Significance
Post-translational modification is crucial for regulating protein function. Acetylation of N-terminal residues is a major form of post-translational modification, but the functions of N-terminal acetyltransferases in vivo are not well understood. Here, we report a Drosophila NAT, which we name Mnat9, that plays an essential role in development. Loss of Mnat9 leads to the abnormal activation of JNK signaling, causing cell death in developing organs. Additionally, Mnat9 directly binds and stabilizes microtubules. We show that these Mnat9 functions are independent of its acetylation activity. Furthermore, Mnat9 and human NAT9 are functionally conserved. This study shows the developmental function of a microtubule-associated NAT in JNK signaling, providing insight into the function of human NAT9.
Keywords: JNK, microtubule stability, cell death, Drosophila development, NAT (N-terminal acetyltransferase)
Abstract
Regulation of microtubule stability is crucial for the maintenance of cell structure and function. While the acetylation of α-tubulin lysine 40 by acetylase has been implicated in the regulation of microtubule stability, the in vivo functions of N-terminal acetyltransferases (NATs) involved in the acetylation of N-terminal amino acids are not well known. Here, we identify an N-terminal acetyltransferase, Mnat9, that regulates cell signaling and microtubule stability in Drosophila. Loss of Mnat9 causes severe developmental defects in multiple tissues. In the wing imaginal disc, Mnat9 RNAi leads to the ectopic activation of c-Jun N-terminal kinase (JNK) signaling and apoptotic cell death. These defects are suppressed by reducing the level of JNK signaling. Overexpression of Mnat9 can also inhibit JNK signaling. Mnat9 colocalizes with mitotic spindles, and its loss results in various spindle defects during mitosis in the syncytial embryo. Furthermore, overexpression of Mnat9 enhances microtubule stability. Mnat9 is physically associated with microtubules and shows a catalytic activity in acetylating N-terminal peptides of α- and β-tubulin in vitro. Cell death and tissue loss in Mnat9-depleted wing discs are restored by reducing the severing protein Spastin, suggesting that Mnat9 protects microtubules from its severing activity. Remarkably, Mnat9 mutated in the acetyl-CoA binding site is as functional as its wild-type form. We also find that human NAT9 can rescue Mnat9 RNAi phenotypes in flies, indicating their functional conservation. Taken together, we propose that Mnat9 is required for microtubule stability and regulation of JNK signaling to promote cell survival in developing Drosophila organs.
Microtubules are a major cytoskeleton component involved in various biological processes, such as cell division, cell migration, and development (1, 2). Abnormal functioning of microtubules can cause a variety of developmental and degenerative defects in animals and human (3–5). Thus, it is a central issue to understand how microtubule stability is regulated.
Stability of microtubules is affected by microtubule-associated proteins (MAPs) that promote polymerization or disassembly (6, 7), where groups of proteins such as Eb1 and patronin are required for protecting the plus and minus ends of microtubules (8, 9). Microtubule stability is also influenced by various post-translational modifications of tubulins, such as acetylation and phosphorylation (10, 11). More than 10 acetylation sites have been identified in tubulins via mass spectrometry, but the physiological importance of most of these sites has yet to be confirmed. Among acetylated lysine residues, K40 acetylation of α-tubulin is the most abundant form associated with stable microtubules (12, 13). Tubulin K40 acetylation, which is mediated by conserved acetyltransferase α-Tat1, plays a role in mechanosensation in mice and Caenorhabditis elegans (14). Similarly, Drosophila tubulin acetyltransferase (dTAT) is also involved in mechanosensation by affecting microtubule stability and dynamics (15). All known acetylation sites are located in the internal region of microtubules, with no N-terminal acetylation found yet.
N-terminal acetylation is likely to be the first modification of most proteins as it occurs at the beginning of translation (16). It has been estimated that more than 80% of eukaryotic proteins are subject to N-α-terminal acetylation mediated by N-acetyltransferases (NATs) (17), with eight different kinds of NATs (NatA through NatH) found so far. However, their functions in vivo are not well understood despite their roles in the regulation of many biological processes including protein folding, protein stability, protein–protein interaction, and protein trafficking (16, 18–21).
Also important to microtubule stability are the potential links between microtubule dynamics and cell signaling in developmental events. Loss of morphogenetic protein Decapentaplegic (TGFβ-BMP) leads to epithelial cell extrusion and death in the developing Drosophila wing by an altered cytoskeletal organization (22). Interestingly, defective microtubule spindles during mitosis can activate c-Jun N-terminal kinase (JNK) signaling and cell death in the wing disc (23). These studies suggest that morphogenetic signaling and microtubule organization regulate each other; however, little is known about the relationship between tubulin acetylation and the JNK signaling pathway. It is an intriguing question how defective microtubule acetylation alters morphogenetic or cell stress signaling in developing organs. From an earlier search for genes involved in growth regulation, we became interested in an uncharacterized Drosophila gene, CG11539, that encodes a NAT family protein. This NAT, which we name microtubule-associated Nat9 (Mnat9), does not belong to the eight known NAT classes. Mnat9 is related to human NAT9, which has been vaguely classified as a member of the GCN5 family with no currently known function except that a DNA sequence polymorphism (A699G) on porcine NAT9 affects pig litter size (24).
In this work, we show that Mnat9 is a microtubule-associated NAT protein required for microtubule stability as well as cell survival by regulating JNK signaling in developing organs. These Mnat9 functions are independent of its acetylation activity. Based on our results, this study suggests that Mnat9 is an evolutionarily conserved NAT family protein with functions in microtubule stability and JNK signaling.
Results
Depletion of Mnat9 Causes Developmental Defects.
Mnat9 is a small protein consisting of 200 amino acids with a single N-terminal acetyltransferase domain like other NATs (Fig. 1A). To test whether Mnat9 is required for normal development, we carried out tissue-specific knockdown of Mnat9 using the Gal4 upstream activating sequences (UAS) method (25). Various tissues including eye, abdomen, thorax, and wing showed developmental defects in Mnat9 RNAi conditions driven by different Gal4 lines (SI Appendix, Fig. S1). Among these defects, we decided to further characterize the Mnat9 function in the wing, where Mnat9 RNAi showed the most robust phenotypes.
Fig. 1.
Knockdown of Mnat9 causes apoptotic cell death in the wing disc. (A) Mnat9 consists of 200 amino acids with a single domain named N-acetyltransferase domain. (B, C) Effects of Mnat9 RNAi in the en domain. en > RFP, dcr2 control shows a normal wing (B). Knockdown of Mnat9 using en-Gal4 causes a size reduction and vein defects (red arrows) with blisters in the posterior part of the wing (C). (D, E) Effects of Mnat9 RNAi in the C96 wing margin domain. C96 > dcr2 control shows a normal wing (D). Mnat9 knockdown causes a notched wing (E). (F, G) Effects of Mnat9 RNAi in the ptc domain. ptc > dcr2 control shows a normal wing (F). Mnat9 knockdown causes a reduction in the L3 to L4 area (G). (H) Apoptotic cell death marker cleaved caspase-3 was highly induced in the ptc domain within the wing pouch (WP, red arrow). (I, J) Effects of p35 on en > Mnat9 RNAi effects. Mnat9 RNAi causes a size reduction of the wing with blisters (I). Mnat9 RNAi phenotype is suppressed by p35 (J). (K–M) Effects of p35 on C96 > Mnat9 RNAi effects. Mnat9 RNAi shows a notched wing (K). Mnat9 RNAi phenotype is suppressed by p35 (L). Quantification of Mnat9 RNAi phenotype rescue by p35 overexpression (M). (N, O) Effects of Nat6 RNAi. Wing notching by C96 > Mnat9 RNAi (N). In contrast, C96 > Nat6 RNAi shows no wing-notching phenotype. (Scale bar for the wings: 0.5 mm. Scale bar for the wing disc in H: 50 μm.)
When Mnat9 was knocked down using the posterior-specific engrailed (en)-Gal4, adult wing sizes were reduced due to defects in the targeted region (Fig. 1 B and C), with blisters showing in 90% of the wings examined (n = 165). C96-Gal4 and ptc-Gal4 drive Gal4 expression in the dorsoventral (DV) boundary and the anterior–posterior (AP) boundary region of the wing disc, respectively; as expected, knockdown using C96-Gal4 and ptc-Gal4 correspondingly resulted in notching along the wing margin and a reduction in the AP boundary region between the L3 and L4 veins (Fig. 1 D–G). Four independent RNAi lines (v104497, v49580, v31519, and 11539R-3) showed similar phenotypes. To determine the specificity of the Mnat9 RNAi phenotypes, we checked whether overexpression of normal Mnat9 can rescue the RNAi phenotypes. Since Drosophila Mnat9 is the RNAi target, we used a human NAT9 expression construct that cannot be silenced by Mnat9 RNAi due to sequence variations. The Mnat9 RNAi wing phenotype was rescued by overexpressing human NAT9 (see Fig. 6 for details), and therefore, the Mnat9 RNAi phenotypes are probably not due to off-target effects or artifacts caused by overexpressed RNAs. As the v104497 RNAi line showed intermediate severity, this line was chosen for further experiments.
Fig. 6.
