Significance
Metazoan species optimize the timing of reproduction to maximize fitness. To understand how biological clocks direct reproduction, we investigated the neural substrates that produce oogenesis rhythms in the genetically amenable model organism Drosophila melanogaster. The neuropeptide allatostatin C (AstC) is an insect counterpart of the vertebrate neuropeptide somatostatin, which suppresses gonadotropin production. A subset of the brain circadian pacemaker neurons produces AstC. We have uncovered that these clock-associated AstC neurons generate the circadian oogenesis rhythm via brain insulin-producing cells and the insect gonadotropin juvenile hormone. Identification of a conserved neuropeptide pathway that links female reproduction and the biological clock offers insight into the molecular mechanisms that direct reproductive timing.
Keywords: biological clock, somatostatin, vitellogenesis, insulin, Drosophila
Abstract
The link between the biological clock and reproduction is evident in most metazoans. The fruit fly Drosophila melanogaster, a key model organism in the field of chronobiology because of its well-defined networks of molecular clock genes and pacemaker neurons in the brain, shows a pronounced diurnal rhythmicity in oogenesis. Still, it is unclear how the circadian clock generates this reproductive rhythm. A subset of the group of neurons designated “posterior dorsal neuron 1” (DN1p), which are among the ∼150 pacemaker neurons in the fly brain, produces the neuropeptide allatostatin C (AstC-DN1p). Here, we report that six pairs of AstC-DN1p send inhibitory inputs to the brain insulin-producing cells, which express two AstC receptors, star1 and AICR2. Consistent with the roles of insulin/insulin-like signaling in oogenesis, activation of AstC-DN1p suppresses oogenesis through the insulin-producing cells. We show evidence that AstC-DN1p activity plays a role in generating an oogenesis rhythm by regulating juvenile hormone and vitellogenesis indirectly via insulin/insulin-like signaling. AstC is orthologous to the vertebrate neuropeptide somatostatin (SST). Like AstC, SST inhibits gonadotrophin secretion indirectly through gonadotropin-releasing hormone neurons in the hypothalamus. The functional and structural conservation linking the AstC and SST systems suggest an ancient origin for the neural substrates that generate reproductive rhythms.
To maximize reproductive fitness, every species on earth optimizes the timing of its reproduction (1). Large mammals, such as goats and sheep, which have a gestation period of ∼6 mo, breed in the fall so they can give birth in the spring. Birds and small mammals with shorter gestation periods breed in both the spring and summer. Biological clocks monitor the changes in day length that occur as the seasons progress so they can better direct behavior, but it remains unclear how clocks direct seasonal changes in reproduction.
In the fruit fly Drosophila melanogaster, ∼150 brain pacemaker neurons that express the highly conserved circadian clock genes work together to generate a robust diurnal rhythmicity in locomotion and many other biological processes (2–4). Reproductive activity in Drosophila is also under the control of the circadian clock. Males lacking clock genes, such as period (per) or timeless (tim), release significantly fewer sperm from the testes to the seminal vesicles (5). Female mating activity also shows a robust circadian rhythmicity that is lost in per or tim mutants (6). In addition to mating activity, oogenesis shows a diurnal rhythmicity that is maintained in the absence of environmental circadian cues (i.e., in constant darkness) (7). It remains unclear, though, how the biological clock regulates reproductive behaviors and their associated physiological processes.
In Drosophila, insulin/insulin-like signaling (IIS) from insulin-producing cells (IPCs) located in the pars intercerebralis region of the brain plays a crucial role in oogenesis. The IPCs are a set of 14 median neurosecretory cells that produce Drosophila insulin-like peptides (Dilps) 2, 3, and 5. Brain IIS directly stimulates germline stem cell (GSC) division in the germarium and promotes germline cyst and oocyte development (8). In addition to its direct roles, IIS also regulates the synthesis of the steroid hormone ecdysteroid (20E) and the sesquiterpenoid juvenile hormones (JHs), both of which constitute complex endocrine networks essential for oocyte production (9–11). 20E signaling is critical for the stimulation of GSC proliferation that occurs upon mating (12), and for the maintenance of GSCs in aging females (13, 14). With 20E, the JHs promote vitellogenesis by stimulating yolk protein synthesis by the fat body and then its uptake by developing oocytes (15).
It is clear that JHs are essential for vitellogenesis; ablation of the corpora allata (CA), the sole endocrine organ that produces and secretes JHs, results in a marked loss of vitellogenesis that is reversible upon treatment with JH mimics (16). The many central and peripheral signals that regulate JH can be divided into two groups: Allatotropins activate JH biosynthesis, while allatostatins inhibit it. In Drosophila, IIS promotes JH biosynthesis and secretion through insulin receptor (lnR) in the CA (11). Ecdysis triggering hormones from the peritracheal gland Inka cells stimulate JH production during eclosion and in stress conditions (17). The neuropeptide allatostatin C (AstC) inhibits JH biosynthesis in several species. AstC was first identified in the hawkmoth Manduca sexta (where it was designated Manse-AST) based on its allatostatic activity on isolated CAs (18). Genes encoding AstC have since been identified in the true armyworm Pseudaletia unipuncta (19) and in D. melanogaster (20). The AstC peptide is highly conserved across insect species, with AstC from M. sexta and D. melanogaster differing by only one amino acid residue. AstC expression occurs mainly in the central nervous system and in the enteroendocrine cells of the gastrointestinal system (20, 21). In adult Drosophila, although knockdown of either AstC or its two G protein-coding receptor (GPCR) receptors (star1 and AICR2) increase JH levels (22, 23), the natural contexts in which AstC inhibits JH levels in adults remain unclear.
A subset of posterior dorsal neurons (DN1p) in the brain clock neuron network produces AstC (AstC-DN1p) (24). In this study, we report that AstC from AstC-DN1p generates a circadian oogenesis rhythm. This AstC acts through its two GPCR receptors to suppress vitellogenesis by blocking Dilp secretion from the IPCs. Our results suggest that as AstC-DN1p activity peaks at dawn, it represses IPC activity. As AstC-DN1p activity reaches its lowest point in the afternoon, IPC activity is derepressed. It is this modulation of IPC activity that then modulates vitellogenesis initiation. In addition, AstC is orthologous to the vertebrate somatostatins (SST) that inhibit gonadotrophin releasing hormone (GnRH) secretion. Thus, we expect our discovery of a causal association between AstC-producing clock neurons and oogenesis rhythms in Drosophila will also offer insight into the neural and molecular mechanisms that determine reproduction timing in seasonal breeding animals.
Results
AstC-Gal4 Neurons Regulate Multiple Processes in Female Reproduction.
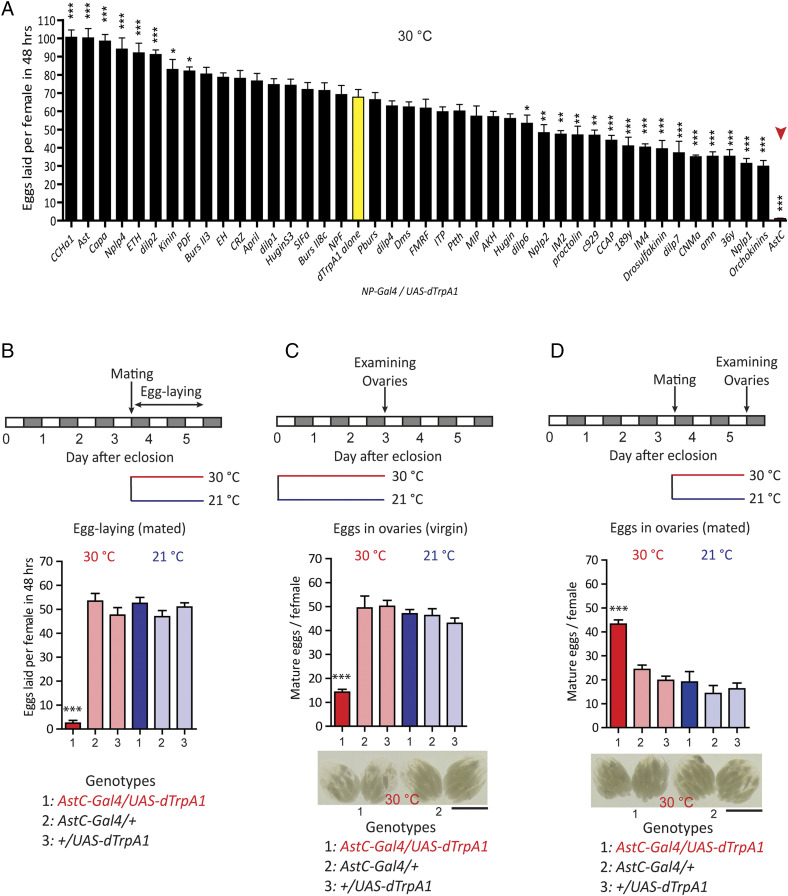
In a search for brain factors that regulate female reproduction, we expressed the warmth-activated cation channel dTrpA1 in select groups of neuromodulator Gal4 neurons and activated them with a temperature shift to 30 °C (25). Females mated individually with wild-type Canton-S (CS) males at 21 °C were allowed to lay eggs for 48 h at 30 °C. Among 42 Gal4 lines, we found activation of AstC-Gal4 neurons produced the strongest suppression of female fecundity (Fig. 1 A and B), although it showed limited effects on mating receptivity (26).
