Significance
The strength of peptide binding to class I MHC proteins is a correlate of immunogenicity. However, many peptides of interest, including shared tumor antigens, bind weakly and are poorly immunogenic. Considerable effort has been spent generating stronger-binding variants of these, with the goal of using them as vaccines to elicit responses against the unmodified peptides. We show that common modifications used to improve peptide binding can unpredictably alter T cell recognition. The alterations arise from structurally silent, “through-protein” effects that are sensed differently by different T cell receptors. Our results have implications for the design and use of modified peptides, as well as efforts to predict the immunogenicity of peptides incorporating similar modifications, such as tumor neoantigens.
Keywords: T cell receptor, MHC, binding, allostery, molecular dynamics
Abstract
Presentation of peptides by class I MHC proteins underlies T cell immune responses to pathogens and cancer. The association between peptide binding affinity and immunogenicity has led to the engineering of modified peptides with improved MHC binding, with the hope that these peptides would be useful for eliciting cross-reactive immune responses directed toward their weak binding, unmodified counterparts. Increasing evidence, however, indicates that T cell receptors (TCRs) can perceive such anchor-modified peptides differently than wild-type (WT) peptides, although the scope of discrimination is unclear. We show here that even modifications at primary anchors that have no discernible structural impact can lead to substantially stronger or weaker T cell recognition depending on the TCR. Surprisingly, the effect of peptide anchor modification can be sensed by a TCR at regions distant from the site of modification, indicating a through-protein mechanism in which the anchor residue serves as an allosteric modulator for TCR binding. Our findings emphasize caution in the use and interpretation of results from anchor-modified peptides and have implications for how anchor modifications are accounted for in other circumstances, such as predicting the immunogenicity of tumor neoantigens. Our data also highlight an important need to better understand the highly tunable dynamic nature of class I MHC proteins and the impact this has on various forms of immune recognition.
T cells direct adaptive immune responses against pathogens and cancer by recognizing antigenic peptides bound and presented by major histocompatibility complex (MHC) proteins. Strong peptide binding to MHC proteins has long been considered a prerequisite for efficient recognition (1), and studies continue to show that peptide affinity for class I MHC proteins is a correlate of immunogenicity (2, 3). This association between peptide/MHC affinity and immunogenicity influenced early approaches to develop reagents and immune therapies that rely on T cell recognition of peptide/MHC complexes. For example, studies on shared tumor antigens found that improving peptide affinity for class I MHC molecules by optimizing anchor residues could improve immunogenicity in vitro (4, 5). Such anchor-modified peptides (sometimes referred to as heteroclitic peptides or mimotopes) were subsequently explored as therapeutic cancer vaccines, with generally disappointing outcomes (6).
Indeed, while they can improve peptide/MHC binding, substitutions at peptide anchors do not always lead to improved immunogenicity. This was noted early in the MART-1 system: although substitution of the position 2 (p2) alanine with leucine for the 26 to 35 decamer improved immunogenicity with several clones, the same substitution in the 27 to 35 nonamer eliminated immunogenicity with others (4). Similar discrimination between wild-type (WT) and anchor-modified variants has been seen with other peptides (7). These in vitro results were consistent with vaccination studies (8–10) and have been hypothesized to contribute, at least in part, to the poor performance of clinical trials with modified shared tumor antigens (7, 11).
The in vitro results with shared tumor antigens indicate that anchor-modified peptides can be perceived as different antigens from their WT counterparts. This is at first surprising, as at least for the p2 position, the sidechain anchoring the peptide to the class I MHC protein is typically fully buried within the p2 pocket and not accessible to T cell receptors (TCRs). Moreover, anchor modification of peptides presented by class I MHC proteins has generally not been associated with peptide conformational changes (12–15).
The different results seen with TCR recognition of WT and anchor-modified peptides are consistent with the TCR’s high sensitivity to subtle changes in the properties of peptide/MHC complexes. In some cases where anchor sensitivity has been observed, mechanistic detail has been provided and relates to how TCRs might differentially sense peptide motion or trigger peptide conformational changes (13, 16, 17). However, deeper insight into how anchor modifications alter peptide/MHC properties and how different TCRs perceive them is warranted, as careful peptide modification may still be a viable strategy for selectively improving immune responses in cancer and infectious disease (11, 18, 19). Moreover, the impact of peptide anchor modification is of heightened interest in the identification of immunologically active neoantigens. Neoantigens with mutations that improve peptide binding to class I MHC proteins are often found to be immunogenic (20), yet the prediction of immunogenic neoantigens based on this observation is hindered by false positives (21, 22).
Recently, we studied the recognition of WT and an anchor-modified variant of the well-known gp100209 tumor antigen presented by the class I MHC protein HLA-A2 (ITDQVPFSV). In examining T cells expressing three different gp100209-specific TCRs, we observed a large variation in functional responses toward the anchor-modified, gp100209–2M variant (IMDQVPFSV) (23). However, we observed few differences when we examined T cell recognition of the WT peptide. Earlier studies with two of the same TCRs showed similar discrimination between the WT and modified gp100209 peptides (24). These results reflected curious differences seen in vaccination studies with the two peptides (9, 10). Thus, over a wide array of experiments in different systems, ranging from in vitro to in vivo, TCRs appear to discriminate between the WT and anchor-modified gp100209 peptides.
Here, we studied recognition of gp100209 anchor-modified peptides in detail. Using a panel of TCRs (23, 24), we found strikingly large differences in how the TCRs perceive anchor modification. Structural data show that sensitivity to anchor modification can arise not from changes in how TCRs interact with the site of the modification, but rather from how modifications alter interactions between the TCR and MHC protein in distant parts of the interface. The data thus reveal a through-protein, allosteric effect whose consequences are TCR dependent and whose strength varies with the size and chemistry of the amino acid in the HLA-A2 p2 pocket. The consequences of anchor modification are associated with changes in the motional properties of the peptide/HLA-A2 complex, which are emerging as highly significant in modulating immune recognition (25). Further results suggest this allosteric effect is not limited to modifications at the first primary anchor but can occur with substitutions at other anchor positions. Our results demonstrate that, separate from generating a more stable peptide/MHC complex, peptide anchor modification can have significant, albeit structurally silent, consequences for TCR recognition that should be considered when assessing T cell-dependent responses to WT and anchor-modified peptides. This includes peptides intentionally modified (e.g., vaccine candidates) as well as those where modifications occur through mutational processes, such as in cancer neoantigens.
Results
The Identity of the gp100209 Anchor Residue Affects TCR Binding in a TCR-Dependent Fashion.
Previously studying the SILv44, R6C12, and T4H2 TCRs (SI Appendix, Table S1), we observed a correlation between TCR affinity toward the anchor-modified gp100209–2M peptide (referred to as T2Met) presented by HLA-A2 and the extent of cytokine release in functional assays, using both transduced Jurkat cells as well as peripheral blood mononuclear cells (23). In those studies, T4H2 recognized the T2Met peptide with the strongest affinity and was most potent, whereas SILv44 recognized T2Met the weakest and was least potent. Surprisingly, much smaller differences were observed between the TCRs when the unmodified WT gp100209 peptide was studied. Earlier experiments with two of the same TCRs showed similar results (24). These observations are curious given our prior work showing the WT and T2Met peptides are structurally indistinguishable when bound to HLA-A2 and that the p2 sidechains are fully buried (12). These findings prompted us to ask how the SILv44, R6C12, and T4H2 TCRs sense differences between the WT and anchor-modified peptides in greater detail.
