Significance
Chemical methods to install histone posttranslational modifications in live cells promise extraordinary potential in epigenome biology and medicine due to the use of endogenous protein levels and lack of genetic manipulation. In this study, we developed a chemical catalyst, PEG-LANA-DSSMe, that binds with nucleosome’s acidic patch and promotes regioselective, synthetic histone acetylation at H2BK120 in living cells. Importantly, the synthetic acetylation suppressed H2B ubiquitination, a critical epigenome mark in cell physiology. This achievement is an example of epigenome manipulation in living cells, entirely relying on the chemical system. In the long term, our catalyst system with further improvements may contribute to both therapy and elucidation of epigenetic mechanisms.
Keywords: catalyst, epigenome, histone, acetylation, ubiquitination
Abstract
Chemical modifications of histones, such as lysine acetylation and ubiquitination, play pivotal roles in epigenetic regulation of gene expression. Methods to alter the epigenome thus hold promise as tools for elucidating epigenetic mechanisms and as therapeutics. However, an entirely chemical method to introduce histone modifications in living cells without genetic manipulation is unprecedented. Here, we developed a chemical catalyst, PEG-LANA-DSSMe 11, that binds with nucleosome’s acidic patch and promotes regioselective, synthetic histone acetylation at H2BK120 in living cells. The size of polyethylene glycol in the catalyst was a critical determinant for its in-cell metabolic stability, binding affinity to histones, and high activity. The synthetic acetylation promoted by 11 without genetic manipulation competed with and suppressed physiological H2B ubiquitination, a mark regulating chromatin functions, such as transcription and DNA damage response. Thus, the chemical catalyst will be a useful tool to manipulate epigenome for unraveling epigenetic mechanisms in living cells.
The eukaryotic epigenome is defined by multiple chemical modifications of biomacromolecules, including DNA methylation and histone posttranslational modifications (PTMs), and plays a pivotal role in regulating gene expressions. Dysregulation of the epigenome is tightly linked to numerous disorders in humans, such as type 2 diabetes, Alzheimer’s disease, and various types of cancer (1–3). Thus, new methods to manipulate the epigenome hold promise for both therapy and elucidation of epigenetic mechanisms. The levels of histone PTMs are regulated by the interplay of two groups of enzymes; writers which introduce PTMs (e.g., histone acetyltransferase, HAT) and erasers which remove them (e.g., histone deacetylase, HDAC). One approach to manipulate histone PTMs is use of small molecule inhibitors of these chromatin-modifying enzymes. So far, several HDAC inhibitors have been approved for the treatment of cancer, and further candidates are under clinical evaluation (4).
In recent years, new chemical biology approaches that introduce histone PTMs entirely through artificial means without reliance on chromatin-modifying enzymes have emerged (5). These include protein semisynthesis using native chemical ligation (6, 7), chemistry-coupled posttranslational mutagenesis (8, 9), and synthetic histone acetylation by chemical catalysis (10, 11). Among these, the chemical catalysis approach is unique because it does not require any genetic manipulations, and thus has great potential for therapeutic use and broad epigenome-related biology studies. So far, two chemical catalyst systems to directly modify histones in nucleosomes have been reported. The first catalyst system is composed of an oligoDMAP-based catalyst (DMAP: 4-dimethylaminopyridine), such as 8DMAP or 16DMAP, and an acetyl donor, such as an N-methoxydiacetamide derivative or a phenyl acetate-derivative (PAc-gly) (10, 12). These oligocationic catalysts bind to nucleosomes through electrostatic interactions with negatively charged DNA. The DMAP moiety of the catalyst activates the acetyl donor to generate an acetyl pyridinium ion intermediate, the active species promoting acetylation of proximate lysine residues on histone tails. Interestingly, synthetic histone acetylation of Xenopus chromatin by 16DMAP with PAc-gly prevented DNA replication in cell-free Xenopus egg extract system, providing a novel link between histone acetylation and cell cycle (12). However, oligoDMAP-based catalysts did not work in live cells, probably due to inactivation of the catalytically active species (i.e., acetyl pyridinium ion) under highly reductive and nucleophilic environment in cells (e.g., the presence of glutathione [GSH]) (13). The second catalyst system composed of LANA-DSH catalyst, in which a DSH catalyst is conjugated with a chromatin-binding LANA (latency-associated nuclear antigen) peptide, binds to the acidic patch of nucleosomes, and efficiently promotes histone- and site-selective acetylation of the proximate H2BK120 residue in both recombinant nucleosomes and endogenous chromatin in isolated nuclei, using acetyl thioesters (e.g., acetyl-CoA) as acetyl donors (11) (Fig. 1A). In contrast to the oligoDMAP-based catalysts, ligand-conjugated DSH catalysts can acetylate target proteins in living cells when the ligands are stable and functional in the cells (14). Biologically significant protein acetylation in living cells, such as histone acetylation, however, was not achieved yet. Thus, we aimed at manipulating epigenome in living cells by a chemical catalyst system without any genetic manipulations. We here disclose a chemical catalyst system, composed of PEG-LANA-DSH and NAC-Ac, enabling regioselective histone acetylation and epigenome manipulation in living cells.
Fig. 1.
