Significance
Axon degeneration is an active program of subcellular self-destruction that drives pathology in the injured and diseased nervous system. SARM1 is an inducible NAD+ hydrolase and the central executioner of axon loss. In healthy axons, the SARM1 NADase is autoinhibited. With injury or disease, this autoinhibition is relieved and SARM1 depletes NAD+, inducing a metabolic crisis and subsequent axon loss. Here we combine peptide screening, cryo-electron microscopy, and site-directed mutagenesis with analysis of axonal metabolomics and axon degeneration to define five domain interactions within and across SARM1 protomers that are required to maintain an inactive SARM1 octamer. These structural insights may enable the development of SARM1 inhibitors that stabilize this autoinhibited conformation and thereby block axon degeneration.
Keywords: neurodegeneration, axonopathy, metabolism, neuropathy, NMN
Abstract
Axon degeneration is an active program of self-destruction mediated by the protein SARM1. In healthy neurons, SARM1 is autoinhibited and, upon injury autoinhibition is relieved, activating the SARM1 enzyme to deplete NAD+ and induce axon degeneration. SARM1 forms a homomultimeric octamer with each monomer composed of an N-terminal autoinhibitory ARM domain, tandem SAM domains that mediate multimerization, and a C-terminal TIR domain encoding the NADase enzyme. Here we discovered multiple intramolecular and intermolecular domain interfaces required for SARM1 autoinhibition using peptide mapping and cryo-electron microscopy (cryo-EM). We identified a candidate autoinhibitory region by screening a panel of peptides derived from the SARM1 ARM domain, identifying a peptide mediating high-affinity inhibition of the SARM1 NADase. Mutation of residues in full-length SARM1 within the region encompassed by the peptide led to loss of autoinhibition, rendering SARM1 constitutively active and inducing spontaneous NAD+ and axon loss. The cryo-EM structure of SARM1 revealed 1) a compact autoinhibited SARM1 octamer in which the TIR domains are isolated and prevented from oligomerization and enzymatic activation and 2) multiple candidate autoinhibitory interfaces among the domains. Mutational analysis demonstrated that five distinct interfaces are required for autoinhibition, including intramolecular and intermolecular ARM-SAM interfaces, an intermolecular ARM-ARM interface, and two ARM-TIR interfaces formed between a single TIR and two distinct ARM domains. These autoinhibitory regions are not redundant, as point mutants in each led to constitutively active SARM1. These studies define the structural basis for SARM1 autoinhibition and may enable the development of SARM1 inhibitors that stabilize the autoinhibited state.
Axon degeneration is a key feature of neurodegeneration in which injured and diseased axons activate a program of subcellular self-destruction (1, 2). Mechanistically, the choice between axon survival and degeneration depends on the balance between axon maintenance and axon destruction proteins (3). SARM1 (sterile α and TIR motif-containing protein 1) is the essential prodegenerative protein in this pathway. Loss of SARM1 is profoundly axoprotective in animal models of nerve injury (4, 5), peripheral neuropathy (6–8), traumatic brain injury (9–11), and glaucoma (12). SARM1 can also drive neuronal cell death. SARM1 knockout (KO) inhibits neuronal loss induced by mitochondrial dysfunction (13) in a mouse model of TDP-43–associated amyotrophic lateral sclerosis (ALS) (14) and in multiple models of photoreceptor neurodegeneration (15, 16). These studies demonstrate that traumatic, metabolic, toxic, genetic, and neuroinflammatory insults can all activate SARM1 and highlight the importance of elucidating the mechanism of SARM1 regulation.
SARM1 functions as an octamer (17, 18) in which each monomer is composed of a mitochondrial targeting sequence, an N-terminal domain with armadillo repeats (ARM), two sterile α-motif (SAM) domains, and a Toll/interleukin-1 receptor (TIR) domain (5). While TIR domains are canonically known for scaffolding functions (19), the SARM1 TIR domain is the founding member of an evolutionarily conserved family of NAD+ hydrolases (NADases) (18, 20–22). SARM1 enzyme activity is necessary for axon degeneration (20), and active SARM1 TIR is sufficient to induce degeneration in otherwise healthy neurons (23). To control this prodegenerative activity, SARM1 regulation must incorporate two key features: 1) enzymatic activity must be inhibited in healthy neurons, and 2) injury- or disease-induced prodegenerative signals must relieve this autoinhibition. Autoinhibition is a common mechanism for tightly controlling protein activity, effectively maintaining an “off” state at equilibrium and rapidly responding to stimuli to induce the “on” state (24–26). The N-terminal ARM domain is required for autoinhibition of SARM1, as deletion mutants lacking the entire N terminus are constitutively active, leading to spontaneous axon degeneration (5). Moreover, the N-terminal ARM region physically associates with the enzymatic TIR domain of SARM1, suggesting that the N terminus may mediate autoinhibition via direct inhibition of the TIR NADase (27). However, the detailed molecular basis for this autoinhibition remains elusive.
Here we have combined functional and structural analysis of SARM1 to define the domain interfaces required for autoinhibition. To identify a region in the ARM domain that mediates autoinhibition, we screened a collection of peptides that tile this domain and identified a single peptide that potently inhibits SARM1 NADase enzyme activity. This peptide encompasses an evolutionarily conserved sequence of hydrophobic residues that are required for its inhibitory activity. Accordingly, mutation of these required hydrophobic residues within the full-length SARM1 protein disrupts autoinhibition, resulting in a constitutively active SARM1 enzyme that cleaves NAD+ and induces axon degeneration in the absence of an injury stimulus. Expression of a protein composed of a concatemer of this hydrophobic region in neurons effectively blocks SARM1-induced NAD+ loss and axon degeneration. To understand SARM1 autoinhibition more comprehensively, we solved a cryo-electron microscopy (cryo-EM) structure of the SARM1 octamer and identified five intramolecular and intermolecular interfaces occurring between domains of SARM1 protomers that are required for autoinhibition. These include not only two interfaces between the autoinhibitory ARM domain and enzymatic TIR domain, but also an ARM-ARM interface as well as two distinct ARM-SAM interfaces. Point mutations in any of these interfaces can create a constitutively active SARM1 enzyme, indicating that these autoinhibitory regions do not redundantly maintain SARM1 in an off state. Instead, maintenance of the off state requires both cis and trans interactions among all domains of the SARM1 octamer. The many potential sites for activating mutations suggest that polymorphisms in SARM1 in the human population could be an unappreciated cause of neurodegenerative disorders. As SARM1 promotes axon degeneration in many peripheral and central nervous system diseases, elucidating the mechanism of autoinhibition may aid in the development of strategies to maintain or reestablish autoinhibition and hence preserve neurons and axons in neurodegenerative diseases.
Results
A Peptide Screen Identifies a Region Involved in SARM1 Autoinhibition.
