Significance
The nanoscale organization of ligands and receptors is critical for cellular communication yet inherently challenging to investigate. We have here devised a DNA origami-based biointerface which allows the experimenter to adjust protein distances with nanometer precision as a means to enhance or disturb signaling while being responsive to large-scale reorganization processes during cell activation. Applying this biointerface to study the spatial requirements of T cell activation, we find that the smallest signaling-competent receptor unit consists of two stably ligated T cell receptors (TCRs) within a distance of 20 nanometers. Spatial organization of the physiological ligand pMHC, however, is not a relevant parameter of antigen-mediated T cell activation, as single, well-isolated pMHC molecules efficiently stimulate T cells.
Keywords: DNA origami, nanobiotechnology, T cell activation, pMHC, serial engagement
Abstract
T cells detect with their T cell antigen receptors (TCRs) the presence of rare agonist peptide/MHC complexes (pMHCs) on the surface of antigen-presenting cells (APCs). How extracellular ligand binding triggers intracellular signaling is poorly understood, yet spatial antigen arrangement on the APC surface has been suggested to be a critical factor. To examine this, we engineered a biomimetic interface based on laterally mobile functionalized DNA origami platforms, which allow for nanoscale control over ligand distances without interfering with the cell-intrinsic dynamics of receptor clustering. When targeting TCRs via stably binding monovalent antibody fragments, we found the minimum signaling unit promoting efficient T cell activation to consist of two antibody-ligated TCRs within a distance of 20 nm. In contrast, transiently engaging antigenic pMHCs stimulated T cells robustly as well-isolated entities. These results identify pairs of antibody-bound TCRs as minimal receptor entities for effective TCR triggering yet validate the exceptional stimulatory potency of single isolated pMHC molecules.
T cells are activated by productive T cell antigen receptor (TCR)–peptide/MHC (pMHC) interactions within the immunological synapse, the dynamic area of contact between a T cell and an antigen-presenting cell (APC). Despite demonstrably low TCR–pMHC affinities (1), T cells detect the presence of a few stimulatory antigenic pMHCs on the APC surface, where they are typically vastly outnumbered by nonstimulatory endogenous pMHCs (2, 3). While mechanisms underlying this remarkable sensitivity remain unresolved, spatial arrangement of TCRs and pMHCs and their assembly into organized structures has been regarded by some as a key parameter (4, 5), yet given the differences in experimental systems applied, no consensus has been reached.
An observation, which has often been interpreted in favor of spatial TCR–pMHC arrangements within the immunological synapse, is that upon ligand engagement, monomeric (6, 7) TCRs rapidly reorganize to form TCR microclusters, where protein kinases and adapter proteins become enriched (8–10). In particular, this has led to the notion that preclustered ligands could facilitate T cell activation at low antigen density by promoting TCR clustering (11–14). In two recent studies, T cell activation was reported to require either deliberate preclustering of single ligand molecules at spacings below 50 nm (15) or their stochastic accumulation to small clusters (16). Both studies used high-affinity ligands with long TCR dwell times, such that TCR organization mirrored ligand organization. It therefore appears plausible that certain spatial requirements for the organization of triggered TCRs control TCR-proximal signaling. Whether the same spatial requirements also apply for the organization of the physiological TCR-ligand pMHC is, however, unclear. Indeed, there are indications to the contrary: spatially well-separated pMHC–TCR binding events were observed to elicit T cell activation (17), and even in T cell–induced microclusters, individual pMHCs are not in close molecular proximity (6). In line with this, a single agonist pMHC molecule on an APC surface can trigger T cell activation (18), implying that the remarkable stimulatory potency of agonist pMHCs does not depend on their (pre)clustering.
Based on the above considerations, we hypothesized that proximity of triggered TCRs, but not necessarily of agonist pMHCs, should modulate the sensitivity of the T cell response. To test this in a defined and systematic manner, we engineered an APC-mimicking biointerface with dual functionality: first, it should generate defined exclusion zones around individual ligands to isolate them as they cluster during T cell activation, and second, it should permit the directed preorganization of single ligand molecules with nanometer precision. To this end, we employed rectangular DNA origami platforms anchored to fluid-phase planar supported lipid bilayers (SLB) and functionalized with either one of two different types of TCR ligands: high-affinity, anti-TCR, single-chain antibody fragments (scFV) to serve as templates for the organization of TCRs or low-affinity antigenic pMHCs as the natural ligand (Fig. 1A). In this fashion, our dynamic biointerface afforded experimentally controllable and defined ligand distances on the nanoscale while permitting at the same time the microscopic reorganization of ligands and receptors in the course of T cell activation.
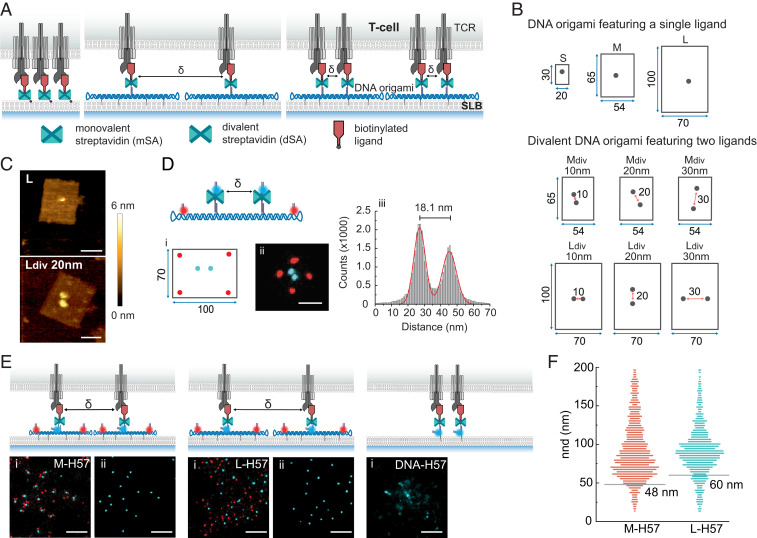
Fig. 1.
Mobile DNA origami platforms for nanoscale ligand organization. (A) Ligands directly anchored to the SLB via mSA can rearrange freely during T cell activation (Left). DNA origami platforms functionalized with ligands at predefined positions and attached to the SLB via cholesterol-modified oligonucleotides set a minimum distance δ between neighboring ligands (Middle). Divalent DNA origami platforms feature two ligands with a predefined δ (Right). (B) DNA origami layouts and nomenclature. DNA origami platforms of different sizes (small, medium, and large) were functionalized with either one (S, M, and L) or two (Mdiv, Ldiv) dSA as indicated in the schemes. Distances are given in nanometers. (C) Representative AFM images of large DNA origami platforms featuring a single ligand or two ligands spaced 20 nm apart. (Scale bar, 50 nm.) (D) Mapping ligand localization on divalent DNA origami platforms via DNA-PAINT superresolution microscopy. (i) Biotinylated ligands were replaced with biotinylated DNA-PAINT docking strands. (ii) Exemplary pseudocolor DNA-PAINT super-resolution image of large DNA origami platforms featuring two ligand attachment sites at 20 nm distance. Ligand (cyan) and platform (red) positions were imaged consecutively by Exchange-PAINT (53) using Cy3B-labeled imager strands. (Scale bar, 50 nm.) (iii) Cross-sectional histogram of ligand positions from summed DNA-PAINT localizations of 100 individual DNA origami platforms. (E) Representative pseudocolor DNA-PAINT super-resolution images of H57-decorated constructs within microclusters showing platform (red) and ligand (cyan) sites (i). Note that due to the position of the imager strand, small discrepancies between the detected position and the actual ligand position are possible. (ii) DNA-PAINT ligand positions after postprocessing. DNA-anchored H57-scFVs free to move without restrictions. DNA-H57 (δ ∼5 nm), M-H57 (δ = 48 nm), and L-H57 (δ = 60 nm) are shown. (Scale bar, 200 nm.) (F) Nnds of ligand positions identified in E. Minimum ligand distances δ are indicated as gray lines. In total, 6.8% and 13.5% of determined nnds were below δ for M and L platforms, respectively. Data are from at least 17 cells recorded in three independent experiments.
When confronting CD4+ effector T cells with this novel biointerface, we found that T cell activation via monovalent anti-TCR scFV required close proximity (≤20 nm) of ligands within units of at least two molecules, which is in line with previous studies using stably binding ligands (15, 16). Intriguingly, pMHCs did not exhibit this requirement but instead stimulated T cells robustly as well-isolated entities. Together, our results indicate that early T cell signaling emerges from small assemblies of triggered TCRs, which can be formed either by stable TCR binding of closely spaced anti-TCR scFV molecules or by repeated short-lived engagement of single pMHC molecules.
Results
Mobile DNA Origami Platforms for Nanoscale Ligand Organization.
We designed rectangular DNA origami tiles of different sizes and ligand occupancies (Fig. 1B and SI Appendix, Figs. S1–S5). At sites chosen for modification, staple strands were elongated and hybridized with biotin-conjugated oligonucleotides. To rule out double ligand occupancy at a single modification site, we used divalent streptavidin (dSA) (19, 20) for the attachment of site-specifically biotinylated TCR ligands (Fig. 1C). dSA distances on DNA origami platforms featuring two ligand attachment sites were verified via DNA points accumulation for imaging in nanoscale topography (DNA-PAINT) (21), a localization-based super-resolution microscopy method using freely diffusing dye-labeled oligonucleotides that transiently bind to their target-bound complements to achieve the necessary target “blinking” (Fig. 1D; for details, refer to SI Appendix, Fig. S5). Two types of ligands (labeled with Alexa Fluor 555, AF555) were used: 1) a single-chain antibody fragment derived from the TCRβ-reactive monoclonal antibody H57-597 (H57-scFV) (22) and 2) the nominal activating ligand of the 5c.c7 TCR, that is, the IEk-embedded moth cytochrome c peptide (pMHC).
