Significance
Endothelial cell types such as HUVECs and HAECs have been used as model systems to study vascular health. This study provides a comprehensive, time-dependent comparison of the transcriptomic responses of HAECs and HUVECs to shear stress by making measurements at 1, 4, and 24 h to capture the responses at early, mid, and late time points after shearing. The results indicate that the responses of HAECs and HUVECs are qualitatively similar for endothelial function-relevant genes and several pathways such as inflammation, oxidative stress, and angiogenesis. The findings also show that HAECs exhibit faster kinetics as compared to HUVECs. This study offers insights into the mechanisms of common and differential stress responses across cell types.
Keywords: atherosclerosis, endothelial cells, systems biology, temporal analysis of flow response, transcriptomics
Abstract
The two main blood flow patterns, namely, pulsatile shear (PS) prevalent in straight segments of arteries and oscillatory shear (OS) observed at branch points, are associated with atheroprotective (healthy) and atheroprone (unhealthy) vascular phenotypes, respectively. The effects of blood flow-induced shear stress on endothelial cells (ECs) and vascular health have generally been studied using human umbilical vein endothelial cells (HUVECs). While there are a few studies comparing the differential roles of PS and OS across different types of ECs at a single time point, there is a paucity of studies comparing the temporal responses between different EC types. In the current study, we measured OS and PS transcriptomic responses in human aortic endothelial cells (HAECs) over 24 h and compared these temporal responses of HAECs with our previous findings on HUVECs. The measurements were made at 1, 4, and 24 h in order to capture the responses at early, mid, and late time points after shearing. The results indicate that the responses of HAECs and HUVECs are qualitatively similar for endothelial function-relevant genes and several important pathways with a few exceptions, thus demonstrating that HUVECs can be used as a model to investigate the effects of shear on arterial ECs, with consideration of the differences. Our findings show that HAECs exhibit an earlier response or faster kinetics as compared to HUVECs. The comparative analysis of HAECs and HUVECs presented here offers insights into the mechanisms of common and disparate shear stress responses across these two major endothelial cell types.
Vascular endothelial cells (ECs) at the blood–vascular interface sense the shear stress exerted by blood flow on the vessel walls to initiate a cascade of intracellular signaling and functional responses. The transcriptional events in this response, which involve the key EC transcription factors Krüppel-like factors 2 and 4 (KLF2 and KLF4, respectively), modulate important vascular functions such as vasodilation through endothelial nitric oxide synthase (eNOS or NOS3), cell proliferation and angiogenesis through VEGF, and inflammation involving immune factors. Arterial ECs experience mainly two types of shear stresses, namely, the pulsatile or laminar shear stress (PS) associated with blood flow with clear direction in the straight parts of arteries, and the oscillatory or disturbed shear stress (OS) without a clear direction at branch points and curvatures. PS results in an atheroprotective phenotype due to cell quiescence, angiogenesis, and absence of inflammation, whereas OS is atheroprone due to increases in inflammation and cell turnover, perturbation of cholesterol metabolism, exacerbation of oxidative stress, and contribution to plaque formation through the interplay of macrophages and platelets with ECs (1–4).
As an EC model to study vascular homeostasis and health, human umbilical vein endothelial cells (HUVECs) have been widely used due to the ease of their availability and culturing. However, because of the unique features of the umbilical vein, which carries blood from the placenta to the fetus at a low pressure and is venous in nature, there have been concerns about the appropriateness of using HUVECs as a surrogate to study the molecular dynamics and functional behaviors of ECs in the arterial system (5–8). In this regard, several other sources of ECs, such as human aortic endothelial cells (HAECs) and human microvascular endothelial cells have been used to study vascular health and diseases, e.g., angiogenesis and atherosclerosis (8). Although there are a few studies comparing basal gene expressions among ECs from different sources under static condition (9, 10), there is a lack of comprehensive investigations that make quantitative and qualitative comparisons among these ECs in terms of their physiological or pathophysiological responses to different patterns of flow. While there have been a few studies comparing the responses of HUVECs and HAECs to perturbations such as adenosine diphosphate (ADP) and cyclic strain (11, 12), their differential responses to the PS and OS flow patterns have not been explored. Furthermore, these studies were typically carried out at one time point, providing only a snapshot of EC cell states (6, 10–12).
Recently, we carried out a comprehensive longitudinal study of the responses of HUVECs to OS and PS spanning 24 h (13). There are other studies in both HAECs and HUVECs, but only at a single time point (14–16). There is a lack of information on the time-dependent responses to shear stress between the widely studied HUVECs with ECs of arterial origin, i.e., HAECs. Comparison of the responses of these two types of ECs would allow an assessment of the utility of the commonly employed HUVECs in studies aiming to elucidate the responses of human arterial ECs to different flow patterns in health and disease. Furthermore, the similarities and differences found between these two types of cells in their responses to different flow patterns would provide insights into the mechanisms under physiological and pathophysiological conditions. Toward these goals, we have examined the temporal transcriptional responses of HAECs to OS and PS flows over 24 h, and compared these results with our prior work on HUVECs (13). Our systematic comparisons reveal that the flow-regulated HAEC and HUVEC responses are qualitatively similar for many key mechanotransduction pathways important for endothelial function and physiology and that there are some differences in the kinetics of the responses and specific genes.
Results and Discussion
Transcriptome-Wide Global Comparison.
The schematic of data analysis is outlined in SI Appendix, Fig. S1. The numbers of OS/PS differentially expressed (DE) genes at 1, 4, and 24 h are shown in SI Appendix, Fig. S2. The overall numbers of DE genes and the temporal trends were similar in both cell types. However, the number of DE genes in HAECs was greater than that in HUVECs at 1 and 4 h. In HAECs, the increase in the number of DE genes was temporally more rapid, e.g., with 11% (369 genes) of all 3,217 DE genes (all three time periods combined) being DE at 1 h, and 51% (1,655 genes) of all DE genes changing at 4 h; in contrast, HUVECs had DE of only about 8% (226 genes) and 35% (947 genes) of all 2,714 DE genes at 1 and 4 h, respectively (SI Appendix, Fig. S2 A and B). These results suggest an earlier response (faster kinetics) in HAECs, although the trends of responses over time were similar. We also analyzed the genes that were commonly and uniquely OS/PS up- or down-regulated in the two types of ECs: For up-regulated genes, HAECs and HUVECs showed more unique than common DE genes at 24 h (SI Appendix, Fig. S2C), while for down-regulated genes, the numbers of common and unique DE genes were comparable at 24 h (SI Appendix, Fig. S2D).