Acetylation activity of Mnat9 is not required for its function in JNK signaling and microtubule stability. (A) Mnat9 has the acetyl-CoA binding motif conserved in all known NATs. (B) Microtubule turbidity assay results show that the addition of either Mnat9 or Mnat9[AcDel] increases the speed of microtubule polymerization. (C–F) Effects of Mna9 overexpression in the notum. Overexpression of wild-type Mnat9 shows a weak thorax cleft (D). Overexpression of mutated Mnat9[AAA] (E) or Mnat9[AcDel] (F) shows a similar thorax cleft phenotype as the wild type. (G–I) Rescue of Mnat9 RNAi phenotype by C96-Gal4. The wing-notching phenotype caused by Mnat9 RNAi (G) is fully suppressed by overexpressing hNAT9 (H, n = 88) or hNAT9[AAA] (I, n = 44). (J–L) Rescue of Mnat9 RNAi phenotype by en-Gal4. The small and wrinkled wing phenotype caused by Mnat9 RNAi (J) is almost fully suppressed by overexpressing hNAT9 (K, n = 60) or hNAT9[AAA] (L, n = 36). (M, N) Effects of Mnat9 overexpression by en-Gal4 in microtubule stability. Treatment of the wing imaginal disc with demecolcine (50 μM) destabilizes the tubulin pattern. The posterior compartment with overexpression of Mnat9 (M) or Mnat9[AcDel] (N) is more resistant to demecolcine than the anterior control region. [Scale bars: 200 μm (C–F), 0.5 mm (G–L), and 50 μm (M, N).]
The efficiency of Mnat9 RNAi was validated with a Western blot experiment using the Mnat9 antibody generated in this study. When RNAi was ubiquitously expressed by hs-Gal4 in the third instar larvae, we observed a reduction of Mnat9 in the whole larval extract (SI Appendix, Fig. S2A). We also tested the effects of Mnat9 RNAi in Drosophila S2 cells by treating the cells with synthesized dsRNAs against Mnat9 (26). After 5 d of incubation, Mnat9 was undetectable in the Western blot experiment (SI Appendix, Fig. S2B). From these results, we verified that the Mnat9 protein level was efficiently reduced by RNAi.
Due to the lack of an Mnat9 mutant fly line, the CRISPR-Cas9 system was used to generate mutants for further confirmation of the RNAi phenotypes. Two different guide RNAs targeting N-terminal and C-terminal parts were expressed using a single vector [(27) and SI Appendix, Fig. S3C)]. Each guide RNA was coexpressed with Cas9 in the germline by using several promoters, including nos promoter. Despite extensive screening efforts, we could not recover the germline mutations in Mnat9 for unknown reasons. However, both guide RNAs with Cas9 expression driven by nub-Gal4 efficiently induced somatic mutant clones (50%, n = 34), resulting in notched wing phenotypes consistent with the Mnat9 RNAi effects (SI Appendix, Fig. S3 A and B).
Mnat9 Is Required for Cell Survival in Developing Organs.
As wing tissues were lost by Mnat9 RNAi and mutations, we tested whether Mnat9 RNAi by ptc-Gal4 causes apoptotic cell death in the ptc domain along the AP boundary (Fig. 1H and SI Appendix, Fig. S4). We found that cleaved-caspases were more strongly induced in the targeted ptc domain within the wing pouch than other regions.
To confirm the cell death effect of Mnat9 depletion, we tested whether the p35 cell death inhibitor (28) can suppress the wing defects. As shown earlier, Mnat9 knockdown using en-Gal4 caused both size reduction and abnormally thin veins in the wing (Fig. 1I). Overexpression of p35 rescued the wing defects almost completely (93% of progenies, n = 33, Fig. 1J), with a similar rescue found in C96-Gal4 cases (Fig. 1 K–M). Notched wings from C96 > Mnat9 RNAi as well were mostly rescued by p35 overexpression. These results suggest that the developmental defects in the wing caused by Mnat9 depletion were mainly due to apoptosis.
To check whether the cell death phenotype was due to the general effects of NAT knockdown, an N-terminal acetyltransferase 6 (Nat6, also known as Naa80, an actin-specific NAT) RNAi line was tested using C96-Gal4 (Fig. 1 N and O). When Nat6 was knocked down, no notching was found in the wing. Similar experiments were done with other known Drosophila NATs including NAT10, Naa15-16, Naa50, deco, and Naa30A, with no notching found by knockdown of any of these genes (SI Appendix, Fig. S5), in contrast with the effects of Mnat9 RNAi that caused wing notching in most of the progeny. This allows us to conclude that the notching phenotype was Mnat9-specific.
Knockdown of Mnat9 Leads to the Activation of JNK Signaling.
Apoptotic cell death is regulated by various signaling pathways including Wnt and Notch (29). To test whether Mnat9 depletion alters JNK signaling, we examined the expression of the puc-lacZ reporter, which is known to reflect activated JNK signaling (30). Our data showed that Mnat9 RNAi by en-Gal4 strongly increased the level of puc-lacZ, indicating elevated JNK signaling activity (Fig. 2 A and A’). We also found that both puc-lacZ and Dcp-1 levels were strongly elevated within the posterior domain of the wing disc (Fig. 2A’’).
Fig. 2.
Depletion of Mnat9 induces JNK signaling activation. (A–A’’’) puc-lacz, a JNK activity reporter, was induced in Mnat9 RNAi areas marked by RFP. (B–B’’) The level of the activated form of JNK (pJNK) was increased in the Mnat9 knockdown region (GFP). (C–C’’) The level of Mmp1, which is a downstream target of pJNK, was elevated in the Mnat9 knockdown region (GFP). (D–F) Wing phenotypes caused by Mnat9 depletion were efficiently rescued by puc overexpression. (G) Knockdown of Mmp1 sufficiently rescued the Mnat9 knockdown phenotype while showing no phenotype in the absence of Mnat9 RNAi. (Scale bar for the wings: 0.5 mm. Scale bar for the wing discs in A and B: 50 μm.)
In addition, the level of phosphorylated JNK (pJNK) was up-regulated by Mnat9 RNAi (Fig. 2 B–B’’), and furthermore, Mnat9 RNAi also led to the ectopic expression of Mmp1 (Fig. 2 C–C’’), a matrix metalloprotease up-regulated by JNK signaling (31). Puc is a major negative regulator for pJNK by acting as a phosphatase to pJNK (30). Thus, we tested whether Mnat9 RNAi phenotypes can be suppressed by puc overexpression. Results demonstrated that overexpression of puc and puc-HA rescued the Mnat9 RNAi phenotypes, showing normal wings in 63% (n = 41) and 56% (n = 46) of adult progeny, respectively (Fig. 2 E and G).
Mmp1 RNAi also rescued the Mnat9 RNAi phenotype in the wing (Fig. 2F), while knockdown of Mmp1 itself did not affect the wing. Similarly, one copy of the Mmp1 mutant (Mmp112/+) rescued the notching phenotype caused by Mnat9 RNAi (SI Appendix, Fig. S6B). Overexpression of Mmp1 resulted in minor notching (SI Appendix, Fig. S6C). When Mmp1 overexpression was combined with Mnat9 depletion, 100% lethality resulted (SI Appendix, Fig. S6D). These data suggest that the ectopic expression of Mmp1 is at least in part responsible for the wing defects caused by Mnat9 RNAi.
In addition to Mnat9 RNAi, we tested the effects of Mnat9 somatic mutation induced by the Cas9 system described earlier (SI Appendix, Fig. S3C). Mnat9 mutation in the wing pouch region by nub > Cas9 led to a strong increase in the pJNK level (SI Appendix, Fig. S3D). Taken together, these results indicate that cell death in the developing wing caused by Mnat9 RNAi is related to increased JNK signaling.
Overexpression of Mnat9 Suppresses JNK Activity.
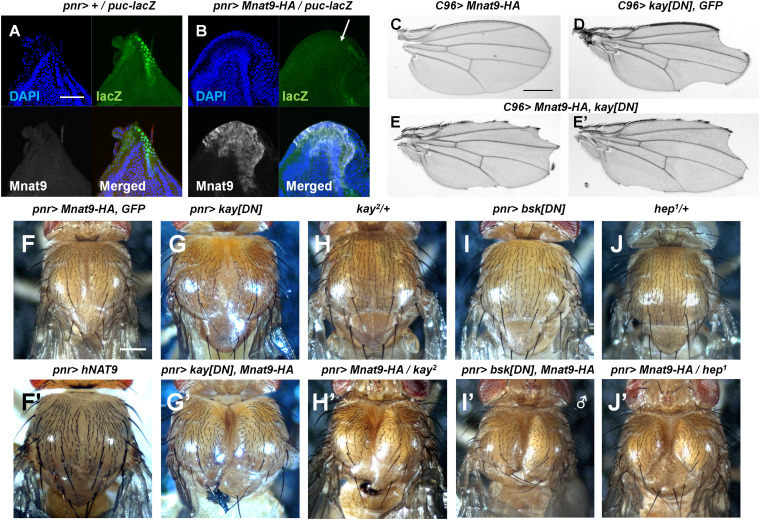
Disruption of cellular homeostasis can provoke JNK signaling as a cellular stress response (32–35). Since Mnat9 RNAi results in JNK activation in the wing disc, Mnat9 may be involved in the suppression of cellular stress or the regulation of JNK signaling or both. To test whether Mnat9 can affect the JNK pathway, we examined whether overexpression of Mnat9 can inhibit JNK activity. Since JNK activation in the notum of the wing disc is required for thorax closure, we overexpressed Mnat9 in the notum using pnr-Gal4. While normal wing discs showed endogenous puc-lacZ expression in the notum, Mnat9 overexpression greatly suppressed it (Fig. 3 A and B), indicating that Mnat9 can act as a negative regulator of JNK signaling.
Fig. 3.
Overexpression of Mnat9 suppresses JNK activity. puc-lacZ expression in the notum region of the wing disc (A) was strongly suppressed by overexpressing Mnat9 (B, white arrow). The overexpressed dominant-negative form of Kay protein causes the notched wing phenotype in adult male wings (D). Ectopic expression of Mnat9 enhances the phenotype (E, E′), while a single expression of Mnat9 shows normal adult wings (C). Overexpression of Mnat9 or human homolog hNAT9 causes a mild thorax closure defect phenotype (F, F′). Mutant or down-regulation of JNK signaling components further enhances the Mnat9 overexpression phenotype (G–J′). [Scale bars: 50 μm (A, B), 0.5 mm (C–E′), and 200 μm (F–J′).]
To further confirm the inhibitory effects of Mnat9 overexpression on JNK signaling, we carried out additional genetic tests with kayak (kay), which encodes the Drosophila Fos, a member of the downstream effector (AP-1) of JNK signaling. Overexpression of Kay[DN] (dominant-negative form) is known to suppress JNK pathway activity (36) and here caused the wing-notching phenotype in adult male flies (Fig. 3D). Mnat9 overexpression by C96-Gal4 did not affect wing development (Fig. 3C) but rather enhanced the notching phenotype of Kay[DN] (Fig. 3 E and E’).
We also used pnr-Gal4 to check the effects of genetic interaction in the notum using UAS-kay[DN], kay2 mutants, UAS-bsk[DN], and hep1 mutants. Overexpression of Mnat9-HA or Kay[DN] showed mild to intermediate levels of the thorax cleft phenotype (Fig. 3 F and G), while on the other hand, coexpression of both Mnat9-HA and Kay[DN] resulted in a strong cleft phenotype with a near-complete split of the thorax (Fig. 3G’). Like Kay[DN], kay2/+ heterozygote mutants also enhanced the thorax cleft phenotype of Mnat9 overexpression (Fig. 3 H and H’); likewise, bsk[DN] and hep1/+ heterozygote mutants also showed similar phenotype enhancement (Fig. 3 I and J’). From these genetic interactions with multiple JNK pathway genes, we confirm that Mnat9 overexpression may inhibit JNK signaling.
Since Mnat9 showed genetic interaction with several components of the JNK signal transduction pathway, including hep, bsk, kay, puc, and Mmp1, we hypothesized that Mnat9 may act upstream of the JNK pathway. To test this hypothesis, we examined the relationship between Mnat9 and eiger (egr, a TNF homolog), which acts as a ligand for its receptor Wengen to trigger JNK signaling. The notched wing phenotype by Mnat9 RNAi was enhanced by Egr overexpression by C96-Gal4. However, because Egr overexpression itself causes mild notching in the wing (SI Appendix, Fig. S7C), we further tested their genetic interaction using egr RNAi. Although egr RNAi by C96-Gal4 did not affect wing development (SI Appendix, Fig. S7A), the wing-notching phenotype by Mnat9 RNAi was strongly suppressed by egr RNAi (SI Appendix, Fig. S7 D–F) and enhanced by Egr overexpression (SI Appendix, Fig. S7G). These results suggest that Mnat9 acts antagonistically to the function of Egr. We performed real-time PCR to test whether Mnat9 RNAi increases egr expression (SI Appendix, Fig. S8) and found that, instead of an increase, Mnat9 RNAi led to a weak decrease (about 20%) in the egr messenger RNA (mRNA) level. Hence, Mnat9 seems to down-regulate egr at the post-transcriptional level.
Mnat9 Stabilizes Microtubules In Vivo and the Loss of Mnat9 Causes Mitotic Defects.
JNK signaling can be activated by multiple mechanisms, including impaired spindle microtubules during mitosis (23, 37). Since microtubules are regulated by acetylation, we figured that ectopic activation of JNK signaling in the Mnat9-depleted wing disc may be due to defects in microtubule stability. To test this idea, we used an antibody against Mnat9 for immunostaining in syncytial embryos where mitotic spindles can be easily detected. The localization of Mnat9 dynamically changed during the processes of mitosis (SI Appendix, Fig. S9). During interphase and prophase, Mnat9 staining was distributed around the nucleus with partial overlap with tubulin staining. However, Mnat9 was highly enriched at the spindle poles and around spindle microtubules during metaphase and anaphase (Fig. 4 A–A’’’ and SI Appendix, Fig. S9). During telophase, Mnat9 staining was enriched around DAPI staining and also colocalized with tubulin in the midbody region containing spindle microtubules. This led to the hypothesis that Mnat9 function might be related to microtubule spindles during mitosis.
Fig. 4.
Depletion of Mnat9 causes mitotic spindle defects in S2 cell and syncytial embryos. (A–A’’’) Mnat9 is localized around the mitotic spindles stained with anti-Tub and enriched in spindle pole regions of Drosophila embryos. (B, C–C’’) Live imaging using constitutively expressed α-tubulin linked with GFP in S2 cells. Unlike control (B), Mnat9 targeted dsRNA treated S2 cells showed abnormal and defective mitotic spindles with 30% penetrance (C–C’’). (D) Wild-type embryo shows normal spindles. (E, F) Mnat9 RNAi using mat-Gal4 causes mitotic defects. Two independent RNAi lines were used (yellow arrows: abnormal DAPI/spindle patterns, white arrow: loss of chromosomes). Demecolcine destabilizes microtubules (G–G′). Treatment with demecolcine for 20 min is sufficient to impair mitotic spindle arrays in the early embryo. (H) Demecolcine-mediated mitotic spindle defects are suppressed when Mnat9 is overexpressed maternally by mat-Gal4. Normal: normal mitotic spindle. Mild: slightly affected mitotic spindle. Intermediate: defective array and abnormal shape of mitotic spindle. Severe: almost no mitotic spindle left. (Scale bar: 20 μm.)
To address this possibility, we knocked down Mnat9 by double-stranded RNA (dsRNA) in the S2 cell line that constitutively expresses the GFP–α-tubulin fusion protein (9). Live imaging after 3 d of dsRNA treatment revealed that nearly 30% of the mitotic spindles (n = 20) were disrupted in Mnat9-depleted S2 cells, while dsRNA untreated cells showed no defects (Fig. 4 B and C’’). These results show that Mnat9 is necessary for maintaining normal mitotic spindle arrays. Next, we tested whether Mnat9 is required for normal mitosis in vivo during nuclear division cycles in syncytial embryos (38). Wild-type embryos showed a normal pattern of spindles at metaphase of nuclear cycle 10 (Fig. 4D), at which stage chromosomes are aligned along the equator. In contrast, embryos expressing Mnat9 RNAi by maternal-Gal4 showed various abnormal mitotic patterns, such as defective chromosome alignments, short spindles, and a loss of chromosomes in 20 to 30% of nuclei (n = 461), indicating that Mnat9 is required for normal mitosis (Fig. 4 E and F).
When we knocked down Mnat9 using en-Gal4, the number of PH3 stained nuclei increased about twofold (SI Appendix, Fig. S10 A–C). Excess PH3 staining can be caused by increased cell proliferation or longer mitosis due to mitotic defects (39). Because Mnat9 knockdown results in smaller wings rather than over-proliferation, the increased PH3 staining is likely to be due to mitotic extension or arrest. This is also supported by the observations that the reduced wing size by Mnat9 RNAi can be restored by the overexpression of p35, but not CycE, that promotes the cell cycle (SI Appendix, Fig. S10 D–F).
Since Mnat9 is enriched at the spindle poles during mitosis (Fig. 4A’), we performed an in vivo microtubule stability assay with syncytial embryos (38). Wild-type embryos showed a normal pattern of spindle arrays at metaphase during nuclear division (Fig. 4G, Mock). When wild-type embryos were treated with a microtubule inhibitor, demecolcine, the pattern of tubulin staining and mitosis at cycle 10 was impaired in almost all nuclei (Fig. 4G’, Demecolcine). Demecolcine treatment impaired 100% of the wild-type embryos, with more than half of them showing severe disruption (Fig. 4H). In contrast, overexpression of Mnat9 strongly suppressed the effects of demecolcine on mitotic spindles (Fig. 4H). These results suggest that Mnat9 is not only required for normal mitosis but also functions to stabilize spindle microtubules in vivo.
Mnat9 Stabilizes Microtubules and Promotes Microtubule Polymerization.
As Mnat9 immunostaining showed overlapping localization with 2mitotic spindles in embryos, we examined whether Mnat9 is physically associated with microtubules via an in vitro microtubule bundling assay (38, 40). In this assay, MAPs can be pelleted in the presence of microtubules after centrifugation at 100,000 g. In the absence of microtubules, GST-Mnat9 was barely detected in the pellet after centrifugation, while conversely, a significant fraction of Mnat9 was pelleted in the presence of microtubules (Fig. 5A). This result indicates that Mnat9 directly binds to microtubules in vitro. We also confirmed the interaction between Mnat9 and microtubules by immunoprecipitation in S2 cultured cells (Fig. 5B), and additionally, a tubulin heterodimer binding assay revealed that GST-Mnat9 can bind to tubulin dimer (αβ) (Fig. 5C). Such findings support that Mnat9 binds to microtubules as well as tubulins, like many other MAPs (41).