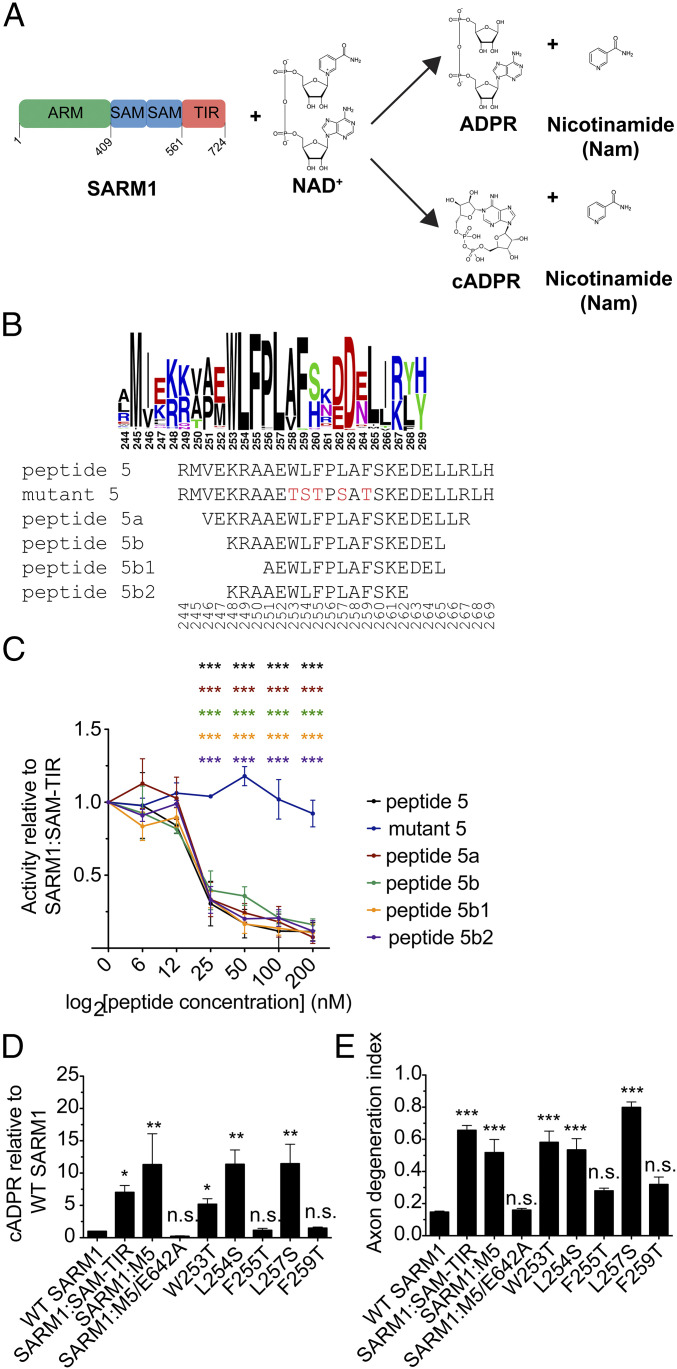
While wild-type SARM1 is autoinhibited, a SARM1 mutant (SARM1:SAM-TIR) lacking the N-terminal ARM motifs is constitutively active (5). The active SARM1 hydrolase converts NAD+ into nicotinamide (Nam) and either ADPR or cyclic ADPR (cADPR) (Fig. 1A). To identify residues or regions within the SARM1 N terminus that mediate autoinhibition of its NADase activity, we designed 12 peptides that tiled the most evolutionarily conserved region of the N terminus from V183 to E450 (SI Appendix, Fig. S1A). We produced human SARM1:SAM-TIR in HEK293T cells and tested whether the peptides could inhibit SARM1 NADase activity (SI Appendix, Fig. S1B). Each peptide was incubated at three concentrations with SARM1:SAM-TIR for 30 min before addition of 2.5 μM NAD+ to start the reaction. After 2 h, reaction products were isolated and measured via high-performance liquid chromatography (HPLC). A representative chromatogram shows their detection using known standards to identify peaks corresponding to NAD+ and the SARM1 hydrolase products ADPR and nicotinamide (SI Appendix, Fig. S1C). Because ADPR is more easily detectable and is an abundant SARM1 hydrolase product in vitro, we quantified its formation as a measure of SARM1:SAM-TIR NADase activity. We evaluated the ability of each peptide to block NADase activity by normalizing to SARM1:SAM-TIR NADase activity in the absence of peptide. At 2 μM of peptide, all peptides showed some inhibition of SARM1:SAM-TIR, suggesting nonspecific inhibition at this highest dose. In contrast, at 0.2 and 1 μM, peptide 5 inhibited NADase activity by more than 80%, whereas none of the other 11 peptides had a significant effect at these lower doses. These results suggested that peptide 5 interacted with SARM1 in a specific manner that led to its functional inhibition (SI Appendix, Fig. S1 C and D).
Fig. 1.
A conserved hydrophobic region of peptide 5 mediates inhibition of SARM1 NADase and prodegenerative activity. (A) Domain organization of SARM1 and the enzymatic reaction that it catalyzes. SARM1 can hydrolyze NAD+ into either ADPR and nicotinamide (Nam) or cyclic ADPR (cADPR) and Nam. (B) Schematic of the N-terminal region encompassed by peptide 5 and sequences of peptides derived from peptide 5. Black letters correspond to hydrophobic residues, green letters to nonpolar residues, magenta letters to polar residues, blue letters to basic residues, and red letters to acidic residues. The size of the residue letter indicates the frequency of that amino acid(s) at that position. (C) Twofold dose–response curves for peptide-mediated inhibition of SARM1:SAM-TIR NADase activity in vitro. Error bars represent ± SEM. n = 4 to 6; two-way ANOVA was used for statistical analysis with ***P < 0.001 with asterisk color denoting a concentration of a given peptide having an effect significantly different from measurements without the peptide. (D) DRG sensory neurons from SARM1 KO mice were infected with variants of SARM1, and metabolites were extracted 3 dpi to measure cADPR levels relative to neurons infected with wild-type SARM1. Error bars represent SEM; n = 4; one-way ANOVA with Dunnett posttest was used for statistical analysis with *P < 0.05 and **P < 0.01. (E) DRG sensory neurons from SARM1 KO mice were infected with variants of SARM1, and axons were imaged 5 d later. Axon fragmentation is quantified using the axon degeneration index (29). Error bars represent SEM, n = 4 to 5; one-way ANOVA with Dunnett posttest was used for statistical analysis with ***P < 0.001 and n.s. meaning not significantly different from control.
The SARM1 peptide 5 extends from arginine at 244 to histidine at 269. The most striking feature of this sequence is a highly evolutionarily conserved region of hydrophobic residues. We used MUSCLE to conduct multiple sequence alignment for 439 metazoan sequences with high homology to human SARM1. This analysis shows the conserved residues at each position where the tall black letters from W253 to F259 indicate these highly evolutionarily conserved hydrophobic residues (Fig. 1B). Reasoning that these residues may be conserved because they are functionally important for inhibition, we designed a mutant variant of the peptide, in which the conserved hydrophobic residues were mutated to hydrophilic residues (W253T/L254S/F255T/L257S/F259T). We compared the mutant and wild-type peptides using the in vitro NADase assay and found that peptide 5 blocked SARM1:SAM-TIR activity in a dose-dependent manner with an IC50 of about 20 nM (Fig. 1C). Strikingly, the mutant peptide 5 lacking the conserved hydrophobic residues failed to block SARM1:SAM-TIR NADase activity, demonstrating that these five residues are necessary for the inhibitory function of the peptide (Fig. 1 B and C). To define the minimal sequence necessary for inhibition, we synthesized variants of peptide 5 that were truncated from both ends (Fig. 1B). All truncated peptides inhibited the SARM1:SAM-TIR NADase with a dose–response similar to that of peptide 5 (Fig. 1C), demonstrating that these flanking residues do not contribute to autoinhibition. This mutational analysis highlights these conserved hydrophobic residues (W253 to F259) as essential for mediating the inhibitory effect of peptide 5 on SARM1:SAM-TIR NADase activity.
Although we initially measured the effects of peptide 5 on SARM1:SAM-TIR to bypass any potential interference from the endogenous SARM1 N terminus, we next sought to determine if peptide 5 would be capable of inhibiting the NADase activity of full-length SARM1. We purified full-length SARM1 protein and found it to be active, indicating that the autoinhibition that occurs in neurons is lost upon purification. Addition of peptide 5 efficiently inhibited full-length SARM1 NADase activity with a dose–response comparable to that of SARM1:SAM-TIR, demonstrating that the presence of the endogenous N terminus does not appreciably interfere with peptide 5 inhibition in vitro (SI Appendix, Fig. S2A). Peptide 5 could inhibit NADase activity by acting either 1) directly on the enzymatic portion of the protein located in the C-terminal TIR domain or 2) on an allosteric site in the SAM domains that then transduces the signal to the enzyme. To distinguish between these possibilities, we purified the SARM1 TIR domain and performed the in vitro NADase assay. Peptide 5 still inhibited the NADase activity of isolated SARM1 TIR domains, while the mutant peptide 5 did not (SI Appendix, Fig. S2B). These findings are consistent with a mechanism of inhibition in which these hydrophobic residues directly interact with the TIR domain of the protein.
