain architectures, additional subclasses of the bifunctional/fusion family were identified. By changing the alignment threshold, distinct clusters of bifunctional enzymes were formed within each major subclass. The analysis provided the ability to identify and define subclasses of the bifunctional family and predict their potential roles within glycoconjugate pathways.
Analysis of Bifunctional/Fusion PGTs.
Accessory domains fused to the PglC-like catalytic domain can be further classified as: sugar-modifying, GT, or regulatory. Furthermore, two clusters include unexpected fusions of the polyPGT and monoPGT superfamilies. The network, colored by type of fusion domain, is shown in Fig. 3. Some domains were not included in these classifications because of their low frequency and/or poor characterization (Fig. 3, light pink nodes and all fused fold types identified are listed in SI Appendix). In general, the accessory domains are consistent with the sugar-modifying and GT functions expected in the overall pathway. Although certain accessory domains (e.g., UDP-sugar acetyltransferase–monoPGT and polyPGT–monoPGT fusions) are specifically fused to either the N-terminus or the C-terminus of the monoPGT domain, others do not show a preference for the fusion order based on their respective function or domain family (e.g., UDP-sugar epimerase–monoPGT fusion). The presence of these fusions suggests either that they occurred as gene fusion events and were operon-architecture–driven or that they were driven by effects such as substrate availability, which do not depend on gene order. Together, the identification and characterization of fused monoPGT proteins is consistent with potential cross-talk and interaction of proteins within the pathway. In the case of the monoPGT–GT fusion enzymes, biochemical evidence corroborates close interaction of these enzymes in the pathway. Reconstitution of the Campylobacter PGT and GTs (PglA, PglJ, PglH, and PglI) in vitro results in complete conversion to the polyprenol diphosphate–linked heptasaccharide, which is the substrate for the oligosaccharyl transferase (10). The failure to detect intermediates also suggests that there is close interaction between these pathway enzymes.
Identification of fused regulatory domains suggests the possibility of pathway modulation. Notably, regulation via phosphorylation of a monoPGT (CapM) in the CPS pathway of Staphylococcus aureus showed that Tyr phosphorylation by the kinase CapB led to an increase in total lipid I production (30). The observation of monoPGT-regulatory domain fusions represented in the SSN suggests that these regulatory domains may serve a similar function in their respective pathways.
Sugar-Modifying Enzymes.
A variety of enzymes catalyze NDP-sugar transformations, and genes encoding these enzymes are often found in operons for glycoconjugate biosynthesis to provide low-abundance building blocks such as N,N′-diacetyl bacillosamine (diNAcBac), N-acetylquinovosamine (QuiNAc), or N-acetylfucosamine (FucNAc). Common transformations include redox reactions, dehydration, amino-transfer, and N- or O-acylation. These processes allow for a variety of novel NDP-sugars to be biosynthetically accessible from common starting NDP-sugars.
Within the context of the monoPGT superfamily, there are 10 unique families of sugar-modifying enzymes fused to the monoPGT catalytic core at either the N or the C terminus (Fig. 4A). Of these, eight can be functionally assigned with confidence based on sequence conservation (summarized in SI Appendix, Table S1). The well-characterized, NADPH-dependent C4′,C6′–dehydratase enzymes PglF (Campylobacter N-linked glycosylation) and CapD (S. aureus CPS), share a conserved three-residue motif: Thr395, Asp396, and Lys397 (numbering for C. jejuni PglF) (31, 32). In contrast, PseB, the NADPH-dependent dehydratase involved in pseudaminic acid biosynthesis for flagellin posttranslational modification, shows a different stereochemical outcome although it retains the Thr, Asp, and Lys motif. However, the enzyme does not utilize the Thr in the dehydration of the sugar moiety but instead includes a Tyr in a position nonhomologous to any in the triad (33). Enzymatic function of sugar-modifying domains for members of both classes of dehydratases were inferred by using these sequence markers in conjunction with overall sequence identity and superfamily identifiers. Altogether, 89 C4′,C6′–dehydratase fusions with monoPGTs were identified (SI Appendix, Table S1).
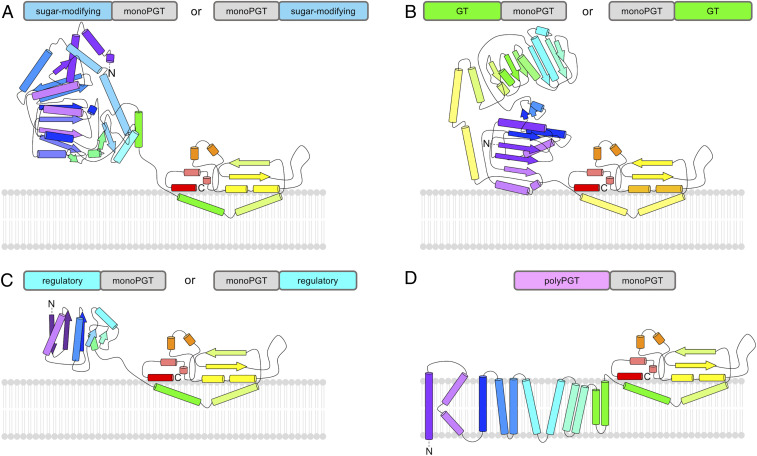
Fig. 4.