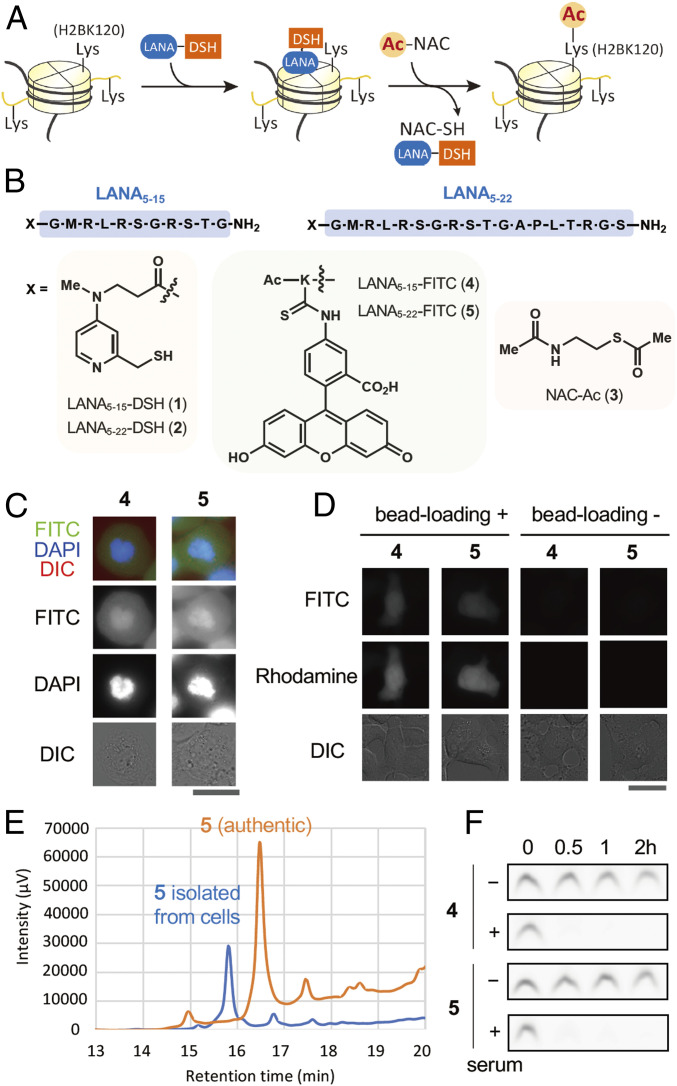
Histone-acetylation catalyst LANA-DSH is unstable in living cells. (A) Schematics of site-selective histone acetylation by LANA-DSH. LANA-DSH binds to nucleosomes, activates the acetyl donor (NAC-Ac), and acetylates H2BK120. (B) Structures of LANA5–15-DSH 1, LANA5–22-DSH 2, NAC-Ac 3, LANA5–15-FITC 4, and LANA5–22-FITC 5. (C) Representative images of LANA-FITC (4 and 5) with DAPI and DIC in fixed HeLa S3 cells. (Scale bar, 25 µm.) (D) Representative microscopic images of LANA-FITC (4 and 5) in living HeLa S3 cells. (Left) LANA-FITC (100 μM) was introduced with dextran-rhodamine (loading control) by bead loading. (Right) ANA-FITC (50 μM) was treated with cells for 30 min. (Scale bar, 25 μm.) (E) HPLC traces of chart of 5 with fluorescence detection (ex = 494 nm, em = 518 nm). Authentic peptide 5 (orange), and recovered peptide from the cells (blue). (F) Serum stability assay of 4 and 5. Each peptide was incubated with or without 25% human-serum-containing medium at 37 °C for indicated time.
Results
Histone-Acetylation Catalyst LANA-DSH Is Unstable in Living Cells.
We first examined whether LANA-DSH can be applied for in-cell histone acetylation. We used two types of LANA-DSH catalysts, LANA5–15-DSH 1 and LANA5–22-DSH 2. Because 1 and 2 (and other LANA-conjugated compounds 4–10 used in this study) were not cell permeable, we used bead-loading method (15) to introduce the compounds into human cultured cells. NAC-Ac 3 was used as a cell-permeable acetyl donor (14) (Fig. 1B). As reported previously (11), these catalysts efficiently promoted acetylation of H2BK120 in recombinant nucleosomes in test tubes (SI Appendix, Fig. S1). However, we could not detect H2BK120 acetylation by the catalyst system in living cells. Since LANA is composed of natural amino acids, we hypothesized that LANA-DSH catalysts were rapidly decomposed in living cells. To examine in-cell stability of LANA, we synthesized LANA5–15-FITC 4 and LANA5–22-FITC 5 (Fig. 1B). Using fixed mitotic cells, we confirmed that both compounds have affinity to chromatin (Fig. 1C). However, when these compounds were introduced into living cells, they did not show any chromatin localization (Fig. 1D). We analyzed 5 recovered from living cells by HPLC and found that the compound was almost completely decomposed (Fig. 1E). Further, both 4 and 5 were immediately decomposed in the presence of human serum in vitro (Fig. 1F). These data indicate that LANA-DSH catalysts are decomposed by proteases in living cells.
Conjugation of PEG Stabilizes LANA in Living Cells.
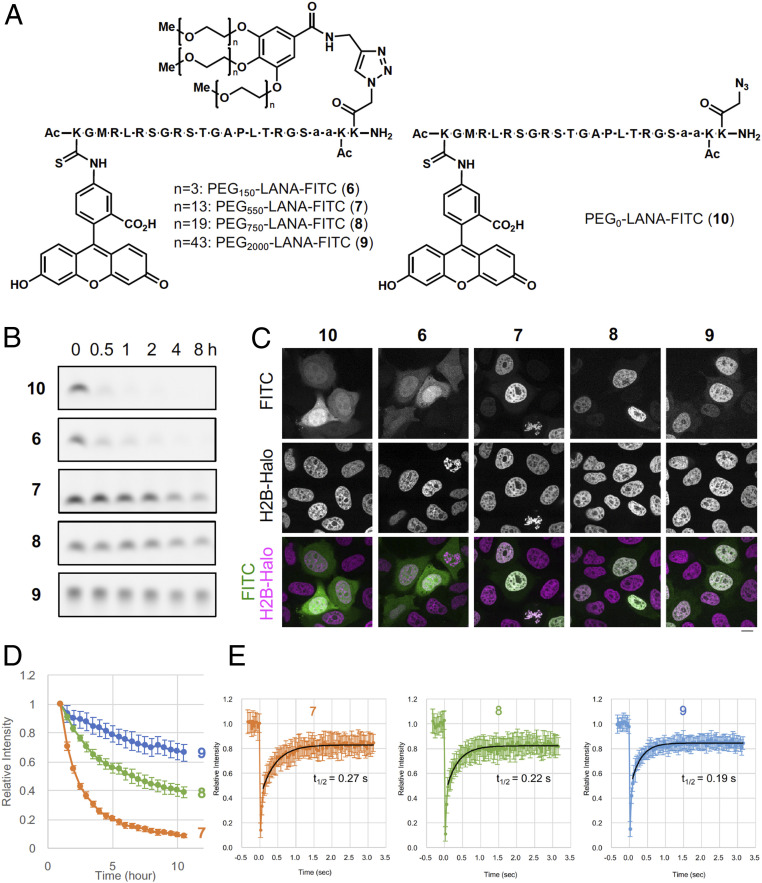
Since conjugation with poly(ethylene glycol) [PEG] moieties (i.e., pegylation) has been used to improve the pharmacokinetic and pharmacodynamic properties of peptides and proteins (16), we next expected that conjugation of a PEG moiety with a proper size to LANA should increase stability toward proteolysis. We thus synthesized LANA5–22-FITC conjugated with three PEG chains, whose molecular weight was ∼150 (PEG150-LANA-FITC 6), 550 (PEG550-LANA-FITC 7), 750 (PEG750-LANA-FITC 8), or 2,000 (PEG2,000-LANA-FITC 9) (Fig. 2A). As a negative control, we also synthesized PEG0-LANA-FITC 10 without the PEG chains (Fig. 2A). We compared stability of these compounds in the presence of human serum. While 10 and 6 were decomposed within 30 min, 7, 8, and 9 were much more stable (Fig. 2B). We then examined the stability of compounds 6–10 in HeLa cells expressing H2B-HaloTag. The localizations of the compounds were analyzed by fluorescent microscopy. If the compounds are stable in living cells, they bind with nucleosomes via the LANA moiety and localize to the chromatin regions, which are visualized by H2B-HaloTag signals. Consistent with the serum stability assay, 10 and 6 did not localize on chromatin, indicating that these two compounds were immediately decomposed in living cells (Fig. 2C). In contrast, the signals of 7, 8, and 9 colocalized with that of H2B-HaloTag, indicating that these compounds are sufficiently stable in living cells and retain binding affinity of the LANA moiety to chromatin histones (Fig. 2C). Time-lapse imaging showed that attachment of longer PEG chains conferred greater in-cell stability to LANA-FITC (Fig. 2D). The approximate half-lives of 7, 8, and 9 were calculated to be 1.2, 5.3, and 20 h, respectively. We further examined residence times of the compounds on chromatin in living cells by fluorescence recovery after photobleaching (FRAP). In cells loaded with PEG-LANA-FITC, a small spot was photobleached and the rate of fluorescence recovery was measured (Fig. 2E). The recovery t1/2 corresponding to the residence time of 7 was 0.27 s, indicating that 7 bound with chromatin transiently. Residence time of 8 (t1/2 = 0.22 s) or 9 (t1/2 = 0.19 s) was shorter than that of 7, suggesting that conjugation with longer PEG may reduce affinity of PEG-LANA-FITC with chromatin. Consistently, electrophoretic mobility shift assay of 7–10 showed that the conjugation with longer PEG chains weakened the binding to recombinant nucleosomes (SI Appendix, Fig. S2). Judging from the results of the FRAP experiments with 7, we estimated the dissociation constant (Kd) of 7 in living cells to be ∼100 µM (SI Appendix, Fig. S3 and SI Appendix). This value was ∼60× higher than that in test tubes, suggesting that in-cell reaction may require higher concentrations of the catalysts than the reactions in test tubes. Taken together, these data demonstrate that conjugation of a longer PEG chain with the LANA ligand is advantageous for in-cell stability, but disadvantageous for LANA-mediated nucleosome binding.