Fig. 1.
Activation of AstC-Gal4 neurons inhibits both egg production and egg laying. (A) Number of eggs laid per female (n = 40 to 60) of the indicated neuropeptide-Gal4 (NP-Gal4) genotype crossed with UAS-dTrpA1 over 48 h at 30 °C (hence activation of dTrpA1). The means ± SEM of the data are shown. Dunnett’s test; *P < 0.05, **P < 0.005, ***P < 0.001 for comparisons against UAS-dTrpA1 alone (yellow bar). The red arrowhead highlights AstC-Gal4/UAS-dTrpA1. (B, Upper) Behavioral protocol for measuring mated female egg laying used in A and B. (Lower) Number of eggs laid per female (n = 40 to 60) of the indicated genotype over 48 h at the indicated temperature (30 °C, red and 21 °C, blue). The means ± SEM of the data are shown. One-way ANOVA followed by Tukey’s test for multiple comparisons; ***P < 0.001 for comparisons of AstC-Gal4/UAS-dTrpA1 against the appropriate parental (light red bars) and temperature controls (blue bars). (C, Top) Protocol for measuring virgin female egg production. (Middle) Number of mature eggs per virgin female (n = 40 to 60) of the indicated genotype kept for 3 d posteclosion at the indicated temperature (30 °C, red and 21 °C, blue). The means ± SEM of the data are shown. One-way ANOVA followed by Tukey’s test for multiple comparisons; ***P < 0.001 for comparisons of AstC-Gal4/UAS-dTrpA1 against the appropriate parental and temperature controls. (Bottom) Ovaries of 3-d-old virgin females of the indicated genotype kept for 3 d posteclosion at 30 °C. (Scale bar, 1 mm.) (D, Top) Protocol for measuring mated female oviposition. (Middle) Number of mature eggs per mated female (n = 40 to 60) of the indicated genotype 2 d postcopulation at the indicated temperature (30 °C and 21 °C). The means ± SEM of the data are shown. One-way ANOVA followed by Tukey’s test for multiple comparisons; ***P < 0.001 for comparisons of AstC-Gal4/UAS-dTrpA1 against the appropriate parental and temperature controls. (Bottom) ovaries of mated females of the indicated genotype kept at 30 °C for 2 d postcopulation. (Scale bar, 1 mm.)
We measured female fecundity by measuring egg-laying activity. Since egg-laying is the final outcome of oogenesis, ovulation, and oviposition, we examined each of these processes in the presence or absence of AstC-Gal4 neuron activation. We measured oogenesis by counting the number of fully mature stage 14 oocytes (hereafter, mature eggs) in the ovaries of 3-d-old virgin females (Fig. 1C). We examined oogenesis only in virgin females, because mating increases ovulation and oviposition rates, both of which reduce the number of eggs in the ovaries. After a 3-d posteclosion incubation at 30 °C, we found control females carrying either AstC-Gal4 or UAS-dTrpA1 averaged 45 ± 5 mature eggs in their ovaries. In contrast, females carrying both the AstC-Gal4 and UAS-dTrpA1 transgenes with AstC neuron activation showed ∼70% fewer mature eggs in their ovaries, compared to controls (Fig. 1C), indicating that AstC-Gal4 neuron activity inhibits oogenesis.
We next evaluated the function of AstC-Gal4 neurons in ovulation and oviposition by counting the numbers of eggs in the ovaries and uterus of mated females, respectively. We reasoned that eggs should build up in the ovaries or uterus, respectively, after a disruption of ovulation or oviposition in mated females. First, we mated 3- to 4-d-old females with CS males at 21 °C and then transferred them to 31 °C for 48 h. While control females carrying either the Gal4 or UAS transgene retained 19 ± 5 mature eggs in their ovaries, those with AstC-Gal4 neuron activation retained 40 ± 5 mature eggs in their ovaries (Fig. 1D). Blocking either ovulation or oviposition seems to cause mated females to hold more mature eggs in their ovaries because AstC-Gal4 activation also reduces egg production. Ovulation, which is the transfer of an egg from the ovary to the uterus is harder to quantify in flies, but AstC-Gal4 neuron activation seems to have a limited effect on ovulation. In contrast, knockdown of tyrosine β-hydroxylase, which suppresses the synthesis of the ovulation-inducing neurotransmitter octopamine produces a profound reduction in ovulation (27) (SI Appendix, Fig. S1). Together, these results implicate AstC-Gal4 neurons specifically in oogenesis and oviposition.
AstC Neuropeptide Regulates Oogenesis, but Not Oviposition.
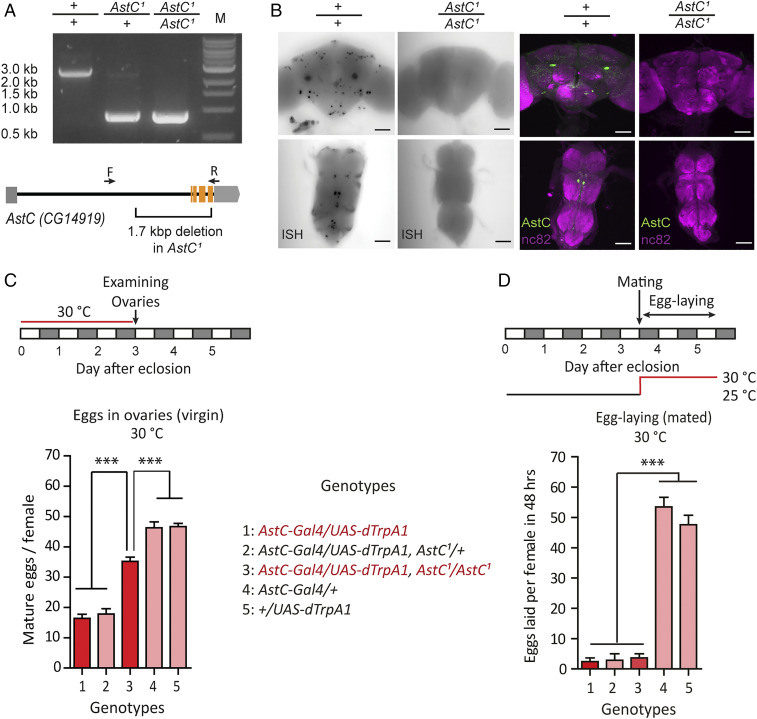
To further clarify the role of AstC in female fecundity, we generated an AstC null-mutant allele (AstC1) with a 1.7-kb deletion in the AstC gene using the CRISPR/Cas9 gene-editing system (28) (Fig. 2A). We verified via mRNA in situ hybridization and anti-AstC labeling that AstC mutants do not produce AstC transcripts or protein in the central nervous system (CNS) (Fig. 2B). We were unable to detect a significant difference in egg-laying after mating between the mutant and controls with a standard 48-h assay. This is probably because postmating fecundity is already near its maximum, particularly in young females, generating a ceiling effect. Thus, we quantified cumulative egg-laying for 6 d (SI Appendix, Fig. S2A) and found AstC-deficient females (AstC1/AstC1) laid 18% more eggs than heterozygous controls. This suggests that AstC deficiency would impact fecundity more strongly in older females. Mutant flies expressing AstC in AstC-Gal4 cells showed even lower fecundity than controls, confirming that AstC inhibits female fecundity (SI Appendix, Fig. S2A). We also found that nervous system-specific knockdown of AstC via RNAi (nSyb-Gal4/AstC-RNAi) increased female fecundity when measured 48 h after mating, confirming the importance of neuronal AstC (SI Appendix, Fig. S2B).
Fig. 2.
AstC neuropeptide modulates egg production. (A) PCR confirmation of the 1.7-kb deletion in AstC homozygote mutant female flies compared with w1118 and AstC heterozygous mutants. Gene span indicates three exons involved in the deletion area. Arrows indicate positions of primers used for PCR confirmation (Materials and Methods). (B) AstC mRNA (Left) and AstC peptide expression levels (Right) in wild-type and AstC mutants were confirmed through in situ hybridization (ISH) and anti-AstC immunostaining (green, anti-AstC; magenta, anti-nc82). The anti-nc82 visualizes Bruchpilot protein, which strongly labels neuropil. (Scale bars, 50 μm.) (C, Upper) Protocol for measuring virgin female egg production. (Lower) Number of mature eggs per virgin female (n = 40 to 60) of the indicated genotype kept for 3 d posteclosion at the indicated temperature at 30 °C. The means ± SEM of the data are shown (C and D). One-way ANOVA followed by Tukey’s test for multiple comparisons; ***P < 0.001 for comparisons between indicated genotypes or groups (C and D). (D, Upper) Protocol for measuring mated female egg laying. (Lower) Number of eggs laid per female (n = 40 to 60) of the indicated genotype for 48 h at 30 °C.
