Significance
The same mutation can have different effects among individuals, and one reason for this is the presence of additional mutations, a relationship known as epistasis. Epistatic effects are difficult to predict. We report a rare example of positive epistasis between a silent mutation (that changes nucleotide but not the encoded amino acid) and missense mutation(s) (that change amino acid and cause cystic fibrosis disease). Individually, each mutation has a detrimental effect, but they are beneficial when present together. The underlying molecular effect is in their ability to markedly modulate the ribosomal speed on mRNA in a synergistic fashion. In a broader setting of human diseases, our results suggest that epistasis with silent mutation broadens the spectrum of disease outcome.
Keywords: synonymous SNP, translation, cotranslational folding, mutations, CFTR
Abstract
Epistasis refers to the dependence of a mutation on other mutation(s) and the genetic context in general. In the context of human disorders, epistasis complicates the spectrum of disease symptoms and has been proposed as a major contributor to variations in disease outcome. The nonadditive relationship between mutations and the lack of complete understanding of the underlying physiological effects limit our ability to predict phenotypic outcome. Here, we report positive epistasis between intragenic mutations in the cystic fibrosis transmembrane conductance regulator (CFTR)—the gene responsible for cystic fibrosis (CF) pathology. We identified a synonymous single-nucleotide polymorphism (sSNP) that is invariant for the CFTR amino acid sequence but inverts translation speed at the affected codon. This sSNP in cis exhibits positive epistatic effects on some CF disease–causing missense mutations. Individually, both mutations alter CFTR structure and function, yet when combined, they lead to enhanced protein expression and activity. The most robust effect was observed when the sSNP was present in combination with missense mutations that, along with the primary amino acid change, also alter the speed of translation at the affected codon. Functional studies revealed that synergistic alteration in ribosomal velocity is the underlying mechanism; alteration of translation speed likely increases the time window for establishing crucial domain–domain interactions that are otherwise perturbed by each individual mutation.
Across individuals, the effect of a mutation may vary because of additional genetic variations, known as epistasis—a concept introduced by Bateson to describe the masking effect of one genetic variant on another (1). Genetic interactions can alter the magnitude of the mutational effect (i.e., antagonistic or synergistic epistasis) or completely change the sign of the effect (positive or negative sign-changing epistasis). Recent developments in genetic high-throughput analyses and emergent deep mutational scan technologies have revealed high prevalence of both intragenic (i.e., interactions between pairs of mutations within an individual gene) or intergenic (i.e., pairs of mutations in different genes) epistasis (2). In the context of human diseases, epistasis complicates the spectrum of clinical manifestations and broadens heterogeneity of disease outcomes (3, 4), and it is likely to influence theratyping strategies (5, 6). The nonadditive relationship between mutations and different layers on which a mutation can exert an effect (e.g., transcription, translation, protein folding and stability, and cellular fitness) (2, 7) limit our ability to predict the outcome of combined inter- and/or intragenic mutations, even given the knowledge of the individual effects of those mutations.
Studies on intragenic epistasis have a rich history in the framework of protein structure–function relationships, where thermodynamic stability or protein–ligand interactions are used as key parameters to identify whether additional mutations alleviate or aggravate the defect of individual mutations (8, 9). However, these studies have considered only mutations that change the amino acid sequence (i.e., missense mutations), leaving out synonymous or silent single-nucleotide polymorphisms (sSNPs), that alter a nucleotide sequence or a codon but not the encoded amino acid. Growing evidence over the past decade suggests that sSNPs are pervasive and may alter protein structure and function (10–15). Moreover, genome-wide association studies link sSNPs with more than 50 human disorders, highlighting the wide-ranging impact of sSNPs as driver mutations, including individual response to therapeutic intervention (16–20). Yet, the association of sSNPs with epistasis and their relationship to other mutations remains elusive.
In this study, we addressed the epistatic effect of a common sSNP on various missense mutations with causal links to cystic fibrosis (CF)—a common inherited lethal disease, caused by abnormalities in the CF transmembrane conductance regulator (CFTR). Over 2,000 variants have been reported in the CFTR gene (https://cftr2.org/). The majority of them (more than 90%) are SNPs (i.e., change of one nucleotide), comprising 40% missense variants (i.e., amino acid change), 10% nonsense mutations (i.e., premature termination codon), and 15% frameshift defects (i.e., nucleotide insertions or deletions) (21, 22). Among these, 408 have been confirmed as CF disease-causing (http://www.genet.sickkids.on.ca/), while the remainder are of unclear clinical significance. Furthermore, effects of concomitantly occurring CF-causing and non–CF-causing variants at the same locus (i.e., so-called complex alleles), particularly with regard to sSNPs, remains a largely understudied area. CFTR is naturally rich in sSNPs (22, 23), and an analysis of the 1000 Genomes Project data (24) revealed that four sSNPs (c.1584G > A, c.2562T > G, c.2280G > A, and c.3870A > G) are the most prevalent. The allele variance is highest at position 2562 (48% with 2,400 G-alleles from a total of 5,008 alleles; Fig. 1A). In previous work, we showed that the c.2562T > G sSNP exchanges codon ACT (read by a high-abundance transfer RNA [tRNA]) to codon ACG (pairs to a low-abundance tRNA), and this codon velocity inversion is deleterious for CFTR cotranslational folding, stability, and ion transport (13). Here, we address the epistatic effects of the c.2562T > G sSNP on CF-causing missense variants, in which the amino acid change alone markedly destabilizes CFTR channel conformation and activity. Strikingly, for some CF-causing missense mutations, we observed a clear positive epistatic effect of the c.2562T > G sSNP. We performed functional analyses and provide molecular explanation for the synergistic effects of the sSNP on clinically relevant missense mutations. Our data offer evidence for sign-changing positive epistasis with potentiation of the effects for each mutation at the translational level.