Fig. 5.
Mnat9 binds and stabilizes microtubules in vitro and in vivo and can acetylate tubulin N terminus. (A) Microtubule-binding assay results show that Mnat9 can bind with microtubules in vitro. (B) Immunoprecipitation results show that Mnat9 can bind with microtubules in vivo. (C) Tubulin heterodimer and MBP-Mnat9 show binding. (D) Dilution stress destabilizes microtubules in the absence of Mnat9 (right red arrow). (D') Quantification of (D). In the presence of Mnat9, microtubules are more resistant to the dilution stress (left red arrow). (E) The addition of Mnat9 suppresses microtubule dissociation by cold stress. (F) Microtubule polymerization is greatly enhanced in the presence of Mnat9. (G) DTNB assay results show that Mnat9 has NAT activity on N-terminal peptides of both α-84D and β-56D tubulin.
Most MAPs can be classified into three major groups, namely microtubule stabilizers, microtubule destabilizers, and motor proteins (38, 42–46). As Mnat9 is a small protein with no conserved motor domain, it is unlikely to be a motor protein. Based on the mitotic spindle defects of Mnat9 RNAi in S2 cells and syncytial embryos (Fig. 4 B–H), it is possible that Mnat9 may affect microtubule stability by direct interaction. To test this possibility, we utilized the phenomenon that microtubules become unstable when taxol concentration decreases with dilution (47) and examined whether Mnat9 can affect microtubule stability under dilution stress (Fig. 5 D and D’). After the in vitro polymerization of microtubules from tubulin dimers, taxol and microtubules were serially diluted to ratios of 1:2, 1:5, 1:20, and 1:50 with reaction buffer. We observed that the microtubule levels were decreased at the 1:50 dilution in the absence of Mnat9 (right red arrow in Fig. 5D); following Mnat9 addition though, microtubules became more resistant to dilution stress (left red arrow in Fig. 5D). We also checked whether Mnat9 can protect microtubules from cold stress. From a microtubule turbidity assay, we observed that microtubules were more resistant to cold stress when Mnat9 was added (Fig. 5E). These results suggest that Mnat9 increases microtubule stability in vitro.
The dynamic instability of microtubules is dependent on polymerization as well as depolymerization. As some microtubule-binding proteins act not only as microtubule stabilizers but also polymerization enhancers (38), we tested whether Mnat9 is involved in microtubule polymerization using an in vitro assay in which the amount of polymerized microtubules was measured after high-speed centrifugation (100,000 g) of the polymerization reaction mixtures. Results from this assay indicated that microtubules were greatly enriched when Mnat9 was added during the polymerization step (Fig. 5F). Results from these assays indicate that Mnat9 regulates microtubule stability by inhibiting depolymerization and promoting polymerization. Since Mnat9 is a putative N-terminal acetyltransferase that can bind to microtubules, we tested whether it has NAT enzyme activity toward tubulin using N-terminal peptides with the 5,5′-dithiobis-(2-nitrobenzoic acid) (DTNB) assay (48). Our data showed that Mnat9 can acetylate the N-terminal peptides of both α-tubulin (α-84D) and β-tubulin (β-56D) (Fig. 5G). However, αβ-tubulin dimers were not acetylated by Mnat9. This suggests that Mnat9 cotranslationally acetylates substrates like other NATs including NatA, B, C, D, and E(16).
The Acetylation Function of Mnat9 Might Be Dispensable for Microtubule Stabilization or JNK Signaling.
Our acetyltransferase assay above indicates that Mnat9 can acetylate the N-terminal peptides of α- and β-tubulin. An important question is whether the acetyltransferase activity of Mnat9 is required for its biological function. Interestingly, our in vitro assays shown in Fig. 5 indicate that Mnat9 can stabilize microtubules in the absence of acetyl-CoA. Therefore, the acetyltransferase activity of Mnat9 may not be essential for its function in stabilizing microtubules. To test this possibility, we generated mutant forms of Mnat9 defective in acetylation activity. Most GNAT family enzymes and all NATs have a conserved acetyl-CoA binding motif, Q/RxxGxG/A (x can be any amino acid), that is essential for their acetyltransferase activity (49–55). We found that Mnat9 also has this acetyl-CoA binding motif (Fig. 6A). Hence, we mutated the acetyl-CoA binding motif to produce two different mutant constructs—Mnat9[AAA] that has alanine substitutions AxxAxA in the RxxGxG acetyl-CoA binding site and Mnat9[AcDel] that has a deletion of six amino acids in the RxxGxG site (RGKGFG).
Next, we tested whether the acetyl-CoA binding site of Mnat9 is essential for microtubule polymerization. For quantitative analysis, we performed a microtubule polymerization turbidity assay. In the absence of MBP-Mnat9, the rate of microtubule polymerization was very slow (Vmax: 0.2 mOD/min). Under this condition, the addition of MBP-Mnat9 strongly facilitated the rate of microtubule polymerization (Vmax: 8.4 mOD/min) (Fig. 6B). Although the Mnat9[AcDel] protein also showed a weakly reduced microtubule polymerization activity, it showed a significantly high level of activity with 70% of wild-type Vmax (Vmax: 5.9 mOD/min), as shown in Fig. 6B. This result suggests that acetyl-CoA binding might not be essential for Mnat9’s function.
We then tested the effects of mutant forms of Mnat9 in developing animals. Since overexpression of wild-type Mnat9 by pnr-Gal4 resulted in a mild defect in thorax closure (Fig. 6D), first we checked whether overexpression of mutated Mnat9 can show a similar thorax defect. Remarkably, overexpression of Mnat9[AAA] or Mnat9[AcDel] showed similar phenotypes (Fig. 6 C–F), suggesting that the overexpression phenotype might be independent of Mnat9 NAT activity.
Next, we tested whether acetyl-CoA mutant forms of Mnat9 can rescue Mnat9 RNAi phenotypes. Since both wild-type and mutated Mnat9 transgenes are targets for Mnat9 RNAi, we used human Mnat9 (hNAT9), which as previously mentioned is not affected by RNAi due to DNA sequence variations; hNAT9 and Mnat9 show 49% protein identity and 54% DNA sequence identity, respectively (SI Appendix, Fig. S11). We generated transgenic fly lines carrying wild-type UAS-hNAT9 or a mutant form UAS-hNAT9[AAA]. Wing phenotypes caused by Mnat9 RNAi with C96-Gal4 or en-Gal4 were almost completely rescued when hNAT9 was overexpressed (Fig. 6 G, H, J, and K). These data support that Mnat9 and hNAT9 are conserved in structure and function. The rescue of Mnat9 RNAi phenotypes by hNAT9 also confirms that Mnat9 RNAi phenotypes are not off-target effects. Importantly, overexpression of wild-type hNAT9 or mutated hNAT9[AAA] showed a similar rescue of Mnat9 RNAi phenotypes (Fig. 6 I and L), strongly suggesting that the N-terminal acetylation activity of Mnat9 might not be critical for its function in wing development.
To test whether the N-terminal acetylation function of Mnat9 affects microtubule stability, we performed an ex vivo microtubule stability assay with wing discs. The wing discs were dissected from third instar larvae and cultured in medium supplemented with insulin (6.2 μg/mL). During cultivation, a high level of demecolcine (50 μM) was applied to en > Mnat9-HA, GFP discs for 75 min. After fixation, discs were stained with SiR-Tubulin, a fluorogenic chemical that strongly binds to the microtubules (56, 57). The anterior compartment used as control showed a low level of SiR-Tublin, while in contrast, Mnat9 overexpression strongly elevated the levels of microtubules in the posterior compartment (Fig. 6M). As demecolcine destabilizes microtubules, this result supports that Mnat9 promotes microtubule stability. Overexpression of Mnat9[AcDel] also showed strongly enhanced levels of SiR-Tubulin in the posterior region, indicating that the acetyl-CoA binding site is not essential for the microtubule stabilization function of Mnat9 (Fig. 6N). We also obtained similar results from an ex vivo microtubule stabilization assay performed live without tissue fixation (SI Appendix, Fig. S12). These observations illustrate that the N-terminal acetylation activity of Mnat9 might not be critical for its function in wing development and microtubule stabilization.
Mnat9 RNAi Wing Phenotypes Are Related to Microtubule Stability.
Our data above suggest that Mnat9 has functions in wing development, that is, protection of wing tissues from JNK-induced cell death and stabilization of microtubules. While it has been reported that the stability of the cytoskeleton affects the pJNK level (58), the underlying mechanism remains unclear. Based on our data, we hypothesized that apoptotic cell death caused by the loss of Mnat9 may also be affected by decreased microtubule stability. To verify, we examined whether the Mnat9 function is related to the Eb1 microtubule stabilizer (59). We found that Eb1 RNAi alone did not induce the notched wing phenotype (Fig. 7C), but when combined with Mnat9 RNAi, Eb1 RNAi considerably enhanced the notched wing phenotype (Fig. 7 D and J).
Fig. 7.
Mnat9 RNAi phenotypes and cell death are modified by microtubule regulators. Compared to control (A), Mnat9 RNAi phenotypes (B) are enhanced by eb-1 RNAi (D) but rescued by spastin RNAi (F). Eb-1 is a microtubule stabilizer, whereas spastin is a microtubule destabilizer. Eb-1 or spastin RNAi shows no notching phenotype in normal Mnat9 conditions (C, E). Quantitative data are shown in J. (G, G′) Mnat9 RNAi by C96-Gal4 shows weak staining for cleaved Cas3 in the DV boundary. (H, H′) eb-1 RNAi strongly enhances the level of cleaved Cas3 by Mnat9 RNAi. (I) Spastin RNAi suppresses the level of cleaved Cas3 by Mnat9 RNAi. [Scale bars: 0.5 mm (A–F) and 50 μm (G–I′).] Dashed line: DV boundary.
To further confirm the relationship between Mnat9 and microtubule stability, we checked if Mnat9 RNAi phenotypes can be altered by Spastin, a microtubule destabilizer (60). Here, while spastin knockdown itself showed no visible wing-notching phenotype, it could rescue the Mnat9 RNAi phenotype (Fig. 7 E, F, and J). We also looked at whether microtubule stability affects cell death in Mnat9-depleted wing discs; results showed that Eb1 RNAi strongly elevated the cleaved caspase-3 level, while spastin RNAi decreased it (Fig. 7 G–I’). These data suggest that the Mnat9 function in the wing can be affected by the altered stability of microtubules.
Discussion
In this work, we have discovered biochemical and biological functions of Mnat9 N-acetyltransferase in Drosophila. Mnat9 is essential for protecting wing tissues from ectopic activation of JNK signaling and cell death during development, and additionally, Mnat9 is associated with microtubules and is required for their stability. The N-terminal acetylation function of Mnat9, interestingly, is not required for the roles of Mnat9 in microtubule stability or JNK signaling, although Mnat9 can acetylate the N-termini of both α-tubulin (α-84D) and β-tubulin (β-56D) in vitro.
Targeted knockdown of Mnat9 resulted in developmental defects in various body structures, indicating the critical developmental functions of Mnat9. The wing disc is an epithelial primordium for the adult wing and associated tissues, including the notum. Consistent with this, Mnat9 RNAi specifically in the DV boundary region of the wing disc led to notching in the wing margin via ectopic JNK signaling and cell death. Overexpressing p35 but not CycE was able to revert Mnat9 RNAi wing phenotypes, indicating that the induced cell death was due to Drice- and Dcp-1–mediated apoptosis (28). We found that reduced Mmp1 can partially suppress the Mnat9 RNAi phenotype; since Mmp1 is one of the downstream targets of pJNK, our data suggest a causal relationship between ectopic JNK signaling and cell death.
This study presents evidence suggesting that Mnat9 is closely associated with microtubules and has various functions related to them. In the syncytial embryo, Mnat9 showed overlapping localization with spindle poles; consistent with this, the knockdown of Mnat9 in Drosophila embryos and S2 cells led to abnormal spindles and a disruption of mitosis. Our data also show that Mnat9 physically interacts with microtubules and is required for normal mitosis in S2 cells. Furthermore, the disruption of mitotic spindles by demecolcine in embryos was suppressed by Mnat9 expression, providing in vivo evidence for its role in microtubule stability (Fig. 4H). Our ex vivo experiments with wing discs also support that Mnat9 expression increases microtubule levels as detected by SiR-Tubulin. In terms of polymerization, our data from in vitro microtubule assays demonstrate that the rate of polymerization is enhanced more than 10 times by the addition of Mnat9, while conversely, the rate of depolymerization decreases, both of which point to the effect of Mnat9 on microtubule stability.
Despite extensive studies on microtubule stability and JNK signaling, not much is known about the relationship between these two processes. Interestingly, it has been reported that microtubule-interfering chemical agents increase c-Jun levels in cancer cells (58) and elevate pJNK in mouse DRG neurons (61). Recent studies have shown that microtubule destabilization leads to the activation of JNK signaling in different cellular contexts, suggesting a causal relationship between microtubule stability and JNK signaling (58, 62). In developing Drosophila wing discs, it has also been shown that an abnormal spindle assembly can activate JNK cell stress signaling to drive apoptotic cell death (23). These studies are consistent with our findings that Mnat9 affects not only microtubule stability but also JNK signaling. An important question then becomes whether the loss of Mnat9 leads to the activation of JNK signaling as a secondary consequence of cellular stress induced by microtubule defects. There seem to be two possibilities in this case: either Mnat9 is not necessarily involved in the inhibition of JNK signaling, or, alternatively, Mnat9 acts as a negative regulator of JNK signaling in addition to its role in microtubule stability. We prefer the latter possibility since Mnat9 was shown in this work to be required to suppress JNK signaling, and its overexpression inactivates puc-lacZ expression in the notum and enhances the phenotypes of heterozygous mutations in the JNK pathway (Fig. 3).
Based on the physical interaction between Mnat9 and microtubules, we also propose that Mnat9 is directly required for microtubule stability by antagonizing the activities of microtubule severing factors. Genetic interaction between Mnat9 and spastin suggests a mechanism of how Mnat9 enhances microtubule stability in vivo, as follows. Spastin depolymerizes microtubules by severing (63); in this work, reduced levels of Spastin rescued wing-notching phenotypes caused by Mnat9 RNAi, suggesting that Mnat9 may act to antagonize the severing activity of Spastin. In addition, Mnat9 may promote Eb1 activity to stabilize microtubules (Fig. 7). Since Mnat9 has a catalytic activity to acetylate α-and β-tubulin in vitro, it is possible that N-terminal acetylated microtubules may be more resistant to the severing activity of Spastin. Remarkably, however, our data suggest that Mnat9 regulates microtubule stability independent of its acetylation activity. First, microtubule polymerization can be enhanced without acetylation in the presence of Mnat9 in vitro (Fig. 5 F and G). Second, Mnat9 mutated in the Acetyl-CoA binding site still has a high level of microtubule polymerization activity in vitro and functional in vivo (Fig. 6 B–F). Third, acetylation-defective hNAT9 is also as functional as wild-type hNAT9 in vivo (Fig. 6 G–L). These data suggest that the role of Mnat9 in microtubule stability is not mediated by the acetylation of tubulins or other MAPs. Instead, Mnat9 may modulate microtubule regulators like Spastin and Eb1 in an acetylation-independent manner. Although we mutated the putative acetyl-CoA binding sites highly conserved in most acetyltransferases, we cannot rule out the possibility that Mnat9 may have an alternative site for its acetylation function. It is also possible that the acetylation activity of Mnat9 may be involved in specific processes that were not explored in this study.
Based on the results of this work, Mnat9 is likely the first NAT family protein with the functions of microtubule stability and JNK signaling during animal development. Our data also provide in vivo evidence for the functional conservation between Mnat9 and human NAT9. It would be interesting to see whether hNAT9 plays similar roles in microtubule stability and JNK signaling in human development and physiology.
Materials and Methods
Strains and Crosses.
All genetic crosses were carried out at 29 °C in standard Bloomington medium unless otherwise specified. The following Drosophila strains were provided by the Bloomington stock center: en-Gal4 (#30564), UAS-dcr2; en-Gal4, UAS-EGFP (#25752), UAS-dcr2; en > RFP,NRE-EGFP (#30730), UAS-dcr2, C96-Gal4 (#25757), UAS-dcr2 (#24646, #24650, and #24651), nub-Gal4; UAS-cas9 (#67086), UAS-cas9/CyO; C96-Gal4 (#67076), UAS-Naa80-RNAi (#63663), mmp112/Cyo (#58709), UAS-mmp1 (#58700), UAS-kay[DN] (#7215), kay2 (#42217), UAS-bsk[DN] (#6409), hep1 (#58779), pnr-Gal4 (#3039), UAS-Eb1-RNAi (#36680), and UAS-spastin-RNAi (#53331).
The following Drosophila strains were provided by the Vienna Drosophila Resource Center UAS-Mnat9-RNAi (#104497, #49580, and #31519), UAS-NAT10-RNAi (#13479), UAS-Naa15-16-RNAi (#17571), UAS-san-RNAi (#31741), UAS-deco-RNAi (#35982), and UAS-Naa30A-RNAi (#101769). The FlyORF stock center provided the UAS-Mnat9-HA (F002403), UAS-puc (F001556), and UAS-puc-HA (F001235), and the Kyoto Stock Center provided the esg-Gal4 lines (#14863 and #109126) and the puc-lacZ (#109029). The female wing or thorax was used for image data acquisition unless otherwise specified.
Generation of Transgenic Lines.
We generated UAS-Mnat9[AAA] and UAS-Mnat9[AcDel] transgenic flies using oligo synthesis service (Twist) and UAS-hNAT9 and UAS-hNAT9[AAA] transgenic flies using human hNAT9 complementary DNA (cDNA) (Genscript). Injection service was done by BestGene.
Immunohistochemistry.