Fig. 2.
Conjugation of PEG stabilizes LANA in living cells. (A) Structures of PEG-LANA-FITC (6–10). (B) Serum stability assay of 6–10. Each peptide was incubated with 25% human-serum-containing medium at 37 °C for indicated times. (C) Representative images of PEG-LANA-FITC 6–10 in living cells. HeLa cells expressing H2B-HaloTag were stained with HaloTag TMR Ligand and loaded with PEG-LANA-FITC. (Scale bar, 10 µm.) (D) Stability of 7–9 in living HeLa cells. Fluorescence images of PEG-LANA-FITC-loaded cells were acquired every 30 min and nuclear intensities were measured. Graphs represent relative fluorescence intensities (averages with SDs; n = 12 each). (E) Photobleaching was performed using HeLa cells loaded with 7–9. Average intensities with SDs (n = 20) are shown with curves after fitting (black lines). Half-time recoveries are also indicated.
PEG-LANA-DSH Catalysts Promotes Histone H2BK120-Selective Acetylation in Test Tube.
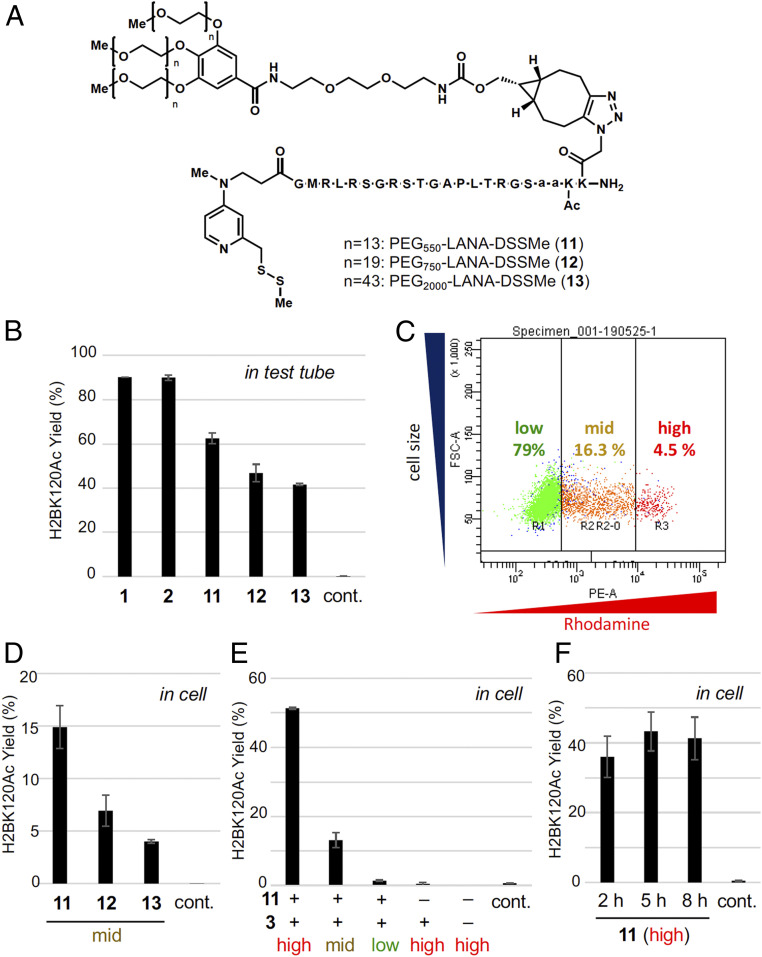
We then synthesized chemical catalysts conjugated with PEG-LANA ligands (Fig. 3A). As the catalyst moiety, we used DSSMe, a prodrugged DSH catalyst (14), which is reduced to the active DSH form by biocompatible reductants, such as GSH or Tris(2-carboxyethyl)phosphine (TCEP), prior to the reaction. The acetylation activity of the three catalysts (PEG550-LANA-DSSMe 11, PEG750-LANA-DSSMe 12, and PEG2,000-LANA-DSSMe 13) was first examined in test tubes containing recombinant nucleosomes as substrates. All the catalysts promoted acetylation of H2BK120 in recombinant nucleosomes, although the activity of PEG-LANA-DSSMe 11–13 was slightly weaker than that of LANA-DSH 1 and 2 (Fig. 3B).
Fig. 3.
Histone H2BK120 acetylation by PEG-LANA-DSH in living cells. (A) Structures of PEG-LANA-DSSMe (11–13). (B) LC-MS/MS analysis of acetylation yield for H2BK120 on a recombinant nucleosome (0.37 µM) treated with 3 (10 mM) and TCEP (0.1 mM) in the presence of 1, 2, or 11–13 (2 µM) or in the absence of any catalysts (control) at 37 °C for 5 h in a test tube. Error bars represent the data range of two independent experiments. (C) Representative dot plot of rhodamine signals (horizontal) and cell size (vertical). HeLa S3 cells were loaded with PEG-LANA-DSSMe and dextran-rhodamine, and analyzed by flow cytometry. Judging from the strength of rhodamine signals, high, mid, and low fractions were defined and separated. (D–F) H2BK120 acetylation yield in living cells determined by LC-MS/MS analysis. HeLa S3 cells pretreated with BSO (100 µM) were loaded with 11, 12, or 13 (500 µM), TCEP (2 mM), dextran-rhodamine (0.5 mg/mL), and/or 3 (10 mM), and incubated with growth medium containing 3 (30 mM) at 37 °C for 8 h or indicated time. Acetylation yield of high, mid, or low fractions are shown. Control indicates acetylation yield without any treatment. Error bars represent the data range of two independent experiments.