Representative subfamilies of bifunctional monoPGT enzymes. Predicted topologies of fusions of the monoPGT core domain modeled with (A) sugar-modifying enzymes (aminotransferase WbpE as model), (B) glycosyl transferases (GT-B fold, WbnH as model), (C) regulatory domains (SpoIIAA as model), and (D) polyPGTs (MraY as model). In all cases the PGT domain is represented by PglC at the C terminus, and the catalytic domain is located on the cytoplasmic face of the membrane. Topology diagrams were generated based on experimentally determined structures using the PDB ID 5W7L for the monoPGT domain and the following PDB IDs for the fused domain: (A) 3NU8, (B) 4XYW, (C) 1TIL, and (D) 4J72. Diagrams are color-ramped purple to red from N terminus to C terminus.
The N. gonorrhoeae PglB includes a monoPGT catalytic domain, which is fused at the C terminus to an N-acetyltransferase domain. The acetyltransferase catalyzes acylation of a C4′amino-sugar precursor, affording UDP-diNAcBac, which is the substrate for the monoPGT (25). All N-acetyltransferase–containing bifunctional enzymes in the InterPro family IPR041561 bear the acetyltransferase domain at the C terminus of the fusion protein. Additionally, these bifunctional enzymes are not found solely in protein glycosylation pathways. Other predicted pathways include exopolysaccharide, colanic acid, and CPS biosynthesis. The gene fusion event could have occurred in an ancient progenitor which was then retained in differing glycosylation pathways of similar organisms. In each of the respective pathways, there are still examples without the monoPGT–acetyltransferase fusion, and, therefore, retention of the fusion would be specific to distinct species. Alternatively, the fusion events may have occurred independently from each other.
Further sugar-modifying elaborations include reductase, O-acetyltransferase, dehydrogenase, and aminotransferase domains. Based on InterPro accession and sequence identity, a total of 15 enzymes have been identified from these classes (SI Appendix, Table S1). However, functional information on each of these respective domains is limited, especially with respect to active-site conservation. Because of their low redundancy within the superfamily, we consider these fusions to be outliers of the sugar-modifying subclass.
Glycosyl Transferases.
Bifunctional enzymes including monoPGT and GT domains represent ∼2% of the total monoPGT superfamily and 46% of all monoPGT fusions (Fig. 4B). GTs elaborate the initial Pren-PP-sugar product of the PGT reaction at the membrane interface. At the time of writing, there are 110 GT families annotated in the CAZy database (34) (http://www.cazy.org/GlycosylTransferases.html). The predominant GT architectures in the monoPGT SSN include GT-1, GT-2, and GT-4 families adopting either a GT-A (GT-2) or a GT-B (GT-1 and GT-4) fold. Both GT-A and GT-B comprise two Rossmann-fold (α/β/α) domains with divergence at the insertion point of one domain into the other (35). Individual GT clusters (SI Appendix, Fig. S1) were annotated based on conservation of catalytic residues and genomic context (genome neighborhood). Substrate specificity for GTs is driven both by the NDP-activated sugar donor and the acceptor substrate. Using sequence-guided approaches, 9 of 12 clusters of GT–monoPGT fusions have predicted GTs based on homology as well as CAZy and InterPro annotations. The functions of the remaining three clusters are unknown. GT fusion enzymes, separated by cluster and annotated by GT fold and predicted homologs, are summarized in SI Appendix, Fig. S3. There is currently not enough functional information about the donor and acceptor specificity of the GTs to discern whether all members in a particular GT-fusion cluster have the same or similar substrate specificity based on sequence alone.
In many members of clusters 1 and 2, the catalytic Asp-Glu dyad, conserved across most monoPGT enzymes, is replaced by Asn-Arg, and the canonical Pro-Arg-Pro motif proposed to be involved in nucleotide binding is replaced by Pro-Glu-Leu (22). However, the GT domain of cluster 1 shares sequence identity with WbbL (23 to 38% ID), a rhamnosyl transferase that uses thymidine diphosphate-activated rhamnose rather than the UDP-activated sugars used in most pathways (36, 37). The function of the monoPGT domain in these fusions is unknown.
Notably, three clusters (2, 3, and 11; SI Appendix, Fig. S3) include GT domains from the WecB/TagA/CpsF InterPro family (IPR004629). TagA enzymes exhibit a novel GT-E fold described by Clubb and coworkers (38) which contains only one Rossmann-fold domain rather than the two observed in GT-A and GT-B folds. There is a second, smaller domain in place of the Rossmann fold, which comprises three α-helices and a β-hairpin. Clusters 2 and 3 are the largest clusters including TagA-like fusions and are connected through one linkage between two representative nodes (UniProt IDs: A0A433I5J1 and A0KWR7). The divergence is based on their domain architecture and conservation of the catalytic dyad. Cluster 2 lacks the Asp-Glu and Pro-Arg-Pro motifs in most members, and the GT domain is at the C terminus. In contrast, Cluster 3 enzymes retain the Asp-Glu and Pro-Arg-Pro motifs, and the GT domain is at the N terminus. This diversity of N-to-C domain architecture in the case of the GT-monoPGT bifunctional enzymes is consistent with fusion events in the monoPGT superfamily that are generally not driven by operon architecture.
