Abstract
Mycobacterium simiae is an emerging and spreading pathogen in Iran and little data about its drug susceptibility test (DST) and no standard treatment regimen are available. We report a case of multidrug‐resistant M. simiae respiratory infection in a 65‐year‐old woman with a history of previous Mycobacterium tuberculosis infection. The patient was treated with clarithromycin, levofloxacin, and cotrimoxazole for one year and eventually died while still suffering from respiratory problems. For DST, broth microdilution method was used according to the Clinical and Laboratory Standards Institute guidelines as well as molecular DST in clinical isolate. Mycobacterium simiae was resistant to streptomycin, moxifloxacin, clarithromycin, and cotrimoxazole antibiotics and was sensitive to clofazimine and amikacin antibiotics. Inappropriate use of antibiotics without determining the pattern of antibiotic resistance increases the likelihood of resistance and, for resistant specimens, the need to review the treatment protocol and replace antibiotics. Effectiveness based on antibiotic resistance pattern is essential.
Keywords: Case report, multidrug resistance, Mycobacterium simiae
Mycobacterium simiae is an emerging and spreading pathogen in Iran and little data about its drug susceptibility test (DST) and no standard treatment regimen are available. We report a case of multidrug‐resistant M. simiae respiratory infection in a 65‐year‐old woman with a history of previous Mycobacterium tuberculosis infection.

Introduction
Infections caused by non‐tuberculous mycobacteria (NTM) is an emerging public health concern in many countries, especially in developing countries [1]. In Iran, Mycobacterium fortuitum, Mycobacterium kansasii, and Mycobacterium simiae are the most common causes of NTM infections. Mycobacterium simiae is a slow‐growing photochromogenic Mycobacterium that was first identified in 1965. It is rarely associated with human infections. It causes lung infections in the elderly with lung abnormality or history of previous tuberculosis (TB), and also in immunocompromised patients. According to the American Thoracic Society (ATS) and Infectious Disease Society of America (IDSA) guidelines released in 2007, radiological, microbiological, and clinical findings are required to identify NTM respiratory infection [2]. There is limited information to discuss the drug susceptibility of M. simiae in vitro and the response to drug therapy in vivo. Mycobacterium simiae is usually resistant to first‐line anti‐TB drugs and has a completely different treatment regimen than TB, including moxifloxacin, clarithromycin, and trimethoprim‐sulfamethoxazole [3]. In this study, we identified M. simiae that was resistant to common treatment, and this is also the first study on M. simiae resistance to treatment in Mashhad.
Case Report
A 65‐year‐old woman was referred to Ghaem Hospital affiliated to Mashhad University of Medical Sciences in July 2018 with symptoms of severe cough, sputum, and chest pain [4]. The patient had a history of TB in the previous five years and had received TB drugs including isoniazid, ethambutol, pyrazinamide, and rifampin for six months and showed improvement and as well as negative acid‐fast bacilli (AFB) smear and sputum culture. A chest X‐ray (CXR) in Anteroposterior (AP) position revealed multiple scattered nodules in both lung fields, especially the right with upper and middle preference. Opacity was seen with the formation of a fluid surface in the lower zone of the left lung, which could indicate a cavitary lesion (Fig. 1A). High‐resolution computed tomography (HRCT) revealed hydropneumothorax at the base of the left lung. Multiple nodules in lung tissue with several cavitary foci were observed in the right and left lung fields. A scattered consolidation patch was also evident in the lung field (Fig. 1B, C). In the patient's blood test, white blood cell count was 11,800/mm3, with 77% neutrophils, 20% lymphocytes, 3% monocytes, and erythrocyte sedimentation rate (ESR) in the first hour 88. Laboratory findings combined with the clinical signs of the patient raised the possibility of mycobacterial infection. Therefore, direct smear microscopy for AFB and mycobacterial culture were performed on the patient's sputum sample. All three samples obtained from the patient were smear‐positive for AFB using the Ziehl–Neelsen method. Also, mycobacterial culture of the patient's samples on Lowenstein–Jensen medium resulted positive for AFB 12 days after inoculation. Mycobacterium simiae was identified using the native reverse line probe assay [5]. This result was also confirmed by sequencing of the ITS (16S‐23S) rRNA region with accession number (MN124510). Therefore, treatment with levofloxacin 1000 mg, clarithromycin 1000 mg, and cotrimoxazole 800 mg orally was initiated. The patient was treated with these drugs for one year, but still had clinical symptoms (such as severe cough and sputum) and chest radiographic findings. Unfortunately, the patient died while still suffering from respiratory problems.