To quantitatively address histone selectivity of the catalyst system, we performed acetylation reactions using a mixture of recombinant nucleosomes and Escherichia coli dihydrofolate reductase (eDHFR) as a substrate, and quantified acetylation yield of 16 lysines in nucleosome and 6 lysines in eDHFR using liquid chromatography-tandem mass spectrometry (LC-MS/MS) analyses. Donor treatment only did not significantly increase acetylation (<5% yield) in all the lysine residues in nucleosome and eDHFR (SI Appendix, Fig. S4). Addition of catalyst 11, however, significantly increased H2BK120 acetylation (81% yield) with slight increase of acetylation of H2BK116 (10% yield), which is the second nearest lysine to the catalyst position, and did not significantly increase acetylation in other lysine residues in nucleosome and eDHFR (<6% yield, SI Appendix, Fig. S4).
The Optimized Catalyst 11 Promotes H2BK120 Acetylation in up to 51% Yield in Living Cells.
Next, we addressed whether PEG-LANA-DSSMe catalysts can promote H2BK120 acetylation in living cells. Cells were pretreated with l-buthionine-sulfoximine (BSO) to reduce the intracellular concentration of GSH, because we previously showed that the activity of DSH catalyst slightly decreased in the presence of high concentration of GSH (11). We confirmed that BSO treatment did not affect HDAC/HAT activities or H2BK120 ubiquitination level (SI Appendix, Fig. S5). Then, PEG-LANA-DSSMe catalysts 11‒13 were reduced by TCEP and loaded into HeLa S3 cells by bead loading. Here, we added dextran-rhodamine as a loading control. The levels of dextran-rhodamine in cells represented the intracellular concentration of catalysts. After the acetylation reaction was conducted by adding acetyl donor 3, cells were stained with 4′,6-diamidine-2′-phenylindole dihydrochloride (DAPI) to exclude potential artifacts caused by contaminated dead cells. We sorted DAPI-negative cells (i.e., living cells) with different levels of rhodamine signals (high, mid, or low) by flow cytometry (Fig. 3C), and acetylation yield was determined by LC-MS/MS analysis. Overall, the percentage of DAPI-negative cells was more than 90% (SI Appendix, Fig. S6). The same experiments using 7, instead of 11‒13, suggested that the “high” fraction may contain ∼3.2 × 10−16 mol 7 in a cell, which corresponds to 427 µM effective concentration in nuclei provided that all the catalysts loaded into the cells existed inside the nuclei, while the “mid” and “low” fractions may contain 0.67 × 10−16 and 0.054 × 10−16 mol 7/cell compounds, respectively (SI Appendix, Fig. S7).
We quantified the yield of H2BK120 acetylation by three PEG-LANA-DSSMe catalysts 11–13 in the "mid" live cells. All catalysts promoted H2BK120 acetylation in living cells (Fig. 3D and SI Appendix, Fig. S8). Since the acetylation yield by 11 was higher than that by 12 or 13, we determined 11 to be the optimum chemical catalyst for in-cell reaction, and used catalyst 11 in the following studies. We performed two negative control experiments (SI Appendix, Fig. S9). First, we synthesized PEG-LANAmut-DSSMe 14, containing a mutant form of the LANA peptide that cannot bind to the nucleosome and examined its acetylation activity. 14 did not promote H2BK120 acetylation, indicating the importance of targeting the catalysts to nucleosome in cells. Second, we separately treated two populations of cells with either catalyst 11 alone or acetyl donor 3 alone, then mixed those two populations of the cells, and analyzed the acetylation level. H2BK120 acetylation was not observed in this case, confirming that 11 promoted acetylation in cells. For the cells containing a high level of 11, H2BK120 was 51% acetylated (Fig. 3E). A time-course analysis revealed that the conversion plateaued within 5 h (Fig. 3F). This reaction profile was consistent with the 1.2-h half-life of 7 in cells as determined from the time-lapse experiments (Fig. 2D). In cells containing middle level of 11, 13% H2BK120 was acetylated (Fig. 3E), indicating that efficiency of the acetylation reaction was dependent on the catalyst concentration in cells.
Synthetic Histone Acetylation by Catalyst 11 Blocks H2B Ubiquitination.
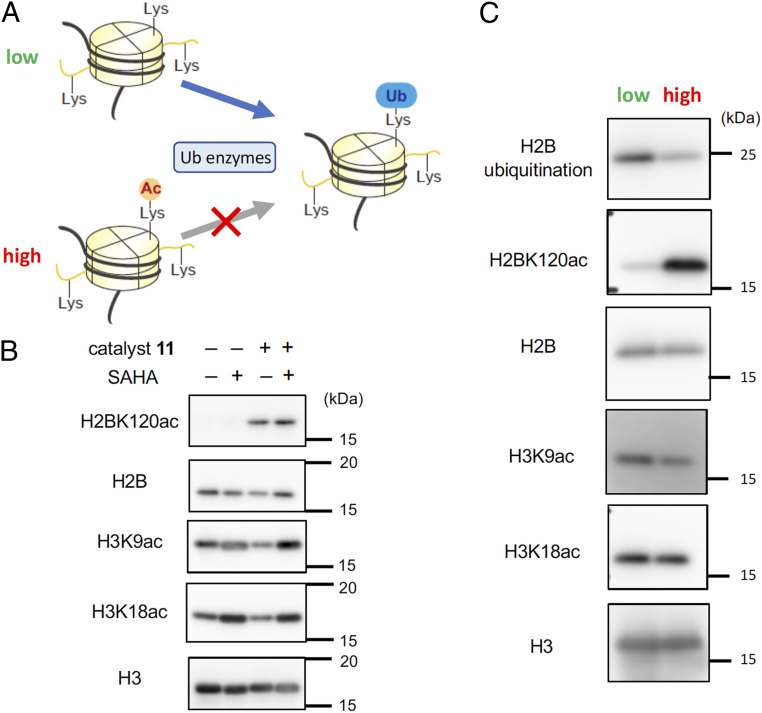
Finally, to investigate the impact of the synthetically introduced H2BK120 acetylation on the epigenome of living cells, we examined the level of H2BK120 ubiquitination, a mark regulating various chromatin functions, such as transcription and DNA damage response (17). We hypothesized that the acetyl group at H2BK120 would function as a protecting group, thus inhibiting the physiological ubiquitination reaction at the same residue mediated by ubiquitin ligases (Fig. 4A). We first addressed whether H2BK120 acetylation is removed by HDAC in cells. While H3 tail acetylation level was significantly increased by treatment of suberoylanilide hydroxamic acid (SAHA), an HDAC inhibitor, H2BK120 acetylation level was not changed by the treatment (Fig. 4B), indicating that H2BK120 is not the main target of HDACs in living cells. Thus, synthetically introduced H2BK120Ac is likely to be stable in cells. We then compared the H2BK120 ubiquitination level in cells containing high or low level of H2BK120 acetylation and found that H2BK120 ubiquitination level was significantly reduced in cells with high-level H2BK120 acetylation (Fig. 4C and SI Appendix, Figs. S10 and S11). In contrast, the acetylation level at histone tail was not affected by high-level H2BK120 acetylation (Fig. 4C). These data showcase that synthetic H2BK120 acetylation in living cells mediated by the chemical catalyst system can work as an inhibitor of H2BK120 ubiquitination and perturb the epigenetic states.