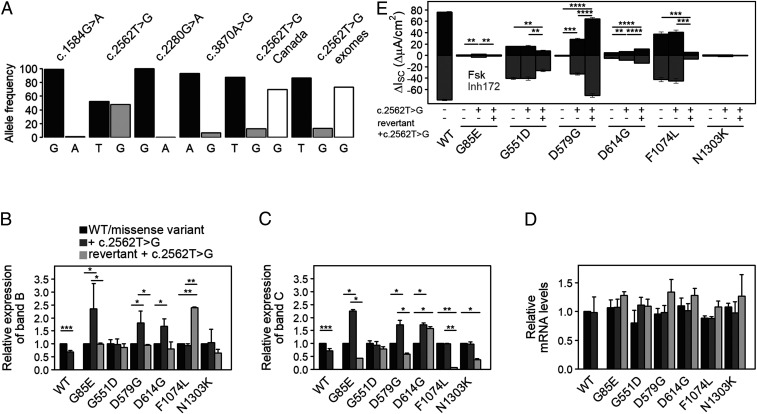
Fig. 1.
c.2562T > G is a common sSNP in the CFTR gene and augments functional expression of specific disease-causing variants. (A) Allele frequencies of CFTR sSNPs within the general population obtained from the 1000 Genomes Project (24), as well as c.2562T > G (T, black; G, gray) in CF patient cohorts from Canada and the US patients’ exomes (dbGaP, NCBI). Excluding p.delF508, the frequency of the G-allele (white bars) rises. (B and C) Quantification of the expression of CFTR band B (B) and band C (C) from immunoblots (SI Appendix, Fig. S1C) normalized to both NPT (encoded on the same plasmid and used as transfection control) and endogenous ACTB (serving as loading control). Expression levels of each variant with the missense mutation alone were set to 1. Data are mean ± SEM (n = 6 to 9). *P < 0.0046 and **P < 0.00856 (unequal variance t test on log2-transformed data with post hoc Bonferroni’s correction). (D) Steady-state CFTR mRNA levels were measured by qRT-PCR and normalized to the NPT transcript. Values obtained for WT-CFTR were set to 1. Data are mean ± SEM (n = 3 to 6). (E) Quantification of changes in short-circuit currents (ΔIsc) measured in FRT monolayers transiently transfected with CFTR variants. Transepithelial ion transport was induced with the CFTR agonist forskolin (Fsk), followed by arrest with CFTR inhibitor172 in the presence of a basolateral-to-apical chloride gradient. Revertant refers to a variant with engineered silent codon substitution at the missense codon. Data are mean ± SEM (n = 3 to 5). *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001; Student’s t test.
Results
The c.2562T > G sSNP Enhances Functional Expression of Certain Disease-Associated CFTR Missense Variants.
In the general population, c.2565T > G is the most common sSNP in CFTR (Fig. 1A), and some studies report its prevalence among patients with CFTR-related disorders (25–28). We evaluated the frequency of c.2565T > G in two North American cohorts, the Canadian CF gene modifier study (2,967 patients (29)), and the US CF patients’ exomes (database of Genotypes and Phenotypes [dbGaP], National Center for Biotechnology Information [NCBI]). The G-allele occurs in 12.6% of the Canadian CF cohort and 13.3% of the US CF exomes (Fig. 1A), predominantly on haplotypes different from those that carry the most common CFTR defect, p.delF508. The G-allele is depleted in individuals who are homozygous for p.delF508 (i.e., 0.3% in Canadian patients and 1.1% in US CF exomes), indicating that c.2562T > G is not typically associated in cis with the p.delF508-CFTR haplotype. Since in both CF patient cohorts individuals with p.delF508 dominate, restricting the analysis to patients who are either heterozygous or homozygous for CF-causing mutations other than p.delF508 (and assuming that the frequency of c.2562T > G in cis with p.delF508 is essentially zero), the G-allele displays higher frequency than in the general population (Fig. 1A, white bars). This group comprises a variety of CF-causing mutations (e.g., splicing defects, nonsense, and missense mutations), all of which are rare with a frequency below a few percent (for example, the p.G551D allele exhibits the highest frequency in the Canadian cohort of 1.75%).
To elucidate the effect of c.2562T > G in cis, we considered only missense SNPs; splicing and nonsense mutations were eliminated, as they can alter length and stability of the messenger RNA (mRNA). We chose six different missense mutations with varying CF clinical outcomes (p.G85E, p.G551D, p.D579G, p.D614G, p.F1074L, and p.N1303K), which are located upstream and downstream of the c.2562T > G sSNP in the primary CFTR sequence. We transiently transfected these six missense variants in immortalized CF bronchial epithelial (CFBE41o−) cells and monitored their effects on expression of the complex glycosylated CFTR (band C) and core glycosylated, endoplasmic reticulum (ER)-retained CFTR (band B). With the exception of p.G551D, all missense SNPs conferred severe defects on CFTR maturational efficiency as evidenced by decreased yields of both band C and band B (SI Appendix, Fig. S1 A and B).
We next introduced the c.2562T > G sSNP (p.T854T) in cis with these six missense mutations and examined effects on CFTR mRNA and protein expression. Following transient transfection in CFBE41o− cells, c.2562T > G alone reduced the amount of both mature and immature CFTR (band C and band B, respectively) (Fig. 1 B and C and SI Appendix, Fig. S1C). In contrast, the c.2562T > G sSNP significantly enhanced levels of bands C and B of G85E-, D579G-, and D614G-CFTR (i.e., “amplifier” effect), but did not alter CFTR expression levels when combined with p.G551D, p.F1074L, or p.N1303K (Fig. 1 B and C and SI Appendix, Fig. S1C). Notably, the c.2562T > G sSNP caused no significant changes in mRNA expression of any variant including wild-type (WT)-CFTR (Fig. 1D), suggesting that the observed effect is solely at the level of translation.
To address any cis-acting effects elicited by c.2562T > G on CFTR channel activity, we utilized short-circuit current measurements (Ussing chamber) to quantify transepithelial ion transport in Fischer rat thyroid (FRT) cells, which represent a standard model for polarizing epithelia expressing apical CFTR and are viewed by the US Food and Drug Administration as informative concerning drug label expansion for CFTR modulators (30). Our previous study on the effect of c.2562T > G alone showed that this sSNP causes subtle structural alterations in CFTR, constricting channel openings by approximately 30% and decreasing conductance by about half of that of WT-CFTR (13). The presence of c.2562T > G with p.G85E, p.D579G, or p.D614G significantly augmented mutant CFTR activity in FRT cells (Fig. 1E), which correlated with higher levels of complex glycosylated active CFTR (band C; Fig. 1C). The c.2562T > G sSNP only marginally influenced the ion transport phenotype of G551D-, N1303K-, or F1407L-CFTR (Fig. 1E). Notably, the three missense variants (p.G85E, p.D579G, or p.D614G), whose protein expression levels and function were augmented by c.2562T > G, were located upstream of the sSNP.
Missense Mutations, Along with the Amino Acid Substitution, Also Alter Codon Speed.
In a previous study, we observed that c.2562T > G (p.T854T) modifies translation velocity at the Thr codon (13); hence, we reasoned that the effect of this sSNP on the CF-causing missense mutation might also be connected with ribosome velocity. The primary effect of any missense mutation is the change in the encoded amino acid, which alone is deleterious for CFTR folding and channel function (31, 32). An additionally significant yet underappreciated area that contributes to translation dynamics is the effect of the codon speed alterations together with amino acid substitution. Thus, we asked whether the codon change that occurs with the missense mutation alters translation velocity at the affected codon.