We also compared the global gene expression differences quantitatively. SI Appendix, Fig. S3 shows a scatterplot of OS/PS log2 fold changes (FCs) in HUVECs vs. HAECs for all genes DE in both cell types at respective time points and max read counts of 10 or more at any time point (1, 4, and 24 h). As time progressed, although the correlation of OS/PS log2 FCs between HAECs and HUVECs decreased slightly and the number of genes eligible for comparison increased, the P values became more significant statistically.
Broad EC Relevant Functions.
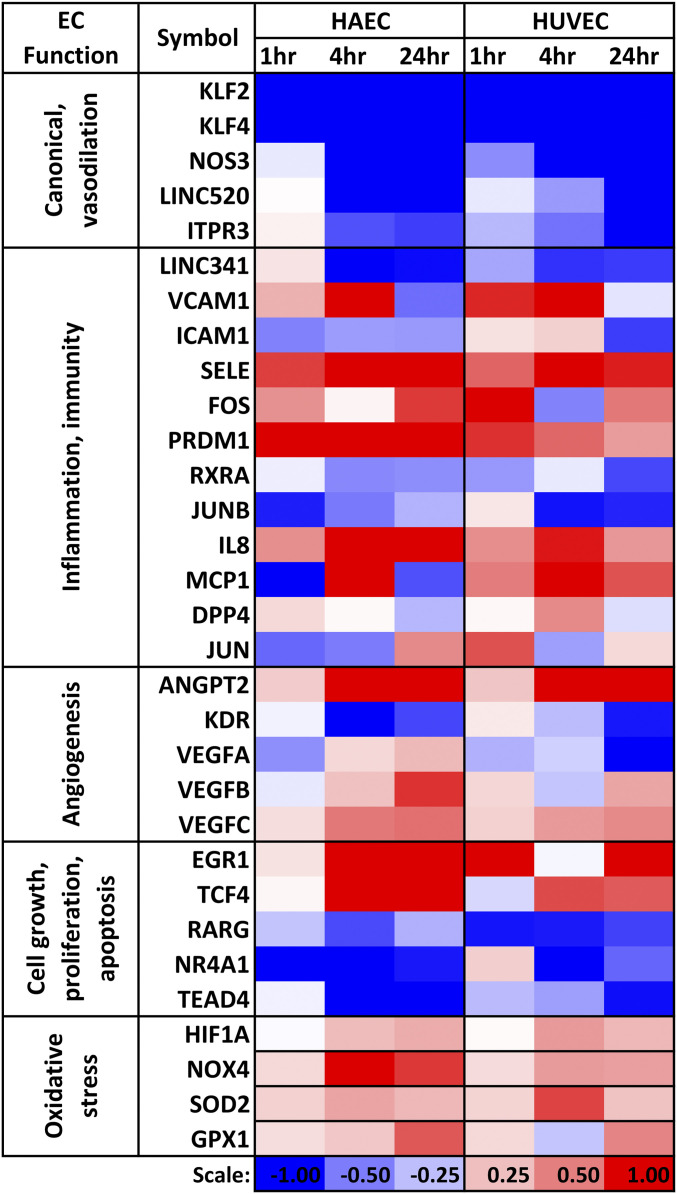
ECs help maintain vascular homeostasis and participate in angiogenesis and immune defense responses such as inflammation and wound healing (17). Facilitation of vasodilation and regulation of blood pressure are important elements in vascular homeostasis. The key genes relevant to EC function include the canonical lineage transcription factors (TFs) KLF2 and KLF4 (18–22), NOS3 (a vasodilator/anticoagulant), and proinflammatory proteins VCAM1 and ICAM1. Elevated expression of NOS3 leads to increased production of nitric oxide and consequently the vasodilation phenotype. In our HAECs vs. HUVECs comparison of OS/PS results, the expression levels of KLF2, KLF4, and NOS3 were down-regulated significantly in both EC types (Fig. 1). VCAM1, ICAM1, and SELE play important roles in leukocyte attachment to ECs. VCAM1 was up-regulated by OS/PS early on in both cell types. ICAM1 was down-regulated in HAECs over all three time points, but only at 24 h in HUVECs. E-selectin (SELE) was strongly up-regulated by OS/PS in both cell types over all three time points. In addition to protein-coding genes, there were also similar trends in HAECs and HUVECs for OS vs. PS differential expression of EC function-relevant long noncoding (lnc) RNAs. For example, LINC00520 (C14orf34) and LINC00341 (C14orf49), the two lncRNA genes we previously identified to be PS inducible (13, 23, 24), were strongly and similarly regulated in both cell types (Fig. 1).
Fig. 1.
Heatmap of log2(OS/PS fold change) for select EC function-relevant genes in HAECs and HUVECs across time. Dataset S1 (“OSbyPS_L2FC”) provides the log2 fold change for all the genes with raw read count >10.
We also observed similarities between the two EC types in the OS/PS responses of regulators of angiogenesis such as ANGPT2 and several OS up-regulated TFs such as EGR1, HIF1A, FOS, PRDM1, and TCF4 (Fig. 1). Among OS down-regulated TFs, we identified RXRA, RARG, JUNB, NR4A1, and TEAD4, besides KLF2 and KLF4. The opposite trend of EGR1 (a target of retinoic acid receptors) to that of RXRA and RARG was similar in HAECs and HUVECs (13). The up-regulation of HIF1A in both types of ECs was consistent with its regulation by EGR1 (13). NR4A1, which is abundantly expressed in ECs and protects against activation by TNF-α and IL-1β through transcriptional up-regulation of IκBα (25), was down-regulated by OS in both cell types. Thus, NR4A1 acts as an antiinflammatory molecule under PS in both cell types.
TCF4 (Entrez gene ID 6925) was up-regulated in both cell types (Fig. 1). TCF4 has a role in WNT3A and β-catenin-mediated proliferation of ECs (26). It also appears to play a role in regulating epithelial–mesenchymal transition (27), and recent genome-wide association studies have pointed to its role in several diseases, including Fuchs’ endothelial corneal dystrophy. PRDM1, whose levels are increased upon virus induction, was up-regulated in OS vs. PS in both cell types at 1 and 4 h. While PRDM1 levels have been shown to be regulated by VEGF signaling in some cell types (28), its levels may possibly be regulated in a VEGF-independent manner in ECs, since VEGF A/B/C and VEGFR (KDR) levels were either down-regulated or only slightly up-regulated at 1 and 4 h in our data in both cell types (Fig. 1).
Transcriptional Regulation of Gene Expression.
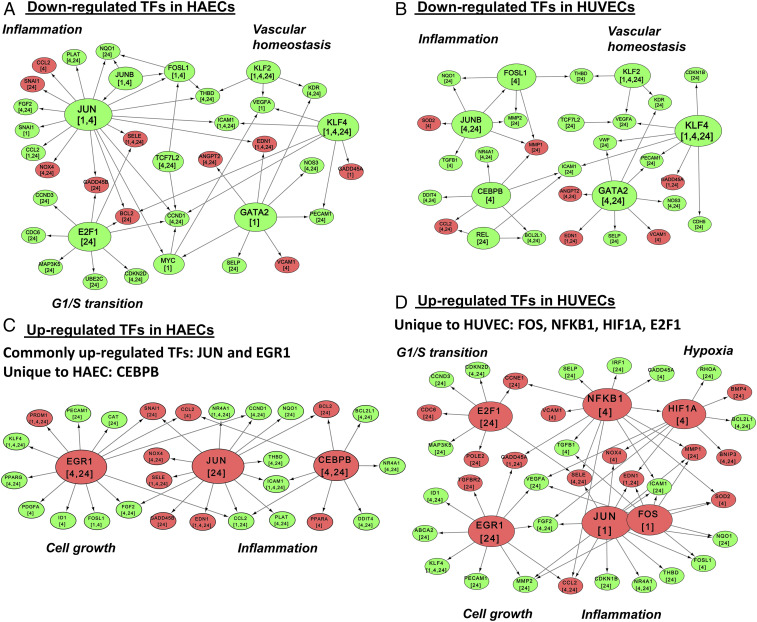
To assess the similarity/difference in the regulation of gene expression between the two cell types, we carried out TF-target network analysis using the TRANSFAC database, as explained in SI Appendix, Methods. Briefly, a network of all OS/PS DE TFs and their DE targets at the same or future time points was developed separately for HAECs and HUVECs. These networks were built for OS/PS down-regulated TFs (Fig. 2 A and B) and OS/PS up-regulated TFs (Fig. 2 C and D). Several highly connected TFs such as KLF2, KLF4, and GATA2 in the down-regulated networks (Fig. 2 A and B and SI Appendix, Fig. S4G), as well as JUN and EGR1 in the up-regulated networks (Fig. 2 C and D and SI Appendix, Fig. S4F), were common between the two cell types. Among the highly connected TFs, KLF2, one of the key regulatory genes for EC function, showed similar response with similar targets in both cell types at all time points (Fig. 2 A and B). However, KLF4 showed stronger down-regulation (with a similar number of DE targets) in HAECs than in HUVECs. EGR1 showed stronger up-regulation with a larger number of DE targets in HAECs than in HUVECs (Fig. 2 C and D).
Fig. 2.
Comparison of transcriptional regulatory networks under OS and PS conditions. Shown are important targets of down-regulated TFs in HAECs (A) and HUVECs (B), and important targets of up-regulated TFs in HAECs (C) and HUVECs (D). The TF target relationships were obtained from the TRANSFAC database. DE TFs were connected to their DE targets when the target was DE at the same or a later time point. The green and red colors for the TF nodes indicate down-regulation and up-regulation, respectively. The same color coding is used for the target nodes.
Many TF genes showed time-dependent and cell-type-specific responses. For example, in HAECs, JUN was down-regulated at 1 and 4 h (Fig. 2A) and became up-regulated at 24 h (Fig. 2C). However, in HUVECs, JUN was up-regulated at 1 h (Fig. 2D) and became down-regulated at 4 h (Fig. 1). JUNB was down-regulated at 1 and 4 h in HAECs (Fig. 2A and SI Appendix, Fig. S4G), and 4 and 24 h in HUVECs (Fig. 2B). Also, the targets for E2F1, which is important for cell cycle progression from G1 to S phase, were down-regulated in HAECs (Fig. 2A) and up-regulated in HUVECs at 24 h (Fig. 2D). CEBPB, which is involved in immune and inflammatory responses, was up-regulated at 4 and 24 h (Fig. 2C) in HAECs and down-regulated at 4 h in HUVECs (Fig. 2B). The TFs FOS, NFκB1, HIF1A, and E2F1 were up-regulated (with their targets DE at the same or at later time points) only in HUVECs (Fig. 2D). In HAECS, even though HIF1A appeared slightly up-regulated at 4 and 24 h (Fig. 1), its log2 FCs were below the cutoff of log2(1.3). Hence, it does not appear in the network of up-regulated TFs in HAECs (Fig. 2C). As shown in SI Appendix, Fig. S4H, several HIF1A targets are similarly regulated in both HAECs and HUVECs, examples include up-regulated genes CXCR4, BMP4, PTGS2, CHKA (choline kinase alpha), NOX4, and EDN1 (endothelin 1, a vasoconstrictor), and down-regulated genes KLF8, FGF2, ADAMTS1, IL11, VEGFA, PTGS2, and CHKA. We observe that the glycolytic metabolism is similarly altered between HUVECs and HAECs in OS/PS, whereas there are some differences in oxidative metabolism. Wu et al. (29) have recently shown that HIF1α is involved in metabolic reprogramming in HAECs. We observe a similar trend in both HAECs and HUVECs in terms of changes associated with HIF1α target genes (SI Appendix, Fig. S4H).
Overall, we were able to identify common key regulators, such as KLF2, KLF4, and EGR1, which are important for endothelial function, as well as cell-type-specific TFs such as E2F1 and CEBPB.
The similarities may be due to the underlying mechanisms involving master EC lineage-dependent transcription factors (LDTFs) coordinating with signal dependent transcription factors (SDTFs) to determine the transcriptional regulation under flow.
Aberrant EC Function Pathways.
The comparisons of longitudinal responses of HAECs and HUVECs provided significant insights into the mechanisms that contribute to the functional responses of endothelium to shear stress. While there were some differences, especially in the kinetics of responses, the majority of the stress-response mechanisms were similar in the two types of ECs, pointing to similar endpoint EC biology. This enabled us to present here detailed mechanisms that are responsive to shear stress in both cell types.
Cell cycle.
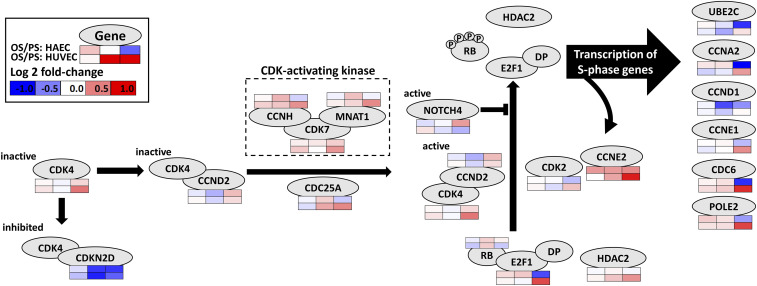
Cell cycle genes exhibited differential expression in OS/PS over the 24 h of study in both HAECs (SI Appendix, Figs. S4A and S5A) and HUVECs (SI Appendix, Fig. S5B). Genes such as CCNE2 and GADD45A/B were up-regulated in both cell types, whereas CCKN1B and CCKN2D were down-regulated in both (SI Appendix, Fig. S4A). E2F1, a key gene in regulating G1/S transition, was OS/PS differently expressed for both HAECs and HUVECs, and the same is true for its several targets such as CCNA2 and CCNE1. The role of E2F1 in cell cycle transition is discussed in detail below.
Fig. 3 provides a detailed view of the key mechanisms of G1/S transition in the cell cycle. Under resting condition, CDK4 is inactive and inhibited by CDKN2D. The up-regulation of CDK-activating kinases such as CDK7 activates CDK4 (bound to CCND2), which in turn phosphorylates RB. During the G1 phase, the transcription factor E2F1 is bound to RB. Once RB is phosphorylated, it releases E2F1, which then moves into the nucleus to regulate the expression of its target genes (30). RB phosphorylation is also modulated by the CDK2-CCNE2 complex. CDK4, CDK7, CCND2, and CCNE2 levels are increased in both cell types (in some cases, below log2 FC threshold), especially at 24 h. Thus, RB is likely phosphorylated in both cell types, although the moderate up-regulation of NOTCH4 in HAECs at 24 h (SI Appendix, Fig. S4A) may counteract this effect (31). OS/PS E2F1 levels are down-regulated in HAECs and up-regulated in HUVECs at 24 h. Thus, in HAECs, the net effect on S-phase genes seems to be mixed. In our data, most S-phase gene targets of E2F1 are OS/PS down-regulated at 24 h; the degree of down-regulation of E2F1 in HAECs is much greater (log2 FC of −0.72) than the up-regulation of CDK2 (0.15) and CCND2 (0.26). In HUVECs, log2 FC for E2F1 is 0.72, consistent with the up-regulation of its targets such as CDC6 and POLE2. The E2F1-target CCNE1 in HUVECs is also slightly up-regulated in OS vs. PS. CCNE1 can further phosphorylate RB, thus establishing a positive feedback (32). CCNA2, another target of E2F1 (33), is also down- and up-regulated in HAECs and HUVECs, respectively, consistent with the changes in E2F1.
Fig. 3.
Commonalities and differences in the G1/S transition in HAECs and HUVECs. The temporal gene expression changes are shown as heat maps Below or Above the gene nodes (oval shapes). The up-regulation of CDK-activating kinases such as CDK7 activates CDK4 (bound to CCND2), which phosphorylates RB. In G1 phase, the TF E2F1 is bound to RB. The phosphorylation of RB causes the release of E2F1, which can then move into the nucleus to regulate the expression of its target genes. RB phosphorylation is also promoted by the binding of CDK2 to CCNE2. CDK4, CDK7, CCND2, and CCNE2 levels are slightly to moderately increased in both cell types, especially at 24 h. Thus, RB is getting phosphorylated in both cell types. However, E2F1 levels are down-regulated in HAECs and up-regulated in HUVECs at 24 h. Thus, in HAECs, the net effect on S-phase genes may be mixed. In our data, most S-phase gene targets of E2F1 are indeed down-regulated at 24 h in HAECs. In HUVECs, E2F1 and its targets such as CDC6 and POLE2 are up-regulated at 24 h.
Inflammation.
Among inflammation-related genes, VCAM1, SELE, and IL8 were up-regulated by OS/PS in both cell types at 1 and 4 h (Fig. 1). At 24 h, VCAM1 was substantially down-regulated in HAECs, but showed no change in HUVECs. NFκB1/2 levels were almost unchanged in HAECs, but moderately up-regulated in HUVECs (see Dataset S1, “OSbyPS_L2FC”) (34). MCP1 (CCL2) was up-regulated by OS/PS at all time points in HUVECs, but only at 4 h in HAECs. Overall, OS/PS effects on inflammation appear to persist over the 24-h period in both cell types, although VCAM1 was DE at 24 h only in HAECs and it was DE at 1 h only in HUVECs.
Oxidative stress.
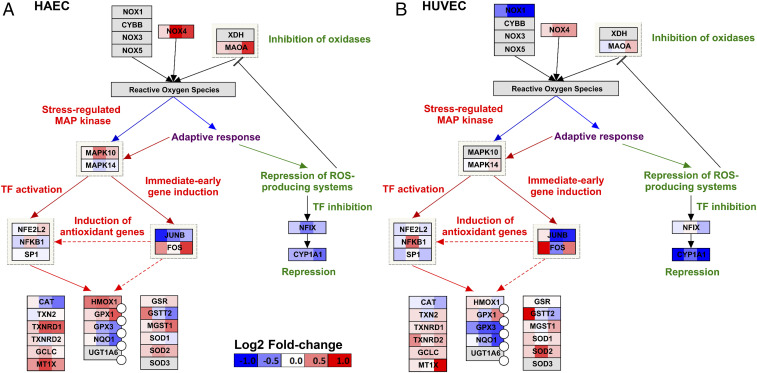
Oxidative stress response to OS/PS persisted throughout the 24 h in both cell types (Fig. 4 and SI Appendix, Fig. S4B). Among oxidative stress-related genes, SOD2, NOX4, and HIF1A were up-regulated in both cell types, although the magnitude and the time of response of up-regulation differ: NOX4 was significantly up-regulated at 4 and 24 h in HAECs, but only moderately in HUVECs; SOD2 was significantly up-regulated at 4 h in HUVECs, but not in HAECs. NQO1 became increasingly down-regulated in both cell types with time.
Fig. 4.
Comparison of OS vs. PS differential expression response of (A) HAECs and (B) HUVECs for Oxidative Stress pathway (WikiPathways)-related genes. Color scale of log2 fold changes: −1 (blue) to 0 (white) to +1 (red). The overall response at the pathway level is similar for the two cell types, although some genes show stronger fold changes in HAECs vs. HUVECs (e.g., NOX4) or weaker fold changes in HAECs vs. HUVECs (e.g., CYP1A1 or SOD2). For some genes, e.g., JUNB, the difference is temporal.
Angiogenesis.
The expressions of angiogenesis pathway genes in response to OS/PS were similar in HAECs and HUVECs across time. Several genes were down-regulated, including KDR and FGF2 (Fig. 1 and SI Appendix, Fig. S6). On the other hand, VEGF A/B/C showed moderate up- or down-regulation, but statistical significance was achieved only at some time points. One of the key genes, ANGPT2, was significantly up-regulated in both cell types across time, but the effect was stronger in HAECs (Fig. 1). PDGFB and PDGFRA were up-regulated in both cell types, although PDGFB FCs were stronger in HAECs and appeared earlier in time (SI Appendix, Fig. S6).
TGF-β signaling.
TGF-β signaling pathway shows similarities between the two cell types. For example, genes such as TGF-β 1 (TGFB1) and ID1 were down-regulated early; e.g., TGFB1 at 1 h in HAECs (below threshold), and 4 h in HUVECs; ID1 at 1 and 4 h in HAECs, and 4 h in HUVECs (SI Appendix, Fig. S4C). SMAD6 was significantly down-regulated at 1 h in both cell types. Several genes were significantly DE at both 4 h and 24 h in HAECs, but not altered or DE only at 24 h in HUVECs; examples include HDAC1, CDKN2B, KLF10, TGFBR3, and MEF2A. cMYC was down-regulated in HAECs, but slightly up-regulated in HUVECs at 1 h. The levels of TGFBR 1/2 showed similar trends throughout the 24-h period. SMAD 6/7/9 showed different trends between HAECs and HUVECs, e.g., SMAD9 was up-regulated at 4 h in HAECs, but down-regulated at 1 h in HUVECs (SI Appendix, Fig. S4C). BMP2 was down-regulated only in HAECs at 4 and 24 h, and BMP4 was up-regulated in both cell types across the time course.
Lysosome and autophagy.
In the lysosome pathway, the DE genes include CTSK, CTSS, NAGA, ACP5, LAMP3, SLC11A1, ABCA2, IGF2R, LITAF, and HYAL1 (SI Appendix, Fig. S4 D and E). These genes also show some temporal differences between the two cell types. Among the ATP6V family of genes, ATP6V1A was up-regulated in both cell types. Among autophagy-related genes, IRS2 and DAPK2 were up-regulated in both cell types (Dataset S1, “OSbyPS_L2FC”), and DDIT4 and ATG9B were down-regulated in both cell types (SI Appendix, Fig. S4 D and E). Thus, autophagy function is regulated similarly in the two cell types. However, it is important to note that there were a few genes which showed significant differences between the two cell types in terms of temporal extent or magnitude of change. The genes with magnitude differences include PIK3CD and BCL2, which are up-regulated, and PIK3R3, MRAS, and BCL2L1, which are down-regulated.
The similarity of HAEC and HUVEC responses to OS/PS was also apparent in the pathway/functional enrichment results shown in SI Appendix, Table S1 obtained using DAVID version 6.8 (35, 36) for Kyoto Encyclopedia of Genes and Genomes (KEGG) (37), Reactome, and Biocarta pathways. SI Appendix, Table S1 lists the –log10 (enrichment P value) for a select set of pathways/processes relevant to EC functions. Several pathways such as PI3K-Akt signaling, RAS signaling, and TNF signaling show similar trends across time in the two cell types. For some pathways, statistical significance is achieved earlier in HAECs than HUVECs. These include focal adhesion, cell cycle, ECM–receptor interaction, HIF-1 signaling, and VEGF signaling, suggesting a faster kinetic response in HAECs.
Kinetics of Shear Stress Responses in HAECs and HUVECs.
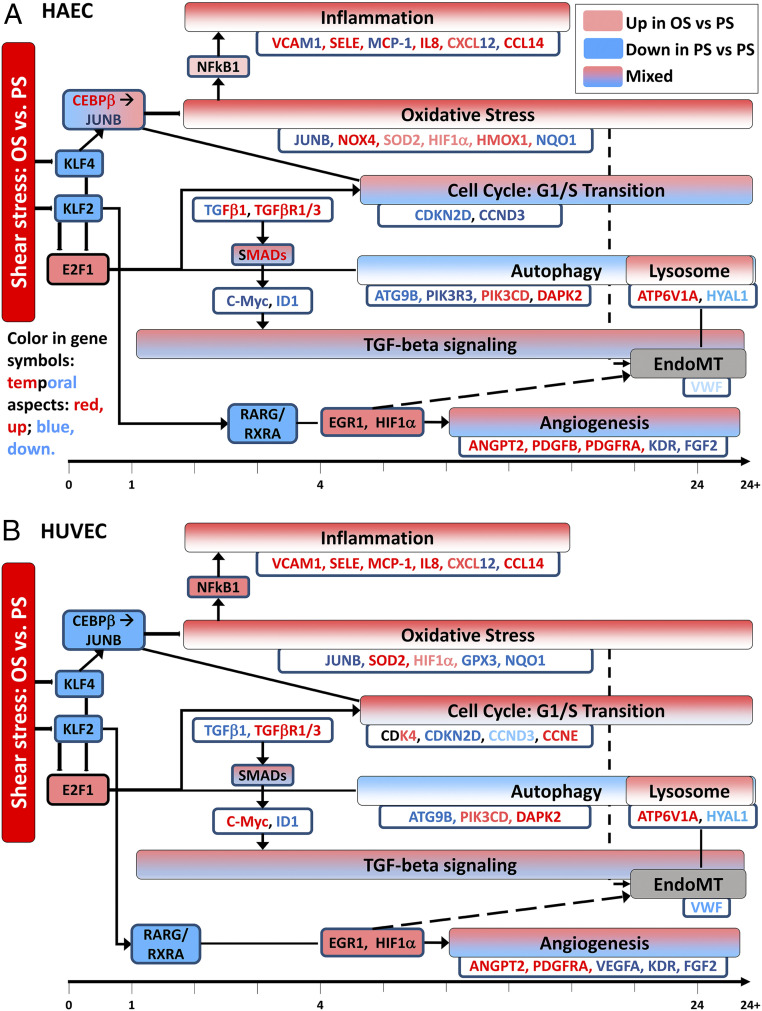
In our prior work comparing OS vs. PS responses in HUVECs, we developed a temporal map of changes in the EC function-related pathways such as inflammation, oxidative stress, cell cycle, TGF-β signaling, and lysosomal trafficking [Fig. 5B, based on and modified from Figure 3 of Ajami et al. (13)]. Here we have developed such a temporal functional map for HAECs (Fig. 5A). Several pathways show overall similar responses in HAECs and HUVECs, providing evidence for common phenotypic mechanisms. Some of the pathways such as oxidative stress and inflammation are overall up-regulated by OS vs. PS, i.e., most genes in these pathways are up-regulated. Some of these findings are in agreement with other previous results on HUVECs, e.g., Sun et al. (38) have shown that OS increased gene and protein expressions for VCAM1 and SELE and levels of mitochondrial ROS, and Hsu et al. (39) have reported similar findings. Gene expressions of VCAM1, ICAM1, and MCP1 have been shown to increase in OS vs. PS in HAECs [Brooks et al. (40)] and HUVECs [Fan et al. (41)]. Autophagy is overall down-regulated (42), whereas TGF-β signaling and angiogenesis pathways show a mix of up- and down-regulated genes in both cell types. Cell cycle pathways, particularly the G1/S transition pathway, show mixed responses in HAECs (Fig. 5A), whereas it is largely up-regulated (though only mildly in terms of FC) in HUVECs (Fig. 5B). Inflammation-related processes and oxidative-stress response pathway show generally similar responses in both cell types; NFκB is up-regulated slightly in HAECs and moderately (DE at 4 h) in HUVECs, and JUNB and NQO1 are down-regulated in both cell types. SOD2, one of the key genes related to oxidative stress, is up-regulated significantly in HUVECs, but only slightly in HAECs, although the response persists throughout the 24-h period. HIF1A is slightly up-regulated in both cell types; Nayak et al. (43) have shown that KLF2 overexpression causes HIF1A inhibition in ECs, and Feng et al. (44) found up-regulation of HIF1A in low shear (mimicking OS) vs. high shear (mimicking PS) conditions in HUVECs. While angiogenesis pathway and its genes show mixed responses in both cell types, the responses are generally similar [e.g., ANGP2 is up-regulated in both cell types, as also observed by others (45)]; in terms of kinetics, the changes appear earlier in HAECs than in HUVECs. Autophagy (42, 46) and lysosome pathways are broadly down- and up-regulated, respectively, in both cell types, although there are some differences in the differential expressions of several genes. For example, VWF, which has a role in blood clotting, is significantly down-regulated at 24 h only in HUVECs. ABCA2, a transporter protein, is significantly down-regulated at 4 and 24 h only in HUVECs. BCL2, which is involved in apoptosis, is significantly up-regulated at 24 h in HAECs only.
Fig. 5.
Temporal map of changes in EC function-relevant pathways in HAECs (A) and HUVECs (B). Color-filled boxes represent pathways. Pathways and genes in red are up-regulated (up) by OS vs. PS, while those in blue are down-regulated (down) by OS vs. PS. Some pathways such as TGF-β signaling pathway and angiogenesis are shown in “mixed” color (gradient) due to mixed response (both up- and down-regulated). Such mixed response is generally observed across the time course. For the cell cycle pathway, CCND3 is down-regulated strongly in HAECs, but only moderately in HUVECs. CDK4 is moderately up-regulated only in HUVECs. Inflammation-related processes and oxidative stress response pathway show overall similar response in both cell types, although there are some temporal differences in specific genes. NFκB is slightly and moderately up-regulated in HAECs and HUVECs, respectively, and JUNB and NQO1 are down-regulated in both cell types. SOD2, one of the key genes related to oxidative stress, is up-regulated significantly in HUVECs, but only slightly in HAECs, although the response persists across time. HIF1α is slightly up-regulated in both cell types. While the angiogenesis pathway and its genes show mixed responses in both cell types and the responses are broadly similar, the changes appear earlier in HAECs than HUVECs. Autophagy and lysosome pathways are overall down- and up-regulated, respectively, in both cell types, although there are minor differences in the differential expression of several genes. Angiogenesis exhibits mixed response in both cell types.
Atherogenic and Atheroprotective Responses.
SI Appendix, Fig. S7 shows temporal evolution of OS/PS differential expression for atheroprotective genes (KLF2, KLF4, eNOS [NOS3] and NQO1) and atherogenic genes (VCAM1, ICAM1, E-Sel [SELE], and MCP1 [CCL2]) in HAECs and HUVECs. This signaling-regulatory map is based, in part, on the KEGG pathway “Fluid shear stress and atherosclerosis—Homo sapiens (human)” (hsa05418) (37, 47, 48). OS and PS are sensed by some common proteins such as integrins and VEGFR by mechanisms not yet fully understood (49, 50). The expression level of integrins (e.g., ITGA2) (SI Appendix, Fig. S7) is up-regulated in OS/PS and that of VEGFR2 (KDR) is down-regulated. From a signaling perspective, MEF2 appears to be affected by only PS (49), and its gene expression shows down-regulation by OS/PS in both cell types. VEGFR signaling activates AMPK and eNOS (NOS3; also positively regulated by AMPK). In turn, AMPK and MEF2 regulate the expression of KLF2, whereas KLF4 is primarily regulated by AMPK. On one hand, OS/PS down-regulates the levels of KLF2 and KLF4 in both HAECs and HUVECs to result in the down-regulation of eNOS and ASS (thus leading to reduced vasodilation and increased inflammation) and the antioxidants NQO1, GPX3, GSTO2 (a representative of glutathione S-transferases), and GSTT2 (Fig. 4 and SI Appendix, Figs. S4B and S7). On the other hand, OS/PS slightly up-regulates NFκB to result in the inductions of VCAM1, SELE, and MCP1 (CCL2) in both HAECs and HUVECs. Overall, inflammation is increased in OS vs. PS in both HAECs and HUVECs. In summary, in both HAECs and HUVECs, the OS vs. PS response is down-regulated for genes contributing to atheroprotection and up-regulated for those contributing to atherogenesis.
Conclusions
We report a systematic comparison of longitudinal responses to OS and PS between HAECs and HUVECs. At 24 h, HAECs and HUVECs had more distinct than common up-regulated DE genes, but the numbers of distinct and common down-regulated DE genes were comparable between these two types of ECs. At a functional level, these two cell types had similar differential OS/PS expressions for several EC function- and pathway-relevant genes. The functionally relevant genes with similar responses include KLF2, KLF4, NOS3, and ITPR3 (canonical functions), VCAM1 and SELE (inflammation), and ANGPT2 (angiogenesis). However, several functionally relevant genes exhibit temporal or directional (up- vs. down-regulated) differences between HAECs and HUVECs. These genes include LINC520 (regulation of NOS3), ICAM1, JUN, JUNB, FOS, and MCP1 (inflammation), VEGFR2 (KDR) and VEGF A/B (angiogenesis), EGR1 (cell growth), and NOX4 and SOD2 (oxidative stress). Temporally, HAECs exhibit an earlier shear stress response, or faster kinetics, as compared to HUVECs. For example, genes involved in focal adhesion, cell cycle, and ECM–receptor interaction achieve statistical significance earlier in HAECs than HUVECs. The similarities in the responses across the EC types suggest common underlying mechanisms that involve master EC LDTFs, which coordinate with SDTFs to determine the transcriptional regulation under flow. The master LDTFs such as KLF2 and KLF4 exhibit very similar OS/PS responses between the two EC types. The same is true for some of the SDTFs such as JUN, GATA2, and EGR1, albeit with some temporal differences. The genetic heterogeneities between HAECs and HUVECs in conjunction with differences in the responses of some of the SDTFs such as CEBPB and E2F1 may be driving the differences in transcriptional responses between the two EC types. As an example, for cell-cycle, several E2F1 target genes such as CCND1, CCNE1, and CD6 exhibit directional, temporal, and/or magnitude differences between HAECs and HUVECs.
Our analysis demonstrates that HUVECs, despite the unique features of the umbilical vein, can serve as an in vitro model to study physiological and pathological responses of vascular endothelium and understand the underlying mechanisms in health and disease, in addition to the bona fide model of HAECs. While the response of OS/PS is similar overall at the functional level, the directional and kinetic differences between HAECs and HUVECs, especially at the gene level can vary and should be considered when interpreting and extrapolating the results from HUVECs. Further investigations with longitudinal comparisons among other mammalian model systems, similar to the present study, would shed more light on the mechanisms associated with pathological responses and offer temporally relevant therapeutic interventions. In addition, as highlighted in some recent studies (51, 52), we recognize that genetic variations may contribute to functional changes and the atherosclerotic phenotype.
Materials and Methods
Culture conditions of HUVECs and shear stress experiments for OS and PS were as previously described (53, 54). In the published work on HUVECs, the measurements were made at 10 time points between 1 and 24 h (13). In the present study on HAECs, measurements were made at 3 time points (1, 4, and 24 h). The comparison was made with respect to the data at these 3 common time points, i.e., 1, 4, and 24 h. Please refer to SI Appendix, Methods for specific details of RNA sequencing and data analyses (SI Appendix, Fig. S1).
Supplementary Material
Acknowledgments
This work was supported in part by NIH Research Grants R01HL106579 and HL108735 (to S.C., S.S., J.Y.-J.S., and Z.B.C) R01LM012595 (to S.S.), and U19AI090023 (to S.S.), and NSF Research Grant CCF0939370 (to S.S.).
Footnotes
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2023236118/-/DCSupplemental.
Data Availability.
The new sequence data on HAECs reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE160611). The HUVEC data used in this study are available at GEO (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE103672).
References
- 1.Chiu J. J., Chien S., Effects of disturbed flow on vascular endothelium: Pathophysiological basis and clinical perspectives. Physiol. Rev. 91, 327–387 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chistiakov D. A., Orekhov A. N., Bobryshev Y. V., Effects of shear stress on endothelial cells: Go with the flow. Acta Physiol. (Oxf.) 219, 382–408 (2017). [DOI] [PubMed] [Google Scholar]
- 3.Baeyens N., Bandyopadhyay C., Coon B. G., Yun S., Schwartz M. A., Endothelial fluid shear stress sensing in vascular health and disease. J. Clin. Invest. 126, 821–828 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dai G., et al. , Distinct endothelial phenotypes evoked by arterial waveforms derived from atherosclerosis-susceptible and -resistant regions of human vasculature. Proc. Natl. Acad. Sci. U.S.A. 101, 14871–14876 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Patel H., Chen J., Das K. C., Kavdia M., Hyperglycemia induces differential change in oxidative stress at gene expression and functional levels in HUVEC and HMVEC. Cardiovasc. Diabetol. 12, 142 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McCall M. N., et al. , MicroRNA profiling of diverse endothelial cell types. BMC Med. Genomics 4, 78 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Staton C. A., Reed M. W., Brown N. J., A critical analysis of current in vitro and in vivo angiogenesis assays. Int. J. Exp. Pathol. 90, 195–221 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Onat D., Brillon D., Colombo P. C., Schmidt A. M., Human vascular endothelial cells: A model system for studying vascular inflammation in diabetes and atherosclerosis. Curr. Diab. Rep. 11, 193–202 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chi J. T., et al. , Endothelial cell diversity revealed by global expression profiling. Proc. Natl. Acad. Sci. U.S.A. 100, 10623–10628 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bhasin M., et al. , Bioinformatic identification and characterization of human endothelial cell-restricted genes. BMC Genomics 11, 342 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang W., et al. , Nucleolin down-regulation is involved in ADP-induced cell cycle arrest in S phase and cell apoptosis in vascular endothelial cells. PLoS One 9, e110101 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sung H. J., Yee A., Eskin S. G., McIntire L. V., Cyclic strain and motion control produce opposite oxidative responses in two human endothelial cell types. Am. J. Physiol. Cell Physiol. 293, C87–C94 (2007). [DOI] [PubMed] [Google Scholar]
- 13.Ajami N. E., et al. , Systems biology analysis of longitudinal functional response of endothelial cells to shear stress. Proc. Natl. Acad. Sci. U.S.A. 114, 10990–10995 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Casella G., et al. , Transcriptome signature of cellular senescence. Nucleic Acids Res. 47, 11476 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sawada M., Yamamoto H., Ogasahara A., Tanaka Y., Kihara S., β-aminoisobutyric acid protects against vascular inflammation through PGC-1β-induced antioxidative properties. Biochem. Biophys. Res. Commun. 516, 963–968 (2019). [DOI] [PubMed] [Google Scholar]
- 16.Wang H. S., Li F., Runge M. S., Chaikof E. L., Endothelial cells exhibit differential chemokinetic and mitogenic responsiveness to alpha-thrombin. J. Surg. Res. 68, 139–144 (1997). [DOI] [PubMed] [Google Scholar]
- 17.Félétou M., “Multiple functions of the Endothelial cells” in Colloquium Series in Integrated Systems Physiology: From Molecule to Function, Granger D. N., Granger J. P., Eds. (Morgan & Claypool Life Sciences, San Rafael, CA, 2011), pp. 3–17. [Google Scholar]
- 18.Sangwung P., et al. , KLF2 and KLF4 control endothelial identity and vascular integrity. JCI Insight 2, e91700 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Atkins G. B., Jain M. K., Role of Krüppel-like transcription factors in endothelial biology. Circ. Res. 100, 1686–1695 (2007). [DOI] [PubMed] [Google Scholar]
- 20.Kuebler W. M., The flow-dependent transcription factor KLF2 protects lung vascular barrier function in acute respiratory distress syndrome. Am. J. Respir. Crit. Care Med. 195, 553–555 (2017). [DOI] [PubMed] [Google Scholar]