We next asked whether the AstC neuropeptide is responsible for the oogenesis inhibition induced by activation of AstC-Gal4 neurons. We found that AstC-deficiency rescued the reduced oogenesis caused by AstC-Gal4 neuron activation (Fig. 2C), confirming that the AstC gene product (i.e., the AstC neuropeptide) produced by AstC-Gal4 neurons is responsible for oogenesis inhibition. We found, however, that AstC-Gal4 neuron activation still showed a partial reduction in oogenesis even in the absence of AstC. This suggests AstC-Gal4 neurons contain at least one additional unknown oogenesis inhibitor. Finally, we found AstC-deficiency did not relieve the oviposition blockade caused by AstC-Gal4 neuron activation (Fig. 2D). Thus, it is not AstC that is responsible for the oviposition phenotype. Together, we conclude that AstC-deficient females show increased fecundity due to increased oogenesis.
AstC from a Subset of DN1p Circadian Clock Neurons Regulates Oogenesis.
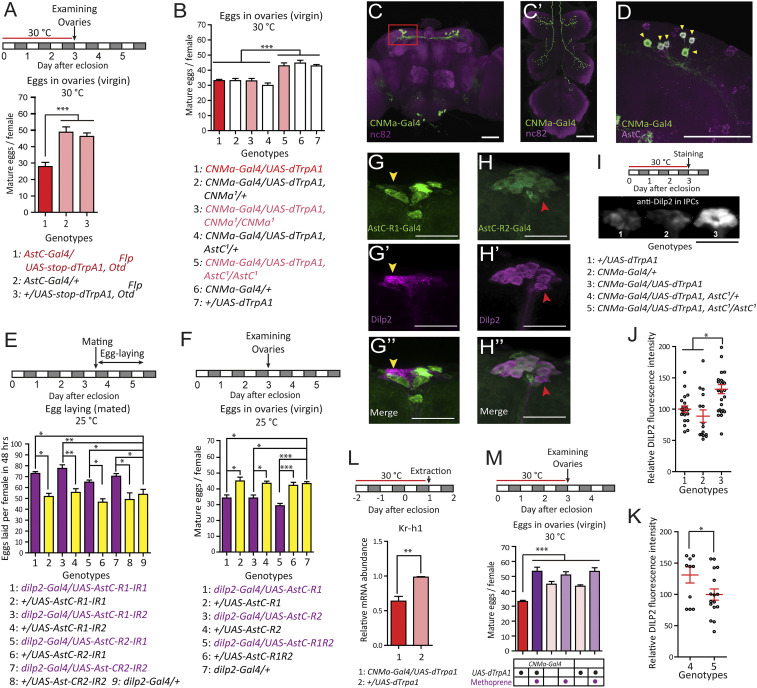
We detected AstC-Gal4 expression in ∼140 brain cells and ∼80 ventral nerve cord (VNC) cells, ∼40 cells of which were positive for anti-AstC (SI Appendix, Fig. S3). To map the AstC neurons that regulate oogenesis, we examined the function of the brain and VNC neurons separately by restricting dTrpA1 expression in brain neurons using a brain-specific OtdFLP (29). We found thermal activation of brain AstC neurons still inhibited oogenesis, but to a lesser extent (a 72% vs. 42% reduction) than activation of all AstC-Gal4 neurons (Fig. 1C vs. Fig. 3A). This suggests brain neurons explain about 58% of the oogenesis phenotype, with the rest likely being attributable to AstC neurons in the VNC. To further map the brain AstC neurons relevant for oogenesis, we reassessed the CNMa-Gal4 driver, which we had found reduced fecundity in our initial screen (Fig. 1A and SI Appendix, Fig. S4). We did this because CNMa and AstC transcripts colocalize in clock-associated DN1s (30). Thermal activation of CNMa-Gal4 neurons reduced oogenesis to a level comparable to what activation of the brain AstC-Gal4 neurons did (Fig. 3B). The somas of CNMa-Gal4 neurons occur exclusively in the brain, with six pairs in the dorsal brain and two in the subesophageal zone (SOZ) (Fig. 3C). All of the dorsal CNMa-Gal4 neurons express AstC (arrowheads in Fig. 3D), but the SOZ neurons, which project into the VNC, do not. We further characterized the brain CNMa-Gal4 neurons with various antisera and found that the six dorsal pairs also express the clock protein PERIOD (PER) and Diuretic hormone 31 (Dh31) (SI Appendix, Fig. S5 A and B). Furthermore, Clk4.1M-Gal4, which labels the DN1ps (4, 31), showed colocalization with anti-AstC (SI Appendix, Fig. S5C). Together, these observations indicate that the dorsal CNMa-Gal4 neurons are a subset of DN1ps, which are core clock neurons that regulate diurnal rhythms of rest/activity in Drosophila (24). Hereafter, we refer to this group of DN1p neurons that express AstC, CNMa, and Dh31 as AstC-DN1p. The DN1p cluster is divided into two subpopulations based on their morphological characteristics: Anterior-projecting neurons (a-DN1p) that innervate the anterior optic tubercle (AOTU) and ventro-contralateral–projecting neurons (vc-DN1p) that do not (32). Notably, AstC-DN1p showed projections to AOTU, indicating it comprises a-DN1p neurons (SI Appendix, Fig. S5D).
Fig. 3.
AstC from AstC-DN1p inhibits JH-dependent oogenesis progression via IPCs. (A) Above, protocol for measuring virgin female egg production used in A and B. (Lower) number of mature eggs per virgin female (n = 40 to 60) of the indicated genotype kept 3 d posteclosion at 30 °C. The means ± SEM of the data are shown (A, B, E, F, and J–M). One-way ANOVA followed by Tukey’s test for multiple comparisons; *P < 0.05; **P < 0.005; ***P < 0.001 for comparisons between indicated genotypes or groups (A, B, E, F, and M). (B) Number of mature eggs per virgin female (n = 30 to 40) of the indicated genotype kept 3 d posteclosion at 30 °C. (C) The brain (C) and VNC (C′) of a 4-d-old virgin female of the CNMa-Gal4/UAS-mCD8-EGFP genotype stained with the nc82 (magenta) and anti-GFP (green) antibodies. (Scale bars, 50 μm.) (D) Dorso-lateral area of the brain indicated by the red box in C from a CNMa-Gal4/UAS-mCD8-EGFP female CNS stained with anti-AstC (magenta) and anti-GFP (green) antibodies. Yellow arrowheads indicate six cells positive for both anti-AstC and anti-GFP in one hemisphere of the brain. (Scale bar, 50 μm.) (E, Upper) Behavioral protocol for measuring mated female egg laying. (Lower) Number of eggs laid per female (n = 40 to 60) of the indicated genotype for 48 h after mating at 25 °C. (F, Upper) Protocol for measuring virgin female egg production. (Lower) Number of mature eggs per virgin female (n = 40 to 60) of the indicated genotype kept 3 d posteclosion at 25 °C. (G) IPCs of a 4-d-old virgin AstC-R1-Gal4/UAS-mCD8-EGFP female stained with anti-Dilp2 (magenta) and anti-GFP (green) antibodies. (Scale bars, 50 μm.) This specific preparation has one IPC that is positive for anti-GFP (yellow arrowhead). (H) IPCs of a 4-d-old virgin AstC-R2-Gal4/UAS-mCD8-EGFP female stained with anti-Dilp2 (magenta) and anti-GFP (green) antibodies. (Scale bars, 50 μm.) Usually, 13 of 14 anti-Dilp2-positive IPCs are positive for anti-GFP. A red arrowhead indicates one IPC that lacks anti-GFP labeling. (I, Upper) Protocol for Dilp2 accumulation experiment in IPCs. (Lower) Images of IPCs from virgin females of the indicated genotype and condition stained with an anti-Dilp2 antibody. (Scale bar, 100 μm.) (J and K) Relative fluorescence intensity of anti-Dilp2 staining of IPCs from females of the indicated genotype in I, kept for 3 d posteclosion at 30 °C. Each circle indicates a relative fluorescence intensity from all IPC somas of a brain. The relative fluorescence intensity is calculated by setting the average of (genotype-1 brains in J and genotype-5 brains in K) as 100%. Unpaired t test; *P < 0.05 for comparisons between indicated genotypes. (L, Upper) Protocol for fly preparation for quantitative RT-PCR. (Lower) Kr-h1 transcript levels in 1-d-old adult females of the indicated genotypes kept for 3 d at 30 °C. Unpaired t test; **P < 0.005. (M, Upper) Protocol for measuring virgin female egg production. (Lower) Number of mature eggs per virgin female (n = 20 to 40) of the indicated genotype kept for 3 d posteclosion at 30 °C. Magenta circle indicates methoprene treatment.