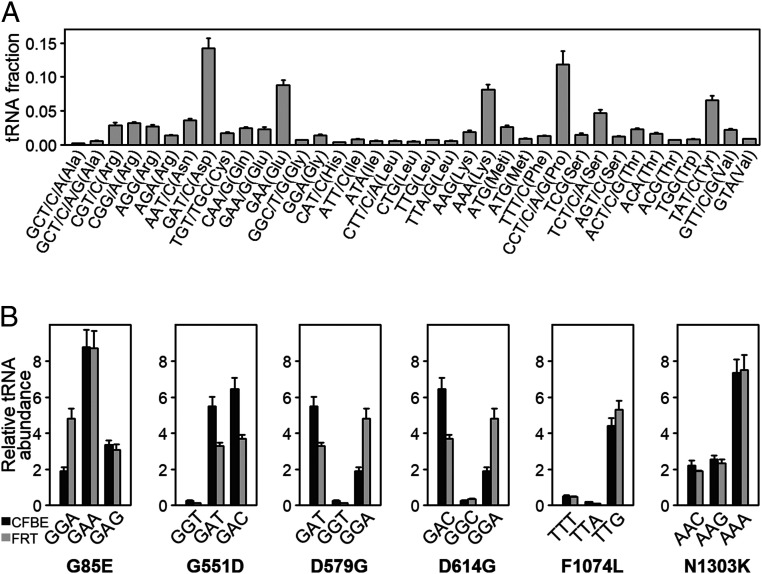
The concentration of the cognate tRNA is one major determinant of ribosome velocity at any given codon (33, 34). To elucidate the effect of the missense mutation at the codon level due to tRNA abundance, we used tRNA-tailored microarrays to determine the tRNA concentration in CFBE41o− (Fig. 2A). The missense variants p.G85E and p.G551D exchange GGA (Gly) to GAA (Glu) and GGT (Gly) to GAT (Asp), respectively (Table 1). Both tRNAs reading Glu (GAA) and Asp (GAT) codons are more abundant than the tRNAs pairing to Gly codons in WT-CFTR (GGA at G85 or GGT at G551, respectively). Consequently, the mutant codons are expected to be read faster compared to the codons in WT-CFTR (Table 1). In contrast, missense mutations p.D579G, p.D614G, and p.F1074L introduced a codon pairing to low-abundance tRNAs, and consequently, would be translated with slower velocity than the codon in WT-CFTR (Table 1). For p.D579G and p.D614G, the Gly-encoding codons GGT or GGC, respectively, are read by the same relatively rare tRNA isoacceptor (Fig. 2A). The p.N1303K is not expected to change codon speed, as abundance of the tRNAs reading the AAC (Asn) codon in WT-CFTR is nearly equal to that of the mutated AAG (Lys) codon (Table 1).
Fig. 2.
tRNA concentrations in CFBE41o− and FRT cells. (A) Absolute tRNA concentrations in CFBE41o− cells determined from tRNA microarrays. tRNAs are designated by cognate codon(s) and amino acid. Data are mean ± SEM (n = 4). (B) Comparisons of the abundance of tRNA isoacceptors pairing to native WT-CFTR sequence (first codon), missense mutation (second codon), and revertant (third codon) in CFBE41o− (black) and FRT (gray) cells. For example, for p.G85E, GGA is the WT codon, GAA is the missense codon, and GAG is the revertant codon. Data are mean ± SEM (n = 3 to 4). Abundance was determined using comparative tRNA microarrays to HeLa tRNAs, and isoacceptors’ concentrations are presented relative to levels found in HeLa cells.
Table 1.
Difference in tRNA abundance among CFTR missense variants and engineered synonymous revertants
| CFTR disease mutation | WT codon | Mutated codon | tRNA concentration* | Change in tRNA concentration |
| p.G85E | GGA (Gly) | GAA (Glu) | 1.89→8.77 | 4-fold increase |
| GAG (Glu) | 1.89→ 3.35 | 2-fold increase | ||
| p.G551D | GGT (Gly) | GAT (Asp) | 0.27 → 5.5 | 20-fold increase |
| GAC (Asp) | 0.27 → 6.45 | 24-fold increase | ||
| p.D579G | GAT (Asp) | GGT (Gly) | 5.5 → 0.27 | 20-fold decrease |
| GGA (Gly) | 5.5 → 1.89 | 2-fold decrease | ||
| p.D614G | GAC (Asp) | GGC (Gly) | 6.45 → 0.56 | 12-fold decrease |
| GGA (Gly) | 6.45 → 1.89 | 3-fold decrease | ||
| p.F1074L | TTT (Phe) | TTA (Leu) | 0.52 → 0.19 | 3-fold decrease |
| TTG (Leu) | 0.52 → 4.4 | 8-fold increase | ||
| p.N1303K | AAC (Asn) | AAG (Lys) | 2.22 → 2.54 | No change |
| AAA (Lys) | 2.22 → 7.34 | 3-fold increase |
Codons designating the disease mutation appear in lightface font; those of the revertant mutants appear in boldface font.
tRNA concentrations for specific codons in CFBE41o− cells are given here, whereas the concentrations for all isoacceptors are summarized in Fig. 2. The first number indicates the concentration of the tRNA that reads the WT codon (column 2), and the second value designates the concentration of the tRNA pairing to the mutated codon (column 3). The synonymous SNP at the missense codon in the revertants is designated in bold.
In order to elucidate whether the amplifier effect of c.2562T > G could be linked to changes in translation kinetics, we engineered synonymous substitutions at the missense codon that do not modify the amino acid identity, but instead alter the tRNA concentration or the speed at which the codon is read. For example, in the engineered p.D579G and p.D614G codons, the tRNA concentration changed only 2-fold as compared to 20-fold or 12-fold increases in the original missense mutations, respectively (Table 1). Hence, these engineered silent codons would exhibit velocities similar to the WT codons (D579 or D614, respectively). For p.G551D, p.F1074L, or p.N1303K, the engineered codons pair to tRNAs with higher abundance (Table 1), thus further increasing the speed differential compared to the original G551, F1074, or N1303 codons, respectively. These engineered silent codon substitutions are referred to as “revertant” mutants, and they all contain the c.2562T > G sSNP. Of note, options for constructing revertants were restricted by the limited number of synonymous codons possible for individual amino acids (E, D, G, L, and K, respectively; Table 1).
Strikingly, the “amplifier” effect of the c.2562T > G sSNP on G85E-, D579G-, and D614G-CFTR revertants was diminished as indicated by lower protein expression in CFBE41o− cells (Fig. 1 B and C). For the F1074L and N1303K revertants, we observed a decrease in mature CFTR (band C; Fig. 1C), which was accompanied by either no changes in the ER-residing glycoform (N130K revertant) or marked increase in band B expression (F1074L revertant) (Fig. 1B). For the G551D revertant, no effect was detected (Fig. 1 B and C). Importantly, none of the engineered synonymous codon substitutions altered mRNA steady-state levels of the revertants (Fig. 1D), suggesting that the observed effect is solely at the level of translation.