Fig. 4.
Synthetic histone acetylation by PEG-LANA-DSH blocks H2B ubiquitination. (A) Schematics of possible effects of synthetic H2BK120 acetylation. (B) HeLa S3 cells pretreated with BSO (100 μM) were loaded with 11 (500 μM), TCEP (2 mM), dextran-rhodamine (0.5 mg/mL), and 3 (10 mM), and incubated with growth medium containing 3 (30 mM) at 37 °C for 8 h. After further incubating the cells with growth medium without the catalyst and donor at 37 °C for 36 h, the cells were treated with or without SAHA (1 μM) for 6 h at 37 °C. Cells were recovered without sorting by flow cytometer, and histones were purified. The levels of H2BK120ac and H3-tail acetylation (H3K9ac and H3K18ac) were examined by Western blotting. Representative data of two independent experiments are shown. (C) Levels of H2BK120ac, H2BK120 ubiquitination, and H3-tail acetylation (H3K9ac and H3K18ac) in H2BK120-acetylated or nonacetylated cells. HeLa S3 cells pretreated with BSO (100 µM) were loaded with 11 (500 µM), TCEP (2 mM), dextran-rhodamine (0.5 mg/mL), and 3 (10 mM), incubated with growth medium containing 3 (30 mM) at 37 °C for 8 h for the chemical acetylation reaction and further incubated with growth medium in the absence of catalysts and donors at 37 °C for 36 h for equilibrating the ubiquitination/deubiquitination processes mediated by the physiological enzymes in living cellular machinery. Histones were purified from high or low cell fraction obtained through sorting by flow cytometry, and analyzed by Western blotting. Representative data of two independent experiments are shown.
Discussion
Chemical methods to install PTMs in live cells promise extraordinary potential in biology and medicine due to the use of endogenous protein levels and lack of genetic manipulation. In this study, we developed a chemical system comprising a protease-resistant nucleosome-binding catalyst (PEG-LANA-DSH) and cell-permeable acetyl donor (NAC-Ac), enabling regioselective lysine acetylation of chromatin histones in living cells. Increasing the size of PEG conferred the catalyst with greater in-cell stability, but reduced its acetylation activity, indicating that there is an optimum PEG length balancing the stability and catalyst activity. We applied bead loading to deliver an active catalyst into live cells and subsequent sorting to analyze the successfully introduced cells, which can bypass the common and highly problematic endocytic entrapment of catalyst cargo. The optimized catalyst 11 promoted H2BK120 acetylation in up to 51% yield in living cells in a catalyst-concentration dependent manner.
We provided quantitative chemo-physical parameters, such as half-lives and binding constants of the pegylated catalysts in living cells. It is noteworthy that the dissociation constant (Kd) of 7 in living cells was ca. 60 times higher than that in test tubes. This is probably due to the presence of many competing acidic patch-binding proteins in living cells, such as Dot1L (18) and RCC1 (19, 20). Interestingly, those proteins often exert bivalent binding with nucleosome. For example, Dot1L binds with acidic patch and DNA via its catalytic domain and DNA-binding domain, respectively. It was reported that the presence of DNA binding domain lowered the Kd value of Dot1L catalytic domain by ∼60 times (18). Therefore, imparting a DNA-binding motif, such as octa-arginine, to the catalyst would improve its nucleosome-binding affinity, which may enable more efficient synthetic acetylation reaction in living cells with lower catalyst loading.
We also demonstrated that the chemical catalyst system modulated epigenome in living cells without genetic manipulation: synthetic H2BK120 acetylation suppressed H2B ubiquitination, a critical epigenome mark in cell physiology (17). This is the first example of epigenome manipulation in living cells entirely relying on the chemical system. Our data indicate that synthetic H2BK120 acetylation is not removed by HDACs, and thus it can continuously block H2BK120 from ubiquitination enzymes. This mode of inhibitory action is conceptually analogous to regioselective protection of a functional group existing in a small organic molecule, a frequently-used strategy in organic synthesis. This function of H2BK120ac may be somewhat inconsistent with the report claiming that H2BK120ac precedes H2B ubiquitination deposition (21). Further analysis for the functions of H2BK120 acetylation is needed. It is known that the presence of H2B ubiquitination on nucleosomes can enhance the activity of Dot1L catalyzing H3K79 methylation (22), and both Dot1L and the H2B E3 ubiquitin ligase ring finger protein 20 (RNF20) are required for MLL-rearranged leukemia (23, 24). Therefore, in the long term, our catalyst system may be developed as a unique H2B ubiquitination inhibitor, which may be applicable for anti-MLL-rearranged leukemia, although it requires further improvement of the system, such as improving the method of introducing the catalyst or the cell membrane permeability of the catalyst.
Materials and Methods
In Vitro Histone Acetylation and Sample Preparation for LC-MS/MS Analysis.
Recombinant nucleosomes and eDHFR-GFP were prepared as described previously (10, 14). LANA-DSH or PEG-LANA-DSSMe was preincubated with NAC-Ac in 20 mM Tris⋅HCl (pH 7.5) with TCEP at room temperature (r.t.) for 1 h, and recombinant nucleosomes (0.37 μM for DNA/histone octamer concentration), or recombinant nucleosomes and eDHFR-GFP (1 µM) were added. The mixture was incubated at 37 °C for 5 h. The nucleosomes and eDHFR-GFP were precipitated by trichloroacetic acid (16.6%). After DNA was digested by DNase I (Takara, 2270A) for 30 min at 37 °C, the samples were mixed with acetone (74%). After overnight incubation at −30 °C, the proteins were collected by centrifugation, air dried, and dissolved in Milli-Q water (MQ). To the solution, 50 mM aqueous ammonium bicarbonate (NH4HCO3 aq.) and 25% propionic anhydride solution (methanol/propionic anhydride, 3:1 (vol/vol)) were added, and pH was adjusted to 8 by adding ammonia solution. After 30-min incubation at r.t., the solvents were removed by Speed-Vac evaporator. The samples were resuspended in 50 mM NH4HCO3 aq. with 0.1% ProteaseMAX (Promega, V2072) and digested with 10 ng/μL Trypsin Gold (Promega, V5280), 10 ng/μL Glu-C (Promega, V1651) in 50 mM NH4HCO3 aq. with 0.02% ProteaseMAX at 37 °C for 3 h or overnight. Then 5% aqueous formic acid (vol/vol) was added, and the solvents were removed by Speed-Vac evaporator to obtain dried digested samples, which were dissolved in 0.1% aqueous formic acid (vol/vol). After centrifugation (15,000 rpm, 5 min), the supernatant was used for LC-MS/MS analysis.