Our goal was to create a nanostructured biointerface that allows for independent control of overall ligand surface density as well as nanoscale spacing while permitting the large-scale reorganization of ligated receptors to microclusters in the course of immunological synapse formation and T cell activation. For this, ligand-decorated DNA origami were anchored via cholesterol-modified oligonucleotides (23) to SLBs containing His-tagged adhesion (ICAM-1) and costimulatory (B7-1) molecules at a density of 100 molecules per square micrometer each. As shown in Fig. 1B, we designed DNA origami platforms of three different sizes: 30 × 20 nm (small), 65 × 54 nm (medium), and 100 × 70 nm (large). SLBs featuring ligands attached either via a His-tagged monovalent streptavidin (mSA) or a DNA/dSA anchor were used here as control, as they do not impose any external spatial constraints on ligand organization. The diffusion coefficients of all SLB-anchored DNA origami platforms were about half of that of mSA-anchored H57-scFVs, with Dorigami-scFv ∼0.25 µm2 ⋅ s−1 and DmSA-scFv = 0.6 µm2 ⋅ s−1 (SI Appendix, Fig. S6 and Table S1). Ligand occupancies of DNA origami platforms were determined via single-molecule microscopy of the biointerfaces and by comparing the platform brightness to the brightness of a single fluorescent ligand molecule (SI Appendix, Fig. S7). For DNA origami platforms with a single modification site, functionalization efficiency was ∼60%; platforms with two sites for modification yielded a mixed population featuring two ligands (∼40%), one ligand (∼45%), and no ligand (∼15%) (SI Appendix, Table S2). Note that our functionalization strategy was optimized to strictly avoid the presence of two ligands at a single modification site since double occupancies at less than 1% of modification sites may very well bias our results. For each experiment, the surface density of ligands on SLBs was assessed by relating the average fluorescence signal per area to the brightness of a single fluorescent ligand molecule.
To verify that DNA origami platforms were effective in physically separating ligands, we used DNA-PAINT super-resolution imaging to determine nearest neighbor distances (nnds) of ligands within T cell–induced microclusters (Fig. 1E and SI Appendix, Figs. S8 and S9). For this, T cell blasts isolated from CD4+ 5c.c7 TCR-transgenic mice were incubated for 10 min on biointerfaces featuring H57-scFV–functionalized DNA origami platforms at ligand densities of ∼4 µm−2 and fixed afterward. We analyzed medium (M-H57) and large (L-H57) DNA origami platforms as well as H57-scFVs directly anchored to the bilayer, which lacked DNA origami–induced exclusion zones (DNA-H57). T cells on all biointerfaces showed characteristic microcluster formation. For M-H57 and L-H57, nnds between individual ligands measured within microclusters corresponded well with minimum ligand distances δ as permitted by the DNA origami platforms (48 and 60 nm, respectively; see Fig. 1F). Note that for DNA-H57, ligand positions that could not be assigned as ligands were spaced below the resolution limit of DNA-PAINT at the experimental conditions applied (∼15 nm); here, quantitative PAINT analysis (24) yielded an estimated average ligand spacing of ∼8 nm.
Physical Separation of scFV-Ligated TCRs in Microclusters Compromises T Cell Activation.
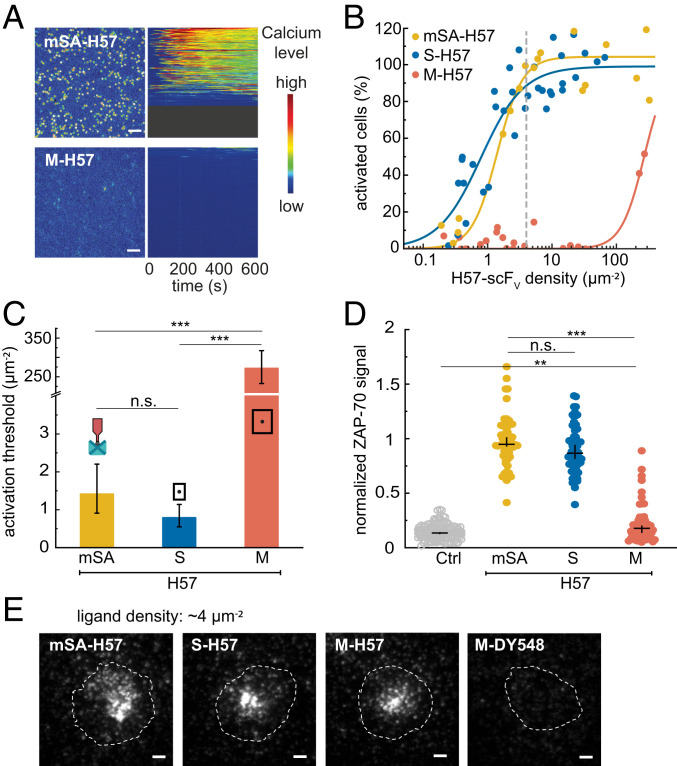
Upon TCR engagement with stimulatory ligands, TCR-proximal signaling results in increased levels of intracellular calcium acting as second messenger to promote T cell activation. We therefore conducted live-cell ratiometric calcium imaging using the calcium-sensitive dye Fura-2 AM to monitor activation levels of T cells (Fig. 2A; for details on the analysis of calcium traces, please refer to Materials and Methods and SI Appendix, Figs. S10–S12). The percentage of activated cells was determined for each biointerface at different ligand densities. Data were plotted as dose–response curves (Fig. 2B) and fitted with Eq. 6 (see Materials and Methods) to determine the ligand densities at half-maximum response, hereafter referred to as “activation threshold.” All fit parameters are listed in SI Appendix, Table S3.
Fig. 2.
H57-scFV-induced T cell activation depends on ligand spacing. (A) Pseudo-colored images of T cells recorded in a typical calcium experiment for mSA-H57 and M-H57 at a ligand density of ∼4 µm−2 (Left). Corresponding pseudo-colored calcium flux traces of >100 cells per region (Right). Cell traces (each horizontal line is one cell) are ranked by their integrated calcium flux in descending order. Calcium was imaged every 1 s. (Scale bar, 4 µm.) (B) Dose–response curves for T cell activation at different ligand surface densities. Each data point corresponds to the percentage of activated cells determined in an individual experiment at a specific ligand density. Data were normalized to a positive control (=100%) containing His-tagged ICAM-1 and B7-1 at 100 µm−2 and His-tagged pMHC (His-pMHC) at 150 µm−2. Dose–response curves were fitted with Eq. 6 (see Materials and Methods) to extract activation thresholds (C). For each dose–response curve, data are from at least three independent experiments and three different mice. For details, refer to SI Appendix, Table S3. Error bars represent the 95% CI. ***P < 0.001; n.s., not significant. A matrix containing the results of significance tests for all combinations of ligands is shown in Dataset S1. (D) ZAP-70–recruitment to the T cell plasma membrane at a ligand density of ∼50 µm−2. T cells were fixed 10 min after cell seeding and immunostained for ZAP-70. The signal was analyzed and normalized using a positive control (as in B) as a reference. Cells on bilayers containing only ICAM-1 and B7-1 at 100 µm−2 are shown as a negative control. Data are shown as medians ± CI, with each data point representing a single cell. n > 50 cells from at least three independent experiments and three different mice. ***P < 0.001; **P < 0.05; n.s., not significant. A matrix containing the results of significance tests for all combinations of ligands is shown in Dataset S2. (E) Representative TIRF images for the different constructs featuring AF555-labeled ligands at densities of ∼4 µm−2 (indicated by a vertical dashed gray line in B). The cell outline is indicated by a dashed white contour line. Images were recorded 10 min after seeding. Dy548-labeled DNA origami platforms without ligands at ∼4 µm−2 are shown for comparison. (Scale bar, 2 µm.)
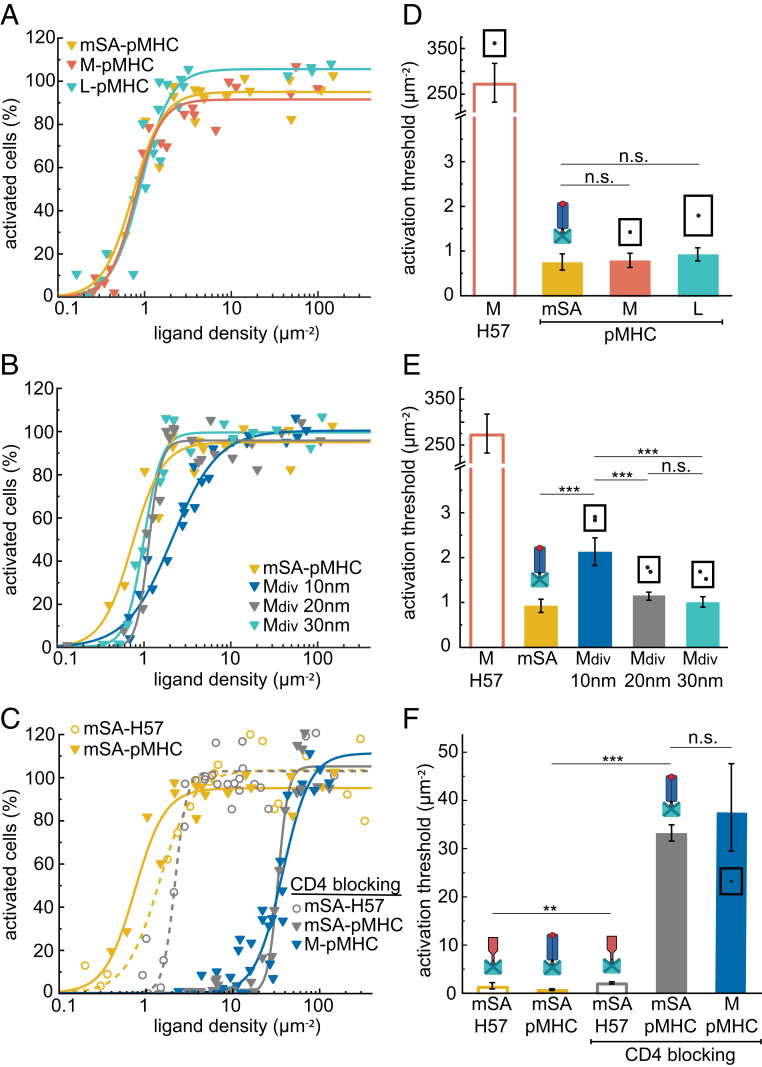
H57-scFV anchored to SLBs via either mSA (Fig. 2 B and C) or DNA/dSA (SI Appendix, Fig. S13) activated T cells at thresholds amounting to ∼1 molecule per square micrometer, similar to nominal SLB-resident pMHCs (see Fig. 4). Attachment of H57-scFV to small DNA origami platforms (S-H57), which allow for close packing of ligands down to distances δ of 20 nm (SI Appendix, Table S4), did not affect recorded activation thresholds. However, for M-H57, which prevents ligand packing below δ = 48 nm, the activation threshold increased by more than two orders of magnitude. Even at surface densities of ∼200 µm−2, at which the SLB was almost fully covered by M-H57, T cell activation was inefficient. Of note, the presence of high densities of ligand-free DNA origami platforms on SLBs (∼50 µm−2) neither activated T cells on bilayers without ligands nor impeded T cell activation driven by bilayer-anchored His-pMHC (SI Appendix, Fig. S14), indicating that the mere presence of DNA origami platforms does not interfere with T cell activation. Activation trends observed at 24 °C were maintained at 37 °C yet with lower activation thresholds (SI Appendix, Fig. S15 A and B).
Fig. 4.