We next addressed whether the engineered revertants influence CFTR channel activity in the setting of c.2562T > G. In polarized FRT cells, the engineered synonymous codon substitutions either reduced or did not affect transepithelial ion transport exhibited by p.G85E, p.G551D, p.F1074L, or p.N1303K (Fig. 1E)—findings which generally agree with trends in band C levels observed for the same revertants expressed in CFBE41o− cells (Fig. 1C). In contrast, the D579G and D614G revertants exhibited a marked enhancement in short-circuit currents (Fig. 1E), which was not anticipated given the decreased band C expression levels detected in CFBE41o− (Fig. 1C). Since CFTR expression was measured in CFBE41o− cells and ion transport in FRT cells, we postulated that this could be explained by distinct tRNAGly and tRNAAsp concentrations in the two cell lines (i.e., those tRNAs pairing to the engineered codons (GGA) versus missense codons, GAT or GAC in p.D579G or p.D614G, respectively) (Table 1). Using tRNA-tailored microarrays, we detected strong differences in the concentrations of tRNAGly and tRNAAsp isoacceptors between CFBE41o− and FRT cells (Fig. 2B). In FRT cells, the concentration of tRNAsAsp (pairing to codons GAT or GAC) is higher than that of tRNAGly reading the revertant GGA codon, while in CFBE41o− cells, the concentration of tRNAGly is much lower than that of tRNAsAsp (Fig. 2B). Indeed, the protein expression levels of D579G and D614G revertants were augmented in FRT cells (SI Appendix, Fig. S2), indicating that within the same cell line, functional and biochemical data corroborate one another, and tRNA concentrations likely explain differences among cell lines. Notably, for p.G85E, p.F1074L, and p.N1303K, the concentrations of tRNAs reading the missense and revertant codons follow the same trend of decrease or increase in both cell lines (Fig. 2B), which explains the observed congruence in functional and biochemical results. Taken together, these findings indicate that the epistatic effect of the c.2562T > G sSNP depends on consequences of the missense mutation caused by the codon change. Since host tRNA concentrations modulate codon speed, the missense mutation may alter codon velocity when concentrations differ between cognate tRNAs decoding the new mutated codon versus the WT codon.
The c.2562T > G sSNP Causes Subtle Structural Rearrangements in the CFTR Variants.
In the context of WT-CFTR, c.2562T > G mildly alters domain–domain contacts and enhances protease resistance, but with no discernible changes in the cotranslational folding of the single domains (13). Thus, we next investigated whether presence of the c.2562T > G sSNP in cis with missense variants also influences mutant CFTR structure. Using limited proteolysis directly in permeabilized CFBE41o− cells (35), we found that c.2562T > G modestly but reproducibly increases protease resistance of D579G- and D614G-CFTR, but has no effect on N1303K-CFTR (Fig. 3). Notably, c.2562T > G did not alter gross proteolytic pattern of each single mutant (D579G- or D614G-CFTR). These results, together with observed similar susceptibilities of both complex (band C) and ER-retained (band B) forms, argue against c.2562T > G causing large structural rearrangements, and suggest that c.2562T > G induces more subtle, local conformational changes and exerts effects on the overall topology of CFTR, which is in accordance with the effect of c.2562T > G alone on WT-CFTR (13). Revertants showed similar (p.N1303K) or lower (p.D579G and p.D614G) proteolytic susceptibility compared to their missense mutant counterparts, but strikingly, overall proteolytic patterns changed (Fig. 3), which is indicative of new protease-resistant fragments and/or conformations. These results support the notion that the c.2562T > G sSNP stabilizes the structure of the D579G- and D614G-CFTR in synergy with the p.D579G or p.D614G missense mutations.
Fig. 3.
The c.2562T > G sSNP influences proteolytic stability of some CFTR variants. Representative immunoblots of limited trypsin digestion of CFTR mutants expressed in permeabilized CFBE41o− cells and probed with an anti-CFTR NBD2 (#596) antibody (Left). Band B and band C designate immature and mature CFTR glycoforms, respectively. NBD2 denotes the nucleotide-binding domain 2–containing fragments. Quantification of bands B and C of CFTR relative to untreated samples (set to 1) (Right). Data are mean ± SD (n = 2 to 4).
Effect of the c.2562T > G sSNP Is Mediated through Changes in Ribosomal Velocity.
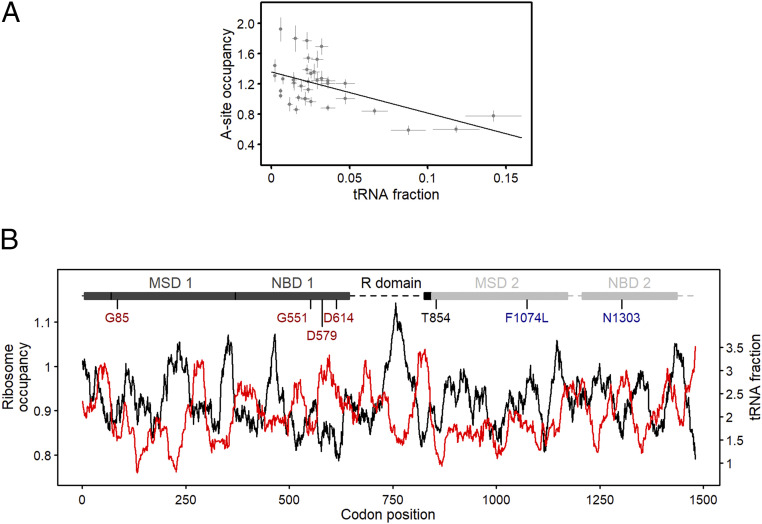
For several CFTR missense variants, tRNA abundance links the c.2562T > G “amplifier” effect on CFTR functional enhancement to speed alterations at the missense codon (i.e., when the concentration differs among tRNAs decoding the new mutated codon compared to the WT codon) (Fig. 2 and Table 1), suggesting that underlying mechanism(s) of the epistasis occur through effects on translation. We therefore employed ribosome profiling (Ribo-Seq) in CFBE41o− cells to determine ribosomal dwelling occupancy at A-site codons (the site that accepts the aminoacyl-tRNA), which correlates with the translational speed of any given codon (36). Thereby, slowly translated codons exhibit larger ribosomal occupancy than fast translated codons. To address whether ribosome occupancy at the A-site codon correlates with the concentration of the cognate tRNA, we calibrated ribosome-protected fragments on the A-site using the 5′ ends of the sequencing reads across all mRNAs (see Materials and Methods) and compared the A-site occupancy to the abundance of each tRNA in CFBE41o− cells determined from the tRNA arrays (Fig. 4A). Codons with high ribosomal A-site codon occupancy are decoded by low-abundance tRNAs, and, vice versa, codons pairing to high-abundance tRNAs exhibited low A-site codon occupancy (Fig. 4A). The very good correlation supports the notion that tRNA concentration is a good predictor of the codon speed.
