Significance
Over the past two decades, natural protein-based hydrogel biomaterials have catapulted cell biology into the third dimension, becoming the modern workhorses of 3D cell and organoid culture. Although these systems mimic many important aspects of native tissue while permitting matrix remodeling and biological integration, spatiotemporal control over cell fate within these materials has not been demonstrated. Here, we introduce a generalizable strategy to covalently decorate cell-laden natural hydrogels with bioactive proteins including growth factors and then use these methods to anisotropically govern cell fates in ways currently unobtainable in synthetic biomaterials. Enabling 4D biochemical tunability within natural protein-based gels, these approaches are likely to find great utility in probing and directing biological functions and in engineering heterogenous functional tissues.
Keywords: biomaterials, photochemistry, hydrogel, protein, 4D biology
Abstract
Hydrogel biomaterials derived from natural biopolymers (e.g., fibrin, collagen, decellularized extracellular matrix) are regularly utilized in three-dimensional (3D) cell culture and tissue engineering. In contrast to those based on synthetic polymers, natural materials permit enhanced cytocompatibility, matrix remodeling, and biological integration. Despite these advantages, natural protein-based gels have lagged behind synthetic alternatives in their tunability; methods to selectively modulate the biochemical properties of these networks in a user-defined and heterogeneous fashion that can drive encapsulated cell function have not yet been established. Here, we report a generalizable strategy utilizing a photomediated oxime ligation to covalently decorate naturally derived hydrogels with bioactive proteins including growth factors. This bioorthogonal photofunctionalization is readily amenable to mask-based and laser-scanning lithographic patterning, enabling full four-dimensional (4D) control over protein immobilization within virtually any natural protein-based biomaterial. Such versatility affords exciting opportunities to probe and direct advanced cell fates inaccessible using purely synthetic approaches in response to anisotropic environmental signaling.
Over the past several decades, hydrogels have found widespread utility as biomaterial constructs for three-dimensional (3D) cell culture and tissue engineering (1–5). Fulfilling a role similar to the extracellular matrix (ECM) in native tissue, gel biomaterials can provide encapsulated cells with mechanical support in a defined geometry, anchors for adhesion and migration, and bioactive chemical signals to guide complex cellular functions (e.g., proliferation, differentiation) (6–8). Classically, such materials are categorized as “synthetic” or “natural” based on the origin of their underlying components (9). Synthetic polymer-based gels, including those composed of poly(ethylene glycol) (PEG) and polyacrylamide, have gained significant traction within the biomaterials community, owing to tunable control over their initial network mechanics, chemical composition, and biodegradability (10–12). Since these systems are synthetically defined, stimuli-responsive functional handles can be installed during formulation to afford constructs with exogenously modifiable properties. Though synthetic gels have been engineered to be responsive to many external stimuli (e.g., pH, temperature, enzyme) (13–15), those sensitive to light can be uniquely modulated in four dimensions (4D, comprising 3D space and time) through lithographically defined irradiation (16, 17). In addition to regulating viscoelastic properties of these materials (18–22), photochemical methodologies have been developed to immobilize bioactive peptides and proteins in user-defined patterns within cell-laden gels (23–30). Such spatiotemporal control over network biofunctionalization offers exciting opportunities in recapitulating the dynamic biochemical heterogeneity characteristic to native tissues.
Despite the historically superior tunability of synthetic hydrogel materials, many cell biologists and tissue engineers alike continue to gravitate toward natural protein-based systems, including those derived from fibrin, collagen I, gelatin, and decellularized extracellular matrix (dECM) (31). These materials generally exhibit higher biocompatibility and enhanced matrix remodeling compared with synthetic alternatives. Moreover, natural biomaterials innately provide encapsulated cells with many of the same biochemical cues present in the native ECM, giving rise to greater cell–material integration and functional engineered tissue (9). With the goal of gaining 4D control over natural biomaterial properties, recent efforts have sought to functionalize protein-based materials with photosensitive moieties. Installation of alkene functionality (e.g., methacrylate, acrylate) on reaction components has permitted photopolymerization of natural gels (32–34), just as chemical cross-linking with photoscissile moieties (e.g., nitrobenzyl) has enabled their photodegradation (35, 36). Though existing methods offer spatiotemporal control over the mechanical properties of natural biopolymer-based hydrogels, strategies to selectively modulate the biochemical aspects of these systems in a user-defined and heterogeneous fashion remain largely unexplored. When properly developed, these advances would further solidify natural gels as choice materials for fundamental cell studies and translational applications.
A recent report from the Zenobi-Wong group was the first to demonstrate photoimmobilization of proteins within natural hydrogels (30). Relying on a sortase transpeptidase-mediated coupling of a chemically tagged streptavidin onto a photouncaged polyglycine peptide grafted to a backbone material, biotinylated proteins of interest could be noncovalently immobilized with spatial control within natural gels. While the group beautifully demonstrated controlled axon guidance using nerve growth factor within an engineered hyaluronan-based matrix (37), photopatterned cell function on or within protein-based biomaterials was not demonstrated. Seeking to fill this important void, we postulated that the photomediated oxime ligation—a chemistry recently developed and reported by our group for photopatterning of synthetic polymer-based materials (26, 38)—could be used to spatially modify natural gels, but with additional benefits associated with covalent protein immobilization, chemical accessibility, design simplicity, and overall ease of use. As this chemistry represents one of exceptionally few bioorthogonal reactions that can be photochemically triggered, thereby enabling user-defined control over when and where ligation occurs with high specificity in the presence of living systems, we identified the photomediated oxime ligation as uniquely capable of spatiotemporally regulating cell function on/in protein-based gels.
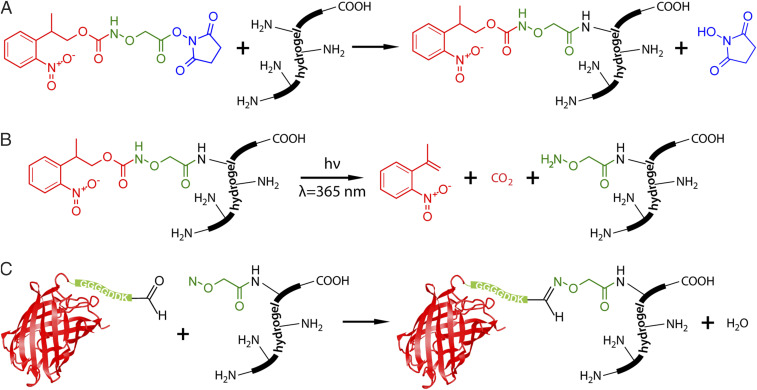
In this paper, we introduce a generalizable strategy to covalently decorate naturally derived biomaterials with bioactive proteins including growth factors to spatially control encapsulated cell fate. Taking advantage of primary amines ubiquitously present on proteins (both at their N terminus and on lysine side chains), we minimally functionalize natural gel precursors with a 2-(2-nitrophenyl) propoxycarbonyl (NPPOC)-photocaged alkoxyamine (H2NO-) through reaction with an activated N-hydroxysuccinimide ester (-OSu), a modification chemistry chosen due to its comparatively long-term reagent stability, synthetic accessibility, and validated use for installing functional handles onto proteins (Fig. 1A). The trifunctional NPPOC-HNO-OSu small molecule can be produced on gram scale through readily accessible chemistry (SI Appendix, Method S1) and used to modify a wide variety of natural biomaterials. Upon exposure of the functionalized gel to cytocompatible near-ultraviolet (UV) light (λ = 365 nm), the photocage is cleaved in a dose-dependent manner, liberating the alkoxyamine and permitting localized condensation with aldehyde-functionalized proteins (Fig. 1 B and C). Through mask-based and laser-scanning lithographic activation of this photomediated oxime ligation, we achieve full 4D control over protein immobilization within three distinct natural biopolymer-based hydrogels. Highlighting the versatility of the approach, we pattern two responses that—despite considerable effort by our laboratory and others—have not been achieved using purely synthetic materials, namely, two-dimensional (2D) primary rat hepatocyte proliferation on collagen gels using immobilized epidermal growth factor (EGF) and 3D U2OS Notch signaling activation within fibrin gels decorated with tethered Delta-1.
Fig. 1.
Hydrogel modification and photopatterning through photomediated oxime ligation. (A) Conjugation of NPPOC-HNO-OSu to primary amines present on the protein-based hydrogel. (B) A reactive alkoxyamine is liberated upon photocleavage of the NPPOC cage with cytocompatible light (λ = 365 nm). (C) Aldehyde-modified proteins are covalently immobilized within the hydrogel via oxime ligation.
Results and Discussion
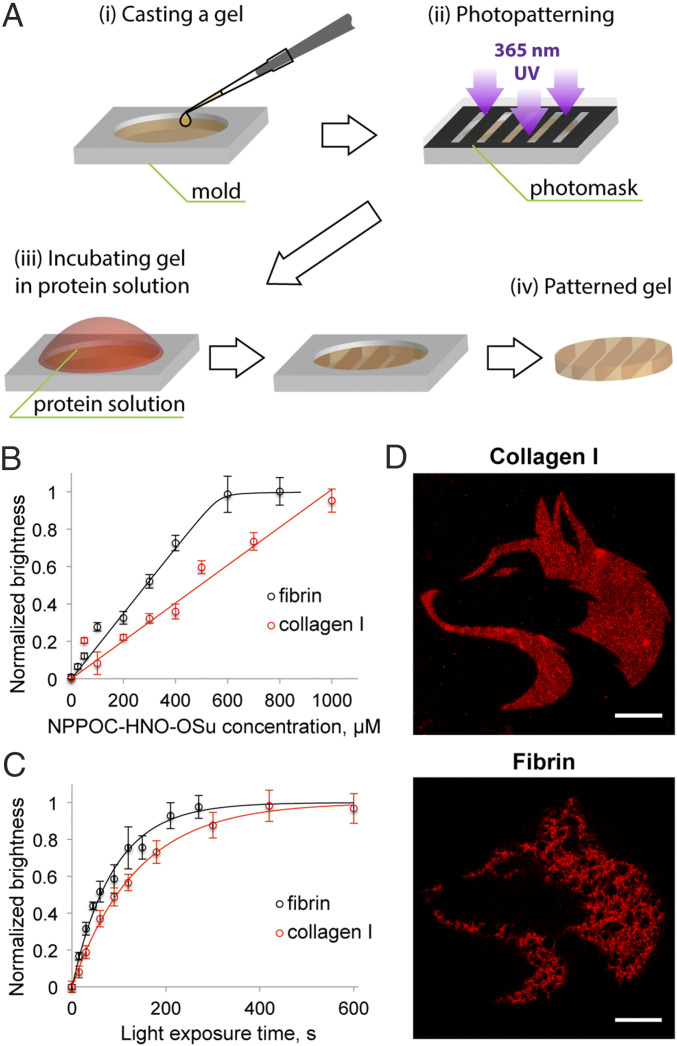
To test our workflow and verify its compatibility with different natural biomaterials, we biochemically decorated gels based on collagen I (4 mg mL−1) or fibrin (10 mg mL−1) with full-length proteins. Prior to gelation via conventional methodologies, collagen I and fibrin gel precursors were incubated with varying amounts of NPPOC-HNO-OSu (0 to 1 mM) to create constructs bearing different concentrations of the photocaged alkoxyamine. Through theoretical calculations and experimental measurements, we found that well under 1% of collagen and fibrin’s primary amines were modified with the photocaged alkoxyamine following all treatment conditions; this small extent of labeling (<1 modification on average per protein) was anticipated to have minimal impact on ECM protein function (SI Appendix, Methods S2–S4 and Fig. S1). For each protein-based gel system, transparent hydrogels were formed in cylindrical molds (diameter = 3.2 mm, height = 1 mm) and exposed to near-UV light (λ = 365 nm, 10 mW cm−2, 10 min). Based on previously reported NPPOC photouncaging kinetics (26, 39), irradiation conditions were expected to yield complete alkoxyamine photoliberation in a manner that is fully cytocompatible. After light exposure, gels were swollen in a solution containing an aldehyde-functionalized mCherry (mCherry-CHO), a red fluorescent protein site-specifically modified with the reactive carbonyl at its C terminus through sortase-tag enhanced protein ligation (STEPL) (27, 40) (Fig. 2A and SI Appendix, Methods S5–S7). After gel–protein conjugation by oxime ligation, gels were washed to remove unbound protein prior to analysis. Taking advantage of the patterned protein’s inherent fluorescence, we quantified the extent of mCherry immobilization via fluorescent confocal microscopy (Fig. 2B). As expected, we observed a linear correlation between the amount of NPPOC-HNO-OSu employed and the total amount of protein phototethered within each gel. The variance in patterning slope between the collagen I and fibrin systems was attributed to differences in the total amine content of the gel precursors, resulting in a scaled functionalization for a given activated ester concentration. Control gels reacted with any amount of NPPOC-HNO-OSu but never exposed to light yielded no mCherry immobilization, indicating that the alkoxyamine remained fully caged throughout gel formation and successfully inhibited protein ligation and that nonspecific protein fouling of the gel did not occur (SI Appendix, Fig. S2).
Fig. 2.
(A) Mask-based lithographic patterning of natural hydrogels. Gels are selectively irradiated with near-UV light prior to incubation with aldehyde-modified proteins of interest and subsequently washed to remove unconjugated species. (B) Immobilized mCherry fluorescence brightness as a function of NPPOC-HNO-OSu labeling concentration following light treatment (λ = 365 nm, 10 mW cm−2, 10 min). (C) Immobilized mCherry fluorescence brightness in collagen I and fibrin gels as a function of UV exposure time. Fluorescence brightness values are normalized between 0 (no fluorescence) and 1 (max observed fluorescence for the given experiment). The best-fit solid line assumes first-order photocleavage kinetics of NPPOC and complete oxime ligation. Error bars correspond to ±1 SD about the mean for n = 3 experimental replicates. (D) Mask-based lithographic patterning of mCherry (red) into collagen I and fibrin gels in arbitrary 2D patterns. (Scale bars: 50 µm.)
Encouraged that our synthetic workflow could be used to functionalize natural gels with full-length proteins, we sought to evaluate the effects of different light irradiation conditions on gel patterning. Hydrogels based on fibrin or collagen I were modified with NPPOC-HNO-OSu (500 and 300 µM, respectively), cast as thin cylindrical constructs, and exposed to near-UV light (λ = 365 nm, 10 mW cm−2) for varied amounts of time (0 to 10 min). Following incubation with mCherry-CHO (0.5 mg mL−1) and fluorescent quantification by confocal microscopy, we observed an exponential dose-dependent response predicted by NPPOC photocleavage kinetics and consistent with previous patterning studies of synthetic PEG gels (26) (Fig. 2C). Specifically, the NPPOC photodeprotection process was found to follow first-order reaction kinetics with decay constants of k = 0.011 ± 0.002 s−1 in fibrin and 0.007 ± 0.001 s−1 in collagen I for λ = 365 nm at 10 mW cm−2. The minor variation in dose responsiveness between the two gel systems was attributed to subtle differences in the chemical microenvironment within each protein that yielded slightly varied photouncaging kinetics.
After demonstrating dose-dependent immobilization of full-length proteins in natural materials using flood illumination, we next employed mask-based lithographic techniques to pattern NPPOC uncaging in arbitrary 2D geometries extending throughout the full thickness of each gel. Successful patterning was again observed in collagen I- and fibrin-based systems, as indicated by the spatially controlled immobilization of mCherry-CHO in the shape of the University of Washington’s former logo within gels (Fig. 2D). Similar results were observed for studies involving gels derived from decellularized cardiac ECM (SI Appendix, Fig. S3). Though micrometer-scale patterning resolution was achieved in each material type, patterning results in fibrin and dECM reflect the fibrous structure characteristic of these natural materials.
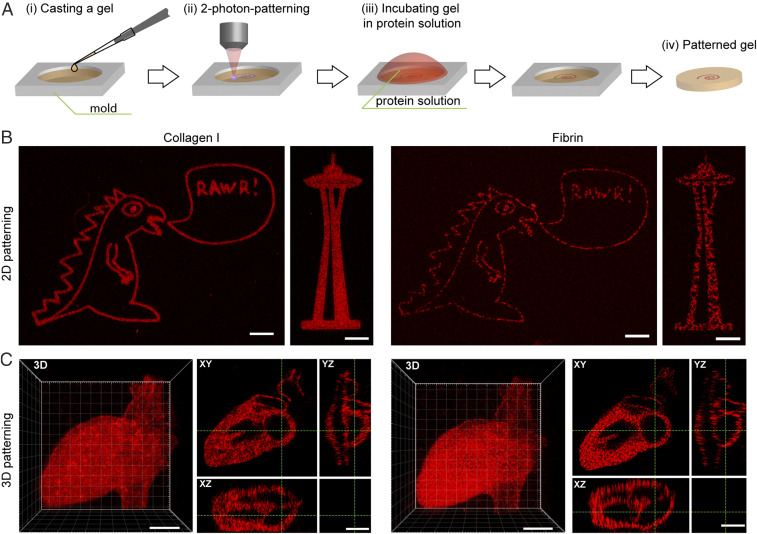
While photomask-based lithography is readily employed to immobilize proteins within large gel volumes in a geometrically regulated and scalable manner, such methods are fundamentally limited in that patterns can be specified in only two of the three spatial dimensions (i.e., x-y but not in z). Previous efforts by our group and others have established multiphoton laser-scanning lithographic patterning as an effective technique to locally modify user-defined hydrogel subvolumes with 4D control (23, 41–44). Using these methodologies, photochemical reactions are confined to the focal point of a femtosecond pulsed laser with high precision (submicrometer control over activation in x and y dimensions; 2 to 3 µm in z); raster scanning of the laser focal point within the sample results in localized material alteration in custom geometries (Fig. 3A). Here, we employed these techniques to immobilize mCherry-CHO within NPPOC-HNO-OSu-functionalized collagen I- and fibrin-based gels in arbitrary shapes. Controlling the laser raster pattern in the x and y dimensions and stepping this activation through thin z sections, 2D “slice” patterns were generated in shapes including the Seattle Space Needle and a vocal dinosaur (Fig. 3B). Utilizing different laser scan 2D geometries at each z location, fully 3D patterns were created in the shape of an anatomically correct human heart (Fig. 3C). Uniform 3D patterning with micrometer-scale patterning resolution was observed for both natural protein-based gel systems.
Fig. 3.
Multiphoton-based lithographic patterning of mCherry-CHO within collagen I and fibrin gels. (A) After casting, gels are exposed to a femtosecond pulsed laser (λ = 740 nm) to activate NPPOC cleavage and liberate the alkoxyamine for oxime ligation with aldehyde-functionalized proteins in user-defined patterns. (B) Two-dimensional control over biomolecule immobilization is achieved within 3D gels, with speckled fluorescence highlighting the fibril nature of assembled fibrin. (C) Three-dimensional patterning of an anatomical heart model is achieved in each natural protein-based gel, showcased with 3D and cross-sectional views. mCherry-CHO is shown in red. (Scale bars: 50 µm.)
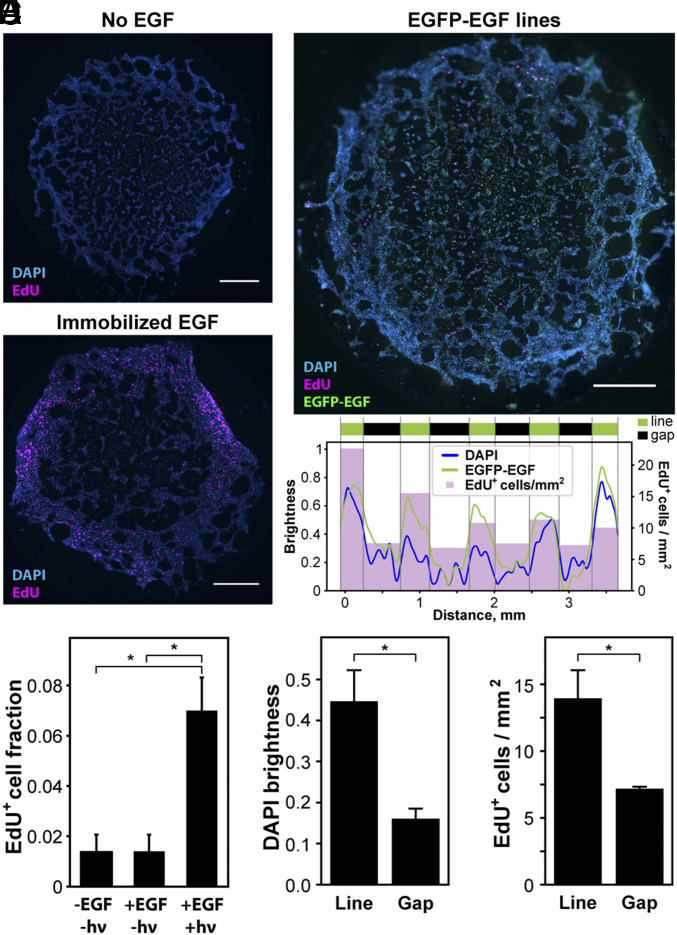
Heartened by our ability to immobilize site-specifically modified fluorescent proteins within natural hydrogel biomaterials, we next sought to employ these methods to spatially direct complex biological fates using growth factors. We turned our initial attention to EGF, a bioactive protein that stimulates proliferation of many cell types, including primary hepatocytes isolated from the liver (45, 46). Collagen I gels modified with NPPOC-HNO-OSu (250 µM) were photochemically decorated with an aldehyde-functionalized EGF (EGF-CHO) that had been site-specifically tagged using STEPL. As hoped, freshly harvested primary rat hepatocytes seeded on EGF-modified gel surfaces exhibited a statistically significant increase in DNA synthesis (P < 0.001, one-way ANOVA with Tukey’s post hoc) after 6 d in culture relative to control gels lacking EGF (Fig. 4 A and B and SI Appendix, Fig. S4), indicating that the tethered EGF remained bioactive. We next exploited mask-based lithography to photopattern collagen gels with lines (500 µm wide) of EGFP-EGF-CHO, a chimeric fusion of enhanced green fluorescent protein and the STEPL-tagged EGF-CHO that could be fluorescently visualized after immobilization. Samples were imaged after 1 and 6 d in culture on the protein-patterned surfaces, where we observed nonuniform hepatocyte coverage on the patterned surfaces only at the later time point; cell density and the number of cells synthesizing DNA indicated by 5-ethynyl-2′-deoxyuridine (EdU) incorporation/staining were statistically higher on day 6 (P = 0.023 for DAPI, P = 0.039 for EdU, t test) but not on day 1 in gel regions functionalized with EGFP-EGF-CHO compared with those without the immobilized growth factor (Fig. 4 C and E and SI Appendix, Figs. S5 and S6). This study is significant for several reasons: 1) It demonstrates patterned regulation of a primary cell type using a photoactivatable hydrogel, a feat that is strikingly absent from the literature likely due to difficulties typically associated with culturing such cells on/in synthetic gels. 2) Hepatocytes are notoriously nonproliferative cells, particularly in vitro, and these results appear to be a promising step toward promoting their expansion in culture. 3) Utilization of the photomediated oxime ligation in conjunction with the site-specifically modified EGF allows it to remain bioactive, despite its identity as a fragile growth factor.
Fig. 4.
Patterned cellular behavior on collagen I gels. (A) Primary rat hepatocytes exhibited enhanced DNA synthesis when seeded on collagen I gels containing tethered EGF. (B) Quantification of EdU-positive cell fraction indicates statistically significant increase in DNA synthesis for hepatocytes on EGF-modified gels. (C) Hepatocytes on collagen I gels patterned with EGFP-EGF-CHO in a 500 µm line pattern. Mean DAPI brightness and EdU+ cell density showing local cell density and proliferation correlate with local EGFP-EGF-CHO concentration. (D and E) EGFP-EGF-CHO-patterned line regions support higher cell counts and higher extents of DNA synthesis than those in the interspaced gap regions. EdU staining is shown in pink, nuclei are in blue, and EGFP-EGF-CHO is in green. All analyses were performed on samples 6 d after seeding. Error bars correspond to ±1 SE about the mean for n ≥ 3 experimental replicates. (Scale bars: 1 mm.) *P < 0.05.
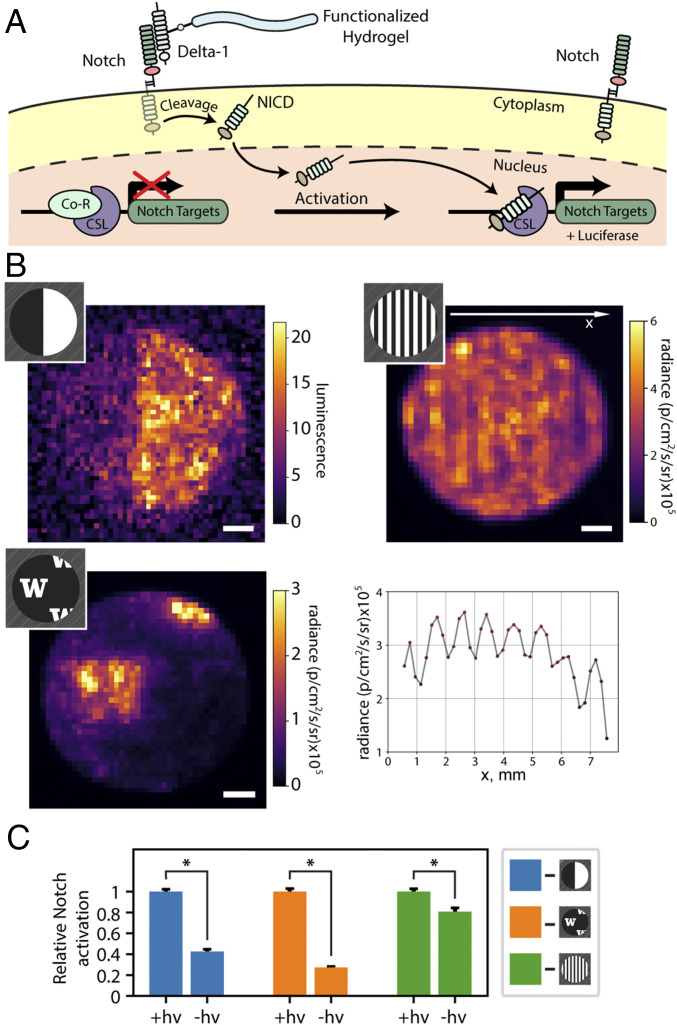
Having demonstrated the ability to direct cellular behavior in response to immobilized protein cues on gel surfaces, we next sought to exploit the inherent bioorthogonality of the photomediated oxime ligation to biochemically pattern natural hydrogels in the presence of encapsulated cells and to subsequently direct their fate in 3D. Specifically, we patterned Delta-1 protein to activate the Notch pathway (Fig. 5A), an evolutionary conserved signaling system required for normal embryonic development, regulation of tissue homeostasis, and maintenance of stem cells in adults (47, 48) that has proven unattainable in synthetic gels. Of the techniques typically employed to assess Notch activation (e.g., RT-PCR, RNA microarray, Western blot analysis, with luciferase reporter cells), luciferase is the only method that can readily provide spatial readout, essential in evaluating gel patterning fidelity. Since luciferase activity cannot be easily resolved in three dimensions using conventional fluorescent imaging techniques (3D samples are imaged as 2D projections, as done using IVIS® imaging for in vivo analysis), we turned our focus to 2D patterns of Delta-1 extending uniformly throughout the gel thickness (SI Appendix, Fig. S7). For these studies, recombinant Delta-1 was expressed in HEK293F mammalian cells, purified, and modified statistically with 2-5-dioxopyrrolidin-1-yl 4-formylbenzoate. This aldehyde-modified protein (Delta-1-CHO) was immobilized within cell-laden hydrogels in various geometries using mask-based photolithography. To quantify and visualize Notch activation in response to patterned Delta-1, U2OS osteosarcoma reporter cells expressing renilla luciferase constitutively and firefly luciferase upon Notch activation were encapsulated within fibrin gels. Whole-gel luminescent imaging on day 7 revealed statistically significant (P < 0.001, t test) enhanced Notch signaling in gel regions matching mask geometry (Fig. 5 B and C), as evidenced by higher firefly luciferase-catalyzed luminescence in the light-exposed biomaterial subvolumes. Critically, luminescence originating from renilla luciferase was constant for cells throughout the entire gel (SI Appendix, Fig. S8), indicating that the protein patterning itself does not affect cell viability; these results are consistent with recent studies demonstrating that the cellular proteome is unaltered in response to the near-UV light treatments employed here (49). Since full 3D control over Delta-1 immobilization within protein-based hydrogels can be achieved using laser-scanning lithographic patterning (SI Appendix, Fig. S9), we anticipate future opportunities in directing Notch and other complex signaling in 3D gels.
Fig. 5.
Patterned Notch activation within fibrin gels. (A) The schematic of Notch signaling in U2OS osteosarcoma CSL/luciferase Notch reporter cells. Firefly luciferase is expressed in response to the up-regulation of Notch signaling caused by the interaction with immobilized Delta-1. (B) Fibrin gel-encapsulated U2OS Notch reporter cells show localized up-regulation of firefly luciferase expression in response to different patterns of immobilized Delta-1-CHO. The half-gel pattern was imaged using Chemidoc XRS+ (Bio-Rad), while 500 µm wide lines and the W pattern were imaged using an IVIS® Spectrum. Line pattern analysis shows modulation of luciferase expression matching regions of immobilized Delta-1. (C) Notch signaling is significantly enhanced for cells within the patterned Delta-1 regions (+hν) compared with that outside (−hν) for each mask geometry (indicated by color). Average luminescence values for functionalized gel regions of each geometry were normalized to 1. Error bars correspond to ±1 SE about the mean. All analyses were performed on samples 7 d after encapsulation/photopatterning. (Scale bars: 1 mm.) *P < 0.001.
Like all scientific strategies, the material-based approaches presented here are not without limitation. Since the bioactive protein is exposed to cells throughout patterning, these techniques are best applied to applications in which 1) the protein’s tethered and soluble forms have inherently different bioactivities, 2) biological function is to be specified on timescales that exceed that of the patterning process (e.g., proliferation, differentiation, migration, morphogenesis), 3) protein immobilization can be performed in the absence of cells, or 4) patterning can be accomplished comparatively fast in the presence of encapsulated living cells, potentially using thin gels. Moreover, while nonspecific adsorption of the selected proteins was not observed in the gel systems employed in this report, background “fouling” should be evaluated for each patterned protein/gel combination of interest. Despite these potential limitations, the ability to spatially control complex cellular functions within the protein-based biomaterials most commonly exploited for 3D cell culture is a significant and powerful advance.
Conclusion
In this article, we have introduced a robust and versatile synthetic workflow to immobilize bioactive proteins site specifically and with spatiotemporal control within natural hydrogel biomaterials. Relying on a photomediated oxime ligation that is bioorthogonal and compatible with common lithographic patterning techniques, gel functionalization can be controlled with micrometer-scale resolution in a dose-dependent manner and in the presence of living cells. Enabling 4D biochemical tunability within biomaterial platforms that have proven the modern workhorses of 3D cell and organoid culture, we anticipate that these approaches will find great utility in probing and directing biological functions and in engineering heterogenous functional tissues.
Materials and Methods
Synthesis of NPPOC-HNO-OSu.
NPPOC-HNO-OSu was synthesized on a gram scale via a two-step reaction from commercially available precursors with ∼80% overall yield. The complete experimental details are given in SI Appendix, Method S1.
Natural Protein-Based Hydrogel Functionalization with NPPOC-HNO-OSu.
For collagen I functionalization.
Collagen I (1 mL of rat tail collagen I at 4 mg mL−1 in 0.02 N acetic acid, Corning) solution was brought to pH 7 to 7.5 on ice through the addition of phosphate buffered saline (PBS)-buffered NaOH, quickly mixed with NPPOC-HNO-OSu (25 mM in dimethyl sulfoxide [DMSO], 0 to 20 µL), and cast in molds. Formed gels were protected from light and washed in PBS overnight prior to patterning.
For fibrinogen functionalization.
Stock fibrinogen solution (1 mL of 50 mg mL−1 in Hank’s balanced salt solution) was quickly mixed with NPPOC-HNO-OSu (25 mM in DMSO, 0 to 50 µL) and incubated (1 h) at room temperature protected from light. To make gels, the functionalized fibrinogen solution was diluted to 10 mg mL−1 through addition of PBS and thrombin (0.5 to 1.0 unit mL−1) prior to casting in molds. Formed gels were protected from light and washed in PBS overnight prior to patterning.
Synthesis of Aldehyde-Modified Polyglycine Probe for STEPL.
An aldehyde-modified polyglycine peptide was synthesized through standard fluorenylmethoxycarbonyl (Fmoc)-based solid-phase methodologies involving a butyloxycarbonyl-protected N-terminal glycine and a 4-methyltrityl-protected C-terminal lysine reside. After selective deprotection of 4-methyltrityl (1% trifluoroacetic acid [TFA] in dichloromethane) on resin, aldehyde functionality was installed through condensation with 4-formylbenzoic and the ε-amino group of the C-terminal lysine. Resin was washed (dimethylformamide three times and dichloromethane three times) prior to peptide cleavage/deprotection (95:5 TFA:H2O, 20 mL, 2 h) and precipitation (diethyl ether, 180 mL, 0 °C, two times). The crude peptide was purified via semipreparative reversed-phase high-performance liquid chromatography using a 55-min gradient (5 to 100% of acetonitrile and 0.1% TFA in H2O) and lyophilized to give the final product [denoted H-GGGGDDK(CHO)-NH2]. Peptide purity was confirmed by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. The complete experimental details are given in SI Appendix, Method S5.
Aldehyde Functionalization of mCherry, EGF, and EGFP-EGF Proteins via STEPL.
STEPL plasmids for mCherry, EGF, and EGFP-EGF were constructed using standard cloning techniques (SI Appendix, Method S6) and transformed into BL21(DE3) Escherichia coli (Thermo Fisher). For protein expression, transformants were grown at 37 °C in a lysogeny broth that contained ampicillin (100 μg mL−1) until an optical density of 0.6 (λ = 600 nm). Expression was induced with isopropyl β-d-1-thiogalactopyranoside (0.5 mM) and then agitated overnight at 18 °C. Cells were collected via centrifugation and lysed by sonication. The clarified lysate was loaded onto HisPur Ni-NTA resin (ThermoFisher), which was washed (20 mM Tris, 50 mM NaCl, and 20 mM imidazole) to remove unbound proteins. After treatment of the resin with H-GGGGDDK(CHO)-NH2 (20×, 4 h, 37 °C), aldehyde-tagged proteins were eluted and purified by dialysis (molecular weight cutoff, ∼10 kDa). The protein identity and purity were confirmed by liquid chromatography–tandem mass spectrometry and sodium dodecyl sulfate polyacrylamide gel electrophoretic analysis. The protein concentrations were determined by UV absorption (λ = 280 nm) prior to use. STEPL-modified proteins were denoted as mCherry-CHO, EGF-CHO, and EGFP-EGF-CHO. The complete experimental details are given in SI Appendix, Method S7.
Aldehyde Functionalization of Delta-1 Protein.
Delta-1 protein was recombinantly expressed in HEK293F mammalian cells (ThermoFisher) and purified by immobilized metal affinity chromatography (SI Appendix, Method S8); 2-5-dioxopyrrolidin-1-yl 4-formylbenzoate (SI Appendix, Method S2, 40 mM in DMSO, 5× molar excess) was added to Delta-1 (1 mg mL−1 in PBS), incubated at room temperature (2 h), dialyzed in PBS at 4 °C, and sterile filtered (0.2 µm syringe filter). This product (denoted Delta-1-CHO) was used without additional purification.
Hydrogel Patterning and Imaging.
For photomask patterning, NPPOC-HNO-OSu-modified natural hydrogels were exposed to collimated UV light (λ = 365 nm, 10 mW cm−2, 0 to 10 min) through a patterned chrome photomask (Photo Sciences) using a Lumen Dynamics OmniCure S1500 Spot UV Curing system equipped with an internal 365 nm band-pass filter and a second in-line 360 nm cut-on long-pass filter. For the multiphoton laser-scanning lithography patterning experiments, an Olympus FV1000 MPE BX61 Multiphoton Microscope with a 20× objective was used. Gels were scanned in x-y scanned regions of interest (ROIs) at different z positions throughout the gel thickness. Each 2D ROI was scanned 16 to 63 times with pulsed laser light (λ = 740 nm, 0 to 100 laser power) with a 1.2 µm z interval to generate 3D patterns. After NPPOC cleavage, gels were incubated for ∼10 h with the aldehyde-tagged protein (0 to 100 µM in PBS) at 4 °C and protected from light. To wash away any unreacted protein, gels were gently agitated in PBS (16 h). Experiments involving patterned immobilization of fluorescent proteins were visualized by fluorescent, confocal, or multiphoton microscopy using standard imaging parameters.
Modulating Hepatocyte Survival and Proliferation Rate in Response to Patterned EGF.
Collagen I gels (30 µL, 2 mg mL−1) were functionalized with NPPOC-HNO-OSu (250 µM) as previously described. Gels were subsequently exposed to UV light (λ = 365 nm, 10 mW cm−2, 10 min) either directly or through a chrome mask patterned with 500 µm wide lines. Gels were then incubated with EGF-CHO or EGFP-EGF-CHO (0.1 mg mL−1 in Tris buffer, 10 h) prior to extensive washing in PBS (three times, 8 h). Primary hepatocytes were isolated from female Lewis/SsNHsd rats (Envigo) based on a previously described protocol (50) approved by the Institutional Animal Use and Care Committee at the University of Washington, then seeded (80,000 cells cm−2) on gels in Dulbecco’s modified Eagle’s medium (supplemented with 10% fetal bovine serum, 1% penicillin–streptomycin, 15 mM Hepes, 0.04 µg mL−1 dexamethasone, 70 µg mL−1 glucagon, and 1% insulin-transferrin-sodium selenite supplement, Sigma-Aldrich). Cells were cultured for 6 d with media changes on days 1, 3, 4, and 5. On days 4 and 5 media were supplemented with EdU (1:1,000, Invitrogen). After 1 and 6 d of culture, samples were fixed (4% paraformaldehyde) prior to staining for nuclei (Hoechst 33342, 1:2,000) and EdU (Click-iT EdU Imaging Kit, Invitrogen C10340). Whole samples were fluorescently imaged (Nikon Eclipse Ti high-resolution fluorescent widefield imaging system). The fraction of EdU-positive nuclei was determined using a custom MATLAB script measuring the ratio of EdU-positive to Hoechst-positive pixels for each image. Line/gap edges were ascribed to low- to high-intensity transitions in the EGFP-EGF-CHO fluorescence. The line/gap cell density was assessed for patterned gels by measuring the mean nuclei fluorescence along the direction of the line. EdU+ cell analysis was performed with the aid of a custom MATLAB segmentation algorithm which detected and determined locations of EdU+ nuclei.
Patterned Notch Activation of U2OS Cells in Response to Immobilized Delta-1.
U2OS osteosarcoma CSL/luciferase Notch reporter cells (106 cell mL−1) were encapsulated in collagen I gels (30 µL, 10 mg mL−1) functionalized with NPPOC-HNO-OSu (250 µM). Cell-laden gels were cast in the presence of Delta-1-CHO (0.05 mg mL−1) and irradiated with light (λ = 365 nm, 10 mW cm−2, 10 min) through various photomasks (half on; array of W’s; 500 µm wide lines) to pattern aminooxy uncaging. Two hours after light exposure, gels were placed in and washed with medium (Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum, 1% penicillin–streptomycin, 2 µg mL−1 aprotinin; changed daily for 6 d). Luciferase expression was determined on day 7 using a Promega Dual-Glo Luciferase Assay per manufacturer’s instruction, with d-luciferin (150 µg mL−1) added to the medium prior to analysis. Gels were imaged using a ChemiDoc XRS+ (BioRad) and In Vivo Imaging System Spectrum (PerkinElmer IVIS®). Overlaying the aligned photomask image file onto each of the patterned gels, Notch activation was quantified for cells within the Delta-modified gel subvolumes and compared with those in the unfunctionalized regions. Statistical analyses were performed using a standard t test.
Supplementary Material
Acknowledgments
We thank J. Davis, D.-H. Kim, N. Sniadecki, Y. Zheng, and all other members of the Gree Research Scholars group for helpful discussion; D. Hailey of the University of Washington (UW) Garvey Imaging Center for his ongoing support and advice; I. Bernstein of the Fred Hutchinson Cancer Research Center for providing plasmid for Delta-1 expression; C. Murry of the UW for providing the U2OS Notch reporter cells used in this work; and D. Corbett for his assistance with IVIS® imaging. We acknowledge support from S. Edgar at the UW Mass Spectrometry Center as well as that from the NIH and N. Peters at the UW W. M. Keck Microscopy Center (Grant S10 OD016240). This work was supported by a generous donation from Gree Real Estate, a Faculty Early Career Development (CAREER) Award (DMR 1652141 to C.A.D.) from the NSF, as well as a Maximizing Investigators’ Research Award (R35GM138036 to C.A.D.) and a Director’s New Innovator Award (DP2HL137188, K.R.S.) from NIH.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2014194118/-/DCSupplemental.
Data Availability.
All study data are included in the article and SI Appendix.
References
- 1.Lee K. Y., Mooney D. J., Hydrogels for tissue engineering. Chem. Rev. 101, 1869–1879 (2001). [DOI] [PubMed] [Google Scholar]
- 2.Peppas N. A., Hilt J. Z., Khademhosseini A., Langer R., Hydrogels in biology and medicine: From molecular principles to bionanotechnology. Adv. Mater. 18, 1345–1360 (2006). [Google Scholar]
- 3.Tibbitt M. W., Anseth K. S., Hydrogels as extracellular matrix mimics for 3D cell culture. Biotechnol. Bioeng. 103, 655–663 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Seliktar D., Designing cell-compatible hydrogels for biomedical applications. Science 336, 1124–1128 (2012). [DOI] [PubMed] [Google Scholar]
- 5.Zhang Y. S., Khademhosseini A., Advances in engineering hydrogels. Science 356, eaaf3627 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.DeForest C. A., Anseth K. S., Advances in bioactive hydrogels to probe and direct cell fate. Annu. Rev. Chem. Biomol. Eng. 3, 421–444 (2012). [DOI] [PubMed] [Google Scholar]
- 7.Burdick J. A., Murphy W. L., Moving from static to dynamic complexity in hydrogel design. Nat. Commun. 3, 1269 (2012). [DOI] [PubMed] [Google Scholar]
- 8.Daly A. C., Riley L., Segura T., Burdick J. A., Hydrogel microparticles for biomedical applications. Nat. Rev. Mater. 5, 20–43 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Caliari S. R., Burdick J. A., A practical guide to hydrogels for cell culture. Nat. Methods 13, 405–414 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lutolf M. P., Gilbert P. M., Blau H. M., Designing materials to direct stem-cell fate. Nature 462, 433–441 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kratochvil M. J., et al. , Engineered materials for organoid systems. Nat. Rev. Mater. 4, 606–622 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aisenbrey E. A., Murphy W. L., Synthetic alternatives to Matrigel. Nat. Rev. Mater. 5, 539–551 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lu Y., Aimetti A. A., Langer R., Gu Z., Bioresponsive materials. Nat. Rev. Mater. 1, 16075 (2016). [Google Scholar]
- 14.Tam R. Y., Smith L. J., Shoichet M. S., Engineering cellular microenvironments with photo- and enzymatically responsive hydrogels: Toward biomimetic 3D cell culture models. Acc. Chem. Res. 50, 703–713 (2017). [DOI] [PubMed] [Google Scholar]
- 15.Badeau B. A., DeForest C. A., Programming stimuli-responsive behavior into biomaterials. Annu. Rev. Biomed. Eng. 21, 241–265 (2019). [DOI] [PubMed] [Google Scholar]
- 16.Ruskowitz E. R., DeForest C. A., Photoresponsive biomaterials for targeted drug delivery and 4D cell culture. Nat. Rev. Mater. 3, 17087 (2018). [Google Scholar]
- 17.Rapp T. L., DeForest C. A., Visible light-responsive dynamic biomaterials: Going deeper and triggering more. Adv. Healthc. Mater. 9, e1901553 (2020). [DOI] [PubMed] [Google Scholar]
