Significance
Nucleosomes, the fundamental packaging unit of eukaryotic chromosomes, affect gene expression profiles based on their positioning. By shifting nucleosomes along genomic DNA, chromatin remodelers are key reorganizing factors that are often mutated in cancers and developmental disorders. Here, we demonstrate how one remodeler class, called Chd1, uses autoinhibitory elements to control the direction of nucleosome sliding. We show that, through positioning of its DNA-binding domain, the autoinhibitory elements of Chd1 can block an initial stage of DNA distortion required for sliding. This interference by autoinhibitory domains weakens remodeler–nucleosome interactions, transiently preventing subsequent binding and hydrolysis of ATP from ratcheting nucleosomes along DNA.
Keywords: chromatin remodeling, nucleosome sliding, Snf2 ATPase, asymmetric histone core, allostery
Abstract
Chromatin remodelers are ATP (adenosine triphosphate)-powered motors that reposition nucleosomes throughout eukaryotic chromosomes. Remodelers possess autoinhibitory elements that control the direction of nucleosome sliding, but underlying mechanisms of inhibition have been unclear. Here, we show that autoinhibitory elements of the yeast Chd1 remodeler block nucleosome sliding by preventing initiation of twist defects. We show that two autoinhibitory elements—the chromodomains and bridge—reinforce each other to block sliding when the DNA-binding domain is not bound to entry-side DNA. Our data support a model where the chromodomains and bridge target nucleotide-free and ADP-bound states of the ATPase motor, favoring a partially disengaged state of the ATPase motor on the nucleosome. By bypassing distortions of nucleosomal DNA prior to ATP binding, we propose that autoinhibitory elements uncouple the ATP binding/hydrolysis cycle from DNA translocation around the histone core.
Chromatin remodelers are ATP (adenosine triphosphate)-dependent motors that dynamically alter the occupancy, composition, and positioning of nucleosomes throughout the genome. A fundamental question is how chromatin remodelers are regulated to achieve specific outcomes. Along with ISWI (imitation switch) remodelers, Chd1 is responsible for establishing evenly spaced nucleosome arrays that block cryptic transcription within gene coding regions (1, 2). Chd1 and a number of ISWI remodelers are sensitive to the amount of DNA outside the nucleosome core, with faster sliding achieved when more DNA is available on the entry side and correspondingly little or no DNA is on the exit side (3–6). Both Chd1 and ISWI possess a DNA-binding domain that binds to DNA flanking the nucleosome (7–9), yet it has been unclear how this domain, binding to DNA either entering or exiting the nucleosome, controls action of the ATPase motor.
Recent work suggests that remodelers shift nucleosomes using a twist defect mechanism (10–13). Like other remodelers, the ATPase motor of Chd1 shifts nucleosomes by engaging DNA at an internal location on the nucleosome called superhelix location 2 (SHL2) (9). At this site, the ATPase motor begins shifting DNA on the entry side prior to ATP binding (10), with the ADP (adenosine diphosphate)-bound and nucleotide-free states of the motor pulling the tracking strand of the DNA duplex toward itself by 1 nt (11, 12). We refer to this DNA distortion, where only the tracking but not guide strand of nucleosomal DNA has been shifted, as the tracking strand bulge. According to the model, the tracking strand bulge transitions to a twist defect when the ATPase motor binds ATP, which enables the motor to further distort the DNA duplex and pull in an entire base pair (bp) of DNA at SHL2 (13). For nucleosome repositioning, the twist defect must shift further onto the nucleosome, which is proposed to occur through a corkscrew shift of DNA toward the dyad that transforms the distorted DNA back to canonical DNA at SHL2. Twist defects therefore provide a means for ratcheting DNA around the histone core in a discontinuous process, initiated when the ATPase motor engages with nucleosomal DNA without ATP.
Chd1 has two autoregulatory elements that flank the ATPase motor. One regulatory element is a pair of N-terminal chromodomains that can interfere with ATPase action (14). The ATPase motor has two RecA-type domains that must jointly grip DNA to stimulate DNA translocation. As shown in a Chd1 crystal structure, the chromodomains can simultaneously bind to both ATPase lobes and inhibit the motor with an acidic helix that packs against and occludes the DNA-binding surface of ATPase lobe 2 (14). To accommodate this binding mode of the chromodomains, the two lobes of the ATPase motor must be widely separated, which prevents them from achieving the tight interface required for ATP hydrolysis. Supported by increased rates of ATP hydrolysis when the chromodomain/lobe 2 interface was disrupted, this inhibitory interface of the chromodomains was proposed to help discriminate nucleosomes from naked DNA (14).
The other regulatory element identified in the Chd1 crystal structure was an extended peptide segment immediately following the ATPase motor, which we named the bridge. Packing against both ATPase lobes, the bridge has the potential to directly influence ATPase activity, and, based on sequence homology, this element was also identified in ISWI remodelers (14). Subsequent work suggested that this segment was an inhibitory element in ISWI, termed NegC, that blocks sliding in the absence of the DNA-binding domain (15). The bridge/NegC element appears linked to DNA sensing, as both Chd1 and ISWI could still slide nucleosomes but lost centering activity when this element was replaced with flexible Gly/Ser-rich linkers (16, 17). How the bridge/NegC element inhibits the ATPase motor has not been investigated.
In Chd1-nucleosome complexes trapped with ADP•BeF3–, the DNA-binding domain was shown to simultaneously bind to exit DNA while contacting the chromo-ATPase unit bound at SHL1 and SHL2 (9, 18, 19). We have hypothesized that such a domain arrangement should be inhibitory, since the DNA-binding domain on exit DNA would make the remodeler insensitive to whether or not DNA is available on the entry side (9). However, apparently counter to this idea, the Chd1 cryo-electron microscopy (cryo-EM) structures have revealed an ATPase motor in a hydrolysis-competent state, fully engaged with nucleosomal DNA (18, 19). A central unanswered question is whether or not nucleosome sliding is stimulated when the DNA-binding domain is bound to exit DNA and, if not, how the organization observed in cryo-EM structures can be understood.
Here, we describe how regulatory elements of the Chd1 remodeler interrupt the twist defect mechanism to determine the direction of nucleosome sliding. To clarify the contributions of entry and exit DNA, we used asymmetrically blocked nucleosomes that can only be shifted in one direction. With this system, we show that yeast Chd1 is autoinhibited when there is no entry DNA available for its DNA-binding domain. Full autoinhibition requires cooperative contributions from both the chromodomains and bridge and is relieved when the DNA-binding domain can bind entry-side DNA. The chromodomains and bridge weaken interactions of the remodeler to nucleosomes in nucleotide-free and ADP-bound states, and this disruption destabilizes the tracking strand bulge in the absence of entry-side DNA. Our model integrates autoregulation of the ATPase motor with the twist defect cycle, providing a mechanistic basis for how chromatin remodeling can be directed by DNA availability outside the nucleosome.
Results
Asymmetrically Blocked Nucleosomes Provide Direct Evidence of Remodeler Autoinhibition.
A major challenge in understanding how remodelers directionally slide nucleosomes is that every nucleosome presents two substrates. Not only is each half of the nucleosome related—entry DNA for a remodeler on one face of the nucleosome is exit DNA for a remodeler on the other face—but remodeler action on one face changes the availability of DNA on both sides. Moreover, since sliding on each side shifts DNA in opposite directions, remodeler action on one side eliminates remodeling products created from action on the other side. Thus, when one side of the nucleosome is highly stimulating, it is impossible in bulk to directly measure remodeler activity on the more poorly stimulating side. We have proposed that Chd1 is autoinhibited when its DNA-binding domain is on the exit side (9, 17). However, the inability to directly study remodeler action on the slower acting side of the nucleosome has prevented confirmation of an autoinhibitory state on the nucleosome.