LC-MS/MS Analysis and Quantification of the Stoichiometry of Acetylation.
LC-MS/MS analysis was conducted as described previously with some modifications (10). For eDHFR, LC was performed as follows: linear gradient of 2–90% acetonitrile with 0.1% formic acid (vol/vol) versus water with 0.1% formic acid (vol/vol) over 17 min at 40 °C with a flow rate of 20 µL min−1 after 1 min equilibration at 2%. Targeted precursor ions and collision energies for H2B114-125 and eDHFR are described in SI Appendix, Tables S1 and S2. Data analysis was carried out using PeakView software (AB Sciex, Version 1.2.0.3) as described previously (10), and the average values were obtained from duplicated measurements. The stoichiometry of acetylated lysines was calculated as a percentage of the total peak area of the extracted ion chromatogram for acetylated peptides in the sum of those for acetylated peptides and propionylated peptides. For histones except H2B114-125 peptide, selected fragment ions are the same as previously described (10). For H2B114-125 peptide and eDHFR, selected fragment ions are shown in SI Appendix, Tables S1 and S2.
Cell Culture and Treatment.
HeLa S3 cells were grown in Dulbecco’s modified Eagle’s medium (DMEM; Gibco, 12430) with 10% fetal bovine serum (FBS; Gibco), 1× GlutaMAX (Gibco), 100 U/mL penicillin, and 100 µg/mL streptomycin (Gibco) at 37 °C in an atmosphere of 5% CO2. Cells were treated with 1 µM SAHA (Sigma, SML0061) and/or 100 µM BSO (Sigma, B2515) in growth medium.
Fluorescent Microscopy for Fixed Cells.
To observe the subcellular distribution of FITC-labeled compounds in fixed cells, HeLa S3 cells were cultured in glass-bottom dishes and washed with PBS, followed by fixation with 4% paraformaldehyde in PBS at r.t. for 10 min. Fixed cells were washed twice with TBS-TX (25 mM Tris⋅HCl (pH7.4), 137 mM NaCl, 2.7 mM KCl, 0.1% Triton X-100), incubated for 5 min in TBS-TX, and treated with 3% Bovine Serum Albumin (BSA; Sigma, 10735086001) in TBST [20 mM Tris⋅HCl (pH7.5), 150 mM NaCl, 0.1% Tween20] at r.t. for 30 min. Then, the cells were incubated with 80-µM FITC-labeled compounds in TBST with 1% BSA for 1 h, washed three times with TBS-TX, and imaged by fluorescent microscopy (DMi8; Leica) in TBST containing 1 µg/mL DAPI (Dojindo Molecular Technologies).
Fluorescent Microscopy for Living Cells.
To observe the subcellular distribution of fluorescein isothiocyanate (FITC)-labeled compounds in living cells, HeLa S3 cells were cultured in glass-bottom dishes and treated with the 50-µM FITC-labeled compounds in Opti-MEM (Gibco) for 30 min. When the compounds were introduced by bead loading into living cells, HeLa S3 cells were cultured in glass-bottom dishes and 8 μL of 100-μM FITC-labeled compounds and 1 mg/mL Dextran-tetramethylrhodamine (Thermo Fisher Scientific, D3307) in PBS was introduced by bead loading (15).Then, the cells were washed three times with FluoroBrite DMEM (Gibco) containing 10% FBS, GlutaMAX, 100 U/mL penicillin, 100 mg/mL streptomycin, and 1 mg/mL heparin sodium (Wako, 081–00136) and then imaged by fluorescent microscopy (DMi8; Leica).
High Performance Liquid Chromatography (HPLC) Analysis of Compounds Isolated from Cells.
First, 100 µL of 0.5-mM compounds in PBS were introduced by bead loading (15) into HeLa S3 cells and the cells were incubated in the growth medium for 60 min. The cells were washed with ice-cold PBS, harvested by cell scraper, washed three times with PBS, and lysed with CRB buffer [50 mM Tris⋅HCl (pH 7.5), 300 mM NaCl, 0.3% Triton X-100] supplemented with 2 mM dithiothreitol (DTT), 0.25 U/μL Benzonase (Millipore, 70664), complete protease inhibitor mixture (Roche, 34185500), and 1 mM phenylmethylsulfonyl fluoride (PMSF) on ice for 30 min. After centrifugation (15,000 rpm, 5 min), the supernatants went through ethanol precipitation at −80 °C for 1 h and centrifugation (10,000 rpm, 15 min) to remove proteins. The solvents of the supernatants were removed by Speed-Vac evaporator to obtain dried samples, which were dissolved in MQ and analyzed by HPLC at 40 °C with a gradient of acetonitrile in 0.1% aqueous TFA/acetic acid (pH 10.5) (method: 2% acetonitrile for 5 min, followed by 10% acetonitrile for 5 min, and a linear gradient of 10–80% acetonitrile over 30 min. TMC Triart C18, ex = 494 nm, em = 518 nm). For the authentic sample, 1-mM compounds mixed with the same volume of HeLa S3 cell lysate, and the mixture went through ethanol precipitation and centrifugation. Then, HPLC sample was prepared as described above and analyzed by HPLC.
Serum Stability Assay.
First, 1 µL of 100-µM FITC-labeled compounds were mixed with 19 µL of 25% Human serum (Sigma-Aldrich, H4522)/RPMI 1640 (Gibco) and incubated at 37 °C for 0, 30, 60, 120, 240, 480 min. After incubation, 6 µL of 5× sample buffer [250 mM Tris⋅HCl (pH6.8), 10% SDS, 30% glycerol, 0.05% bromophenol blue] and 3 µL of 1 M DTT were added and boiled at 95 °C for 5 min. The samples were separated by Novex 16% Tricine Protein Gels (Life Technologies), and FITC signals were detected by ImageQuant LAS 4010 (GE Healthcare).
HeLa Cell Expressing H2B-HaloTag.
PB533-H2B-Halo vector was constructed using H2B and Halo fragments amplified from pBOS-H2B (25) and Halo Tag vector (Promega), respectively. The fragments were inserted into PB533 PiggyBac vector (System Biosciences). HeLa cells were routinely maintained in DMEM (Nacalai Tesque) supplemented with l-glutamine/penicillin/streptomycin (2 mM l-glutamine, 100 U/mL penicillin, 0.1 mg/mL streptomycin; Sigma-Aldrich) and 10% FBS (Thermo Fisher Scientific) at 37 °C under 5% CO2 atmosphere. To establish a HeLa cell line that stably expresses H2B-Halo, cells were transfected with PB533-H2B-Halo and Super PiggyBac Transposase expression vector (System Biosciences) using FuGene HD transfection reagent (Promega) according to the instruction manual, and 1 mg/mL G418 (Nacalai Tesque) was added 24 h after transfection. One week later, single colonies were isolated, and H2B-Halo–positive colonies were further selected by staining with 0.5 μM HaloTag TMR Ligand (Promega).
