Significance
The mTORC1 eukaryotic cell growth regulator dynamically responds to changes in environmental nutrient levels. While many upstream regulators are known to modulate mTORC1 activity, the full complement of these is unknown. Here, using a genome-wide FACS-based CRISPR-Cas9 screen, we identify almost all known positive regulators of mTORC1 as well as many others. The results of our screens highlighted the importance of mitochondrial health in the regulation of mTORC1, which led us to investigate how mitochondrial stress impinges on mTORC1 signaling. Ultimately, we found that two kinases, AMPK and HRI, together signal mitochondrial distress to mTORC1. Furthermore, full inhibition of mTORC1 by mitochondrial stress requires the ATF4-dependent up-regulation of two mTORC1 pathway inhibitors, Sestrin2 and Redd1.
Keywords: mTORC1, CRISPR-Cas9 screen, mitochondria
Abstract
In mammalian cells, nutrients and growth factors signal through an array of upstream proteins to regulate the mTORC1 growth control pathway. Because the full complement of these proteins has not been systematically identified, we developed a FACS-based CRISPR-Cas9 genetic screening strategy to pinpoint genes that regulate mTORC1 activity. Along with almost all known positive components of the mTORC1 pathway, we identified many genes that impact mTORC1 activity, including DCAF7, CSNK2B, SRSF2, IRS4, CCDC43, and HSD17B10. Using the genome-wide screening data, we generated a focused sublibrary containing single guide RNAs (sgRNAs) targeting hundreds of genes and carried out epistasis screens in cells lacking nutrient- and stress-responsive mTORC1 modulators, including GATOR1, AMPK, GCN2, and ATF4. From these data, we pinpointed mitochondrial function as a particularly important input into mTORC1 signaling. While it is well appreciated that mitochondria signal to mTORC1, the mechanisms are not completely clear. We find that the kinases AMPK and HRI signal, with varying kinetics, mitochondrial distress to mTORC1, and that HRI acts through the ATF4-dependent up-regulation of both Sestrin2 and Redd1. Loss of both AMPK and HRI is sufficient to render mTORC1 signaling largely resistant to mitochondrial dysfunction induced by the ATP synthase inhibitor oligomycin as well as the electron transport chain inhibitors piericidin and antimycin. Taken together, our data reveal a catalog of genes that impact the mTORC1 pathway and clarify the multifaceted ways in which mTORC1 senses mitochondrial dysfunction.
The mechanistic target of rapamycin complex 1 (mTORC1) is a eukaryotic cell growth regulator that responds to nutrient and growth factor availability. Under nutrient-replete conditions, mTORC1 licenses anabolic processes while inhibiting catabolic ones. Given the myriad of stimuli that mTORC1 responds to, it is no surprise that a diverse set of proteins, many as part of large complexes, act in a coordinated manner to regulate mTORC1 activity.
The heterodimeric Rag GTPases (RagA/B and RagC/D) play a central role in the control of mTORC1 by nutrients. In response to amino acids, as well as glucose and cholesterol, GTP-bound RagA/B and GDP-bound RagC/D mediate the recruitment of mTORC1 to the lysosomal surface (1–5). Once at the lysosome, GTP-bound Rheb, which is under the control of growth factors through the TSC complex pathway, binds to mTORC1 and stimulates its kinase activity (6–13). Together, Rheb and the Rags form a GTPase-based coincidence detector at the lysosomal surface that ensures that mTORC1 becomes activated only when nutrient and growth factor conditions are optimal. Given that the Rag and Rheb GTPases are central arbiters of mTORC1 activation, the regulation of their respective nucleotide states is of great interest.
Dozens of proteins have been shown to modulate mTORC1 activity, many acting indirectly through one of several key effectors. However, the relative contributions of these proteins to the regulation of mTORC1 activity has not been systematically interrogated. Additionally, the majority of the proteins that regulate mTORC1 were identified using proteomic approaches. While fruitful, these studies leave open the possibility that proteins that play a role in mTORC1 regulation through transient or indirect interactions with pathway components, or are not easily detected with mass spectrometry-based proteomics, have not been identified.
Advances in CRISPR-Cas9-based screening have generated large catalogs of gene essentiality data in numerous cell lines, which can be leveraged to identify genes that are coessential with those encoding components of the mTORC1 pathway (14, 15). Though this type of analysis reveals many established mTORC1 regulators, a caveat is that it relies on cell fitness rather than mTORC1 activity as a readout. A recent study utilized a gene-trap approach to identify mTORC1 regulators in haploid cells and define new relationships among established components (16). The CRISPR-screening strategy we present here expands this toolbox by enabling screening in a large set of genetically diverse cell lines of different lineages and allows for the identification of genes that regulate mTORC1 signaling but whose loss is not tolerated long term. We carried out a genome-wide CRISPR-Cas9 screen and a series of focused sublibrary screens to identify positive regulators of mTORC1. The hits from these screens ultimately led us to study how mTORC1 senses mitochondrial dysfunction. We find that two kinases (AMPK and HRI) act in a coordinated fashion to mediate the inhibition of mTORC1 caused by mitochondrial stress.
Results
A FACS-Based Genetic Screen to Identify mTORC1 Regulators.
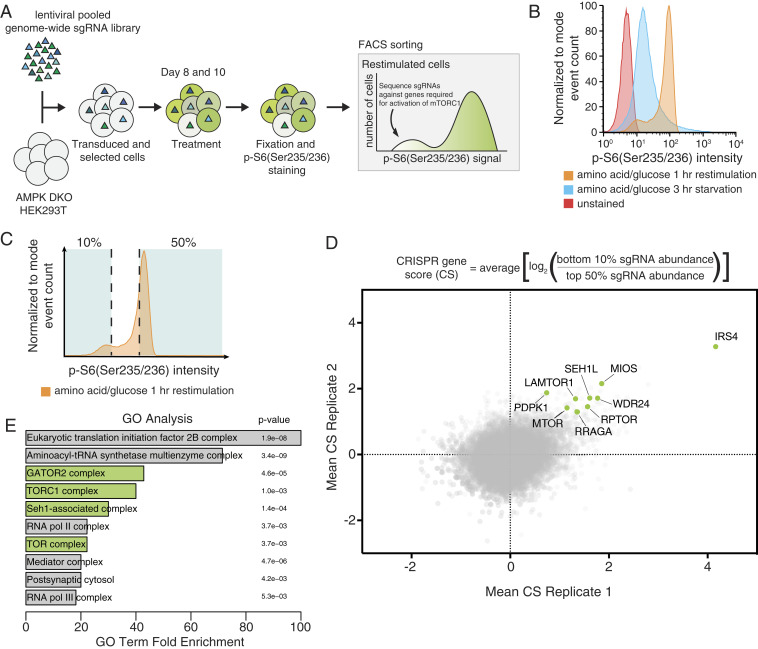
To identify genes that contribute to mTORC1 activation, we devised a fluorescence-activated cell sorting (FACS)-based CRISPR-Cas9 screening strategy that uses the phosphorylation of rpS6, a well-established marker of mTORC1 activity, as a readout (Fig. 1A). We chose the HEK293T cell line for the screen because of its robust regulation of mTORC1 activity in response to amino acids and glucose starvation and restimulation (Fig. 1B) and undertook initial screens in cells lacking both AMPKα1 and AMPKα2 (hereafter AMPK double knockout [DKO] cells) in order to more easily identify regulators that act independently of energetic stress (3). Generation of high-quality screening data from this strategy required careful consideration of downstream processing steps, so cell fixation, immunostaining, and DNA extraction protocols were extensively optimized.
Fig. 1.