Regulatory Domains.
The two most highly represented classes of regulatory domains in monoPGT fusions (Fig. 4C) are the signal transduction response regulators (IPR001789) and the sulfate transporter and anti-sigma factor antagonist (STAS) family (IPR002645), consisting of 49 and 29 representative nodes, respectively (SI Appendix, Table S2). Both of these domains are involved in phospho-regulation of their effector protein targets. Response regulatory domains, such as CheY, are receiver domains in a two-component response regulation system, where the receiver domain is fused to the protein it regulates. Paired with a kinase, often a histidine kinase, the response regulation domain becomes phosphorylated at a conserved aspartic acid (Asp57 in Escherichia coli CheY) (39) which triggers a downstream signal, commonly through up-regulation of genes or through activation of enzymatic activity (40). MonoPGT fusions containing the response regulatory domain show conservation of the CheY residue Asp57 (Glu in some members) that forms the phosphoaspartyl group. Other residues that are involved in coordinating the catalytic magnesium ion (Asp12 and Asp13 E. coli CheY numbering) and phosphorylation-dependent conformational change (Lys109) are not conserved. It is thus possible that these regulatory fusion domains are not functional or have a different mechanism of action.
Members of the STAS domain include anti-anti sigma factors such as SpoIIAA from Bacillus subtilis (UniProt ID: P10727), which is involved in regulating transcription factor σ and, therefore, expression of sporulation genes in B. subtilis (41). All monoPGT–STAS fusion enzymes contain a conserved serine residue in the STAS domain, which in SpoIIAA is phosphorylated by the kinase SpoIIAB. Together, observation of these fusion enzymes further supports the premise that regulation of bacterial glycosylation may occur at the monoPGT step.
Poly–MonoPGT Fusions.
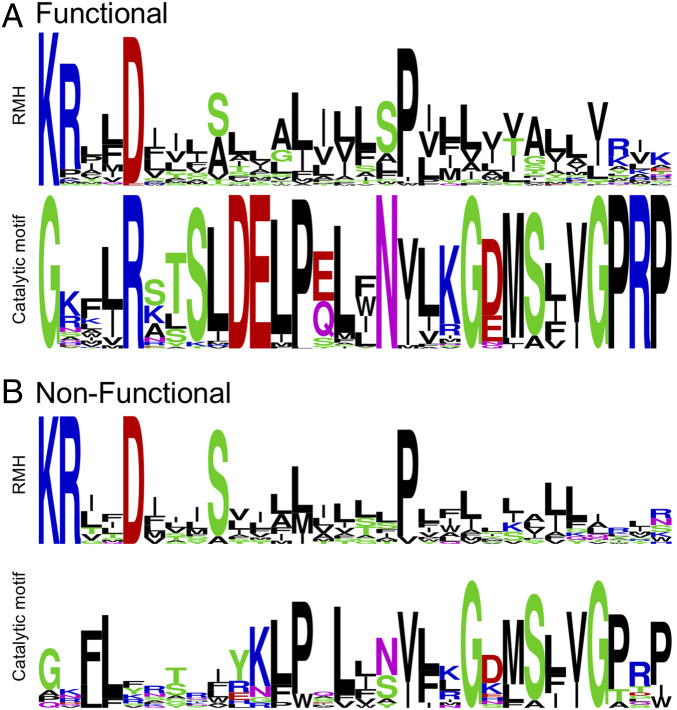
An unexpected fusion that was identified contains both a polyPGT (IPR018480) domain and a monoPGT (IPR003362) domain (from either the small or large monoPGT family) (Fig. 4D). There are two distinct clusters of these fusions within the network (Fig. 3, magenta nodes). In one cluster, the catalytic residues of the sequences are retained in some fusions and, therefore, assumed to encode functional polyPGT and monoPGT domains (polyPGTF–monoPGTF). In the second cluster, only the polyPGT catalytic residues are conserved and the monoPGT catalytic dyad is not retained (polyPGTF–monoPGTNF). A comparison of the sequence conservation of polyPGTF–monoPGTNF to C. concisus PglC homologs at the catalytic dyad shows divergence of the nonfunctional monoPGT domain (Fig. 5). The pseudoenzyme domain of polyPGTF–monoPGTNF enzymes retains sequence similarity in the RMH motif. In these pseudoenzymes, the KXXXD motif at the start of the RMH is strictly conserved, and the general pattern of aliphatic residues flanking a proline at the kink of the RMH is also conserved (Fig. 5).
Fig. 5.
Sequence logos of the RMH motif and catalytic motif for (A) C. concisus PglC homolog sequences and (B) polyPGTF–monoPGTNF sequences. Sequence logos were generated using WebLogo (65).