The following primary antibodies were used: rabbit anti-Mnat9 (generated with full sequence Mnat9 protein by Abclonal, 1:150), mouse anti-MMP1 (Developmental Studies Hybridoma Bank [DSHB], 1:1:1 mixture with three antibodies, 1:100), mouse anti–β-gal (DSHB, 1:100), chicken anti–β-gal (Abcam, 1:100), rabbit anti-pJNK (Promega, 1:500), rabbit anti–cleaved caspase-3 (Cell Signaling, 1:100), rabbit anti–Dcp-1 (Cell Signaling, 1:100), mouse anti-tubulin (Sigma-Aldrich, 1:100), rat anti-tubulin (Merck Millipore, 1:200), and chicken anti-GFP (Invitrogen, 1:500). The following secondary antibodies were used: donkey fluorescein isothiocyanate, Cy3, and Cy5 (Jackson ImmunoResearch, 1:300).
Third instar larvae were dissected in phosphate-buffered saline (PBS). Samples were fixed in 4% paraformaldehyde fixative for 20 min at room temperature with agitation. Fixed samples were washed for 10 min in PBS and permeabilized by incubating for 30 min in 0.3 to 0.5% Triton X-100 in PBS (PBT). Next, they were blocked for at least 60 min in 5% normal goat serum, 0.3% PBT solution. Primary antibodies were diluted in blocking solution and samples were incubated for 1 h at room temperature or overnight at 4 °C with agitation. After incubation, samples were washed three times for 20 min in 0.3% PBT and then incubated with secondary antibodies and DAPI for 1 h at room temperature. They were then washed four times for 20 min before mounting on slides for imaging. Vectashield (Vector Laboratories, H-1000) medium was used for mounting, and an LSM 710 confocal microscope was used for imaging.
Embryos were collected via standard protocol (64) and fixed with methanol. Immunostaining was carried out using the same protocol as larval disc staining, except that saponin was used as the detergent instead of Triton X-100.
Western Blot.
Drosophila S2 cells, or third instar larvae, were collected and extracted with radioimmunoprecipitation assay buffer or 0.5% (3-((3-cholamidopropyl) dimethylammonio)-1-propanesulfonate) (CHAPS) buffer in Hepes. Protease mixture (cOmplete, Roche) and PhosSTOP (Roche) were added to the lysis buffer right before the experiment. Cells or tissues were homogenized and the protein concentration was calculated using the Bradford assay.
Gradient sodium dodecyl sulphate (SDS) gels of 10%, 12%, or 4 to 20% were used to separate the proteins. Electrophoresis and wet transfer were done in a Bio-Rad electrophoresis chamber.
The following primary antibodies were used: rabbit anti-Mnat9 (Abclonal/1:1,500), mouse anti-H2A (Santa Cruz/1:1,000), rabbit anti-H3 (Merck Millipore/1:3,000), mouse anti-MBP (NEB/1:2,000), and mouse anti-Tubulin (Sigma-Aldrich/1:1,000). Secondary antibodies were from Thermo Fisher Scientific and Sigma-Aldrich.
Immunoprecipitation.
Transfection was carried out with Effectene (Qiagen) according to the manufacturer’s instructions. A total of 1 to 2 μg DNA was used for each transfection. For immunoprecipitation, cells were lysed using CHAPS buffer (0.1 to 0.5% CHAPS/115 mM NaCl/20 mM Hepes/1.2 mM CaCl2/1.2 mM MgCl2/2.4 mM K2HPO4 and Protease Inhibitor mixture [Roche]), and the lysates were precleared by incubating with protein A magnetic beads (Surebeads/Bio-Rad) for 1 h at 4 °C. The immunoprecipitates captured by the protein A magnetic beads were incubated with the clear lysates overnight at 4 °C. Immunoprecipitates were washed and boiled at 94 °C for 5 min and then subjected to SDS-polyacrylamide gel electrophoresis (PAGE).
Cell Maintenance.
Drosophila S2 cells were utilized for all cell experiments, including immunoprecipitation. The cell line was provided by the Drosophila Genomics Resource Center (DGRC) in Indiana and Thermo Fisher Scientific. For S2 cell live imaging, the S2-act-GFP-alphaTub84B cell line (DGRC/#170) was used, and 10% fetal bovine serum (HyClone) in Schneider’s medium (Sigma-Aldrich) was used for cell maintenance and experiments.
CRISPR Vector.
The Mnat9 guide RNA vector was generated using the pCFD4 vector. Cloning was done by utilizing long primers as previously reported (27). Two different guide RNAs targeted the coding regions of Mnat9 to produce mutations. A fly line established by BestGene was utilized to induce tissue-specific somatic knockout by crossing with fly lines carrying the Cas9 transgene.
DTNB Assay.
The DTNB protocol was used in the N-acetyltransferase assay (65). DTNB (Ellman’s Reagent, 22582) was purchased from Thermo Fisher Scientific. A positive control experiment was done using human Naa50 protein and its known substrates. The oligopeptides used in this study were custom made (Genscript): the peptides contained seven specific amino acids at their N termini (α-tubulin 84D: MRECISI/β-tubulin 56D: MLIGARP) followed by 17 amino acids identical to the adrenocorticotropic hormone peptide sequence (RWGRPVGRRRRPVRVYP), while the internal lysines were replaced with arginines to avoid interference caused by internal acetylation (Nε).
Real-Time PCR.
Before getting heat shock, larvae were raised at 18 °C. Larvae were incubated in a 37 °C water chamber for 1 h and then transferred to a 29 °C incubator for 12 h. After incubation, the total RNA of the larvae was extracted using TRIzol (Thermo) following manufacturer’s instruction. cDNA synthesis and genomic DNA (gDNA) removal steps were carried out using PrimeScript RT Reagent Kit with gDNA Eraser (Takara). Normalized cDNA samples were used for further application. The teal-time PCR experiment was conducted and analyzed using a Bio-Rad CFX96 with QuantiTect SYBR Green PCR Kits (Qiagen). Primers with high primer efficiency (>95%) were used for amplification and sequence information as follows: 1) RPL32 (control), F-ATGCTAAGCTGTCGCACAAATG/R-GTTCGATCCGTAACCGATGT; 2) Eiger 1, F- AATCTCGCTCGATTGCCGAT/R-TGCAGGGCTCTCTTTGGAAG; and 3) Eiger 2, F-TTTGGGGTTCATCGGTCTGG/R-GTCGATGAGGGCATTCTCGT. All primers were purified with the high-performance liquid chromatography method.
Protein Expression and Purification.
pGEX-4T1 (Amersham) was used for cloning and expressing GST-Mnat9, and pMAL-c4X (NEB) was used for expressing MBP-Mnat9. In brief, Rosett (DE3) chemically competent cells (Enzynomics) were used for plasmid transformation. One liter of Luria agar culture was grown to an OD600 of 0.6 to 0.7 at 37 °C, followed by transfer to 15 °C with isopropyl β-D-1-thiogalactopyranoside added to a final concentration of 0.3 mM. Cells were incubated at 15 °C in a shaker at 220 rpm for 16 h and harvested by centrifugation at 12,000 g for 30 min at 4 °C. Cell pellets were resuspended in 30 mL of lysis buffer (200 mM NaCl, 20 mM TrisHCl pH 7.4, 1 mM ethylenediaminetetraacetic acid (EDTA), one tablet of cOmplete EDTA-free Protease Inhibitor mixture [Roche]) and incubated at 4 °C with 10 mg of lysozyme and benzonase (100 unit, Enzynomics) for 30 min. After mild sonication (Branson, microtip 10% amplitude, 1 min), lysates were centrifuged for 20 min at 13,000 g. The supernatant containing the soluble protein fraction was added to desired column (for GST-tagged proteins: Sepharose 4B [GE Healthcare], and for MBP-tagged proteins: Amylose Sepharose resin [NEB]). After incubation at 4 °C for 2 h, tubes were centrifuged at 2,000 g for resin separation. After three times washing with lysis buffer, elution buffer was added (10 mM maltose in lysis buffer for MBP-tagged protein elution and 10 mM reduced glutathione [Sigma-Aldrich G4251] in 50 mM Tris HCl for GST-tagged protein elution) and was rocked at room temperature for 3 min. The supernatant was saved after spin down at 2,000 g, and the elution steps were repeated three times. For protein purity confirmation, purified proteins were loaded on SDS-PAGE gel and stained with Coomassie staining. Protein concentration was checked by absorbance measurements at 260 and 280 nm. Proteins with high purity (>90%) were used for further application.
PH3 Staining Quantification.
The number of PH3 stained nuclei and the posterior and anterior wing disc areas were measured using the imageJ program. The number of PH3 stained nuclei per unit area was then calculated to get the relative ratio. Five wing discs were used for each genotype.
Microtubule-Binding Assay.
Microtubule assembly.
Porcine (MT002) or bovine (MT001) tubulin dimers from Cytoskeleton were utilized for the preparation of microtubules. A 5 mg/mL tubulin solution was dissolved in BRB80 buffer with 1 mM GTP (Cytoskeleton). The solution (20 μL) was then mixed with cushion buffer (5 μL, 80 mM Pipes, 1 mM MgCl2, 1 mM EDTA, and 60% glycerol, pH 7.0) and then incubated at 35 °C for 20 min. After the incubation, 200 μL BRB80 buffer and 2 μL taxol (2 mM, Sigma-Aldrich) were added to the mixture.
Microtubule binding.