CRISPR-Cas9 screen identifies positive regulators of mTORC1. (A) Schematic of the CRISPR-Cas9 FACS-based genome-wide screen. (B) Representative flow cytometry histogram of wild-type HEK293T cells not exposed to the primary antibody (red), or immunostained after starvation of amino acids and glucose (blue), or starved of and restimulated with both (orange). (C) Schematic for FACS collection on a representative starved and restimulated sample of cells. (D) Equation for determining CRISPR scores (CSs). Known mTORC1 regulators are highlighted in green. Positive CRISPR scores indicate genes that when lost prevented cells from fully reactivating mTORC1 upon combined glucose and amino acid restimulation as readout by p-S6 levels. Mean CS from two biological replicates of genome-wide screens in AMPK DKO HEK293T cells. (E) GO analysis shows enrichment of mTORC1-related complexes in an unbiased manner. Analysis was performed on the 106 top scoring genes, which had an FDR of <0.05.
AMPK DKO HEK293T cells were transduced with a lentiviral single guide RNA (sgRNA) library targeting ∼18,000 genes and screened 8 and 10 d after to minimize the loss of sgRNAs targeting essential genes (Fig. 1A). After immunostaining with a phospho-rpS6 antibody, cells were separated using a flow cytometry sorter into two fractions: those with the 10% lowest and 50% highest phospho-rpS6 signals (Fig. 1C). For each gene, a “CRISPR score” (CS) was generated by calculating the mean log2 fold change in the abundances of sgRNAs targeting the gene between the two fractions. Replicate screens were compared directly to assess overlap and also subjected to model-based analysis of genome-wide CRISPR-Cas9 knockout (MAGeCK) (17). In this dataset, genes encoding central components of the mTORC1 pathway scored, including genes for the mTOR kinase (MTOR) and its associated protein raptor (RPTOR), along with established upstream regulators of mTORC1, including proteins in the nutrient-sensing pathway such as the RagA GTPase (RRAGA) and Ragulator and GATOR2 subunits (LAMTOR1, MIOS, SEH1L, and WDR24) (Fig. 1D). Furthermore, gene ontology (GO) analysis confirmed in an unbiased fashion that the dataset is enriched for gene sets related to mTORC1 and amino acid signaling (Fig. 1E) (18). As expected because of their connection to the GCN2 pathway, which becomes activated when the levels of cytosolic uncharged tRNAs increase, genes encoding tRNA synthetases also scored highly in this analysis (19).
Overall, using a false discovery rate (FDR) cutoff of <0.1, our screening approach allowed us to identify 173 genes as potential regulators of mTORC1 activity, 77 of which have been previously defined as core fitness genes in the reference set from Hart et al. (20). This suggests that by performing the screens not long after library transduction, we were able to capture interactions between essential processes and mTORC1 signaling.
Genes Scoring in the Genome-Wide Screen Positively Regulate mTORC1 Signaling.
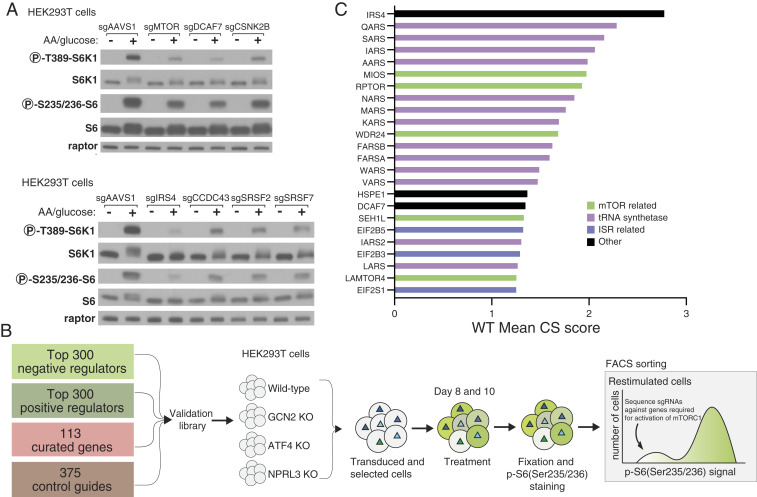
For validation of hit genes from the screen, we used CRISPR-Cas9 to generate individual cell lines, each lacking a high scoring gene with a previously unknown or underappreciated link to mTORC1 signaling. Consistent with the screen, disruption of DCAF7, CSNK2B, IRS4, CCDC43, and SRSF2/7 decreased phosphorylated rpS6 without impacting total rpS6 levels. To assess whether these genes act upstream of mTORC1, rather than impacting rpS6 phosphorylation through another mechanism, such as regulating an rpS6 phosphatase, we also examined the phosphorylation of S6 Kinase 1 (S6K1), which is a direct substrate of the mTORC1 kinase. Gratifyingly, when compared to the AAVS1 control sgRNA, loss of the hit genes decreased the phosphorylation of S6K1, suggesting that they likely act upstream of mTORC1 activity (Fig. 2A).
Fig. 2.
Generation of focused sublibrary and validation of individual gene hits. (A) Validation of select genes from screen. Immunoblot analysis shows mTORC1 signaling is blunted, as detected by decreased phosphorylation of the direct mTORC1 substrate S6K1, in HEK293T cells expressing sgRNAs targeting indicated genes. Signaling was assayed 8 d posttransduction with the indicated sgRNA as described for the primary screen. (B) Categories of genes included in the focused sublibrary along with a schematic of how the focused screens were performed. (C) CS scores for the 24 top scoring genes from a focused sublibrary screen in wild-type HEK293T cells.
In order to validate hits from the primary screen en masse, we generated a focused sublibrary of sgRNAs targeting ∼700 genes identified from 1) the positive regulator screen described here, 2) prior screens aimed at discovering negative regulators of the mTORC1 pathway, and 3) known regulators of the mTORC1 pathway that eluded detection in both screening strategies mentioned above (Fig. 2B). Screens with the focused sublibrary in wild-type (WT) HEK293T cells gave results that largely recapitulated those from the primary screens performed in the AMPK DKO cells, indicating that the screening strategy is robust and that the hits are not dependent on the loss of AMPK. Importantly, genes encoding known positive regulators of the mTORC1 pathway, including MIOS, RPTOR, WDR24, SEH1L, LAMTOR2/4, RHEB, RRAGA, and MTOR, all score within the top 24 genes (Fig. 2C).
Focused Sublibrary Screens Define Epistatic Relationships.
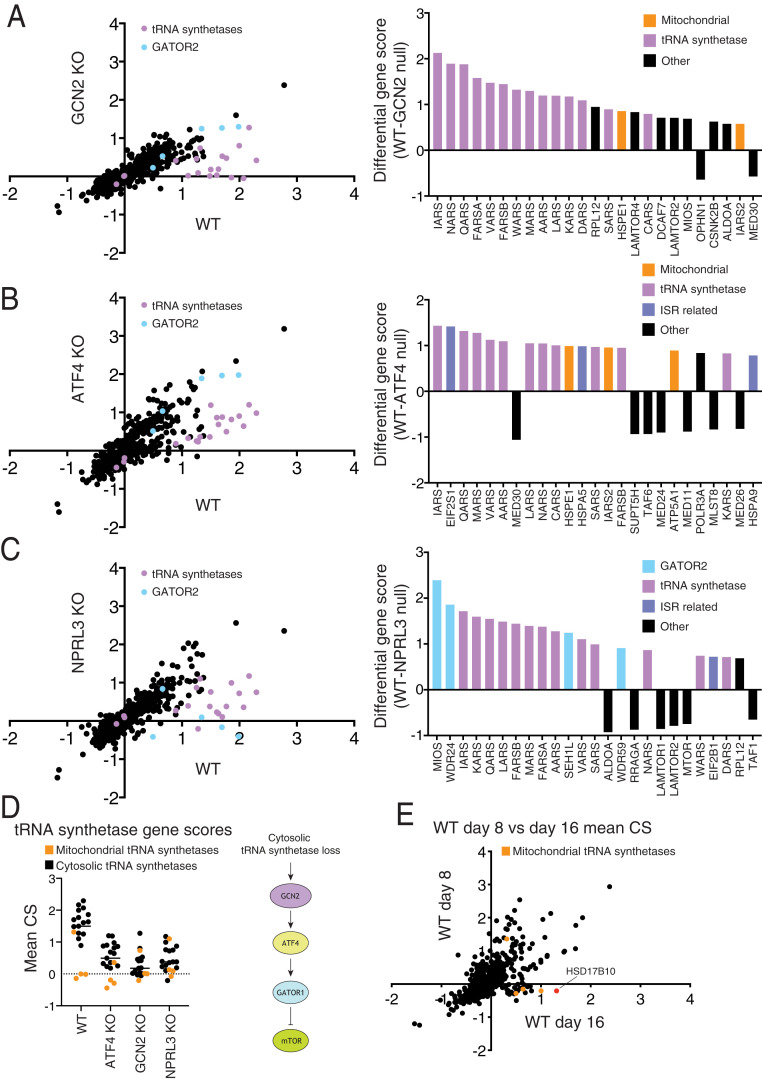
In small-scale screens using the focused sublibrary, we asked whether hit genes impact mTORC1 through the integrated stress response (ISR) or the Rag-based amino acid nutrient-sensing pathway. To interrogate the role of the ISR, we used HEK293T cells lacking the GCN2 kinase, which is activated by uncharged tRNAs that accumulate upon amino acid starvation, or the ATF4 transcription factor, which can be induced by a number of kinases, including GCN2 (19, 21–24). To define genes acting through the Rag pathway, we generated HEK293T cells deficient in NPRL3, a component of GATOR1, a negative regulator of RagA/B whose loss renders mTORC1 signaling insensitive to amino acid or glucose starvation (25–27).
Comparisons of hits from sublibrary screens in wild-type HEK293T cells and those deficient in GCN2 or ATF4 showed that many of the tRNA synthetase genes did not score in the GCN2 or ATF4 KO cells (Fig. 3 A and B), supporting a previous report that defects in tRNA charging impinge upon mTORC1 through GCN2 and ATF4 (28). Screens in the NPRL3-null HEK293T cells revealed two classes of genes that require GATOR1 to impact mTORC1 (Fig. 3C). The first consists of components of GATOR2, a known positive regulator of the pathway that likely acts upstream of GATOR1 to inhibit its function (25). The second consists of the aforementioned tRNA synthetases, supporting a previously proposed model in which defects in tRNA charging, drive ATF4-dependent up-regulation of Sestrin2, an inhibitor of GATOR2 (28–31).
Fig. 3.
Focused sublibrary screens in cells deficient in known stress and amino acid sensing pathways. (A–C) Comparisons of CRISPR scores (CSs) from focused sublibrary screens in wild-type HEK293T to those in cells lacking GCN2, ATF4, or NPRL3. The top 25 differential CSs for each screening pair are included in an adjacent bar graph. The tRNA synthetase genes targeted in the library are highlighted in purple. Genes encoding GATOR2 components are highlighted in light blue. (D) Scores for tRNA synthetase genes present in the focused sublibrary in screens across all indicated cell lines. (E) Focused screens performed at 8 versus 16 d after transduction, highlighting genes whose scores are time dependent.
Given that a variety of mitochondrial electron transport chain (ETC) and adenosine triphosphate (ATP) synthase inhibitors are known to dampen mTORC1 signaling (32–39), it was surprising that only the sgRNAs targeting the cytosolic, but not mitochondrial, tRNA synthetases scored as inhibiting mTORC1 (Fig. 3D). A potential explanation for this discrepancy is that it takes a substantial amount of time for a defect in mitochondrial translation to reduce mitochondrially encoded components of the electron transport chain to levels sufficient to cause mitochondrial dysfunction. Indeed, in focused sublibrary screens performed at longer time points after sgRNA transduction (16 d), the sgRNAs targeting the mitochondrial tRNA synthetase genes did score, as did sgRNAs targeting the HSD17B10 gene, which encodes a key component of the mitochondrial RNase P complex required for tRNA processing (Fig. 3E) (40–42).
Disruption of Mitochondrial Function Inhibits mTORC1 Signaling through AMPK and ATF4.
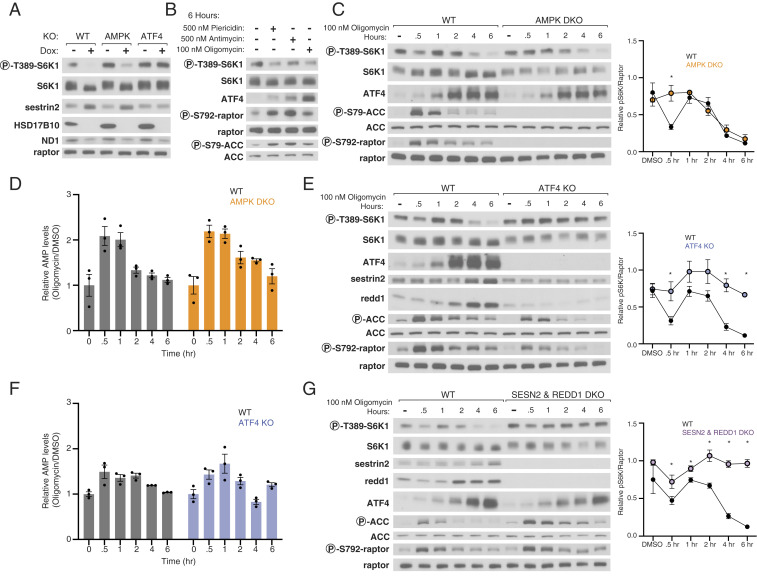
Given that HSD17B10 was among the strongest hits from our focused screens, we asked how its loss might inhibit mTORC1. To validate it, we developed a conditional knockout system, in which doxycycline (dox) suppressed the expression of a cDNA encoding HSD17B10 in HEK293T cells lacking the endogenous HSD17B10 gene (HSD17B10 dox-off cells). Gratifyingly, suppression of HSD17B10 strongly reduced mTORC1 signaling as readout by S6K1 phosphorylation (Fig. 4A). As expected, given its function, its loss also substantially decreased the levels of MT-ND1, which is encoded by the mitochondrial genome and thus depends on mitochondrial translation for its expression (Fig. 4A). In ATF4 KO but not AMPK DKO cells, mTORC1 was completely resistant to HSD17B10 loss (Fig. 4A), suggesting that chronic inhibition of mitochondrial protein synthesis suppresses mTORC1 signaling through ATF4 via the ISR.
Fig. 4.