We first confirmed the impact of p2 anchor modification on the binding of the gp100209 peptide to HLA-A2, using differential scanning fluorimetry (DSF) to measure the melting temperature (Tm) of the peptide/HLA-A2 complex. Consistent with prior measurements (26), we found the T2Met complex was 5 °C more stable than the WT complex (SI Appendix, Fig. S1). This reflects a moderate, but not substantial, improvement in the affinity of the T2Met peptide for HLA-A2 (for comparison, we previously found a 16 °C difference between the WT and p2 anchor-modified MART-1 decamer) (27).
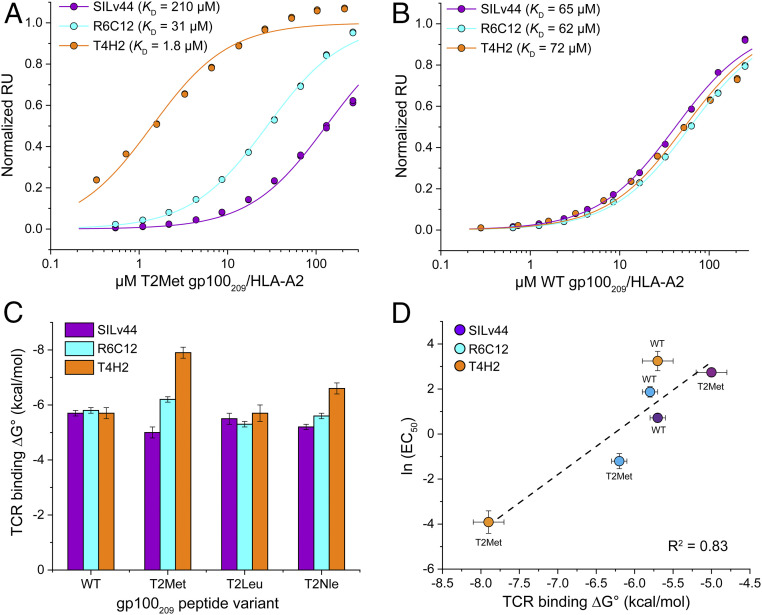
We then repeated our earlier surface plasmon resonance (SPR) binding experiments, measuring the affinity SILv44, R6C12, and T4H2 possess toward the WT and anchor-modified T2Met peptides presented by HLA-A2. We used a global data acquisition and analysis approach that enhanced accuracy with low-affinity interactions (28). The results were nearly identical to our previous findings: with the T2Met peptide, SILv44 bound with a weak KD of 210 μM, R6C12 with an intermediate KD of 31 μM, and T4H2 with a strong KD of 1.8 μM (Fig. 1A and SI Appendix, Table S2 and Fig. S2A). However, when performing the same experiment with the WT peptide we measured nearly identical, moderate affinities in the range of 60 to 70 μM (Fig. 1B and SI Appendix, Table S2). Thus, even though the WT and T2Met peptide/HLA-A2 complexes are structurally indistinguishable (12), the three TCRs sense the peptide threonine-to-methionine anchor modification. Moreover, they do so in a TCR-dependent manner: SILv44 recognizes T2Met with an ∼3-fold weaker KD than WT, R6C12 with an ∼2-fold stronger KD, and T4H2 with a remarkable 40-fold stronger KD (Fig. 1C).
Fig. 1.
The SILv44, R6C12, and T4H2 TCRs sense anchor modification in the gp100209 peptide differently. (A) SPR titrations for the three TCRs binding the T2Met/HLA-A2 complex. The TCRs show an ∼120-fold range in affinity, with T4H2 binding strongest and SILv44 TCR weakest. Data points show representative titrations with duplicate injections. To aid in presentation, data points and fitted curves were normalized to maximum response values determined from the global analysis of multiple datasets as described in Methods. The color scheme for the TCRs is maintained throughout the figure. (B) As in A but binding to the WT peptide/HLA-A2 complex. The TCRs bind with almost identical affinities. (C) Free energy changes (ΔG°) for the TCRs binding the WT, T2Met, T2Leu, and T2Nle peptide/HLA-A2 complexes. Values and error bars are from SI Appendix, Table S2, propagated from the average and SDs of the KD values. (D) EC50 values from the functional titrations in SI Appendix, Fig. S3 are well correlated with the binding free energies. Error bars are from SI Appendix, Table S2 (ΔG°) or propagated from the errors in EC50 measurements in SI Appendix, Fig. S3.
The differential sensitivities to anchor modifications were mirrored in experiments measuring cytokine production, using CD8+ TCR-transduced Jurkat cells cocultured with antigen-pulsed T2 cells (SI Appendix, Fig. S3). The EC50 values determined from these functional experiments correlated well with the in vitro binding results (Fig. 1D). They also confirm the results are not an artifact due to altered peptide binding to HLA-A2 stemming from anchor modification as in the case of SILv44, the better HLA-A2 binding peptide is nonetheless a less potent antigen.
TCR Anchor Sensitivity Is Influenced by Both Structural and Chemical Properties of the gp100209 p2 Anchor Residue.
We next investigated how the characteristics of the p2 anchor residue affect TCR binding. Leucine is a preferred anchor residue for HLA-A2. Although there is no structure of the gp100209 variant with a leucine at p2 bound to HLA-A2, we hypothesized that the SILv44, R6C12, and T4H2 TCRs would recognize this peptide (referred to as T2Leu) similarly to T2Met. Once again, however, the TCRs showed varying levels of sensitivity to this modification (Fig. 1C and SI Appendix, Table S2). In SPR binding experiments, the R6C12 TCR recognized T2Leu fourfold weaker than T2Met. SILv44 recognized T2Leu with an affinity intermediate between WT and T2Met. T4H2 recognized T2Leu with an affinity identical to that of WT.
To further explore the sensitivity of the TCRs, we studied a gp100209 variant with norleucine at position 2 (referred to as T2Nle). Norleucine is isosteric with methionine but replaces the sulfur with a methylene carbon. The SILv44 TCR recognized T2Nle with an affinity slightly stronger than T2Met, but weaker than T2Leu, while R6C12 and T4H2 showed the opposite behavior (Fig. 1C and SI Appendix, Table S2). By DSF, the Tm values of the T2Nle and T2Leu complexes were indistinguishable from that of T2Met (SI Appendix, Fig. S1). Remarkably then, the TCRs thus sense both the shape and chemistry of the p2 sidechain in the gp100209 peptide, independent of the stability of the peptide/HLA-A2 complex.
The Anchor Sensitivity of the T4H2 TCR Is Driven by Dramatic Differences in TCR Binding Thermodynamics and Kinetics.