- 18.Kloxin A. M., Kasko A. M., Salinas C. N., Anseth K. S., Photodegradable hydrogels for dynamic tuning of physical and chemical properties. Science 324, 59–63 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Khetan S., et al. , Degradation-mediated cellular traction directs stem cell fate in covalently crosslinked three-dimensional hydrogels. Nat. Mater. 12, 458–465 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rosales A. M., Mabry K. M., Nehls E. M., Anseth K. S., Photoresponsive elastic properties of azobenzene-containing poly(ethylene-glycol)-based hydrogels. Biomacromolecules 16, 798–806 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arakawa C. K., Badeau B. A., Zheng Y., DeForest C. A., Multicellular vascularized engineered tissues through user-programmable biomaterial photodegradation. Adv. Mater. 29, 1703156 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu L., et al. , Cyclic stiffness modulation of cell‐laden protein–polymer hydrogels in response to user‐specified stimuli including light. Adv. Biosyst. 2, 1800240 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.DeForest C. A., Polizzotti B. D., Anseth K. S., Sequential click reactions for synthesizing and patterning three-dimensional cell microenvironments. Nat. Mater. 8, 659–664 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.DeForest C. A., Anseth K. S., Cytocompatible click-based hydrogels with dynamically tunable properties through orthogonal photoconjugation and photocleavage reactions. Nat. Chem. 3, 925–931 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mosiewicz K. A., et al. , In situ cell manipulation through enzymatic hydrogel photopatterning. Nat. Mater. 12, 1072–1078 (2013). [DOI] [PubMed] [Google Scholar]
- 26.DeForest C. A., Tirrell D. A., A photoreversible protein-patterning approach for guiding stem cell fate in three-dimensional gels. Nat. Mater. 14, 523–531 (2015). [DOI] [PubMed] [Google Scholar]
- 27.Shadish J. A., Benuska G. M., DeForest C. A., Bioactive site-specifically modified proteins for 4D patterning of gel biomaterials. Nat. Mater. 18, 1005–1014 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shadish J. A., Strange A. C., DeForest C. A., Genetically encoded photocleavable linkers for patterned protein release from biomaterials. J. Am. Chem. Soc. 141, 15619–15625 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zheng Y., et al. , 4D hydrogel for dynamic cell culture with orthogonal, wavelength-dependent mechanical and biochemical cues. Mater. Horiz. 7, 111–116 (2020). [Google Scholar]
- 30.Broguiere N., et al. , Morphogenesis guided by 3D patterning of growth factors in biological matrices. Adv. Mater. 32, e1908299 (2020). [DOI] [PubMed] [Google Scholar]
- 31.Hoffman A. S., Hydrogels for biomedical applications. Adv. Drug Deliv. Rev. 64, 18–23 (2012). [DOI] [PubMed] [Google Scholar]
- 32.Nguyen K. T., West J. L., Photopolymerizable hydrogels for tissue engineering applications. Biomaterials 23, 4307–4314 (2002). [DOI] [PubMed] [Google Scholar]
- 33.Burdick J. A., Chung C., Jia X., Randolph M. A., Langer R., Controlled degradation and mechanical behavior of photopolymerized hyaluronic acid networks. Biomacromolecules 6, 386–391 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Benton J. A., DeForest C. A., Vivekanandan V., Anseth K. S., Photocrosslinking of gelatin macromers to synthesize porous hydrogels that promote valvular interstitial cell function. Tissue Eng. Part A 15, 3221–3230 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peng K., et al. , Dextran based photodegradable hydrogels formed via a Michael addition. Soft Matter 7, 4881–4887 (2011). [Google Scholar]
- 36.Tamura M., et al. , Click-crosslinkable and photodegradable gelatin hydrogels for cytocompatible optical cell manipulation in natural environment. Sci. Rep. 5, 15060 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Broguiere N., Isenmann L., Zenobi-Wong M., Novel enzymatically cross-linked hyaluronan hydrogels support the formation of 3D neuronal networks. Biomaterials 99, 47–55 (2016). [DOI] [PubMed] [Google Scholar]
- 38.Farahani P. E., Adelmund S. M., Shadish J. A., DeForest C. A., Photomediated oxime ligation as a bioorthogonal tool for spatiotemporally-controlled hydrogel formation and modification. J. Mater. Chem. B Mater. Biol. Med. 5, 4435–4442 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Adelmund S. M., Ruskowitz E. R., Farahani P. E., Wolfe J. V., DeForest C. A., Light-activated proteomic labeling via photocaged bioorthogonal non-canonical amino acids. ACS Chem. Biol. 13, 573–577 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Warden-Rothman R., Caturegli I., Popik V., Tsourkas A., Sortase-tag expressed protein ligation: Combining protein purification and site-specific bioconjugation into a single step. Anal. Chem. 85, 11090–11097 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Luo Y., Shoichet M. S., A photolabile hydrogel for guided three-dimensional cell growth and migration. Nat. Mater. 3, 249–253 (2004). [DOI] [PubMed] [Google Scholar]
- 42.Hahn M. S., Miller J. S., West J. L., Laser scanning lithography for surface micropatterning on hydrogels. Adv. Mater. 17, 2939–2942 (2005). [Google Scholar]
- 43.DeForest C. A., Anseth K. S., Photoreversible patterning of biomolecules within click-based hydrogels. Angew. Chem. Int. Ed. Engl. 51, 1816–1819 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Arakawa C., et al. , Biophysical and biomolecular interactions of malaria-infected erythrocytes in engineered human capillaries. Sci. Adv. 6, eaay7243 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Barreiros A. P., et al. , EGF and HGF levels are increased during active HBV infection and enhance survival signaling through extracellular matrix interactions in primary human hepatocytes. Int. J. Cancer 124, 120–129 (2009). [DOI] [PubMed] [Google Scholar]
- 46.Bowen W. C., et al. , Development of a chemically defined medium and discovery of new mitogenic growth factors for mouse hepatocytes: Mitogenic effects of FGF1/2 and PDGF. PLoS One 9, e95487 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.D’Souza B., Miyamoto A., Weinmaster G., The many facets of Notch ligands. Oncogene 27, 5148–5167 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Varnum-Finney B., et al. , Immobilization of Notch ligand, Delta-1, is required for induction of notch signaling. J. Cell Sci. 113, 4313–4318 (2000). [DOI] [PubMed] [Google Scholar]
- 49.Ruskowitz E. R., DeForest C. A., Proteome-wide analysis of cellular response to ultraviolet light for biomaterial synthesis and modification. ACS Biomater. Sci. Eng. 5, 2111–2116 (2019). [DOI] [PubMed] [Google Scholar]
- 50.Seglen P. O., Preparation of isolated rat liver cells. Methods Cell Biol. 13, 29–83 (1976). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and SI Appendix.