Residues in the ARM Domain Corresponding to Peptide 5 Are Required for SARM1 Autoinhibition.
Peptide 5 inhibits SARM1 NADase activity in vitro, suggesting that the residues encompassing peptide 5 may be required for autoinhibition of full-length SARM1 in neurons. Moreover, the mutant peptide 5 does not inhibit the SARM1 NADase, highlighting these residues as mediators of the autoinhibitory interaction. Mutations in residues required for autoinhibition are predicted to result in a constitutively active enzyme. To test this hypothesis, we introduced the W253T/L254S/F255T/L257S/F259T mutation into full-length SARM1 (termed SARM1:M5) and assessed enzymatic and prodegenerative activity in primary dorsal root ganglion (DRG) sensory neurons. To avoid confounds from endogenous SARM1 protein, we cultured DRG neurons prepared from SARM1 knockout mice and infected them with lentivirus expressing either wild-type or mutant SARM1:M5. We extracted metabolites 3 d post infection (dpi), a time when soma and axons appeared morphologically normal. Using liquid chromatography-tandem mass spectrometry (LC-MS/MS), we previously showed that, in DRG neurons, the NAD+ hydrolase product cADPR is a reliable biomarker of SARM1 activity (28). We found that cADPR levels were elevated in neurons expressing SARM1:M5 to a similar degree as those expressing the constitutively active SARM1:SAM-TIR (5). To confirm that the cADPR generation was due to direct SARM1 enzymatic activity, we generated a double mutant of SARM1 including both the M5 mutations in the putative autoinhibitory domain and a mutation in the catalytic glutamate at position 642 (E642A) that renders SARM1 enzymatically dead (20). In contrast to SARM1:M5, SARM1:M5/E642A does not increase neuronal cADPR levels (Fig. 1D). Consistent with the generation of cADPR, both SARM1:SAM-TIR and SARM1:M5 reduced the levels of neuronal NAD+ compared to neurons expressing wild-type SARM1, whereas SARM1:M5/E642A had no effect on NAD+ levels (SI Appendix, Fig. S2C). Taken together, these data indicate that SARM1:M5 has lost autoinhibitory regulation and therefore functions as a constitutively active enzyme.
Finally, we assessed the requirements for each of the five hydrophobic residues by mutating them individually within the SARM1 full-length protein. These five mutants were expressed in DRG neurons, and their activity was examined by monitoring cADPR levels. The expression of SARM1 (W253T), SARM1 (L254S), and SARM1 (L257S) resulted in constitutive activity similar to SARM1:M5, whereas SARM1 (F255T) and SARM1 (F259T) expression did not affect cADPR levels (Fig. 1D). The results were similar when measuring NAD+ loss (SI Appendix, Fig. S2C). Hence, we conclude that W253, L254, and L257 are each required for autoinhibition of SARM1 enzymatic activity.
SARM1:M5 lacks autoinhibitory capability and is therefore a constitutively active NADase, and so we hypothesized that this mutant might stimulate axon loss even in the absence of injury. We transduced SARM1 knockout neurons with lentivirus expressing wild-type SARM1 or SARM1:M5 and imaged axons to observe their morphological integrity 5 d later (5 dpi) (SI Appendix, Fig. S2D). Axon fragmentation was quantified via an axon degeneration index (29), where higher numbers correlate with increased axon fragmentation (Fig. 1E). Expression of wild-type SARM1 did not disrupt axon morphology; however, expression of SARM1:M5 induced axon degeneration to a similar extent as the constitutively active SARM1:SAM-TIR protein, leaving only a few axons that were continuous and intact (SI Appendix, Fig. S2D and Fig. 1E). This prodegenerative activity of SARM1:M5 was completely abolished by the additional mutation of the critical NADase catalytic residue E642, demonstrating that the degenerative activity requires enzymatic activity. The single mutations that caused constitutive SARM1 NADase activity and the resulting cADPR production (W253T, L254S, and L257S) also caused similar levels of axon degeneration. Conversely, those mutants that had no effect on cADPR levels (F255T and F259T) did not cause axon degeneration. Thus, SARM1:M5 as well as the single SARM1 mutants W253T, L254S, and L257S each demonstrate the key features of a lack of autoinhibition: constitutive activity that induces cADPR production and NAD+ depletion along with axon degeneration in otherwise healthy neurons.
The Peptide 5 Region Is Required for Interaction of the ARM Domain with the TIR Domain.
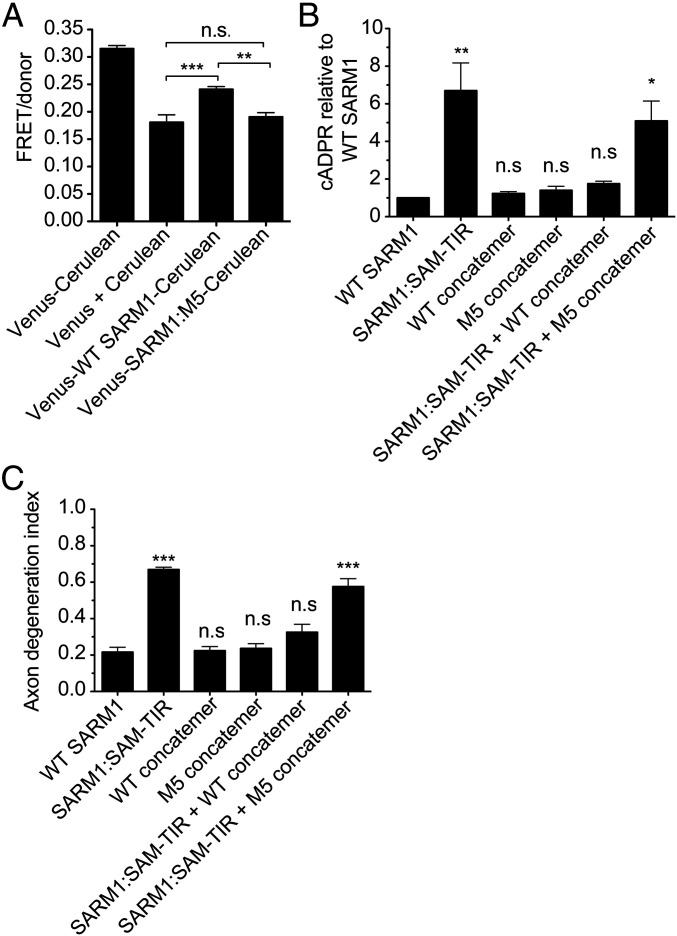
The observations that peptide 5 inhibited enzymatic activity of the isolated TIR domain and that SARM1:M5 is constitutively active suggest that this region of the protein contacts the TIR domain to keep SARM1 in an off state. We previously generated a SARM1 molecule with Cerulean and Venus fluorescent proteins on the N terminus and C terminus, respectively, and used it in fluorescence resonance energy transfer (FRET) studies to detect a close association between the N-terminal ARM region and TIR domain of SARM1 (27). We generated a similar Cerulean/Venus construct using SARM1:M5. Both wild-type SARM1 and SARM1:M5 FRET constructs were expressed in HEK293T cells. We found that wild-type SARM1 fused to Cerulean and Venus produces a FRET signal 33% above the background levels observed when the Cerulean and Venus fluorophores are expressed in trans. In contrast, SARM1:M5 fused to Cerulean and Venus gave a FRET signal that was indistinguishable from background (Fig. 2A). Thus, we show that the exogenous peptide 5 acts directly on the SARM1 TIR domain in vitro, and mutation of the corresponding hydrophobic residues in SARM1 disrupts the interaction of the N-terminal and TIR domains as measured by FRET. These data support the model that these hydrophobic residues in the N-terminal region interact with the TIR domain.