Figure 1.
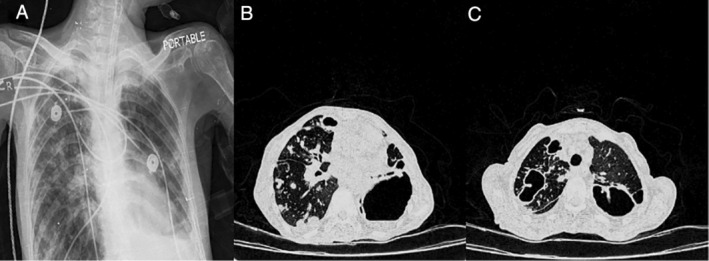
A 65‐year‐old‐woman with Mycobacterium simiae respiratory infection. (A) Multiple scattered nodules were seen in both lung fields, especially the right with upper and middle preference. Opacity image was seen with the formation of a fluid surface in the lower zone of the left lung, which indicates a cavitary lesion. (B, C) Hydropneumothorax at the base of the left lung. Multiple nodules in lung tissue with several cavitary foci were observed in the right and left lung fields. A scattered consolidation patch was also evident in the lung field.
The evaluation of antibiotic resistance pattern by manual method (broth microdilution) using resazurin reagent was based on the Clinical and Laboratory Standards Institute (CLSI) guidelines (M24‐2A) for moxifloxacin, clarithromycin, streptomycin, clofazimine, amikacin, and cotrimoxazole antibiotics. Clofazimine and amikacin were purchased from Sigma Chemical Co. (USA), and the remaining antibiotics were supplied by different pharmaceutical laboratories (clarithromycin, Solarbio, China; streptomycin, Pharmacia‐Upjohn, USA; moxifloxacin, Bayer, Germany; cotrimoxazole, ACS Chemical, Inc., India). The causative agent was cultured in Middlebrook 7H9 Broth base medium (Fluka, Switzerland) for four days to reach the logarithmic growth phase and then the concentration [1.5 × 105 colony forming unit (CFU)/mL] was prepared for inoculation into the wells, 100 μL of 7H9 medium with 5% Oleic acid, Albumin, Dextrose, Catalase (OADC) into each well. The concentration of each antibiotic was placed inside the wells according to Figure 2. We inoculated 100 μL of the prepared bacterial concentration in each well and completely covered the micro‐plates with paraffin to prevent drying. After seven days of incubation at 37°C, 20 μL of resazurin solution was added into each well and incubated overnight at 37°C. The lowest concentration of antibiotic that prevents discolouration from blue to pink is considered as minimal inhibitory concentration (MIC). The MIC of each antibiotic and their interpretation were determined according to Table 1. The MIC determination for each antibiotic was repeated three times. Results showed that M. simiae was resistant to streptomycin, moxifloxacin, clarithromycin, and cotrimoxazole antibiotics and was sensitive to clofazimine and amikacin antibiotics (Fig. 2).
Figure 2.
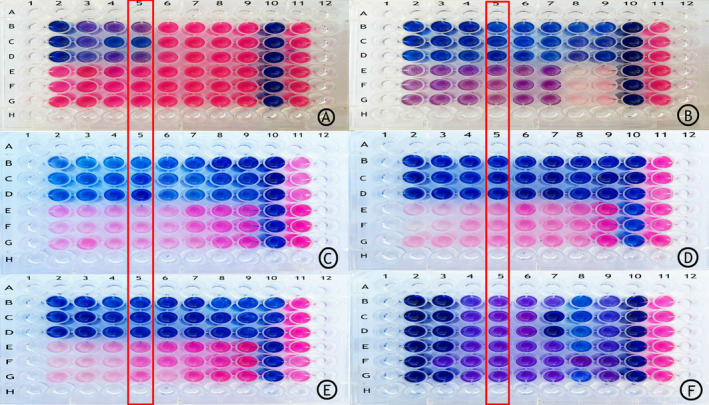
Antibiotic resistance pattern using manual method (broth microdilution) using resazurin reagent: rows B, C, and D in all plates are related to standard Mycobacterium simiae (JCM12377) and rows E, F, and G in all plates are related to the clinical isolation of M. simiae. To ensure the accuracy of the results of each antibiotic for standard sample and clinical isolation, it was performed three times. The concentration of antibiotics was reduced from column 2 to column 9. Plate A contains the antibiotic moxifloxacin (0.25–32 μg/mL). Plate B contains the antibiotic clofazimine (0.015–2 μg/mL). Plate C contains the antibiotic streptomycin (0.5–64 μg/mL). Plate D contains the antibiotic clarithromycin (2–256 μg/mL). Plate E contains the antibiotic cotrimoxazole (0.03–4 μg/mL). Plate F contains the antibiotic amikacin (0.5–64 μg/mL). Columns 10 and 11 represent sterility control and growth control of each plate, respectively. In columns 1 and 12 and rows A and H, sterile distilled water is added to prevent the plates from drying out.
Table 1.
Antibiotics with tested concentrations and suggested interpretation.
| Compound | Concentrations (μg/mL) | Susceptible | Intermediate | Resistant |
|---|---|---|---|---|
| Clarithromycin | 2–256 | ≤4.0 | 8.0 | ≥32.0 |
| Amikacin | 0.5–64 | ≤2 | 4.0 | ≥8.0 |
| Streptomycin | 0.5–64 | ≤2.0 | 4.0 | ≥8.0 |
| Moxifloxacin | 0.25–32 | ≤0.5 | 1.0, 2.0 | ≥4.0 |
| Clofazimine | 0.015–2 | ≤0.06 | 0.12 | ≥0.25 |
| Cotrimoxazole | 0.03–4 | ≤0.12 | 0.25 | ≥0.5 |
For each antibiotic resistance, polymerase chain reaction (PCR) analysis was performed with primers according to Table 2. The PCR products were sent for sequencing using Sanger sequencing with ABI 3730xl DNA analyser (Applied Biosystems, USA).
Table 2.
Primers used to determine antibiotic resistance.
| Resistance | Locus | Reference | Primer (5 → 3) |
|---|---|---|---|
| CLR | rrl | [6] |
Fw: CGGGATTCGGTCGCAGAAAC Rev: CCAGGTCTGGCCTATCGAAC |
| STR | rpsL | [6] |
Fw: ATTCCGAGGCAGGGCATAAC Rev: TTGCGCGGCATCAGCTCTTC |
|
MOX |
gyr A |
[6] |
Fw: ATTCTGCCGAACGGATCGAG Rev: CGACCGCGTTATCCGAATTG |
| gyr B |
Fw: TGGGCAACACCGAGGTGAAG Rev: ACGGGTCCATGGTGGTTTCC |
||
| SMX‐TMP | Ful p1 | [7] |
Fw: AGTCATAGGTGTCGGCCAAG Rev: GCGGACTGTTCAAAACCAAT |
CLR, clarithromycin; Fw, forward; MOX, moxifloxacin; Rev, reverse; SMX‐TMP, cotrimoxazole; STR, streptomycin.
In gyr A gene, a deletion was performed in the bases of 1148, 1149, and 1150, and the amino acid phenylalanine was removed and replaced with a stop codon. At position 1066, the amino acid glutamate was converted to serine (GAG is converted to TCG) (based on genomic positions in the M. simiae reference strain JCM12377, GenBank accession number AP022568.1). In the gyr B gene, an insertion in the base 442 to which base C has been added and the frame shift has occurred. In the rrl gene in the clinical sample, adenine was deleted at position 217, and a point mutation was made at the base 63 (in standard sample C (cytosine) and in clinical sample A (adenine)). In the rpsl gene at position 115, a point mutation occurred, and the base T (thymine) in the clinical specimen replaced C (cytosine) in the standard sample. Also, at position 289, G (guanine) in the clinical specimen replaced A in the standard sample, and in position 290 in the clinical sample, base A was inserted. In the fol p1 region, there was a point mutation at positions 60 and 715, and guanine and adenine bases in the standard sample replaced cytosine and guanine bases in the clinical specimen, respectively.
Discussion
In recent years, isolation of M. simiae from clinical specimens has been reported in different parts of the world such as Europe, America, and the Middle East. In Iran, M. simiae is endemic and accounts for more than 40% of NTM infections [2]. Successful treatment regimen for M. simiae infection is still a challenge and has not been standardized. Various treatment regimens have been proposed, including clarithromycin, moxifloxacin, and trimethoprim sulfamethoxazole, which have been reported to be effective in some reports [8]. Hamieh et al. founded that 100% of M. simiae isolates in vitro were resistant to streptomycin, 81% to cotrimoxazole, and 70% to moxifloxacin. These results were consistent with the results of our study, which showed resistance to the antibiotics streptomycin, moxifloxacin, and cotrimoxazole [9]. In the study of Coolen‐Allou et al., M. simiae was sensitive to amikacin, moxifloxacin, ciprofloxacin, and clarithromycin, while in our study M. simiae was resistant to moxifloxacin and clarithromycin [10]. Cowman et al. showed that 100% of the M. simiae isolates were sensitive to clofazimine and 89% to amikacin. In our study, M. simiae isolate was also sensitive to these antibiotics [11]. Baghaei et al. used a combination of clarithromycin, ofloxacin, and cotrimoxazole to treat patients with M. simiae respiratory infection, which was a successful treatment, contrary to the results of our study [12]. Van Ingen et al. showed that M. simiae was sensitive to clofazimine and cycloserine and resistant to the antibiotics streptomycin, amikacin, ciprofloxacin (72%), and clarithromycin (84%), while in our study M. simiae was sensitive to amikacin [13]. These results show the differences in the drug susceptibility test (DST) of M. simiae in different geographical areas (Table 3) and emphasize the need to perform the DST before starting treatment and help to choose a successful treatment regimen. It is possible that the increased rate of M. simiae infections in Iran and inappropriate use of antibiotics without specifying the DST has caused resistance to some antibiotics in the clinical sample compared to the standard strain.
Table 3.
Drug susceptibility test of Mycobacterium simiae in different geographical areas and treatment regimens of pulmonary infection M. simiae.
| Number of patients | Susceptibility tests | Treatment | Outcome | |
|---|---|---|---|---|
| Hamieh, 2018 [9]Lebanon | 51 | Susceptible to: (88%) AMK, (19%) SMX‐TMP, and (30%) MOX, two of the isolates that were resistant to all the tested antibiotics, except CLR, were tested against CLO and found to be susceptible | CLR, SMX‐TMP or MOX. CLR with CLO was used in two patients |
Six to 24 months. Four patients noted improvement. Two patients received a combination of CLO and CLR improvement |
|
Coolen‐Allou, 2018 [10] France |
97 |
Susceptible to: AMK, MOX, CIP, and CLR Resistant to: R, E, and H |
CLR, E, MOX, CLO, and AMK | Treatment failure in two patients, other patient found no relapse with M. simiae |
|
Cowman, 2016 [11] United Kingdom |
55 Retrospective study |
Susceptible to: CLO, CYC, and AMK Resistant to: H, E, RB, Ri, STR, and CLR |
— | — |
|
Baghaei, 2012 [12] Iran |
26 |
Resistant to: H, R, E, Z, and STR |
CLR, OFX, and SMX‐TMP | 12 months, 24 patients were cured and two patients failed the treatment |
|
Van Ingen, 2008 [13] The Netherlands |
6 |
Susceptible to: CLO, CYC, and (76%) PRO Resistant to: Ri, E, H, STR, AMK, CIP (72%), and CLR (84%) |
R, E, CIP, and CLR | One of them was cured, one relapsed, and one died |
|
This study 2020 |
1 |
Susceptible to: CLO and AMK Resistant to: STR, CLR, SMX‐TMP, and MOX |
LVX, CLR, and SMX‐TMP | Death |
AMK, amikacin; CIP, ciprofloxacin; CLO, clofazimine; CLR, clarithromycin; CYC, cycloserine; E, ethambutol; H, isoniazid; LVX, levofloxacin; MOX, moxifloxacin; OFX, ofloxacin; PRO, protionamide; R, rifampin; RB, rifabutin; Ri, rifampicin; SMX‐TMP, cotrimoxazole; STR, streptomycin; Z, pyrazinamide.
In conclusion, due to the high prevalence of infections caused by M. simiae in Iran, inappropriate use of antibiotics without determining the pattern of antibiotic resistance has increased the likelihood of resistance and, for resistant specimens, the need to review the treatment protocol and replace antibiotics. Effective determination of antibiotic resistance pattern prior to treatment is essential.