T cell activation is independent of pMHC spacing. Dose–response curves of monovalent (A) and divalent (B) DNA origami platforms featuring pMHC measured at 24 °C and after blocking of CD4 with anti-CD4 Fab (C). Each data point corresponds to the percentage of activated cells determined in an individual experiment at a specific ligand density. For each construct, data are from at least three independent experiments and three different mice. Data for mSA-H57 and M-H57 are shown as references. (D–F) Activation thresholds determined from fits of data from A–C. Error bars represent the 95% CI. ***P < 0.001; **P < 0.05; n.s., not significant. A matrix containing the results of significance tests for all combinations of ligands is shown in Dataset S1.
Calcium signaling is initiated downstream of a tyrosine kinase cascade involving the TCR and TCR-proximal phosphorylation targets. The cytoplasmic tyrosine kinase ZAP-70 binds to phosphorylated immunoreceptor tyrosine-based activation motifs present on the cytoplasmic tails of the TCR-associated CD3 subunits (25). To determine whether early T cell signaling events, such as ZAP-70 recruitment, are subject to minimal ligand distance requirements, we performed immunostaining experiments on T cells, which had been fixed 10 min after their encounter with the biointerface at a ligand density of ∼50 µm−2. As shown in Fig. 2D, ZAP-70 recruitment from the cytoplasm was observed for mSA-H57 and S-H57 but not M-H57, in agreement with the calcium response recorded for H57-scFV attached to the different constructs. This implies that distancing of H57-scFVs affects TCR signaling in its earliest stages. Of note, clustering was observed for all constructs at low activating ligand densities of ∼4 µm−2, even for M-H57, which failed to promote calcium signaling (Fig. 2E and SI Appendix, Fig. S16). The fluid nature of the SLB interface allows ligands to accumulate at the T cell–SLB interface. To test whether the decreased potency of M-H57 was caused by a lower degree of ligand accumulation, we quantified the ligand density and detected a ∼3.5-fold ligand enrichment at the T cell–SLB interface, with no significant differences between constructs (SI Appendix, Fig. S17).
Pairs of Closely Spaced Ligated TCRs Are Sufficient Units for T Cell Activation.
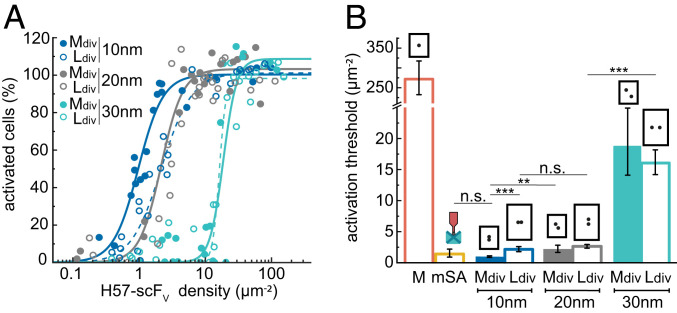
To assess the geometric requirements for efficient T cell activation, we attached two H57-scFVs to 65 × 54 nm DNA origami to generate mobile divalent platforms (Mdiv-H57) with defined distances between individual H57-scFVs of 10, 20, and 30 nm (Fig. 1 B and D). While monovalent platforms of this size no longer supported efficient stimulation (Fig. 2 A and B), divalent platforms with δ = 10 nm and δ = 20 nm ligand spacing were highly potent activators (Fig. 3 A and B). Activation thresholds for Mdiv-H57 with δ = 30 nm were one order of magnitude higher but still significantly below those measured for monovalent M-H57. Importantly, together with our data on M-H57, these results imply that platform-enforced ligand distancing rather than other platform-associated factors such as increased drag forces on the TCR-ligand complex or limited interaction with coreceptors determines the efficiency of activation.
Fig. 3.
Isolated pairs of closely spaced H57-scFVs are sufficient units for T cell activation. (A) Dose–response curves for divalent DNA origami platforms featuring two H57-scFVs at different distances. Each data point corresponds to the percentage of activated cells determined in an individual experiment at a specific ligand density. Fits for Ldiv-H57 using Eq. 6 are shown as dashed lines. For each construct, data are from at least three independent experiments and three different mice. (B) Activation thresholds determined from fits of data from A. Data for mSA-H57 are shown as reference. Error bars represent the 95% CI. ***P < 0.001; **P < 0.05; n.s., not significant. A matrix containing the results of significance tests for all combinations of ligands is shown in Dataset S1.
Although unlikely, we cannot completely rule out that two neighboring Mdiv-H57 platforms contributed to signaling. In fact, the smallest possible distance between two ligands on two different adjacent Mdiv-H57 platforms is ∼40 nm for all layouts (SI Appendix, Table S4), which is slightly below the minimum ligand separation on monovalent M-DNA origami platforms. To increase the minimal distance between two ligand pairs placed on two distinct DNA origami platforms, we increased the platform size to 100 × 70 nm (Ldiv-H57). As shown in Fig. 3, activation thresholds were not markedly affected by this; specifically, for Ldiv-H57 with 10 and 20 nm ligand spacing, for which the minimal interplatform distance between individual ligands amounted to ∼60 nm, we determined similar activation thresholds when compared to those measured for mSA-H57. These observations imply that isolated H57-scFV ligand pairs are by themselves sufficiently stimulatory to activate T cells.
Due to incomplete functionalization, Mdiv and Ldiv DNA origami platforms contained fractions of platforms carrying either two, one, or no ligands. Adding an even 10-fold molar excess of monovalent M-H57 to Mdiv-H5710nm, however, did not affect the activation threshold (SI Appendix, Fig. S18). Hence, when using a mixture of DNA origami platforms featuring two, one, or no H57scFV, only those decorated with two scFVs contributed to the signaling response. We observed efficient ZAP-70 recruitment (SI Appendix, Fig. S19A) and ligand clustering (SI Appendix, Fig. S11 D–I) for all divalent platforms.
Well-Isolated Monomeric Agonist pMHCs Efficiently Activate T Cells.
Results obtained using stably binding TCR-reactive antibodies provide information concerning spatial TCR arrangements and stoichiometries required for productive signaling. It may be unjustified, however, to conclude on similar spatial constraints underlying T cell stimulation via the physiological TCR-ligand pMHC. After all, pMHCs bind TCRs transiently with lifetimes of seconds rather than tens of minutes in a docking angle that is fairly conserved among stimulatory TCR–pMHC pairs. Furthermore, pMHCs offer a second epitope for coreceptor engagement, which is known to sensitize T cells for antigen by a factor of 10 to 50 (2). We therefore assessed whether T cell stimulation via nominal pMHCs followed similar proximity requirements as H57-scFV–mediated T cell activation.
Intriguingly, activation thresholds for nominal pMHCs did not depend on the manner of ligand presentation: pMHCs tethered to mSA (mSA-pMHC) and medium-sized DNA origami platforms (M-pMHC) gave rise to similar dose–response curves at 24 °C (Fig. 4 A and D) and at 37 °C (SI Appendix, Fig. S15 C and D). Even when placing pMHCs on large DNA origami platforms with δ = 60 nm (L-pMHC), the efficiency of T cell triggering was unperturbed.
Furthermore, preorganization of two pMHC molecules on divalent DNA origami platforms did not enhance their stimulatory potency. On the contrary, activation thresholds were slightly increased at a spacing of 10 nm, possibly due to steric hindrance between adjacent pMHCs (Fig. 4 B and E). This effect may even be underrepresented in our study since T cell activation triggered by DNA origami platforms featuring one instead of two pMHCs could not be accounted for. All pMHC constructs produced high levels of ZAP-70 recruitment (SI Appendix, Fig. S19B) and showed clustering on the SLB (SI Appendix, Fig. S20).
To test whether CD4-binding to pMHC affected the different spatial requirements for TCR triggering observed for pMHC and H57-scFV, we blocked CD4 with an anti-CD4 Fab prior to cell seeding. While this markedly increased activation thresholds, the observed effect was similar for pMHC anchored directly to the SLB or attached to DNA origami. In control experiments, T cell activation thresholds for SLB-anchored H57-scFV were only slightly increased by CD4 blocking (Fig. 4 C and F).
Discussion
The molecular organization of membrane-bound ligands and receptors at cell–cell interfaces is critical for cellular communication (26–29) yet presents invariably a formidable challenge to investigators. On the one hand, experimental approaches involving molecularly defined biochemical or structural biology techniques inherently remove molecular players from the live cell context. On the other hand, even state-of-the art imaging reaches its limits given the spatial and temporal resolution necessary to follow nanoscale reorganization processes in situ. Antigen recognition by T cells illustrates this conundrum: while central to adaptive immunity and with most molecular players already identified, knowledge on its operational principles is still limited. The key to identifying molecular mechanisms are cellular interventions that allow for manipulating the organization of molecules in a live cell environment.
With this in mind, we devised a DNA origami-based biointerface, which allowed for experimentally controllable and defined adjustment of protein distances while supporting at the same time the cell-driven spatial reorganization of ligand and receptor molecules in the course of cell activation. By imposing steric constraints to cell-mediated clustering processes, as well as by prearranging clusters of ligands, we were able to identify ligand arrangements for productive TCR triggering. While efficient T cell activation via monovalent TCRβ-reactive scFVs required close proximity of ligands within units of at least two molecules, such a requirement was absent for the natural ligand, that is, nominal pMHC (Fig. 5A), which stimulated T cells effectively when present as individual, well-separated entities. This disparity is likely rooted in the fundamentally different nature by which scFVs and pMHCs interact with the TCR.
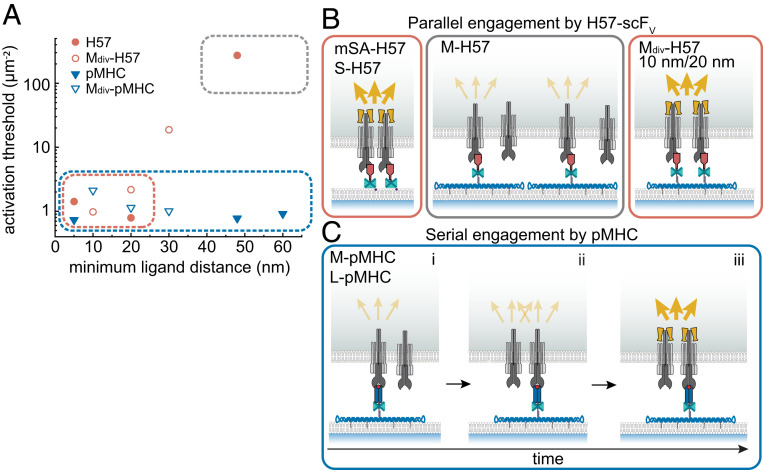
Fig. 5.