Preassembled stable microtubules were further incubated with or without bovine serum albumin, GST, MBP, GST-Mnat9, or MBP-hNAT9 for 10 min at 35 °C. Cushion buffer was half-filled in a hard wall centrifuge tube, and the microtubule–protein mixture was carefully located on top of the cushion buffer to fill the tube. Tubes were centrifuged at 100,000 × g for 1 h at 25 °C. Supernatant and pellet samples were carefully separated to other tubes and labeled. Both were analyzed by SDS-PAGE and Coomassie staining.
In Vitro and In Vivo Microtubule Stability Assay.
The in vitro microtubule stability assay was performed as previously described (38). Briefly, preassembled microtubules (50 μM) were incubated with or without purified recombinant GST-Mnat9 protein (10 μM) for 30 min at 35 °C. After the incubation, the solution was diluted with different folds and incubated for 30 min at 35 °C. The mixtures were then centrifuged at 100,000 × g for 30 min at 20 °C. Pellets were resuspended in SDS sample buffer and electrophoresed. Samples were analyzed by Western blot for quantification.
The in vivo microtubule stability assay was performed in the following steps: first, Drosophila embryos were collected using a grape juice agar plate. Embryos were washed and then gathered with nylon mesh. A 50% bleach solution was used for dechorionation, with the embryos carefully monitored during the dechorionation step to prevent overexposure to bleach. When more than 80% of the embryos were dechorionated, bleaching was quenched and they were incubated in M3 media containing 0.3 μM demecolcine (Sigma-Aldrich) for 20 min. After the incubation, embryos were quickly fixed using the methanol fixation method and immunostained. The number of nuclei was counted for quantification.
Ex Vivo Microtubule Stability Assay.
Drosophila wing imaginal discs were cultivated as previously described (66–68). For live imaging, early third instar larvae were dissected in insect medium (Express Five SFM from Thermo Fisher Scientific, insulin supplement 6.2 μg/mL from Sigma-Aldrich) and incubated for 1 h with SiR700-tubulin (1:500, Spirochrome) and verapamil (10 μM, Spirochrome). After briefly washing two times with PBS solution, the discs were separated in insect medium supplemented with demecolcine (50 μM). Dissected discs were mounted using insect medium and then analyzed with a confocal microscope. For fixed sample preparation, early third instar larvae were dissected in insect medium and incubated for 75 min with demecolcine (50 μM). After incubation, discs were washed two times with insect medium. Then, discs were fixed using ethylene glycol bis(succinimidyl succinate) (10 mM in PBS) for 15 min at room temperature. After three times washing using PBS, discs were further incubated with SiR700-tubulin (1:500, Spirochrome) for 1 h. Discs were then briefly washed with PBS and mounted on the slide glass using Vectashield.
Microtubule Turbidity Assay.
The microtubule turbidity assay was performed as previously described (69). Microtubule polymerization or depolymerization was monitored via spectrophotometer (DU-730, Beckman). Absorbance at 340 nm was continuously recorded as previously described (59). For the microtubule polymerization assay, the cuvettes and spectrophotometer were preincubated at 37 °C for at least 10 min. The reaction mixture (1XBRB80, 1 mM GTP, 5% glycerol, 4 mg/mL tubulin, and 2 mg/mL of the desired protein) was immediately applied to the prewarmed cuvette and recorded. For the microtubule depolymerization assay, the spectrophotometer was preincubated at 6 °C for 30 min before the experiment. The reaction mixture was incubated at 37 °C for 30 min and then immediately transferred to the chilled spectrophotometer and recorded.
Tubulin Heterodimer Binding Assay.
Purified tubulin heterodimer (Cytoskeleton) was incubated with MBP-Mnat9 protein at room temperature for 30 min in MBP column buffer (200 mM NaCl, 20 mM Tris HCl pH 7.4, and 1 mM EDTA with Protease Inhibitor mixture). During incubation, amylose resin (New England BioLabs) was precleared and equilibrated with column buffer. After the incubation, the tubulin and MBP-Mnat9 protein mixture was further incubated with the equilibrated resin for 1 h at 4 °C on the rotator. After four times washing, the resin was eluted using 20 mM maltose in the column buffer. Eluted proteins were analyzed by SDS-PAGE and Western blot.
Supplementary Material
Acknowledgments
We thank T. Igaki, J. Chung, Vienna Drosophila Resource Center, Bloomington stock center, National Institute of Genetics stock center, and FlyORF for fly stocks; DGRC for providing Drosophila cell lines; Flybase for data sets and services; K.O. Cho, S.-T. Hong, and H. Chung for helpful discussions; Ok-Kyung Lee for fly stock maintenance; DSHB for providing monoclonal antibodies; and Addgene for providing vectors. This research was supported by grants NRF-2014K1A1A2042982 and NRF-2017R1A2B3007516 from the National Research Foundation of Korea funded by the Ministry of Education, Science and Technology, Republic of Korea.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2010140118/-/DCSupplemental.
Data Availability.
All study data are included in the article and SI Appendix.
References
- 1.Forth S., Kapoor T. M., The mechanics of microtubule networks in cell division. J. Cell Biol. 216, 1525–1531 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Muroyama A., Lechler T., Microtubule organization, dynamics and functions in differentiated cells. Development 144, 3012–3021 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gartz Hanson M., et al. , Novel α-tubulin mutation disrupts neural development and tubulin proteostasis. Dev. Biol. 409, 406–419 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Abdollahi M. R., et al. , Mutation of the variant α-tubulin TUBA8 results in polymicrogyria with optic nerve hypoplasia. Am. J. Hum. Genet. 85, 737–744 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bahi-Buisson N. et al.; LIS-Tubulinopathies Consortium , The wide spectrum of tubulinopathies: What are the key features for the diagnosis? Brain 137, 1676–1700 (2014). [DOI] [PubMed] [Google Scholar]
- 6.Hirokawa N., Microtubule organization and dynamics dependent on microtubule-associated proteins. Curr. Opin. Cell Biol. 6, 74–81 (1994). [DOI] [PubMed] [Google Scholar]
- 7.Drewes G., Ebneth A., Mandelkow E.-M., MAPs, MARKs and microtubule dynamics. Trends Biochem. Sci. 23, 307–311 (1998). [DOI] [PubMed] [Google Scholar]
- 8.Akhmanova A., Steinmetz M. O., Control of microtubule organization and dynamics: Two ends in the limelight. Nat. Rev. Mol. Cell Biol. 16, 711–726 (2015). [DOI] [PubMed] [Google Scholar]
- 9.Rogers S. L., Rogers G. C., Sharp D. J., Vale R. D., Drosophila EB1 is important for proper assembly, dynamics, and positioning of the mitotic spindle. J. Cell Biol. 158, 873–884 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Janke C., Bulinski J. C., Post-translational regulation of the microtubule cytoskeleton: Mechanisms and functions. Nat. Rev. Mol. Cell Biol. 12, 773–786 (2011). [DOI] [PubMed] [Google Scholar]
- 11.Wloga D., Gaertig J., Post-translational modifications of microtubules. J. Cell Sci. 123, 3447–3455 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Al-Bassam J., Corbett K. D., α-Tubulin acetylation from the inside out. Proc. Natl. Acad. Sci. U.S.A. 109, 19515–19516 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eshun-Wilson L., et al. , Effects of α-tubulin acetylation on microtubule structure and stability. Proc. Natl. Acad. Sci. U.S.A. 116, 10366–10371 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shida T., Cueva J. G., Xu Z., Goodman M. B., Nachury M. V., The major alpha-tubulin K40 acetyltransferase alphaTAT1 promotes rapid ciliogenesis and efficient mechanosensation. Proc. Natl. Acad. Sci. U.S.A. 107, 21517–21522 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yan C., et al. , Microtubule acetylation is required for mechanosensation in Drosophila. Cell Rep. 25, 1051–1065.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aksnes H., Drazic A., Marie M., Arnesen T., First things first: Vital protein marks by N-terminal acetyltransferases. Trends Biochem. Sci. 41, 746–760 (2016). [DOI] [PubMed] [Google Scholar]
- 17.Arnesen T., Towards a functional understanding of protein N-terminal acetylation. PLoS Biol. 9, e1001074 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goetze S., et al. , Identification and functional characterization of N-terminally acetylated proteins in Drosophila melanogaster. PLoS Biol. 7, e1000236 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Scott D. C., Monda J. K., Bennett E. J., Harper J. W., Schulman B. A., N-terminal acetylation acts as an avidity enhancer within an interconnected multiprotein complex. Science 334, 674–678 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hwang C.-S., Shemorry A., Varshavsky A., N-terminal acetylation of cellular proteins creates specific degradation signals. Science 327, 973–977 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Drazic A., et al. , NAA80 is actin’s N-terminal acetyltransferase and regulates cytoskeleton assembly and cell motility. Proc. Natl. Acad. Sci. U.S.A. 115, 4399–4404 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gibson M. C., Perrimon N., Extrusion and death of DPP/BMP-compromised epithelial cells in the developing Drosophila wing. Science 307, 1785–1789 (2005). [DOI] [PubMed] [Google Scholar]