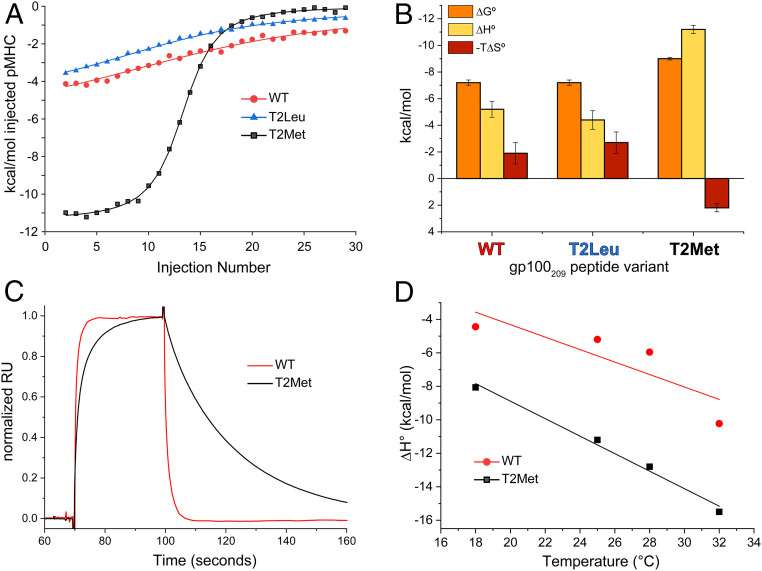
To better understand the sensitivity of TCRs to different p2 anchors in the gp100209 peptide, we studied the thermodynamics of T4H2 TCR recognition of the WT, T2Leu, and T2Met peptide/HLA-A2 complexes via isothermal titration calorimetry (ITC) (Fig. 2A). We restricted these studies to T4H2 as it showed the greatest sensitivity to anchor modification. We again used a global data acquisition and analysis approach that enhanced accuracy with weak interactions. From ITC we obtained KD values slightly stronger than with SPR (SI Appendix, Table S3), consistent with previous work with TCR-peptide/MHC interactions (29–31). Crucially though, the relative differences in binding affinity and thus free energy were the same between the ITC and SPR data, with T4H2 again recognizing the T2Met peptide substantially stronger than both the WT and T2Leu peptides (SI Appendix, Table S3).
Fig. 2.
The T4H2 TCR recognizes the T2Met peptide/HLA-A2 complex with a distinct thermodynamic and kinetic profile. (A) Calorimetric titrations of peptide/HLA-A2 complexes with the T4H2 TCR. The WT and T2Leu complexes are recognized similarly, but recognition of T2Met is strikingly more endothermic. Multiple datasets were analyzed globally with shared stoichiometries and local thermodynamics, facilitating accurate assessments across a range of c values as described in Methods. (B) Breakdown of binding thermodynamics for the data in A. Compared to WT and T2Leu, T2Met is recognized with a much more favorable enthalpy change and an unfavorable entropy change. Values are the average and SDs from the indicated number of titrations in SI Appendix, Table S3. (C) T4H2 binds the T2Met complex with slower association and dissociation rates compared to the WT complex. Data are from the injection of 18 μM of peptide/HLA-A2 complex over the same TCR sensor surface, normalized to the maximum response point (prior to injection spikes) to facilitate comparison of kinetic phases. The slower association and dissociation phases for the T2Met complex are evident (SI Appendix, Fig. S2B). (D) Changes in heat capacity for T4H2 recognition of the WT and T2Met peptides calculated from the binding ΔH° measured at different temperatures.
The thermodynamic data from ITC indicated that the T4H2 TCR recognized the WT and T2Leu peptides with similar, moderately favorable enthalpy (ΔH°) and entropy (ΔS°) values. However, the improved affinity toward the T2Met peptide was driven by strikingly different binding thermodynamics, notably a much more favorable ΔH° offset by an unfavorable change in ΔS° (Fig. 2B and SI Appendix, Table S3). The improved ΔH° is consistent with the formation of stronger interatomic interactions between the TCR and peptide/HLA-A2 when methionine is at p2 of the gp100209 peptide, whereas the more unfavorable ΔS° is consistent with a greater loss of molecular flexibility upon TCR binding. This suggests that the dynamics of the peptide/HLA-A2 protein are different when methionine vs. threonine is present at p2.
To compare with the thermodynamic data, we assessed the kinetics of T4H2 recognition of the WT and T2Met complexes. Dissociation rates for the WT complex were rapid as frequently seen for moderate-to-low affinity TCR-peptide/MHC interactions, precluding quantification of kinetics by SPR. Binding to the T2Met complex, on the other hand, was clearly driven by both slower association and dissociation rates (Fig. 2C and SI Appendix, Fig. S2B). The impact on kinetics is consistent with the binding thermodynamics: a more dynamic peptide/HLA-A2 complex results in slower binding and a higher entropic penalty, whereas a more stably formed interface results in slower dissociation and a more favorable binding enthalpy change.
Lastly, we measured heat capacity changes (ΔCp) for T4H2 recognition of the WT and T2Met complexes (Fig. 2D and SI Appendix, Table S3). Both were recognized with moderately negative ΔCp values, consistent with measurements on other TCR-peptide/MHC systems (32). The ΔCp for recognition of the T2Met peptide, however, was ∼30% larger, suggesting T4H2 forms a more hydrophobic interface with T2Met compared to the WT peptide, consistent with the kinetic and other thermodynamic measurements that indicate a more stably formed TCR interface (33).
The Moderate Sensitivity of the SILv44 TCR to Anchor Modification Is Not Correlated with Structural Differences in the TCR-Peptide/HLA-A2 Interface.
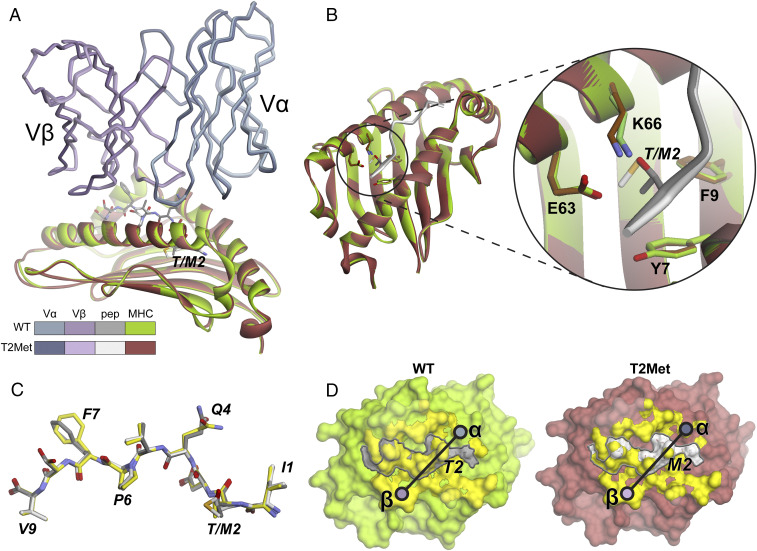
To explore how the TCRs perceived the differences in peptides, we crystallized and solved the structures of select TCR-peptide/HLA-A2 complexes. Although we were unable to crystallize complexes with the R6C12 TCR, we determined structures for T4H2 bound to the WT, T2Met, and T2Leu peptide/HLA-A2 complexes, as well as SILv44 bound to the WT peptide/HLA-A2 complex (SI Appendix, Table S4). The structure of SILv44 bound to the T2Met complex has been described separately (34).
For SILv44, the crystals with the WT peptide were isomorphic with those with the T2Met peptide, and the structure was solved at the same resolution. The complex with the WT peptide has clear density at the TCR-peptide/HLA-A2 interface (SI Appendix, Fig. S4A). The two structures display characteristics typical of TCRs bound to HLA-A2 (35, 36). Superimposing the two complexes by aligning the HLA-A2 binding grooves reveals that SILv44 binds both ligands essentially identically (Fig. 3A and SI Appendix, Table S5). There are no obvious differences in the regions of HLA-A2 surrounding the p2 pocket (Fig. 3B). The peptide conformations in the two complexes are likewise nearly identical (Fig. 3C and SI Appendix, Table S5). Additionally, the conformations of the peptides in the two SILv44 complexes are nearly the same as the peptides in the unbound gp100209 peptide/HLA-A2 crystal structures, with only small differences in the Gln4 and Phe7 sidechain rotamers.
Fig. 3.
The SILv44 TCR binds the WT and T2Met peptide/HLA-A2 complexes with no apparent structural differences. (A) Structural overview showing the TCR variable domains and HLA-A2 peptide binding domains aligned by superimposing residues 1 to 180 of HLA-A2. The color scheme is maintained throughout the figure. (B) Zoomed-in region of the HLA-A2 p2 pocket in the structures. (C) The conformations of the WT and T2Met peptides in the TCR-bound structures are the same and nearly identical to that of the peptide from the unbound WT gp100209/HLA-A2 complex (yellow). (D) Footprints of the TCR over the peptide/HLA-A2 complexes. Circles indicate centers of mass of the TCR variable domains. Yellow surface is from atoms with ≥0.1 Å2 of surface area buried upon TCR binding. Peptide surface is outlined by the gray line.
The SILv44 TCR buries essentially the same amount of solvent accessible surface area in each complex (1,845 Å2 and 1,828 Å2 for WT and T2Met). In both cases, 37% of the buried surface area is attributed to the α-chain and 63% to the β-chain. Most interfacial contacts are thus made by the β-chain, away from the p2 position of the peptide (SI Appendix, Fig. S5). The diagonal crossing angle over the center of the peptide is 27° to 28° (Fig. 3D). Overall then, the two SILv44 structures are essentially identical and thus do not indicate a mechanism for the threefold difference in binding affinity between the WT and T2Met peptides.
The Significant Sensitivity of the T4H2 TCR to Anchor Modification Is Correlated with Changes in TCR/MHC Interactions at a Site Distant from the Peptide p2 Anchor.
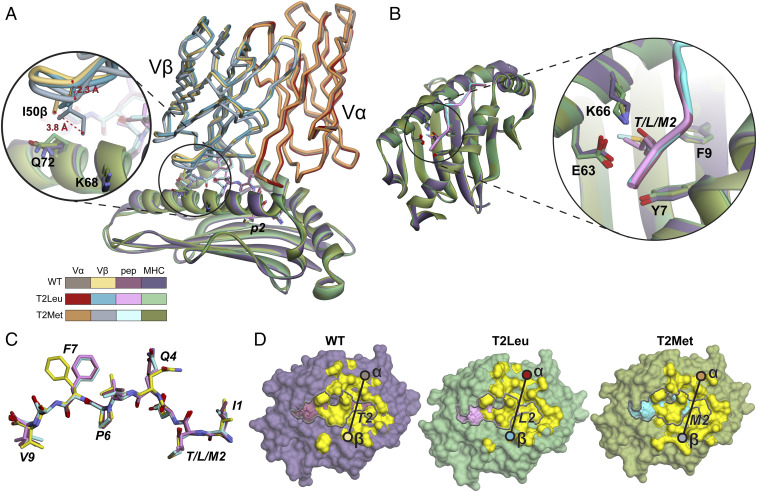
For the T4H2 TCR, the WT, T2Leu, and T2Met crystals were also isomorphous and the structures solved at similar resolutions (SI Appendix, Table S4). All structures have clear density at the TCR-peptide/MHC interface (SI Appendix, Fig. S4B). The structures again display common characteristics of TCR complexes with HLA-A2 (35, 36), although T4H2 binds more orthogonally than SILv44 (crossing angles of 51° to 53° for T4H2).
The structures for T4H2 with the WT and T2Leu peptides are nearly identical (Fig. 4A and SI Appendix, Table S5). However, the structure of T4H2 with the T2Met peptide differs from these due to a systematic displacement in the TCR β-chain. This displacement involves multiple strands of the Vβ framework; when the peptide binding domains are superimposed, the T2Met Vβ domain differs from the WT with a Cα root mean square (RMS) deviation of 1.4 Å, compared to a value of 0.3 Å for the T2Leu/WT Vβ comparison (Fig. 4A and SI Appendix, Fig. S6). The displacement of the T2Met Vβ domain brings the CDR2β loop 2 to 4 Å closer to the HLA-A2 α1 helix in the T2Met structure (Fig. 4A, Inset). The closer approach of CDR2β results in new/altered contacts between the TCR and HLA-A2 α1 helix (SI Appendix, Fig. S7). This translates into greater burial of total and hydrophobic surface: the WT and T2Leu complexes bury a total of 1,750 Å2 and 1,788 Å2 of solvent accessible surface area, respectively, of which 885 Å2 and 912 Å2 are hydrophobic, while the T2Met complex buries a total of 1,935 Å2 of solvent accessible surface area, of which 975 Å2 is hydrophobic. The burial of more hydrophobic surface area with the T2Met complex is consistent with the more negative binding ΔCp, more favorable binding ΔH°, and slower dissociation rate for T4H2 recognition of the T2Met peptide.
Fig. 4.
The T4H2 TCR binds the T2Met peptide/HLA-A2 complex with subtle differences compared to the WT and T2Leu complexes. (A) Structural overview showing the TCR variable domains and HLA-A2 peptide binding domains aligned by superimposing residues 1 through 180 of HLA-A2. The color scheme is maintained throughout the figure. The Vβ domain of the T2Met complex is shifted relative to that of the WT and T2Leu complexes (SI Appendix, Fig. S6). The zoomed-in section highlights the closer approach of CDR2β in the T2Met structure. (B) Zoomed-in region of the HLA-A2 p2 pocket in the structures. (C) The conformations of the WT, T2Leu, and T2Met peptides in the TCR-bound structures are again nearly identical to that of the peptide from the unbound WT gp100209/HLA-A2 complex (yellow), differing predominantly by rotamer shifts in Gln4 and Phe7. (D) Footprints of the TCR over the peptide/HLA-A2 complexes. Circles indicate centers of mass of the TCR variable domains. Yellow surface is from atoms with ≥0.1 Å2 of surface area buried upon TCR binding. Peptide surface is outlined by the gray line.
The p2 pocket regions of HLA-A2 are again very similar in the three T4H2 structures (Fig. 4B). There is however a small, systematic rigid body displacement of the center of the HLA-A2 α1 helix in the structure with the T2Met peptide in the region that interfaces with the CDR2β loop (∼1 Å measured at the backbone of Lys68; Fig. 4A, Inset and SI Appendix, Fig. S8A). This displacement is not seen in the structures of the two unbound peptide/HLA-A2 complexes or in the T2Leu structure (SI Appendix, Fig. S8 B and C). The differences between the structures do not extend to the peptide, as peptide conformations are again nearly identical in the three structures. As with the SILv44 complexes, these are nearly identical to the structure of the peptides from the unbound peptide/HLA-A2 complexes, differing only in sidechain rotamer conformations for Gln4 and Phe7 (Fig. 4C and SI Appendix, Table S5).
Unlike SILv44, T4H2 uses more of the α- than the β-chain to engage the peptide/HLA-A2 complexes. In the complexes with the WT and T2Leu peptides, 68% of the buried surface area is contributed by Vα and 32% by Vβ. Owing to the closer approach of the CDR2β loop, the numbers differ slightly for the complex with the T2Met peptide: 62% is contributed by Vα and 38% by Vβ. Due to its greater utilization of the Vα domain upon binding, the T4H2 TCR is positioned closer to the N-terminal region of the peptide near the HLA-A2 p2 pocket compared to SILv44 (Fig. 4D; for a direct comparison see SI Appendix, Fig. S9).
Peptide-Dependent Consequences of Mutations Confirm the Structural Differences with the T4H2 TCR and Indicate the p2 Residue Is an Allosteric Modulator of TCR Binding.
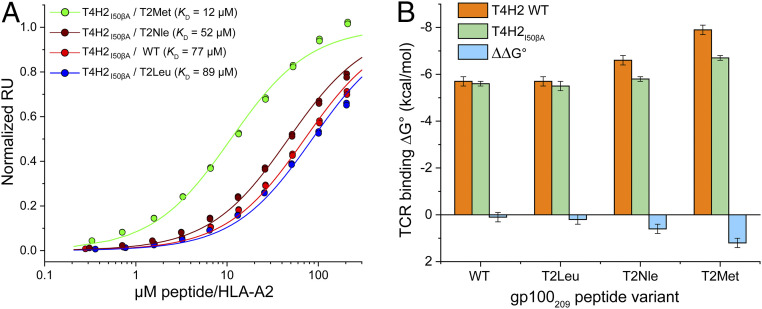
The structural and thermodynamic data indicate that for the T4H2 TCR, the T2Met peptide allows the CDR2β loop to rest closer to the α1 helix of HLA-A2, contributing to the much improved affinity T4H2 has for the T2Met peptide compared to WT. Accordingly, we reasoned that mutations in the T4H2 CDR2β loop should be sensitive to the peptide. We thus mutated Ile50 within CDR2β to alanine and asked how the effects of this mutation on TCR binding varied with peptide. We selected Ile50β as it is a CDR2β hydrophobic loop residue most proximal to the α1 helix, where it packs against the sidechains of Lys68 and Gln72 (SI Appendix, Fig. S7). Notably, Ile50β is nearly 20 Å away from the p2 residue, whereas Lys68 and Gln72 are 12 to 16 Å away (SI Appendix, Fig. S10). The results of the Ile50β→Ala mutation mirrored how the TCR sensed changes to the p2 anchor residue (Fig. 5A and SI Appendix, Table S2). The impact of the mutation on recognition of WT and T2Leu was negligible (e.g., for the WT peptide, the KD moved imperceptibly from 72 to 77 μM). The impact on recognition of T2Nle and T2Met on the other hand was more substantial, with the KD for T2Met dropping from 1.8 to 12 μM (Fig. 5B). These results support our conclusions that the T2Met modification in the gp100209 peptide leads to stronger interactions between the T4H2 TCR and the HLA-A2 α1 helix. The through-protein “action at a distance” revealed by this experiment indicates that the gp100209 p2 anchor is an allosteric modulator of TCR recognition.
Fig. 5.
Peptide-dependent consequences of a mutation in the T4H2 CDR2β loop confirms the structural variances in the T4H2 structures. (A) SPR titrations for the T4H2 TCR with the Ile50β→Ala mutation binding the WT, T2Nle, T2Leu, and T2Met peptide/HLA-A2 complexes. Data points show representative single titrations with duplicate injections. (B) Comparison of binding free energy changes with the WT T4H2 TCR and the Ile50β→Ala mutant. The effect of the mutation, ∼20-Å distant from the p2 residue, is negligible for recognition of the WT peptide, but increases upon p2 anchor modification, to a maximum amount observed with the T2Met peptide. Values and error bars are from SI Appendix, Table S2, propagated from the average and SDs of the KD values.
Peptide Modification Results in Dynamical Differences in gp100209 Peptide/HLA-A2 Complexes.
Although the gp100209 p2 anchor residue allosterically modulates the binding of the SILv44, R6C12, and T4H2 TCRs to various extents, the crystallographic structures do not clearly indicate an underlying mechanism beyond the displacement in Vβ and systematic shift in the HLA-A2 α1 helix seen for T4H2 recognition of T2Met. However, as noted above, the more unfavorable ΔS° and slower association kinetics for T4H2 binding the T2Met compared to the WT complex led us to hypothesize that changes in the flexibility of the peptide/HLA-A2 complexes underlie how the TCRs perceive peptide anchor modification. Indeed, we and others have shown that mutations in peptides presented by class I MHC proteins can modulate peptide and MHC motional properties in ways that influence binding by a range of proteins (16, 37–39). Moreover, it is now recognized that through-protein allosteric effects such as those seen here frequently occur via alterations in protein motions (40).
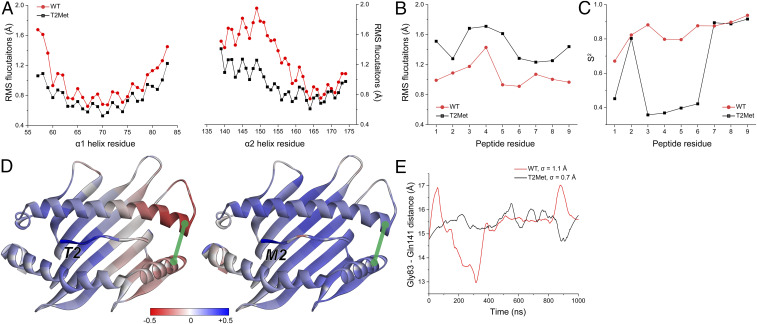
Recently, we described a large library of molecular dynamics (MD) simulations of peptide/HLA-A2 complexes, with more than 50 different peptide/HLA-A2 complexes simulated for 1 μs in explicit solvent (41). We previously used this library to assess variances in peptide dynamics and how dynamical information is communicated through the protein (42). This library included the WT and T2Met gp100209/HLA-A2 complexes, using their available crystal structures as starting coordinates (12). We interrogated these simulations here, computing Cα RMS fluctuations for the α1 and α2 helices of the HLA-A2 protein and the peptide. We observed no clear peptide-dependent differences in the α1 helix, including those amino acids that neighbor the peptide p2 residue. We did observe greater fluctuations of the short arm of the α2 helix (residues 142 to 152; also referred to as the α2–1 helix) in the simulation with the WT peptide (Fig. 6A), consistent with prior findings showing that this region of class I MHC proteins is sensitive to the nature of the bound peptide (38, 43–46).
Fig. 6.
Anchor modification alters the motional properties of the unbound peptide/HLA-A2 complex. (A) The RMS fluctuations for Cα atoms of the HLA-A2 α1 and α2 helix when the WT or T2Met peptide is bound. A clear difference is apparent in the C-terminal short arm of the α2 helix. (B) The RMS fluctuations for Cα atoms of the peptides bound to HLA-A2. T2Met peptide fluctuates more than the WT peptide in the binding groove. (C) Order parameters (S2), calculated from vectors defined between the α-carbons and β-carbons of each residue. Consistent with the fluctuation data, the WT peptide is more ordered in the binding groove. (D) Correlations between the p2 Cα atom and the Cα atoms of the rest of the WT and T2Met peptide/HLA-A2 complexes. Red/blue colors indicate anticorrelated/correlated motion according to the indicated scale. Replacing Thr with Met at p2 shifts motion in the peptide binding groove from anticorrelated to correlated. Green line represents the distance between Gly83 and Gln141. (E) Variance in Gly83–Gln141 distance over the course of the simulations when the WT or T2Met peptides are bound. The WT complex displays more breathing between the helices.
In examining the peptides in the simulations, we observed the opposite behavior: the T2Met peptide exhibited larger fluctuations than the WT peptide across its entire length (Fig. 6B). We also computed order parameters, which revealed more rapid motions in the T2Met peptide (Fig. 6C). These results are consistent with our previous observations showing that p2 “anchor fixing” of the MART-1 nonamer peptide leads to greater peptide mobility (13, 16). A more mobile T2Met peptide in the free peptide/HLA-A2 complex could contribute to the higher entropic penalty and slower association rate for binding by the T4H2 TCR than seen with the WT peptide.
We next computed Cα dynamic cross-correlation matrices for the entire peptide/HLA-A2 complexes. When examining correlations between the p2 residue and the rest of the peptide binding groove, we observed a striking distinction between the WT and T2Met complexes. Whereas motions between p2 and the rest of the groove were all positively correlated with the T2Met peptide, the motions of the WT complex were anticorrelated in the region of HLA-A2 near the C-terminal end of the peptide (Fig. 6D). We hypothesized this could emerge from greater “breathing” of the HLA-A2 binding groove near the C-terminal end in the WT peptide, perhaps driven by the α2 helix motion indicated by the fluctuation data. Indeed, we found that the variance of the distance between residues 83 and 141 at the end of the binding groove was ∼50% greater in the WT than the T2Met complex (Fig. 6D and E). As with the different peptide dynamics, changes in correlated motion of the HLA-A2 protein in response to anchor modification could impact TCR recognition.
Lastly, we asked if anchor modification at the position 9 (p9) primary anchor could have consequences similar to what we have seen at p2. The NY-ESO-1157 shared tumor antigen has been studied extensively, and variants with substitutions for the suboptimal p9 cysteine have been described that improve binding to HLA-A2. Once again, p9 modification in NY-ESO-1157 has not been found to result in structural perturbations in the unbound peptide/HLA-A2 complex (14, 15). Yet, T cells respond to NY-ESO-1157 variants differently (47), and the affinity of NY-ESO-1157-specific TCRs can change with p9 anchor modification (48). As our MD simulation library included the WT NY-ESO-1157 peptide along with variants that had been crystallized, we asked if similar variations in peptide/HLA-A2 motion occurred when the p9 residue was altered. Examining the simulations of the NY-ESO-1157 WT complex and the complex in which the p9 cysteine was replaced with alanine (C9Ala), we again observed differences in correlated motion, although with less divergent fluctuations between the ends of the HLA-A2 α-helices and less impact on the peptide (SI Appendix, Fig. S11).
Discussion
The capacity for subtle modifications in peptides bound and presented by class I MHC proteins to influence TCR recognition is well established. Surprisingly, this extends to modifications at buried primary anchor residues, even when the substitutions lead to imperceptible structural changes in the TCR-exposed surface. However, the scope and scale of TCR sensitivity to anchor modification remain poorly studied. Here, we studied a panel of three TCRs that distinguish between the WT and p2-modified variants of the gp100209 shared tumor antigen presented by the class I MHC protein HLA-A2. We observed a remarkably wide range of sensitivity to anchor modification: the SILv44 TCR bound the T2Met variant 3-fold weaker than the WT peptide, whereas the R6C12 TCR bound the T2Met variant 2-fold more tightly. Although in opposing directions, these variances are on the order of previous descriptions of TCRs that sense anchor-modified peptides (16, 17, 48). Remarkably though, the T4H2 TCR bound the T2Met variant 40-fold more tightly than the WT peptide. Our observations thus reveal a dramatic and unpredictable TCR-dependent sensitivity to peptide anchor modification.
How does anchor modification of the gp100209 peptide impact TCR binding? Although the detailed mechanism is not fully clear, the T4H2 data demonstrate the contribution of a through-protein effect in the peptide/HLA-A2 complex: the nature of the amino acid filling the p2 pocket influences how the TCR interacts with HLA-A2 in distant regions. This is a classical action at a distance, i.e., allosteric effect, in which the action of binding at one site influences binding at another. Notably, allostery here is occurring in the absence of conformational change, as the crystal structures of the WT and T2Met gp100209 peptides bound to HLA-A2 are indistinguishable (12). While it is possible that future structural work could uncover currently unrecognized structural differences, the potential for allostery to occur in the absence of conformational change has long been recognized (49) and can arise from changes in protein motions that occur in response to ligand binding but are not visible structurally (50, 51). This process, termed dynamic allostery, has been linked to a variety of biological processes, including signaling through the TCR (52–54). With class I MHC proteins, dynamic allostery has been suggested to influence the binding of TCRs and other proteins, including elements of the peptide-loading machinery (16, 37–39, 55, 56).
Consistent with dynamic allostery, our simulations show differences in peptide and HLA-A2 motions depending on which gp100209 peptide is bound. For binding of the T4H2 TCR, we also saw clear thermodynamic and kinetic signatures of dynamic allostery. Exactly why the T4H2, SILv44, and R6C12 TCRs respond differently to the motional differences imparted by peptide modification is not yet clear but must ultimately be attributable to features unique to each TCR, such as binding geometry, the physicochemical properties of their various binding loops, and the contacts formed across the interface. In support of this, the SILv44 and T4H2 TCRs bind with distinct footprints, using different amino acids to form different sets of contacts, particularly near the C-terminal end of the peptide and the flanking HLA-A2 α-helices. The SILv44 TCR, which is only moderately sensitive to anchor modification, senses these differences without apparent structural consequences. In the case of the T4H2 TCR, however, the receptor is able to co-opt the motional differences to make stronger interactions with the HLA-A2 protein and bury additional surface, leading to a substantially stronger binding affinity with the T2Met peptide.
Curiously, the allosteric effect of anchor modification in HLA-A2 is not limited to the size of the p2 sidechain, but also its chemistry, as the three TCRs also discriminated between methionine and norleucine. This is an important mechanistic clue, as although both amino acids are hydrophobic, norleucine is more so, and a role for hydrophobicity in influencing motions that underlie dynamic allostery has been demonstrated in other systems (57, 58).
Although our data speak most clearly to the effect of p2 anchor modification, data with the NY-ESO-1157 peptide suggest similar, albeit smaller, consequences for modification at the C terminus. As with gp100209 and other shared tumor antigens, a similar effect of p9 modification could help explain earlier TCR-dependent recognition of variant NY-ESO-1157 peptides even though the structures of the unbound peptide/HLA-A2 complexes are essentially identical (14, 15).