Live-Cell Imaging.
For live-cell imaging in Fig. 2 C and D, H2B-Halo-expressing HeLa cells were plated on a 35-mm glass-bottom dish (IWAKI). On the next day, 0.5 μM TMR-Halo Ligand was added 1 h before loading PEG-LANA-FITC into cells; 5 μL of 100 μM PEG-LANA-FITC was used for bead loading (15). The medium was replaced with FluoroBrite DMEM containing l-glutamine/penicillin/streptomycin and FBS. Thirty to sixty min after the loading, samples were set onto a heating unit (Tokai Hit; 37 °C) with a CO2-control system (Tokken) on a spinning-disk confocal microscope (Nikon Ti-E attached with Yokogawa CSU-W1, operated under NIS Elements Version 4.30). Fluorescent images were acquired using a Plan Apo VC 100× Oil DIC N2 (numerical aperture, NA, 1.4) objective lens, a Nikon LU-N4 Laser unit, and an Andor iXon3 electronmultiplying CCD (EMCCD). Snapshot images in Fig. 2C were acquired using a high excitation power condition (50% of 20-mW 488-nm and 5% of 20-mW 561-nm laser lines, each 100-ms exposure time). For Fig. 2D, cells were imaged every 30 min using a relatively low power condition to keep cells alive (5% of 488-nm and 2% of 561-nm laser lines, each 200 ms). The nuclear areas of individual cells were selected using H2B-Halo signals for thresholding and PEG-LANA-FITC intensities were measured using Fiji/ImageJ Version 1.51f. For FRAP analysis in Fig. 2E, HeLa cells loaded with PEG-LANA-FITC were used. FRAP was performed using a confocal microscope (Olympus FV1000) operated by built-in FV1000 software (ver. 4.2), equipped with a heating unit (Tokai Hit; 37 °C), a CO2-control system (Tokken), and a 60× PlanApoN (NA 1.40) Oil lens. One hundred images were collected using a main scanner (0.3% 488-nm laser transmission; 0.5 μs/pixel; 256 × 64 pixels; pinhole 800 μm; 8× zoom; ∼35.2 ms/frame), and after collecting 10 images, 1-μm-diameter spot was bleached using a second scanner (100% 405-nm laser transmission; 14 ms). The fluorescence intensities of the bleached area were measured, and after background subtraction, the relative intensities to the averages before bleaching were obtained. Using Fiji/ImageJ Version 1.51f (“Curve Fitting” Tool), the recovery curves (from 3 frame after bleaching) were fit with the exponential association kinetics [I = A*(1 − exp(−k*t) + B, where I is the relative intensity; A is the recovered fraction; k is the binding coefficient; t, time (s); and B is the baseline (unbleached fraction)] (26).
In-Cell Histone Acetylation.
HeLa S3 cells were treated with 100 µM BSO in growth medium at 37 °C for 1 d; 0.5 mM PEG-LANA-DSSMe was preincubated with 10 mM NAC-Ac, 2 mM TCEP, and 0.5 mg/mL Dextran-tetramethylrhodamine in PBS for 10 min. The preincubation mixture was incorporated into the cells by bead loading (15), and the cells were incubated in growth medium containing 30 mM NAC-Ac and 100 µM BSO at 37 °C for indicated time. The cells harvested with accutase (Innovative Cell Technologies) were incubated with 1 µg/mL DAPI in growth medium at r.t. for 15 min to stain dead cells, and sorted by FACS Arial II or III (BD Biosciences). The living cells (DAPI negative) were classified into three groups with their tetramethylrhodamine signals: low population have the same signal as nontreated cells, high population have the highest about 5% intensity of the sample, and remaining cells were classified as mid population. Histones were isolated with Histone Purification Kit (Active Motif) according to the manufacturer’s instructions. In brief, histones were extracted with acid, and the H2A/H2B and H3/H4 fractions were isolated by subsequent elution. For LC-MS/MS analysis, the histones were dissolved in 20 µL MQ. To the histone solution, 20 μL of 200 mM NH4HCO3 aq., 40 μL of propionic anhydride solution was added, and pH was adjusted to 8 by adding ammonia solution. After 30-min incubation at r.t., the solvents were removed by Speed-Vac evaporator. Then, the samples digested by Trypsin Gold and Glu-C as described in In Vitro Histone Acetylation and Sample Preparation for LC-MS/MS Analysis in Materials and Methods and analyzed by LC-MS/MS.
Sample Preparation for Western Blotting.
Histones were isolated from the cells by Histone Purification Kit according to the manufacturer’s instructions or by acid extraction. The histones were dissolved in MQ and pH was adjusted by adding 1 M Tris⋅HCl (pH 7.5). To equalize the concentrations of the histones, the samples were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) followed by oriole staining (Bio-Rad), and the signals were detected by BioDoc-It Imaging System and measured by Fiji/ImageJ Version 1.51h. After equalizing the histone concentration, the histones were analyzed by Western blotting.
Antibodies.
Rabbit polyclonal antibodies against H3 (abcam, ab1791), H3K9ac (Merck, 07–352), H3K18ac (abcam, ab1181), and H2B (abcam, ab1790), rabbit monoclonal antibodies against H2B ubiquitination (Cell signaling technology, 5546S), and a mouse monoclonal antibody against H2B (abcam, ab52484), and H2BK120ac were used for Western blotting. For generating monoclonal antibody against H2BK120ac, H2BK120ac-containing synthetic peptide (NH2-CSEGTKAVTK(Ac)YTSSK-CONH2) was coupled to keyhole limpet hemocyanin and used to immunize mice (27); after generating hybridomas, clones were screened by enzyme-linked immunosorbent assay (ELISA) using plates coated with the modified or unmodified peptide conjugated with BSA. After recloning, supernatants from clones reacting with the H2BK120ac-containing synthetic peptide were used to probe blots using recombinant nucleosomes, in which H2BK120 was acetylated by LANA-DSH and acetyl-CoA (11), and the ones giving a single band at the size of histone H2B were selected. Hybridoma cells were routinely grown in GIT medium (Fujifilm, 637–25715), supplemented with 1 ng/mL IL-6 (FunaKoshi, 206-IL-010) and 100 μg/mL Gentamicin (Sigma, G1397-10 mL), at 37 °C in an atmosphere of 5% CO2. For antibody purification, the supernatant of hybridoma cell culture was applied to 1 mL Protein A Sepharose (GE Healthcare, 17–5280-01), which were preequilibrated with 20 mM phosphate buffer (pH7.0). After washing the column with 20 mM phosphate buffer (pH7.0), antibodies were eluted with 5 mL 100 mM Glycine (pH 3.0) and the buffer pH was immediately neutralized using 1.5 M Tris⋅HCl (pH 8.8). The relevant fractions were concentrated in PBS (Nacalai, 27575–31) using Amicon Ultra-4 Centrifugal Filter Units (Millipore, UFC803024).