Next, we asked whether AstC-DN1p acts via the AstC neuropeptide by inducing thermal activation of AstC-DN1p (i.e., CNMa-Gal4 neurons) in AstC or CNMa-deficient mutants. We found that the reduced oogenesis phenotype we observed upon CNMa-Gal4 neuron activation disappeared in AstC-deficient females, but not in CNMa-deficient females (Fig. 3B). Furthermore, restoring AstC expression specifically in AstC-DN1p blocked the fecundity increase caused by AstC-deficiency, reducing it to levels lower than the controls. Notably, these results were indistinguishable from those of rescue experiments using AstC-Gal4 (SI Appendix, Fig. S2A).
AstC from AstC-DN1p Inhibits IPC Secretory Activity.
IIS is closely linked to oogenesis (33), modulating reproduction according to nutritional state. The circadian DN1 neurons, including AstC-DN1p and others, make synaptic contacts with IPCs in the brain pars intercerebralis (34). Thus, we speculated that activation of AstC-DN1p reduces oogenesis by inhibiting the IPCs and IIS. We tested this idea by first examining the anatomical interactions between AstC-DN1p and the IPCs. AstC-DN1p processes labeled with the presynaptic marker nSyb-EGFP indeed project to the necks of the IPCs, to a point between their somas and axons (SI Appendix, Fig. S6A). This is also where DN1 neurons make physical contact with the IPCs (34). These connections likely represent functional synapses between AstC-DN1p and the IPCs, because trans-Tango, which labels functionally connected postsynaptic neurons (35), driven by the DN1p-targeting Clk4.1M-Gal4 produces strong trans-Tango labeling in the pars intercerebralis (36).
There are two GPCRs that reportedly act as AstC receptors in Drosophila: AstC-R1 (also known as star1 or CG7285) and AstC-R2 (also known as AICR2 or CG13702) (37). To examine the function of these AstC receptors in the IPCs, we performed an RNAi-based knockdown of each receptor using the IPC-specific dilp2-Gal4. As we observed in AstC-deficient mutants and in pan-neural AstC-RNAi, AstC receptor knockdown increases egg-laying in mated females (Fig. 3E). This suggests both receptors are required for normal IPC function. We also overexpressed either AstC-R1 or AstC-R2 in the IPCs. As with AstC-DN1p activation, we found that both AstC-R1 and AstC-R2 overexpression in IPCs reduced oogenesis in virgin females (Fig. 3F). Our RNAi results indicate that the IPCs express both AstC receptors. For further confirmation of this result, we examined AstC-R1-Gal4 and AstC-R2-Gal4, each of which carries an extra exon from the TA-Gal4 transgene in its respective AstC receptor locus (38). As expected, the IPCs were labeled by both receptor-Gal4 lines (Fig. 3 G and H). We found AstC-R2-Gal4 labeled 13 of 14 IPCs (n = 4), whereas AstC-R1-Gal4 labeled a more variable number of IPCs (one brain showed 14 of 14 AstC-R1-Gal4+ IPCs, two brains had only one AstC-R1-Gal4+ IPC, and one brain had no AstC-R1-Gal4+ IPCs). This variability in AstC-R1-Gal4 labeling may reflect transient AstC-R1 expression in IPCs. Nevertheless, our evidence suggests AstC neuropeptide from AstC-DN1p acts on IPCs to modulate female reproductive activity.
Having shown that AstC from AstC-DN1p reduces fecundity through the IPCs, we predicted that AstC inhibits insulin secretion from the IPCs. We examined the effect of AstC-DN1p activation on insulin levels in IPCs using anti-Dilp2 staining. Three days of constant AstC-DN1p activation posteclosion induced a marked accumulation of Dilp2 in the IPCs (Fig. 3 I and J). We found AstC-deficiency, on the other hand, led to reduced Dilp2 accumulation in the IPCs (Fig. 3K). These results are consistent with the hypothesis that AstC from AstC-DN1p is a negative regulator of IPC function.
AstC Inhibits JH Biosynthesis Likely via IPCs.
IIS induces oocyte development indirectly by stimulating JH biosynthesis and secretion. In many insects, JH stimulates vitellogenesis in which the yolk proteins are produced by the fat body and sequestered by developing oocytes (15). Thus, we reasoned that inhibition of IPC activity by AstC-DN1p activation would reduce JH biosynthesis leading to reduced vitellogenesis. First, we evaluated JH activity by quantifying mRNA levels of Krüppel homolog 1 (Kr-h1), the transcription of which is directly induced by JHs (39, 40). We asked whether Kr-h1 transcription reflects JH levels by examining three different developmental stages: Pupa S8 (2 d before eclosion), pupa S15 (1 d before eclosion), and adults 1 d after eclosion. According to previous reports, JHs are almost absent at pupa S8, increasing before pupa S15, and peaking 1 d posteclosion (41). Mirroring JH levels, Kr-h1 transcription is lowest at pupa S8, rising to intermediate levels at pupa S15, and peaking in 1-d-old adults (SI Appendix, Fig. S7). Next, we activated AstC-DN1p beginning at pupa S8 for 3 d and then examined Kr-h1 mRNA levels in 1-d-old females (Fig. 3L). We found AstC-DN1p activation reduced Kr-h1 transcription by ∼25%, indicating that AstC-DN1p can alter JH levels. Finally, we asked whether the JH mimic methoprene can rescue the reduction in oogenesis caused by AstC-DN1p activation (Fig. 3M). Indeed, we found methoprene treatment restored oogenesis to control levels. These results suggest AstC-DN1p regulates oocyte development by controlling JH biosynthesis.
AstC-DN1p Generates Circadian Vitellogenesis Rhythm via IPCs.
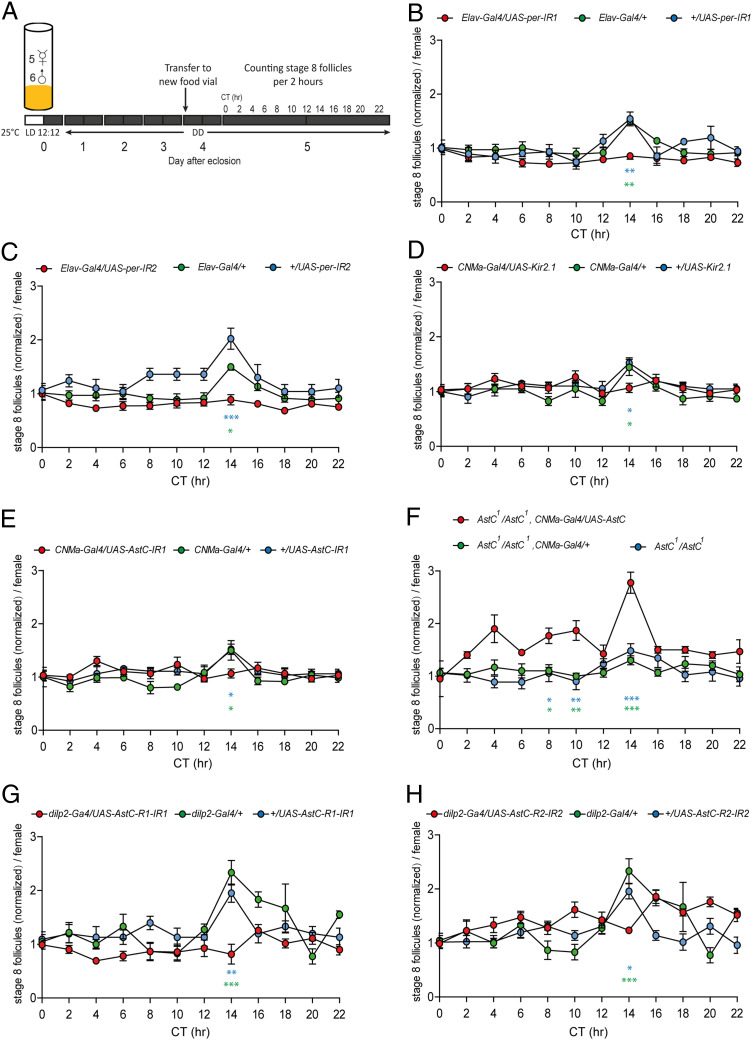
The finding that AstC-DN1p, a subset of the circadian pacemaker neuron network, regulates oogenesis, raised the intriguing hypothesis that it may coordinate oogenesis progression according to the endogenous biological rhythm. Mated females show circadian rhythmicity in oogenesis progression under a photoperiodic condition (light:dark, LD 12:12) (42) and after a transition to constant darkness (DD) (7). But it remains unclear how and whether the endogenous clock generates this oogenesis rhythm. Because oviposition has a strong dependency on light cues and may potentially stimulate oogenesis (42), we studied the oogenesis rhythm under a DD condition. First, we examined the control strain w1118. Virgin females were cultured under a LD 12:12 cycle and transferred to DD together with naïve CS males 1 d after eclosion (SI Appendix, Fig. S8A). Over 5 d in DD (i.e., DD1, DD2, DD3, and DD5), the ovaries from individual females were examined every 2 h. We counted the number of stage 8 follicles in the ovaries because vitellogenesis initiation (i.e., the developmental transition from stage 7 to stage 8 follicles) is an important oogenesis control point that requires JH functions like the stimulation of yolk protein synthesis and uptake by the ovary (43, 44). For the first day under DD (DD1), the ovaries showed more stage 8 follicles and a higher baseline than those in following days (SI Appendix, Fig. S8B). To simplify comparisons between days, we normalized the data to the value at CT0 for each day. We observed circadian changes in the number of stage 8 follicles with an acute peak at CT14 on DD2, DD3, and DD5, but not on DD1 (SI Appendix, Fig. S8C). Notably, we did not see any quantitative or qualitative difference among the DD2, DD3, and DD5 rhythms. The higher baseline rate of vitellogenesis in DD1 suggests a stronger reproductive drive early in pregnancy, which would produce a ceiling effect and mask the circadian vitellogenesis rhythm. Using a cosinor analysis, we detected a rhythm with a 24 h-period in DD2, DD3, and DD5, but not in DD1. We failed to detect a rhythm in several other control groups (SI Appendix, Table S1). Nevertheless, a circadian vitellogenesis rhythm clearly exists, because w1118 and all tested controls showed a clear peak at CT14 that is absent in the test groups (i.e., AstC- or AstC-R1-RNAis). It seems the cosinor analysis simply has difficulty detecting the vitellogenesis rhythm because it shows such strong phase synchronization, producing only a single sharp peak within a single 2-h time bin.