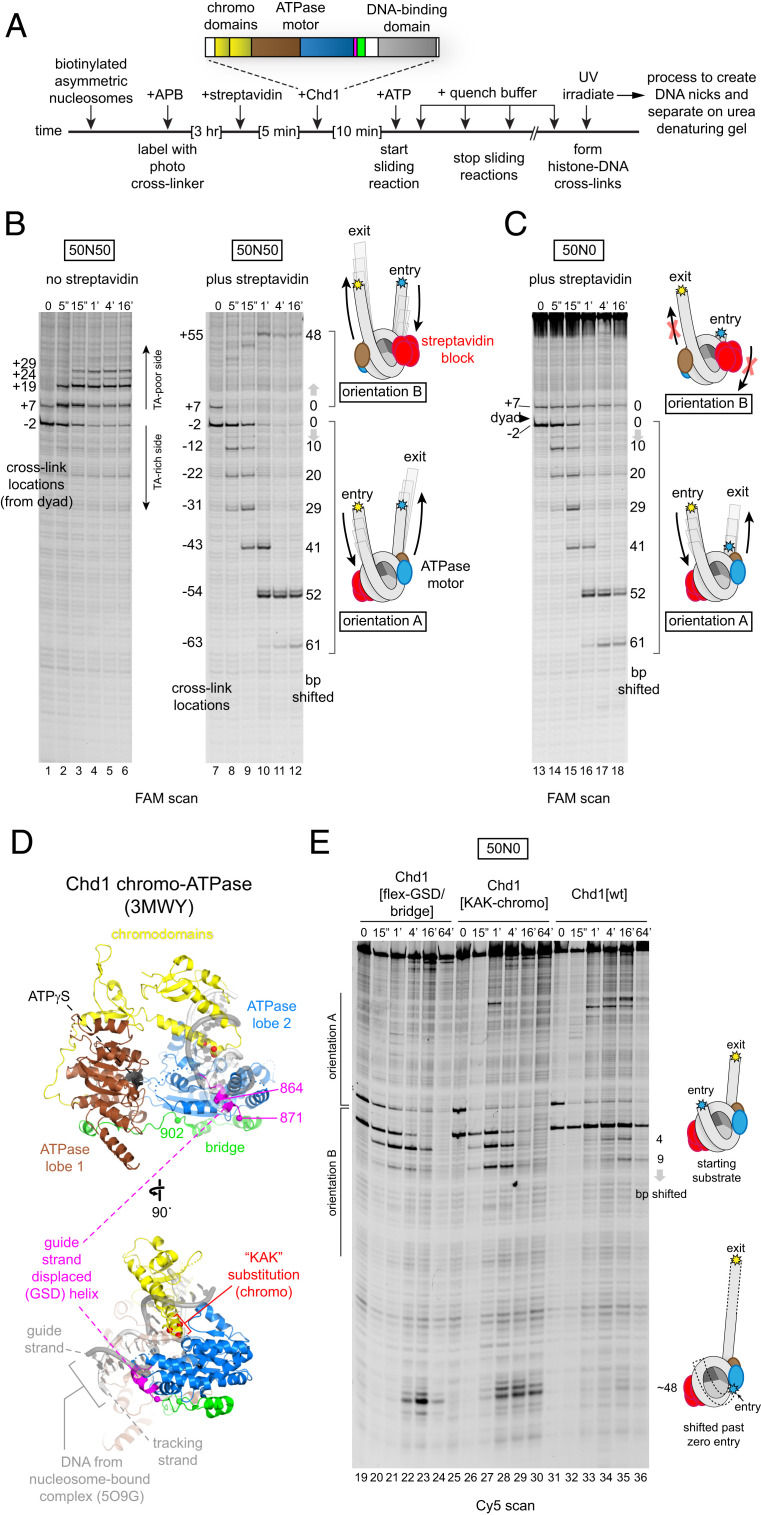
To isolate remodeling reactions on each side of the nucleosome, we built on the design by Rando and Kaufman for creating an asymmetric H3/H4 tetramer (20, 21). In this design, two variants of H3, called H3X and H3Y, have unique amino acid substitutions at the H3–H3 interface that enforce hetero-oligomerization, such that an H3X/H4 dimer can only form a tetramer with an H3Y/H4 dimer and vice versa (20). To block one side, we introduced an H3X(Q76C) substitution for attaching a biotin moiety, where binding of a streptavidin should sterically interfere with remodeler action (SI Appendix, Fig. S1). Since the histone core can deposit on DNA in either orientation, we also introduced an H3Y(M120C) substitution for cross-linking DNA at the nucleosome dyad (10, 22). Cross-linking with 4-azidophenacyl bromide (APB) site-specifically nicks DNA, allowing us to distinguish between the two orientations of the histone core (called “A” and “B,” SI Appendix, Fig. S1).
To test this design, we generated 50N50 nucleosomes using the asymmetric H3X/H3Y tetramer and either the canonical Widom 601 positioning sequence (23) or a 601 variant (601[swap SHL2.5/3.5]) (10, 24) and performed ultraviolet (UV) cross-linking before and after sliding by Chd1 (Fig. 1A). As we previously reported, Chd1 preferentially shifts the histone octamer toward the thymidine-adenosine dinucleotide (TA)-poor side of the 601 sequence (24). Here, in the absence of streptavidin, Chd1 showed the same behavior with both M120C cross-linking sites shifting up the gel and away from the TA-rich side of the 601 sequence (Fig. 1B, lanes 1 to 6). Strikingly, Chd1 produced a unique pattern of M120C cross-linking in the presence of streptavidin. Upon addition of ATP, Chd1 shifted the two distinct nucleosome orientations toward opposite DNA ends: the lower M120C cross-link (representing orientation A) moved down the gel (toward the 6-carboxyfluorescein [FAM] label), whereas the upper M120C cross-link (representing orientation B) shifted higher up in the gel (away from the FAM label; lanes 7 to 12). For both orientations, the majority of M120C cross-links shifted ∼50 nt away from their starting locations, corresponding to pulling all of the 50 bp of entry DNA onto the histone core. Thus, for both orientations of H3X/H3Y, Chd1 moved these streptavidin/biotin-H3X/H3Y nucleosomes unidirectionally, and sliding slowed down or stopped when no more entry-side DNA was available.
Fig. 1.
Disruption of autoregulatory elements allows nucleosome sliding in the absence of entry-side DNA. (A) Schematic of nucleosome sliding assay, where the direction of histone octamer movement is detected via photo–cross-linking APB-labeled H3Y(M120C). Magenta represents the GSD helix and green represents the bridge. (B) The biotin-H3X/H3Y nucleosome design produces two populations of nucleosomes that shift in opposite directions in the presence of streptavidin. After labeling with APB, 50N50 nucleosomes (150 nM) made with the canonical Widom 601 sequence were incubated with 1 µM Chd1 in either absence or presence of 10 µM streptavidin, and nucleosome sliding was initiated with the addition of 2 mM ATP. Each time point was quenched with EDTA, UV-irradiated, and processed to produce DNA nicks that reveal cross-linking from H3Y[M120C]. The samples were analyzed by a urea denaturing gel. Note that upper and lower cross-links migrate in opposite directions upon the addition of the streptavidin block, indicative of the asymmetric H3X/H3Y tetramer assembled in either orientation A or orientation B as described in SI Appendix, Fig. S1. (C) Chd1 cannot shift streptavidin/biotin-H3X/H3Y nucleosomes that lack entry-side DNA. Nucleosome sliding reactions were carried out as in A but here with 50N0 nucleosomes also made with the canonical Widom 601 sequence and lacking flanking DNA on the TA-poor side. Extended gels and Cy5 scan are given in SI Appendix, Fig. S2. (D) The crystal structure of the chromo–ATPase-bridge portion of Chd1 in an inhibited conformation (14). Superimposed is a DNA duplex from the Chd1-nucleosome complex (18) based on its location relative to lobe 2. (E) The GSD helix/bridge and chromodomains are required to block Chd1 sliding when entry-side DNA is absent. Nucleosome sliding reactions contained 150 nM 50N0 streptavidin/biotin-H3X/H3Y nucleosomes made with the 601[swap SHL2.5/3.5] sequence and 1 µM Chd1 variants incubated in the presence of 10 µM streptavidin and 2 mM ATP.
To investigate how reduced availability of entry or exit DNA affected sliding, Chd1 remodeling experiments were performed with 50N0 biotin-H3X/H3Y nucleosomes. In the presence of streptavidin, 50N0 gave two distinct remodeling behaviors: the lower M120C cross-link (orientation A) shifted down the gel as observed for 50N50 nucleosomes, whereas the upper M120C cross-link (orientation B) remained in the same location throughout the remodeling reaction (Fig. 1C). For 50N0 nucleosomes, orientation B possessed 50 bp on the exit side and zero bp on the entry side. These experiments therefore show that yeast Chd1 cannot effectively reposition nucleosomes without entry-side DNA.
To see if the absence of entry-side DNA affected an ATP-bound state of the remodeler in solution, we used two site-specific cross-linking variants, N459C and V721C, to monitor the association of the two ATPase lobes with nucleosomal DNA (9) (SI Appendix, Fig. S3). Cross-linking of the Chd1 ATPase motor was performed with 50N0 and 50N50 streptavidin/biotin-H3X/H3Y nucleosomes (using the canonical 601) either after incubation with ATP, to allow for sliding, or with nonhydrolyzable ATP analogs (AMP-PNP or ATPγS). In all cases where ATP or an ATP analog was present, both N459C and V721C yielded strong cross-links at the SHL2 site where zero-entry DNA was available for 50N0 nucleosomes (SI Appendix, Fig. S3). Therefore, the ATP-bound state of the ATPase motor appeared able to fully engage at SHL2 even when entry-side DNA was lacking. Yet despite this engagement, yeast Chd1 was unable to reposition nucleosomes without entry-side DNA.
The Chd1 Chromodomains and Guide-Strand–Displaced Helix/Bridge Block Nucleosome Sliding in the Absence of Entry-Side DNA.
We hypothesized that nucleosomes without entry DNA were unable to be effectively moved by Chd1 due to autoinhibition. Previous work showed that disruption of the Chd1 bridge, like NegC of ISWI, eliminated nucleosome centering (16, 17). We disrupted the bridge by substituting residues 864 to 902 with a Gly/Ser-rich linker (called Chd1[flex-GSD/bridge]). This segment encompasses the first half of the bridge, which contacts ATPase lobe 2, and a preceding helix that we refer to as the guide-strand–displaced (GSD) helix because it is sterically incompatible with the DNA guide strand of the duplex in Chd1-nucleosome complexes (Fig. 1D).
We tested nucleosome sliding of 50N0 biotin-H3X/H3Y nucleosomes in the presence of streptavidin, and unlike wild type, Chd1[flex-GSD/bridge] robustly shifted orientation B nucleosomes, which lacked entry-side DNA (Fig. 1E, lanes 19 to 24). For these nucleosomes, Chd1[flex-GSD/bridge] shifted DNA ∼48 bp such that the DNA end was shifted to the internal SHL2 site where the ATPase motor acts.
To see if the chromodomains also contributed to autoinhibition in the absence of entry DNA, we introduced a triple mutation into the chromodomains—E265K, D266A, and E268K—called Chd1[KAK-chromo], which removes acidic residues that contact the DNA-binding surface of ATPase lobe 2 in the crystal structure (Fig. 1D) (14). In a native gel sliding assay, disruption of the chromodomains, unlike the bridge, did not appear to alter nucleosome centering (SI Appendix, Fig. S4). However, Chd1[KAK-chromo] also shifted the zero entry of 50N0 streptavidin/biotin-H3X/H3Y nucleosomes to the internal SHL2 site (Fig. 1E, lanes 25 to 30), indicating that this disruption reduces the dependence of Chd1 on entry DNA. Although similar off-the-end products were also created by the wild-type construct (Chd1[wt]), these were minor species with the dominant band being the unshifted 50N0 substrate for orientation B (lanes 31 to 36). These results therefore show that these regulatory elements—the chromodomains and GSD helix/bridge—participate in slowing or stopping nucleosome sliding by yeast Chd1 when entry-side DNA is unavailable.
Activation and Inhibition of Nucleosome Sliding Depends on the Location but Not Direct Contacts of the DNA-Binding Domain.
Entry-side DNA is presumably detected by the DNA-binding domain. As shown by cross-linking and cryo-EM structures, however, the Chd1 DNA-binding domain can also reside on exit-side DNA (9, 18, 19). While bound to exit DNA, we expect that the remodeler would not be able to sense entry DNA and thus be inhibited. On exit DNA, the DNA-binding domain uses conserved residues to contact the chromodomains, and we wondered whether these direct contacts were necessary for autoinhibition.
To see if direct contacts conveyed an inhibitory signal to the ATPase motor, we generated several Chd1 variants with mutations at and adjacent to the DNA-binding domain/chromodomain interface (SI Appendix, Fig. S5A). Deletion of SANT or SLIDE domains weakened sliding activity, and it was inconclusive whether entry/exit DNA sensing was affected. For three clusters of highly conserved residues on the DNA-binding domain—Chd1[D1033A/E1034A/D1038A], Chd1[E1178A/E1179A], and Chd1[D1201A/P1202A]—nucleosomes were shifted similarly as Chd1[wt] with the majority of orientation A nucleosomes shifting until most or all entry DNA was pulled onto the nucleosome but not further (SI Appendix, Fig. S5B). For orientation B 50N0 nucleosomes, where zero bp DNA was available on the entry side, no appreciable movement from the starting position was observed. These wild-type–like behaviors suggest that disrupting this interface did not relieve inhibition of sliding in the absence of entry-side DNA, as observed for Chd1[KAK-chromo] and Chd1[flex-GSD/bridge].
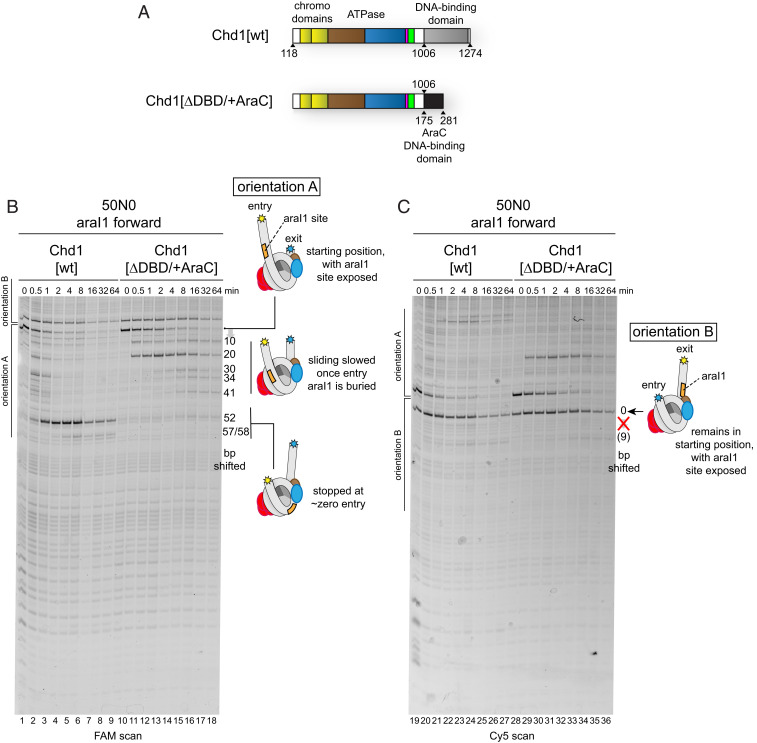
Since removing highly conserved residues at the interface between the DNA-binding domain and chromodomains appeared to have little effect on relieving inhibition, we suspected that autoinhibitory control instead resulted from the location of the DNA-binding domain—on entry versus exit DNA—without direct interdomain contacts. To test this idea, we turned to a chimeric fusion remodeler where the native Chd1 DNA-binding domain was replaced with the DNA-binding domain of the Escherichia coli AraC transcription factor, called Chd1[∆DBD/+AraC] (Fig. 2A). Our previous work showed that this and other DNA-binding domain substitutions can target chimeric Chd1 remodelers to nucleosomes containing specific binding sites recognized by the foreign domain, stimulating the remodeler to reposition these nucleosomes (25–28). As a foreign domain, the AraC DNA-binding domain should clarify whether a specific feature of the native Chd1 DNA-binding domain is required for inhibition when bound to the exit side.
Fig. 2.
Chd1 sliding activity is controlled by the location of the DNA-binding domain on entry or exit DNA. (A) Domain organization of Chd1[wt] and the chimeric Chd1[∆DBD/+AraC] fusion remodeler. Magenta represents the GSD helix, and green represents the bridge. (B) Sliding experiments using 50N0 streptavidin/biotin-H3X/H3Y nucleosomes, made with 601[swap SHL2.5/3.5], containing the araI1 sequence on extranucleosomal DNA. Sliding experiments were carried out with 150 nM 50N0 nucleosomes, 1 µM Chd1 variant, 10 µM streptavidin, and 2 mM ATP. (C) The Cy5 scan for experiments shown in B, which focuses on orientation B nucleosomes. Cartoon schematics highlight the positioning of the extranucleosomal DNA and araI1 sequence (orange) for different cross-linked products.
On 50N0 streptavidin/biotin-H3X/H3Y nucleosomes, we inserted the 17-bp DNA-binding site for AraC, called araI1, on the 50-bp linker located 3 to 20 bp from the nucleosome edge. For these experiments, due to the two orientations of the H3X/H3Y biotin–streptavidin block, the araI1 binding site will therefore either be on entry-side DNA (orientation A) or exit-side DNA (orientation B). For orientation A, the 50N0 streptavidin/biotin-H3X/H3Y nucleosomes were shifted by both Chd1[wt] and Chd1[∆DBD/+AraC] (Fig. 2B). Unlike the ∼50-bp shift observed for Chd1[wt], the majority of 50N0 nucleosomes containing the araI1 sequence were only shifted ∼20 bp by Chd1[∆DBD/+AraC], which is consistent with reduced sliding due to burial of the araI1 site. As a control, a 50N0 nucleosome lacking the araI1 site was also tested, which showed shifts in the same direction by Chd1[∆DBD/+AraC], though at a slower rate (SI Appendix, Fig. S6).
In stark contrast to entry-side targeting, Chd1[∆DBD/+AraC] was unable to significantly shift nucleosomes when its binding site was on the exit side (orientation B; Fig. 2C, lanes 28 to 36, and SI Appendix, Fig. S6). To monitor binding, we introduced the N459C variant into Chd1[∆DBD/+AraC] and performed site-specific cross-linking. Additionally, we detected a cross-link to the araI1 binding site, likely arising from a native cysteine in the AraC DNA-binding domain (SI Appendix, Fig. S7, lanes 8, 9, 13, and 14). The N459C site showed robust cross-linking to SHL2 on both sides of a 50N0 nucleosome, demonstrating that the inability to shift orientation B nucleosomes was not due to poor engagement of the motor (lanes 4, 9, 14, and 19). These results demonstrate that direct interactions with the DNA-binding domain are not required to inhibit nucleosome sliding activity. Instead, sliding activity depends on the location of the DNA-binding domain. The regulatory element most likely affected by the placement of the DNA-binding domain is the bridge. The bridge is physically coupled to the DNA-binding domain through a linker, and as we have shown here, the bridge is required for blocking nucleosome sliding when entry-side DNA is not available.
We questioned whether the bridge might be sufficient for blocking nucleosome sliding when the AraC DNA-binding domain is targeted to exit DNA. To test this, we introduced the KAK substitution into the Chd1-AraC fusion remodeler, called Chd1[KAK/∆DBD/+AraC]. On orientation B nucleosomes, with the araI1 on the exit side, the KAK substitution enabled the Chd1-AraC remodeler to pull nucleosomal DNA farther onto the histone core (SI Appendix, Fig. S8, compare lanes 41 to 43 with 45 to 48 and 50 to 53 with 55 to 58). As with the native DNA-binding domain, the KAK substitution allowed the remodeler to robustly shift the DNA end all the way to the internal SHL2 site where the ATPase motor acts (lanes 55 to 58). Importantly, Chd1[KAK/∆DBD/+AraC] shifted nucleosomes containing the araI1 site faster than those without the araI1 site (SI Appendix, Fig. S8, lanes 65 to 68), which indicates that the specific tethering via the AraC domain to exit DNA stimulated sliding. These results suggest that the bridge by itself is insufficient for preventing Chd1 sliding activity when the DNA-binding domain is on the exit side. Thus, the bridge and chromodomains work together to reinforce an autoinhibited state of the remodeler on the nucleosome.
On nucleosomes lacking entry DNA presented so far, DNA was always available on the exit side. To determine if exit-side DNA was necessary for sliding inhibition, we tested sliding of Chd1[wt] on 0N0 biotin-H3X/H3Y nucleosomes. Chd1[wt] was unable to appreciably shift these 0N0 nucleosomes (SI Appendix, Fig. S9, lanes 1 to 6 and 19 to 24). In contrast, both Chd1[flex-GSD/bridge] and Chd1[KAK-chromo] shifted these nucleosomes ≥50 bp (lanes 7 to 18 and 25 to 36), indicating that the inability for Chd1[wt] to shift 0N0 nucleosomes is therefore due to autoinhibition.
As a control, we used cross-linking of the Chd1[N459C] variant to monitor binding to nucleosomes. Chd1[wt] binds weakly to 0N0 nucleosomes (9, 17), which likely contributes to the inability to shift these nucleosomes. While Chd1[N459C] cross-linking was weaker for 0N0, it was still significant in the presence of AMP-PNP (SI Appendix, Fig. S10). Notably, cross-linking for Chd1[N459C] was extremely weak on 50N0 at the SHL2 where DNA was only available on the entry side (lanes 1 to 4). Therefore, a limitation of using N459C to monitor ATPase binding is that it appears to reflect species that prefer the presence of DNA on the exit rather than entry side of the nucleosome.
Taken together, these results suggest that, although the remodeler is inhibited when its DNA-binding domain is engaged with exit DNA, an autoinhibited state is favored when the DNA-binding domain is unable to bind to entry-side DNA.
The Chromodomains, GSD Helix, and Bridge Inhibit DNA Distortion at SHL2 in the Absence of Entry DNA.
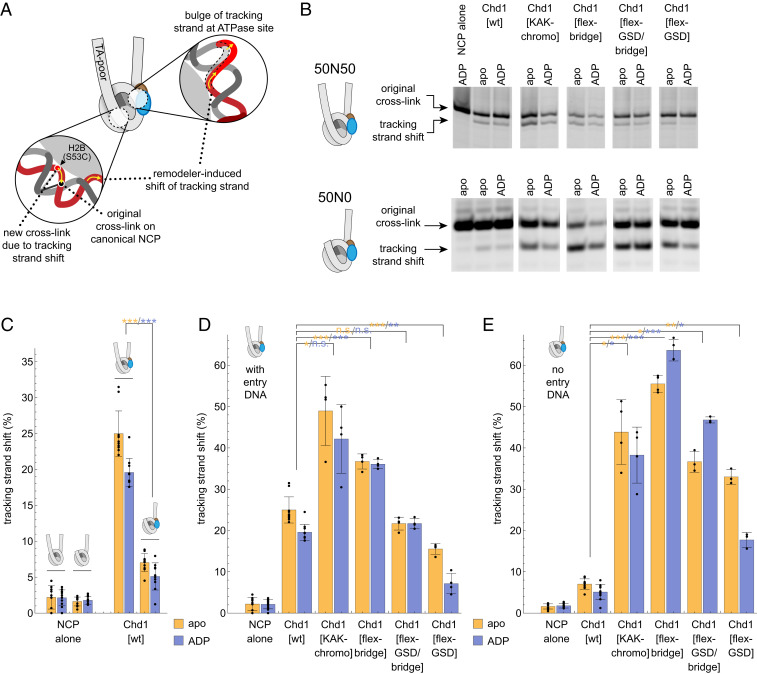
Distortion of the DNA tracking strand at SHL2 is believed to be the first step in nucleosome sliding. To see whether a tracking strand bulge requires entry DNA, we monitored Chd1’s ability to shift the DNA tracking strand on the TA-poor side of the Widom 601 by UV cross-linking to APB-labeled histone H2B(S53C) (Fig. 3A) (10). The tracking strand bulge is specifically catalyzed by the remodeler ATPase in nucleotide-free (apo) and ADP-bound states (10–12). For Chd1[wt], the cross-link corresponding to movement of the tracking strand was much stronger for 50N50 nucleosomes, which had entry DNA on the TA-poor side, compared with 50N0 nucleosomes, which lacked entry DNA (Fig. 3 B and C). This result shows that when entry DNA is unavailable, the Chd1 ATPase motor is significantly less effective in stabilizing this important first step in nucleosome sliding.
Fig. 3.
Regulatory elements of Chd1 antagonize the ability to shift the DNA-tracking strand in the absence of entry-side DNA. (A) Schematic showing how a tracking strand shift by the ATPase motor can be detected ∼30 nt away from the SHL2 binding site with H2B(S53C) cross-linking. (B) Representative gels showing H2B(S53C) cross-linking on 50N0 and 50N50 nucleosomes, made with the Widom 601 sequence. (C) The ability of Chd1[wt] to form the tracking strand shift is significantly compromised without entry-side DNA. The percent tracking strand shift was calculated as the intensity of the cross-link due to the tracking strand shift (lower band) divided by the total intensity of both cross-link bands. (D and E) Comparison of ability to induce the tracking strand shift is shown for Chd1[wt] and variants on nucleosomes with entry-side DNA (50N50, D) or lacking entry-side DNA (50N0, E). Bar graphs in C, D, and E show average values with SDs. *P < 0.01, **P < 0.0005, ***P < 0.00001, or P > 0.01 (n.s.) for comparison of each variant to the same nucleotide state of Chd1[wt].
Inhibitory elements of Chd1 appear to be responsible for the reduction in movement of the tracking strand in the absence of entry DNA. When no entry DNA was available, both Chd1[KAK-chromo] and Chd1[flex-GSD/bridge] increased the fraction of 50N0 nucleosomes showing a shift in the tracking strand (Fig. 3 B–E). To distinguish the contributions of the GSD helix and bridge, Chd1 variants with these disruptions were tested separately. Both Chd1[flex-GSD] and Chd1[flex-bridge] showed a significant increase in tracking strand movement when entry DNA was absent. For the Chd1[flex-GSD] variant, the shift in cross-linking was consistently stronger in nucleotide-free compared with ADP conditions, yet the significance of this difference is unclear since these two states yield similar structures (11, 12). Interestingly, Chd1[flex-bridge] gave a stronger signal for the tracking strand shift than Chd1[wt] even when entry DNA was present (Fig. 3D). This stronger response suggests that inhibition still occurs even in the presence of entry DNA.
Autoinhibitory Elements Weaken Remodeler-Nucleosome Interactions in the Nucleotide-Free and ADP-Bound States.
The crystal structure of the Chd1 chromo-ATPase in the absence of the nucleosome (14) provides a model for how autoinhibitory elements may block the ATPase motor from distorting DNA on the nucleosome. As shown by cross-linking and cryo-EM, when Chd1 is bound to the nucleosome, its chromodomains contact DNA at SHL1, adjacent to the ATPase motor (9, 18, 19). Comparison of these structures suggests that Chd1 could potentially bind to the nucleosome in an autoinhibited conformation, as seen in the crystal structure (SI Appendix, Fig. S11). In this model, ATPase lobe 1 and the chromodomains would bind nucleosomal DNA similarly to that observed in the Chd1-nucleosome complexes. With an inhibited conformation as observed in the crystal structure, ATPase lobe 2 would be detached from nucleosomal DNA, instead interacting with the chromodomains, GSD helix, and bridge.
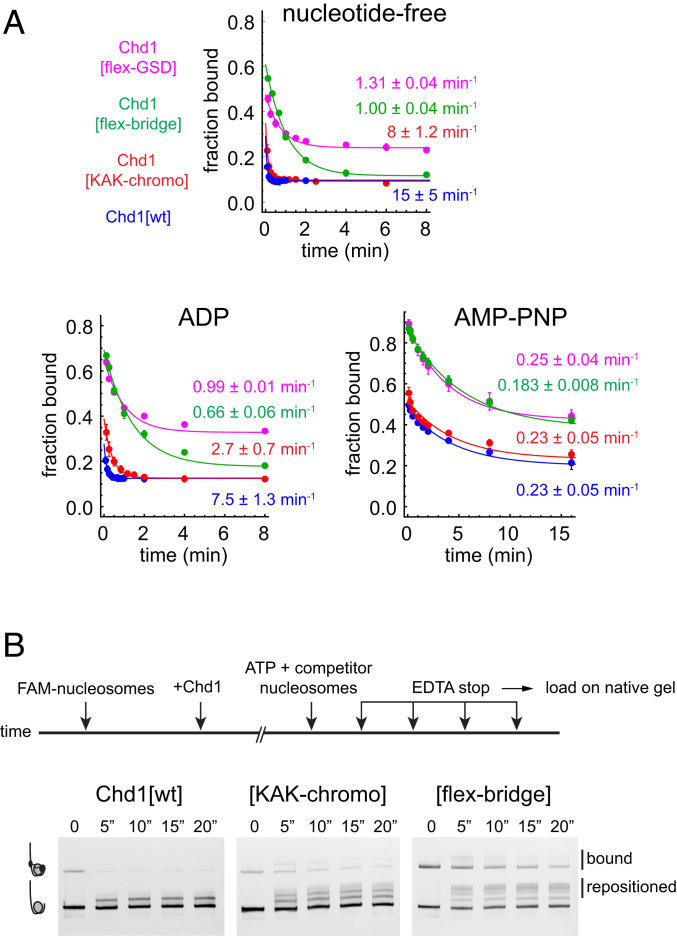
Since this partially detached configuration of Chd1 on the nucleosome would be accompanied by a loss of contacts between ATPase lobe 2 and nucleosomal DNA, we would expect such an inhibited conformation of the remodeler to be less stable on the nucleosome. To examine how inhibitory elements affect stability of Chd1-nucleosome complexes, we measured the dissociation rates of wild-type and variant Chd1 from 40N40 nucleosomes using a competition assay (29). By challenging preincubated Chd1-nucleosome complexes with a 200-fold excess of unlabeled nucleosomes, dissociation rates were calculated by monitoring the disappearance of Chd1-nucleosome complexes over time. For wild type, dissociation rates were strongly affected by nucleotide state: Chd1[wt] rapidly dissociated in nucleotide-free (15 ± 5 min−1) and ADP conditions (7.5 ± 1.3 min−1) yet was much more stable with the ATP mimic AMP-PNP (0.23 ± 0.05 min−1) (Fig. 4A).
Fig. 4.
Chd1 autoregulatory elements stimulate Chd1 dissociation from the nucleosome. (A) Disruption of the GSD helix, chromodomains, and bridge stabilize Chd1-nucleosome complexes in nucleotide-free and ADP conditions. Plots represent quantification of nucleosome dissociation data from native PAGE gels, where preincubated mixtures of 80 nM Chd1 and 20 nM FAM-labeled 40N40 nucleosomes (canonical 601 sequence) were challenged with 4 µM unlabeled 61N2 nucleosomes at time 0. Reactions contained either no nucleotide, 1 mM ADP, or 1 mM AMP-PNP. Note that since the dissociation rate for Chd1[wt] was extremely rapid, nucleotide-free and ADP experiments were conducted at 4 °C for all constructs. For each Chd1 variant and nucleotide condition, each dataset was individually fit to a single exponential decay. Overall rates and SDs were obtained by averaging fits from multiple independent experiments with the following numbers of datasets: Chd1[wt] n = 5 (apo), n = 4 (ADP), and n = 7 (AMP-PNP); Chd1[flex-GSD] n = 2 (apo), n = 2 (ADP), and n = 4 (AMP-PNP); Chd1[flex-bridge] n = 4 (apo), n = 4 (ADP), and n = 4 (AMP-PNP); and Chd1[KAK-chromo] n = 6 (apo), n = 6 (ADP), and n = 6 (AMP-PNP). The progress curves shown are fits to averaged data. (B) The chromodomains and bridge interfere with Chd1 processivity during nucleosome mobilization. Schematic and data for sliding processivity experiments using Chd1 variants. Time points are in seconds. See also SI Appendix, Fig. S14.
Interestingly, disruption of autoinhibitory elements slowed dissociation in nucleotide-free and ADP-bound states yet did not alter rates in the presence of AMP-PNP (Fig. 4A). Disruption of the GSD helix and bridge had the most dramatic effect, slowing dissociation ≥10-fold in nucleotide-free and ADP conditions. Both of these variants also showed a larger fraction of bound nucleosomes in the earliest time points, which may reflect a faster on-rate. For Chd1[KAK-chromo], dissociation was ∼threefold slower compared to Chd1[wt] in ADP conditions. In nucleotide-free conditions, the dissociation rates for Chd1[wt] were at the limit of what this assay could measure and, given the high error, were not statistically different from Chd1[KAK-chromo]. However, compared to Chd1[wt], Chd1[KAK-chromo] consistently showed a stronger signal of nucleosome-bound Chd1 persisting for the first few time points, suggestive of slower off-rates, faster on-rates, or both.
In the crystal structure, the GSD helix blocks part of the DNA-binding surface of lobe 2, whereas there is no obvious interference from the bridge. The similar dissociation rates of Chd1[flex-GSD] and Chd1[flex-bridge] could arise from a dependency of the GSD helix on the bridge to block the DNA-binding surface. However, in addition to interference, the GSD helix has a positive role as well, independently from the bridge, since disruption of the GSD helix alone (residues 864 to 871) severely impaired nucleosome sliding activity (SI Appendix, Fig. S12). Consistent with these differences in sliding activity, disruptions of the GSD helix and bridge yielded distinct profiles for steady state ATP hydrolysis rates: Chd1[flex-bridge] showed similar behavior as Chd1[wt], whereas Chd1[flex-GSD], unlike Chd1[wt], showed no significant ATPase stimulation in the presence of nucleosomes over naked DNA substrates (SI Appendix, Fig. S13). In the autoinhibited structure, the GSD helix packs against a characteristic Snf2-type insertion called the gating helix (11). We found that deletion of 634 to 653, encompassing the gating helix N terminus, crippled sliding activity (SI Appendix, Fig. S12), consistent with a similar deletion previously shown to block nucleosome sliding (19). In addition to the GSD and gating helices being critical for nucleosome sliding, both mutants also showed a similarly increased basal rate of ATP hydrolysis compared to Chd1[wt] (SI Appendix, Fig. S13), perhaps due to a functional connection of these neighboring elements.
These results show that the GSD helix and bridge have distinct, nonoverlapping roles and that the nucleotide states that allow the ATPase to distort tracking strand DNA are those that are most sensitive to autoinhibitory elements.
Autoinhibitory Elements Promote Remodeler Dissociation during Nucleosome Sliding.
Given that these autoinhibitory elements can weaken Chd1-nucleosome interactions, we questioned whether they might destabilize nucleosome binding during sliding, therefore lessening processivity. To assay for processivity, nucleosome sliding reactions were carried out in the presence and absence of unlabeled competitor nucleosomes, where competitor was added either before or concurrently with ATP. In the absence of competitor, Chd1[wt] produced several shifted species of 0N80 nucleosomes (SI Appendix, Fig. S14). As a control, competitor and labeled nucleosomes were mixed with remodeler prior to ATP addition, which blocked the sliding of the labeled 0N80 nucleosomes (SI Appendix, Fig. S14). To measure processivity, Chd1 variants were preincubated with labeled 0N80 nucleosomes, and then sliding was initiated by introducing a mixture of ATP with competitor nucleosomes. Under these conditions, Chd1[wt] only produced one new species of shifted 0N80 nucleosomes, estimated to be shifted by ∼10 bp (Fig. 4B). In contrast, Chd1[KAK-chromo] and Chd1[flex-bridge] shifted nucleosomes tens of bp under competitive conditions, as indicated by multiple nucleosome species. These experiments indicate that the autoinhibitory elements can stimulate complete release of the remodeler during the nucleosome sliding reaction.
We wondered whether the interruption of sliding processivity of Chd1[wt] arose from the creation of exit DNA during the sliding reaction. To test this, we carried out processivity experiments using 15N80 nucleosomes. With these substrates, Chd1[wt] created a similar pattern of shifted nucleosomes as for 0N80 nucleosomes (SI Appendix, Fig. S14). Therefore, initiation of nucleosome sliding appeared to be relatively unaffected by exit DNA. Previous single molecule experiments have shown that Chd1 and ISWI remodelers shift nucleosomes in multi-bp bursts (30–33). The limited processivity that we observe here for Chd1[wt] may reflect such a burst translocation phase with autoinhibition affecting the pauses between bursts, as proposed for ISWI (34).
Discussion
As directly demonstrated using asymmetrically blocked nucleosomes, we show here that yeast Chd1 requires entry DNA for robust nucleosome sliding (Fig. 1). We propose that the dependence on entry DNA arises from regulation of the tracking strand bulge, the first step in nucleosome sliding. In the absence of entry DNA, Chd1’s ability to shift the tracking strand was significantly diminished (Fig. 3C). We show that the tracking strand bulge is antagonized by the chromodomains, GSD helix, and bridge, as this DNA shift was much stronger when these regulatory elements were disrupted (Fig. 3E). We further show that the chromodomains, GSD helix, and bridge weaken Chd1-nucleosome interactions in ADP-bound and nucleotide-free states (Fig. 4A), the stages in the remodeling cycle when the ATPase motor creates a distortion in the DNA-tracking strand (10–12).
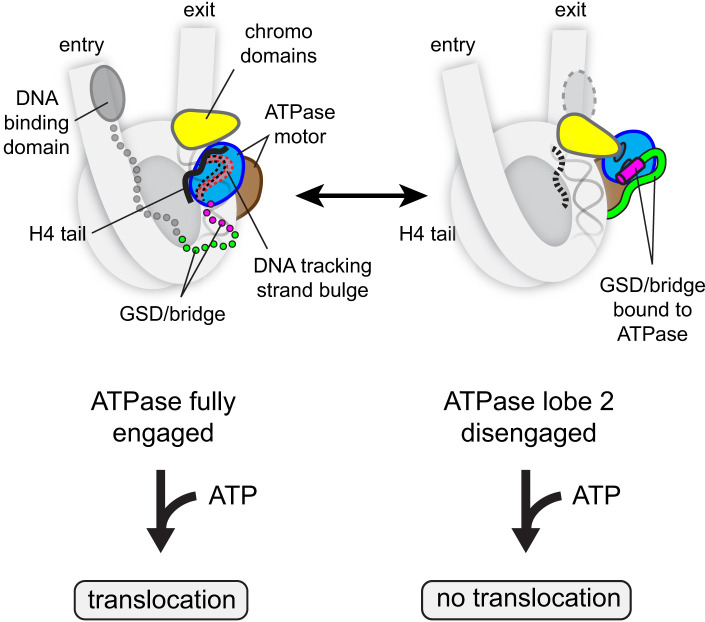
We propose that autoinhibitory elements block nucleosome sliding by promoting an inactive state of the ATPase motor on the nucleosome (Fig. 5). An inhibited conformation of the Chd1 ATPase motor, with ATPase lobe 2 bound by an acidic helix of the chromodomains, GSD helix, and the bridge (14) (Fig. 1D), is compatible with the chromodomains and ATPase lobe 1 binding to the nucleosome (SI Appendix, Fig. S11). A key point of this model is that ATPase lobe 2 can adopt multiple positions when the remodeler is bound to the nucleosome, where it can be fully engaged with nucleosomal DNA or instead bound to autoinhibitory elements (Fig. 5).
Fig. 5.
A model for allosteric regulation of the Chd1 ATPase motor on the nucleosome. The ATPase motor can sample active (Left) and autoinhibited states (Right) on the nucleosome. These states differ primarily by the location of ATPase lobe 2, which is fully engaged with nucleosomal DNA when active or held away from DNA in the inhibited form. The H4 tail (black) stabilizes the active form, whereas the chromodomains and GSD/bridge reinforce the inhibited form. The location of the DNA-binding domain allosterically regulates the GSD helix/bridge, favoring activation on entry DNA. By blocking full engagement with DNA, autoinhibitory elements cause the ATPase to bypass the twist defect cycle and therefore prevents the next ATP binding and hydrolysis event from stimulating DNA translocation.
This inhibited state has two important consequences. One is that it weakens interactions of the remodeler with the nucleosome and promotes dissociation (Fig. 4). Second, while bound to the nucleosome, the ATPase motor is unable to distort nucleosomal DNA in nucleotide-free and ADP-bound states. Given the similar dissociation rates in AMP-PNP (Fig. 4A), the ATPase motor appears relatively unaffected by autoinhibitory elements in the ATP-bound state. This suggests that autoinhibitory elements do not directly destabilize the ATPase motor when fully engaged with nucleosomal DNA. Instead, they interfere once ATPase lobe 2 releases DNA, which occurs more readily in a post-ATP hydrolysis state. Thus, by trapping ATPase lobe 2 away from DNA, autoinhibitory elements can prevent DNA distortions in the nucleotide-free and ADP-bound states of the motor. We propose that maturation of twist defects from nucleotide-free to ATP-bound states is critical for DNA translocation and that by bypassing the first stage, inhibitory elements divert the ATPase motor to a nonproductive pathway where ATP binding and hydrolysis are unable to stimulate DNA movement.
This autoinhibitory mechanism suggests how ATP hydrolysis may be uncoupled from DNA translocation (15). In addition, we expect that disruption of ATPase–DNA interactions at any stage of creating twist defects should prevent DNA translocation. Such destabilization could explain a range of so-called ATPase coupling mutants found in Chd1 and other remodelers, where sliding activity is poor despite significant levels of ATP hydrolysis (SI Appendix, Figs. S12 and S13) (11, 12, 27, 35, 36).
Like other remodelers, ATPase lobe 2 of Chd1 binds to the H4 tail when fully engaged with nucleosomal DNA (18, 19). Since nucleotide-free and ADP-bound states of the remodeler bind more weakly to nucleosomes, the H4 tail could help tip the balance toward a DNA-engaged state for lobe 2, promoting DNA distortion prior to ATP binding. Consistent with this idea, Chd1 slides nucleosomes lacking the H4 tail more slowly (37) yet can be significantly rescued by disrupting the inhibitory interface of the chromodomains (14).
Given that disruption of either the chromodomains or GSD/bridge allows for nucleosome sliding without entry DNA, these elements appear to reinforce each other. Such reinforcement likely arises from each element binding to distinct patches of the lobe 2 DNA-binding surface (Figs. 1D and 5). For the chromodomains and GSD helix, the binding surface of lobe 2 is only available when it detaches from nucleosomal DNA. By stabilizing the swung-out conformation of lobe 2 away from nucleosomal DNA, each element should increase the likelihood that the other will bind, and together, they should lengthen the lifetime of a partially disengaged state.
Autoinhibition biases the direction of nucleosome sliding based on the location of the DNA-binding domain. Although a conserved surface of the Chd1 DNA-binding domain can contact the chromodomains (18, 19) and dampen ATP hydrolysis activity when bound to exit DNA (9), our experiments with Chd1[∆DBD/+AraC] show that specific contacts with the DNA-binding domain are not required to block nucleosome sliding (Fig. 2 and SI Appendix, Fig. S6). Thus, rather than through direct contacts, sliding activity of Chd1 appears to be controlled through the location of its DNA-binding domain on the nucleosome. These results support the idea that, with the DNA-binding domain on exit DNA, Chd1-nucleosome complexes captured with ADP•BeF3– and visualized by cryo-EM (18, 19) would not be competent for DNA translocation. Thus, whereas these structures may represent an initial engagement of Chd1 with the nucleosome, robust sliding activity would require repositioning of the DNA-binding domain or ATPase motor on the nucleosome.
An attractive means by which the location of the DNA-binding domain could regulate sliding is by biasing conformational sampling of the bridge. Comparison of Chd1 and ISWI remodeler structures suggests that the bridge can exist in at least three states: fully extended and bound to both lobes of the ATPase motor, as seen for an autoinhibited state of Chd1 (14) (Fig. 1D); in a more compact state, bound only to lobe 2, as observed for ISWI (38) (SI Appendix, Fig. S15); and disordered, as in ISWI- and Chd1-nucleosome complexes with their ATPase motors fully engaged with nucleosomal DNA (12, 18, 19, 39, 40). We propose that the bridge, sampling different bound and unbound states, would be biased by the location of the DNA-binding domain (Fig. 5).
Although the GSD helix and bridge are directly connected and likely affect each other, each also plays unique roles. Disruption of the bridge alone relieves inhibition of the tracking strand bulge, more so than isolated disruption of the GSD helix (Fig. 3 D and E). A possible explanation could be an influence of the bridge on biasing the dynamics of the ATPase lobes, whose central cleft opens and closes during the hydrolysis cycle. The bridge could also exert an inhibitory effect by spatially restricting residues C-terminal to the bridge that have been shown to be important for coupling ATP hydrolysis with nucleosome sliding (35). The GSD helix, in addition to its ability to antagonize stability of Chd1-nucleosome complexes (Fig. 4A), also has a positive role in nucleosome sliding (SI Appendix, Fig. S12). Although its precise role is presently unclear, we note that the GSD helix can pack against the gating helix, which we and others have shown is also important for nucleosome sliding (SI Appendix, Fig. S12) (19), supporting the notion that these helices may jointly assist in the nucleosome sliding process.
Finally, although some aspects are specific for CHD-family remodelers, the regulatory controls we propose here also add mechanistic understanding to models describing behavior of ISWI remodelers. The ISWI family shares several architectural features with Chd1, including a C-terminal DNA-binding domain (41), the bridge (14), and the GSD helix (SI Appendix, Fig. S15). Unlike yeast Chd1, ISWI is strongly activated by binding to the entry side acidic patch, which can stimulate sliding independently of entry DNA (42, 43). Since the acidic patch binding motif of ISWI immediately precedes the DNA-binding domain (43), it appears likely that binding to the entry-side acidic patch can allosterically activate the ATPase motor analogously to the DNA-binding domain on entry DNA.
Given the continuous DNA contact likely required for translocation throughout the ATP hydrolysis cycle, we expect that for ISWI and other remodelers, capturing partially disengaged states of the ATPase motor will be a common theme for how autoregulatory elements interrupt and therefore control action of the ATPase motor.
Materials and Methods
Protein Constructs.
Primers for generating Chd1 variants are given in SI Appendix, Table S1. All amino acid substitutions were generated through PCR-based mutagenesis, using Phusion (New England Biolabs). Histone H3 amino acid changes for the asymmetric H3X/H3Y tetramer are listed in SI Appendix, Fig. S1. All other nucleosomes contained histone H3 with only the C110A substitution. For twist defect experiments, H2B[S53C] provided an attachment site for the photo-cross linker.
For experiments with streptavidin/biotin-H3X/H3Y nucleosomes, “Streptavidin-Alive” was used, which contains a C-terminal 6xHis tag in a pET21a expression vector (44). To purify biotin-H3X/H4, streptavidin binding affinity was weakened by introducing two mutations into Streptavidin-Alive as described (45), corresponding to S16A and G37T of Streptavidin-Alive. Note that Streptavidin-Alive[S16A/G37T] retains relatively tight binding to biotin.