Supplementary Material
Acknowledgments
We thank Dr. Kana Tanabe, Ms. Yuki Kobayashi, and Ms. Junko Kato for assistance with experiments, Mr. Christopher W. Adamson for reading the manuscript and helpful comments, and MAB Institute, Inc. for generating anti-H2BK120Ac monoclonal antibody. This work was supported by Ministry of Education, Culture, Sports, Science and Technology (MEXT)/Japan Society for the Promotion of Science (JSPS) KAKENHI [JP17H01522/JP17K19479/JP20H00489 (M.K.), JP19KK0179 (M.K., K.Y., and S.A.K.), JP18H04536 (K.Y.), JP18H05534 (H. Kurumizaka), JP17H01417 and JP18H05527 (H. Kimura)], the Platform Project for Supporting Drug Discovery and Life Science Research (BINDS) from Japan Agency for Medical Research and Development (AMED) (JP20am0101076, H. Kurumizaka), Japan Science and Technology Agency (JST) Exploratory Research for Advanced Technology (ERATO) (JPMJER1901m, H. Kurumizaka), and the “Epigenome Manipulation Project” of the All-RIKEN Projects supported by RIKEN.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. T.W.M. is a guest editor invited by the Editorial Board.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2019554118/-/DCSupplemental.
Data Availability.
All study data are included in the article and/or SI Appendix.
References
- 1.Hyun K., Jeon J., Park K., Kim J., Writing, erasing and reading histone lysine methylations. Exp. Mol. Med. 49, e324 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mirabella A. C., Foster B. M., Bartke T., Chromatin deregulation in disease. Chromosoma 125, 75–93 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kundaje A. et al.; Roadmap Epigenomics Consortium , Integrative analysis of 111 reference human epigenomes. Nature 518, 317–330 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eckschlager T., Plch J., Stiborova M., Hrabeta J., Histone deacetylase inhibitors as anticancer drugs. Int. J. Mol. Sci. 18, 1414 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yamatsugu K., Kawashima S. A., Kanai M., Leading approaches in synthetic epigenetics for novel therapeutic strategies. Curr. Opin. Chem. Biol. 46, 10–17 (2018). [DOI] [PubMed] [Google Scholar]
- 6.Müller M. M., Muir T. W., Histones: At the crossroads of peptide and protein chemistry. Chem. Rev. 115, 2296–2349 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.David Y., Vila-Perelló M., Verma S., Muir T. W., Chemical tagging and customizing of cellular chromatin states using ultrafast trans-splicing inteins. Nat. Chem. 7, 394–402 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wright T. H., et al. , Posttranslational mutagenesis: A chemical strategy for exploring protein side-chain diversity. Science 354, aag1465 (2016). [DOI] [PubMed] [Google Scholar]
- 9.Yang A., et al. , A chemical biology route to site-specific authentic protein modifications. Science 354, 623–626 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ishiguro T., et al. , Synthetic chromatin acylation by an artificial catalyst system. Chem 2, 840–859 (2017). [Google Scholar]
- 11.Amamoto Y., et al. , Synthetic posttranslational modifications: Chemical catalyst-driven regioselective histone acylation of native chromatin. J. Am. Chem. Soc. 139, 7568–7576 (2017). [DOI] [PubMed] [Google Scholar]
- 12.Kajino H., et al. , Synthetic hyperacetylation of nucleosomal histones. RSC Chemical Biology 1, 56–59 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mizumoto S., et al. , Hydroxamic acid-piperidine conjugate is an activated catalyst for lysine acetylation under physiological conditions. Chem. Asian J. 15, 833–839 (2020). [DOI] [PubMed] [Google Scholar]
- 14.Hamajima W., et al. , Site-selective synthetic acylation of a target protein in living cells promoted by a chemical catalyst/donor system. ACS Chem. Biol. 14, 1102–1109 (2019). [DOI] [PubMed] [Google Scholar]
- 15.Sato Y., Stasevich T. J., Kimura H., Visualizing the dynamics of inactive X chromosomes in living cells using antibody-based fluorescent probes. Methods Mol. Biol. 1861, 91–102 (2018). [DOI] [PubMed] [Google Scholar]
- 16.Harris J. M., Martin N. E., Modi M., Pegylation: A novel process for modifying pharmacokinetics. Clin. Pharmacokinet. 40, 539–551 (2001). [DOI] [PubMed] [Google Scholar]
- 17.Meas R., Mao P., Histone ubiquitylation and its roles in transcription and DNA damage response. DNA Repair (Amst.) 36, 36–42 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Worden E. J., Hoffmann N. A., Hicks C. W., Wolberger C., Mechanism of cross-talk between H2B ubiquitination and H3 methylation by Dot1L. Cell 176, 1490–1501.e12 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Makde R. D., England J. R., Yennawar H. P., Tan S., Structure of RCC1 chromatin factor bound to the nucleosome core particle. Nature 467, 562–566 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.England J. R., Huang J., Jennings M. J., Makde R. D., Tan S., RCC1 uses a conformationally diverse loop region to interact with the nucleosome: A model for the RCC1-nucleosome complex. J. Mol. Biol. 398, 518–529 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gatta R., et al. , An acetylation-mono-ubiquitination switch on lysine 120 of H2B. Epigenetics 6, 630–637 (2011). [DOI] [PubMed] [Google Scholar]
- 22.McGinty R. K., Kim J., Chatterjee C., Roeder R. G., Muir T. W., Chemically ubiquitylated histone H2B stimulates hDot1L-mediated intranucleosomal methylation. Nature 453, 812–816 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Daigle S. R., et al. , Selective killing of mixed lineage leukemia cells by a potent small-molecule DOT1L inhibitor. Cancer Cell 20, 53–65 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang E., et al. , Histone H2B ubiquitin ligase RNF20 is required for MLL-rearranged leukemia. Proc. Natl. Acad. Sci. U.S.A. 110, 3901–3906 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kanda T., Sullivan K. F., Wahl G. M., Histone-GFP fusion protein enables sensitive analysis of chromosome dynamics in living mammalian cells. Curr. Biol. 8, 377–385 (1998). [DOI] [PubMed] [Google Scholar]
- 26.Kimura H., Cook P. R., Kinetics of core histones in living human cells: Little exchange of H3 and H4 and some rapid exchange of H2B. J. Cell Biol. 153, 1341–1353 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hayashi-Takanaka Y., et al. , Distribution of histone H4 modifications as revealed by a panel of specific monoclonal antibodies. Chromosome Res. 23, 753–766 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix.