Significance
A central question in biology is whether trait differences are the result of variation in gene number, sequence, or regulation. Snake venoms are an excellent system for addressing this question because of their genetic tractability, contributions to fitness, and high evolutionary rates. We sequenced and assembled the genome of the Tiger Rattlesnake to determine whether the simplest rattlesnake venom was the product of a simple or complex genotype. The number of venom genes greatly exceeded the number of venom proteins producing the simple phenotype, indicating regulatory mechanisms were responsible for the production of the simplest, but most toxic, rattlesnake venom. We suggest that the retention of genomic complexity may be the result of shared regulatory elements among gene-family members.
Keywords: genotype–phenotype, gene regulation, methylation, chromatin
Abstract
Variation in gene regulation is ubiquitous, yet identifying the mechanisms producing such variation, especially for complex traits, is challenging. Snake venoms provide a model system for studying the phenotypic impacts of regulatory variation in complex traits because of their genetic tractability. Here, we sequence the genome of the Tiger Rattlesnake, which possesses the simplest and most toxic venom of any rattlesnake species, to determine whether the simple venom phenotype is the result of a simple genotype through gene loss or a complex genotype mediated through regulatory mechanisms. We generate the most contiguous snake-genome assembly to date and use this genome to show that gene loss, chromatin accessibility, and methylation levels all contribute to the production of the simplest, most toxic rattlesnake venom. We provide the most complete characterization of the venom gene-regulatory network to date and identify key mechanisms mediating phenotypic variation across a polygenic regulatory network.
Although nearly every cell within a eukaryotic organism possesses the same genome, variation in gene regulation allows this single genome to produce different types of cells with unique functions (1). Divergence in gene regulation is pervasive at the cellular, organismal, population, and species levels and can affect phenotypic divergence (2–4). Discerning the phenotypic impacts of regulatory variation, however, requires understanding how this variation transmits through gene-regulatory networks to affect particular traits (5). Although several examples of adaptive expression variation in natural populations exist for traits with relatively simple genetic bases (i.e., one to several genes; refs. 6 and 7), most traits are complex products of poorly characterized developmental pathways involving many loci (8). As a result, linking regulatory change to phenotypic divergence in polygenic, complex traits remains a challenge.
Snake venoms have emerged as a system for studying the adaptive impacts and phenotypic consequences of regulatory variation in polygenic traits because of their genetic tractability (9, 10), contributions to fitness (11, 12), and high evolutionary rates (13, 14). Snake venoms comprise 5 to 25 toxin-gene families that show extreme levels of expression divergence among toxin genes at all phylogenetic scales (15–17). Because venoms are secretions, no complicating developmental processes are interposed between the expressed genes and their final product (10), allowing variation in toxin expression to directly alter the venom phenotype in a tractable manner; differences in toxin expression have been shown to be correlated with differences in venom function and toxicity (18–20).
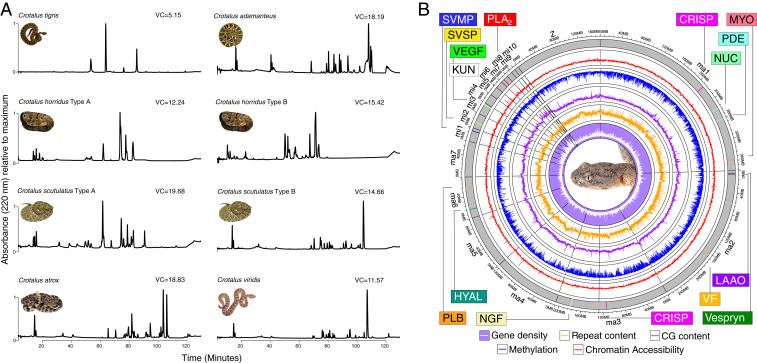
The largest axis of phenotypic variation in rattlesnake venoms is associated with differences in neurotoxic activities across individuals (18) known as the A–B dichotomy (21). Here, simple type A venoms have high neurotoxicity resulting from potent phospholipase () neurotoxins and low levels of tissue-damaging snake-venom metalloproteinases (SVMPs). More complex type B venoms, on the other hand, lack neurotoxic s (but possess other paralogs) and express high levels of SVMPs. The A–B dichotomy has been documented both within (14) and between species (18) and is summarized in Fig. 1A. Recent work in rattlesnake venoms showed that gene presence–absence explained some, but not all, of this variation, and variation within venom types was also present (Fig. 1A; refs. 22 and 23). Among rattlesnakes, the Tiger Rattlesnake (Crotalus tigris) possesses the simplest, but most toxic, venom of any species (venom complexity [VC] estimates are shown in Fig. 1A; SI Appendix, Table S1). Calvete et al. (24) found that four toxic proteins comprised 93% of the C. tigris venom proteome, whereas most viper venoms analyzed by similar techniques possessed tens to hundreds of toxic proteins. Whether this phenotypic simplicity is the result of a simple genotype through gene loss or a complex genotype mediated through gene-regulatory mechanisms, however, is unclear.
Fig. 1.
A complex genetic architecture underlies the simple venom phenotype of the Tiger Rattlesnake. (A) Comparative proteomics shows the simplicity of the Tiger Rattlesnake (C. tigris) venom phenotype. RP-HPLC of whole venoms for eight individuals across six species of rattlesnake. Chromatographic peaks represent a particular toxic protein or set of proteins. Estimates of VC, our proxy for phenotypic complexity, are shown next to each venom profile, and the Tiger Rattlesnake (VC = 5.15) possessed a significantly simpler venom than all other rattlesnakes shown here (see SI Appendix, Table S1 for details). For species polymorphic for the A–B venom dichotomy, where type A venoms are simple and neurotoxic and type B venoms are complex and hemotoxic, venom type is listed next to species name. C. tigris possesses a type A venom, whereas Crotalus adamanteus, C. atrox, and C. viridis possess type B venoms. See SI Appendix, Fig. S4 for transcriptome–proteome relationship in C. tigris. (B) Circos plot of the RaGOO assembly displaying gene density, repeat content, GC content, proportion of methylated cytosines in the venom gland, and chromatin accessibility in the venom gland across 100-kb windows at chromosome scale. Venom toxins that were reliably mapped to chromosome scaffolds are shown as colored vertical lines and labeled accordingly. CRISP, cysteine-rich secretory protein; HYAL, hyaluronidase; KUN, Kunitz-type toxin; LAAO, L-amino acid oxidase; MYO, myotoxin; NGF, nerve growth factor; NUC, nucleotidase; PDE, phosphodiesterase; PLB, phospholipase B; VEGF, vascular endothelial growth factor; VF, venom factor. Snake image credits: M.P.H. and Travis Fisher (photographer).
To characterize the genetic architecture underlying the simplest rattlesnake venom, we sequenced and assembled the Tiger Rattlesnake genome. We then used this genome, along with RNA-sequencing (RNA-seq), Assay for Transposase-Accessible Chromatin using sequencing (ATAC-seq), and whole-genome bisulfite sequencing (WGBS), to determine whether 1) the simplest venom phenotype was matched by a simple genetic architecture or 2) molecular and evolutionary mechanisms mediated the production of a simple phenotype from a more complex genotype. Here, an approximate one-to-one mapping of genotype to phenotype (i.e., venom gene to venom protein) would represent a simple genotype. A complex genotype would be characterized by a many-to-one mapping of genotype to phenotype, where many functional, but silenced, venom genes would be present in the genome, consistent with definitions from other systems (reviewed in ref. 25). We found evidence that gene loss, chromatin accessibility, and methylation levels all contributed to the production of the simplest, most toxic rattlesnake venom. Although deletion events reduced genomic complexity in particular toxin-gene tandem arrays, other genomic regions maintained complexity similar to other rattlesnakes with more complex venom phenotypes. In these regions, chromatin accessibility and methylation levels were significant predictors of toxin-gene expression and, therefore, putatively enabled a complex genetic architecture to produce a simple phenotype. Overall, we provided the most complete genomic characterization of the venom-gene regulatory network to date and identified key mechanisms generating phenotypic variation across a polygenic regulatory network.
Results
De Novo Genome Assembly and Annotation.
We sequenced the Tiger Rattlesnake genome using PacBio ( coverage; N50 of 25.73 kb; SI Appendix, Fig. S1) and Illumina ( coverage; 150 paired-end [PE]) sequencing technologies. The hybrid de novo assembly produced 4,228 scaffolds with a N50 scaffold/contig length of 2.11 Mb. Although the final assembly was 1.61 Gb in size, similar to other venomous snake genome assemblies (26–29), the N50 contig length was order(s)-of-magnitude larger in our assembly relative to other assemblies (Table 1). We assessed the completeness of the assembly using the BUSCO (version [v] 3) (30) vertebrata gene set () and found that the assembly was 95.8% complete (93.9% single-copy, 1.9% duplicated, 2.0% fragmented, and 2.2% missing), similar to other recent squamate genomes (e.g., Komodo Dragon 95.7%; ref. 31).
Table 1.
Tiger Rattlesnake genome assembly statistics and comparisons to other snake genome assemblies
| Tiger Rattlesnake | Prairie Rattlesnake | Five-Pace Viper | King Cobra | Indian Cobra | |
| Assembly size, Gb | 1.61/1.59 | 1.34 | 1.47 | 1.66 | 1.79 |
| Number of scaffolds | 4,228/160 | 7,043 | 160,256 | 296,399 | 1,897 |
| Scaffold N50, Mb | 2.11/207.72 | 179.89 | 2.12 | 0.23 | 223.35 |
| Number of contigs | 4,228 | 166,667 | 17,322 | 816,633 | 13,066 |
| Contig N50, Mb | 2.110 | 0.015 | 0.220 | 0.004 | 0.300 |
| GC content, % | 39.9/39.8 | — | — | 40.6 | 40.5 |
| Protein-coding genes | 18,240 | 17,352 | 21,194 | 18,506 | 23,248 |
| Putative venom protein-coding genes | 51 | 92 | 262 | 232 | 139 |
For the Tiger Rattlesnake, de novo assembly statistics are shown on the left, and RaGOO assembly statistics are shown on the right (where appropriate). The 160 RaGOO scaffolds do not include the 220 scaffolds assigned to chromosome 0 (160 RaGOO scaffolds and 220 unassigned de novo scaffolds in chromosome 0; 380 scaffolds total).
We next used RaGOO (32) to scaffold our de novo assembly to the Prairie Rattlesnake genome assembly (28). The scaffolded assembly reduced the total number of scaffolds from 4,228 to 380 (160 new scaffolds and 220 unassigned de novo scaffolds) and increased the scaffold N50 from 2.11 to 207.72 MB (Table 1), producing a chromosomal-level assembly; all 18 chromosomes (7 macro-, 10 micro-, Z chromosome) identified in the Prairie Rattlesnake genome assembly (28) were assembled here (Fig. 1B). Genomic content in the Tiger Rattlesnake genome was comparable to other recent snake genomes. For the de novo assembly, CG content, repeat content, and gene density were 39.9% (39.8% RaGOO assembly), 42.8% (44.0% RaGOO assembly), and 1.17 genes per 100 kb (1.13 genes per 100 kb RaGOO assembly), respectively. We performed RNA-seq on 25 tissues from the genome individual (SI Appendix, Fig. S2 and Table S2) and used MAKER (33), FGENESH (34), and manual assessment to annotate 18,240 protein-coding genes, 51 of which were homologous to putative toxins from other venomous snake species. To determine the chromosomal location of the 51 putative toxin genes in the Tiger Rattlesnake genome, we analyzed the mapping results of the scaffolded assembly and compared the locations of Tiger Rattlesnake toxin genes to the locations of orthologous toxin genes in the Prairie Rattlesnake (Fig. 1B; Dataset S1). Most major toxin-gene families (e.g., and SVMP) shared similar chromosomal locations in the Tiger and Prairie Rattlesnake (28) genomes, although other toxin-gene families (e.g., C-type lectins [CTLs] and vascular endothelial growth factor [VEGF]) mapped to different chromosomal locations (Fig. 1B; Dataset S1).
The Genetic Basis of the Simplest Venom.
We identified significantly fewer toxin genes in the Tiger Rattlesnake genome () than found in the Prairie Rattlesnake genome (; < 0.001) and other venomous snake species (Table 1), indicating that gene loss likely played a role in the production of the simplest rattlesnake venom. Not all of these genes, however, were likely to be expressed and directly contribute to the venom phenotype. We next used RNA-seq to determine which of these 51 genes were actively expressed in the venom gland of the Tiger Rattlesnake. We identified 20 toxin genes that were highly expressed (transcripts per million [TPM] >1,000 [1 k]) in both venom glands of the genome individual and an additional six toxin genes with TPM > 500, suggesting that approximately half of the putative toxin genes identified in the genome were appreciably expressed in the venom-gland transcriptome; results were largely consistent across seven individuals (SI Appendix, Fig. S3 and Dataset S1). Fifteen toxins were confirmed by quantitative mass spectrometry to be present in the venom of the genome animal (SI Appendix, Fig. S4), although proteomic detection thresholds likely precluded the detection of other toxins (10). Overall, we confirmed that 15 to 26 of the 51 identified putative toxin genes contributed to the venom phenotype and, therefore, that 25 to 36 of the identified putative toxin genes did not.
Gene Loss Contributes to the Evolution of Simple Venoms.
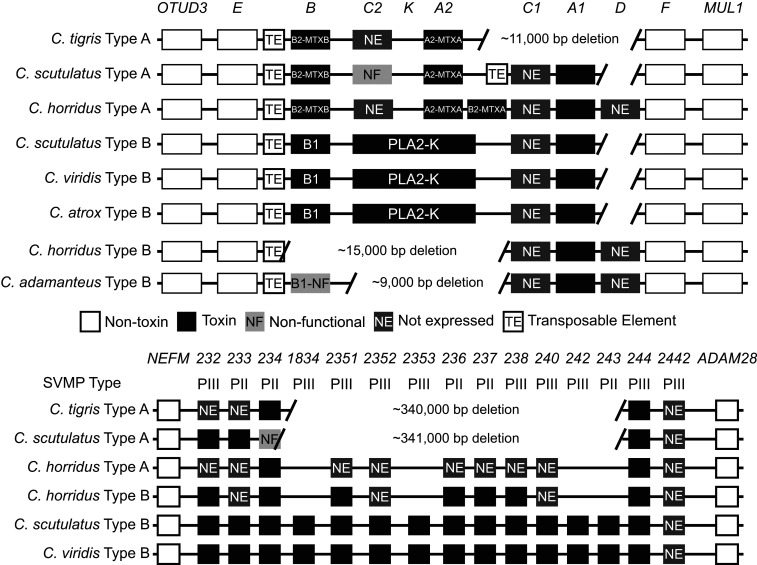
To identify specific toxin-gene losses in the Tiger Rattlesnake, we compared the Tiger Rattlesnake genome to several other rattlesnake species using the Prairie Rattlesnake genome (28) as well as the data of Dowell et al. (22, 23); Dowell and colleagues used bacterial artificial chromosome cloning to investigate the genetic architecture of the and SVMP toxin-gene families across several species and venom types. We found large deletions on both microchromosome 7 (s) and microchromosome 1 (SVMPs) in the Tiger Rattlesnake genome (Fig. 2).
Fig. 2.
Gene losses in two major toxin-gene families partly facilitate the evolution of a simple venom phenotype in the Tiger Rattlesnake (C. tigris). Genomic position for the (Upper) and SVMP (Lower) toxin tandem arrays across several rattlesnake species. Toxin genes are shown as black boxes, and toxin-gene nomenclature is taken from Dowell et al. (22, 23). Flanking nontoxin genes (e.g., OTUD3) are shown as white boxes. The Tiger Rattlesnake genome has large deletions in both toxin tandem arrays, although deletions do not completely explain the simplest venom phenotype; for example, five functional SVMP genes are retained in the genome, but only two are expressed. PII, type II SVMP; PIII, type III SVMP.
On microchromosome 7, we found an 11-kb deletion in the Tiger Rattlesnake genome that resulted in the deletion of three venom genes: -C1, -A1, and -D (Fig. 2). At least two of these three genes have been found in all other rattlesnakes examined to date, suggesting that this deletion event was unique to the Tiger Rattlesnake (although other deletion events in this region were found in other species; e.g., -K; Fig. 2). The mean number of functional venom genes across all species in this region was four (range three to seven), although no single paralog was shared across all species, reflecting the dynamic evolutionary history of this genomic region (22). The Tiger Rattlesnake possessed three venom genes: the basic (-B2-MTXB) and acidic (-A2-MTXA) subunits of the defining neurotoxin of type A venoms (18, 21) and -C2, which was not expressed. Although we found less than average genomic complexity and a large deletion event in the Tiger Rattlesnake genome, complexity was not different in the Tiger Rattlesnake relative to other rattlesnake species with much more complex venoms (e.g., Crotalus horridus type B).
On microchromosome 1, we found a large deletion event (340 kb) in the Tiger Rattlesnake genome that resulted in the deletion of 10 SVMP genes. Unlike microchromosome 7, however, the deletion event on microchromosome 1 was shared with the Mojave Rattlesnake (Crotalus scutulatus) with type A venoms (Fig. 2); more recent work (not included in Fig. 2) identified 30 SVMP paralogs in the Western Diamondback Rattlesnake (Crotalus atrox) genome (35). Including these data, the mean number of functional SVMP venom genes across all rattlesnake species investigated to date was 13 (range 4 to 30). The Tiger Rattlesnake possessed five SVMP venom genes, but only two of these genes, SVMP-234 and SVMP-244, were expressed. Despite gene deletions playing a larger role in the SVMP genomic region of the Tiger Rattlesnake genome relative to the genomic region, gene loss alone could not completely explain the production of the simplest venom phenotype. We found similar deletion events on both microchromosomes in species with more complex phenotypes, as well as the retention of functional paralogs that were not expressed in the Tiger Rattlesnake genome (e.g., -C2 and SVMP-233), indicating that regulatory mechanisms were also enabling the production of a simple phenotype from a more complex genotype.
Methylation Levels Predict Toxin-Gene Expression.