Disclosure Statement
Appropriate written informed consent was obtained for publication of this case report and accompanying images.
Author Contribution Statement
Hadi Lotfi, Hadi Farsiani, and Amir Hooshang Alvandi carried out the laboratory experiments and were involved in the acquisition of data. Hadi Lotfi, Mojtaba Sankian, and Ehsan Aryan wrote the first draft of the manuscript. Ahmad Khalifeh Soltani critically revised the article for important intellectual content. Zahra Meshkat, Hadi Lotfi, and Ehsan Aryan collaboratively contributed to the concept of the original research reported in this study. All authors reviewed and approved the final version of the manuscript.
Acknowledgment
The authors received financial support from the Mashhad University of Medical Sciences (grant number 961176).
Lotfi, H , Aryan, E , Sankian, M , et al. (2021) A case of multidrug‐resistant Mycobacterium simiae in an elderly woman. Respirology Case Reports, 9(3), e00715 10.1002/rcr2.715
Associate Editor: Semra Bilaceroglu
References
- 1. Piersimoni C, and Scarparo C. 2008. Pulmonary infections associated with non‐tuberculous mycobacteria in immunocompetent patients. Lancet Infect. Dis. 8(5):323–334. [DOI] [PubMed] [Google Scholar]
- 2. Hashemi‐Shahraki A, Darban‐Sarokhalil D, Heidarieh P, et al. 2013. Mycobacterium simiae: a possible emerging pathogen in Iran. Jpn. J. Infect. Dis. 66(6):475–479. [DOI] [PubMed] [Google Scholar]
- 3. Heidarieh P, Mirsaeidi M, Hashemzadeh M, et al. 2016. In vitro antimicrobial susceptibility of nontuberculous mycobacteria in Iran. Microb. Drug Resist. 22(2):172–178. [DOI] [PubMed] [Google Scholar]
- 4. Lotfi H, Sankian M, Meshkat Z, et al. 2021. Mycobacterium simiae pulmonary infection: a case series and literature review. Respirol. Case Rep. (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. kamali Kakhki R, Aryan E, Meshkat Z, et al. 2020. Development of a cost‐effective line probe assay for rapid detection and differentiation of Mycobacterium species: a pilot study. Rep. Biochem. Mol. Biol. 8(4):383–393. [PMC free article] [PubMed] [Google Scholar]
- 6. Bakuła Z, Modrzejewska M, Pennings L, et al. 2018. Drug susceptibility profiling and genetic determinants of drug resistance in Mycobacterium kansasii . Antimicrob. Agents Chemother. 62(4):e01788–e01717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ameen SM, and Drancourt M. 2013. In vitro susceptibility of Mycobacterium tuberculosis to trimethoprim and sulfonamides in France. Antimicrob. Agents Chemother. 57(12):6370–6371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Griffith DE, Aksamit T, Brown‐Elliott BA, et al. 2007. An official ATS/IDSA statement: diagnosis, treatment, and prevention of nontuberculous mycobacterial diseases. Am. J. Respir. Crit. Care Med. 175(4):367–416. [DOI] [PubMed] [Google Scholar]
- 9. Hamieh A, Tayyar R, Tabaja H, et al. 2018. Emergence of Mycobacterium simiae: a retrospective study from a tertiary care center in Lebanon. PLoS One. 13(4):e0195390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Coolen‐Allou N, Touron T, Belmonte O, et al. 2018. Clinical, radiological, and microbiological characteristics of Mycobacterium simiae infection in 97 patients. Antimicrob. Agents Chemother. 62(7):e00395–e00318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cowman S, Burns K, Benson S, et al. 2016. The antimicrobial susceptibility of non‐tuberculous mycobacteria. J. Infect. 72(3):324–331. [DOI] [PubMed] [Google Scholar]
- 12. Baghaei P, Tabarsi P, Farnia P, et al. 2012. Pulmonary disease caused by Mycobacterium simiae in Iran's national referral center for tuberculosis. J. Infect. Dev. Ctries 6(1):23–28. [DOI] [PubMed] [Google Scholar]
- 13. Van Ingen J, Boeree M, Dekhuijzen P, et al. 2008. Clinical relevance of Mycobacterium simiae in pulmonary samples. Eur. Respir. J. 31(1):106–109. [DOI] [PubMed] [Google Scholar]