To determine whether the vitellogenesis rhythm is under the control of the biological clock, we knocked down the expression of PER, a core molecular clock protein in the nervous system. PER-immunostaining confirmed that two independent pan-neuronal per-RNAi lines showed almost no anti-PER labeling in the brains examined at ZT23, when the control groups showed robust anti-PER labeling (SI Appendix, Fig. S9). Remarkably, the CT14 peak was completely absent in both per-RNAi lines, highlighting a clear link between the biological clock and the vitellogenesis rhythm. Next, we examined the role of the AstC-DN1p clock neurons in the vitellogenesis rhythm. As with the controls mentioned above, CNM-Gal4 or UAS-Kir2.1 (i.e., inward-rectifying K+ channel that silences neurons) alone showed the peak at CT14 in the number of stage 8 follicles. When these two lines were combined (i.e., CNMa-Gal4/UAS-Kir2.1) to induce silencing of the AstC-DN1p neurons, however, the peak was absent. Similarly, AstC-DN1p–specific expression of AstC-RNAi also suppressed the CT14 peak (Fig. 4C). Conversely, the restoration of AstC expression in AstC-DN1p rescued the circadian vitellogenesis rhythm in AstC-deficient mutant females (AstC1/1) by restoring the CT14 peak (Fig. 4D).
Fig. 4.
AstC signaling from AstC-DN1p to IPCs generates the circadian vitellogenesis rhythm. (A) Protocol for measuring circadian vitellogenesis rhythm in B–H. Groups of five virgin females of the indicated genotypes and six CS males cultured under a 12:12 LD cycle were placed in food vials upon eclosion and then transferred to constant darkness on day 2. On day 6, the ovaries were dissected every 2 h and stage 8 follicles were counted. (B–E) The normalized number of stage 8 follicles from the indicated genotypes at the indicated circadian time. For normalization, the number of follicles at each circadian time point was divided by the number of follicles at CT0. The means ± SEM of the data are shown. In each circle, n = 6 to 10. One-way ANOVA followed by Tukey’s test for multiple comparisons among genotypes at each time point; ***P < 0.001; **P < 0.005; *P < 0.05; no labeling, P > 0.05. The symbol for P value is color-coded according to the control group used for comparison. UAS-RNAi alone controls in B, C, E, G, and H, all genotypes in D, and AstC1/AstC1, CNMa-GAL4/+ in F were from a separate experimental cohort. For a better comparison between genotypes, Elav-GAL4 and dilp2-Gal4 control data are presented twice in B and C and G and H, respectively.
Since our evidence suggests AstC-DN1p supplies an inhibitory input to the IPCs, attenuating IIS, we asked whether the vitellogenesis rhythm requires IPC-specific function of the AstC receptors. We did this by performing an IPC-specific knockdown of either AstC-R1 or -R2 (Fig. 4 G and H). As before, we found the number of stage 8 follicles in control ovaries peaked at CT14. Similar to AstC-RNAi and the AstC mutant, RNAi-mediated knockdown of either AstC receptor in the IPCs dampened the circadian vitellogenesis rhythm by suppressing the peak at CT14. From these results, we conclude that the AstC signaling axis from AstC-DN1p to the IPCs is critical for the circadian vitellogenesis rhythm.
Discussion
In this study, we discovered that six pairs of DN1p neurons that are part of the circadian pacemaker neuron network in the brain make functional inhibitory connections to the brain IPCs. The IPCs are endocrine sensors that link the organism’s nutritional status with anabolic processes, such as those associated with growth in developmental stages and with reproduction in adults (45). In juvenile stages, activation of IIS through the InR results in larger flies, whereas inhibition of this pathway produces smaller flies (46). Consistent with this, we also found that forced activation of the AstC-DN1p (i.e., CNMa-Gal4/UAS-NaChBac) during development resulted in 12% smaller adults, confirming their role as a negative regulator of the IPCs (SI Appendix, Fig. S6 B and C). In adults, the IPCs are associated with many physiological and behavioral processes, such as feeding (47), glycaemic homeostasis (48), sleep (49), lifespan, and stress resistance (50). As such, the IPCs receive a variety of modulatory inputs from both central and peripheral sources, such as sNPF, corazonin, tachykinins, limostatin, allatostatin A, adipokinetic hormone, GABA, serotonin, and octopamine (51, 52). Regarding reproduction, IIS directed by the IPCs stimulates GSC proliferation and vitellogenesis. Our results also indicate that AstC from AstC-DN1p suppresses the secretory activity of the IPCs and JH-dependent oocyte development (i.e., vitellogenesis). Indeed, we found that the JH mimic methoprene can rescue the suppression of oogenesis induced by AstC-DN1p activation. From these results we conclude that IPCs are inhibited by AstC released by AstC-DN1p. A similar link between IIS and the circadian clock has also been reported in mammals (53–55), but the mechanism remains unclear (34).
Although our genetic evidence supporting the inhibitory action of AstC-DN1p on IPCs is compelling, it is also puzzling because a previous study found forced activation of 8 to 10 pairs of DN1p neurons (i.e., Clk4.1-LexA+ neurons) induced Ca2+ transients in IPCs. This study also found that, under LD 12:12 conditions, the IPCs showed electrical activity early in the morning when DN1p neurons are also active (34). The same study, however, reported that, under DD conditions, the IPCs showed no bursting activity in the morning (i.e., CT0 to -4). Instead, they showed bursting activity in the late afternoon (i.e., CT8 to -12) when DN1 activity falls (see below). Furthermore, DN1p activation evokes varying levels of Ca2+ transients from individual IPCs, some of which produce no detectable Ca2+ transient. Thus, like mammalian pancreatic β-cells, the IPCs in Drosophila seem to comprise a heterogeneous cell population (56). We noted that individual IPCs show highly variable AstC-R1 expression, which would also lead to individual IPCs showing variable responses to AstC.
In D. melanogaster, the LD cycle generates an egg-laying rhythm by influencing oogenesis and oviposition. Oviposition depends on light cues, whereas oogenesis cycles with the circadian rhythm that itself continues to run in DD conditions (7). In live-brain Ca2+ imaging experiments, DN1 neurons show a circadian Ca2+ activity rhythm that peaks around CT19 and reaches its lowest point between CT6 and CT8 (57). This DN1 activity rhythm correlates well with the rhythm of vitellogenesis initiation we observed in this study. In our model, the lowest point in DN1 Ca2+ activity between CT6 and CT8 leads to a significant attenuation of AstC secretion. This leads to a derepression of IPC activity, which eventually induces JH biosynthesis and vitellogenesis initiation. The 6-h delay required for previtellogenic stage 7 follicles to develop into vitellogenic stage 8 follicles (58) would result in a peak in the number of stage 8 follicles between CT12 and CT14. Notably, the ovaries of the AstC-deficient mutant showed similar numbers of stage 8 oocytes at all examined circadian time points, indicating that any other JH- or vitellogenesis-regulating factors play only minor roles in producing the circadian vitellogenesis rhythm.