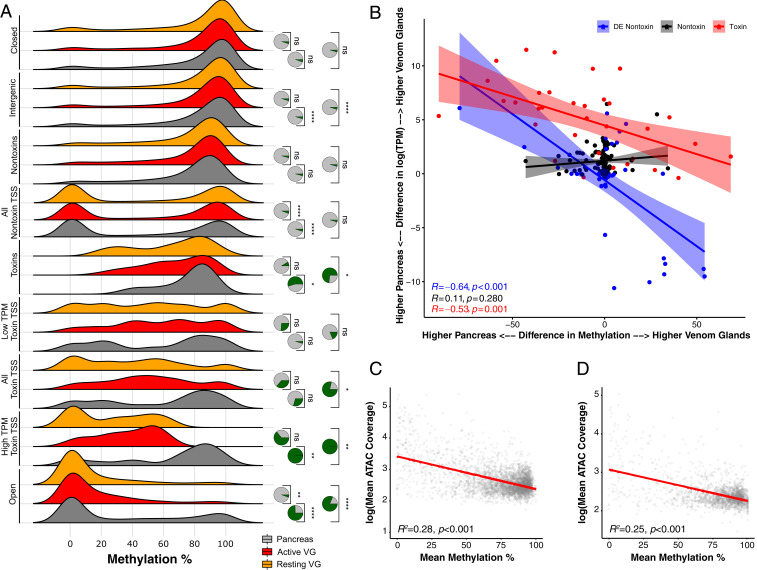
We performed WGBS on three tissues, two venom glands (one milked at 4 d and indicated as “Active,” one milked at >30 d and indicated as “Resting”) and the pancreas (control; Fig. 3; SI Appendix, Table S3). We recovered high genome-wide CpG methylation across all tissues (77%), consistent with other vertebrates (36, 37), and found decreased methylation near transcription start sites (TSSs) for all annotated protein-coding genes across all samples (SI Appendix, Fig. S5). Methylation and expression levels were significantly negatively correlated for all genes across all tissues ( < 0.001), as were toxin-specific methylation and expression levels in the venom glands (; SI Appendix, Fig. S6). To determine if differences in methylation predicted differences in gene expression across venom glands and pancreas, we performed a linear regression and found significant negative correlations in toxin genes () and differentially expressed nontoxin genes ( < 0.001), but not in similarly expressed nontoxins (; Fig. 3B); these results indicated that toxin and nontoxin transcripts that were differentially expressed across pancreas and venom glands exhibited significant, correlated changes in methylation level, whereas nontoxin transcripts that were similarly expressed across pancreas and venom glands were also similarly methylated. Highly expressed toxin genes were demethylated in the venom gland and heavily methylated in the pancreas (Fig. 3B; SI Appendix, Fig. S6), as expected, suggesting that methylation played a role in venom-gene expression. Gene Ontology (GO)-enrichment analysis of the differentially expressed nontoxin genes () showed that these genes were involved with protein production, particularly the ribosomal machinery of the venom-gland cells; roughly a third of the terms were directly related to ribosomal activity/components (SI Appendix, Fig. S7). Given that these genes were differentially expressed and differentially methylated across venom glands and pancreas (Fig. 3B), methylation appeared to also play a role in regulating the nonvenom components associated with venom translation/secretion and, therefore, in the overall production of the simple venom.
Fig. 3.
Methylation levels and chromatin accessibility jointly regulate gene expression in the venom glands of the Tiger Rattlesnake. (A) Distributions of methylation levels for pancreas, active venom glands (VG), and resting VGs across venom-gland-specific chromatin-accessible regions (Open), high-expression toxin-gene TSSs (High TPM Toxin TSS), all toxin-gene TSSs, low-expression (Low TPM) toxin-gene TSSs, entire toxin-genic regions including introns (Toxin), all nontoxin-gene TSSs, entire nontoxin-genic regions including introns (Nontoxin), intergenic regions, and inaccessible chromatin regions (Closed). TSS regions included bp around the estimated start site. High TPM toxins represent the 20 most highly expressed toxins (TPM > 1,000). Low TPM toxins represent the 32 transcripts with TPM < 1,000. Nontoxin genes represent genes actively expressed in the VGs that do not produce toxic proteins. Statistical significance was assessed by using t tests. Pie charts represent the proportion of 10,000 bootstrapped t tests that were significant ( < 0.05) after subsampling; green represents significant tests, and gray represents nonsignificant tests. (B) Difference in methylation level for TSS ( bp) across pancreas and VGs significantly predicts expression differences across the same tissues. Each point represents an individual transcript. Blue points represent nontoxins that were significantly differentially expressed (DE) across VGs and pancreas (mean TPM > 300; Padj < 0.01). Black points represent nontoxins that were not significantly DE across VGs and pancreas (mean TPM > 300; Padj > 0.1). Red points represent toxins. The y axis shows the difference in expression between VGs and pancreas; positive values represent transcripts more highly expressed in the VGs, whereas negative values represent transcripts more highly expressed in the pancreas. The x axis shows the difference in methylation level between VGs and pancreas; positive values represent transcripts more highly methylated in the VGs, whereas negative values represent transcripts more highly methylated in the pancreas. Overall, transcripts that exhibit DE across pancreas and VGs also exhibit differences in methylation levels; transcripts that are more highly expressed in the VGs have reduced methylation levels in the VG and vice versa. (C and D) Chromatin accessibility and methylation level are significantly negatively correlated across the (C) and SVMP (D) genomic regions. The y axis represents log-transformed ATAC-seq mean coverage estimates across 2-kb bins for all three VGs. The x axis represents mean methylation percentage across 2-kb bins for both VGs sequenced.
Chromatin Accessibility Predicts Toxin-Gene Expression.
We performed ATAC-seq on nine tissues, six venom glands (right and left venom glands from three individuals) and three control tissues (Harderian gland, labial gland, and pancreas; SI Appendix, Table S4). We found increased chromatin accessibility near TSS for all nontoxin genes across all control tissues with little to no accessibility near toxin TSS (SI Appendix, Fig. S8). In the venom glands, we found increased chromatin accessibility near toxin TSSs and transcription end sites (TESs) relative to nontoxins in the venom gland; this pattern was even more pronounced for the 20 most highly expressed toxins described above. Highly expressed toxins exhibited much higher levels of chromatin accessibility relative to lowly expressed toxins in the venom glands, where low-expression toxin-gene accessibility was much more similar to nontoxin-gene accessibility (SI Appendix, Figs. S8 and S9). Highly expressed toxin genes were also unique in having increased accessibility near TESs in the venom gland; low-expression toxin genes and nontoxin genes did not have accessible TESs in any tissue (SI Appendix, Figs. S8 and S9). Overall, chromatin accessibility, similar to methylation, was correlated with toxin-gene expression levels.
Chromatin Accessibility and Methylation Jointly Allow a Complex Genotype to Produce a Simple Venom Phenotype.
To assess the relationship between chromatin accessibility and methylation, as well as their potential joint role in co-regulating toxin-gene expression, we compared the distributions of methylation levels for pancreas and both venom glands across the following genomic regions: 1) venom-gland-specific chromatin-accessible regions (Open; see Materials and Methods for details), 2) high-expression toxin gene (High TPM Toxin) TSSs, 3) all toxin-gene TSSs, 4) low-expression (Low TPM) toxin-gene TSSs, 5) entire toxin-genic regions including introns (Toxin), 6) all nontoxin-gene TSSs, 7) entire nontoxin-genic regions including introns (Nontoxin), 8) intergenic regions, and 9) inaccessible chromatin regions (Closed; Fig. 3A; SI Appendix, Fig. S10). We found significant reductions in venom-gland methylation levels relative to the pancreas for at least one comparison across the Open ( < 0.001; see right tails of distributions), High TPM Toxin TSSs (0.001 0.009), All Toxin TSSs (), and Toxin (0.011 0.038) regions, indicating that chromatin accessibility and methylation were significantly and negatively correlated; both putatively contributed, in tandem, to the regulation of toxin-gene expression in the Tiger Rattlesnake. Indeed, chromatin accessibility and methylation level were significantly negatively correlated across both the (Fig. 3C) and SVMP (Fig. 3D) genomic regions ( < 0.001). Intergenic regions were also significantly different ( < 0.001), although our bootstrapping analysis showed less confidence in this result (Fig. 3A; SI Appendix, Fig. S10). For nontoxin TSSs, methylation was significantly greater in the active venom gland relative to both the pancreas and resting venom gland ( < 0.001). Low TPM Toxin TSSs (0.440 0.910), Nontoxins (0.083 0.160), and Closed (0.120 0.950) regions were not significantly different in methylation level across pancreas and either venom gland (Fig. 3A), as expected. The significant result in High TPM Toxin TSS regions and nonsignificant result in Low TPM Toxin TSS regions showed that chromatin accessibility and methylation were directly related to toxin-gene expression level (Fig. 3A; SI Appendix, Fig. S8).
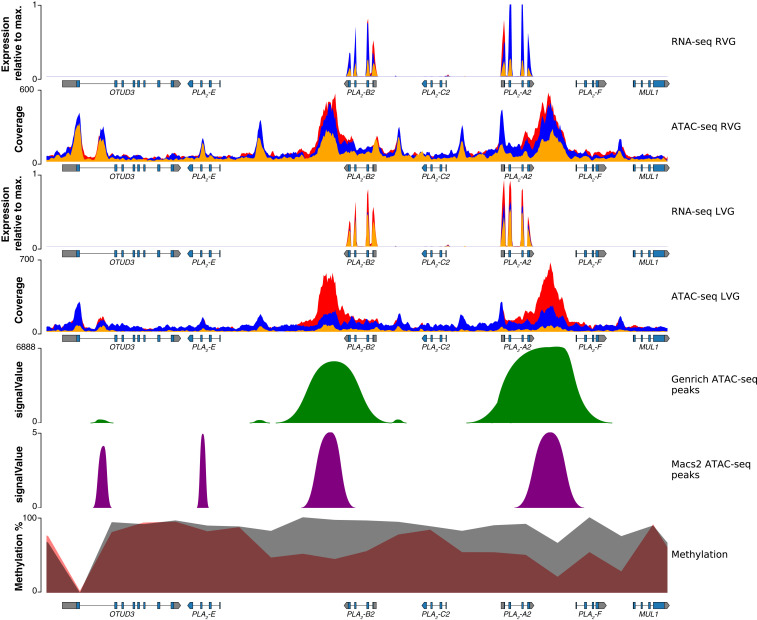
We next investigated venom-gland-specific chromatin-accessible regions (which represent putative venom-specific promoters and enhancers; see Materials and Methods for details), methylation levels, and expression levels for all genomic regions containing toxin genes, regardless of expression level (Figs. 4 and 5; SI Appendix, Figs. S11–S29). For the region on microchromosome 7, the Tiger Rattlesnake possessed three venom genes: the basic (-B2-MTXB) and acidic (-A2-MTXA) neurotoxin subunits, which were the most highly expressed toxin genes in the venom-gland transcriptome, and -C2, which was not expressed (Fig. 2; SI Appendix, Fig. S3). We found increased accessibility and reduced methylation near -B2-MTXB and -A2-MTXA and reduced accessibility and increased methylation near -C2 (Fig. 4). Venom-gland-specific chromatin-accessible regions were identified near the active genes, but not -C2, as expected, and these results were largely consistent across all toxin genes (SI Appendix, Figs. S11–29).