AMPK and ATF4 signal mitochondrial distress to mTORC1. (A) Loss of HSD17B10 inhibits mTORC1 activation by amino acids and glucose in an ATF4-dependent manner. HSD17B10 knockout HEK293T cells with dox-off conditional HSD17B10 expression were cultured with or without doxycycline (dox) for 8 d and then starved of amino acids and glucose for 1 h and restimulated with both for 30 min. Immunoblot analyses of mTORC1 signaling and HSD17B10 expression. ND1 levels are expected to decrease upon loss of mitochondrial translation. (B) Inhibition of complex I, complex III, or the ATP synthase suppresses mTORC1 signaling. mTORC1 activity was assayed in response to a 6-h treatment with vehicle (DMSO), piericidin (500 nM), antimycin (500 nM), or oligomycin (100 nM). (C) AMPK mediates the first phase of mTORC1 inhibition in response to mitochondrial distress. Immunoblot analyses of mTORC1 signaling over the course of a 6-h treatment with 100 nM oligomycin in wild-type and AMPK DKO HEK293T cells. The levels of S6K1 phosphorylation were quantified using densitometry with ImageJ. Values shown are the mean ± SEM for n = 3 biologically independent experiments. P values were determined using a two-sided Student’s t test. *P < 0.05 (D) AMP levels increase acutely upon treatment with oligomycin and subsequently return to baseline levels. Metabolite extracts were analyzed by LC-MS and relative AMP levels are shown as the mean ± SEM for n = 3 biologically independent experiments. (E and F) ATF4 mediates the second phase of mTORC1 signaling in response to mitochondrial distress. Time course experiments were performed and analyzed as in C and D. (G) Sestrin2 and Redd1 are required for the ISR to inhibit mTORC1 signaling in response to mitochondrial distress. Time course experiments were performed and analyzed as in C.
Because mitochondrially encoded proteins are components of the ETC, we asked how its acute inhibition impacts mTORC1 signaling. We treated wild-type HEK293T cells with an inhibitor of complex I (piericidin), complex III (antimycin), or ATP synthase (oligomycin) for 6 h. As expected, these treatments reduced the rate of mitochondrial oxygen consumption (SI Appendix, Fig. S1). The inhibitors also dampened mTORC1 signaling, activated AMPK, and induced ATF4 expression, with piericidin and antimycin having milder effects than oligomycin (Fig. 4B). Given this, we carried out future experiments with oligomycin.
Over a 6-h time course experiment in wild-type cells, oligomycin inhibited mTORC1 signaling in two distinct phases. A decrease in mTORC1 signaling first occurred after 30 min of oligomycin treatment, which correlated with an increase in AMP levels as well as AMPK activity, as detected by the phosphorylation of its substrates ACC and Raptor (Fig. 4 C and D). After 1 h of oligomycin treatment, mTORC1 signaling was restored to pretreatment levels, but starting at 2 to 4 h after oligomycin addition, mTORC1 was again inhibited but in this case in the absence of a substantial increase in AMP levels. Instead, at these later time points, the inhibition of mTORC1 correlated with an induction of the ISR as detected by an increase in ATF4 expression (Fig. 4C).
To understand the contributions of AMPK and ATF4 to the oligomycin-induced inhibition of mTORC1, we carried out oligomycin treatment time course experiments in the AMPK DKO and ATF4 KO cells. Although, at short time points AMP levels increased in both WT and AMPK DKO cells (Fig. 4D), in the absence of AMPK, mTORC1 signaling was not inhibited 30 min after addition of oligomycin (Fig. 4C). However, mTORC1 signaling in AMPK DKO cells remained sensitive to inhibition at longer time points (Fig. 4C), suggesting an AMPK-independent response was being activated.
In cells lacking ATF4, oligomycin did not inhibit mTORC1 signaling at either short or long time points after its addition (Fig. 4E). Indeed, in these cells oligomycin mildly increased mTORC1 signaling at the shorter time points, perhaps explaining why the AMPK-dependent inhibition of mTORC1 seen at 30 min after oligomycin addition in wild-type cells was absent in ATF4 KO cells. Importantly, loss of ATF4 did not greatly alter the impact of oligomycin on AMP levels or the phosphorylation of AMPK substrates (Fig. 4 E and F). Thus, inhibition of the ATP synthase with oligomycin treatment inhibits mTORC1 in two phases, the first of which is dependent on AMPK and the second on ATF4.
Both Sestrin2 and Redd1 Are Required for Oligomycin to Inhibit mTORC1 Signaling.
Given the critical role of ATF4 in mediating the inhibition of mTORC1 caused by oligomycin as well as loss of HSD17B10, we sought to understand the downstream mechanisms through which it acts. Mining of RNA sequencing (RNAseq) datasets from cells with activated ATF4 revealed increases in the mRNAs for Sestrin2 and Redd1 (DDIT4), which encoded two well-known repressors of mTORC1 signaling (43–45). Sestrin2 is a leucine sensor that inhibits Rag-based nutrient signaling to mTORC1 by binding to GATOR2, while Redd1 inhibits growth factor signaling to mTORC1 via the tuberous sclerosis complex (TSC) pathway (29, 30, 46, 47). It is well-appreciated that ISR induction via GCN2 activation up-regulates Sestrin2, and Redd1 has been shown to be up-regulated by PERK activation, which is also a component of the ISR (28, 48). In our hands, oligomycin increased Sestrin2 and Redd1 expression in a fashion that correlated with ATF4 levels and was dependent on ATF4 (Fig. 4E).
To test the roles of Sestrin2 and Redd1 in mediating the effects of oligomycin on mTORC1, we generated HEK293T cells lacking Sestrin2 or Redd1 or both. The individual loss of either Sestrin2 or Redd1 did not significantly impact the capacity of oligomycin to inhibit mTORC1 (SI Appendix, Fig. S2 A and B). In the Sestrin2 and Redd1 DKO cells (Fig. 4G), baseline mTORC1 signaling was slightly boosted, but it was still inhibited at 30 min of oligomycin treatment. At the later time points, however, loss of both Sestrin2 and Redd1, like that of ATF4, prevented oligomycin from inhibiting mTORC1. We conclude that the induction of both Sestrin2 and Redd1 is required to inhibit mTORC1 signaling in response to oligomycin treatment at long time points. Moreover, there must be additional ATF4 targets that account for the boost in mTORC1 signaling seen in the ATF4 KO cells treated at early time points with oligomycin, as this effect was not prevented by the loss of Sestrin2 and Redd1.
mTORC1 Is Largely Insensitive to Mitochondrial Dysfunction in Cells Lacking the AMPK and HRI Kinases.
We next asked how oligomycin treatment induces ATF4 and inhibits mTORC1. ATF4 is induced at the level of its translation in a fashion that depends on the phosphorylation of the α-subunit of the translation initiation factor eIF2 (49, 50). Multiple kinases can phosphorylate eIF2α, including GCN2 in response to amino acid starvation and HRI in response to mitochondrial stress caused by oligomycin (19, 43, 45). In GCN2 KO cells, oligomycin still induced ATF4 and its targets Sestrin2 and Redd1, and also inhibited mTORC1 (SI Appendix, Fig. S3). Thus, the relatively small impact that oligomycin has on levels of amino acids like aspartate (SI Appendix, Fig. S4) is likely not sufficient to suppress tRNA charging and thereby activate GCN2.