This fusion of PGTs is particularly striking in that the polyPGTs and monoPGTs most commonly function in different glycan and glycoconjugate biosynthetic pathways. Therefore, typically the genes are localized in different operons. To look more closely at these pathways, a genome neighborhood network and diagram were constructed. The genome neighborhood that includes a functional monoPGT and a polyPGTF–monoPGTNF does not show similarity to the known monoPGT or polyPGT glycosylation pathways (SI Appendix, Fig. S4A). The functional monoPGT gene and the polyPGTF–monoPGTNF gene share a common ancestor (vide infra). Other than this single example, there is not a functional monoPGT localized in any of the genome neighborhoods that include polyPGTF–monoPGTNF fusions. Furthermore, these genome neighborhoods (SI Appendix, Fig. S4 D–F) include enzymes in peptidoglycan biosynthesis, such as MurJ, MurC, and MurM which are associated with polyPGT pathways. A subset of polyPGTF–monoPGTNF fusions have a second nonfunctional monoPGT gene in the genome neighborhood, which is fused to a GT (SI Appendix, Fig. S4 A–D). These PGT–GT fusion enzymes are found in GT cluster 1 (vide supra; SI Appendix, Fig. S3) with highest sequence identity to the rhamnosyl transferase WbbL in the GT domain which is in the mycobacterial mycolylarabinogalactan–peptidoglycan pathway. PG pathways commonly use a polyPGT, and, therefore, it is reasonable to assume that these polyPGTF–monoPGTNF enzymes also act in polyPGT-dependent pathways.
Phylogenetic Reconstruction.
A representative phylogenetic tree was generated by curating a set of representative sequences from each of the largest 100 clusters in addition to four of the largest sugar-modifying and GT-fusion clusters, as well as the characterized monoPGT enzymes E. coli WcaJ (24, 42), Salmonella enterica WbaP (43), S. aureus CapM (30), N. gonorrhoeae PglB (25), C. jejuni PglC (23, 44), and C. concisus PglC (21, 22) (Fig. 6). Members from each family (small, large, and bifunctional/fusion) are represented in the reconstruction. A complete list of all UniProt IDs used in this analysis is provided in SI Appendix, Table S3. This analysis provides complementary information to the SSN, supporting substantial divergence in the superfamily. Rather than dependence on pairwise alignments between each member, phylogenetic reconstructions depend on multiple sequence alignments and include all sequences comparatively. The pattern, described as “radial burst” evolution by Babbitt and coworkers (45), shows several early divergence events, followed by subsequent branching in a radial manner. Notably, the phylogenetic reconstruction supports a model in which the distinct monoPGT architectures were reinvented in different clades to generate the same enzyme architecture. Specifically, the small monoPGT architecture has branched from a large monoPGT in multiple clades in the phylogenetic tree, indicating that the small “PglC-like” architecture is the most modern version of this superfamily.
Fig. 6.
Phylogenetic tree of the monoPGT superfamily. The reconstruction is colored by the following families and subclasses of enzymes: small “PglC-like” (blue), large “WcaJ-like” (green), sugar-modifying bifunctional (red), glycosyl transferase bifunctional (yellow), regulatory bifunctional (cyan), polyPGT–monoPGT fusions (magenta), and uncharacterized members (black). A full list of UniProt IDs used in the reconstruction is provided in SI Appendix, Table S3 (listed in a clockwise direction starting from the sequence marked by *).
Discussion
Protein fusions comprise at least 4% of the monoPGT superfamily, the remaining sequences being small and large monoPGTs (38% and 47%, respectively) and sequences which could not be categorized into families based on sequence alone (11%). A recent comprehensive study of fusion event frequency across genomes from all three domains of life has revealed that, although there is a wide range among the 90 genomes characterized, on average, bacterial genomes include 14% fusion proteins (46). Additionally, the frequency of fusion proteins is higher for metabolic enzymes, possibly supporting shuttling of pathway intermediates. However, comparison of the number of fusion proteins between superfamilies, especially integral membrane protein superfamilies, is not well documented (47).
The identification and classification of fusions of monoPGTs has provided a broader understanding of their roles in bacterial glycan biosynthesis pathways. The prevalence of sugar-modifying and GT bifunctional enzymes is consistent with the existence of protein–protein interactions facilitating product transfers within the respective pathways. Fusions with regulatory domains point to possible regulation of glycan biosynthesis at the PGT reaction specifically, allowing for conservative use of the limited supply of PrenP (48), the common substrate in these glycoconjugate biosynthesis pathways.
Within a single organism, there can be multiple distinct pathways responsible for the diversity of glycoconjugates, each using PrenP as the lipid carrier (49). Sugar-modifying–monoPGT bifunctional enzymes impart a unique advantage to the respective pathways, especially if the sugar-modifying domain catalyzes the final step before the monoPGT transfers the sugar phosphate to PrenP. Such bifunctional enzymes could result in an advantageous increase in the local concentration of a modified soluble sugar substrate.
Similarly, the GT–monoPGT bifunctional enzymes might enhance flux through the pathway by overcoming a potentially unfavorable equilibrium of the PGT (21, 30). Regulation of glycoconjugate biosynthetic pathways at the PGT reaction may be the most efficient means for tuning pathway flux, in that it is the first membrane-committed step in the pathway, and the reactions catalyzed by PGTs, unlike those catalyzed by GTs, are isoenergetic. Regulation of the PGT reaction conserves PrenP and allows for triaging between the pathways within the bacterial cell. Such questions can now be addressed by biochemical analysis of monoPGT–regulatory domain fusions.
The most unexpected finding is the presence of fusions containing a domain from each of the distinct monoPGT and polyPGT superfamilies. The structural and mechanistic divergence of these two superfamilies is indicative of independent evolution of fold and function, also referred to as nonhomologous isofunctional enzymes (50, 51). The evolution of these isofunctional enzymes may have been driven by the necessity to bind and transform the membrane-embedded PrenP substrate. The monoPGTs and polyPGTs have distinct folds. The monoPGTs have no known structural homologs that carry out functions other than phosphoglycosyl transfer, whereas the polyPGTs, exemplified by MraY, do bear structural similarity to membrane proteins of the resistance-nodulation–cell division permease family which transports lipid components across the cell membrane (52–55). These analogous monoPGT and polyPGT enzymes are involved in different glycoconjugate biosynthesis pathways, and thus, the genes are not encoded in the same operon. Therefore, fusion proteins including the catalytic domains from the polyPGT and monoPGT superfamilies are not likely to have occurred as a result of gene fusion events. Rather, it is possible that these polyPGTF–monoPGTNF fusion enzymes resemble an ancestral enzyme. Notably, the monoPGT–core fold, exemplified by C. jejuni PglC, has been shown to increase the local membrane concentration of the PrenP substrate (48). Such an enrichment could increase catalytic throughput of the fused polyPGT enzyme and confer a selective advantage.
Conclusion.
Herein, we have illustrated the architectural diversity of monoPGT enzymes, highlighting modifications that augment their ability to support pathway function. In addition to the three original classifications in the monoPGT superfamily, we have applied network analysis that reveals unanticipated fusions in the bifunctional family to include sugar-modifying enzymes, GTs, and regulatory proteins. Fusion subclasses were identified for which the functional role is not yet understood. These include the regulatory domain–monoPGT and polyPGT–monoPGT fusion enzymes. Annotating these domain architectures in the context of the sequence similarity network allows a greater understanding of this diverse superfamily. Future target identification and functional analyses of these distinct subclasses in the monoPGT superfamily are enabled through their identification in the SSN. Altogether, the identification and bioinformatic analyses of fusion enzymes described herein allows for better characterization of the monoPGT superfamily within the context of bacterial glycosylation pathways.
Materials and Methods
Sequence Dataset Curation and SSN Generation.
Sequence dataset curation for the SSN was conducted following the methods described by Copp et al. (27). Briefly, all sequences in the InterPro family IPR003362 (63,152 sequences at the time of writing) were downloaded from UniProt (http://www.uniprot.org/) and converted to FASTA format using Galaxy (56). This includes one Protein Data Bank (PDB) entry (C. concisus PglC, 5W7L) and nine biochemically characterized enzymes (SI Appendix, Table S4) (10, 21, 23, 25, 30, 42, 43, 57, 58). The dataset was reduced using the Cluster Database at High Identity with Tolerance webserver (59) (http://weizhongli-lab.org/cdhit_suite/cgi-bin/index.cgi) with two sequential percent-identity cuts (90% and 70%). The 70% identity reduction dataset reduced the number of sequences from 63,152 to 19,221 representative sequences. Of the 63,152 sequences, 38,878 are nonredundant sequences, based on a 95% identity cutoff. All putative eukaryotic sequences (28) were removed following manual inspection of codon usage which indicated that they were most likely the result of contaminating prokaryotic DNA incorporated in the sequencing sample.
All-by-all Basic Local Alignment Search Tool (BLAST) calculations were run using the EFI-EST (https://efi.igb.illinois.edu/efi-est/) (28) with an alignment score threshold of 90 and further filtering by sequence length with a minimum and maximum requirement of 180 and 1,000 amino acids, respectively (note that 180 residues is of sufficient length to include the catalytic core). To facilitate visualization, the final network generated was 40% representative, collapsing sequences of 40% identity into representative nodes (8,927 nodes and 58,663 edges), and was visualized using Cytoscape (29). The alignment threshold and representative node percentage were selected based on empirical analysis. Lower thresholds yielded more edges, but the main cluster became too populated and did not provide distinct clustering (SI Appendix, Fig. S1 A and B). Alternatively, increasing the percent identity used as the cutoff to generate the representative node network increased the total number of singletons (SI Appendix, Fig. S1 C and D).
Genome Neighborhood Network and Diagram.
The genome neighborhood network and diagram was generated using the Enzyme Function Initiative-Genome Neighborhood Tool (60) (EFI-GNT; https://efi.igb.illinois.edu/efi-gnt/) with the SSN generated above (BLAST E-value of 1 × 10−90). The neighborhood reading frame was set to 20 frames and the minimal co-occurrence was set to 20%. Additional genome neighborhood diagrams were generated using the FASTA search option on the EFI-GNT webtool (60).
Phylogenetic Reconstruction.
The representative node, comprising the largest number of sequences, was selected from each of the largest 100 clusters (based on the total number of representative nodes in the cluster). Biochemically or structurally characterized members were included (E. coli WcaJ, S. enterica WbaP, S. aureus CapM, N. gonorrhoeae PglB, C. jejuni PglC, and C. concisus PglC). Lastly, curated sequences of additional bifunctional sugar-modifying or glycosyl transferase enzymes localized to smaller clusters, not represented in the largest 100 clusters, were included. A multiple sequence alignment and phylogenetic reconstruction of the curated sequences was generated using ClustalOmega (61). The phylogenetic reconstruction was visualized using the Interactive Tree of Life webserver (62) (https://itol.embl.de/). The UniProt IDs contained in the reconstruction (Fig. 6) are listed in SI Appendix, Table S3.
Supplementary Material
Acknowledgments
We thank Dr. Gregory J. Dodge, Dr. Margarita Tararina, and Hannah Bernstein for valuable input. We also thank Professors Yu (Brandon) Xia and Christian Whitman for reading and helpful discussions of the manuscript. Financial support for this work was provided by the NIH (R01 GM131627 to B.I. and K.N.A.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2018289118/-/DCSupplemental.
Data Availability.
Bioinformatic datasets in .xgmml and .excel format have been deposited in Mendeley (https://dx.doi.org/10.17632/zcx42s9mzf.1). All other study data are included in the article and/or SI Appendix.
References
- 1.Tytgat H. L., Lebeer S., The sweet tooth of bacteria: Common themes in bacterial glycoconjugates. Microbiol. Mol. Biol. Rev. 78, 372–417 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Price N. P., Momany F. A., Modeling bacterial UDP-HexNAc: polyprenol-P HexNAc-1-P transferases. Glycobiology 15, 29R–42R (2005). [DOI] [PubMed] [Google Scholar]
- 3.Lukose V., Walvoort M. T. C., Imperiali B., Bacterial phosphoglycosyl transferases: Initiators of glycan biosynthesis at the membrane interface. Glycobiology 27, 820–833 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bouhss A., Trunkfield A. E., Bugg T. D., Mengin-Lecreulx D., The biosynthesis of peptidoglycan lipid-linked intermediates. FEMS Microbiol. Rev. 32, 208–233 (2008). [DOI] [PubMed] [Google Scholar]
- 5.Neuhaus F. C., Baddiley J., A continuum of anionic charge: Structures and functions of D-alanyl-teichoic acids in gram-positive bacteria. Microbiol. Mol. Biol. Rev. 67, 686–723 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nothaft H., Szymanski C. M., Protein glycosylation in bacteria: Sweeter than ever. Nat. Rev. Microbiol. 8, 765–778 (2010). [DOI] [PubMed] [Google Scholar]
- 7.Whitfield C., Trent M. S., Biosynthesis and export of bacterial lipopolysaccharides. Annu. Rev. Biochem. 83, 99–128 (2014). [DOI] [PubMed] [Google Scholar]
- 8.Whitfield C., Biosynthesis and assembly of capsular polysaccharides in Escherichia coli. Annu. Rev. Biochem. 75, 39–68 (2006). [DOI] [PubMed] [Google Scholar]
- 9.O’Riordan K., Lee J. C., Staphylococcus aureus capsular polysaccharides. Clin. Microbiol. Rev. 17, 218–234 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Glover K. J., Weerapana E., Imperiali B., In vitro assembly of the undecaprenylpyrophosphate-linked heptasaccharide for prokaryotic N-linked glycosylation. Proc. Natl. Acad. Sci. U.S.A. 102, 14255–14259 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fry B. N., et al. , The lipopolysaccharide biosynthesis locus of Campylobacter jejuni 81116. Microbiology (Reading) 144, 2049–2061 (1998). [DOI] [PubMed] [Google Scholar]
- 12.Jaffee M. B., Imperiali B., Optimized protocol for expression and purification of membrane-bound PglB, a bacterial oligosaccharyl transferase. Protein Expr. Purif. 89, 241–250 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jaffee M. B., Imperiali B., Exploiting topological constraints to reveal buried sequence motifs in the membrane-bound N-linked oligosaccharyl transferases. Biochemistry 50, 7557–7567 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boyle D. S., Donachie W. D., mraY is an essential gene for cell growth in Escherichia coli. J. Bacteriol. 180, 6429–6432 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lehrer J., Vigeant K. A., Tatar L. D., Valvano M. A., Functional characterization and membrane topology of Escherichia coli WecA, a sugar-phosphate transferase initiating the biosynthesis of enterobacterial common antigen and O-antigen lipopolysaccharide. J. Bacteriol. 189, 2618–2628 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Soldo B., Lazarevic V., Karamata D., tagO is involved in the synthesis of all anionic cell-wall polymers in Bacillus subtilis 168. Microbiology (Reading) 148, 2079–2087 (2002). [DOI] [PubMed] [Google Scholar]
- 17.Chung B. C., et al. , Crystal structure of MraY, an essential membrane enzyme for bacterial cell wall synthesis. Science 341, 1012–1016 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hakulinen J. K., et al. , MraY-antibiotic complex reveals details of tunicamycin mode of action. Nat. Chem. Biol. 13, 265–267 (2017). [DOI] [PubMed] [Google Scholar]
- 19.Al-Dabbagh B., et al. , Catalytic mechanism of MraY and WecA, two paralogues of the polyprenyl-phosphate N-acetylhexosamine 1-phosphate transferase superfamily. Biochimie 127, 249–257 (2016). [DOI] [PubMed] [Google Scholar]