Fig. 2.
The M5 region inhibits SARM1 activation in cells. (A) Wild-type SARM1 with Cerulean attached to the N terminus and Venus attached to the C terminus exhibited a positive FRET/donor ratio in HEK293T cells, but similarly tagged SARM1:M5 did not. Error bars represent SEM; n = 4; one-way ANOVA with Bonferroni posttest was used for statistical analysis with **P < 0.01 and ***P < 0.001. (B) DRG sensory neurons from SARM1 KO mice were infected with SARM1:SAM-TIR mutant along with constructs expressing concatemers of either the wild-type peptide 5 region or the peptide 5 M5 mutant region. Metabolites were extracted at 3 d after infection to measure cADPR levels. Metabolite levels are shown as compared to those observed in neurons infected with wild-type SARM1 alone. Error bars represent SEM; n = 4; one-way ANOVA with Dunnett posttest was used for statistical analysis with *P < 0.05 and **P < 0.01. (C) DRG sensory neurons from SARM KO mice were imaged at 5 dpi to measure axon degeneration. Error bars represent SEM; n = 4; one-way ANOVA with Dunnett posttest was used for statistical analysis with ***P < 0.001 and n.s. meaning not significantly different from control.
Having characterized the function of this autoinhibitory sequence, we next used this knowledge to develop a SARM1 inhibitor that functions in DRG neurons. Peptide 5 is a potent inhibitor in vitro, but our efforts to add cell permeant tags to peptide 5 to introduce this peptide into neurons were unsuccessful. As an alternative, we created a lentiviral vector expressing a concatemer of four repeats of the wild-type peptide 5 (R244-H269) or the mutant peptide 5 (conserved hydrophobic residues mutated), each separated by a short linker. We tested whether this concatemer could inhibit the constitutively active SARM1:SAM-TIR, which produces cADPR and promotes axonal degeneration when expressed in SARM1 knockout neurons. Coexpression of the wild-type concatemer with SARM1:SAM-TIR blocked cADPR generation and axon degeneration in neurons. In contrast, the mutant concatemer did not block either of these phenotypes (Fig. 2 B and C). Thus, the peptide 5 region blocks SARM1 NADase activity in neurons, likely via these conserved hydrophobic residues, and rescues axons from degeneration.
Overall Structure and the Five Interfaces in the Autoinhibited SARM1.
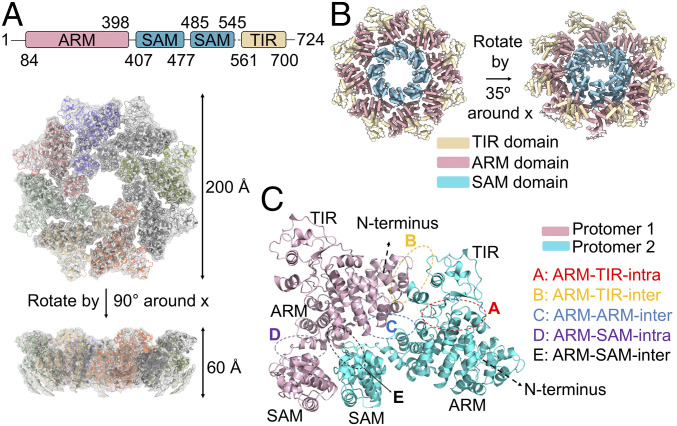
To further elucidate the molecular basis of autoinhibition, we set out to determine the SARM1 structure using cryo-EM (SI Appendix, Figs. S3–S5). We expressed SARM1 in SF9 insect cells as a maltose-binding protein (MBP) fusion (MBP-SARM1) and purified it to homogeneity. The elution volume of SARM1 from a Superdex 200 gel filtration column indicated a preformed oligomer (SI Appendix, Fig. S3A), consistent with the octameric ring assembly seen previously (17, 18). The cryo-EM data (SI Appendix, Fig. S3B) were collected using a Titan Krios microscope operating at 300 keV equipped with a K3 direct electron detection camera. They were processed in Relion 3.1 (30) to obtain a final map at 3.4-Å resolution based on gold-standard Fourier shell correlation between half maps (SI Appendix, Fig. S3 C–F, S4, and S5 and Table S1). The local resolution for the majority of the central SAM domain and ARM domain ranges between 3.0 and 3.5 Å (SI Appendix, Fig. S3F). The local resolution of the peripheral TIR domain was relatively lower (∼5 to 6 Å) in the initial reconstruction. By symmetry expansion, density subtraction, and focused classification, we improved the resolution of the TIR domain to ∼4 Å (SI Appendix, Fig. S3F), leading to unambiguous identification of the two critical interfaces between ARM and TIR domains.
The octameric ring of SARM1 is highly compact and has a diameter of ∼200 Å and a height of ∼60 Å (Fig. 3A). In the center of the ring, the two SAM domains stack on top of each other and oligomerize to form the central hub. The eight ARM domains form the next layer of the ring outside the central hub; each ARM domain packs tightly with its own SAM domain (intramolecular interaction) and with a neighboring SAM domain (intermolecular interaction) (Fig. 3 A–C). Strikingly, the TIR domains do not contact each other, but are rather spaced out at the periphery of the octamer, sandwiched between two adjacent ARM domains, but have no clear interaction with any SAM domains (Fig. 3B). Although the linker between SAM and TIR domains is flexible and invisible in our cryo-EM map, we assigned the ARM-SAM and TIR domains in direct contact as shown within one protomer (Fig. 3C). This linker is also disordered in the recently reported SARM1 cryo-EM structures (31, 32), which, however, used a different designation of the protomer; in this designation, a TIR domain does not contact the rest of the domains in the same protomer (SI Appendix, Fig. S6 A–D). While our assignment of the protomer is arbitrary, as in the published structures (31, 32), and the distances between the SAM domain C terminus and the TIR domain N terminus are similar between the two assignments, we deem more probable that during protein translation and folding, a TIR domain would interact with its own ARM domain first. Our assignment also allows easier designation of the two interfaces between ARM and TIR as intracellular and intermolecular ARM-TIR interfaces.
Fig. 3.
Overall structure of human SARM1. (A) Domain arrangement of SARM1 and the octameric model fitted into the composite EM map combining the C8 symmetry map and the TIR-focused refined C1 map. The dimensions of the octamer are indicated. (B) Ribbon diagram of the octameric model, color coded by domains. ARM: light pink; SAM: sky blue; TIR: wheat. (C) Five intramolecular and intermolecular interfaces between SARM1 domains are shown on two extracted protomers.
By extracting two adjacent protomers from the homo-octamer, we identified five unique interfaces in addition to the previously identified SAM-SAM interfaces (17): ARM-TIR intramolecular (ARM-TIR I), ARM-TIR intermolecular (ARM-TIR II), ARM-ARM, ARM-SAM intramolecular (ARM-SAM I), and ARM-SAM intermolecular (ARM-SAM II) (Fig. 3C). Among them, the ARM-TIR I interface covers the region of the ARM domain in peptide 5 identified in our peptide screening, which buries ∼880 Å2 surface area per surface, the largest among all interfaces. This knowledge, the isolated TIR domains in the octamer, and further structure-based mutagenesis (see below) indicated that the observed homo-octamer represents the autoinhibited state of SARM1. In comparison with the published SARM1 cryo-EM structures (31, 32), our SARM1 structure is highly similar (SI Appendix, Fig. S6A). However, our TIR domain has an ordered BB loop, in contrast to the published structures (SI Appendix, Fig. S6E), and thus exhibits a better-specified ARM-TIR II interface involving the BB loop. Of note, in addition to the evidence provided by the peptide studies that the ARM-TIR I interface is functionally important, our mutational analysis substantiates the role of the ARM-TIR II interface (see the following section) in keeping SARM1 inactive.
Detailed Interactions and Mutagenesis in the Five Interfaces of the SARM1 Octameric Ring.
To examine the importance of all five observed interfaces, we scrutinized the surfaces involved and identified potential key interacting residues. We then generated SARM1 constructs with mutations in these residues, introduced them into SARM1 KO neurons, and measured their activity by monitoring neuronal cADPR levels.
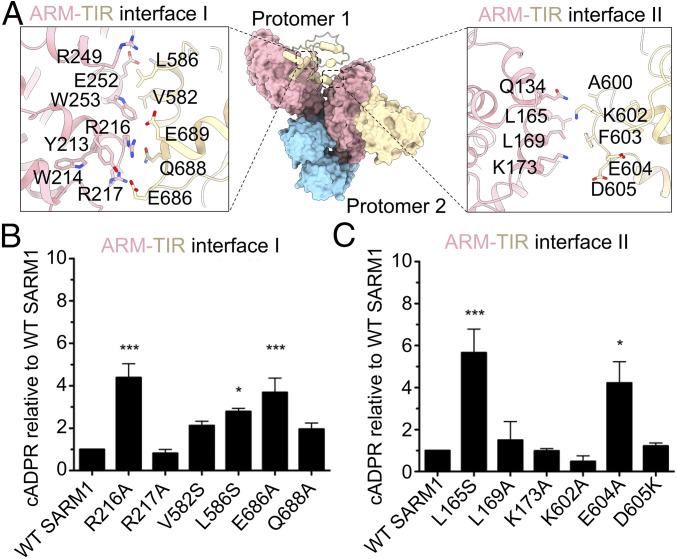
The ARM-TIR I interface covers the α10-α11 junction and α13 of the ARM domain and αA and the C-terminal αE of the TIR domain (Figs. 3C and 4 A, Left, and SI Appendix, Fig. S7). In line with the peptide-screening result, a core region of the interface is the hydrophobic interaction between W253 of ARM, whose mutation led to constitutive SARM1 activation (Fig. 1 D and E and SI Appendix, Fig. S2 C and D), and V582 and L586 of TIR (Fig. 4 A, Left). Mutations V582S and L586S correspondingly resulted in constitutive SARM1 activity as measured by cADPR production, further implicating this ARM-TIR interface in the control of SARM1 autoinhibition (Fig. 4B). The L254 and L257 residues highlighted by our peptide inhibitor (Fig. 1 D and E and SI Appendix, Fig. S2 C and D) are not localized at the ARM-TIR interface, but are instead buried within the ARM domain itself. Mutations in these residues may therefore affect the ARM domain folding (SI Appendix, Fig. S8A), thereby perturbing the ARM-TIR I interaction. The ARM-TIR I interface is further stabilized by peripheral electrostatic and hydrogen-bonding interactions. The side chains of R216 and R217 of the ARM domain form salt bridges with the side chains of E686 and E689 of the TIR domain, and Q688 of the TIR domain potentially further contributes to the interaction networks (Fig. 4 A, Left). On the ARM domain side, the R216A mutation resulted in high cADPR and loss of autoinhibition, while the R217A mutation had no effect on basal SARM1 activity (Fig. 4B). On the TIR domain side of the interface, both E686A and Q688A resulted in constitutive SARM1 activity (Fig. 4B).
Fig. 4.
Structural and functional analyses of the ARM-TIR interfaces in the octamer. (A) Interactions involved in ARM-TIR interface I (Left) and ARM-TIR interface II (Right). (B and C) Functional effects of mutations at these two interfaces. DRG sensory neurons from SARM1 KO mice were infected with various SARM1 constructs with indicated mutations in ARM-TIR interface I (B) or ARM-TIR interface II (C). Metabolites were extracted 3 dpi to measure cADPR levels relative to neurons infected with wild-type SARM1. Error bars represent SEM; n = 3 to 4. One-way ANOVA with Dunnett posttest was used for statistical analysis with *P < 0.05 and ***P < 0.001.
At another region of the ARM-TIR I interface, R249 and E252 of the ARM create a hydrogen-bonding triad with Q585 of TIR, and two aromatic residues, F255 and F259, line up near the critical W253 residue (SI Appendix, Fig. S8B). The role of these two Phe residues is still unclear, with conflicting data. While we found that F255T and F259T mutants showed minimal effects on SARM1 autoinhibition in neurons (SI Appendix, Fig. S2 C and D and Fig. 1 D and E), others have shown that overexpression of a F255R/P256R SARM1 double mutant decreased viability of HEK293T cells, providing indirect evidence of SARM1 activation (33). It is possible that the Thr mutations were less disruptive than the Arg mutations or that disrupting the Pro is the primary cause of the phenotype in this double mutant. Collectively, these structural and mutational analyses confirmed and extended the peptide-mapping studies.
The ARM-TIR II interface, with a buried surface area of ∼141.5 Å2 per partner, is much smaller and involves the α7 (residues L165-K173) and α4-α5 hinge (the loop contributing Q134) of the ARM domain and the BB loop (residues A600-D605) of the TIR domain (Fig. 4 A, Right, and SI Appendix, Fig. S7). Q134 of the ARM domain forms a van der Waals interaction with A600 and K602 of the TIR domain. L165 and L169 of the ARM domain establish hydrophobic contact with F603 of the TIR domain, while K173 has the potential to make salt bridges with E604 and D605 (Fig. 4 A, Right). The SARM1 mutant L165S resulted in increased cADPR consistent with constitutive SARM1 activity, whereas the mutations L169A and K173A did not alter autoinhibition (Fig. 4C). We also generated mutants on the TIR side of the interface at the BB loop. We found that the E604A mutant was constitutively active, the D605K mutant showed an activity similar to wild-type SARM1, and K602A exhibited extremely low cADPR levels and NADase activity (Fig. 4C). However, interpretation of these mutants is complicated by the implicated role of the BB loop in mediating NADase activity of the TIR domain (18, 27), whereby relief from autoinhibition by K602A and D605K mutations could be offset by their effect on TIR NAD+ catalysis itself. Nonetheless, the observed hyperactivity of the E604A mutant supports the role of this interface in autoinhibition.
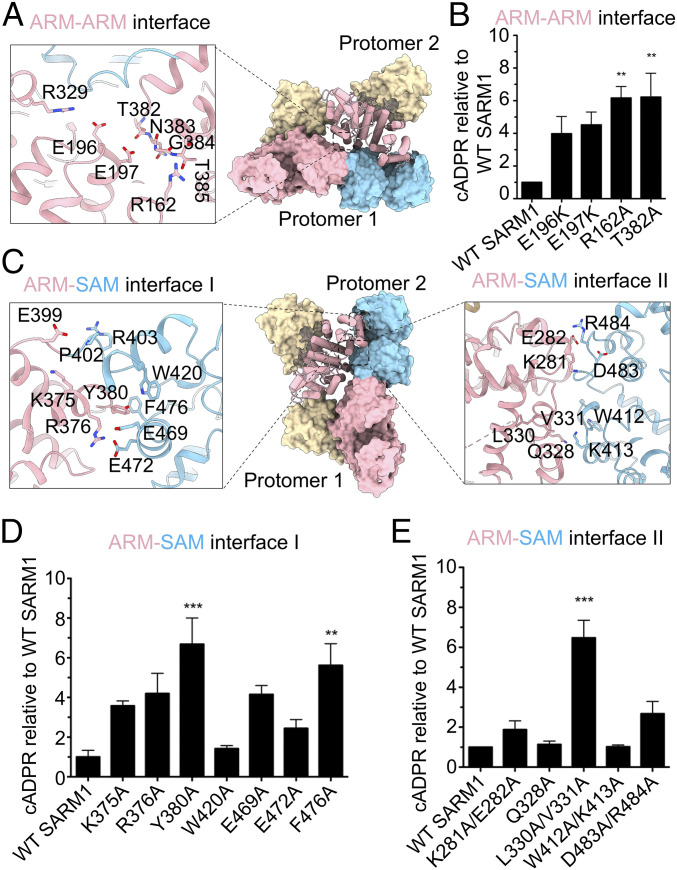
In the ARM-ARM interface, with a buried surface area of ∼96.2 Å2 per partner, E197 on α9 and R162 on α6 of one ARM domain interact with the adjacent ARM domain at a loop connecting α20 and α21 at residues T382-T385 (Fig. 5A and SI Appendix, Fig. S7). A major interaction comes from the side chain of E197 and the backbone amide nitrogen of G384, which stabilizes the hinge formed by residues T382, N383, G384, and T385 of the adjacent ARM domain (Fig. 5A). There is no obvious interaction between E196 and the T382-T385 loop. However, E196 is within the distance for a salt bridge with R329 of the same ARM domain to stabilize the folding of the ARM domain itself (Fig. 5A). We generated a number of mutants (E196K, E197K, R162A, and T382A) on both sides of this interface and found that all of them had at least a fourfold increase in cADPR levels, signifying constitutive activation (Fig. 5B). Thus, these ARM-ARM interactions occurring within the SARM1 octamer appear to be a critical regulator of SARM1 activity.
Fig. 5.
Structural and functional analyses of the ARM-ARM and ARM-SAM interfaces. (A) Interactions involved in the ARM-ARM interface. (B) Functional effects of mutations at the ARM-ARM interface. DRG sensory neurons from SARM1 KO mice were infected with SARM1 constructs with mutations in the ARM-ARM interface. Metabolites were extracted 3 dpi to measure cADPR levels relative to neurons infected with wild-type SARM1. (C) Interactions involved in ARM-SAM interface I (Left) and ARM-SAM interface II (Right). (D and E) Functional effects of mutations at ARM-SAM interfaces. DRG sensory neurons from SARM1 KO mice were infected with SARM1 constructs with mutations in ARM-SAM interface I (D) or ARM-SAM interface II (E). Metabolites were extracted 3 dpi to measure cADPR levels relative to neurons infected with wild-type SARM1. Error bars represent SEM; n = 3 to 4; one-way ANOVA with Dunnett posttest was used for statistical analysis with **P < 0.01 and ***P < 0.001.
The intramolecular ARM-SAM I interface (∼482.8 Å2 buried surface area per partner) involves residues K375-Y380 of the ARM domain and two regions of the connected SAM domain (Fig. 5 C, Left, and SI Appendix, Fig. S7). An electrostatic interaction between K375 and E399 of ARM appears to restrain the flexibility of the ARM-SAM linker (Fig. 5 C, Left). R376 forms hydrogen bonds with both E469 and E472 of the SAM domain, while Y380 is cradled by the hydrophobic residues W420 and F476 of the SAM domain (Fig. 5 C, Left). The K375A, R376A, and Y380A mutations in the ARM component of this interface each resulted in an over threefold increase in cADPR levels, signifying constitutive SARM1 activity (Fig. 5D). Analysis of mutants in the SAM side of this interface showed that E469A and F476A mutations resulted in constitutive SARM1 activity, and mutations at W420A and E472A had little effect (Fig. 5D).
The intermolecular ARM-SAM II interface (∼645.8 Å2 buried surface area per partner) involves α17 (D325-D335) on the ARM domain and residues P410-E416 at the N-terminal region of the SAM domain (Fig. 5 C, Right, and SI Appendix, Fig. S7). V331 of ARM establishes van der Waals interaction with W412 and K413 of the SAM domain in the adjacent protomer (Fig. 5 C, Right). The double mutation L330A/V331A in the ARM domain resulted in constitutive activity, while the Q328A mutation on a nearby residue or the opposing SAM domain double mutant W412A/K413A did not change SARM1 basal activity (Fig. 5E). Because L330 is buried within the ARM domain, the L330A/V331A double mutation may also interrupt the interior folding of the ARM domain to indirectly affect the interaction with the SAM domain, similar to how mutations L254S and L257S on buried residues of another ARM domain overcome SARM1 autoinhibition (Fig. 1 C and D and SI Appendix, Fig. S2 C and D). A second region of this interface occurs between α15 (K281-S290) of the ARM domain and a loop (F476-N486) connecting the two SAM domains. The charged residues K281/E282 of the ARM domain may have the potential to establish salt bridges with D483/R484 on the SAM domain (Fig. 5 C, Right). Disrupting this interface with the double mutation D483A/R484A on the SAM domain or the double mutation K281A/E282A in the ARM domain did not significantly change basal SARM1 activity (Fig. 5E). Taken together, these structural and functional studies reveal the metastability of the SARM1 autoinhibited state that requires all five interfaces in the octamer to maintain the off state. Disruption of any of these interfaces can result in loss of autoinhibition and lead to constitutive activation of the SARM1 NADase and neurodegeneration.
Discussion
Multisite Regulation of SARM1 Autoinhibition: Implications for Human Genetic Variations and Drug Discovery.
SARM1 is an inducible NADase and the major executioner of axon degeneration responsible for both Wallerian and dying-back axon loss in injury and disease (1, 2). Here we have investigated the mechanism by which SARM1 is held in an autoinhibited state in healthy neurons. Using peptide mapping, we first identified an evolutionarily conserved hydrophobic region of the ARM domain that is necessary and sufficient for SARM1 autoinhibition. A peptide encompassing this region (peptide 5) and the protein itself are inhibitors of the SARM1 NADase, while mutations of key residues in this region render SARM1 a constitutively active NADase capable of triggering degeneration in otherwise healthy neurons. Residues in this region promote the association of the SARM1 ARM domain with the enzymatic TIR domain, consistent with the model that these residues participate in the binding of the autoinhibitory ARM domain to the TIR domain.
We then solved the cryo-EM structure of full-length SARM1, which provided evidence that the preformed octameric assembly is an autoinhibited form and depicted an overview of the SARM1 autoinhibition state composed of five interdomain interfaces that are both intramolecular and intermolecular. It is intriguing that recombinant full-length SARM1 has enzymatic activity, yet we obtained its structure in the autoinhibited form. Most likely, the sample is a mixture of active and inactive conformations, and the inactive state is more stable and thus preferentially extracted in the cryo-EM analysis. In line with the identified important role of peptide 5, this region of the ARM domain, including the key residue W253, is a critical element mediating the ARM-TIR I interface through mainly hydrophobic interactions. Intriguingly, systematic mutagenesis of the structure-informed interdomain interfaces suggests that all these interfaces regulate the activity of SARM1 nonredundantly. That is, interactions at multiple interfaces together coordinate SARM1 autoinhibition, and disruption of any one of the interfaces relieves this inhibition and causes SARM1 activation. There may be two mechanisms by which the observed octameric ring structure inhibits TIR domain activity. First, previous studies suggested that TIR domains need to oligomerize to activate their NADase activity (18); thus, the octamer may restrain the TIR domain catalytic activity by holding the TIR domains in isolated states and preventing their oligomerization. Second, the BB-loop region of the TIR domain is in direct contact with the ARM domain as observed in the ARM-TIR II interface, which may keep the BB-loop in a conformation that is incompatible with catalysis.
The “multi-site” regulation in SARM1 autoinhibition also implies that SARM1 may be activated through different site-specific stimuli, in which the TIR activity is tightly controlled through some distal allosteric site. With an exquisite sensitivity of SARM1 to perturbations, human genetic variation in amino acids required for SARM1 autoinhibition would be predicted to generate constitutively active SARM1 proteins that could promote axon loss and be an unrecognized contributor to human neurodegenerative disease. On the other hand, these findings suggest that high-affinity SARM1 inhibitors can be derived from the natural autoinhibitory mechanism. Screens for small molecules that mimic or strengthen the autoinhibitory interactions will be a booming field for the future mechanistic study or therapeutic intervention of axon degeneration.
Allosteric Inhibition and the Mechanism of SARM1 Activation.
Not only must SARM1 be autoinhibited in healthy neurons, but this autoinhibition must be relieved in order to activate SARM1. Our findings here in conjunction with prior studies (27, 34) suggest that relief of autoinhibition likely entails the release of the TIR domain from the interaction with ARM domains intramolecularly and intermolecularly. Posttranslational modifications and ligand binding are rapid and local mechanisms to relieve autoinhibition in other proteins (24–26); however, the hydrophobic nature of the major ARM region that suppresses the TIR domain through direct interaction makes this region a poor candidate for modification or even ligand binding. Instead, we hypothesize that sensing of an injury-induced cue occurs at a site away from the direct ARM-TIR interfaces.
In two recently published papers (32, 33), the NAD+ substrate of SARM1 was shown to directly bind to the N-terminal ARM domain (SI Appendix, Fig. S8C), which was proposed to allosterically promote the interaction of the ARM domain with the TIR domain and to promote inhibition of the SARM1 NADase. Armadillo-repeat domains are typical scaffold domains initially identified in β-catenin, which are often responsible for the recognition of different substrates or ligands (35). Identification of NAD+ as a SARM1 ligand raises the probability that SARM1 may act as a pattern recognition receptor for sensing a different agonist or antagonist for NADase activity and downstream signaling. Similar cases can be found in inflammasomes in which the nucleotide-binding domain and leucine-rich-repeat domain sense ligands in different NLR proteins (36, 37). Allosteric inhibition by NAD+ at the ARM domain suggests that the same site may also transmit an allosteric activating signal to the TIR. Indeed, we previously demonstrated that the K193 residue involved in NAD+ interaction (32, 33) is required for SARM1 activation (38), consistent with the model that an injury sensor region including K193 transduces the injury signal to regions of the ARM domain in direct contact with the TIR domain to release the TIR for oligomerization and activation of enzymatic activity. What might this injury signal be? Upon axon injury, the NAD+ biosynthetic enzyme NMNAT2 is lost (39), leading to loss of NAD+ and an increase in the levels of its precursor NMN. Hence, either or both of these changes are candidate injury signals. Indeed, NMN may function through SARM1 to promote axon degeneration (40, 41), and recently it was shown that NMN and an NMN mimetic can enhance activity of the SARM1 enzyme (31, 34). Hence, NMN is a candidate injury signal for relief of SARM1 autoinhibition. Integration of our discovery of the SARM1 autoinhibition mechanism, work on SARM1 activation, and future structural studies should enable a comprehensive understanding of SARM1 regulation in both the healthy and diseased nervous systems.
Materials and Methods
Peptide Design and Synthesis.
Peptides were designed to be roughly 24 amino acids long with the goal of keeping evolutionarily conserved regions intact. Peptides were synthesized by GenScript to >80% purity.
Protein Expression and Purification.
For in vitro NADase activity assay, human SARM1 proteins were expressed and purified from HEK293T cells as previously described (20). For cryo-EM studies, N-terminal tobacco etch virus-cleavable MBP-tagged SARM1 (amino acids 20 to 700) was expressed in baculovirus-infected insect cells and purified by amylose affinity and gel filtration chromatography.
In Vitro NADase Assay, Metabolite Extraction, and HPLC.
Purified SARM1 protein on beads was incubated with peptides for 30 min at 37 °C. NAD+ (2.5 μM) was added, and the reaction was incubated at 37 °C for 2 h. For each purified SARM1 construct, we used sufficient SARM1 attached to beads to consume roughly half the added NAD+ in the absence of peptide. Metabolites were extracted using chloroform/methanol, lyophilized, and measured by HPLC as previously described (20).
Culture of Dorsal Root Ganglion Neurons.
Primary DRG cells were isolated from embryonic day 13.5 wild-type or SARM−/− embryos as previously described (29). Lentiviral particles containing SARM1 variants were created as previously described (29), and after 1 d in vitro DRG cultures were transduced with lentivirus.
Metabolite Extraction from DRG Neurons and Measurement by LC-MS/MS.
After 6 d in vitro, metabolites from DRG neuron cultures were collected and measured by LC-MS/MS as previously described (42).
Measurement of Axon Fragmentation.
DRGs were seeded into 96-well plates and distal axons were imaged using a high-content Operetta imager (PerkinElmer) 5 dpi. A previously designed ImageJ macro (29) was used to quantify axon degeneration from bright-field images by calculating a ratio of fragmented axon area to total axon area. For each experiment, this axon degeneration index was averaged over nine fields per well and four wells per condition.
Sensitized Emission FRET.
Measurements of FRET intensity of wild-type or M5 mutant SARM1-FRET constructs were performed as previously described (27).
The cryo-EM Data Collection, Processing, and Model Refinement.
We used an FEI Titan Krios microscope with a K3 direct detection camera to collect 4,776 micrographs of the SARM1 octamer with a pixel size of 0.825 Å. Two-dimensional (2D) and three-dimensional (3D) classifications, 3D refinement, symmetry expansion, and focused classification were conducted in Relion (30). The atomic model was built and refined in Coot (43) and PHENIX (44), respectively, and displayed using ChimeraX (http://www.rbvi.ucsf.edu/chimerax/) and Pymol (45).
Supplementary Material
Acknowledgments
This work was supported by NIH Grants R01CA219866 and RO1NS087632 (to A.D. and J.M.), RF1AG013730 (to J.M.), and AI050872 (to H.W.). We thank T. Fahrner, K. Simburger, X. Sun, and A. Neiner for technical assistance and members of the J.M., A.D., and H.W. laboratories for discussions. The cryo-EM data were collected with the assistance of Richard Walsh and Sarah Sterling at the Harvard Medical School cryo-Electron Microscopy Center supported by NIH National Institute of General Medical Sciences Grant GM103310. C.S. is supported by a Cancer Research Institute Irvington Postdoctoral Fellowship.
Footnotes
Competing interest statement: A.D. and J.M. are cofounders, scientific advisory board members, and shareholders of Disarm Therapeutics, a wholly owned subsidiary of Eli Lilly & Co. Y.S. is a consultant to Disarm Therapeutics.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2023151118/-/DCSupplemental.
Data Availability.
All study data are included in the article and/or supporting information. The atomic coordinates and cryo-EM map have been deposited in the Protein Data Bank (PDB), http://www.rcsb.org/ (PDB ID code 7KNQ) (46), and the EM Data Resource, https://www.emdataresource.org/ (ID code EMD-22954) (47).
References
- 1.Figley M. D., DiAntonio A., The SARM1 axon degeneration pathway: Control of the NAD+ metabolome regulates axon survival in health and disease. Curr. Opin. Neurobiol. 63, 59–66 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Coleman M. P., Höke A., Programmed axon degeneration: From mouse to mechanism to medicine. Nat. Rev. Neurosci. 21, 183–196 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.DiAntonio A., Axon degeneration: Mechanistic insights lead to therapeutic opportunities for the prevention and treatment of peripheral neuropathy. Pain 160 (suppl. 1), S17–S22 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Osterloh J. M., et al. , dSarm/Sarm1 is required for activation of an injury-induced axon death pathway. Science 337, 481–484 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gerdts J., Summers D. W., Sasaki Y., DiAntonio A., Milbrandt J., Sarm1-mediated axon degeneration requires both SAM and TIR interactions. J. Neurosci. 33, 13569–13580 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Geisler S., et al. , Vincristine and bortezomib use distinct upstream mechanisms to activate a common SARM1-dependent axon degeneration program. JCI Insight, 4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Geisler S., et al. , Prevention of vincristine-induced peripheral neuropathy by genetic deletion of SARM1 in mice. Brain 139, 3092–3108 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Turkiew E., Falconer D., Reed N., Höke A., Deletion of Sarm1 gene is neuroprotective in two models of peripheral neuropathy. J. Peripher. Nerv. Syst. 22, 162–171 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Henninger N., et al. , Attenuated traumatic axonal injury and improved functional outcome after traumatic brain injury in mice lacking Sarm1. Brain 139, 1094–1105 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ziogas N. K., Koliatsos V. E., Primary traumatic axonopathy in mice subjected to impact acceleration: A reappraisal of pathology and mechanisms with high-resolution anatomical methods. J. Neurosci. 38, 4031–4047 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marion C. M., McDaniel D. P., Armstrong R. C., Sarm1 deletion reduces axon damage, demyelination, and white matter atrophy after experimental traumatic brain injury. Exp. Neurol. 321, 113040 (2019). [DOI] [PubMed] [Google Scholar]
- 12.Ko K. W., Milbrandt J., DiAntonio A., SARM1 acts downstream of neuroinflammatory and necroptotic signaling to induce axon degeneration. J. Cell Biol. 219, e201912047 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Summers D. W., DiAntonio A., Milbrandt J., Mitochondrial dysfunction induces Sarm1-dependent cell death in sensory neurons. J. Neurosci. 34, 9338–9350 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.White M. A., et al. , Sarm1 deletion suppresses TDP-43-linked motor neuron degeneration and cortical spine loss. Acta Neuropathol. Commun. 7, 166 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ozaki E., et al. , SARM1 deficiency promotes rod and cone photoreceptor cell survival in a model of retinal degeneration. Life Sci. Alliance 3, 1–13 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sasaki Y., et al. , SARM1 depletion rescues NMNAT1 dependent photoreceptor cell death and retinal degeneration. bioRxiv, 1–19 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sporny M., et al. , Structural evidence for an octameric ring arrangement of SARM1. J. Mol. Biol. 431, 3591–3605 (2019). [DOI] [PubMed] [Google Scholar]
- 18.Horsefield S., et al. , NAD+ cleavage activity by animal and plant TIR domains in cell death pathways. Science 365, 793–799 (2019). [DOI] [PubMed] [Google Scholar]
- 19.Narayanan K. B., Park H. H., Toll/interleukin-1 receptor (TIR) domain-mediated cellular signaling pathways. Apoptosis 20, 196–209 (2015). [DOI] [PubMed] [Google Scholar]
- 20.Essuman K., et al. , The SARM1 toll/interleukin-1 receptor domain possesses intrinsic NAD+ cleavage activity that promotes pathological axonal degeneration. Neuron 93, 1334–1343.e5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Essuman K., et al. , TIR domain proteins are an ancient family of NAD+-consuming enzymes. Curr. Biol. 28, 421–430.e4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wan L., et al. , TIR domains of plant immune receptors are NAD+-cleaving enzymes that promote cell death. Science 365, 799–803 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gerdts J., Brace E. J., Sasaki Y., DiAntonio A., Milbrandt J., SARM1 activation triggers axon degeneration locally via NAD+ destruction. Science 348, 453–457 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nussinov R., et al. , Autoinhibition in Ras effectors Raf, PI3Kα, and RASSF5: A comprehensive review underscoring the challenges in pharmacological intervention. Biophys. Rev. 10, 1263–1282 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nussinov R., Tsai C. J., Jang H., Autoinhibition can identify rare driver mutations and advise pharmacology. FASEB J. 34, 16–29 (2020). [DOI] [PubMed] [Google Scholar]
- 26.Khan R. B., Goult B. T., Adhesions assemble! Autoinhibition as a major regulatory mechanism of integrin-mediated adhesion. Front. Mol. Biosci. 6, 144 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Summers D. W., Gibson D. A., DiAntonio A., Milbrandt J., SARM1-specific motifs in the TIR domain enable NAD+ loss and regulate injury-induced SARM1 activation. Proc. Natl. Acad. Sci. U.S.A. 113, E6271–E6280 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sasaki Y., et al. , cADPR is a gene dosage-sensitive biomarker of SARM1 activity in healthy, compromised, and degenerating axons. Exp. Neurol. 329, 113252 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sasaki Y., Vohra B. P. S., Lund F. E., Milbrandt J., Nicotinamide mononucleotide adenylyl transferase-mediated axonal protection requires enzymatic activity but not increased levels of neuronal nicotinamide adenine dinucleotide. J. Neurosci. 29, 5525–5535 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Scheres S. H. W., RELION: Implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bratkowski M., et al. , Structural and mechanistic regulation of the pro-degenerative NAD hydrolase SARM1. Cell Rep. 32, 107999 (2020). [DOI] [PubMed] [Google Scholar]
- 32.Jiang Y., et al. , The NAD+-mediated self-inhibition mechanism of pro-neurodegenerative SARM1. Nature 588, 658–663 (2020). [DOI] [PubMed] [Google Scholar]
- 33.Sporny M., et al. , Structural basis for SARM1 inhibition and activation under energetic stress. eLife 9, 1–25 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao Z. Y., et al. , A cell-permeant mimetic of NMN activates SARM1 to produce cyclic ADP-Ribose and induce non-apoptotic cell death. iScience 15, 452–466 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Valenta T., Hausmann G., Basler K., The many faces and functions of β-catenin. EMBO J. 31, 2714–2736 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sharif H., et al. , Structural mechanism for NEK7-licensed activation of NLRP3 inflammasome. Nature 570, 338–343 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tenthorey J. L., et al. , The structural basis of flagellin detection by NAIP5: A strategy to limit pathogen immune evasion. Science 358, 888–893 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Geisler S., et al. , Gene therapy targeting SARM1 blocks pathological axon degeneration in mice. J. Exp. Med. 216, 294–303 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gilley J., Coleman M. P., Endogenous Nmnat2 is an essential survival factor for maintenance of healthy axons. PLoS Biol. 8, e1000300 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gilley J., Orsomando G., Nascimento-Ferreira I., Coleman M. P., Absence of SARM1 rescues development and survival of NMNAT2-deficient axons. Cell Rep. 10, 1974–1981 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Di Stefano M., et al. , NMN deamidase delays Wallerian degeneration and rescues axonal defects caused by NMNAT2 deficiency in vivo. Curr. Biol. 27, 784–794 (2017). [DOI] [PubMed] [Google Scholar]
- 42.Sasaki Y., Nakagawa T., Mao X., DiAntonio A., Milbrandt J., NMNAT1 inhibits axon degeneration via blockade of SARM1-mediated NAD+ depletion. eLife 5, 1–15 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Emsley P., Cowtan K., Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004). [DOI] [PubMed] [Google Scholar]
- 44.Adams P. D., et al. , PHENIX: A comprehensive python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.DeLano W. L., The PyMOL Molecular Graphics System (Version 1, Schrödinger LLC, 2002).
- 46.Shen C., Wu H., SARM1 octamer. Protein Data Bank. https://www.rcsb.org/structure/7KNQ. Deposited 5 November 2020.
- 47.Shen C., Wu H., SARM1 octamer. EMDataBank. https://www.emdataresource.org/EMD-22954. Deposited 5 November 2020.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or supporting information. The atomic coordinates and cryo-EM map have been deposited in the Protein Data Bank (PDB), http://www.rcsb.org/ (PDB ID code 7KNQ) (46), and the EM Data Resource, https://www.emdataresource.org/ (ID code EMD-22954) (47).