The impact of anchor modification on TCR recognition has previously been discussed as a potential contributor to mixed performance of clinical trials relying on modified versions of the MART-1 shared tumor antigen (7, 11, 17). Although caution is warranted when extrapolating from biochemical to clinical results, our findings can also explain the heterogeneous outcomes when the gp100209 antigen has been used in vivo (9, 10). The confluence of these data, together with our observation of varied TCR responses and the data with the NY-ESO-1157 antigen, suggest that, in general, T cell sensitivity to anchor modification is broad and unpredictable. This should be considered when interpreting outcomes, both clinical and basic, that rely on the presumption that modified and WT peptides are recognized similarly by responding TCRs.
Our findings also have implications for the burgeoning field of neoantigens. Although the potential for neoantigens as therapeutic cancer vaccines is now frequently discussed, predicting those that are immunologically active remains a challenge, due in part to a high rate of false positives. Some neoantigen prediction approaches consider how mutations alter the peptide relative to its WT counterpart, including how mutations are predicted to improve MHC binding (21). The most significant of these would be neoantigens incorporating mutations at anchor positions. The complex consequences from anchor modification demonstrated here—in some cases resulting in stronger T cell recognition, in other cases weaker, with the degree of sensitivity varying by TCR—could complicate efforts to develop neoantigen prediction algorithms. Addressing this problem will require a deeper understanding of how class I MHC proteins are influenced by anchor modifications and how different TCRs perceive them.
In conclusion, we have demonstrated that modification of the anchor positions of peptides presented by class I MHC proteins can have dramatic, TCR-dependent consequences on T cell recognition, even when the modification does not lead to changes in the structure of the peptide/MHC complex. Our results implicate a through-protein mechanism in which the identity of the anchor residue serves as an allosteric modulator of TCR binding. Allostery is linked to changes in both peptide and protein motions in the peptide/MHC complex, with TCR-dependent variances attributable to properties unique to individual receptors. While our studies emphasize the p2 position of the gp100209 peptide presented by HLA-A2, our findings appear generalizable to other peptides and anchor positions. Our results emphasize the need for caution in the use of anchor-modified peptides and reveal further complexities for predicting the consequences of mutations in peptides presented by class I MHC proteins, such as efforts to identify immunogenic tumor neoantigens. Lastly, our results emphasize the need to better understand the highly tunable dynamic nature of peptide/MHC complexes and the consequences this has for immunobiology.
Methods
Proteins and Peptides.
Recombinant, soluble TCRs and pMHCs were generated as previously described (29). Briefly, TCR α- and β-chains, the HLA-A2 heavy chain, and β2m were expressed as inclusion bodies in Escherichia coli and were dissolved in 8 M urea following purification. TCR α- and β-chains with the α-chain in 20% molar excess were rapidly diluted into 50 mM Tris⋅HCl (pH 8.3), 2.5 M urea, 6.3 mM cysteamine, 3.7 mM cystamine, 2 mM EDTA, and 0.2 mM PMSF with the addition of 400 mM ʟ-arginine for SILv44 and R6C12. HLA-A2 heavy chain and β2m in a 1:3 molar ratio with excess peptide were rapidly diluted into 100 mM Tris⋅HCl (pH 8.3), 400 mM ʟ-arginine, 6.3 mM cysteamine, 3.7 mM cystamine, 2 mM EDTA, and 0.2 mM PMSF. TCR and pMHC were incubated in refold buffers for 12 h at 4 °C followed by dialysis against 10 mM Tris⋅HCl (pH 8.3) for 48 h. TCRs and pMHCs were purified by anion exchange followed by size-exclusion chromatography. Peptides were commercially synthesized by AAPTEC or Genscript. The Ile50β→Ala mutation in the T4H2 TCR was generated using PCR-based mutagenesis and confirmed by sequencing.
Surface Plasmon Resonance.
SPR experiments were performed using a Biacore T200 instrument in 10 mM Hepes (pH 7.4), 150 mM NaCl, 3 mM EDTA and 0.005% surfactant P-20 at 25 °C. TCRs were coupled to a Biacore CM5 sensor chip using EDC-NHS amine coupling. Approximate coupling RUs were 1,500 to 3,000 for steady-state measurements and 500 for kinetic measurements. Increasing concentrations of peptide/HLA-A2 complexes were flowed over immobilized TCR at a rate of 5 μL/min for steady-state measurements or 100 μL/min for kinetic measurements. Each concentration was injected in duplicate for each experiment. For determining KD values, steady-state responses were determined by averaging the final 10 s of each injection and subtracting the response values from identical injections over a blank surface. To enhance accuracy and precision, we used a previously described global analysis approach (28) where each experiment (i.e., an n = 1) consisted of injecting two to four different peptide/HLA-A2 complexes (WT, T2Met, T2Leu, and/or T2Nle) over the same TCR sensor surface. These data, consisting of RU vs. concentration from duplicate injections of different peptide/HLA-A2 complexes over the same TCR surface, were then globally fit to a 1:1 binding model in OriginPro 2018, with local KD values for each individual peptide/HLA-A2 series and a shared maximum response. KD values for each peptide/HLA-A2 experiment were averaged, with errors reported as the SDs as shown in SI Appendix, Table S2. For kinetic measurements with the T2Met peptide, the reference-subtracted dissociation phases of the sensorgrams were globally fit to a single exponential decay model in OriginPro 2018, with a shared koff between the responses from the different injected concentrations.
Protein Crystallography.
Crystals of the SILv44-WT/HLA-A2 complex were grown in 16% PEG-3350 and 236 mM ammonium citrate dibasic at a protein concentration of 6 mg/mL. Crystals of the T4H2-WT/HLA-A2, T4H2-T2Met/HLA-A2, and T4H2-T2Leu/HLA-A2 complexes were grown in 18% PEG-10000, 16% glycerol, 100 to 250 mM Tris⋅HCl pH 8.5, and 60 to 100 mM NaCl at protein concentrations of 6 to 9 mg/mL. The T4H2-T2Met/HLA-A2 complex crystals were improved by seeding with initial, crushed crystals of the same complex. All crystals were grown by hanging drop vapor diffusion at 23 °C. Crystals were cryoprotected by soaking in 16 μL mother liquor supplemented with 4 μL 75% glycerol for ∼30 s prior to flash freezing in liquid nitrogen. Diffraction data were collected on the 22ID or 23ID-D beamlines at the Advanced Photon Source at Argonne National Laboratories. Data were indexed, integrated, and scaled in HKL2000. The SILv44-WT peptide/HLA-A2 structure was solved using the Phaser molecular replacement module in PHENIX using the TCR and peptide/HLA-A2 from the SILv44-T2Met/HLA-A2 structure as separate search models (34). The initial search model for the T4H2 TCR was built with Sculptor in PHENIX using the α-chain of Protein Data Bank (PDB) 3QEU and the β-chain of PDB 4C56 (59, 60). The search model for the peptide/HLA-A2 complex was PDB 1TVH (12). The T4H2-WT/HLA-A2 structure was solved by molecular replacement using these search models in Phaser with the peptide and complementarity determining region (CDR) loops of the TCR removed. The T4H2 TCR and peptide/HLA-A2 from the T4H2-WT/HLA-A2 structure were used as search models for the T4H2-T2Met/HLA-A2 and T4H2-T2Leu/HLA-A2 structures, again with the peptide and CDR loops removed. Following molecular replacement, all models were rebuilt using PHENIX Autobuild. Multiple rounds of restrained refinement were performed using PHENIX Refine. Evaluation of models and map fitting were performed using COOT. Structures were evaluated by MolProbity during and after refinement. Structures were visualized using PyMOL and Discovery Studio. Solvent accessible surface areas were measured using a probe radius of 1.4 Å. The PDB IDs for the structures determined here are 6VMC (T4H2-T2Leu/A2), 6VM9 (T4H2-T2Met/A2), 6VMA (T4H2-WT/A2), and 6VM7 (SILv44-WT/A2). The PDB ID of the separately determined SILv44-T2Met/A2 structure used for comparison is 6VM8 (34).