Fig. 4.
CFTR translation is nonuniform, and mutations alter translation speed along with amino acid exchange. (A) Ribosomal A-site codon occupancy from Ribo-Seq correlates with tRNA abundance determined by tRNA microarrays (Fig. 2A) in CFBE 41o− cells (R = 0.52; Pearson correlation coefficient). Data are mean ± SEM (n = 3 for Ribo-Seq and n = 4 for the tRNA microarrays). (B) Ribosomal A-site dwelling occupancy (black, right axis) and tRNA concentrations (red, right axis) as a function of CFTR mRNA nucleotide position. Both curves were smoothed with a window of 30 codons and are inversely correlated (P < 2.22 × 10−16; two-sample Kolmogorov-Smirnov test). Independent of the smoothing window size, both curves strongly inversely correlate (SI Appendix, Fig. S3). A schematic of CFTR domains is shown at the top of the sequence. MSD, membrane-spanning domain; NBD, nucleotide-binding domain; R, regulatory. The largest part of the R domain is unstructured (dashed line). Missense mutations upstream (red) and downstream (blue) of the c.2562T > G (Thr854, black) are color coded.
We next graphed the tRNA concentration versus A-site occupancy for each codon along the length of CFTR mRNA (Fig. 4B and SI Appendix, Fig. S3). mRNA regions with codons pairing to low-abundance tRNAs appeared as valleys in the tRNA profiles and inversely correlated with regions of high ribosome occupancy as evidenced by peaks indicating low translation (Fig. 4B). By contrast, valleys in the ribosome profiling spectrum (i.e., fast translating regions) inversely correlated with peaks in the tRNA abundance profiles, or regions of codons pairing to high-abundance tRNAs (Fig. 4B).
The overall translation profile of CFTR is nonuniform. Interestingly, both the D579 and D614 residues localize to a region that is rapidly translated (i.e., low ribosome occupancy) (Fig. 4B), and the tRNAsAsp pairing to both WT codons (GAT and GAC, respectively) are highly abundant (Fig. 2). However, the tRNAGly reading the missense codons of p.D579G and p.D614G (GGT or GGC, respectively) is among the rarest tRNAs in CFBE41o− cells (Fig. 2A), which would markedly reduce the translational velocity at these positions. Moreover, reversal of codon speed in the engineered D579G and D614G revertants would likely change the average velocity of the entire region (Fig. 4B) and consequently alter cotranslational CFTR folding, which explains changes in proteolytic susceptibility of the revertants (Fig. 3). The G85, G551, and N1303 residues are located in regions of average ribosomal occupancy, which indicates moderate translation rates at these codons (Fig. 4B). For p.G85E, the mutated codon is read by a tRNA with fourfold higher abundance (Table 1), thereby modestly elevating ribosomal velocity. The mutated codon for p.G551D (GAT) pairs to a 20-fold higher abundance tRNA (Table 1), which would increase translational speed at this position. The p.N1303K variant is not anticipated to significantly alter translational speed, since the native codon (AAC) and mutated codon (AAG) pair to tRNAs with relatively the same abundance (Table 1). Conversely, the F1074 residue localizes to a region of moderately high ribosome occupancy (Fig. 4B). The mutated codon for p.F1074L (TTA) is read by a tRNA with tree-fold lower abundance (Table 1), and thus, would further decrease ribosomal velocity (Fig. 4B).
Together, our results suggest that c.2562T > G exerts robust positive effects and partially rescues the processing and function of CFTR variants with missense mutation located 5′ upstream of the sSNP through a mechanism dependent on translation speed. In addition to effects caused by the primary amino acid substitution, changes in velocity at the missense codon appear to play a crucial role (i.e., greater alterations in codon speed correlate with larger rescuing effects of the c.2562T > G sSNP).
Discussion
Our results demonstrate that an intragenic sSNP, which alone is deleterious, exerts a large positive epistatic effect on disease-causing missense mutations and alleviates CFTR folding and aberrant ion transport activity. The largest effect we observed was in combination with missense mutations that, along with the amino acid substitution, affected translation rate at the mutated codon (e.g., p.D579G and p.D614G). The synergistic alteration in ribosomal velocity by the sSNP, together with the missense mutation, likely increases the time window for cotranslational folding and facilitates establishment of crucial interactions that are otherwise perturbed by each individual mutation.
Overall, the positive effect of c.2562T > G is exerted on missense variants that localize 5′ upstream of the sSNP. The larger the effect of a codon velocity change through the missense substitution (usually an order of magnitude or higher), the higher the rescuing effect of the c.2562T > G sSNP on CFTR activity and expression. Larger velocity changes at a single codon are more likely to influence the ribosomal speed of larger mRNA regions (37). The c.2562T > G sSNP decreases the speed of translation at the Thr854 codon (13), which, combined with the translational effect of the missense mutation, likely mimics the effects of global translational perturbations. Along that line, low growth temperature (37), mild inhibition of translation by emetine (38), or mutations of specific ribosomal proteins (39), which globally reduce elongation speed, facilitate the folding and function of mutant proteins.
CFTR folding is cotranslational (35) and orchestrated by the translation kinetics (40). Mutation-based local alterations of programmed ribosome velocity or changes in the components of translational machinery that reduce the elongation speed impact CFTR biogenesis and function (13, 39, 40). The c.2562T > G sSNP resides in a segment near the predicted edge of the R-domain at an interface between the N-terminal “lasso” motif and membrane spanning domain-2 (MSD2), which modulates interdomain interactions and stabilizes the final three-dimensional topology of CFTR (41). Thr854 is therefore located at a strategic point crucial for establishing domain–domain interactions. The p.D579G and p.D614G defects severely compromise such interdomain contacts and overall CFTR assembly (42). In the context of these missense variants, c.2562T > G–induced reduction in translational velocity may provide an additional time window for lasso–R-domain–MSD2 interfaces to properly align. Support for this hypothesis is found in the proteolytic patterns of D579G- and D614G-CFTR in combination with c.2562T > G, showing slightly enhanced proteolytic resistance but WT-like signatures (Fig. 3).
In summary, our findings describe a rare occurrence of positive epistasis with a participation of sSNP, by which the detrimental effects of the individual sSNP and missense mutation switch sign when both mutations are present together. Moreover, to our knowledge, this is the first study that provides evidence that missense mutations, in addition to effects caused by the primary amino acid substitution, also modulate translation velocity through the altered codon. Clearly, the amino acid change dominates the phenotype of any particular missense mutation; however, additional effects on ribosome velocity through the codon substitution are the underlying mechanisms of the positive epistatic interactions with the nearby sSNP. Since changes in codon speed correlate with the cognate tRNA abundance, information on the cellular tRNA concentrations could be used to predict combined translational effect(s) of synonymous and nonsynonymous variants. Predicting epistatic effects among mutations is likely to reveal molecular signatures that contribute to disease heterogeneity, mechanisms underlying genotype–phenotype relationships, and the likelihood of individual response to precision medicine therapies.
Materials and Methods
Cell Culture and Transfection.
Parental CFBE41o− cells (a kind gift from Karl Kunzelmann, University of Regensburg, Regensburg, Germany; Dieter Gruenert, University of California, San Francisco, CA) were maintained in Earle’s minimal essential medium (Biochrom), supplemented with 2 mM l-glutamine (Gibco) and 10% fetal calf serum (PAN-Biotech). Parental FRT cells were a kind gift from Michael Welsh, University of Iowa, Iowa City, IA, and cultured in F12 Ham Coon’s modified nutrient mixture (Sigma, F6636) supplemented with 2.68 g sodium bicarbonate, 850 μL 2N hydrochloric acid (pH 7.3), and 5% fetal bovine serum as described previously (39, 43).
WT or missense variant CFTRs cDNAs—alone or in combination with the c.2562T > G sSNP—were cloned into the pcDNA3 vector (Invitrogen) and transiently transfected using either polyethylenimine (linear, molecular weight 40,000 Da, Polysciences) for CFBE41o− cells or Lipofectamine 3000 (Invitrogen, L3000015) for FRT.
Antibodies and Immunoblotting.
The CFTR antibody employed here was mouse monoclonal anti-NBD2 (596; dilution 1:2,000), which was purchased from John R. Riordan and Tim Jensen, University of North Carolina, Chapel Hill, NC, through the Cystic Fibrosis Foundation Therapeutics Antibody Distribution Program (Bethesda, MD). Additional antibodies utilized included mouse anti–β-actin (anti-ACTB; dilution 1:4,000; no. A228, Sigma-Aldrich), anti-tubulin mouse monoclonal (dilution 1:10,000; no. 302211, Synaptic Systems), rabbit anti–neomycin phosphotransferase (NPT) (dilution 1:1,000; no. H06-747, Millipore), goat anti–mouse-horseradish peroxidase (HRP) (for CFBE41o−: dilution 1:10,000, no. 170-5047, BioRad; for FRT: dilution 1:10,000, no. P0447, Agilent); and goat anti–rabbit-HRP (for CFBE41o−: dilution 1:3,000, no. 170-5046, BioRad; for FRT: dilution 1:10,000, no. 31460, Thermo Fisher).
Western blots were performed as described previously (13). Densities of CFTR bands B and C were normalized to 1) NPT, which is expressed within the pcDNA3 backbone and therefore provides a measure of transfection efficiency, and 2) β-actin or α-tubulin, which serve as sample loading controls.
Partial Permeabilization of the Cells and Limited Proteolysis.
Permeabilized CFBE41o− cells were prepared as previously described (35). Briefly, cells were harvested by scraping after twice washing with ice-cold 1× phosphate-buffered saline (PBS; pH 7.4). Cells were centrifuged at 225 × g for 2 min at 4 °C and resuspended in 6 mL ice-cold buffer A (110 mM KOAc, 20 mM Hepes-KOH pH 7.2, 2 mM MgOAc) containing 2 µL/mL digitonin (40 mg/mL stock dissolved in dimethyl sulfoxide, Merck). Following exactly 5 min incubation on ice, permeabilization was stopped by adding 8 mL ice-cold buffer A. Permeabilized cells were recovered by centrifugation at 225 × g for 2 min at 4 °C, resuspended in 200 µL buffer A, and then immediately used or stored at −80 °C. The stability of individual CFTR domains was probed by limited proteolysis. Equal amounts of permeabilized cells were incubated 15 min with trypsin (tosyl phenylalanyl chloromethyl ketone-treated, dissolved in 1 mM HCl to 10 mg/mL stock; Sigma-Aldrich) in concentrations ranging from 38 to 0.3 µg/mL. Proteolysis was arrested by adding 2.5× complete protease inhibitor, 12.5 U benzonase nuclease (Novagen), and 8 µL 5× SDS loading buffer. CFTR-derived fragments were detected by immunoblotting using anti-NBD2 antibody (#596).
Short-Circuit Current Measurements.
CFTR-dependent ion transport studies were performed at 37°C as previously described (39). Briefly, FRT cells transiently transfected with CFTR variants were grown 4 d on permeable supports at air–liquid interface and mounted in P2300 Ussing chambers (MC8 apparatus, Physiologic Instruments). Cells were equilibrated 5 min in a basal-to-apical chloride gradient (i.e., basal regular ringer and apical low chloride ringer). Following sodium channel inactivation with amiloride (100 µM) and 5-min stabilization, the CFTR agonist forskolin (Fsk, 10 μM; activator of protein kinase A) was added and tracings stabilized for at least 5 min. At the end of experiment, CFTR activity was ablated by the CFTR Inhibitor172, (Inh172, 10 μM). Changes in CFTR-mediated transepithelial current (∆Isc) were calculated from the highest level of stable plateaus achieved.
RNA Extraction, Quantitative RT-PCR, and tRNA Microarrays.
Steady-state mRNA levels were measured by qRT-PCR. Total RNA was isolated using TriReagent (Sigma-Aldrich) and then DNase I (Thermo Fisher) treated and reverse transcribed using oligo-(dT)18 primers and Revert Aid H Minus Reverse Transcriptase (Thermo Fisher). PCR was performed in clear 96-well plates (Sarstedt) sealed with adhesive tape (Sarstedt) in a T Professional thermocycler (Biometra) employing 2× SensiFAST SYBR Hi-ROX Mix (BioLine). CFTR and the NPT references were amplified with the following primer pairs (5′→3′): CFTR forward, CCTATGTCAACCCTCAACACG, and reverse, ACTATCACTGGCACTGTTGC; and NPT forward, TGCTCCTGCCGAGAAAGTAT, and reverse, GCTCTTCGTCCAGATCATCC. The final concentration of primers was 300 nM, and relative levels of product were calculated using the ΔΔCT method. Nontemplate and nonreverse transcribed samples served as controls. qRT-PCR assays displayed a linear range over five (R2 = 0.99969, efficiency = 91%) or four (R2 = 0.998; efficiency = 93%) orders of magnitude for CFTR or NPT, respectively.
tRNA microarrays were performed as previously described (13, 44). Fluorescent stem-loop RNA/DNA oligonucleotide bearing a Cy3 fluorescent dye (Microsynth) was ligated to total tRNA from CFBE41o− or FRT cells and compared to HeLa tRNA labeled with Atto647 stem-loop RNA/DNA oligonucleotide. Quantification was performed by normalizing the median of the Cy3-tRNA signal for each tRNA isoacceptor from CFBE41o− or FRT to the corresponding Atto647-labeled HeLa tRNA signal. The full set of tRNA isoacceptors for FRT cells is published in ref. 44. The tRNAs abundance is given relative to the corresponding HeLa isoacceptor (Fig. 2B) (i.e., ratio >1 means tRNA in CFBE41o− or FRT is higher than the HeLa isoacceptor, and vice versa). The relative signal for each tRNA isoacceptor comparing CFBE41o− to HeLa was converted into absolute value (Fig. 2A) using the absolute values of HeLa tRNA set as quantified in ref. 13.
Ribo-Seq and Data Analysis.
Ribo-Seq data sets from three independent biological replicates were generated in ref. 13. The triplicates correlate very well with Pearson correlation coefficients of 0.94, 0.90, and 0.92 for replicate 1 versus 2, for replicate 2 versus 3, and for replicate 1 versus 3, respectively.
These data were used to calculate the genome-wide codon occupancy (13, 45). The majority of the sequencing reads (ribosome-protected fragments, RPFs) ranged from 27 to 29 nucleotides in length. To precisely determine the position of A- and P-site codons in each RPF, we followed the procedure described in refs. 46 and 47 using the calibration tool (https://github.com/AlexanderBartholomaeus/MiMB_ribosome_profiling). Briefly, RPFs were binned into groups of equal read length, and each group was aligned to start codons. The RNase I used to generate RPFs generates fragments with precise 5′ ends that enable reliable alignment using 5′ ends (47). Ribosomes located at the start codon accommodate AUG at the P-site, which causes a characteristic drop in read density upstream of the start codon. For each read length, we used this feature to define the offset between the 5′-RPF end and the codon at the P-site. The A-site codon coverage was determined by an additional offset of three nucleotides. The A- and P-site codons are located approximately in the center of each read as opposed to the 5′- and 3′-RPF ends, which are bias-free and on a metagenome scale, report on genuine codon elongation events (48, 49). Single-transcript profiles (Fig. 4B and SI Appendix, Fig. S3) were smoothed, which additionally removes some intrinsic bias in the RPF library generation. Biases in the 5′- and 3′-RPF ends originate from the experimental protocol in the Ribo-Seq datasets (e.g., nuclease cleavage, adapter ligation) (36, 50). The A-site occupancy in these triplicates correlate better than the RPFs, with Pearson correlation coefficients of 0.97, 0.94, and 0.94 for replicate 1 versus 2, for replicate 2 versus 3, and for replicate 1 versus 3, respectively.
Genome Data Analysis.
US CF patient exome data were downloaded from the dbGaP (NCBI) under accession numbers phs000254.v2.p1.c1 and phs000254.v2.p1.c2.
Supplementary Material
Acknowledgments
This work was supported by Deutsche Forschungsgemeinschaft (FOR1805 to Z.I.), muko e.V. German Cystic Fibrosis Foundation (project 1603 to Z.I.), NIH (R01HL136414 to E.J.S. and Z.I.; K99HL151965 to K.E.O.), US Cystic Fibrosis Foundation (IGNATO17XXO to Z.I.; SORSCH13XXO and SORSCH14XXO to E.J.S.; and OLIVER17F0 to K.E.O.), and Burroughs Wellcome Fund (Collaborative Research Travel Grant to K.E.O.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2010612118/-/DCSupplemental.
Data Availability.
Ribosome profiling and tRNA microarray data have been deposited within Gene Expression Omnibus under accession numbers GSE74365 and GSE53991, respectively. All other study data are included in the article and/or supporting information.
References
- 1.Bateson, Discussion on the influence of heredity on disease, with special reference to tuberculosis, cancer, and diseases of the nervous system: Introductory address. Proc. R. Soc. Med. 2, 22–30 (1909). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Domingo J., Baeza-Centurion P., Lehner B., The causes and consequences of genetic interactions (epistasis). Annu. Rev. Genomics Hum. Genet. 20, 433–460 (2019). [DOI] [PubMed] [Google Scholar]
- 3.Fu W., O’Connor T. D., Akey J. M., Genetic architecture of quantitative traits and complex diseases. Curr. Opin. Genet. Dev. 23, 678–683 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang X., Fu A. Q., McNerney M. E., White K. P., Widespread genetic epistasis among cancer genes. Nat. Commun. 5, 4828 (2014). [DOI] [PubMed] [Google Scholar]
- 5.Joshi D., Ehrhardt A., Hong J. S., Sorscher E. J., Cystic fibrosis precision therapeutics: Emerging considerations. Pediatr. Pulmonol. 54 (suppl. 3), S13–S17 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Touat M., et al. , Mechanisms and therapeutic implications of hypermutation in gliomas. Nature 580, 517–523 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Poelwijk F. J., Krishna V., Ranganathan R., The context-dependence of mutations: A linkage of formalisms. PLoS Comput. Biol. 12, e1004771 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bank C., Hietpas R. T., Jensen J. D., Bolon D. N., A systematic survey of an intragenic epistatic landscape. Mol. Biol. Evol. 32, 229–238 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Olson C. A., Wu N. C., Sun R., A comprehensive biophysical description of pairwise epistasis throughout an entire protein domain. Curr. Biol. 24, 2643–2651 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bartoszewski R. A., et al. , A synonymous single nucleotide polymorphism in DeltaF508 CFTR alters the secondary structure of the mRNA and the expression of the mutant protein. J. Biol. Chem. 285, 28741–28748 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Canale A. S., et al. , Synonymous mutations at the beginning of the Influenza A virus hemagglutinin gene impact experimental fitness. J. Mol. Biol. 430, 1098–1115 (2018). [DOI] [PubMed] [Google Scholar]
- 12.Kimchi-Sarfaty C., et al. , A “silent” polymorphism in the MDR1 gene changes substrate specificity. Science 315, 525–528 (2007). [DOI] [PubMed] [Google Scholar]
- 13.Kirchner S., et al. , Alteration of protein function by a silent polymorphism linked to tRNA abundance. PLoS Biol. 15, e2000779 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu Y., et al. , Non-optimal codon usage is a mechanism to achieve circadian clock conditionality. Nature 495, 116–120 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou M., et al. , Non-optimal codon usage affects expression, structure and function of clock protein FRQ. Nature 495, 111–115 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bin Y., et al. , Development of an immunochromatographic strip test for rapid detection of citrus yellow vein clearing virus. Arch. Virol. 163, 349–357 (2018). [DOI] [PubMed] [Google Scholar]
- 17.Chu D., Wei L., Nonsynonymous, synonymous and nonsense mutations in human cancer-related genes undergo stronger purifying selections than expectation. BMC Cancer 19, 359 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gartner J. J. et al.; NISC Comparative Sequencing Program , Whole-genome sequencing identifies a recurrent functional synonymous mutation in melanoma. Proc. Natl. Acad. Sci. U.S.A. 110, 13481–13486 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schutz F. A., et al. , Single nucleotide polymorphisms and risk of recurrence of renal-cell carcinoma: A cohort study. Lancet Oncol. 14, 81–87 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Supek F., Miñana B., Valcárcel J., Gabaldón T., Lehner B., Synonymous mutations frequently act as driver mutations in human cancers. Cell 156, 1324–1335 (2014). [DOI] [PubMed] [Google Scholar]