Like the IPCs, the DN1p cluster is also heterogeneous. A subset of the DN1p neurons is most active at dawn and promotes wakefulness (59). Another subset of the DN1p cluster (also known as, spl-gDN1) promotes sleep (36, 60). The DN1p cluster comprises two morphologically distinct subpopulations, a-DN1p and vc-DN1p (32). The a-DN1p subcluster promotes wakefulness by inhibiting sleep promoting neurons, whereas the vc-DN1p subcluster resembles the sleep-promoting spl-gDN1. Our results indicate AstC-DN1p are a-DN1p neurons that project to the AOTU. Although we could not rule out the possibility that AstC-DN1p is also heterogeneous and includes some vc-DN1p neurons, the wake-promoting role of a-DN1p aligns well with the circadian vitellogenesis rhythm that requires the secretory activity of AstC-DN1p to be lowest in the afternoon and highest at dawn. Furthermore, AstC-DN1p neurons express Dh31. Dh31-expressing DN1 clock neurons are intrinsically wake-promoting and Dh31-DN1p activity in the late night or early morning suppresses sleep (61). Again, this is consistent with our observation that AstC-DN1p are also wake-promoting a-DN1p. We speculate Dh31 plays a limited role in oogenesis regulation, because unlike AstC, RNAi-mediated knockdown of Dh31 had a negligible impact on female fecundity (SI Appendix, Fig. S2C).
Besides AstC-DN1p, the female brain has many additional AstC neurons. However, it seems unlikely that other AstC neurons contribute to the circadian vitellogenesis rhythm. This is because restoring AstC expression specifically in AstC-DN1p almost completely restored the vitellogenesis rhythm in AstC-deficient mutants. It is feasible, however, that other AstC neurons contribute to different aspects of female reproduction. Indeed, we noted a sizable difference in the final oogenesis outcome between AstC-Gal4 neuron activation and brain-specific AstC-Gal4 neuron activation. This suggests AstC cells outside of the brain also regulate oogenesis probably in other physiological contexts, such as the postmating responses.
AstC receptors are orthologous to mammalian SST receptors (sstr1-5). SST is a brain neuropeptide that was originally identified as an inhibitor of growth hormone (GH) secretion in the anterior pituitary (62). Thus, our observation that AstC inhibits IIS from IPCs, a major endocrine signal that promotes growth in Drosophila, suggests remarkable structural and functional conservation between the invertebrate AstC and vertebrate SST systems. In addition, SST inhibits the hypothalamic neuropeptide GnRH, which stimulates the anterior pituitary’s production of follicle-stimulating hormone (FSH) and luteinizing hormone (LH) (63). FSH stimulates clutches of immature follicles to initiate follicular development, while LH stimulates ovulation. Thus, both AstC and SST regulate the secretion of gonadotropins (JH in insects, FSH and LH in mammals) indirectly through the IPCs in insects and through the hypothalamic GnRH neurons in mammals. This functional conservation between AstC and SST is also evident in the immune system. AstC inhibits the innate immune system in insects, while SST inhibits inflammation in mammals (64).
In many seasonal breeders, the changing photoperiod as the seasons progress acts as an environmental cue for the biological clock system, which would then direct any necessary physiological changes (65, 66). During the winter, Drosophila females enter a form of reproductive dormancy characterized by a pronounced suppression of vitellogenesis (67). A winter-like condition (i.e., short-day length, low temperature, and food shortage) down-regulates neural activity in the IPCs (68). But the IPCs are not equipped with a cell-autonomous clock, so they must receive seasonal information from the brain clock neuron network. Indeed, two clock related neuropeptides—pigment dispersing factor and short neuropeptide F—from circadian morning pacemaker or M-cells have been implicated in regulating reproductive dormancy (68, 69). Intriguingly, AstC-DN1p neurons are the DN1p subset that receives pigment dispersing factor signals from these M-cells. Furthermore, DN1p can process light and temperature information for the circadian regulation of behavior (31, 70). Finally, our finding that AstC-DN1p generates the circadian vitellogenesis rhythm via the IPCs makes AstC-DN1p neurons the prime candidates for integrating the seasonal cues that control the entrance, maintenance, or exit from reproductive dormancy. Considering the functional and structural conservation between the AstC and SST systems, the SST system may also link the brain clock, GnRH, and/or its downstream reproductive pathways in controlling seasonal reproductive patterns in vertebrates.
Materials and Methods
Fly Stocks.
The following stocks were reported previously or obtained from the Bloomington Drosophila Stock Center (BDSC), the Vienna Drosophila RNAi Center (VDRC), or the Korean Drosophila Resource Center (KDRC): AstC-R1-Gal4 (38), AstC-R2-Gal4 (38), AstC-IR1 (VDRC #13772), AstC-IR2 (VDRC #102735), Dh31-IR1 (VDRC #50293), Dh31-IR2 (VDRC #37763), Dh31-IR3 (VDRC #37764), per-IR1 (BDSC #40878), per-IR2 (BDSC #31285), AstC-R1-IR1 (VDRC #13560), AstC-R1-IR2 (BDSC #JF02656), AstC-R2-IR1 (BDSC #JF01960), AstC-R2-IR2 (VDRC #50000), and AstC-GAL4 (KDRC #10012). Gal4 stocks used for RNAi carried UAS-Dicer2 (VDRC #60007). The complete list of fly stocks used in this study can be found in SI Appendix, Supplementary Materials and Methods. We generated CNMa-Gal4 by inserting the Gal4 ORF into the CNMa ATG start site in a recombineering-ready, premapped genomic BAC clone (Clone ID, CH321-34F01) from the P[acman] library (71). The CNMa-Gal4 construct was inserted into a specific site on the third chromosome (VIE-49b, a gift from Barry J. Dickson, Janelia Research Campus, Ashburn, VA). UAS-AstC was generated by cloning the NotI-AstC-ORF-KpnI fragment amplified from the cDNA clone RH36507 (AY070699) into the SST13 vector, and then inserting it into a specific site on the third chromosome (VIE-49b). The AstC and CNMa mutant alleles were generated via the CRISPR/Cas9 system. All primers or oligonucleotides used in this study are listed in SI Appendix, Table S2.
Bioassays.
Flies were raised on normal fly food at 25 °C in a fly room under a 12-h:12-h, light:dark cycle. For the egg-laying assay, we modified a published protocol (72). For the egg-production assay, freshly eclosed females were placed individually in vials for 3 d. Then, their ovaries were dissected in phosphate buffered saline (PBS) and the mature eggs (stage 14) in both ovaries were counted under a stereomicroscope. To examine the circadian vitellogenesis rhythm, we modified methods described by Allemand (42). The ovarioles were separated and the stages of the vitellogenic follicles were determined according to the method used by Jia et al. (58). For the oviposition assay, the reproductive organs of postmating females were dissected and the mature eggs (stage 14) from both ovaries were counted. For methoprene treatment, flies were fed food with 1.04 μL/mL methoprene (Sigma-Aldrich, catalog number 40596-69-8) or 1 μL/mL 95% ethanol (vehicle control) individually for 3 d after eclosion. All assays were repeated on at least 2 different days.
Immunohistochemistry.
Antibodies used included rabbit anti-GFP antibody (1:1,000; Invitrogen, A11122), rabbit anti-AstC antibody (73), anti-Dh31 (74) (a gift from Jan Veenstra from Université de Bordeaux, France), anti-PER (75) (a gift from Eun Young Kim, Ajou University School of Medicine, Gyeonggi-do, Republic of Korea), anti-Dilp2 (76) (a gift from Yu Kweon from Korea Research Institute of Bioscience and Biotechnology, Daejon, South Korea), mouse anti-nc82 antibody (1:50; Developmental Studies Hybridoma Bank), Alexa Flour 488-labeled goat anti-rabbit IgG (1:1,000; Invitrogen, A11008), and Alexa Fluor 568 goat anti-mouse IgG (1:1,000; Invitrogen, A11004). To quantify anti-Dilp2 fluorescence, maximum intensity z-projections of 15 consecutive confocal stacks (5-μm-thick each) covering all somas of 14 IPCs were merged with ImageJ. Then, the relative anti-Dilp2 fluorescence intensity of each brain was calculated by setting the average of control brains (UAS-dTrpA1/+) (for Fig. 3J) or CNMa-Gal4/UAS-dTrpA1, AstC1/AstC1 brains (for Fig. 3K) as 100%.
Quantitative RT-PCR.
Total RNA was extracted from whole adult female bodies (n = 15) using TRIzol (Takara) according to the manufacturer’s instructions. RNA (1 μg) was reverse transcribed with oligo(dT) primers (Promega) and Accupower RT premix (Bioneer). Quantitative RT-PCR reactions were performed using an IQTM5 real-time PCR detection system (Bio-Rad) with SYBR Premix ExTaq (Takara) according to the manufacturer’s instructions. The gene-specific primers for Rp49 and Kr-h1 described previously (77) were used (SI Appendix, Table S2).
Supplementary Material
Acknowledgments
We thank S.-I. Kang, H.-S. Yoon, J. Mun, and J. Song for excellent technical assistance; B. J. Dickson (Janelia Research Campus), J. Veenstra (Université de Bordeaux), Y. Kwon (Korea Research Institute of Bioscience and Biotechnology), and E. Y. Kim (Ajou University School of Medicine) for reagents; and C. Lim (Ulsan National Institute of Science and Technology) for a cosinor analysis. This work was supported by the Gwangju Institute of Science and Technology (GIST) Research Institute grants funded by the GIST in 2020; Basic Science Research Programs (NRF-2015K2A1B8046794, NRF-2018R1A2A1A05079359, NRF-2019R1A4A1029724) through the National Research Foundation of Korea (NRF) funded by Ministry of Science and Information and Communication Technology, Republic of Korea; and by Slovak Research and Development Agency (Grants APVV-16-0395 and APVV-18-0201) and Vedecká grantová agentúra MŠVVaŠ SR a SAV (Grant VEGA-2/0080/18). Stocks were obtained from the Korea Drosophila Resource Center (NRF-2017M3A9B8069650), the Bloomington Drosophila Stock Center and the Vienna Drosophila Resource Center.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2016878118/-/DCSupplemental.
Data Availability.
All study data are included in the article and supporting information.
References
- 1.Boden M. J., Kennaway D. J., Circadian rhythms and reproduction. Reproduction 132, 379–392 (2006). [DOI] [PubMed] [Google Scholar]
- 2.Kaneko M., Hall J. C., Neuroanatomy of cells expressing clock genes in Drosophila: Transgenic manipulation of the period and timeless genes to mark the perikarya of circadian pacemaker neurons and their projections. J. Comp. Neurol. 422, 66–94 (2000). [DOI] [PubMed] [Google Scholar]
- 3.Helfrich-Förster C., The neuroarchitecture of the circadian clock in the brain of Drosophila melanogaster. Microsc. Res. Tech. 62, 94–102 (2003). [DOI] [PubMed] [Google Scholar]
- 4.Shafer O. T., Helfrich-Förster C., Renn S. C., Taghert P. H., Reevaluation of Drosophila melanogaster’s neuronal circadian pacemakers reveals new neuronal classes. J. Comp. Neurol. 498, 180–193 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beaver L. M., et al. , Loss of circadian clock function decreases reproductive fitness in males of Drosophila melanogaster. Proc. Natl. Acad. Sci. U.S.A. 99, 2134–2139 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sakai T., Ishida N., Circadian rhythms of female mating activity governed by clock genes in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 98, 9221–9225 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Allemand R., Influence de modifications des conditions lumineuses sur les rythmes circadiens de vitellogenese et d’ovulation chez Drosophila melanogaster. J. Insect Physiol. 22, 1075–1080 (1976). [DOI] [PubMed] [Google Scholar]
- 8.LaFever L., Drummond-Barbosa D., Direct control of germline stem cell division and cyst growth by neural insulin in Drosophila. Science 309, 1071–1073 (2005). [DOI] [PubMed] [Google Scholar]
- 9.Tatar M., Yin C., Slow aging during insect reproductive diapause: Why butterflies, grasshoppers and flies are like worms. Exp. Gerontol. 36, 723–738 (2001). [DOI] [PubMed] [Google Scholar]
- 10.Tu M. P., Yin C. M., Tatar M., Impaired ovarian ecdysone synthesis of Drosophila melanogaster insulin receptor mutants. Aging Cell 1, 158–160 (2002). [DOI] [PubMed] [Google Scholar]
- 11.Tu M.-P., Yin C.-M., Tatar M., Mutations in insulin signaling pathway alter juvenile hormone synthesis in Drosophila melanogaster. Gen. Comp. Endocrinol. 142, 347–356 (2005). [DOI] [PubMed] [Google Scholar]
- 12.Ameku T., Niwa R., Mating-induced increase in germline stem cells via the neuroendocrine system in female Drosophila. PLoS Genet. 12, e1006123 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ables E. T., Drummond-Barbosa D., The steroid hormone ecdysone functions with intrinsic chromatin remodeling factors to control female germline stem cells in Drosophila. Cell Stem Cell 7, 581–592 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morris L. X., Spradling A. C., Steroid signaling within Drosophila ovarian epithelial cells sex-specifically modulates early germ cell development and meiotic entry. PLoS One 7, e46109 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Swevers L., Raikhel A. S., Sappington T. W., Shirk P., Iatrou K., “Vitellogenesis and post-vitellogenic maturation of the insect ovarian follicle” in Comprehensive Molecular Insect Science, Gilbert L. I., Gill S., Iatrou K., Eds. (Elsevier, 2005), vol. 1, pp. 87–155. [Google Scholar]
- 16.Gruntenko N. E., et al. , Altered juvenile hormone metabolism, reproduction and stress response in Drosophila adults with genetic ablation of the corpus allatum cells. Insect Biochem. Mol. Biol. 40, 891–897 (2010). [DOI] [PubMed] [Google Scholar]
- 17.Meiselman M., et al. , Endocrine network essential for reproductive success in Drosophila melanogaster. Proc. Natl. Acad. Sci. U.S.A. 114, E3849–E3858 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kramer S. J., et al. , Identification of an allatostatin from the tobacco hornworm Manduca sexta. Proc. Natl. Acad. Sci. U.S.A. 88, 9458–9462 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jansons I. S., Cusson M., McNeil J. N., Tobe S. S., Bendena W. G., Molecular characterization of a cDNA from Pseudaletia unipuncta encoding the Manduca sexta allatostatin peptide (Mas-AST). Insect Biochem. Mol. Biol. 26, 767–773 (1996). [DOI] [PubMed] [Google Scholar]
- 20.Williamson M., Lenz C., Winther Å. M. E., Nässel D. R., Grimmelikhuijzen C. J. P., Molecular cloning, genomic organization, and expression of a C-type (Manduca sexta-type) allatostatin preprohormone from Drosophila melanogaster. Biochem. Biophys. Res. Commun. 282, 124–130 (2001). [DOI] [PubMed] [Google Scholar]
- 21.Žitňan D., Šauman I., Sehnal F., Peptidergic innervation and endocrine cells of insect midgut. Arch. Insect Biochem. Physiol. 22, 113–132 (1993). [Google Scholar]
- 22.Wang C., Zhang J., Tobe S. S., Bendena W. G., Defining the contribution of select neuropeptides and their receptors in regulating sesquiterpenoid biosynthesis by Drosophila melanogaster ring gland/corpus allatum through RNAi analysis. Gen. Comp. Endocrinol. 176, 347–353 (2012). [DOI] [PubMed] [Google Scholar]
- 23.Bendena W. G., Zhang J., Burtenshaw S. M., Tobe S. S., Evidence for differential biosynthesis of juvenile hormone (and related) sesquiterpenoids in Drosophila melanogaster. Gen. Comp. Endocrinol. 172, 56–61 (2011). [DOI] [PubMed] [Google Scholar]
- 24.Díaz M. M., Schlichting M., Abruzzi K. C., Long X., Rosbash M., Allatostatin-C/AstC-R2 is a novel pathway to modulate the circadian activity pattern in Drosophila. Curr. Biol. 29, 13–22.e3 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hamada F. N., et al. , An internal thermal sensor controlling temperature preference in Drosophila. Nature 454, 217–220 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jang Y. H., Chae H. S., Kim Y. J., Female-specific myoinhibitory peptide neurons regulate mating receptivity in Drosophila melanogaster. Nat. Commun. 8, 1630 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Monastirioti M., Distinct octopamine cell population residing in the CNS abdominal ganglion controls ovulation in Drosophila melanogaster. Dev. Biol. 264, 38–49 (2003). [DOI] [PubMed] [Google Scholar]
- 28.Barrangou R., et al. , CRISPR provides acquired resistance against viruses in prokaryotes. Science 315, 1709–1712 (2007). [DOI] [PubMed] [Google Scholar]
- 29.Asahina K., et al. , Tachykinin-expressing neurons control male-specific aggressive arousal in Drosophila. Cell 156, 221–235 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Abruzzi K. C., et al. , RNA-seq analysis of Drosophila clock and non-clock neurons reveals neuron-specific cycling and novel candidate neuropeptides. PLoS Genet. 13, e1006613 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang L., et al. , DN1(p) circadian neurons coordinate acute light and PDF inputs to produce robust daily behavior in Drosophila. Curr. Biol. 20, 591–599 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lamaze A., Krätschmer P., Chen K. F., Lowe S., Jepson J. E. C., A wake-promoting circadian output circuit in Drosophila. Curr. Biol. 28, 3098–3105.e3 (2018). [DOI] [PubMed] [Google Scholar]
- 33.Ikeya T., Galic M., Belawat P., Nairz K., Hafen E., Nutrient-dependent expression of insulin-like peptides from neuroendocrine cells in the CNS contributes to growth regulation in Drosophila. Curr. Biol. 12, 1293–1300 (2002). [DOI] [PubMed] [Google Scholar]
- 34.Barber A. F., Erion R., Holmes T. C., Sehgal A., Circadian and feeding cues integrate to drive rhythms of physiology in Drosophila insulin-producing cells. Genes Dev. 30, 2596–2606 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Talay M., et al. , Transsynaptic mapping of second-order taste neurons in flies by trans-Tango. Neuron 96, 783–795.e4 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guo F., Holla M., Díaz M. M., Rosbash M., A circadian output circuit controls sleep-wake arousal in Drosophila. Neuron 100, 624–635.e4 (2018). [DOI] [PubMed] [Google Scholar]