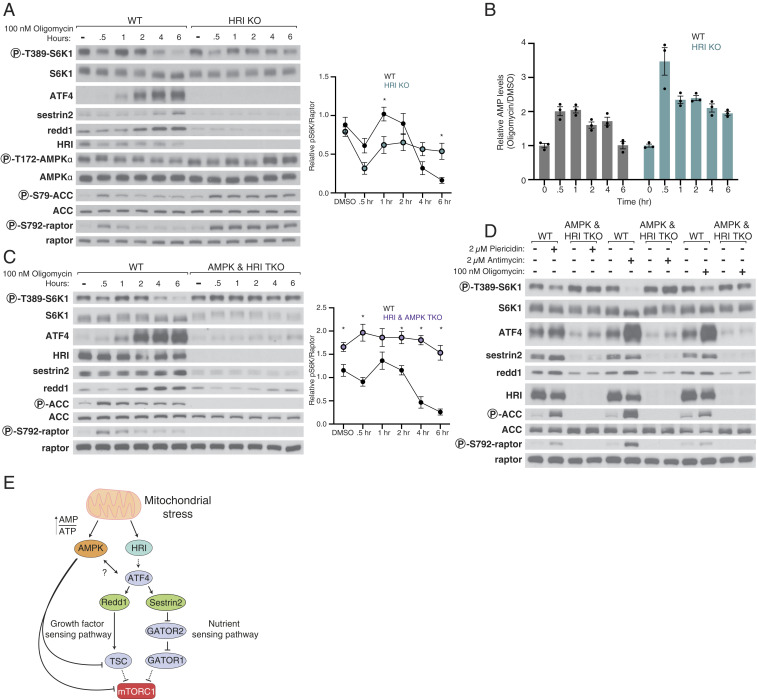
In HRI KO HEK293T cells, mTORC1 remained sensitive to oligomycin at 30 min after its addition but was largely resistant to oligomycin at the longer time points (Fig. 5A). Consistent with this observation, HRI loss also prevented induction by oligomycin of ATF4 and its targets Sestrin2 and Redd1. Interestingly, in the HRI KO HEK293T cells, oligomycin increased the phosphorylation of AMPK itself and AMPK signaling to a similar extent as in wild-type cells, but unlike in the wild-type cells, where it was quickly turned off, it remained active for the duration of the time course (Fig. 5A). This sustained AMPK signaling correlated with an inability of the HRI KO cells to restore normal levels of AMP following oligomycin treatment, indicating that maintenance of energy homeostasis upon inhibition of ATP synthesis requires not only AMPK but also HRI (Fig. 5B). Consistent with our finding that two kinases, AMPK and HRI, participate in the oligomycin-dependent inhibition of mTORC1, in HEK293T cells lacking both of them, mTORC1 was almost completely resistant to oligomycin treatment (Fig. 5C). While loss of AMPK did not prevent oligomycin-induced cell death, loss of ATF4 did reduce it at 48, but not 96 h after oligomycin treatment, whether rapamycin was also added or not (SI Appendix, Fig. S5).
Fig. 5.
Together, AMPK and HRI signal mitochondrial dysfunction to mTORC1. (A) Loss of HRI prevents activation of the ISR in response to mitochondrial distress and renders mTORC1 signaling resistant to the second phase of inhibition caused by oligomycin. Cell lysates were analyzed by immunoblotting and the levels of S6K1 phosphorylation quantified as in Fig. 4. Values are mean ± SEM for n = 3 biologically independent experiments. P values were determined using a two-sided Student’s t test. (B) HRI KO HEK293T cells have increased AMPK activity, which corresponds to an increase in AMP levels upon treatment with oligomycin. Metabolite extracts were analyzed by liquid chromatography-mass spectrometry (LC-MS) and relative AMP levels are shown as the mean ± SEM for n = 3 biologically independent experiments. (C) Loss of both HRI and AMPK prevents inhibition of mTORC1 by oligomycin. Time course experiments were performed and analyzed as in A in cells lacking AMPK and HRI (AMPK and HRI TKO). (D) In HEK293T cells lacking AMPK and HRI, mTORC1 signaling is resistant to the inhibition normally caused by a 6-h treatment with piericidin, antimycin, or oligomycin. (E) Model for pathway leading to mTORC1 inhibition in response to mitochondrial distress.
Finally, we revisited how the electron transport chain inhibitors piericidin and antimycin impact mTORC1. Gratifyingly, in HEK293T cells lacking HRI and AMPK, mTORC1 signaling was largely insensitive to not only oligomycin but also piercidin and antimycin (Fig. 5D). While in these cells oligomycin no longer induced ATF4, piercidin and antimycin still caused a slight increase in ATF4 levels, suggesting that they activate another eIF2α kinase besides HRI, most likely GCN2 (51). Overall, our results show that HRI and AMPK mediate mTORC1 inhibition caused by various forms of mitochondrial dysfunction.
Discussion
Many upstream regulators control mTORC1 and facilitate its dynamic regulation by growth factors and nutrients. Given that dysregulation of the pathway is implicated in human diseases, such as cancer and neurodevelopmental disorders, there has been great interest in understanding how the pathway is regulated (52). The FACS-based CRISPR screens described here provide a systematic interrogation of the mTORC1 pathway in a human cell line in response to changes in nutrient levels. The genes identified in these screens will serve as a resource for future work and complement existing protein–protein interaction datasets.
In addition to identifying known positive regulators, we validated that the suppression of a number of genes not previously connected to mTORC1 can inhibit its activity, and these genes serve as leads for future investigation. For example, how does loss of SRSF2 or SRSF7, which encode splicing factors, impact mTORC1 signaling? A member of the same splicing factor family, SRSF1, has been previously implicated in mTORC1 signaling, but exactly how remains unclear (53). Other gene products that we identified, such as the kinase CSNK2B, might play roles in signaling cascades not previously connected to mTORC1 regulation. Our finding that loss of IRS4 strongly inhibits mTORC1 activity is consistent with IRS4, rather than IRS1/2, being the main transducer of insulin signaling in HEK293-based cell lines, as previously reported (54). It is interesting to consider whether its tissue-specific expression might differentially modulate regulation of mTORC1 by insulin in vivo. Our focused sublibrary approach allowed us to efficiently perform many screens in different genetic backgrounds and to thus systematically define epistatic relationships between hundreds of potential hit genes and core stress pathways.
Finally, our study led us to concentrate on the relationship between mitochondrial stress and mTORC1. We find that two kinases, AMPK and HRI, but not GCN2, are necessary for signaling oligomycin-induced mitochondrial dysfunction to mTORC1 (Fig. 5E). AMPK inhibits mTORC1 at short time periods after mitochondrial inhibition while HRI does so at later times, so that loss of both kinases makes mTORC1 largely refractory to the effects of oligomycin. HRI acts through the transcriptional activation of two established mTORC1 pathway inhibitors, Sestrin2 and Redd1, via the ATF4 transcription factor. Indeed, loss of Sestrin2 and Redd1 together, but not of either alone, is sufficient to render mTORC1 resistant to the long-term inhibitory effects of oligomycin. As several studies demonstrate that mTORC1 regulates ATF4 levels under nonstress conditions, our results reveal further complexity to the relationship between mTORC1 and ATF4 (44, 55). Our findings also reveal that HRI is necessary for restoring normal levels of AMP upon prolonged oligomycin treatment and thus for inactivation of AMPK. This function of HRI is unlikely to require ATF4, as in cells lacking this transcription factor, AMP levels and AMPK activity responded largely normally to oligomycin treatment. Taken in sum, our work builds upon previous evidence that mitochondria “talk to” mTORC1 to bring clarity to the mechanisms involved.
Materials and Methods
Cell Culture.
HEK293T cells were cultured in Dulbecco's modified Eagle's medium (DMEM) (Gibco) supplemented with 10% heat-inactivated fetal calf serum (FCS) (Gibco) and penicillin-streptomycin. Cell lines were maintained at 37 °C and 5% CO2. The mitochondrial response to stress is dependent on the cellular metabolomic state, so care should be taken to ensure cells are not overgrown or nutrient starved ahead of experiments. All cell lines were obtained from American Type Culture Collection (ATCC) and tested for mycoplasma.
Antibodies.
The raptor antibody was from EMD Millipore (09-217); S6K1 pT389 (9234), S6K1 (2708), rpS6 pS235/S236 (2211), rpS6 (2217), Sestrin-2 (8487), ATF-4 (11815), raptor pS792 (2083), acetyl-CoA carboxylase (ACC) (3662), ACC pS79 (3661), AMPKα (5831), AMPKα pT172 (2535), and GCN2 (3302) antibodies were from Cell Signaling Technology; ND1 (19703-1-AP), Sestrin2 (10795-1-AP), and Redd1 (10638-1-AP) antibodies were from Proteintech; HRI (MBS2538144) antibody was from MyBioSource; and the HSD17B10 (TA500724) antibody was from Life Technologies.
sgRNA Cloning, Lentiviral Production, and Lentiviral Transduction.