- 20.Dong Y. Y., et al. , Structures of DPAGT1 explain glycosylation disease mechanisms and advance TB antibiotic design. Cell 175, 1045–1058.e16 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Das D., Kuzmic P., Imperiali B., Analysis of a dual domain phosphoglycosyl transferase reveals a ping-pong mechanism with a covalent enzyme intermediate. Proc. Natl. Acad. Sci. U.S.A. 114, 7019–7024 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ray L. C., et al. , Membrane association of monotopic phosphoglycosyl transferase underpins function. Nat. Chem. Biol. 14, 538–541 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lukose V., et al. , Conservation and covariance in small bacterial phosphoglycosyltransferases identify the functional catalytic core. Biochemistry 54, 7326–7334 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Furlong S. E., Ford A., Albarnez-Rodriguez L., Valvano M. A., Topological analysis of the Escherichia coli WcaJ protein reveals a new conserved configuration for the polyisoprenyl-phosphate hexose-1-phosphate transferase family. Sci. Rep. 5, 9178 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hartley M. D., et al. , Biochemical characterization of the O-linked glycosylation pathway in Neisseria gonorrhoeae responsible for biosynthesis of protein glycans containing N,N′-diacetylbacillosamine. Biochemistry 50, 4936–4948 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Entova S., Billod J. M., Swiecicki J. M., Martín-Santamaría S., Imperiali B., Insights into the key determinants of membrane protein topology enable the identification of new monotopic folds. eLife 7, e40889 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Copp J. N., Anderson D. W., Akiva E., Babbitt P. C., Tokuriki N., Exploring the sequence, function, and evolutionary space of protein superfamilies using sequence similarity networks and phylogenetic reconstructions. Methods Enzymol. 620, 315–347 (2019). [DOI] [PubMed] [Google Scholar]
- 28.Gerlt J. A., et al. , Enzyme function initiative-enzyme similarity tool (EFI-EST): A web tool for generating protein sequence similarity networks. Biochim. Biophys. Acta 1854, 1019–1037 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shannon P., et al. , Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rausch M., et al. , Coordination of capsule assembly and cell wall biosynthesis in Staphylococcus aureus. Nat. Commun. 10, 1404 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Riegert A. S., et al. , Structural and biochemical investigation of PglF from campylobacter jejuni reveals a new mechanism for a member of the short chain dehydrogenase/reductase superfamily. Biochemistry 56, 6030–6040 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li W., et al. , Analysis of the Staphylococcus aureus capsule biosynthesis pathway in vitro: Characterization of the UDP-GlcNAc C6 dehydratases CapD and CapE and identification of enzyme inhibitors. Int. J. Med. Microbiol. 304, 958–969 (2014). [DOI] [PubMed] [Google Scholar]
- 33.Ishiyama N., et al. , Structural studies of FlaA1 from Helicobacter pylori reveal the mechanism for inverting 4,6-dehydratase activity. J. Biol. Chem. 281, 24489–24495 (2006). [DOI] [PubMed] [Google Scholar]
- 34.Lombard V., Golaconda Ramulu H., Drula E., Coutinho P. M., Henrissat B., The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 42, D490–D495 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lairson L. L., Henrissat B., Davies G. J., Withers S. G., Glycosyltransferases: Structures, functions, and mechanisms. Annu. Rev. Biochem. 77, 521–555 (2008). [DOI] [PubMed] [Google Scholar]
- 36.Mills J. A., et al. , Inactivation of the mycobacterial rhamnosyltransferase, which is needed for the formation of the arabinogalactan-peptidoglycan linker, leads to irreversible loss of viability. J. Biol. Chem. 279, 43540–43546 (2004). [DOI] [PubMed] [Google Scholar]
- 37.Grzegorzewicz A. E., et al. , Development of a microtitre plate-based assay for lipid-linked glycosyltransferase products using the mycobacterial cell wall rhamnosyltransferase WbbL. Microbiology (Reading) 154, 3724–3730 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kattke M. D., et al. , Structure and mechanism of TagA, a novel membrane-associated glycosyltransferase that produces wall teichoic acids in pathogenic bacteria. PLoS Pathog. 15, e1007723 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sanders D. A., Gillece-Castro B. L., Stock A. M., Burlingame A. L., Koshland D. E. Jr, Identification of the site of phosphorylation of the chemotaxis response regulator protein, CheY. J. Biol. Chem. 264, 21770–21778 (1989). [PubMed] [Google Scholar]
- 40.Stock A. M., Robinson V. L., Goudreau P. N., Two-component signal transduction. Annu. Rev. Biochem. 69, 183–215 (2000). [DOI] [PubMed] [Google Scholar]
- 41.Masuda S., et al. , Crystal structures of the ADP and ATP bound forms of the Bacillus anti-sigma factor SpoIIAB in complex with the anti-anti-sigma SpoIIAA. J. Mol. Biol. 340, 941–956 (2004). [DOI] [PubMed] [Google Scholar]