- 21.Ferec C., Cutting G. R., Assessing the disease-liability of mutations in CFTR. Cold Spring Harb. Perspect. Med. 2, a009480 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tsui L. C., Dorfman R., The cystic fibrosis gene: A molecular genetic perspective. Cold Spring Harb. Perspect. Med. 3, a009472 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hill A. E., et al. , Longevity and plasticity of CFTR provide an argument for noncanonical SNP organization in hominid DNA. PLoS One 9, e109186 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Auton A. et al.; 1000 Genomes Project Consortium , A global reference for human genetic variation. Nature 526, 68–74 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bombieri C., et al. , Recommendations for the classification of diseases as CFTR-related disorders. J. Cyst. Fibros. 10 (suppl. 2), S86–S102 (2011). [DOI] [PubMed] [Google Scholar]
- 26.de Cid R., et al. , Independent contribution of common CFTR variants to chronic pancreatitis. Pancreas 39, 209–215 (2010). [DOI] [PubMed] [Google Scholar]
- 27.Steiner B., Truninger K., Sanz J., Schaller A., Gallati S., The role of common single-nucleotide polymorphisms on exon 9 and exon 12 skipping in nonmutated CFTR alleles. Hum. Mutat. 24, 120–129 (2004). [DOI] [PubMed] [Google Scholar]
- 28.Tzetis M., et al. , CFTR gene mutations–Including three novel nucleotide substitutions–And haplotype background in patients with asthma, disseminated bronchiectasis and chronic obstructive pulmonary disease. Hum. Genet. 108, 216–221 (2001). [DOI] [PubMed] [Google Scholar]
- 29.Li W., et al. , Unraveling the complex genetic model for cystic fibrosis: Pleiotropic effects of modifier genes on early cystic fibrosis-related morbidities. Hum. Genet. 133, 151–161 (2014). [DOI] [PubMed] [Google Scholar]
- 30.Durmowicz A. G., Lim R., Rogers H., Rosebraugh C. J., Chowdhury B. A., The U.S. food and drug administration’s experience with Ivacaftor in cystic fibrosis. Establishing efficacy using in vitro data in lieu of a clinical trial. Ann. Am. Thorac. Soc. 15, 1–2 (2018). [DOI] [PubMed] [Google Scholar]
- 31.Bose S. J., et al. , Towards next generation therapies for cystic fibrosis: Folding, function and pharmacology of CFTR. J. Cyst. Fibros. 19 (suppl. 1), S25–S32 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Manfredi C., Tindall J. M., Hong J. S., Sorscher E. J., Making precision medicine personal for cystic fibrosis. Science 365, 220–221 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kirchner S., Ignatova Z., Emerging roles of tRNA in adaptive translation, signalling dynamics and disease. Nat. Rev. Genet. 16, 98–112 (2015). [DOI] [PubMed] [Google Scholar]
- 34.Zhang G., Ignatova Z., Folding at the birth of the nascent chain: Coordinating translation with co-translational folding. Curr. Opin. Struct. Biol. 21, 25–31 (2011). [DOI] [PubMed] [Google Scholar]
- 35.Kleizen B., van Vlijmen T., de Jonge H. R., Braakman I., Folding of CFTR is predominantly cotranslational. Mol. Cell 20, 277–287 (2005). [DOI] [PubMed] [Google Scholar]
- 36.Ingolia N. T., Ribosome profiling: New views of translation, from single codons to genome scale. Nat. Rev. Genet. 15, 205–213 (2014). [DOI] [PubMed] [Google Scholar]
- 37.Zhang G., Hubalewska M., Ignatova Z., Transient ribosomal attenuation coordinates protein synthesis and co-translational folding. Nat. Struct. Mol. Biol. 16, 274–280 (2009). [DOI] [PubMed] [Google Scholar]
- 38.Meriin A. B., et al. , A novel approach to recovery of function of mutant proteins by slowing down translation. J. Biol. Chem. 287, 34264–34272 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oliver K. E., et al. , Slowing ribosome velocity restores folding and function of mutant CFTR. J. Clin. Invest. 129, 5236–5253 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim S. J., et al. , Protein folding. Translational tuning optimizes nascent protein folding in cells. Science 348, 444–448 (2015). [DOI] [PubMed] [Google Scholar]
- 41.Zhang Z., Chen J., Atomic structure of the cystic fibrosis transmembrane conductance regulator. Cell 167, 1586–1597.e9 (2016). [DOI] [PubMed] [Google Scholar]
- 42.Thibodeau P. H., et al. , The cystic fibrosis-causing mutation deltaF508 affects multiple steps in cystic fibrosis transmembrane conductance regulator biogenesis. J. Biol. Chem. 285, 35825–35835 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sabusap C. M., et al. , Analysis of cystic fibrosis-associated P67L CFTR illustrates barriers to personalized therapeutics for orphan diseases. JCI Insight 1, e86581 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Polte C., et al. , Assessing cell-specific effects of genetic variations using tRNA microarrays. BMC Genomics 20 (suppl. 8), 549 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lareau L. F., Hite D. H., Hogan G. J., Brown P. O., Distinct stages of the translation elongation cycle revealed by sequencing ribosome-protected mRNA fragments. elife 3, e01257 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bartholomaus A., Ignatova Z., “Codon resolution analysis of ribosome profiling data” in Ribosome Profiling: Methods and Protocols, Labunskyy V. M., Ed. (Methods in Molecular Biology, Springer, 2021) in press. [DOI] [PubMed] [Google Scholar]
- 47.Mohammad F., Woolstenhulme C. J., Green R., Buskirk A. R., Clarifying the translational pausing landscape in bacteria by ribosome profiling. Cell Rep. 14, 686–694 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.O’Connor P. B., Andreev D. E., Baranov P. V., Comparative survey of the relative impact of mRNA features on local ribosome profiling read density. Nat. Commun. 7, 12915 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tunney R., et al. , Accurate design of translational output by a neural network model of ribosome distribution. Nat. Struct. Mol. Biol. 25, 577–582 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bartholomäus A., Del Campo C., Ignatova Z., Mapping the non-standardized biases of ribosome profiling. Biol. Chem. 397, 23–35 (2016). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Ribosome profiling and tRNA microarray data have been deposited within Gene Expression Omnibus under accession numbers GSE74365 and GSE53991, respectively. All other study data are included in the article and/or supporting information.