Individual sgRNAs (SI Appendix, Dataset S3) were cloned into lentiCRISPRv2-Opti (Addgene #163126) at the BsmBI site as described by the depositor. Virus was generated as in ref. 56. Briefly, 750,000 HEK293T cells were plated in DMEM supplemented with 20% FCS. At 12 h postseeding, VSV-G envelope (Addgene #8454) and psPAX2 (Addgene #12260) were cotransfected with lentiCRISPRv2-Opti containing the indicated sgRNA sequences or pCW57 with the indicated cDNAs with XTremeGene 9 Transfection Reagent (Roche). After 12 h, the culture medium was refreshed. At 28 h posttransfection, supernatant-containing virus was collected and passed through a 0.45-μm filter. Virus was stored at −80 °C until use. For lentiviral transduction, cells were spinfected at 1,200 × g for 45 min at 37 °C with 8 μg/mL polybrene and virus-containing medium. After 12 h, the culture medium was refreshed, and after 24 h, cells were selected with puromycin or blasticidin for 4 d.
Pooled Genome-Wide CRISPR-Cas9 Screens.
The pooled genome-wide lentiviral sgRNA library (Addgene #1000000100) was prepared and screens were performed as described in ref. 57 with slight modifications. Briefly, 600 million HEK293T cells were transduced with the viral pool to achieve 1,000-fold library coverage after puromycin selection. After 72 h of puromycin selection, 200 million cells were passaged and expanded every 2 d until day 7 when 1 billion cells were plated into fibronectin-coated 15-cm dishes. On days 8 and 10, the cells were washed three times with phosphate-buffered saline (PBS) supplemented with 1 mM CaCl2 and 500 μM MgCl2 and placed in RPMI lacking amino acids and glucose for 3 h. After 3 h, amino acids and glucose were added for 1 h and the cells harvested. Cells were collected by washing once with ice-cold PBS, after which Accumax solution (Sigma) was added, and plates were nutated at 4 °C for 10 min. After 10 min, cells were collected into a 50-mL centrifuge tube (Celltreat) and pelleted via centrifugation at 4 °C at 300 × g for 3 min. Subsequently, cells were fixed by resuspension in formalin for 5 min at room temperature. At 5 min cells were pelleted via centrifugation at 4 °C at 300 × g for 3 min. Finally, cells were permeabilized by resuspension in 90% methanol:PBS. After permeabilization cells were stored at −20 °C until staining.
Cell Staining and FACS.
A total of 500 million previously fixed and permeabilized cells were pelleted by centrifugation at 4 °C at 300 × g for 3 min and then washed once with ice-cold PBS. Cells were then blocked with 5% bovine serum albumin (BSA) in PBS for 1 h at room temperature. For primary antibody staining, cells were incubated with primary antibody diluted (1:100) in 0.05% BSA in PBS at room temperature for 1 h. After one wash with 0.05% BSA in PBS, cells were resuspended in secondary antibody diluted (1:200) in 0.05% BSA in PBS and incubated for 1 h. Finally, cells were washed twice in 0.05% BSA in PBS and filtered through a cell strainer prior to FACS sorting. Cells were sorted using a BD FACSAria III. Gates were drawn to collect the low intensity fraction at 10% and the high intensity fraction at 50%. Sorting was preformed until ∼50 million cells were obtained for the lower fraction. Data were analyzed using FlowJo software (TreeStar).
Analyses of CRISPR-Cas9 Screens.
Genomic DNA was extracted from sorted cells by first decross-linking cells in a solution of 10 mM Tris pH 7.5 and 10 mg/mL Proteinase K (Roche) in PBS at 55 °C for 24 h. Subsequently, the Qiagen QIAmp DNA Blood Maxi kit was used according to the manufacturer’s instructions. High-throughput sequencing libraries were prepared as described (58, 59). Sequencing reads were aligned to the sgRNA library and quantified. A pseudo count was added to each value and the log2-transformed fold change in abundance was then calculated between low and high fractions. MaGeCK analysis was performed as in ref. 60 and ref. 17.
Focused sgRNA Sublibrary Screens.
The focused sublibrary (SI Appendix, Dataset S1, Addgene #164084) was designed by selecting the top scoring genes in the positive regulator screen described here and prior screens aimed at discovering negative regulators of the mTORC1 pathway. Additionally, a curated list of known mTORC1-related genes and control sgRNAs were included. Ten sgRNAs per gene were selected from Addgene #1000000100 along with 375 control sgRNAs that were selected randomly from this library. The oligonucleotide pool was synthesized by Agilent Technologies and cloned as described in ref. 59. Focused sublibrary screens were carried out as described for genome-wide screens with modifications. Briefly, 60 million wild-type or indicated knockout HEK293T cells were transduced with the viral pool to achieve 1,000-fold library coverage after puromycin selection. After 72 h of puromycin selection, 20 million cells were passaged and expanded every 2 d until day 7, when 100 million cells were plated into fibronectin-coated 15-cm dishes. Screen replicates were analyzed by averaging high and low ratios and then calculating mean CS.
GO Term Enrichment.
GO term enrichment was performed in R using the topGO R package (18). Top scoring positive regulator genes with an FDR less than 5% from MAGeCK analysis were compared to all genes in the genome-wide library using the classic Fisher statistic from the topGO package for GO terms in the cellular component category. GO terms with fewer than three members were filtered out by setting the node size to 3. Terms with P values greater than 0.01 were filtered out and then ranked by fold enrichment.
Starvation, Drug Treatments, and Time Course Experiments.
Cells were seeded in DMEM culture medium at 720,000 cells per well in a fibronectin-coated six-well plate the day before an experiment. For nutrient starvation experiments, cells were washed once with RPMI lacking amino acids and glucose and then starved in 2 mL of the same media. After 1 h, amino acids and glucose were added back for 30 min at which point cells were harvested and lysed. For the drug treatment experiments, cells were washed three times with PBS and then treated for 6 h in RPMI containing dimethylsulfoxide (DMSO), oligomycin (100 nM) (Sigma), antimycin (500 nM or 2 μM) (Sigma), or piericidin (500 nM or 2 μM) (Sigma). Thorough washing of cells cultured in DMEM culture medium prior to treatment with ETC inhibitors in RPMI is important for obtaining consistent inhibition of mTORC1 by the inhibitors. For time course experiments, RPMI containing DMSO or oligomycin (100 nM) was prepared and medium was refreshed at the indicated time points. Cells at all times were collected simultaneously.
Cell Lysis.
Cells were briefly washed with ice-cold PBS and then scraped into a Triton X-100-based lysis buffer (1% Triton X-100, 10 mM beta-glycerol phosphate, 10 mM pyrophosphate, 40 mM Hepes pH 7.4, 2.5 mM MgCl2 with 1 tablet of ethylenediaminetraacetic acid (EDTA)-free protease inhibitor [Roche] per 50 mL buffer). Cell lysates were clarified by centrifugation at 17,000 × g at 4 °C for 10 min.
Generation of the Dox-Off and KO HEK293T-Based Cell Lines.
The cDNA for human HSD17B10 was codon optimized and synthesized by IDT as a gBlock gene fragment. This gene fragment was then cloned into the pCW57.kc1 (Addgene #164076) vector using NEBuilder HiFi DNA Assembly (NEB). Cells were transduced with lentivirus generated with the pCW57.kc1 vector and selected with blasticidin for 3 d. Subsequently, KO cells were generated using either px330 (Addgene #42230) or lentiCRISPRv2-Opti (Addgene #163126). After transfection as described in ref. 3 or transduction, cells were selected with puromycin for 4 d and then allowed to recover for 7 d. Cells were then sorted by a flow cytometer into 96-well plates containing 200 μL DMEM (Gibco) supplemented with 30% heat-inactivated FCS (Gibco) and penicillin-streptomycin in each well. Clones were screened for KO by immunoblotting for proteins encoded by targeted genes. Double and triple KO cell lines were generated with the same process, beginning with SESN2 KO or AMPK DKO. AMPK DKO, SESN2 KO, and ATF4 KO HEK293T cells were generated as described in refs. 3, 30, and 44, respectively.