- 21.SenBanerjee S., et al. , KLF2 Is a novel transcriptional regulator of endothelial proinflammatory activation. J. Exp. Med. 199, 1305–1315 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fontijn R. D., et al. , Expression of nitric oxide-transporting aquaporin-1 is controlled by KLF2 and marks non-activated endothelium in vivo. PLoS One 10, e0145777 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miao Y., et al. , Enhancer-associated long non-coding RNA LEENE regulates endothelial nitric oxide synthase and endothelial function. Nat. Commun. 9, 292 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang T. S., et al. , LINC00341 exerts an anti-inflammatory effect on endothelial cells by repressing VCAM1. Physiol. Genomics 49, 339–345 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.You B., Jiang Y. Y., Chen S., Yan G., Sun J., The orphan nuclear receptor Nur77 suppresses endothelial cell activation through induction of IkappaBalpha expression. Circ. Res. 104, 742–749 (2009). [DOI] [PubMed] [Google Scholar]
- 26.Zhang Y., Chidiac R., Delisle C., Gratton J. P., Endothelial NO synthase-dependent S-nitrosylation of β-catenin prevents its association with TCF4 and inhibits proliferation of endothelial cells stimulated by Wnt3a. Mol. Cell. Biol. 37, e00089-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Forrest M. P., Hill M. J., Quantock A. J., Martin-Rendon E., Blake D. J., The emerging roles of TCF4 in disease and development. Trends Mol. Med. 20, 322–331 (2014). [DOI] [PubMed] [Google Scholar]
- 28.Arulanandam R., et al. , VEGF-mediated induction of PRD1-BF1/Blimp1 expression sensitizes tumor vasculature to oncolytic virus infection. Cancer Cell 28, 210–224 (2015). [DOI] [PubMed] [Google Scholar]
- 29.Wu D., et al. , HIF-1α is required for disturbed flow-induced metabolic reprogramming in human and porcine vascular endothelium. eLife 6, e25217 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang K. C., et al. , Role of microRNA-23b in flow-regulation of Rb phosphorylation and endothelial cell growth. Proc. Natl. Acad. Sci. U.S.A. 107, 3234–3239 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Noseda M., Karsan A., Notch and minichromosome maintenance (MCM) proteins: Integration of two ancestral pathways in cell cycle control. Cell Cycle 5, 2704–2709 (2006). [DOI] [PubMed] [Google Scholar]
- 32.Bertoli C., Skotheim J. M., de Bruin R. A., Control of cell cycle transcription during G1 and S phases. Nat. Rev. Mol. Cell Biol. 14, 518–528 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen T., et al. , The retinoblastoma protein selectively represses E2F1 targets via a TAAC DNA element during cellular senescence. J. Biol. Chem. 287, 37540–37551 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang W., et al. , TLR4 activation induces nontolerant inflammatory response in endothelial cells. Inflammation 34, 509–518 (2011). [DOI] [PubMed] [Google Scholar]
- 35.Huang D. W., Sherman B. T., Lempicki R. A., Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 (2009). [DOI] [PubMed] [Google Scholar]
- 36.Huang D. W., Sherman B. T., Lempicki R. A., Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37, 1–13 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kanehisa M., Furumichi M., Tanabe M., Sato Y., Morishima K., KEGG: New perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 45, D353–D361 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sun Z., et al. , Activation of GPR81 by lactate inhibits oscillatory shear stress-induced endothelial inflammation by activating the expression of KLF2. IUBMB Life 71, 2010–2019 (2019). [DOI] [PubMed] [Google Scholar]
- 39.Hsu P. L., Lin Y. C., Ni H., Mo F. E., Ganoderma triterpenoids exert antiatherogenic effects in mice by alleviating disturbed flow-induced oxidative stress and inflammation. Oxid. Med. Cell. Longev. 2018, 3491703 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brooks A. R., Lelkes P. I., Rubanyi G. M., Gene expression profiling of human aortic endothelial cells exposed to disturbed flow and steady laminar flow. Physiol. Genomics 9, 27–41 (2002). [DOI] [PubMed] [Google Scholar]
- 41.Fan W., et al. , Shear-sensitive microRNA-34a modulates flow-dependent regulation of endothelial inflammation. J. Cell Sci. 128, 70–80 (2015). [DOI] [PubMed] [Google Scholar]
- 42.Lien S. C., et al. , Mechanical regulation of cancer cell apoptosis and autophagy: Roles of bone morphogenetic protein receptor, Smad1/5, and p38 MAPK. Biochim. Biophys. Acta 1833, 3124–3133 (2013). [DOI] [PubMed] [Google Scholar]
- 43.Nayak L., Lin Z., Jain M. K., “Go with the flow”: How Krüppel-like factor 2 regulates the vasoprotective effects of shear stress. Antioxid. Redox Signal. 15, 1449–1461 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Feng S., et al. , Mechanical activation of hypoxia-inducible factor 1α drives endothelial dysfunction at atheroprone sites. Arterioscler. Thromb. Vasc. Biol. 37, 2087–2101 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tressel S. L., Huang R. P., Tomsen N., Jo H., Laminar shear inhibits tubule formation and migration of endothelial cells by an angiopoietin-2 dependent mechanism. Arterioscler. Thromb. Vasc. Biol. 27, 2150–2156 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu J., et al. , Shear stress regulates endothelial cell autophagy via redox regulation and Sirt1 expression. Cell Death Dis. 6, e1827 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Heo K. S., Fujiwara K., Abe J., Disturbed-flow-mediated vascular reactive oxygen species induce endothelial dysfunction. Circ. J. 75, 2722–2730 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nigro P., Abe J., Berk B. C., Flow shear stress and atherosclerosis: A matter of site specificity. Antioxid. Redox Signal. 15, 1405–1414 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bryan M. T., et al. , Mechanoresponsive networks controlling vascular inflammation. Arterioscler. Thromb. Vasc. Biol. 34, 2199–2205 (2014). [DOI] [PubMed] [Google Scholar]
- 50.Zhou J., Li Y. S., Chien S., Shear stress-initiated signaling and its regulation of endothelial function. Arterioscler. Thromb. Vasc. Biol. 34, 2191–2198 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Krause M. D., et al. , Genetic variant at coronary artery disease and ischemic stroke locus 1p32.2 regulates endothelial responses to hemodynamics. Proc. Natl. Acad. Sci. U.S.A. 115, E11349–E11358 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stolze L. K., et al. , Systems genetics in human endothelial cells identifies non-coding variants modifying enhancers, expression, and complex disease traits. Am. J. Hum. Genet. 106, 748–763 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen Z., et al. , Shear stress, SIRT1, and vascular homeostasis. Proc. Natl. Acad. Sci. U.S.A. 107, 10268–10273 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Guo D., Chien S., Shyy J. Y., Regulation of endothelial cell cycle by laminar versus oscillatory flow: Distinct modes of interactions of AMP-activated protein kinase and Akt pathways. Circ. Res. 100, 564–571 (2007). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The new sequence data on HAECs reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE160611). The HUVEC data used in this study are available at GEO (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE103672).