- 37.Kreienkamp H. J., et al. , Functional annotation of two orphan G-protein-coupled receptors, Drostar1 and -2, from Drosophila melanogaster and their ligands by reverse pharmacology. J. Biol. Chem. 277, 39937–39943 (2002). [DOI] [PubMed] [Google Scholar]
- 38.Kondo S., et al. , Neurochemical organization of the Drosophila brain visualized by endogenously tagged neurotransmitter receptors. Cell Rep. 30, 284–297.e5 (2020). [DOI] [PubMed] [Google Scholar]
- 39.Minakuchi C., Zhou X., Riddiford L. M., Krüppel homolog 1 (Kr-h1) mediates juvenile hormone action during metamorphosis of Drosophila melanogaster. Mech. Dev. 125, 91–105 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kayukawa T., et al. , Transcriptional regulation of juvenile hormone-mediated induction of Krüppel homolog 1, a repressor of insect metamorphosis. Proc. Natl. Acad. Sci. U.S.A. 109, 11729–11734 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bownes M., Rembold H., The titre of juvenile hormone during the pupal and adult stages of the life cycle of Drosophila melanogaster. Eur. J. Biochem. 164, 709–712 (1987). [DOI] [PubMed] [Google Scholar]
- 42.Allemand R., [Rhythm of vitellogenesis and ovulation in photoperiod ld 12:12 of drosophila melanogaster]. J. Insect Physiol. 22, 1031–1035 (1976). In French. [DOI] [PubMed] [Google Scholar]
- 43.Postlethwait J. H., Handler A. M., The roles of juvenile hormone and 20-hydroxy-ecdysone during vitellogenesis in isolated abdomens of Drosophila melanogaster. J. Insect Physiol. 25, 455–460 (1979). [Google Scholar]
- 44.Jowett T., Postlethwait J. H., The Regulation of yolk polypeptide synthesis in Drosophila ovaries and fat body by 20-hydroxyecdysone and a juvenile hormone analog. Dev. Biol. 80, 225–234 (1980). [DOI] [PubMed] [Google Scholar]
- 45.Nässel D. R., Insulin-producing cells and their regulation in physiology and behavior of Drosophila 1. Can. J. Zool. 90, 476–488 (2012). [Google Scholar]
- 46.Demontis F., Perrimon N., Integration of Insulin receptor/Foxo signaling and dMyc activity during muscle growth regulates body size in Drosophila. Development 136, 983–993 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cognigni P., Bailey A. P., Miguel-Aliaga I., Enteric neurons and systemic signals couple nutritional and reproductive status with intestinal homeostasis. Cell Metab. 13, 92–104 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Haselton A., et al. , Partial ablation of adult Drosophila insulin-producing neurons modulates glucose homeostasis and extends life span without insulin resistance. Cell Cycle 9, 3063–3071 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Crocker A., Shahidullah M., Levitan I. B., Sehgal A., Identification of a neural circuit that underlies the effects of octopamine on sleep:wake behavior. Neuron 65, 670–681 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Broughton S. J., et al. , Longer lifespan, altered metabolism, and stress resistance in Drosophila from ablation of cells making insulin-like ligands. Proc. Natl. Acad. Sci. U.S.A. 102, 3105–3110 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nässel D. R., Kubrak O. I., Liu Y., Luo J., Lushchak O. V., Factors that regulate insulin producing cells and their output in Drosophila. Front. Physiol. 4, 252 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nässel D. R., Vanden Broeck J., Insulin/IGF signaling in Drosophila and other insects: Factors that regulate production, release and post-release action of the insulin-like peptides. Cell. Mol. Life Sci. 73, 271–290 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Van Cauter E., et al. , Modulation of glucose regulation and insulin secretion by circadian rhythmicity and sleep. J. Clin. Invest. 88, 934–942 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Peschke E., Peschke D., Evidence for a circadian rhythm of insulin release from perifused rat pancreatic islets. Diabetologia 41, 1085–1092 (1998). [DOI] [PubMed] [Google Scholar]
- 55.Boden G., Ruiz J., Urbain J. L., Chen X., Evidence for a circadian rhythm of insulin secretion. Am. J. Physiol. 271, E246–E252 (1996). [DOI] [PubMed] [Google Scholar]
- 56.Bonner-Weir S., Aguayo-Mazzucato C., Physiology: Pancreatic β-cell heterogeneity revisited. Nature 535, 365–366 (2016). [DOI] [PubMed] [Google Scholar]
- 57.Liang X., Holy T. E., Taghert P. H., Synchronous Drosophila circadian pacemakers display nonsynchronous Ca2+ rhythms in vivo. Science 351, 976–981 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jia D., Xu Q., Xie Q., Mio W., Deng W. M., Automatic stage identification of Drosophila egg chamber based on DAPI images. Sci. Rep. 6, 18850 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kust N., et al. , Functional analysis of Drosophila HSP70 promoter with different HSE numbers in human cells. PLoS One 9, e101994 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Guo F., Chen X., Rosbash M., Temporal calcium profiling of specific circadian neurons in freely moving flies. Proc. Natl. Acad. Sci. U.S.A. 114, E8780–E8787 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kunst M., et al. , Calcitonin gene-related peptide neurons mediate sleep-specific circadian output in Drosophila. Curr. Biol. 24, 2652–2664 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Brazeau P., et al. , Hypothalamic polypeptide that inhibits the secretion of immunoreactive pituitary growth hormone. Science 179, 77–79 (1973). [DOI] [PubMed] [Google Scholar]
- 63.Bhattarai J. P., et al. , Somatostatin inhibition of gonadotropin-releasing hormone neurons in female and male mice. Endocrinology 151, 3258–3266 (2010). [DOI] [PubMed] [Google Scholar]
- 64.Pintér E., Helyes Z., Szolcsányi J., Inhibitory effect of somatostatin on inflammation and nociception. Pharmacol. Ther. 112, 440–456 (2006). [DOI] [PubMed] [Google Scholar]
- 65.Whittier J. M., Crews D., “Seasonal reproduction: Patterns and control” in Hormones and Reproduction in Fishes, Amphibians, and Reptiles, Norris D. O., Jones R. E., Eds. (Plenum Press, 1987), pp. 385–409. [Google Scholar]
- 66.Sharp P. J., Photoperiodic regulation of seasonal breeding in birds. Ann. N. Y. Acad. Sci. 1040, 189–199 (2005). [DOI] [PubMed] [Google Scholar]
- 67.Saunders D. S., Henrich V. C., Gilbert L. I., Induction of diapause in Drosophila melanogaster: Photoperiodic regulation and the impact of arrhythmic clock mutations on time measurement. Proc. Natl. Acad. Sci. U.S.A. 86, 3748–3752 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ojima N., Hara Y., Ito H., Yamamoto D., Genetic dissection of stress-induced reproductive arrest in Drosophila melanogaster females. PLoS Genet. 14, e1007434 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nagy D., et al. , Peptidergic signaling from clock neurons regulates reproductive dormancy in Drosophila melanogaster. PLoS Genet. 15, e1008158 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang Y., Liu Y., Bilodeau-Wentworth D., Hardin P. E., Emery P., Light and temperature control the contribution of specific DN1 neurons to Drosophila circadian behavior. Curr. Biol. 20, 600–605 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Venken K. J. T., et al. , Versatile P[acman] BAC libraries for transgenesis studies in Drosophila melanogaster. Nat. Methods 6, 431–434 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yapici N., Kim Y. J., Ribeiro C., Dickson B. J., A receptor that mediates the post-mating switch in Drosophila reproductive behaviour. Nature 451, 33–37 (2008). [DOI] [PubMed] [Google Scholar]
- 73.Žitňan D., Kingan T. G., Kramer S. J., Beckage N. E., Accumulation of neuropeptides in the cerebral neurosecretory system of Manduca sexta larvae parasitized by the braconid wasp Cotesia congregata. J. Comp. Neurol. 356, 83–100 (1995). [DOI] [PubMed] [Google Scholar]
- 74.Park J. H., Schroeder A. J., Helfrich-Förster C., Jackson F. R., Ewer J., Targeted ablation of CCAP neuropeptide-containing neurons of Drosophila causes specific defects in execution and circadian timing of ecdysis behavior. Development 130, 2645–2656 (2003). [DOI] [PubMed] [Google Scholar]
- 75.Kim E. Y., et al. , A role for O-GlcNAcylation in setting circadian clock speed. Genes Dev. 26, 490–502 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kwak S. J., et al. , Drosophila adiponectin receptor in insulin producing cells regulates glucose and lipid metabolism by controlling insulin secretion. PLoS One 8, e68641 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fichelson P., Brigui A., Pichaud F., Orthodenticle and Kruppel homolog 1 regulate Drosophila photoreceptor maturation. Proc. Natl. Acad. Sci. U.S.A. 109, 7893–7898 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and supporting information.