Treatment with Doxycycline.
Doxycycline was prepared as a stock solution (30 μg/mL), and stored at −80 °C. Cells were cultured in DMEM (Gibco) supplemented with 10% heat-inactivated FCS (Gibco) and penicillin-streptomycin and 30 ng/mL doxycycline for 10 d prior to experiments.
Western Blot Quantifications.
Western blot band intensities were measured using ImageJ. Average pixel intensities were collected over equal area boxes for bands and nearby areas without bands as background controls. Pixel densities were then inverted by subtracting 255 from all values. Background controls were then subtracted from corresponding band intensities. Finally, net pS6K values were divided by net raptor values. Because the mobility of S6K changes depending on its phosphorylation state, which can confound analysis, we chose to normalize pS6K levels to the raptor loading control.
Oxygen Consumption Rates.
Oxygen consumption rates (OCRs) were measured using a Seahorse XFe96 Analyzer (Agilent). A total of 25,000 cells were seeded 18 h prior to the experiment in fibronectin-coated Seahorse XFe96 cell culture plates. One hour prior to the experiment, cells were incubated in Seahorse Media (Agilent) supplemented with 10 mM glucose, 2 mM glutamine, and 1 mM pyruvate containing vehicle (DMSO), oligomycin (1 μM), antimycin (500 nM), or piericidin (500 nM). Three OCR measurements were taken for each treatment.
Metabolomics.
Cells were washed twice with ice cold PBS and extracted on dry ice in 0.8 mL 80% methanol containing 500 nM internal standards (Metabolomics Amino Acid Mix Standard: Cambridge Isotope Laboratories, Inc.). Cell extracts were collected using a cell scraper and transferred to a microcentrifuge tube. Samples were vortexed for 10 min at 4 °C and centrifuged at 17,000 × g for 10 min at 4 °C. Supernatants were transferred to new microcentrifuge tubes and evaporated to dryness by vacuum centrifugation. Dried polar extracts were stored at −80 °C until analysis. Polar metabolite profiling was performed as described in ref. 3.
Cell Viability.
Cells were seeded at 750,000 cells per 10-cm dish and refreshed 12 h later with RPMI medium (Gibco) supplemented with 10% dialyzed heat-inactivated FCS (Gibco) and penicillin-streptomycin containing vehicle (DMSO), oligomycin (100 nM), or oligomycin (100 nM) plus rapamycin (20 nM). At 48 or 96 h after, cells were collected, washed once with PBS, and stained with Propidium Iodide ReadyProbes reagent (Thermo Fisher Scientific). Data were collected on a BD LSRFortessa-HTS and analyzed using FlowJo software (TreeStar).
Supplementary Material
Acknowledgments
We thank all members of the D.M.S. laboratory for helpful insights; G. Liu and P. Rosen for critical reading of the manuscript; C. Thoreen (Yale University) for the gift of the ATF4-null HEK293T cell line; and P. Thiru for help with MAGeCK analysis. We thank the Metabolite Profiling Core Facility at the Whitehead Institute for running metabolomics samples and for data analysis. This work was supported by grants to D.M.S. from the NIH (R01 CA103866, R01 CA129105, and R01 AI047389). K.J.C. was supported by an Massachusetts Institute of Technology (MIT) School of Science fellowship in cancer research and an NSF fellowship (2016197106). J.M.O. was supported by a fellowship (F30CA210373) from the National Cancer Institute (NCI) and the Harvard–MIT Medical Student Training Program (MSTP) training grant (T32GM007753) from the National Institute of General Medical Sciences. C.H.A. had fellowship support from the NIH (NRSA F31 CA228241-01). J.B.S. was funded by a NCI F99/K00 predoctoral to postdoctoral transition fellowship (K00CA234839). J.M.R. was supported by a fellowship grant from the NCI of the NIH (F31CA232355). D.M.S. is an investigator of the Howard Hughes Medical Institute and an American Cancer Society research professor.
Footnotes
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2022120118/-/DCSupplemental.
Data Availability.
All study data are included in the article and/or supporting information.
References
- 1.Sancak Y., et al. , The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science 320, 1496–1501 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Efeyan A., et al. , Regulation of mTORC1 by the Rag GTPases is necessary for neonatal autophagy and survival. Nature 493, 679–683 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Orozco J. M., et al. , Dihydroxyacetone phosphate signals glucose availability to mTORC1. Nat. Metab. 2, 893–901 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Castellano B. M., et al. , Lysosomal cholesterol activates mTORC1 via an SLC38A9-Niemann-Pick C1 signaling complex. Science 355, 1306–1311 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sancak Y., et al. , Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell 141, 290–303 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Inoki K., Li Y., Xu T., Guan K. L., Rheb GTPase is a direct target of TSC2 GAP activity and regulates mTOR signaling. Genes Dev. 17, 1829–1834 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Inoki K., Li Y., Zhu T., Wu J., Guan K. L., TSC2 is phosphorylated and inhibited by Akt and suppresses mTOR signalling. Nat. Cell Biol. 4, 648–657 (2002). [DOI] [PubMed] [Google Scholar]
- 8.Long X., Lin Y., Ortiz-Vega S., Yonezawa K., Avruch J., Rheb binds and regulates the mTOR kinase. Curr. Biol. 15, 702–713 (2005). [DOI] [PubMed] [Google Scholar]
- 9.Menon S., et al. , Spatial control of the TSC complex integrates insulin and nutrient regulation of mTORC1 at the lysosome. Cell 156, 771–785 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tee A. R., et al. , Tuberous sclerosis complex-1 and -2 gene products function together to inhibit mammalian target of rapamycin (mTOR)-mediated downstream signaling. Proc. Natl. Acad. Sci. U.S.A. 99, 13571–13576 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tee A. R., Manning B. D., Roux P. P., Cantley L. C., Blenis J., Tuberous sclerosis complex gene products, Tuberin and Hamartin, control mTOR signaling by acting as a GTPase-activating protein complex toward Rheb. Curr. Biol. 13, 1259–1268 (2003). [DOI] [PubMed] [Google Scholar]
- 12.Li Y., Inoki K., Guan K. L., Biochemical and functional characterizations of small GTPase Rheb and TSC2 GAP activity. Mol. Cell. Biol. 24, 7965–7975 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Manning B. D., Tee A. R., Logsdon M. N., Blenis J., Cantley L. C., Identification of the tuberous sclerosis complex-2 tumor suppressor gene product tuberin as a target of the phosphoinositide 3-kinase/akt pathway. Mol. Cell 10, 151–162 (2002). [DOI] [PubMed] [Google Scholar]
- 14.Kim E., et al. , A network of human functional gene interactions from knockout fitness screens in cancer cells. Life Sci. Alliance 2, e201800278 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tsherniak A., et al. , Defining a cancer dependency map. Cell 170, 564–576.e16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.De Zan E., et al. , Quantitative genetic screening reveals a Ragulator-FLCN feedback loop that regulates the mTORC1 pathway. Sci. Signal. 13, eaba5665 (2020). [DOI] [PubMed] [Google Scholar]
- 17.Li W., et al. , MAGeCK enables robust identification of essential genes from genome-scale CRISPR/Cas9 knockout screens. Genome Biol. 15, 554 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rahnenfuhrer J., Alexa A., topGO: Enrichment Analysis for Gene Ontology (R package version 2.40.0, Bioconductor, 2020). DOI: 10.18129/B9.bioc.topGO. [DOI]
- 19.Inglis A. J., et al. , Activation of GCN2 by the ribosomal P-stalk. Proc. Natl. Acad. Sci. U.S.A. 116, 4946–4954 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hart T., et al. , High-resolution CRISPR screens reveal fitness genes and genotype-specific cancer liabilities. Cell 163, 1515–1526 (2015). [DOI] [PubMed] [Google Scholar]