- 23.Poulton J. S., McKay D. J., Peifer M., Centrosome loss triggers a transcriptional program to counter apoptosis-induced oxidative stress. Genetics 212, 187–211 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiugang Z., Jing L., Yonggang L., Characterization, expression profile, polymorphism and association of porcine NAT9 gene. Mol. Biol. Rep. 39, 3137–3142 (2012). [DOI] [PubMed] [Google Scholar]
- 25.Duffy J. B., GAL4 system indrosophila: A fly geneticist’s swiss army knife. Genesis 34, 1–15 (2002). [DOI] [PubMed] [Google Scholar]
- 26.Kao L.-R., Megraw T. L., RNAi in cultured Drosophila cells. Methods Mol. Biol. 247, 443–457 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Port F., Chen H.-M., Lee T., Bullock S. L., Optimized CRISPR/Cas tools for efficient germline and somatic genome engineering in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 111, E2967–E2976 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hay B. A., Wolff T., Rubin G. M., Expression of baculovirus P35 prevents cell death in Drosophila. Development 120, 2121–2129 (1994). [DOI] [PubMed] [Google Scholar]
- 29.Pećina-Slaus N., Wnt signal transduction pathway and apoptosis: A review. Cancer Cell Int. 10, 22 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martín-Blanco E., et al. , Puckered encodes a phosphatase that mediates a feedback loop regulating JNK activity during dorsal closure in Drosophila. Genes Dev. 12, 557–570 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Uhlirova M., Bohmann D., JNK- and Fos-regulated Mmp1 expression cooperates with Ras to induce invasive tumors in Drosophila. EMBO J. 25, 5294–5304 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Essers M. A. G., et al. , FOXO transcription factor activation by oxidative stress mediated by the small GTPase Ral and JNK. EMBO J. 23, 4802–4812 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang M. C., Bohmann D., Jasper H., JNK signaling confers tolerance to oxidative stress and extends lifespan in Drosophila. Dev. Cell 5, 811–816 (2003). [DOI] [PubMed] [Google Scholar]
- 34.Brown M., et al. , An initial phase of JNK activation inhibits cell death early in the endoplasmic reticulum stress response. J. Cell Sci. 129, 2317–2328 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Szegezdi E., Logue S. E., Gorman A. M., Samali A., Mediators of endoplasmic reticulum stress-induced apoptosis. EMBO Rep. 7, 880–885 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rallis A., Moore C., Ng J., Signal strength and signal duration define two distinct aspects of JNK-regulated axon stability. Dev. Biol. 339, 65–77 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gerlach S. U., Eichenlaub T., Herranz H., Yorkie and JNK control tumorigenesis in Drosophila cells with cytokinesis failure. Cell Rep. 23, 1491–1503 (2018). [DOI] [PubMed] [Google Scholar]
- 38.Zhang G., Beati H., Nilsson J., Wodarz A., The Drosophila microtubule-associated protein mars stabilizes mitotic spindles by crosslinking microtubules through its N-terminal region. PLoS One 8, e60596 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bougé A. L., Parmentier M. L., Tau excess impairs mitosis and kinesin-5 function, leading to aneuploidy and cell death. Dis. Model. Mech. 9, 307–319 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhao W. M., Seki A., Fang G., Cep55, a microtubule-bundling protein, associates with centralspindlin to control the midbody integrity and cell abscission during cytokinesis. Mol. Biol. Cell 17, 3881–3896 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fukata Y., et al. , CRMP-2 binds to tubulin heterodimers to promote microtubule assembly. Nat. Cell Biol. 4, 583–591 (2002). [DOI] [PubMed] [Google Scholar]
- 42.Goodson H. V., Jonasson E. M., Microtubules and microtubule-associated proteins. Cold Spring Harb. Perspect. Biol. 10, a022608 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cassimeris L., The oncoprotein 18/stathmin family of microtubule destabilizers. Curr. Opin. Cell Biol. 14, 18–24 (2002). [DOI] [PubMed] [Google Scholar]
- 44.Tangutur A. D., Kumar D., Krishna K. V., Kantevari S., Microtubule targeting agents as cancer chemotherapeutics: An overview of molecular hybrids as stabilizing and destabilizing agents. Curr. Top. Med. Chem. 17, 2523–2537 (2017). [DOI] [PubMed] [Google Scholar]
- 45.Walczak C. E., Microtubule dynamics and tubulin interacting proteins. Curr. Opin. Cell Biol. 12, 52–56 (2000). [DOI] [PubMed] [Google Scholar]
- 46.Akhmanova A., Steinmetz M. O., Tracking the ends: A dynamic protein network controls the fate of microtubule tips. Nat. Rev. Mol. Cell Biol. 9, 309–322 (2008). [DOI] [PubMed] [Google Scholar]
- 47.Mao T., Jin L., Li H., Liu B., Yuan M., Two microtubule-associated proteins of the Arabidopsis MAP65 family function differently on microtubules. Plant Physiol. 138, 654–662 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thompson P. R., et al. , Regulation of the p300 HAT domain via a novel activation loop. Nat. Struct. Mol. Biol. 11, 308–315 (2004). [DOI] [PubMed] [Google Scholar]
- 49.Pourreza A., et al. , Mutagenesis analysis of a conserved region involved in acetyl coenzyme A binding in the aminoglycoside 6′-N-acetyltransferase type Ib encoded by plasmid pJHCMW1. Antimicrob. Agents Chemother. 49, 2979–2982 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhu H.-Y., et al. , In silico identification and characterization of N-Terminal acetyltransferase genes of poplar (Populus trichocarpa). Int. J. Mol. Sci. 15, 1852–1864 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Salah Ud-Din A. I., Tikhomirova A., Roujeinikova A., Structure and functional diversity of GCN5-related N-acetyltransferases (GNAT). Int. J. Mol. Sci. 17, 1018 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Abboud A., Bédoucha P., Byška J., Arnesen T., Reuter N., Dynamics-function relationship in the catalytic domains of N-terminal acetyltransferases. Comput. Struct. Biotechnol. J. 18, 532–547 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lu L., Berkey K. A., Casero R. A. Jr, RGFGIGS is an amino acid sequence required for acetyl coenzyme A binding and activity of human spermidine/spermine N1acetyltransferase. J. Biol. Chem. 271, 18920–18924 (1996). [DOI] [PubMed] [Google Scholar]
- 54.Marmorstein R., Structure of histone acetyltransferases. J. Mol. Biol. 311, 433–444 (2001). [DOI] [PubMed] [Google Scholar]
- 55.Vetting M. W., et al. , Structure and functions of the GNAT superfamily of acetyltransferases. Arch. Biochem. Biophys. 433, 212–226 (2005). [DOI] [PubMed] [Google Scholar]
- 56.Derivery E., et al. , Polarized endosome dynamics by spindle asymmetry during asymmetric cell division. Nature 528, 280–285 (2015). [DOI] [PubMed] [Google Scholar]
- 57.Lukinavičius G., et al. , Fluorogenic probes for live-cell imaging of the cytoskeleton. Nat. Methods 11, 731–733 (2014). [DOI] [PubMed] [Google Scholar]
- 58.Wang T. H., et al. , Microtubule-interfering agents activate c-Jun N-terminal kinase/stress-activated protein kinase through both Ras and apoptosis signal-regulating kinase pathways. J. Biol. Chem. 273, 4928–4936 (1998). [DOI] [PubMed] [Google Scholar]
- 59.Tirnauer J. S., Grego S., Salmon E. D., Mitchison T. J., EB1-microtubule interactions in Xenopus egg extracts: Role of EB1 in microtubule stabilization and mechanisms of targeting to microtubules. Mol. Biol. Cell 13, 3614–3626 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Errico A., Ballabio A., Rugarli E. I., Spastin, the protein mutated in autosomal dominant hereditary spastic paraplegia, is involved in microtubule dynamics. Hum. Mol. Genet. 11, 153–163 (2002). [DOI] [PubMed] [Google Scholar]
- 61.Valakh V., Frey E., Babetto E., Walker L. J., DiAntonio A., Cytoskeletal disruption activates the DLK/JNK pathway, which promotes axonal regeneration and mimics a preconditioning injury. Neurobiol. Dis. 77, 13–25 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Moon D. O., et al. , JNK inhibitor SP600125 promotes the formation of polymerized tubulin, leading to G2/M phase arrest, endoreduplication, and delayed apoptosis. Exp. Mol. Med. 41, 665–677 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Roll-Mecak A., Vale R. D., The Drosophila homologue of the hereditary spastic paraplegia protein, spastin, severs and disassembles microtubules. Curr. Biol. 15, 650–655 (2005). [DOI] [PubMed] [Google Scholar]
- 64.Moritz M., et al. , Three-dimensional structural characterization of centrosomes from early Drosophila embryos. J. Cell Biol. 130, 1149–1159 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ree R., et al. , The N-terminal acetyltransferase Naa10 is essential for zebrafish development. Biosci. Rep. 35, e00249 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Aldaz S., Escudero L. M., Freeman M., Live imaging of Drosophila imaginal disc development. Proc. Natl. Acad. Sci. U.S.A. 107, 14217–14222 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tsao C. K., Ku H. Y., Lee Y. M., Huang Y. F., Sun Y. H., Long term ex vivo culture and live imaging of Drosophila larval imaginal discs. PLoS One 11, e0163744 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Restrepo S., Zartman J. J., Basler K., Cultivation and Live Imaging of Drosophila Imaginal Discs. Methods in Molecular Biology (Humana Press Inc., 2016), pp. 203–213. [DOI] [PubMed] [Google Scholar]
- 69.Gaskin F., Analysis of microtubule assembly kinetics using turbidimetry. Methods Mol. Biol. 777, 99–105 (2011). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and SI Appendix.