All Chd1 constructs were based on an N- and C-terminally truncated form of Saccharomyces cerevisiae Chd1 (residues 118 to 1274), here called Chd1[wt] (25). Chd1[cys-lite] is identical to Chd1[wt], except all five native cysteines have been mutated to alanine (9). Chd1[KAK-chromo] contained three substitutions, E265K, D266A, and E268K (14); Chd1[flex-GSD] had residues 864 to 871 substituted with an 8-residue Gly/Ser linker; Chd1[flex-GSD/bridge] had residues 864 to 902 substituted with a 39-residue Gly/Ser linker; Chd1[flex-bridge] had residues 884 to 902 substituted with a 19-residue Gly/Ser linker; and Chd1[∆Ngating] had residues 634 to 653 replaced by a 5-residue linker (Gly-Ser-Ser-Ser-Gly). For Chd1[∆DBD/+AraC], residues 118 to 1,006 of Chd1 were fused with the DNA-binding domain of AraC (residues 175 to 281). Chd1[∆SLIDE] encompassed residues 118 to 1,161 (25), and Chd1[∆SANT] spanned residues 118 to 1,274 but lacked residues 1,106 to 1,125. Chd1[N459C] and Chd1[V721C] had an otherwise cysteine-free background as described (9). N459C was introduced into Chd1[∆DBD/+AraC] without mutating any native cysteines.
Protein Purification.
Histones were expressed and purified as previously described (9, 46). Chd1 proteins were expressed and purified as described (9, 17). Streptavidin was expressed in BL21 Star (DE3) cells harboring Rosetta 2 in 2xTY media and when OD600 nm = 0.5, cells were induced with 0.35 mM isopropyl-β-ᴅ-thiogalactoside for 4.5 h. Cells were resuspended in 50 mM Tris−HCl, pH 7.5, 100 mM NaCl, 1 mM ethylenediaminetetraacetic acid (EDTA), flash frozen in liquid nitrogen, and stored at −80 °C. Inclusion bodies containing streptavidin were processed in a manner similar to that of histones (46). Thawed cells were briefly incubated with 0.46 mM phenylmethylsulfonyl fluoride, 1 mM benzamidine, 1 mg/mL lysozyme, and then sonicated for three rounds (50 pulses/round). Lysate was clarified via centrifugation at 23,000 × g for 20 min at 4 °C, after which the supernatant was discarded and the pellet was resuspended in resuspension buffer + 1% Triton X-100. Inclusion bodies were spun at 23,000 × g for 10 min at 4 °C, followed by another round of resuspension followed by clarification in the presence of detergent. This was then followed by two more rounds of resuspension and centrifugation in resuspension buffer without Triton X-100. The final pellet was smeared with a spatula in a 50 mL conical tube and stored at −20 °C for further processing. For purification, streptavidin was solubilized in unfolding buffer (7 M guanidinium chloride, 20 mM Tris−HCl, pH 7.5, and 10 mM DTT) by incubating at room temperature with gentle rocking for 1 h. Streptavidin was dripped into ice cold phosphate-buffered saline (PBS) and then loaded onto a 5-mL HisTrap HP column pre-equilibrated in His⋅Bind A buffer (20 mM Tris−HCl pH 7.8, 500 mM NaCl, 10 mM imidazole, 10% glycerol, 5 mM β-mercaptoethanol). Streptavidin was eluted in a 1 M imidazole bump. Fractions were pooled and desalted into PBS over a HiPrep 26/10 Desalting column (Cytiva). Protein was aliquoted and flash frozen.
Histone Preparation.
For making biotinylated nucleosomes, the histone H3X[Q76C] variant was labeled with biotin-maleimide and then refolded with H4 by overnight dialysis against 2 M NaCl and 10 mM Tris−HCl pH 7.5. To isolate the biotinylated species, the H3X[Q76C-biotin]/H4 dimer was applied to a 5-mL HisTrap column to which ∼10 mg of Streptavidin-Alive[S16A/G37T] had been prebound. To remove nonspecifically bound proteins, the column was washed with His⋅Bind A buffer plus 1.5 M guanidinium chloride. After reaching baseline, the biotinylated H3X[Q76C-biotin]/H4 dimer was eluted with His⋅Bind A buffer plus 3 M guanidinium chloride. The H3X[Q76C-biotin]/H4 was added to unfolding buffer with an equimolar amount of H3Y[M120C]/H4 plus two equivalents of histones H2A and H2B. Refolding and purification by size exclusion chromatography was carried out as for the nonbiotinylated octamer (46).
Nucleosome Constructs.
All primers used for making nucleosomal DNA are given in SI Appendix, Table S2. DNA templates were either the canonical Widom 601 sequence (23) or a 60 variant called 601[swap SHL2.5/3.5], which swapped a 16-bp segment (24 to 39 bp from the dyad) between the TA-rich and TA-poor sides (10, 24). The Widom 601 sequence produced a biased orientation of H3X/H3Y with a preference for the H3X-half on the TA-poor side and the H3Y-half on the TA-rich side (compare orientation A [strong] and orientation B [weak] in Fig. 1B, lanes 7 to 12). In contrast, 601[swap SHL2.5/3.5] decreased the barrier for nucleosome sliding and produced a more even distribution of the two H3X/H3Y orientations (SI Appendix, Fig. S16).
Nucleosomes with the canonical Widom 601 were used for experiments shown in Figs. 1 B and C, 3, and 4 and SI Appendix, Figs. S12–S14, whereas 601[swap SHL2.5/3.5] was used for all other experiments. For experiments in Fig. 2 and SI Appendix, Figs. S6, S7, and S8, nucleosomes containing the 17-bp araI1 sequence on the DNA flanking the TA-rich side were prepared in two orientations: araI1 forward 5′ TATGGATAAAAATGCTA and araI1 reverse 5′ TAGCATTTTTATCCATA. All fluorescently labeled nucleosomal DNA fragments were generated by large-scale PCR using 5′-labeled FAM and Cy5 primers (Integrated DNA Technologies). DNA fragments for competitor nucleosomes were digested out of a 34-mer array using EcoRV to generate 61N2 DNA fragments, as previously described (9).
Site-Specific Cross-linking.
All cross-linking experiments were carried out as previously described (9, 10). For tracking strand shift experiments, the ADP stock (2 µmols or 100 mM) was treated with 2 U hexokinase in the presence of 100 mM MgCl2 and 245 mM glucose (14 µmols) for 20 min at room temperature, in 20 µl final volume.
Nucleosome Dissociation Assay.
In a 30-µl reaction volume, Chd1 (100 nM) and FAM-labeled 40N40 nucleosomes (20 nM) were preincubated at room temperature for 10 min in 1X Binding Buffer (10 mM HEPES-KOH, pH 7.6, 80 mM NaCl, 2 mM MgCl2, 1 mM DTT, 0.04 mg/mL bovine serum albumin [BSA], and 5% sucrose) with or without nucleotide (1 mM ADP or AMP-PNP). At time 0, a 200-fold excess of unlabeled 61N2 competitor nucleosomes (4 µM final concentration) was added and rapidly mixed. Native polyacrylamide gels (4 to 4.5%, 60:1 acrylamide:bis) were pre-run in 0.25xTBE for ∼20 min at 100 V. For each time point, a 2-µl aliquot was removed and immediately loaded on actively running gel. ADP and apo time courses were carried out in the cold room, whereas AMP-PNP time courses were carried out at room temperature. For AMP-PNP samples, running buffer of gels were pre-equilibrated at 4 °C and electrophoresis tanks were placed in tubs filled with ice to maintain the chilled temperature during electrophoresis. Gels were scanned on Typhoon 5, Cy2 setting. Images were quantified in ImageJ, and fraction bound was determined by scaling the intensity of Chd1-free and Chd1-bound nucleosome bands by the number of occupied SHL2 sites. Free nucleosomes were 0% bound, 1:1 Chd1-nucleosome complexes were 50% bound, and 2:1 complexes were 100% bound. Dissociation rates were calculated and plotted using Mathematica (Wolfram).
Nucleosome Sliding and Processivity Assays.
Nucleosome sliding assays were performed similarly to previously described (27). Remodeler processivity assays were carried out in sliding buffer (20 mM HEPES pH 7.5, 50 mM KCl, 100 µg/mL BSA, 5 mM MgCl2, 1 mM DTT, and 5% sucrose) at room temperature with 80 nM Chd1 and a final concentration of 30 nM 6-FAM labeled 0–601–80 or 15–601–80 nucleosomes. After a 10-min preincubation, sliding was initiated with 1 mM ATP, plus or minus 0–601–80 competitor nucleosomes (3 µM). For each time point, 1 µL of reaction was transferred to 6 µL quench buffer (20 mM HEPES pH 7.5, 50 mM KCl, 100 µg/mL BSA, 1 mM DTT, 5% sucrose, 25 mM EDTA, and 150 µg/mL salmon sperm DNA) and placed on ice. Native polyacrylamide gels (6%) were scanned on a Typhoon 5.
ATPase Assay.
Nicotinamide adenine dinucleotide-coupled ATP hydrolysis experiments were carried out as previously described (9). Data were fit using Mathematica to the standard Michaelis–Menten equation, velocity = kcat × [Chd1] × [NCP]/(KM + [NCP]) + c, where c is the basal ATPase rate.