Ligand-specific spatial requirements for T cell activation. (A) Minimum ligand distances δ are plotted versus the activation threshold. For DNA origami platforms featuring a single ligand, δ corresponds to the smallest possible distances between two ligands on adjacent platforms assuming quasi-crystalline packing in clusters. For divalent platforms, it corresponds to the predefined distance between the two ligands on an individual platform. The lateral extension of mSA-anchored ligands was approximated with 5 nm. The spatial regimes allowing (red box) and prohibiting (gray box) parallel engagement via H57 as well as those that allow serial engagement via pMHC (blue box) are indicated. (B) Parallel engagement model of antibody-induced triggering. Two or more triggered TCRs within 20 nm form signaling-competent TCR assemblies, resulting in ZAP-70 recruitment (yellow dumbbells) and initiation of calcium signaling (yellow arrows). H57-scFV–triggered T cell activation requires at least two simultaneous ligand–receptor engagements within a distance of 20 nm (“parallel engagement”). This can occur via T cell–induced clustering of mSA-anchored scFVs or preorganization of scFVs on divalent DNA origami platforms (red boxes). H57-scFvs isolated on M-DNA origami platforms at δ = 48 nm fail to stimulate T cells efficiently (gray box). (C) Serial engagement model of pMHC-induced triggering (blue box). A single isolated pMHC molecule can create a signaling-competent TCR assembly by sequentially engaging multiple TCRs. (i) A single pMHC–TCR binding event does not induce efficient downstream signaling. (ii) Via serial, short-lived binding events, a single isolated pMHC molecule may engage several TCRs sequentially. (iii) If two or more TCRs within a distance of 20 nm are triggered in this way, a signaling-competent TCR assembly is created.
With an interaction half-life of ∼50 min at room temperature (22), a H57-scFV molecule can be expected to engage a single TCR and stay bound for the entire duration of the experiment. As a consequence, the specific arrangement of H57-scFV serves as a template for the molecular organization of ligated TCR molecules. In this case, T cell stimulation was only observed under conditions that allowed for close proximity of ligated TCRs in clusters (δ ≤ 20 nm). In agreement with Wind et al. (15), enforcement of minimum ligand distances of 48 nm created a signaling-impaired state. In contrast, isolated TCR pairs enforced by divalent DNA origami platforms were still signaling competent: platforms featuring scFVs at 10 and 20 nm distance and therefore closely mimicking IgG antibodies (30) were most potent.
As has become evident from our experiments involving H57-scFVs for stimulation, parallel engagement of two proximal TCRs is required for T cell activation (Fig. 5B). In case of freely diffusing ligands, the probability for the formation of such parallel engagements depends on the ligand (and receptor) density as well as the ligand–receptor dwell time. Evidently, in our system, this condition was met at a minimum density of approximately one H57-scFV molecule per square micrometer (Fig. 2C), which amounts to 100 or more scFV molecules per T cell synapse. Note that the high affinity of the H57-scFV–TCR interaction impeded the dissociation of any ligand–receptor pairs once they had formed. Similar requirements for productive signaling were previously reported in a chimeric antigen receptor system, where DNA hybridization was used to mimic receptor–ligand interactions (16); in that system, initiation of signaling was found to depend on clustering of ligated receptors, which, in turn, required high ligand densities (≥1 µm−2) and long ligand–receptor dwell times.
In a natural APC–T cell interface, however, neither of these conditions applies: the nominal antigen density on APCs can be as low as one to five antigenic pMHC molecules per cell (31, 32) and dwell times for the TCR–pMHC interaction are short (1.7 s and 100 ms at room temperature and 37 °C, respectively) (22). The probability of stochastic formation of parallel engagements is negligible, which already suggests that pMHC-triggered T cell activation follows different recognition dynamics. We have now validated this notion by actively isolating individual pMHC molecules with the use of DNA origami platforms to prohibit the formation of parallel engagements, which left the efficiency of antigen detection and associated TCR proximal signaling unperturbed. Our data suggest that TCRs triggered by pMHC give rise to similar signaling-competent TCR assemblies as does H57-scFV but without the need for either ligand preorganization or T cell–induced ligand clustering. Instead, the short dwell times allow for sequential binding of the same pMHC molecule to multiple TCRs; as a consequence, a single isolated pMHC molecule may promote an assembly of signaling-competent TCRs by serial engagement of two or more TCRs within a range of 20 nm (Fig. 5C). Groves and colleagues concluded from the activation behavior of transgenic and TCR T cells on SLBs featuring grid barriers that TCR microclusters triggered all or nothing, with two pMHC molecules per cluster substantially increasing triggering probability (33). Together with our data, this indicates that a certain number or local density of TCRs in a signaling-competent arrangement is necessary to trigger a cluster. It seems quite plausible that the environment in a real APC–T cell interface allows efficient triggering of a microcluster by a single pMHC. Indeed, sequential engagement and triggering of up to ∼200 TCRs by a single pMHC molecule has been reported (34) and proposed to enhance the T cell’s sensitivity for antigen, particularly at low antigen densities (35).
Alternatively, triggering TCR-CD3 complexes via pMHC may differ qualitatively from one following antibody binding, possibly allowing effective downstream signaling via a single, well-isolated pMHC-engaged TCR. While our results rule out a specific role for the coreceptor CD4 in such a scenario, it is conceivable that binding-induced changes in TCR-CD3 quaternary structure, which are unique to pMHC engagement, give rise to a different or more pronounced state of CD3 phosphorylation with efficient ZAP70 recruitment and activation following suit.
In conclusion, we have demonstrated that, unlike high-affinity TCR-reactive scFVs, monomeric agonist pMHCs stimulate T cells efficiently as well-isolated entities. We propose that signaling-competent TCR assemblies evolve via parallel binding of closely spaced high-affinity ligands or via serial, rapid, and short-lived engagement by a single nominal pMHC. This concept is in good agreement with recently published findings on TCR triggering by pMHC (6, 17) as well as artificial ligands (15, 16) and reconciles the concept of sensitized T cell antigen detection in the single-molecule regime (18) with functionally relevant TCR clustering (36, 37).
Materials and Methods
Assembly of DNA Origami Platforms.
DNA origami structures were assembled in a single folding reaction carried out in a test tube (AB0620, Thermo Fisher Scientific) with 10 µl folding mixture containing 10 nM M13mp18 scaffold DNA (New England Biolabs), 100 nM unmodified oligonucleotides (Integrated DNA Technologies, Eurofins), 500 nM fluorescently labeled (IBA Lifesciences) or biotinylated oligonucleotides (Biomers), and folding buffer (5 mM Tris [AM9855G, Thermo Fisher Scientific], 50 mM NaCl [AM9759, Thermo Fisher Scientific], 1 mM ethylenediaminetetraacetic acid (EDTA) [AM9260G, Thermo Fisher Scientific], and 12.5 mM MgCl2 [AM9530G, Thermo Fisher Scientific]). Oligonucleotide sequences are shown in SI Appendix, Tables S5–S8. At sites chosen for ligand attachment, staple strands were elongated at their 3′ end with 21 bases. At sites chosen for cholesterol anchor attachment, staple strands were elongated at either their 3′ end (L) or 5′ end (S and M) with 25 bases, respectively. DNA origami were annealed using a thermal protocol (90 °C for 15 min; 90 °C to 4 °C, 1 °C/min; 4 °C for 6 h) and purified using 100 kDa Amicon Ultra centrifugal filters (UFC510096, Merck). DNA origami were stored up to 4 wk at −20 °C. For functionalization, DNA origami were incubated with a 10× molar excess of dSA for 30 min at 24 °C, and excessive dSA was removed using 100 kDa Amicon Ultra centrifugal filters (UFC210024, Merck). As a last step, AF555-conjugated and site-specifically biotinylated TCR ligands (H57-scFV, pMHC) were added at a 10× molar excess for 60 min at 24 °C. Functionalized DNA origami platforms were used for experiments at the same day without further purification.
Agarose Gel Electrophoresis.
DNA origami were mixed with DNA loading buffer (B7025S, New England Biolabs) and subjected to agarose gel electrophoresis (1% agarose [A9539, Sigma-Aldrich], 1× Tris Acetate-EDTA [15558042, Thermo Fisher Scientific], 10 mM MgCl2) to validate correct folding. Agarose gels were run at 24 °C for 75 min at 100V, stained with 1× SYBR Gold Nucleic Acid Stain (S11494, Thermo Fisher Scientific), and visualized with a Gel Doc XR+ (Bio-Rad).
High-Speed Atomic Force Microscopy Imaging.
High-speed atomic force microscopy (HS-AFM) (38–40) (Research Institute of Biomolecule Metrology, Japan) was conducted in tapping mode at 24 °C in AFM imaging buffer (40 mM Tris, 2 mM EDTA, and 12.5 mM MgCl2) with free amplitudes of 1.5 to 2.5 nm and amplitude set points larger than 90%. Silicon nitride cantilevers with electron-beam deposited tips (USC-F1.2-k0.15, NanoWorld AG), nominal spring constants of 0.15 Nm−1, resonance frequencies around 500 kHz, and a quality factor of ∼2 in liquids were used. For sample preparation, 2 µl of 500 µM MgCl2 solution was preincubated on a freshly cleaved mica surface for 5 min followed by a washing step with deionized water. A total of 2 µl purified DNA origami solution (1:10 diluted in AFM imaging buffer) was applied to the mica surface for 5 min. Finally, the sample was washed with AFM imaging buffer.
Preparation of Functionalized Planar SLBs.
Vesicles containing 98% 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine and 2% 1,2-dioleoyl-sn-glycero-3-[N(5-amino-1-carboxypentyl)iminodiaceticacid]succinyl[nickel salt] (Ni-DOGS NTA) (Avanti Polar Lipids) were prepared at a total lipid concentration of 0.5 mg ⋅ ml−1 as described (22) in 10× Dulbecco’s phosphate-buffered saline (PBS) (D1408-500 mL, Sigma-Aldrich). Glass coverslips (#1.5, 24 × 60 mm, Menzel) were plasma cleaned for 10 min and attached with the use of dental imprint silicon putty (Picodent twinsil 22, Picodent) to Lab-Tek 8-well chambers (Thermo Fisher Scientific) from which the glass bottom had been removed (41). Coverslips were incubated with a fivefold diluted vesicle solution for 10 min before they were extensively rinsed with PBS (D1408-500 mL, Sigma-Aldrich). For functionalization, SLBs were first incubated for 60 min with cholesterol oligonucleotides (Integrated DNA Technologies) complementary to the elongated staple strands at the bottom side of the DNA origami and then rinsed with PBS. DNA origami were incubated on SLBs in PBS + 1% bovine serum albumin (BSA) (A9418-10G, Sigma-Aldrich) for 60 min. Finally, His10-tag ICAM-1 (50440-M08H, Sino Biological) (270 ng ⋅ mL−1) and His10-tag B7-1 (50446-M08H, Sino Biological) (130 ng ⋅ mL−1) were incubated for 75 min at 24 °C and then rinsed off with PBS. PBS was replaced with Hank’s Balanced Salt Solution (HBSS) for imaging (H8264-500 mL, Sigma-Aldrich).
Total Internal Reflection Fluorescence Microscopy.
Total internal reflection fluorescence (TIRF) microscopy experiments were performed on a home-built system based on a Zeiss Axiovert 200 microscope equipped with a 100×, numerical aperture (NA) = 1.46, Plan-Apochromat objective (Zeiss). Total internal reflection illumination was achieved by shifting the excitation beam parallel to the optical axis with a mirror mounted on a motorized table. The setup was equipped with a 488-nm optically pumped semiconductor laser (Sapphire, Coherent), a 532-nm diode-pumped solid state (DPSS) laser (Spectra Physics Millennia 6s), and a 640-nm diode laser (iBeam smart 640, TOPTICA). Laser lines were overlaid with an OBIS Galaxy Beam Combiner (Coherent). Acousto-optic modulators (Isomet) were used to modulate intensity (1 to 3 kW/cm2) and timings using an in-house developed package implemented in LabVIEW (National Instruments). A dichroic mirror (Di01-R405/488/532/635-25 × 36, Semrock) was used to separate excitation and emission light. Emitted signals were split into two-color channels using an OptoSplit II Image Splitter (Oxford Instruments) with a dichroic mirror (DD640-FDi01-25×36, Semrock) and emission filters for each color channel (ET 570/60, ET 675/50, Chroma) and imaged on the same back-illuminated Andor iXon Ultra EMCCD camera.
DNA Origami Platform Characterization and Determination of Ligand Surface Densities.
Ligand occupancies of SLB-anchored DNA origami were determined via brightness analysis. In a first step, the dark fraction of DNA origami, bearing no ligand, was determined for each construct separately via two-color colocalization experiments. For this, DNA origami were modified with a single or two biotinylated oligonucleotides labeled with Abberior STAR 635P (Biotin-DNA-AS635P) and prestained with YOYO-1 iodide (YOYO) at a concentration of 1 µg ⋅ ml−1 for 45 min at 24 °C. Excessive YOYO was removed using 100 kDa Amicon Ultra centrifugal filters, and DNA origami-bearing SLBs were produced as described above. Positions of diffraction-limited spots were determined for both color channels and corrected for chromatic aberrations as described (6). Detected signal positions were counted as colocalized if signals were within a distance of 240 nm. The fraction of DNA origami carrying at least one biotin modification, was determined by relating the number of signals in the red color channel (Biotin-DNA-AS635P) that colocalized with a signal in the green color channel (YOYO), Ncoloc_1, to the number of detected green signals, Ntotal_1 (Eq. 1). In a second step, the efficiency of functionalization of existing biotin groups with a ligand, was determined using DNA origami labeled with Biotin-DNA-AS635P and TCR ligands (H57-scFV, pMHC [both labeled with AF555]). For this, the number of green signals (H57-scFV, pMHC) that colocalized with a red signal (Biotin-DNA-AS635P), Ncoloc_2, was divided by the number of red signals (Biotin-DNA-AS635P, Ntotal_2).
| [1] |
The fraction of DNA origami carrying at least one ligand is then given by
| [2] |
For divalent DNA origami modified at two sites, ligand occupancy was evaluated by using a MATLAB (MathWorks)-based maximum-likelihood estimator to determine position, integrated brightness B, full width at half-maximum, and local background of individual signals in the images (42, 43). Briefly, DNA origami were anchored to SLBs, and the integrated brightness B was determined for all recorded positions. Images were taken at multiple different locations. The brightness values B of a monomer reference [bilayer-anchored 2xHis6-tag pMHC-555 (6)] were used to calculate the probability density function (pdf) of monomers, ρ1(B). Because of the independent photon emission process, the corresponding pdfs of N colocalized emitters can be calculated by a series of convolution integrals.
| [3] |
A weighted linear combination of these pdfs was used to calculate the brightness distribution of a mixed population of monomers and oligomers.
| [4] |
Brightness values for each DNA origami construct were pooled and used to calculate . For H57-scFV, brightness values were corrected according to the protein to dye ratio (0.93 to 1.0) of the used H57-scFV preparation (see Protein Expression and Purification). A least-square fit with Eq. 4 was employed to determine the weights of the individual pdfs, αN, with . For all fits, no higher contributions than dimers (α2) were observed. A minimum of ∼700 brightness values was applied to calculate ρ1(B) and ρ2(B). To account for DNA origami carrying no ligand, was determined by multiplying evaluated monomer α1 and dimer α2 contributions with the fraction of DNA origami carrying at least one ligand.
| [5] |
For mobility analysis of DNA origami, ∼10 image sequences were recorded at different locations on the SLB at an illumination time of 3 ms and a time lag of 10 ms. Images were analyzed using in-house algorithms implemented in MATLAB (44). Mean-square displacements (MSDs) were averaged over all trajectories, plotted as a function of time lag, and the diffusion coefficient (D) was determined by fitting the function , where denotes the localization precision; diffusion coefficients were determined from the first two data points of the MSD t-plot.
The average surface densities of AF555-labeled ligand on SLBs were determined by dividing mean intensities per square micrometer by the single-molecule brightness of bilayer-anchored AF555-labeled His-pMHC at eight positions on the SLB.
To determine the levels of nonspecific adsorption of ligands to the SLB, we incubated ligands in the absence of DNA origami platforms directly on the SLB. At a concentration of 50 nM bt-H57 and bt-pMHC (which typically yields a surface density of ∼20 ligands per square micrometer when attached to DNA origami platforms), a surface density of ∼0.1 ligands per square micrometer was detected, indicating ∼0.5% nonspecifically bound ligands. All experiments were carried out at 24 °C, unless stated otherwise.
For experiments with SLBs featuring both Mdiv-H5710nm and M-H57, platforms were functionalized separately as described above. M-H57 platforms were additionally modified with biotin-AS635P to be able to distinguish the two DNA origami platforms on the SLB. To obtain the three different molar ratios of Mdiv-H5710nm:M-H57 (1:0, 1:2, and 1:10), appropriate amounts of M-H57 were added to a SLB containing Mdiv-H5710nm after 15 min, and both constructs were then incubated for 60 min and rinsed with PBS. For imaging, PBS was replaced for HBSS.
Sample Preparation for DNA-PAINT Imaging of Divalent DNA Origami.
On divalent DNA origami platforms (Mdiv, Ldiv), biotinylated TCR ligands were replaced with biotinylated DNA-PAINT docking strands (Biotin-TEG-V-2T-P3*’) to image ligand positions. Additionally, four staple strands at the corners were extended with P1′ DNA-PAINT docking strands for barcoding. Four buffers were used for sample preparation: 1) buffer A (10 mM Tris-HCl, 100 mM NaCl, and 0.05% Tween 20, pH 8.0), 2) buffer B (5 mM Tris-HCl, 10 mM MgCl2, 1 mM EDTA, and 0.05% Tween 20, pH 8.0), 3) buffer C (1× PBS and 500 mM NaCl, pH 7.2), and 4) imaging buffer.
The imaging buffer consisted of buffer C supplemented with 1× protocatechuic acid (PCA) (40× PCA: 154 mg PCA, 10 mL water and NaOH were mixed and adjusted to pH 9.0), 1× protocatechuate 3,4-dioxygenase(PCD) (100× PCD: 9.3 mg PCD in 13.3 mL buffer [100 mM Tris-HCl, pH 8.0, 50 mM KCl, 1 mM EDTA, and 50% glycerol]), and 1× trolox (100× trolox: 100 mg trolox, 430 µl 100% methanol, and 345 µl 1M NaOH in 3.2 mL UP-H2O). Stock solutions were stored at −20 °C.
For sample preparation, a µ-slide VI 0.5 glass bottom (80607, Ibidi) was used with a channel volume of 40 µl. Solutions were always added on one side of the channel and removed from the opposite side. A total of 100 µl biotin-labeled BSA dissolved in buffer A at 1 mg ⋅ ml−1 (A8549-10MG, Sigma-Aldrich) was flushed through the channel five times and incubated for 5 min. After washing with 1 mL buffer A, 100 µl NeutrAvidin (0.5 mg ⋅ ml−1 in buffer A, [31000, Thermo Fisher Scientific]) was flushed through the channel five times and incubated for 5 min. After washing the channel with 3 mL buffer A, a solution of 100 µl biotin-modified DNA strands (Z-TEG-Biotin [M platform], Biotin-TEG-Z [L platform]) in buffer A at 1 µM was flushed through five times and incubated for 15 min. The channel was then washed with 2 mL buffer B before flushing in five times 100 µl the DNA origami in buffer B at a concentration of 500 pM. DNA origami were incubated for 20 min and washed with 1 mL buffer B. A total of 100 µl 90-nm standard gold nanoparticles (G-90-100, Cytodiagnostics) diluted 1:5 in buffer C were added and incubated for 3 min before washing with 2 mL buffer C. Finally, 500 µl the imaging solution containing 8 nM of the imager strand P1-Cy3B was flushed into image barcode positions. After washing the channel with 3 mL buffer C, 500 µl 10-nM imager strand P3-Cy3B was flushed into image ligand positions. Imaging parameters for DNA-PAINT on divalent DNA origami platforms are shown in SI Appendix, Table S9.
Sample Preparation for DNA-PAINT Imaging of DNA Origami Platforms within Microclusters.
For DNA-PAINT imaging, biotinylated oligos were extended with DNA-PAINT docking sites at their 3′ end (P3*’-2T-Z′-4T-TEG-Biotin), and DNA origami platforms were functionalized as described above by attaching biotinylated H57-scFV labeled with AS635P. A DNA-anchored H57-scFV free to move without restrictions (DNA-H57) was assembled by incubating P3*’-2T-Z′-4T-TEG-Biotin with a 10× molar excess of dSA at 24 °C for 15 min, followed by addition of H57-scFV to the second available binding pocket of the dSA. TCR ligands were added at a 10× molar excess over dSA at 24 °C for 60 min. Finally, the construct was anchored to complementary cholesterol oligonucleotides on SLBs. DNA origami without ligands were labeled by hybridizing an Alexa Fluor 647 (AF647)-labeled oligo to an elongated staple strand next to the ligand attachment site. Prior to T cell seeding, ligand surface densities on SLBs were determined as described before. Then, 4.0 × 106 cells per well were seeded onto SLBs and allowed to settle for 10 min at 24 °C. Fixation buffer (PBS, 8% formaldehyde [28908, Thermo Fisher Scientific], 0.2% glutaraldehyde [G7776-10ML, Sigma-Aldrich], 100 mM Na3VO4 [S6508-10G, Sigma-Aldrich], 1 M NaF [S7920-100G, Sigma-Aldrich]) was added 1:1 and incubated for 20 min at 24 °C before samples were rinsed with PBS. Permeabilization buffer (PBS, 100 mM Na3VO4, 1 M NaF, and 0.2% Triton X [85111, Thermo Fisher Scientific]) was added, and after 1 min, samples were washed with PBS before blocking with passivation buffer (PBS, 3% BSA) for 30 min. MgCl2 was added additionally to each buffer to a final concentration of 10 mM. Samples were extensively washed with passivation buffer, and 0.05% sodium azide was added to each well. Samples were kept up to 72 h at 4 °C prior further usage.
Before adding gold fiducials, samples were washed with 100 µl buffer C. Subsequently, 100 µl 90-nm standard gold nanoparticles, diluted 1:2 in buffer C, were added and incubated for 3 min before washing with 2 mL buffer C. Prior to image acquisition, all fluorophores were deactivated by a high-intensity bleach pulse. Imaging parameters for DNA-PAINT cell experiments are detailed in SI Appendix, Table S10. A total of 500 µl imaging buffer containing imager strand P1-Cy3B at 8 nM was flushed into image barcode positions. After washing the channel with 4 mL buffer C, 500 µl 10-nM imager strand P3-Cy3B in imaging buffer was added to image ligand positions.
DNA-PAINT Super-Resolution Microscopy Setup.
Super-resolution microscope 1.
DNA-PAINT imaging was partly carried out using an inverted Nikon Eclipse Ti microscope (Nikon Instruments) and the Perfect Focus System by applying an objective-type TIRF configuration with an oil-immersion objective (Apo SR TIRF 100×, NA = 1.49, oil). Two lasers were used for excitation: 640 nm (150 mW, Toptica iBeam smart) or 561 nm (200 mW, Coherent Sapphire). They were coupled into a single-mode fiber, which was connected to the microscope body via a commercial TIRF Illuminator (Nikon Instruments). The laser beam was passed through cleanup filters (ZET642/20 or ZET561/10, Chroma Technology) and coupled into the microscope objective using a beam splitter (ZT647rdc or ZT561rdc, Chroma Technology). Fluorescence light was spectrally filtered with two emission filters (ET705/72m or ET600/50m, Chroma Technology) and imaged with a sCMOS camera (Andor Zyla 4.2) without further magnification, resulting in an effective pixel size of 130 nm after 2 × 2 binning. Camera readout sensitivity was set to 16-bit, and readout bandwidth was set to 540 MHz.
Super-resolution microscope 2.
DNA-PAINT imaging was partly carried out using an inverted Nikon Eclipse Ti2 microscope (Nikon Instruments) with the Perfect Focus System by applying an objective-type TIRF configuration with an oil-immersion objective (Apo SR TIRF 100×, NA = 1.49, oil). A 561-nm laser (MPB Communications, 2W, DPSS system) was used for excitation and was coupled into a single-mode fiber. The beam was coupled into the microscope body using a commercial TIRF Illuminator (Nikon Instruments). The laser beam was passed through cleanup filters (ZET561/10, Chroma Technology) and coupled into the microscope objective using a beam splitter (ZT561rdc, Chroma Technology). Fluorescence light was spectrally filtered with two emission filters (ET600/50m, Chroma Technology) and imaged with a sCMOS camera (Andor Zyla 4.2) without further magnification, resulting in an effective pixel size of 130 nm after 2 × 2 binning. Camera readout sensitivity was set to 16-bit, and readout bandwidth was set to 540 MHz.
Data Analysis for DNA-PAINT.
Determination of TCR-ligand positions of divalent DNA origami.
Raw imaging data (TIFF image stacks) was subjected to spot finding and subsequent super-resolution reconstruction using the Picasso software package (45). Drift correction was performed with a redundant cross-correlation and gold nanoparticles as fiducials. For the DNA origami averaging and distance calculation, 49 to 100 individual structures were picked and averaged using the Picasso Average tool. The averaged ligand positions were plotted and fitted using the Origin software. The average distance between two potential ligand positions was then calculated using the distance between the maxima of the fit. Localization precision was determined by nearest neighbor analysis (46).
Determination of nnds within microclusters.
Raw imaging data (TIFF image stacks) was subjected to spot finding and subsequent super-resolution reconstruction using the Picasso software package. Drift correction was performed with a redundant cross-correlation and gold nanoparticles as fiducials. Channel alignment from exchange experiments was performed using gold nanoparticles as well. Microclusters were identified either based on the accumulation of localizations of individual P3-Cy3B (for DNA-H57) or P1-Cy3B (for M-H57 and L-H57) imager strand binding events. Individual ligand positions characterized by clouds of localizations were identified within the microclusters. Center positions of localization clouds were determined using a modified Ripley’s K Function, where the number of neighboring localizations within 10 nm was taken into account and the center position, defined as the localization with the highest number of neighbors, was taken as the determined ligand position. True repetitively visited ligand sites were then identified as exhibiting ≥15 localizations within a 10-nm distance of the center position. Furthermore, it was required that the mean frame number of all localizations recorded for an individual site was between 20 and 80% of the total number of frames (SI Appendix, Fig. S9). Undesired imager sticking was filtered out by removing all sites harboring more than 80% of the localizations within 5% of frames of the total acquisition. The resulting detected ligand positions were then used to measure nnds.
Calcium Imaging Experiments and Analysis.
For 20 min at 24 °C, 106 T cells were incubated in T cell media supplemented with 5 µg ⋅ ml−1 Fura-2 AM (11524766, Thermo Fisher Scientific). Excessive Fura-2 AM was removed by washing 3× with HBSS + 2% fetal bovine serum (FBS). T cells were diluted with HBSS + 2% FBS to get a final concentration of 5 × 103 cells ⋅ µl−1. Immediately after T cells landed on the functionalized SLBs, 105 cells were transferred to the Lab-Tek chamber and image acquisition was started. Fura-2 AM was excited using a monochromatic light source (Polychrome V, TILL Photonics), coupled to a Zeiss Axiovert 200M equipped with a 10× objective (Olympus) and 1.6× tube lens, and an Andor iXon Ultra EMCCD camera. A longpass filter (T400lp, Chroma) and an emission filter were used (510/80ET, Chroma). Imaging was performed with excitation at 340 and 380 nm with illumination times of 50 and 10 ms, respectively. The total recording time was 10 min at 1 Hz. Precise temperature control was enabled by an in-house–built incubator equipped with a heating unit. Unless stated otherwise, experiments were carried out at 24 °C.
ImageJ was used to generate ratio and sum images of 340/380 nm. T cells were segmented and tracked via the sum image of both channels using an in-house MATLAB algorithm based on Gao et al. (47). Cellular positions and tracks were stored and used for intensity extraction based on the ratio image. Intensity traces were normalized to the starting value at time point zero. Traces were categorized in “activating” and “nonactivating” based on an arbitrary activation threshold ratio of 0.4. The activation threshold was chosen based on a comparison of individual traces of a positive control (ICAM-1 100 µm−2, B7-1 100 µm−2, and His-pMHC 150 µm−2) and a negative control (ICAM-1 100 µm−2 and B7-1 100 µm−2) (n > 40). For generating dose–response curves, at least 20 calcium measurements (of typically ∼150 to 350 cells in a region of interest) were conducted, with each measurement at a specific ligand density. The percentage of activated cells was evaluated for each measurement and normalized to the positive control. Data were plotted as percent activated cells A as a function of ligand surface densities L to generate dose–response curves and fitted with Eq. 6 to extract the activation threshold TA, the maximum response Amax, and the Hill coefficient n:
| [6] |
All fit parameters and their 95% CIs are summarized in SI Appendix, Table S3. Statistical significance between obtained values TA,1 and TA,2 for two different data sets was determined via a bootstrap ratio test (48, 49) as follows: a bootstrap sample was obtained by drawing n data points (sampling with replacement) from a dose–response curve, where n equals the size of the data set. From each data set, 1,000 bootstrap samples were drawn and fitted via Eq. 6. This yielded activation threshold values and for each of the bootstrap samples from the two different data sets. The ratio () was calculated for each pair of bootstrap samples. If the 100 × (1 − α)% CI of log(TA,1/TA,2) did not contain 0, the null hypothesis of equal TA was rejected at a significance level of α.
T Cell Fixation and Immunostaining.
Prior to T cell seeding, ligand surface densities on SLBs were determined as described above. For 10 min at 24 °C, 2.5 × 106 cells/well were seeded onto SLBs and allowed to settle. Fixation buffer (PBS, 8% formaldehyde [28908, Thermo Fisher Scientific], 0.2% glutaraldehyde [G7776-10ML, Sigma-Aldrich], 100 mM Na3O4V [S6508-10G, Sigma-Aldrich], and 1 M NaF [S7920-100G, Sigma-Aldrich]) was added 1:1 and incubated for 20 min at 24 °C before samples were rinsed with PBS. For immunostaining, permeabilization buffer (PBS, 100 mM Na3O4V, 1 M NaF, and 0.2% Triton X [85111, Thermo Fisher Scientific]) was added, and after 1 min, samples were washed with PBS before blocking with passivation buffer (PBS, 3% BSA) for 30 min. ZAP-70 antibody labeled with AF647 (clone 1E7.2, #51-6695-82, Thermo Fisher Scientific) was added at a final concentration of 1.25 µg ⋅ ml−1 and incubated overnight at 4 °C. For experiments with DNA origami, MgCl2 was added additionally to each buffer at a final concentration of 10 mM to ensure nanoplatform integrity during fixation and immunostaining. Samples were washed extensively with passivation buffer. Imaging was performed in HBSS.
Determination of TCR Surface Densities.
Average TCR surface densities were calculated from T cells in contact with ICAM-1–functionalized SLBs and labeled to saturation with H57-scFV site-specifically labeled with AF555 (6). T cell brightness per square micrometer was then divided by the single-molecule signal of a single bilayer-anchored His-pMHC–AF555.
Tissue Culture.
Primary CD4+ T cells isolated from lymph node or spleen of 5c.c7 TCR-transgenic mice were pulsed with 1 µM moth cytochrome c (MCC) peptide 88–103 peptide (C18-reverse phase HPLC-purified; sequence: ANERADLIAYLKQATK, T cell epitope underlined, Elim Biopharmaceuticals, Inc.) and 50 U ⋅ ml−1 IL-2 (eBioscience) for 7 d to arrive at an antigen-experienced T cell culture (50). T cells were maintained at 37 °C and 5% CO2 in Roswell Park Memorial Institute (RPMI) 1640 Media (Life Technologies) supplemented with 10% FBS (Merck), 100 µg ⋅ ml−1 penicillin (Life Technologies), 100 µg ⋅ ml−1 streptomycin (Life Technologies), 2 mM l-glutamine (Life Technologies), 0.1 mM nonessential amino acids (Lonza), 1 mM sodium pyruvate (Life Technologies), and 50 µM β-mercaptoethanol (Life Technologies). Dead cells were removed on day 6 after T cell isolation by means of density-dependent gradient centrifugation (Histopaque 1119, Sigma). Antigen-experienced T cells were used for experiments on day 7 to 9.
Protein Expression and Purification.
We extended a single-chain antibody fragment of the variable domain (scFV) reactive against the murine TCR-β chain (mAb clone: H57-597) C-terminally with an AviTagTM for site-specific biotinylation followed by a 3C protease cleavable 12× histidine tag. The H57-scFV was expressed in Escherichia coli as inclusion bodies and refolded in vitro as described (22). After refolding, the H57-scFV was concentrated with a stirred 10-kDa ultrafiltration cell (PBGC04310, Merck) and purified by means of gel filtration (Superdex 200 10/300 GE Healthcare Life Sciences) using an ÄKTA pure chromatography system (GE Healthcare Life Sciences). Eluted fractions containing monomeric H57-scFV were concentrated with 10 kDa Amicon Ultra-4 centrifugal filters (Merck), treated with a 3C protease (71493, Merck) to remove the 12× histidine tag, and subjected to site-specific biotinylation with a BirA biotin ligase (ACROBiosystems). 3C protease (containing a 6× histidine tag), BirA biotin ligase (containing a glutathioneS-transferase (GST)-tag), and unprocessed H57-scFV (still containing a 12× histidine tag) were removed by a batch purification with Ni-NTA agarose (Thermo Fischer Scientific) and glutathione agarose (Thermo Fisher Scientific). The supernatant containing biotinylated H57-scFV was further purified via gel filtration using Superdex 75 (10/300 GL, GE Healthcare Life Sciences). Finally, biotinylated H57-scFVs were randomly conjugated on surface-exposed lysines with AF555 carboxylic acid, succinimidyl ester (Thermo Fisher Scientific) or Abberior STAR 635P (AS635P) carboxylic acid, and succinimidyl ester (Abberior) according to the manufacturer’s instructions. To remove excess dye, the AF555- or AS635P-conjugated and biotinylated H57-scFVs were purified by gel filtration using Superdex 75 (10/300 GL, GE Healthcare Life Sciences). Fractions containing monomeric, fluorescently labeled, and biotinylated H57-scFV were again concentrated with 10 kDa Amicon Ultra-4 centrifugal filters (UFC801024, Merck) and stored in PBS supplemented with 50% glycerol at −20 °C. The protein to dye ratio ranged between 0.93 and 1.0 for the AF555-labeled H57-scFV and was 2.0 for the AS635P-labeled H57-scFV as determined by spectrophotometry (280 to 555 nm ratio).
The 2xHis6-tag pMHC-AF555 was produced as described in ref. 6. Biotinylated pMHC was produced as follows: the I-Ek protein subunits I-Ek-biotin and I-Ekβ0 were expressed as inclusion bodies in E. coli. To arrive at I-Ek/MCC(ANP)-biotin complexes, I-Ek-biotin and I-Ekβ0 subunits were refolded in vitro in the presence of MCC(ANP) sequence: ANERADLIAYL[ANP]QATK (Elim Biopharmaceuticals, Inc.) as described (41, 51). Refolded I-Ek/MCC(ANP)-biotin complexes were purified via 14.4.4S mAb-based affinity chromatography (Cyanogen bromide-activated Agarose, Merck) followed by gel filtration (Superdex 200 10/300, GE Healthcare Life Sciences). Eluted protein fractions were site-specifically biotinylated using the BirA (ACROBiosystems) and subjected to a batch purification step with glutathione agarose (Thermo Fisher Scientific) to remove the BirA ligase and gel filtration (Superdex 200 10/300, GE Healthcare Life Sciences). Fractions containing monomeric I-Ek/MCC(ANP)-biotin complexes were concentrated with 10 kDa Amicon Ultra-4 centrifugal filters (UFC801024, Merck), snap frozen in liquid nitrogen, and stored in 1× PBS at −80 °C. The MCC peptide derivative MCC-C (sequence: ANERADLIAYLKQATKGGSC) was purchased from Elim Biopharmaceuticals, purified via reversed phase chromatography (Pursuit XRs C18 column, Agilent), and conjugated to AF555 C2-maleimide (Thermo Fisher Scientific) according to the manufacturer’s instructions. Purity of the MCC-C peptide and efficient fluorophore coupling to AF555 C2-maleimide was verified via matrix-assisted laser desorption/ionization-time-of-flightmass spectrometry (Bruker). For fluorescence labeling of pMHC, we exchanged the placeholder peptide MCC(ANP) for site-specifically AF555-conjugated MCC peptides (MCC-AF555) under acidic conditions (1× PBS and 100 mM citric acid, pH5.1) for 72 h at room temperature. Following peptide exchange, AF555-conjugated I-Ek/MCC complexes (either C-terminally extended with 2xHis6 or biotin) were subjected to gel filtration (Superdex 200 10/300 GE Healthcare Life Sciences) to remove an excess of unreacted MCC-AF555. Fractions containing monomeric AF555-conjugated I-Ek/MCC complexes were concentrated with 10 kDa Amicon Ultra-4 centrifugal filters and stored in 1× PBS supplemented with 50% glycerol at −20 °C. Quantitative I-Ek/MCC[ANP] peptide replacement (>99%) was verified via spectrophotometry (280 to 555 nm ratio).
Trans-dSA was prepared with some adaptions as described (20). The pET21a (+) vectors encoding “alive” (i.e., biotin binding) and “dead” (i.e., biotin nonbinding) streptavidin subunits were kindly provided by Alice Ting (Stanford University). We substituted the 6× histidine tag of the “alive” subunit with a cleavable 6× glutamate tag to allow for purification via cation exchange chromatography preceded by a recognition site of the 3C protease for optional removal of the tag (7). Both “alive” and “dead” streptavidin subunits were expressed in E. coli (BL-21) for 4 h at 37 °C and refolded from inclusion bodies as described (19). After refolding, the streptavidin tetramer mixture was concentrated in a stirred 10 kDa ultrafiltration cell (PBGC04310, Merck). Further concentration and buffer exchange to 20 mM Tris-HCl pH 8.0 were carried out with 10 kDa Amicon Ultra-4 centrifugal filters (Merck). The mixture of tetramers was then purified by anion exchange chromatography (MonoQ 5/50 GE Healthcare Life Sciences) using a column gradient from 0.1 to 0.4 M NaCl. dSA was eluted with 0.25 M NaCl, concentrated again (10k Da Amicon Ultra-4 centrifugal filters), and further purified via gel filtration (Superdex 200 10/300, GE Healthcare Life Sciences). After removal of the poly-E tag with the 3C protease (Merck), the protein was again subjected to gel filtration (Superdex 200 10/300 GE Healthcare Life Sciences). Monomeric fractions of dSA were then concentrated (10 kDa Amicon Ultra-4 centrifugal filters) and stored in PBS supplemented with 50% glycerol at −20 °C.
mSA was produced as described in ref. 52. In brief, the sequence of the “dead” subunit was C-terminally extended with a 3xHis6-tag for attachment to lipid bilayers containing 18:1 Ni-DOGS NTA. After expression in E. coli and refolding from inclusion bodies in vitro, the mixture of tetramers was purified by anion exchange chromatography (MonoQ 5/50 GE Healthcare Life Sciences) using a column gradient from 0.1 to 0.4 M NaCl. mSA was eluted with 0.22 M NaCl, concentrated again (10 kDa Amicon Ultra-4 centrifugal filters), and further purified via gel filtration (Superdex 200 10/300, GE Healthcare Life Sciences). After removal of the poly-E tag located on the “alive” subunit with 3C protease (Merck) followed by a gel filtration step (Superdex 200 10/300 GE Healthcare Life Sciences), the protein was stored in PBS supplemented with 50% glycerol at −20 °C.
Animal Model and Ethical Compliance Statement.
Björn Lillemeier (Salk Institute) kindly gifted us 5c.c7 αβ TCR-transgenic mice bred onto the B10.A background. Animal husbandry, breeding, and euthanization for T cell isolation was evaluated by the ethics committees of the Medical University of Vienna and approved by the Federal Ministry of Science, Research and Economy (BMWFW-66.009/0378-WF/V/3b/2016). They were performed in accordance with Austrian law (Federal Ministry for Science and Research, Vienna, Austria), the guidelines of the ethics committees of the Medical University of Vienna, and the guidelines of the Federation of Laboratory Animal Science Associations (FELASA), which match those of Animal Research: Reporting in vivo Experiments (ARRIVE). Furthermore, animal husbandry, breeding, and euthanization for T cell isolation was conducted under Project License I4BD9B9A8L, which was evaluated by the Animal Welfare and Ethical Review Body of the University of Oxford and approved by the Secretary of State of the UK Home Department. They were performed in accordance with Animals (Scientific Procedures) Act 1986, the guidelines of the ethics committees of the Medical Science of University of Oxford, and the guidelines of FELASA, which match those of ARRIVE. Both male and female mice at 8 to 12 wk old were randomly selected and killed for isolation of T cells from lymph nodes and spleen.
Statistical Analysis.
Analysis of statistical significance for Zap-70 immunostaining and ligand enrichment data were performed via bootstrap testing (48). A bootstrap sample was obtained by drawing n observations with replacement from the original data set of size n. For comparison of two populations, 1,000 bootstrap samples were drawn from each population and the medians and () were determined. The difference between the medians for each pair of bootstrap samples was calculated. If the 100 × (1 − α)% CI of the differences did not contain 0, the null hypothesis of equal medians was rejected at a significance level of α.
Supplementary Material
Acknowledgments
This work was supported by the Austrian Science Fund (FWF projects V538-B26 [E.S. and J.B.H.] and I4662-B [M.B.]; the PhD program Cell Communication in Health and Disease W1205, R.P., J.B.H. and H.S.), the TU Wien doctoral college BioInterface (J.B.H.), the European Fund for Regional Development (EFRE, IWB2020, A.K. and J.P.), the Federal State of Upper Austria (J.P.), the Vienna Science and Technology Fund (WWTF, LS13-030, G.J.S. and J.B.H.), the Boehringer Ingelheim Fonds (R.P.), the German Research Foundation through the Emmy Noether Program (DFG JU 2957/1-1, R.J.), the SFB1032 (project A11, R.J.), the European Research Council through an ERC Starting Grant (MolMap; grant agreement number 680241, R.J.), the Allen Distinguished Investigator Program through The Paul G. Allen Frontiers Group (R.J.), the Danish National Research Foundation (Centre for Cellular Signal Patterns, DNRF135, R.J.), the Human Frontier Science Program through a Young Investigator Grant (HFSP RGY0065), the Max Planck Foundation (R.J.) and the Max Planck Society (R.J.), the International Max Planck Research School for Molecular and Cellular Life Sciences (A.S.E.), the German Research Foundation through the Quantitative Biosciences Munich graduate school (T.S.), and the Wellcome Trust (Principal Research Fellowship 100262 Z/12/Z, E.K.). We thank V. Mühlgrabner for help with tissue culture.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2016857118/-/DCSupplemental.
Data Availability.
All study data are included in the article and supporting information.
References
- 1.Davis M. M., et al. , Ligand recognition by Aβ T cell receptors. Annu. Rev. Immunol. 16, 523–544 (1998). [DOI] [PubMed] [Google Scholar]
- 2.Irvine D. J., Purbhoo M. A., Krogsgaard M., Davis M. M., Direct observation of ligand recognition by T cells. Nature 419, 845–849 (2002). [DOI] [PubMed] [Google Scholar]
- 3.Purbhoo M. A., Irvine D. J., Huppa J. B., Davis M. M., T cell killing does not require the formation of a stable mature immunological synapse. Nat. Immunol. 5, 524–530 (2004). [DOI] [PubMed] [Google Scholar]
- 4.Fooksman D. R., Organizing MHC class II presentation. Front. Immunol. 5, 158 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goyette J., Nieves D. J., Ma Y., Gaus K., How does T cell receptor clustering impact on signal transduction? J. Cell Sci. 132, jcs226423 (2019). [DOI] [PubMed] [Google Scholar]
- 6.Brameshuber M., et al. , Monomeric TCRs drive T cell antigen recognition. Nat. Immunol. 19, 487–496 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rossboth B., et al. , TCRs are randomly distributed on the plasma membrane of resting antigen-experienced T cells. Nat. Immunol. 19, 821–827 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Campi G., Varma R., Dustin M. L., Actin and agonist MHC-peptide complex-dependent T cell receptor microclusters as scaffolds for signaling. J. Exp. Med. 202, 1031–1036 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yokosuka T., et al. , Newly generated T cell receptor microclusters initiate and sustain T cell activation by recruitment of Zap70 and SLP-76. Nat. Immunol. 6, 1253–1262 (2005). [DOI] [PubMed] [Google Scholar]
- 10.Varma R., Campi G., Yokosuka T., Saito T., Dustin M. L., T cell receptor-proximal signals are sustained in peripheral microclusters and terminated in the central supramolecular activation cluster. Immunity 25, 117–127 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cochran J. R., Cameron T. O., Stern L. J., The relationship of MHC-peptide binding and T cell activation probed using chemically defined MHC class II oligomers. Immunity 12, 241–250 (2000). [DOI] [PubMed] [Google Scholar]
- 12.Giannoni F., et al. , Clustering of T cell ligands on artificial APC membranes influences T cell activation and protein kinase C theta translocation to the T cell plasma membrane. J. Immunol. 174, 3204–3211 (2005). [DOI] [PubMed] [Google Scholar]
- 13.Anderson H. A., Hiltbold E. M., Roche P. A., Concentration of MHC class II molecules in lipid rafts facilitates antigen presentation. Nat. Immunol. 1, 156–162 (2000). [DOI] [PubMed] [Google Scholar]
- 14.Gombos I., Detre C., Vámosi G., Matkó J., Rafting MHC-II domains in the APC (presynaptic) plasma membrane and the thresholds for T-cell activation and immunological synapse formation. Immunol. Lett. 92, 117–124 (2004). [DOI] [PubMed] [Google Scholar]
- 15.Cai H., et al. , Full control of ligand positioning reveals spatial thresholds for T cell receptor triggering. Nat. Nanotechnol. 13, 610–617 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Taylor M. J., Husain K., Gartner Z. J., Mayor S., Vale R. D., A DNA-based T cell receptor reveals a role for receptor clustering in ligand discrimination. Cell 169, 108–119.e20 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin J. J. Y., et al. , Mapping the stochastic sequence of individual ligand-receptor binding events to cellular activation: T cells act on the rare events. Sci. Signal. 12, eaat8715 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang J., et al. , A single peptide-major histocompatibility complex ligand triggers digital cytokine secretion in CD4(+) T cells. Immunity 39, 846–857 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Howarth M., et al. , A monovalent streptavidin with a single femtomolar biotin binding site. Nat. Methods 3, 267–273 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fairhead M., Krndija D., Lowe E. D., Howarth M., Plug-and-play pairing via defined divalent streptavidins. J. Mol. Biol. 426, 199–214 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jungmann R., et al. , Single-molecule kinetics and super-resolution microscopy by fluorescence imaging of transient binding on DNA origami. Nano Lett. 10, 4756–4761 (2010). [DOI] [PubMed] [Google Scholar]
- 22.Huppa J. B., et al. , TCR-peptide-MHC interactions in situ show accelerated kinetics and increased affinity. Nature 463, 963–967 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Langecker M., et al. , Synthetic lipid membrane channels formed by designed DNA nanostructures. Science 338, 932–936 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jungmann R., et al. , Quantitative super-resolution imaging with qPAINT. Nat. Methods 13, 439–442 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Trebak M., Kinet J. P., Calcium signalling in T cells. Nat. Rev. Immunol. 19, 154–169 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Casaletto J. B., McClatchey A. I., Spatial regulation of receptor tyrosine kinases in development and cancer. Nat. Rev. Cancer 12, 387–400 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kanchanawong P., et al. , Nanoscale architecture of integrin-based cell adhesions. Nature 468, 580–584 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shaw A., et al. , Spatial control of membrane receptor function using ligand nanocalipers. Nat. Methods 11, 841–846 (2014). [DOI] [PubMed] [Google Scholar]
- 29.Wamhoff E.-C., et al. , Programming structured DNA assemblies to probe biophysical processes. Annu. Rev. Biophys. 48, 395–419 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shaw A., et al. , Binding to nanopatterned antigens is dominated by the spatial tolerance of antibodies. Nat. Nanotechnol. 14, 184–190 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Palmer E., Naeher D., Affinity threshold for thymic selection through a T-cell receptor-co-receptor zipper. Nat. Rev. Immunol. 9, 207–213 (2009). [DOI] [PubMed] [Google Scholar]
- 32.Harding C. V., Unanue E. R., Quantitation of antigen-presenting cell MHC class II/peptide complexes necessary for T-cell stimulation. Nature 346, 574–576 (1990). [DOI] [PubMed] [Google Scholar]
- 33.Manz B. N., Jackson B. L., Petit R. S., Dustin M. L., Groves J., T-cell triggering thresholds are modulated by the number of antigen within individual T-cell receptor clusters. Proc. Natl. Acad. Sci. U.S.A. 108, 9089–9094 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Valitutti S., Müller S., Cella M., Padovan E., Lanzavecchia A., Serial triggering of many T-cell receptors by a few peptide-MHC complexes. Nature 375, 148–151 (1995). [DOI] [PubMed] [Google Scholar]
- 35.González P. A., et al. , T cell receptor binding kinetics required for T cell activation depend on the density of cognate ligand on the antigen-presenting cell. Proc. Natl. Acad. Sci. U.S.A. 102, 4824–4829 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pageon S. V., et al. , Functional role of T-cell receptor nanoclusters in signal initiation and antigen discrimination. Proc. Natl. Acad. Sci. U.S.A. 113, E5454–E5463 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ma Y., et al. , Clustering of the ζ-Chain can initiate T cell receptor signaling. Int. J. Mol. Sci. 21, 3498 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Preiner J., et al. , High-speed AFM images of thermal motion provide stiffness map of interfacial membrane protein moieties. Nano Lett. 15, 759–763 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Karner A., et al. , Tuning membrane protein mobility by confinement into nanodomains. Nat. Nanotechnol. 12, 260–266 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ando T., et al. , A high-speed atomic force microscope for studying biological macromolecules. Proc. Natl. Acad. Sci. U.S.A. 98, 12468–12472 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Axmann M., Schütz G. J., Huppa J. B., Measuring TCR-pMHC binding in situ using a FRET-based microscopy assay. J. Vis. Exp. 104, e53157 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moertelmaier M., Brameshuber M., Linimeier M., Schütz G. J., Stockinger H., Thinning out clusters while conserving stoichiometry of labeling. Appl. Phys. Lett. 87, 1–3 (2005). [Google Scholar]
- 43.Schmidt T., Schutz G. J., Gruber H. J., Schindler H., Local stoichiometries determined by counting individual molecules. Anal. Chem. 68, 4397–4401 (1996). [Google Scholar]
- 44.Wieser S., Moertelmaier M., Fuertbauer E., Stockinger H., Schütz G. J., (Un)confined diffusion of CD59 in the plasma membrane determined by high-resolution single molecule microscopy. Biophys. J. 92, 3719–3728 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schnitzbauer J., Strauss M. T., Schlichthaerle T., Schueder F., Jungmann R., Super-resolution microscopy with DNA-PAINT. Nat. Protoc. 12, 1198–1228 (2017). [DOI] [PubMed] [Google Scholar]
- 46.Endesfelder U., Malkusch S., Fricke F., Heilemann M., A simple method to estimate the average localization precision of a single-molecule localization microscopy experiment. Histochem. Cell Biol. 141, 629–638 (2014). [DOI] [PubMed] [Google Scholar]
- 47.Gao Y., Kilfoil M. L., Accurate detection and complete tracking of large populations of features in three dimensions. Opt. Express 17, 4685–4704 (2009). [DOI] [PubMed] [Google Scholar]
- 48.MacKinnon J. G., “Bootstrap hypothesis testing” in Handbook of Computational Econometrics, Belsley D. A., Kontoghiorghes E. J., Eds. (John Wiley & Sons, Ltd, 2009), pp. 183–213. [Google Scholar]
- 49.Wheeler M. W., Park R. M., Bailer A. J., Comparing median lethal concentration values using confidence interval overlap or ratio tests. Environ. Toxicol. Chem. 25, 1441–1444 (2006). [DOI] [PubMed] [Google Scholar]
- 50.Huppa J. B., Gleimer M., Sumen C., Davis M. M., Continuous T cell receptor signaling required for synapse maintenance and full effector potential. Nat. Immunol. 4, 749–755 (2003). [DOI] [PubMed] [Google Scholar]
- 51.Xie J., et al. , Photocrosslinkable pMHC monomers stain T cells specifically and cause ligand-bound TCRs to be ‘preferentially’ transported to the cSMAC. Nat. Immunol. 13, 674–680 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Platzer R., et al. , Unscrambling fluorophore blinking for comprehensive cluster detection via photoactivated localization microscopy. Nat. Commun. 11, 4993 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jungmann R., et al. , Multiplexed 3D cellular super-resolution imaging with DNA-PAINT and exchange-PAINT. Nat. Methods 11, 313–318 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and supporting information.