- 21.Costa-Mattioli M., et al. , Translational control of hippocampal synaptic plasticity and memory by the eIF2alpha kinase GCN2. Nature 436, 1166–1173 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harding H. P., et al. , Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol. Cell 6, 1099–1108 (2000). [DOI] [PubMed] [Google Scholar]
- 23.Dever T. E., et al. , Phosphorylation of initiation factor 2 alpha by protein kinase GCN2 mediates gene-specific translational control of GCN4 in yeast. Cell 68, 585–596 (1992). [DOI] [PubMed] [Google Scholar]
- 24.Harding H. P., et al. , The ribosomal P-stalk couples amino acid starvation to GCN2 activation in mammalian cells. eLife 8, e50149 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bar-Peled L., et al. , A Tumor suppressor complex with GAP activity for the Rag GTPases that signal amino acid sufficiency to mTORC1. Science 340, 1100–1106 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shen K., et al. , Architecture of the human GATOR1 and GATOR1-Rag GTPases complexes. Nature 556, 64–69 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shen K., Valenstein M. L., Gu X., Sabatini D. M., Arg-78 of Nprl2 catalyzes GATOR1-stimulated GTP hydrolysis by the Rag GTPases. J. Biol. Chem. 294, 2970–2975 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ye J., et al. , GCN2 sustains mTORC1 suppression upon amino acid deprivation by inducing Sestrin2. Genes Dev. 29, 2331–2336 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saxton R. A., et al. , Structural basis for leucine sensing by the Sestrin2-mTORC1 pathway. Science 351, 53–58 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wolfson R. L., et al. , Sestrin2 is a leucine sensor for the mTORC1 pathway. Science 351, 43–48 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Parmigiani A., et al. , Sestrins inhibit mTORC1 kinase activation through the GATOR complex. Cell Rep. 9, 1281–1291 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dennis P. B., et al. , Mammalian TOR: A homeostatic ATP sensor. Science 294, 1102–1105 (2001). [DOI] [PubMed] [Google Scholar]
- 33.Kalender A., et al. , Metformin, independent of AMPK, inhibits mTORC1 in a rag GTPase-dependent manner. Cell Metab. 11, 390–401 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bolster D. R., Crozier S. J., Kimball S. R., Jefferson L. S., AMP-activated protein kinase suppresses protein synthesis in rat skeletal muscle through down-regulated mammalian target of rapamycin (mTOR) signaling. J. Biol. Chem. 277, 23977–23980 (2002). [DOI] [PubMed] [Google Scholar]
- 35.Kim D. H., et al. , mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell 110, 163–175 (2002). [DOI] [PubMed] [Google Scholar]
- 36.Vander Haar E., Lee S. I., Bandhakavi S., Griffin T. J., Kim D. H., Insulin signalling to mTOR mediated by the Akt/PKB substrate PRAS40. Nat. Cell Biol. 9, 316–323 (2007). [DOI] [PubMed] [Google Scholar]
- 37.Inoki K., Zhu T., Guan K.-L., TSC2 mediates cellular energy response to control cell growth and survival. Cell 115, 577–590 (2003). [DOI] [PubMed] [Google Scholar]
- 38.Kim S. G., et al. , Metabolic stress controls mTORC1 lysosomal localization and dimerization by regulating the TTT-RUVBL1/2 complex. Mol. Cell 49, 172–185 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gwinn D. M., et al. , AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol. Cell 30, 214–226 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Oerum S., et al. , Structural insight into the human mitochondrial tRNA purine N1-methyltransferase and ribonuclease P complexes. J. Biol. Chem. 293, 12862–12876 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reinhard L., Sridhara S., Hällberg B. M., The MRPP1/MRPP2 complex is a tRNA-maturation platform in human mitochondria. Nucleic Acids Res. 45, 12469–12480 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vilardo E., et al. , A subcomplex of human mitochondrial RNase P is a bifunctional methyltransferase–extensive moonlighting in mitochondrial tRNA biogenesis. Nucleic Acids Res. 40, 11583–11593 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guo X., et al. , Mitochondrial stress is relayed to the cytosol by an OMA1-DELE1-HRI pathway. Nature 579, 427–432 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Park Y., Reyna-Neyra A., Philippe L., Thoreen C. C., mTORC1 balances cellular amino acid supply with demand for protein synthesis through post-transcriptional control of ATF4. Cell Rep. 19, 1083–1090 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fessler E., et al. , A pathway coordinated by DELE1 relays mitochondrial stress to the cytosol. Nature 579, 433–437 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brugarolas J., et al. , Regulation of mTOR function in response to hypoxia by REDD1 and the TSC1/TSC2 tumor suppressor complex. Genes Dev. 18, 2893–2904 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ellisen L. W., Growth control under stress: mTOR regulation through the REDD1-TSC pathway. Cell Cycle 4, 1500–1502 (2005). [DOI] [PubMed] [Google Scholar]
- 48.Whitney M. L., Jefferson L. S., Kimball S. R., ATF4 is necessary and sufficient for ER stress-induced upregulation of REDD1 expression. Biochem. Biophys. Res. Commun. 379, 451–455 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Costa-Mattioli M., Walter P., The integrated stress response: From mechanism to disease. Science 368, eaat5314 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pakos-Zebrucka K., et al. , The integrated stress response. EMBO Rep. 17, 1374–1395 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mick E., et al. , Distinct mitochondrial defects trigger the integrated stress response depending on the metabolic state of the cell. eLife 9, e49178 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Saxton R. A., Sabatini D. M., mTOR signaling in growth, metabolism, and disease. Cell 169, 361–371 (2017). [DOI] [PubMed] [Google Scholar]
- 53.Michlewski G., Sanford J. R., Cáceres J. F., The splicing factor SF2/ASF regulates translation initiation by enhancing phosphorylation of 4E-BP1. Mol. Cell 30, 179–189 (2008). [DOI] [PubMed] [Google Scholar]
- 54.Fantin V. R., et al. , Characterization of insulin receptor substrate 4 in human embryonic kidney 293 cells. J. Biol. Chem. 273, 10726–10732 (1998). [DOI] [PubMed] [Google Scholar]
- 55.Ben-Sahra I., Hoxhaj G., Ricoult S. J. H., Asara J. M., Manning B. D., mTORC1 induces purine synthesis through control of the mitochondrial tetrahydrofolate cycle. Science 351, 728–733 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang T., Lander E. S., Sabatini D. M., Viral packaging and cell culture for CRISPR-based screens. Cold Spring Harb. Protoc. 2016, pdb.prot090811 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang T., et al. , Identification and characterization of essential genes in the human genome. Science 350, 1096–1101 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang T., Lander E. S., Sabatini D. M., Large-scale single guide RNA library construction and use for CRISPR-Cas9-based genetic screens. Cold Spring Harb. Protoc. 2016, pdb.top086892 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang T., Lander E. S., Sabatini D. M., Single guide RNA library design and construction. Cold Spring Harb. Protoc. 2016, pdb.prot090803 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang B., et al. , Integrative analysis of pooled CRISPR genetic screens using MAGeCKFlute. Nat. Protoc. 14, 756–780 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or supporting information.