Supplementary Material
Acknowledgments
We thank Christian Kaiser for sharing reagents and protocols for streptavidin purification, Bob Schleif for the AraC template and discussions about the araI1 binding site, and Kevin Catalan for subcloning Chd1[∆SANT]. This work was supported by the NIH (R01-GM04192 to G.D.B.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2014498118/-/DCSupplemental.
Data Availability.
All study data are included in the article and SI Appendix.
References
- 1.Gkikopoulos T., et al. , A role for Snf2-related nucleosome-spacing enzymes in genome-wide nucleosome organization. Science 333, 1758–1760 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Smolle M., et al. , Chromatin remodelers Isw1 and Chd1 maintain chromatin structure during transcription by preventing histone exchange. Nat. Struct. Mol. Biol. 19, 884–892 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang J. G., Madrid T. S., Sevastopoulos E., Narlikar G. J., The chromatin-remodeling enzyme ACF is an ATP-dependent DNA length sensor that regulates nucleosome spacing. Nat. Struct. Mol. Biol. 13, 1078–1083 (2006). [DOI] [PubMed] [Google Scholar]
- 4.Stockdale C., Flaus A., Ferreira H., Owen-Hughes T., Analysis of nucleosome repositioning by yeast ISWI and Chd1 chromatin remodeling complexes. J. Biol. Chem. 281, 16279–16288 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kassabov S. R., Henry N. M., Zofall M., Tsukiyama T., Bartholomew B., High-resolution mapping of changes in histone-DNA contacts of nucleosomes remodeled by ISW2. Mol. Cell Biol. 22, 7524–7534 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schwanbeck R., Xiao H., Wu C., Spatial contacts and nucleosome step movements induced by the NURF chromatin remodeling complex. J. Biol. Chem. 279, 39933–39941 (2004). [DOI] [PubMed] [Google Scholar]
- 7.Yamada K., et al. , Structure and mechanism of the chromatin remodelling factor ISW1a. Nature 472, 448–453 (2011). [DOI] [PubMed] [Google Scholar]
- 8.Dang W., Bartholomew B., Domain architecture of the catalytic subunit in the ISW2-nucleosome complex. Mol. Cell. Biol. 27, 8306–8317 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nodelman I. M., et al. , Interdomain communication of the Chd1 chromatin remodeler across the DNA gyres of the nucleosome. Mol. Cell 65, 447–459.e6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Winger J., Nodelman I. M., Levendosky R. F., Bowman G. D., A twist defect mechanism for ATP-dependent translocation of nucleosomal DNA. eLife 7, e34100 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li M., et al. , Mechanism of DNA translocation underlying chromatin remodelling by Snf2. Nature 567, 409–413 (2019). [DOI] [PubMed] [Google Scholar]
- 12.Yan L., Wu H., Li X., Gao N., Chen Z., Structures of the ISWI-nucleosome complex reveal a conserved mechanism of chromatin remodeling. Nat. Struct. Mol. Biol. 26, 258–266 (2019). [DOI] [PubMed] [Google Scholar]
- 13.Bowman G. D., Deindl S., Remodeling the genome with DNA twists. Science 366, 35–36 (2019). [DOI] [PubMed] [Google Scholar]
- 14.Hauk G., McKnight J. N., Nodelman I. M., Bowman G. D., The chromodomains of the Chd1 chromatin remodeler regulate DNA access to the ATPase motor. Mol. Cell 39, 711–723 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clapier C. R., Cairns B. R., Regulation of ISWI involves inhibitory modules antagonized by nucleosomal epitopes. Nature 492, 280–284 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leonard J. D., Narlikar G. J., A nucleotide-driven switch regulates flanking DNA length sensing by a dimeric chromatin remodeler. Mol. Cell 57, 850–859 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nodelman I. M., et al. , The Chd1 chromatin remodeler can sense both entry and exit sides of the nucleosome. Nucleic Acids Res. 44, 7580–7591 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Farnung L., Vos S. M., Wigge C., Cramer P., Nucleosome-Chd1 structure and implications for chromatin remodelling. Nature 550, 539–542 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sundaramoorthy R., et al. , Structure of the chromatin remodelling enzyme Chd1 bound to a ubiquitinylated nucleosome. eLife 7, e35720 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ichikawa Y., et al. , A synthetic biology approach to probing nucleosome symmetry. eLife 6, e28836 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ichikawa Y., Kaufman P. D., Novel genetic tools for probing individual H3 molecules in each nucleosome. Curr. Genet. 65, 371–377 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hota S. K., et al. , Nucleosome mobilization by ISW2 requires the concerted action of the ATPase and SLIDE domains. Nat. Struct. Mol. Biol. 20, 222–229 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lowary P. T., Widom J., New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J. Mol. Biol. 276, 19–42 (1998). [DOI] [PubMed] [Google Scholar]
- 24.Winger J., Bowman G. D., The direction that the Chd1 chromatin remodeler slides nucleosomes can be influenced by DNA sequence. J. Mol. Biol. 429, 808–822 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McKnight J. N., Jenkins K. R., Nodelman I. M., Escobar T., Bowman G. D., Extranucleosomal DNA binding directs nucleosome sliding by Chd1. Mol. Cell. Biol. 31, 4746–4759 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Patel A., et al. , Decoupling nucleosome recognition from DNA binding dramatically alters the properties of the Chd1 chromatin remodeler. Nucleic Acids Res. 41, 1637–1648 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nodelman I. M., Bowman G. D., Nucleosome sliding by Chd1 does not require rigid coupling between DNA-binding and ATPase domains. EMBO Rep. 14, 1098–1103 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McKnight J. N., Tsukiyama T., Bowman G. D., Sequence-targeted nucleosome sliding in vivo by a hybrid Chd1 chromatin remodeler. Genome Res. 26, 693–704 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ren R., Ghassabi Kondalaji S., Bowman G. D., The Chd1 chromatin remodeler forms long-lived complexes with nucleosomes in the presence of ADP·BeF3- and transition state analogs. J. Biol. Chem. 294, 18181–18191 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Blosser T. R., Yang J. G., Stone M. D., Narlikar G. J., Zhuang X., Dynamics of nucleosome remodelling by individual ACF complexes. Nature 462, 1022–1027 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Deindl S., et al. , ISWI remodelers slide nucleosomes with coordinated multi-base-pair entry steps and single-base-pair exit steps. Cell 152, 442–452 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Qiu Y., et al. , The Chd1 chromatin remodeler shifts nucleosomal DNA bidirectionally as a monomer. Mol. Cell 68, 76–88.e6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kirk J., Yeast Chd1p remodels nucleosomes with unique DNA unwrapping and translocation dynamics. bioRxiv [Preprint] (2020). 10.1101/376806 (Accessed 8 January 2021). [DOI]
- 34.Hwang W. L., Deindl S., Harada B. T., Zhuang X., Histone H4 tail mediates allosteric regulation of nucleosome remodelling by linker DNA. Nature 512, 213–217 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Patel A., McKnight J. N., Genzor P., Bowman G. D., Identification of residues in chromodomain helicase DNA-binding protein 1 (Chd1) required for coupling ATP hydrolysis to nucleosome sliding. J. Biol. Chem. 286, 43984–43993 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sen P., Ghosh S., Pugh B. F., Bartholomew B., A new, highly conserved domain in Swi2/Snf2 is required for SWI/SNF remodeling. Nucleic Acids Res. 39, 9155–9166 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ferreira H., Flaus A., Owen-Hughes T., Histone modifications influence the action of Snf2 family remodelling enzymes by different mechanisms. J. Mol. Biol. 374, 563–579 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yan L., Wang L., Tian Y., Xia X., Chen Z., Structure and regulation of the chromatin remodeller ISWI. Nature 540, 466–469 (2016). [DOI] [PubMed] [Google Scholar]
- 39.Chittori S., Hong J., Bai Y., Subramaniam S., Structure of the primed state of the ATPase domain of chromatin remodeling factor ISWI bound to the nucleosome. Nucleic Acids Res. 47, 9400–9409 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Armache J. P., et al. , Cryo-EM structures of remodeler-nucleosome intermediates suggest allosteric control through the nucleosome. eLife 8, e46057 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ryan D. P., Sundaramoorthy R., Martin D., Singh V., Owen-Hughes T., The DNA-binding domain of the Chd1 chromatin-remodelling enzyme contains SANT and SLIDE domains. EMBO J. 30, 2596–2609 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Levendosky R. F., Bowman G. D., Asymmetry between the two acidic patches dictates the direction of nucleosome sliding by the ISWI chromatin remodeler. eLife 8, e45472 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dao H. T., Dul B. E., Dann G. P., Liszczak G. P., Muir T. W., A basic motif anchoring ISWI to nucleosome acidic patch regulates nucleosome spacing. Nat. Chem. Biol. 16, 134–142 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Howarth M., et al. , A monovalent streptavidin with a single femtomolar biotin binding site. Nat. Methods 3, 267–273 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu S. C., Wong S. L., Structure-guided design of an engineered streptavidin with reusability to purify streptavidin-binding peptide tagged proteins or biotinylated proteins. PLoS One 8, e69530 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Luger K., Rechsteiner T. J., Richmond T. J., Preparation of nucleosome core particle from recombinant histones. Methods Enzymol. 304, 3–19 (1999). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and SI Appendix.