Fig. 4.
Regulatory landscape for the toxin-gene family in the Tiger Rattlesnake. Chromatin accessibility and methylation levels regulate the expression of toxin genes with increased accessibility and reduced methylation near active genes (i.e., -B2 and -A2) and reduced accessibility and increased methylation near silenced genes. Rows from top to bottom: right venom-gland (RVG) transcriptome, RVG ATAC-seq, left venom-gland (LVG) transcriptome, LVG ATAC-seq, venom-gland specific ATAC-seq peaks identified by Genrich, venom-gland specific ATAC-seq peaks identified by MACS2, and methylation percentage. RNA-seq y axes represent venom-gland expression levels scaled by the most highly expressed transcript in that genomic region. Red, blue, and orange colors in the RNA-seq and ATAC-seq plots represent the three individuals sequenced, respectively; colors are overlaid, and not all may be visible. Genrich-estimated signalValues represent the area under the peak; MACS2-estimated signalValues represent fold change in coverage at the peak summit. For the methylation track, gray shading represents methylation levels in 2-kb bins in the pancreas (control), whereas red shading represents methylation levels in 2-kb bins in the venom gland. Gene annotations are shown under tracks where appropriate; -B2, -C2, and -A2 are toxin genes. Max., maximum.
Fig. 5.
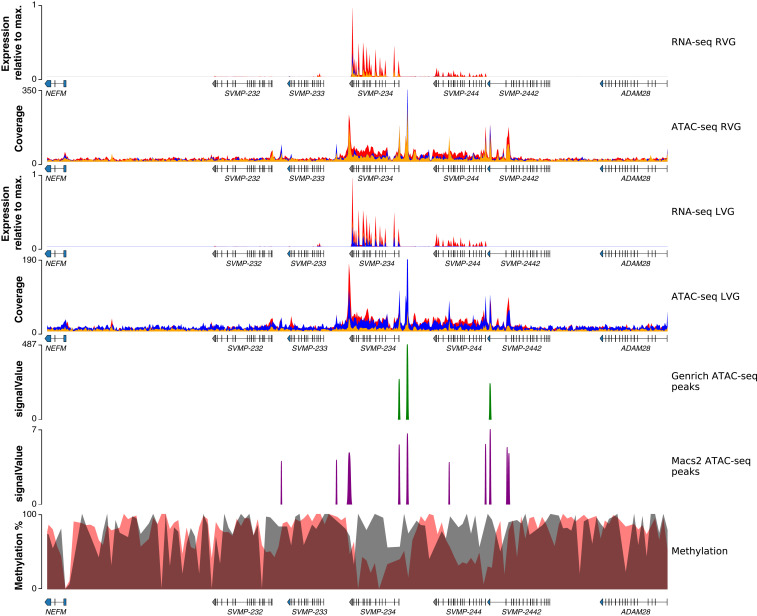
Regulatory landscape for the SVMP toxin-gene family in the Tiger Rattlesnake. Chromatin accessibility and methylation levels regulate the expression of SVMP toxin genes with increased accessibility and reduced methylation near active genes (i.e., SVMP-234 and SVMP-244) and reduced accessibility and increased methylation near silenced genes. Rows from top to bottom: right venom-gland (RVG) transcriptome, RVG ATAC-seq, left venom-gland (LVG) transcriptome, LVG ATAC-seq, venom-gland-specific ATAC-seq peaks identified by Genrich, venom-gland-specific ATAC-seq peaks identified by MACS2, and methylation percentage. RNA-seq y axes represent venom-gland expression levels scaled by the most highly expressed transcript in that genomic region. Red, blue, and orange colors in the RNA-seq and ATAC-seq plots represent the three individuals sequenced, respectively; colors are overlaid, and not all may be visible. Genrich-estimated signalValues represent the area under the peak; MACS2-estimated signalValues represent fold change in coverage at the peak summit. For the methylation track, gray shading represents methylation levels in 2-kb bins in the pancreas (control), whereas red shading represents methylation levels in 2-kb bins in the venom gland. Gene annotations are shown under tracks where appropriate; SVMP-232, SVMP-233, SVMP-234, SVMP-244, and SVMP-2442 are toxin genes. Max., maximum.
For the SVMP region on microchromosome 1, the Tiger Rattlesnake possessed five SVMP venom genes: SVMP-232, SVMP-233, SVMP-234, SVMP-244, and SVMP-2442; only SVMP-234 and SVMP-244 were expressed (Fig. 2; SI Appendix, Fig. S3). We again found increased accessibility and reduced methylation near the two expressed paralogs and reduced accessibility and increased methylation near the three silenced paralogs (Fig. 5). Venom-gland-specific chromatin-accessible regions were mostly identified near the active genes, but not the three silenced paralogs, although several accessible regions were found in the introns of SVMP-2442, which was not expressed (Fig. 5).
Venom-Gene Regulatory Network.
To identify regulatory elements significantly enriched in the venom-gland-specific chromatin-accessible regions described above, we performed a motif discovery analysis using HOMER (38) on both the Genrich (39) and MACS2 (40) datasets independently (see Materials and Methods for details). Using the Genrich dataset, we identified 8 de novo and 13 known motifs that were significantly enriched in venom-gland-specific chromatin-accessible regions; these 21 motifs represented different transcription factors (TFs; Datasets S2 and S3). Using the MACS2 dataset, we identified 24 de novo and 30 known motifs that were significantly enriched in venom-gland-specific chromatin-accessible regions; these 54 motifs represented different TFs (Datasets S4 and S5). Motifs for TFs were shared among the two datasets: seven members of the forkhead class (FOX) of DNA-binding proteins, multiple motifs for the Nuclear Factor I (NFI) family, GRHL2, and SMAD3 (although SMAD3 motifs were not identified in any toxin promotor/TSS; see below).
We identified a full-length NFIC motif and a half-site NFI motif in the promoter/TSS regions of both -B2-MTXB and -A2-MTXA (Datasets S6 and S7), suggesting that these paralogs were co-regulated by the NFI family. Similarly, a FOXA2 motif was identified in the promoter/TSS region of SVMP-234, and a FOXA1 motif was identified in the promoter/TSS region of SVMP-244 (although other motifs were identified in the untranslated region). A GRHL2 motif and a half-site NFI motif were consistently identified across multiple snake venom serine proteinase (SVSP) paralogs. Overall, our motif analysis suggested that 1) paralogs within a toxin-gene family were likely co-regulated by the same (set of) TF(s); and 2) TFs were shared across different toxin-gene families.
To determine if the venom-gland-specific expression of toxin genes occurred through venom-gland-specific TFs, we tested for significant differential expression across venom glands and all other tissues for the 1) 10 candidate TFs described above, and 2) 12 candidate TFs previously identified in the Prairie Rattlesnake (28). Schield et al. (28) identified a set of 12 TFs that were significantly up-regulated in the Prairie Rattlesnake venom gland relative to other tissues, but did not identify specific binding sites to suggest a role in directly regulating venom genes (see Discussion). We found 10 TFs that were significantly differentially expressed across Tiger Rattlesnake venom glands and other tissues. NFIA ( < 0.001), GRHL1 ( < 0.001), and NR4A2 (), all candidates from the previous study, were significantly overexpressed in the Tiger Rattlesnake venom gland relative to all other tissues. FOXA2 (), FOXA3 ( < 0.001), FOXD3 ( < 0.001), FOXO3 ( < 0.001), NFI ( < 0.001), NFIC ( < 0.001), and SMAD3 ( < 0.001) were significantly underexpressed in the venom gland; all other candidates were not differentially expressed across tissue types (Dataset S8).
Discussion
A central question in evolutionary biology is whether major phenotypic differences are largely the result of variation in gene number, gene sequence, or regulation (22, 41, 42). We sequenced and assembled a high-quality genome for the Tiger Rattlesnake to determine whether gene loss or specific regulatory mechanisms produced the simplest rattlesnake venom. The most contiguous snake-genome assembly to date, in terms of contig N50, provided the critical sequence information necessary to infer gene loss (43) and characterize regulatory landscapes across the genome. We used the assembled genome, RNA-seq, ATAC-seq, and WGBS to show that gene deletions, chromatin accessibility, methylation, and specific TFs all contributed to the production of a simple trait from a complex genotype.
Our comparative genomic analyses showed significant reductions in the total number of toxin genes present in the Tiger Rattlesnake genome relative to other rattlesnake species both across the entire genome (28) as well as in specific genomic regions (22, 23, 35). On microchromosome 7, we found a 11-kb deletion in the Tiger Rattlesnake genome that resulted in the deletion of three venom genes, and this deletion event coincided with the location of a known transposable element (TE). Dowell et al. (22) proposed that the TE clusters between genes could facilitate gene duplications and deletions through nonallelic homologous recombination, and our data further support this hypothesis. Evidence of gene duplications and deletions was widespread across the genome. For example, with 15 functional paralogs, SVSPs were the largest toxin-gene family in the Tiger Rattlesnake genome (Dataset S1). Similar to previous work in other toxin genes (29, 35), we found evidence of multiple pseudogenes and orphaned exons on microchromosome 2 (SI Appendix, Figs. S11 and S12), suggesting that past duplications and deletions played a role in generating the current set of SVSP paralogs in the Tiger Rattlesnake genome. Although we found substantial evidence for gene loss, gene deletions alone could not completely explain the simplest venom phenotype, as only half of the identified toxins were expressed at appreciable levels in the venom gland and confirmed in the venom.
To identify regulatory mechanisms governing the expression (or lack thereof) of these genes, we investigated the roles of methylation and chromatin accessibility in toxin-gene regulation. Methylation level and chromatin accessibility were both significant predictors of toxin-gene expression within the venom gland (i.e., high- versus low-expression toxin genes) as well as across tissues, suggesting that each mechanism played a role (perhaps jointly) in the production of the simplest venom phenotype. The demethylation of accessible, venom-specific promoter and enhancer regions was expected; transcription cannot take place unless a gene is both accessible and free of methylation. The redundancy of methylating inaccessible regions, however, was less clear (44, 45). Previous research suggested that methylation was a passive process, by which methyl groups were added following the removal of TFs (44). More recent work, however, has suggested that methylation assists in maintaining chromatin accessibility by facilitating TF binding (46–48). Here, unmethylated sites would recruit methyl-minus TFs to bind and, therefore, prevent chromatin from innately closing. Without the removal of methyl groups, transcription factors could not bind, and chromatin would then rapidly become inaccessible (46–48); this inaccessibility could play a critical role in the prevention of spurious transcription of nearby genes (49), although not all demethylated toxin genes were accessible. For example, toxin genes showed minimal expression in the pancreas despite a range of methylation levels (although the correlation between methylation and expression remained significant; ; SI Appendix, Fig. S6). Toxin genes, however, were not accessible in the pancreas (SI Appendix, Figs. S8 and S9), indicating that methylation level and chromatin accessibility were not always redundant.
Conservative estimates suggest that, in vertebrates, tandemly arrayed genes represent 14% of all genes and 25% of all gene-duplication events, yet little is known about their regulation (50). Toxin genes are known to occur in large tandem arrays (22, 23, 26, 29, 35), and chromatin accessibility can span thousands of base pairs. For example, the two Genrich-identified peaks are 2.9 kb and 3.6 kb, respectively (Fig. 4). Chromatin-accessible regions containing tandemly arrayed genes could, therefore, contain multiple loci, TSSs, and/or TESs (e.g., the broad accessibility of the CTL tandem array; SI Appendix, Fig. S14). High-expression toxin genes were unique in having increased accessibility near both TSSs and TESs. In tandem arrays, TESs and TSSs will often be adjacent, and 72 to 94% of tandemly arrayed genes in vertebrate genomes have been shown to be in parallel transcription orientation (i.e., on the same strand; ref. 50). We found similar patterns in the two largest toxin-gene families here (Fig. 5; SI Appendix, Figs. S11 and S12). Given that toxin paralogs often shared TF binding sites, methylation may have prevented the transcription of nearby, accessible genes that did not need to be expressed in the same array; other highly expressed toxin genes not located in tandem arrays, however, also exhibited increased chromatin accessibility near both TSSs and TESs (SI Appendix, Figs. S15 and S16). To our knowledge, accessible TESs have only been identified once before (51). Further comparative work is required to determine if accessible TSSs and TESs are a signature of highly expressed genes, highly expressed genes in tandem arrays, or venom genes.
TFs also play a central role in transcriptional regulation (52) and phenotypic evolution (53). Although Schield et al. (28) recently found evidence for NFI TF binding sites near of venom genes in the Prairie Rattlesnake genome, suggesting that these TFs may play a role in venom regulation, they did not identify significant enrichment for binding sites associated with any of these TFs in toxin-gene regions (28). Using our ATAC-seq data, we were able to identify a set of regulatory elements significantly enriched in the venom-gland-specific chromatin-accessible regions that corresponded to 10 TFs. Eight of the 10 candidate TFs identified belonged to the FOX or NFI TF families. FOX DNA-binding proteins have been shown to directly interact with chromatin and can also be regulated by members of the NFI family (54), and NFI members have also been shown to affect chromatin structure and activation (55). These results suggest that, for example, the NFI TF family may regulate expression in the Tiger Rattlesnake through chromatin remodeling. Although we identified significant enrichment for these 10 TFs in putative toxin promoter/enhancer regions, none of these TFs were significantly up-regulated in venom glands relative to other tissues in the Tiger Rattlesnake. We did, however, identify three TFs from the Prairie Rattlesnake candidate list that were overexpressed in the venom glands. Our candidate TFs were based on significant enrichment for motifs in venom-gland-specific chromatin-accessible regions, suggesting a regulatory role in toxin-gene expression, whereas the Schield et al. (28) candidate TFs were identified based on expression and lacked significant enrichment for binding sites near toxin genes. Therefore, we propose that the TFs that we identified, such as FOXA1, play a role in directly regulating venom genes while also maintaining important regulatory roles in other tissues, explaining the lack of differential expression across tissue types. The TFs that were overexpressed in the venom gland, on the other hand, may be linked to regulating the nonvenom components associated with venom translation/secretion. Venom production, rather than toxin-gene expression, appears to be linked to a few key venom-specific TFs. Overall, we showed that toxin-gene regulation and venom production were predicted by interactions between chromatin structure, methylation level, and specific sets of TFs.
Structural/copy-number variants, chromatin structure, DNA modifications such as methylation, and TFs have been shown here and elsewhere to play key roles in the generation of complex trait variation, yet other mechanisms, including single nucleotide polymorphisms (SNPs), noncoding RNAs, and posttranscriptional modifications, may also be involved (reviewed in ref. 25). Posttranscriptional modifications have been proposed as a major source of phenotypic variation in snake venoms (56), but more recent work showed, at best, a minor role for such mechanisms (10). microRNAs (miRNAs) have similarly been proposed to underlie differences in venom expression across age classes in rattlesnakes (57), but ontogenetic variation has not been documented in the Tiger Rattlesnake (all individuals included in this study were also adults; Materials and Methods). For any posttranscriptional modifications, including miRNAs, to play a role in generating the simple Tiger Rattlesnake venom, the transcriptome and proteome would need to be discordant. For example, if miRNAs were modulating venom-protein production, the transcriptome and proteome would need to show very different expression levels between toxin transcript and final toxin-protein product. As shown in SI Appendix, Fig. S4, however, this was not observed, suggesting that posttranscriptional modifications did not play a key role in the production of the venom phenotype. SNPs, particularly in the enhancer and promoter regions, on the other hand, likely contribute to the generation of regulatory variation across complex regulatory networks. Unfortunately, a limited sample size () precluded us from addressing SNPs in this study, but determining how sequence variation affects venom regulation across populations and species is central to understanding the evolution of complex regulatory networks; the putative promoters and enhancers we identified should aid in that goal.
The architecture of the genotype–phenotype map can directly influence the rate of adaptation, robustness, and evolvability for a particular trait (58–60). Here, approximately half of the identified venom genes contributed to the venom phenotype. Why half of the venom genes were not expressed and have not undergone pseudogenization, however, was less clear. Simulation studies have suggested that robustness and evolvability can actually increase with the number of genes in a genotype, suggesting that more complex genotype–phenotype maps could be advantageous (61), contrary to cost of complexity theory (62). The SVSP tandem array in the Tiger Rattlesnake genome showed how common pseudogenization can be in tandem gene arrays, further highlighting that functional, but inactive, toxin genes were most likely being maintained by selection. Giorgianni et al. (35) found a similar pattern when examining SVMPs in the Western Diamondback Rattlesnake and posited that functional, but inactive, venom genes may be expressed in other individuals (perhaps in different populations), at different life stages, or in different tissues. We posit an additional hypothesis that some of these paralogs may be retained because of shared/co-opted regulatory elements. For example, SVMP-2442 was not expressed in any of the seven venom-gland transcriptomes sequenced (SI Appendix, Fig. S3), but was retained as a functional paralog in the genome. Giorgianni et al. (35) described this paralog (mdc-1 in their manuscript) as the most likely immediate ancestor of SVMP genes; the locus was also retained, but not expressed, in the Western Diamondback Rattlesnake. We found several venom-specific chromatin-accessible regions in the introns of this gene in the Tiger Rattlesnake genome. These regions likely represented venom-specific enhancer elements (63), suggesting that the maintenance of SVMP-2442 and other functional, but inactive, genes may be the result of purifying selection on shared regulatory elements; recent work has shown that >95% of chromatin-accessible regions likely interact with at least one other regulatory element in the genome (64). Further comparative and functional genomic work will show what role selection on gene-regulatory networks plays in the maintenance of silenced paralogs and genomic complexity.
Conclusion
Although variation in gene expression is ubiquitous, identifying the mechanisms producing such variation, especially for complex phenotypes, is challenging (8). By using the genetic tractability of the venom system, we were able to simultaneously assess the relative contributions of two key mechanisms to the production of the overall phenotype, as well as link these regulatory mechanisms at particular genes to phenotypic divergence in a polygenic trait. Previous work showed that modular regulatory architectures allowed a simple genotype to produce a complex phenotype (2). We show that this modularity may extend to complex genotypes and simple traits, highlighting the potential evolvability of the genotype–phenotype map.
Materials and Methods
High-Molecular-Weight DNA Isolation and PacBio Sequencing.
Blood was extracted from the caudal vein of an adult male C. tigris (52 cm snout-vent length, 56.5 cm total length) collected from Santa Cruz County, Arizona. High-molecular-weight (HMW) genomic DNA (gDNA) was extracted by using a pipette-free protocol (SI Appendix). Single-molecule real-time (SMRT) Cell Sequencing (Sequel) was performed at the University of Delaware Sequencing and Genotyping Center. HMW DNA was sequenced across six SMRT cells and resulted in 3,540,291 filtered subreads (total sequence length of 53,096,151,594 bp; coverage), N50 of 25.73 kb, and a maximum subread length of 164.84 kb (SI Appendix, Fig. S1).
DNA Isolation and Illumina Sequencing.
Blood was extracted from the caudal vein of the adult male C. tigris genome animal. gDNA was extracted for short-read genome-library construction following a standard phenol–chloroform extraction protocol. Approximately 25 L of whole blood was added to 700 L of lysis buffer, 35 L of 10 mg/mL proteinase K, and 1 L of 20 mg/mL ribonuclease A; the solution was incubated at 55 °C for 2 h. One milliliter of phenol–chloroform was added, and the solution was centrifuged at 13,000 rpm for 5 min. The aqueous phase containing DNA was added to 1 mL of chloroform–isoamyl, and the solution was centrifuged at 13,000 rpm for 3 min. Six hundred microliters of the aqueous phase was removed, and 60 L of 3 M sodium acetate and 1,200 L of ice-cold ethanol were added. The solution was placed at −20 °C overnight to allow DNA to precipitate. Precipitate DNA was washed with 70% ethanol and resuspended in 100 L of 10 mM Tris buffer. The genome library was prepared by using the TruSeq DNA PCR-Free Library Prep and sequenced 150 PE on a NovaSeq 6000 at the Translational Science Laboratory in the College of Medicine at Florida State University. Sequencing resulted in coverage.
Genome Assembly.
A hybrid genome assembly was performed by using the Illumina and PacBio data with MaSuRCA (v3.2.8) (65) and default settings on the Clemson University Palmetto Cluster. Genome assembly completeness was assessed by using the BUSCO (v3) (30) vertebrata gene set (). Genome scaffolding was performed by using RaGOO (32); the de novo-assembled genome and the raw Illumina data were provided as input. The Prairie Rattlesnake genome (28) served as the reference. All other parameters were default. For both the de novo and the scaffolded RaGOO assemblies, we calculated gene density, GC content, and repeat content across 100-kb regions using custom python scripts (https://github.com/masonaj157/Ctigris_genome_scripts). For the RaGOO scaffolded assembly, the proportion of methylated cytosines was calculated from the venom-gland WGBS library from the genome individual. For ATAC-seq coverage, venom-gland libraries for the genome individual were aligned to the RaGOO assembly using Bowtie2 (66) as described below. Summary statistics for the RaGOO assembly shown in Fig. 1B were visualized in R using the circlize package (67). See SI Appendix for details.
RNA-Seq.
Total RNA was extracted from 39 individual tissues by using a standard TRIzol method (68); these tissues represent 24 different tissue types across seven individuals (SI Appendix, Table S2). To avoid potentially confounding ontogenetic differences in venom expression common in rattlesnakes (e.g., ref. 16), all individuals sampled were adults (ontogenetic venom variation has also not been documented in C. tigris). To investigate how dependent venom expression was on the last date of venom expulsion, we tested three different stimulation timepoints prior to gland removal: 1) five individuals at 4 d following gland stimulation (standard practice; ref. 69), 2) one individual at 1 d (active state), and 3) one individual >30 d (resting state). Venom expression has also repeatedly been shown to be under genetic, rather than plastic, control (16, 70). Libraries were sequenced 150 PE on a NovaSeq 6000 at the Translational Science Laboratory in the College of Medicine at Florida State University or on a NextSeq 550 at the Clemson University Genomics and Bioinformatics Facility. Low-quality bases were trimmed with Trim Galore! (v0.4.5) (71).
Genome Annotation.
We annotated repeat elements in the de novo-assembled genome using RepeatModeler (72). We then de novo-assembled 25 individual transcriptomes (representing 24 distinct tissues) from the adult male C. tigris genome animal as described (73). We then used MAKER (v2.31.8) (33) to annotate coding sequences in the de novo-assembled genome using the 309,105 de novo-assembled transcripts (SI Appendix), the species-specific repeat library described above, and all protein-coding genes from Anolis carolinensis () (74) and Ophiophagus hannah () (26). After our initial MAKER run, we used BUSCO (v3) (30) and our de novo genome assembly to iteratively train AUGUSTUS (75); three iterations of training were performed. Following the final MAKER iteration, gene IDs were ascribed based on homology, as described (28). Briefly, we used blastp with an e-value threshold of 1 to search a custom UniProt database containing all protein-coding genes from A. carolinensis () (74), O. hannah () (26), and Crotalus (). Because venom-gene families are known to occur in large tandem arrays and MAKER can underestimate the number of paralogs in particular gene families (28), we performed additional annotation steps for venom genes in the C. tigris genome. We used a combination of empirical annotation in FGENESH (34) as described (22, 28), as well as manual annotation using RNA-seq alignments in HiSat2 (v2.1.0) (76); the former identified all genes regardless of expression, whereas the latter was used to explicitly identify expressed toxin genes.
Genomic Alignments.
We used published genome sequences to compare the and SVMP toxin arrays across all rattlesnake species investigated to date. For the genomic region, we aligned sequences from C. tigris (this study), Crotalus viridis (28), and six individuals from Dowell et al. (22): type A and type B C. horridus, type A and type B C. scutulatus, C. atrox, and Crotalus adamanteus. For the SVMP genomic region, we aligned sequences from C. tigris (this study), C. viridis (28), and four individuals from Dowell et al. (23): type A and type B C. horridus and type A and type B C. scutulatus. Alignments were conducted by using MAFFT (v7.407) (77). Given the high rate of gene loss in these regions leading to large differences in length across species, we first anchored the alignments by using the flanking nontoxin genes on either side of the toxin-gene arrays. For the region, we used OTUD3 and MUL1. For the SVMP region, we used ADAM28 and NEFM. We then performed pairwise alignments of each pair of sequences (rather than aligning all at once) to account for the variation in these regions and produce a tractable alignment.
RNA-Seq Analyses.
Transcriptomes were assembled by using the “new Tuxedo” package (78). Trimmed reads were aligned to the de novo-assembled genome by using HiSat2 (v2.1.0) (76). Transcripts were assembled, and expression was quantified by using StringTie (v1.3.4) (79). edgeR (80) was used to calculate differential expression by comparing tissues with replicates (i.e., venom gland, vomeronasal organ, muscle, liver, pancreas, and testes) to the expression of all other tissues. Briefly, we used StringTie and edgeR to measure expression in counts per million (CPM). Only genes with a CPM > 5 in two or more samples were used in downstream analyses. Coverage data were then normalized in edgeR by using default Trimmed Mean of M-values. Genes with a false-discovery rate of <1% and a log-fold change of >2 were considered significantly different. Differentially expressed genes were visualized by using pheatmap in R across all 36 samples (left and right venom-gland transcriptomes were combined for three individuals for plotting) using log2-transformed CPM (SI Appendix, Fig. S2). The 51 putative toxin genes were visualized across all venom-gland samples (SI Appendix, Fig. S3).
ATAC-Seq, Peak Calling, and Motif Analyses.
Nine tissues from three individuals (SI Appendix, Table S4) were dissected immediately following euthanasia and individually prepped for ATAC-seq as described (81) with minor modifications. Approximately 50,000 cells per tissue (i.e., 50 k intact nuclei) were isolated for each library, and nuclei isolation, preparation, and cleanup were carried out as described (81). The Tn5 transposase (Nextera DNA Library Preparation Kit, Illumina) was used for transposition. Libraries were then sequenced 150 PE on a NovaSeq 6000 at the Translational Science Laboratory in the College of Medicine at Florida State University; sequencing depth is summarized in SI Appendix, Table S4. Low-quality bases were trimmed with Trim Galore! (v0.4.5) (71). Overall alignment rates varied from 81.68 to 98.79%. Peaks were independently called for each library by using Genrich (39) and MACS2 (40). To identify venom-gland-specific ATAC-seq peaks, BEDTools (82) was used to 1) identify peaks shared across all six venom-gland libraries (13,655 Genrich peaks and 27,509 MACS2 peaks) and 2) remove any peak identified in a non-venom-gland tissue (pancreas, labial gland, or harderian gland) that overlapped with a peak from the shared venom-gland peak set in (1). This filtering step was performed for the Genrich and MACS2 peak datasets independently and resulted in 2,446 and 5,919 venom-specific peaks in the Genrich and MACS2 datasets, respectively. pyGenomeTracks (83) was used to visualize ATAC-seq data and peak calls. To identify regulatory elements significantly enriched in the venom-specific ATAC-seq peaks relative to the genomic background, we performed a motif-discovery analysis using HOMER (38); analyses were run for the Genrich and MACS2 peak datasets independently. Discovery analyses were run by using “-size 200” and “-mset vertebrates” parameters. Enrichment P values were considered significant, as suggested by the HOMER documentation. Peaks were annotated by using a custom motif file containing all significant de novo and known motifs from either the Genrich or MACS2 run, respectively. To determine if candidate TFs were up-regulated in the venom gland compared to other body tissues, we used DESeq2 (84) to test for significant differential expression. See SI Appendix for details.
WGBS.
Three tissues from two individuals (SI Appendix, Table S4) were dissected immediately following euthanasia and stored in 95% EtOH at 4 °C (85). DNA was extracted 14 d after dissection by using a standard phenol–chloroform procedure, as described above. Library preps were performed 5 to 6 d later by using the Zymo Pico Methyl-Seq Library Prep Kit. Libraries were sequenced 150 PE on a NovaSeq 6000 at the Translational Science Laboratory in the College of Medicine at Florida State University with a targeted sequencing coverage of 20 (86). Low-quality bases were trimmed with Trim Galore! (v0.4.5) (71) with a minimum Phred score of 20. To reduce methylation bias at the 3′ and 5′ ends of reads, we clipped 10 bp on each end from all reads. Using Bismark (87), reads were then mapped to the de novo-assembled genome (–non_directional), and methylation calls were extracted with a minimum observation of 10 methylation states at a given site (–cutoff 10). Non-CpG methylation sites were merged, and the resulting CpG methylation levels were used in downstream analyses. Bismark reports were used to check for M-bias (88) and overall alignment success. BEDTools (82) was used to calculate 1) mean methylation of the two venom glands and 2) mean methylation in 2-kb windows across the de novo-assembled genome for both the venom glands and pancreas. pyGenomeTracks (83) was used to visualize methylation levels (0 to 100%) across particular genomic regions. deepTools (89) was used to plot methylation across all genes as well as 3 kb upstream and downstream of gene start and stop sites. To determine if methylation differed between tissue types in different regions of the genome, we calculated mean methylation of open regions, 1-kb regions around toxin and nontoxin TSS, toxin genes, nontoxin genes, intergenic regions, and closed regions. We used ggpubr and t tests to test for significant differences between the three tissues. To provide confidence in significance estimates, we performed 10,000 bootstrap replicated t tests, subsampling 50 observations ( number of toxin genes) with replacement for each replicate. Finally, to assess the relationship between gene expression, chromatin accessibility, and methylation level, we estimated the degree of correlation between each using ggpubr. To determine if increased methylation resulted in lower gene expression, we measured methylation level of 1-kb regions around TSS and estimated gene expression in TPM with StringTie (78), as described above. We calculated the difference in methylation and gene expression between venom glands and pancreas. We tested three groups of genes for this analysis: toxins, differentially expressed nontoxins (adjusted P [Padj] < 0.01), and nondifferentially expressed nontoxins ( > 0.1). All nontoxins tested had an average TPM greater than 300 across pancreas and venom gland tissues. Differential expression was estimated with DESeq2 (84). To search for enriched GO terms among these differentially expressed nontoxin genes, we used ShinyGO (90) using the Gallus gallus and Rattus norvegicus genomes and a false-discovery rate of 0.05. To determine if chromatin accessibility was correlated with methylation, we calculated mean ATAC-seq coverage in 2-kb windows across the genome to match previously calculated methylation windows. After log-transformation of ATAC-seq coverage, we correlated methylation level and ATAC-seq coverage for the s and SVMPs scaffolds. See SI Appendix for details
Venom Reversed-Phase High-Performance Liquid Chromatography.
Reversed-phase high-performance liquid chromatography (RP-HPLC) was performed on the eight venom samples shown in Fig. 1a. Distinct peaks were identified, Shannon’s diversity index (H) was calculated, and diversity indices were corrected to obtain the effective number of peaks. The 95% CIs were calculated for the effective number of peak estimates for all samples except C. tigris. To determine whether C. tigris possessed a significantly simpler venom phenotype, we determined whether the effective number of peaks for C. tigris fell outside the 95% CI of the distribution. See SI Appendix for details.
Quantitative Venom Proteomics.
We used quantitative mass spectrometry to determine which toxins identified in the genome were present in the venom of the genome individual as described (10). See SI Appendix for details.
Permits and Protocols.
Animals were collected under the following permits: Arizona Game and Fish SP628489/SP673626/SP622613, Texas Parks and Wildlife SPR-0713-098, Florida Fish and Wildlife Conservation Commission LSSC-13-00004, and Jekyll Island Authority. All procedures were approved by Institutional Animal Care and Use Committees at Clemson University (2017-067), the University of Central Florida (13–17W), and Florida State University (1529/1836).
Supplementary Material
Acknowledgments
This work was supported by NSF Grants DEB 1638879 and DEB 1822417 (to C.L.P.) and startup funding provided by Clemson University (C.L.P.). Additional resources were supported by NIH National Institute of General Medical Sciences Institutional Development Award P20GM109094. We thank the Clemson University Genomics and Bioinformatics Facility for computational resources. We thank Bruce Kingham and Olga Shevchenko at the University of Delaware DNA Sequencing & Genotyping Center for assistance on generating high-quality PacBio data; Todd Castoe and Drew Schield for discussions on gene annotation; Travis Fisher for photography; Tim Sackton and the Faculty of Arts and Sciences Informatics Group at Harvard University for advice on ATAC-seq analyses; and Michael Broe for discussions on genome assembly.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2014634118/-/DCSupplemental.
Data Availability.
The de novo genome assembly has been deposited at DNA Data Bank of Japan/European Nucleotide Archive/GenBank (accession no. VORL00000000). Raw PacBio (SRR9915945), Illumina WGS (SRR10189615–26), ATAC-seq (SRR11413274–82), RNA-seq (SRR11524048–77, SRR11545022–24, and SRR11816475–79), and WGBS (SRR11461881–83) data are available under BioProject PRJNA558767. Previously available RNA-seq data used in this study can be found at BioProject PRJNA88989 (accession no. SRR5270853).
References
- 1.Slack J., Metaplasia and transdifferentiation: From pure biology to the clinic. Nat. Rev. Mol. Cell Biol. 8, 369–378 (2007). [DOI] [PubMed] [Google Scholar]
- 2.Van Belleghem S., et al. , Complex modular architecture around a simple toolkit of wing pattern genes. Nat. Ecol. Evol. 1, 0052 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lewis J., Reed R., Genome-wide regulatory adaptation shapes population-level genomic landscapes in Heliconius. Mol. Biol. Evol. 36, 159–173 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sackton T., et al. , Convergent regulatory evolution and loss of flight in paleognathous birds. Science 364, 74–78 (2019). [DOI] [PubMed] [Google Scholar]
- 5.Halfon M., Perspectives on gene regulatory network evolution. Trends Genet. 33, 436–447 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lamichhaney S., et al. , A beak size locus in Darwin’s finches facilitated character displacement during a drought. Science 353, 470–474 (2016). [DOI] [PubMed] [Google Scholar]
- 7.Zhang L., Reed R., Genome editing in butterflies reveals that spalt promotes and Distal-less represses eyespot color patterns. Nat. Commun. 7, 11769 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Savolainen O., Lascoux M., Merila J., Ecological genomics of local adaptation. Nat. Rev. Genet. 14, 807–820 (2013). [DOI] [PubMed] [Google Scholar]
- 9.Margres M., et al. , Linking the transcriptome and proteome to characterize the venom of the eastern diamondback rattlesnake (Crotalus adamanteus). J. Proteomics 96, 145–158 (2014). [DOI] [PubMed] [Google Scholar]
- 10.Rokyta D., Margres M., Calvin K., Post-transcriptional mechanisms contribute little to phenotypic variation in snake venoms. G3 5, 2375–2382 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Holding M., Biardi J., Gibbs H., Coevolution of venom function and venom resistance in a rattlesnake predator and its squirrel prey. Proc. R. Soc. B 283, 20152841 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Margres M., et al. , Quantity, not quality: Rapid adaptation in a polygenic trait proceeded exclusively through expression differentiation. Mol. Biol. Evol. 34, 3099–3110 (2017). [DOI] [PubMed] [Google Scholar]
- 13.Lynch V. J., Inventing an arsenal: Adaptive evolution and neofunctionalization of snake venom phospholipase genes. BMC Evol. Biol. 7, 2 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Strickland J., et al. , Evidence for divergent patterns of local selection driving genome variation in Mojave rattlesnakes (Crotalus scutulatus). Sci. Rep. 8, 17622 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Daltry J. C., Wüster W., Thorpe R. S., Diet and snake venom evolution. Nature 379, 537–540 (1996). [DOI] [PubMed] [Google Scholar]
- 16.Margres M., et al. , Phenotypic integration in the feeding system of the eastern diamondback rattlesnake (Crotalus adamanteus). Mol. Ecol. 24, 3405–3420 (2015). [DOI] [PubMed] [Google Scholar]
- 17.Margres M., et al. , Tipping the scales: The migration-selection balance leans toward selection in snake venoms. Mol. Biol. Evol. 36, 271–282 (2019). [DOI] [PubMed] [Google Scholar]
- 18.Mackessy S. P., Venom composition in rattlesnakes: Trends and biological significance in The Biology of Rattlesnakes, Hayes W. K., Beaman K. R., Cardwell M. D., Bush S. P., Eds. (Loma Linda University Press, Loma Linda, CA, 2008), pp. 495–510. [Google Scholar]
- 19.Calvete J., et al. , Snake venomics of the central American rattlesnake Crotalus simus and the South American Crotalus durissus complex points to neurotoxicity as an adaptive pedomorphic trend along Crotalus dispersal in South America. J. Proteome Res. 9, 528–544 (2010). [DOI] [PubMed] [Google Scholar]
- 20.Margres M., et al. , Functional characterizations of venom phenotypes in the eastern diamondback rattlesnake (Crotalus adamanteus) and evidence for expression-driven divergence in toxic activities among populations. Toxicon 119, 28–38 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Glenn J. L., Straight R. C., Wolf T. B., Regional variation in the presence of canebrake toxin in Crotalus horridus venom. Comp. Biochem. Physiol. C 107, 337–346 (1994). [DOI] [PubMed] [Google Scholar]
- 22.Dowell N., et al. , The deep origin and recent loss of venom toxin genes in rattlesnakes. Curr. Biol. 26, 2434–2445 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dowell N., et al. , Extremely divergent haplotypes in two toxin gene complexes encode alternative venom types within rattlesnake species. Curr. Biol. 28, 1016–1026 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Calvete J. J., Pérez A., Lomonte B., Sánchez E. E., Sanz L., Snake venomics of Crotalus tigris: The minimalist toxin arsenal of the deadliest neartic rattlesnake venom. Evolutionary clues for generating a pan-specific antivenom against crotalid type II venoms. J. Proteome Res. 11, 1382–1390 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marian A., Molecular genetic studies of complex traits. Transl. Res. 159, 64–79 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vonk F., et al. , The king cobra genome reveals dynamic gene evolution and adaptation in the snake venom system. Proc. Natl. Acad. Sci. U.S.A. 110, 20651–20656 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yin W., et al. , Evolutionary trajectories of snake genes and genomes revealed by comparative analyses of five-pacer viper. Nat. Commun. 7, 13107 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schield D., et al. , The origins and evolution of chromosomes, dosage compensation, and mechanisms underlying venom regulation in snakes. Genome Res. 29, 590–601 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Suryamohan K., et al. , The Indian cobra reference genome and transcriptome enables comprehensive identification of venom toxins. Nat. Genet. 52–106–117 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Simao F., Waterhouse R., Ioannidis P., Kriventseva E., Zdobnov E., BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31, 3210–3212 (2015). [DOI] [PubMed] [Google Scholar]
- 31.Lind A., et al. , Genome of the Komodo dragon reveals adaptations in the cardiovascular and chemosensory systems of monitor lizards. Nat. Ecol. Evol. 3, 1241–1252 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alonge M., et al. , RaGOO: Fast and accurate reference-guided scaffolding of draft genomes. Genome Biol. 20, 224 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cantarel B., et al. , Maker: An easy-to-use annotation pipeline designed for emerging model organism genomes. Genome Res. 18, 188–196 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Solovyev V., Kosarev P., Seledsov I., Vorobyev D., Automatic annotation of eukaryotic genes, pseudogenes and promoters. Genome Biol. 7, S10 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Giorgianni M., et al. , The origin and diversification of a novel protein family in venomous snakes. Proc. Natl. Acad. Sci. U.S.A. 117, 10911–10920 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Feng S., et al. , Conservation and divergence of methylation patterning in plants and animals. Proc. Natl. Acad. Sci. U.S.A. 107, 8689–8694 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schmitz R., Lewis Z., Goll M., DNA methylation: Shared and divergent features across eukaryotes. Trends Genet. 35, 818–827 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Heinz S., et al. , Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 3, 576–589 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gaspar J. M., Data from “Genrich.” GitHub. https://github.com/jsh58/Genrich.git. Accessed 1 September 2019.
- 40.Zhang Y., et al. , Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, 1–9 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hoekstra H. E., Coyne J. A., The locus of evolution: Evo devo and the genetics of adaptation. Evolution 61, 995–1016 (2007). [DOI] [PubMed] [Google Scholar]
- 42.Carroll S. B., Evo-devo and an expanding evolutionary synthesis: A genetic theory of morphological evolution. Cell 134, 25–36 (2008). [DOI] [PubMed] [Google Scholar]
- 43.Gordon D., et al. , Long-read sequence assembly of the gorilla genome. Science 352, aae0344 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rea T., The accessible chromatin landscape of the human genome. Nature 489, 75–82 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Clark S., et al. , scNMT-seq enables joint profiling of chromatin accessibility DNA methylation and transcription in single cells. Nat. Commun. 9, 781 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yin Y., et al. , Impact of cytosine methylation on DNA binding specificities of human transcription factors. Science 356, eaaj2239 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lin Q., et al. , Methmotif: An integrative cell specific database of transcription factor binding motifs coupled with DNA methylation profiles. Nucleic Acids Res. 47, D145–D154 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Spektor R., Tippens N., Mimoso C., Soloway P., methyl-ATAC-seq measures DNA methylation at accessible chromatin. Genome Res. 29, 969–977 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Neri F., et al. , Intragenic DNA methylation prevents spurious transcription initiation. Nature 543, 72–77 (2017). [DOI] [PubMed] [Google Scholar]
- 50.Pan D., Zhang L., Tandemly arrayed genes in vertebrate genomes. Comp. Funct. Genomics 2008, 545269 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mieczkowski J., et al. , MNase titration reveals differences between nucleosome occupancy and chromatin accessibility. Nat. Commun. 7, 11485 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.de Mendoza A., et al. , Transcription factor evolution in eukaryotes and the assembly of the regulatory toolkit in multicellular lineages. Proc. Natl. Acad. Sci. U.S.A. 110, E4858–E4866 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wittkopp P., Kalay G., Cis-regulatory elements: Molecular mechanisms and evolutionary processes underlying divergence. Nat. Rev. Genet. 13, 59–69 (2012). [DOI] [PubMed] [Google Scholar]
- 54.Grabowska M., et al. , NFI transcription factors interact with FOXA1 to regulate prostate-specific gene expression. Mol. Endocrinol. 28, 949–964 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fane M., Harris L., Smith A., Piper M., Nuclear factor one transcription factors as epigenetic regulators in cancer. Int. J. Canc. 140, 2634–2641 (2017). [DOI] [PubMed] [Google Scholar]
- 56.Casewell N., et al. , Medically important differences in snake venom composition are dictated by distinct postgenomic mechanisms. Proc. Natl. Acad. Sci. USA 111, 9205–9210 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Durban J., et al. , Integrated “omics” profiling indicates that miRNAs are modulators of the ontogenetic venom composition shift in the central American rattlesnake, Crotalus simus simus. BMC Genomics 14, 234 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cowperthwaite M. C., Bull J. J., Ancel Myers L., Distributions of beneficial fitness effects in RNA. Genetics 170, 1449–1457 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wagner G. P., et al. , Pleiotropic scaling of gene effects and the ‘cost of complexity’. Nature 452, 470–473 (2008). [DOI] [PubMed] [Google Scholar]
- 60.Aguilar-Rodriguesz J., Peel L., Stella M., Wagner A., Payne J., The architecture of an empirical genotype-phenotype map. Evolution 72, 1242–1260 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Catalan P., Wagner A., Manrubia S., Cuesta J., Addings levels of complexity enhances robustness and evolvability in a multilevel genotype-phenotype map. J. R. Soc. Interface 15, 20170516 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Orr H. A., Adaptation and the cost of complexity. Evolution 54, 13–20 (2000). [DOI] [PubMed] [Google Scholar]
- 63.Yan F., Powell D., Curtis D., Wong N., From reads to insight: A hitchhiker’s guide to ATAC-seq data analysis. Genome Biol. 21, 22 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Greenwald W., et al. , Chromatin co-accessibility is highly structured, spans entire chromosomes, and mediates long range regulatory genetic effects. 10.1101/604371 (9 April 2019). [DOI] [Google Scholar]
- 65.Zimin A., et al. , The MaSuRCA genome assembler. Bioinformatics 29, 2669–2677 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Langmead B., Salzberg S. L., Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gu Z., Gu L., Eils R., Schlesner M., Brors B., Circlize implements and enhances circular visualization in R. Bioinformatics 30, 2811–2812 (2014). [DOI] [PubMed] [Google Scholar]
- 68.Rokyta D. R., Lemmon A. R., Margres M. J., Aronow K., The venom-gland transcriptome of the eastern diamondback rattlesnake (Crotalus adamanteus). BMC Genom. 13, 312 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rotenberg D., Bamberger E. S., Kochva E., Studies on ribonucleic acid synthesis in the venom glands of Vipera palaestinae (Ophidia, Reptilia). Biochem. J. 121, 609–612 (1971). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gibbs H. L., Sanz L., Chiucchi J. E., Farrell T. M., Calvete J. J., Proteomic analysis of ontogenetic and diet-related changes in venom composition of juvenile and adult dusky pigmy rattlesnakes (Sistrurus miliarius barbouri). J. Proteomics 74, 2169–2179 (2011). [DOI] [PubMed] [Google Scholar]
- 71.Krueger F., Trim galore. A wrapper tool around cutadapt and fastQC to consistently apply quality and adapter trimming to fastQ files. https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/. Accessed 1 July 2019.
- 72.Smit A., Hubley R., Repeatmodeler. http://www.repeatmasker.org/RepeatModeler/. Accessed 1 February 2019.
- 73.Holding M., Margres M., Mason A., Parkinson C., Rokyta D., Evaluating the performance of de novo assembly methods for venom-gland transcriptomics. Toxins 10, 249 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Alföldi J., et al. , The genome of the green anole lizard and a comparative analysis with birds and mammals. Nature 477, 587–591 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Stanke M., Morgenstern B., Augustus: A web server for gene prediction in eukaryotes that allows user-defined constraints. Nucleic Acids Res. 33, W465–W467 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kim D., Langmead B., Salzberg S., HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Katoh K., Standley D., MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 30, 772–780 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pertea M., Kim D., Perea G., Leek J., Salzberg S., Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 11, 1650–1667 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pertea M., et al. , StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 33, 290–295 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Robinson M., McCarthy D., Smyth G., edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Buenrostro J., Wu B., Chang H., Greenleaf W., ATAC-seq: A method for assaying chromatin accessibility genome-wide. Curr. Protoc Mol. Biol. 109, 21–29 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Quinlan A., Hall I., BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ramirez F., et al. , High-resolution TADs reveal DNA sequences underlying genome organization in flies. Nat. Commun. 9, 189 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Love M., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Schroder C., Steimer W., gDNAa extraction yield and methylation status of blood samples are affected by long-term storage conditions. PloS One 13, e0192414 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ziller M., Hansen K., Meissner A., Aryee M., Coverage recommendations for methylation analysis by whole-genome bisulfite sequencing. Nat. Methods 12, 230–232 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Krueger F., Andrews S., Bismark: A flexible aligner and methylation caller for bisulfite-seq applications. Bioinformatics 27, 1571–1572 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hansen K., Langmead B., Irizarry R., BSmooth: From whole genome bisulfite sequencing reads to differentially methylated regions. Genome Biol. 13, R83 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ramirez F., Dundar F., Diehl S., Gruning B., Manke T., deepTools: A flexible platform for exploring deep-sequencing data. Nucleic Acids Res. 42, W187–W191 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ge S., Jung D., Yao R., ShinyGO: A graphical gene-set enrichment tool for animals and plants. Bioinformatics 36, 2628–2629 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The de novo genome assembly has been deposited at DNA Data Bank of Japan/European Nucleotide Archive/GenBank (accession no. VORL00000000). Raw PacBio (SRR9915945), Illumina WGS (SRR10189615–26), ATAC-seq (SRR11413274–82), RNA-seq (SRR11524048–77, SRR11545022–24, and SRR11816475–79), and WGBS (SRR11461881–83) data are available under BioProject PRJNA558767. Previously available RNA-seq data used in this study can be found at BioProject PRJNA88989 (accession no. SRR5270853).