- 42.Patel K. B., et al. , Functional characterization of UDP-glucose:undecaprenyl-phosphate glucose-1-phosphate transferases of Escherichia coli and Caulobacter crescentus. J. Bacteriol. 194, 2646–2657 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Patel K. B., Ciepichal E., Swiezewska E., Valvano M. A., The C-terminal domain of the Salmonella enterica WbaP (UDP-galactose:Und-P galactose-1-phosphate transferase) is sufficient for catalytic activity and specificity for undecaprenyl monophosphate. Glycobiology 22, 116–122 (2012). [DOI] [PubMed] [Google Scholar]
- 44.Glover K. J., Weerapana E., Chen M. M., Imperiali B., Direct biochemical evidence for the utilization of UDP-bacillosamine by PglC, an essential glycosyl-1-phosphate transferase in the Campylobacter jejuni N-linked glycosylation pathway. Biochemistry 45, 5343–5350 (2006). [DOI] [PubMed] [Google Scholar]
- 45.Akiva E., Copp J. N., Tokuriki N., Babbitt P. C., Evolutionary and molecular foundations of multiple contemporary functions of the nitroreductase superfamily. Proc. Natl. Acad. Sci. U.S.A. 114, E9549–E9558 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ou Y., McInerney J. O., Eukaryote genes are more likely than prokaryote genes to be composites. Genes (Basel) 10, 648 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Enright A. J., Iliopoulos I., Kyrpides N. C., Ouzounis C. A., Protein interaction maps for complete genomes based on gene fusion events. Nature 402, 86–90 (1999). [DOI] [PubMed] [Google Scholar]
- 48.Entova S., Guan Z., Imperiali B., Investigation of the conserved reentrant membrane helix in the monotopic phosphoglycosyl transferase superfamily supports key molecular interactions with polyprenol phosphate substrates. Arch. Biochem. Biophys. 675, 108111 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yother J., Capsules of Streptococcus pneumoniae and other bacteria: Paradigms for polysaccharide biosynthesis and regulation. Annu. Rev. Microbiol. 65, 563–581 (2011). [DOI] [PubMed] [Google Scholar]
- 50.Galperin M. Y., Koonin E. V., Divergence and convergence in enzyme evolution. J. Biol. Chem. 287, 21–28 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Omelchenko M. V., Galperin M. Y., Wolf Y. I., Koonin E. V., Non-homologous isofunctional enzymes: A systematic analysis of alternative solutions in enzyme evolution. Biol. Direct 5, 31 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tseng T. T., et al. , The RND permease superfamily: An ancient, ubiquitous and diverse family that includes human disease and development proteins. J. Mol. Microbiol. Biotechnol. 1, 107–125 (1999). [PubMed] [Google Scholar]
- 53.Viljoen A., et al. , The diverse family of MmpL transporters in mycobacteria: From regulation to antimicrobial developments. Mol. Microbiol. 104, 889–904 (2017). [DOI] [PubMed] [Google Scholar]
- 54.Bernut A., et al. , Insights into the smooth-to-rough transitioning in Mycobacterium bolletii unravels a functional Tyr residue conserved in all mycobacterial MmpL family members. Mol. Microbiol. 99, 866–883 (2016). [DOI] [PubMed] [Google Scholar]
- 55.Seeliger J. C., et al. , Elucidation and chemical modulation of sulfolipid-1 biosynthesis in Mycobacterium tuberculosis. J. Biol. Chem. 287, 7990–8000 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Afgan E., et al. , The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Res. 46, W537–W544 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Katzen F., et al. , Xanthomonas campestris pv. campestris gum mutants: Effects on xanthan biosynthesis and plant virulence. J. Bacteriol. 180, 1607–1617 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Merino S., et al. , A UDP-HexNAc:polyprenol-P GalNAc-1-P transferase (WecP) representing a new subgroup of the enzyme family. J. Bacteriol. 193, 1943–1952 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Huang Y., Niu B., Gao Y., Fu L., Li W., CD-HIT suite: A web server for clustering and comparing biological sequences. Bioinformatics 26, 680–682 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zallot R., Oberg N. O., Gerlt J. A., ‘Democratized’ genomic enzymology web tools for functional assignment. Curr. Opin. Chem. Biol. 47, 77–85 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Madeira F., et al. , The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 47, W636–W641 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Letunic I., Bork P., Interactive tree of life (iTOL): An online tool for phylogenetic tree display and annotation. Bioinformatics 23, 127–128 (2007). [DOI] [PubMed] [Google Scholar]
- 63.Waterhouse A. M., Procter J. B., Martin D. M., Clamp M., Barton G. J., Jalview Version 2–A multiple sequence alignment editor and analysis workbench. Bioinformatics 25, 1189–1191 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lomize M. A., Pogozheva I. D., Joo H., Mosberg H. I., Lomize A. L., OPM database and PPM web server: Resources for positioning of proteins in membranes. Nucleic Acids Res. 40, D370–D376 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Crooks G. E., Hon G., Chandonia J. M., Brenner S. E., WebLogo: A sequence logo generator. Genome Res. 14, 1188–1190 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Bioinformatic datasets in .xgmml and .excel format have been deposited in Mendeley (https://dx.doi.org/10.17632/zcx42s9mzf.1). All other study data are included in the article and/or SI Appendix.