Isothermal Titration Calorimetry.
ITC experiments were performed using a Microcal VP-ITC instrument in 10 mM Hepes (pH 7.4) and 150 mM NaCl at 25 °C. Heat capacity experiments were conducted by performing additional experiments at 18 °C, 28 °C, and 32 °C. Titrations were performed with the TCR in the calorimeter cell and peptide/HLA-A2 in the syringe. Protein concentrations in the cell varied between 15 and 20 μM. Protein concentrations in the syringe were at least 10-fold greater than the protein concentration in the cell. Injection volumes of 10 μL were injected over 20 s spaced 220 s apart for a total of 30 injections. Data were processed and integrated using Origin 7.0 as distributed with the instrument. The first data point was removed due to diffusion across the syringe tip during equilibration as is standard. The c values for the titrations ranged from 3 to 65. While these values are within the accepted range of 1 to 1,000 (61), to improve accuracy at the low end we used a global analysis approach similar to that used with SPR where a single experiment (n = 1) consisted of titrating different peptide/HLA-A2 complexes (WT, T2Met, and T2Leu) into a fresh TCR sample from the same stock. These data were then fit globally to a single-site binding model using NLREG, with ΔH°, KD, and a baseline offset as local parameters and the stoichiometry as a shared, global parameter (62). This approach allowed us to improve accuracy with the low-affinity, low c titrations, as the stoichiometry was constrained by the higher-affinity T2Met titration (63). From these titrations, KD, ΔG°, and ΔS° values were calculated from the multiple measured ΔH° values and equilibrium constants and averaged for each peptide/HLA-A2, with errors reported as the SDs as shown in SI Appendix, Table S2. ΔCp values were determined by linear regression analysis of the ΔH° vs. temperature data, with errors shown as standard fitting errors.
Thermal Stability Measurements.
Thermal stability experiments were measured by differential scanning fluorimetry using a NanoTemper Prometheus instrument collecting backreflection scattering optics. Approximately 10 μL of protein at a concentration of 10 μM was analyzed. The temperature range spanned 20 to 95 °C at a ramp rate of 1 °C/min. Tm values were determined by fitting the temperature derivative of the scattering curve to a Bigaussian function as previously described (27). Measurements were replicated as indicated in SI Appendix, Fig. S1, with values and errors reported as the average and SDs of the multiple measurements.
Molecular Dynamics Simulations Analyses.
Molecular dynamics simulations of the gp100209 and NY-ESO-1157 peptides and variants were previously generated and described (41). For the new analyses presented here, RMS fluctuations, order parameters, binding groove distances, and dynamical cross-correlation matrices were calculated via cpptraj. In displaying interatomic distances, traces were smoothed with a Savitzky–Golay filter using a second-degree polynomial and a 1,000-point window length.
Cells and Functional Analyses.
Jurkat 76 cells stably transduced with the CD8 coreceptor (64) and T2 cells were maintained in RPMI-1640 media supplemented with 10% fetal bovine serum, 100 units/mL penicillin, and 100 μg/mL streptomycin. Jurkat cells were electroporated using the Neon Transfection System (Thermo Fisher) with the pCMV(CAT)T7-SB100 plasmid and the pSBbi-Neo Sleeping Beauty vector bearing either the SILv44, R6C12, or T4H2 TCR α- and β-chains separated by the P2A self-cleaving peptide as previously described (31). Stable transformants were positively selected by culturing cells in complete media containing 1.2 mg/mL of G418. Prior to coculture experiments, TCR-expressing cells were monitored via flow cytometry for TCR cell surface expression by staining cells with anti-human CD3 PE-conjugated antibody (BioLegend). Coculture experiments were conducted with TCR-expressing cells and T2 antigen-presenting cells pulsed with peptide for 2 h at the indicated concentrations, then incubated at a 1:1 ratio at 37 °C for 18 to 20 h. T cell cytokine release was determined by measuring IL-2 cytokine concentration from coculture supernatants via ELISA (BioLegend). Responses were measured in two independent experiments, each consisting of two biological replicates. All four data series were fit together to the Hill equation using MATLAB 2018a to derive EC50 values, with errors reported as standard fitting error as shown in SI Appendix, Fig. S3. pSBbi-Neo was a gift from Eric Kowarz, Goethe University, Frankfurt, Germany, obtained from Addgene (Addgene plasmid #60525). pCMV(CAT)T7-SB100 was a gift from Zsuzsanna Izsvak Max Delbruck Center for Molecular Medicine, Berlin, Germany, obtained from Addgene (Addgene plasmid #34879). Jurkat 76 and T2 cells were a gift from Michael Nishimura, Loyola University Stritch School of Medicine, Chicago, IL.
Supplementary Material
Acknowledgments
This work was supported by grant R35GM118166 from the National Institute of General Medical Sciences, National Institutes of Health (to B.M.B.). A.R.S. was supported by a Walther Cancer Foundation Interdisciplinary Interface Training Project fellowship. X-ray data were collected at the SER-CAT and GM/CA beamlines at the Advanced Photon Source, Argonne National Laboratory. SER-CAT is supported by its member institutions and equipment grants (S10RR25528 and S10RR028976) from the National Institutes of Health. GM/CA has funded in whole or in part with Federal funds from the National Cancer Institute (ACB-12002) and the National Institute of General Medical Sciences (AGM-12006). Use of the Advanced Photon Source was supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. W-31-109-Eng-38.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2018125118/-/DCSupplemental.
Data Availability.
Crystallographic structure data have been deposited in Protein Data Bank (6VM7, 6VMA, 6VM9, and 6VMC). All study data are included in the article and supporting information.
References
- 1.Chen W., Khilko S., Fecondo J., Margulies D. H., McCluskey J., Determinant selection of major histocompatibility complex class I-restricted antigenic peptides is explained by class I-peptide affinity and is strongly influenced by nondominant anchor residues. J. Exp. Med. 180, 1471–1483 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Croft N. P., et al. , Most viral peptides displayed by class I MHC on infected cells are immunogenic. Proc. Natl. Acad. Sci. U.S.A. 116, 3112–3117 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fritsch E. F., et al. , HLA-binding properties of tumor neoepitopes in humans. Cancer Immunol. Res. 2, 522–529 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Valmori D., et al. , Enhanced generation of specific tumor-reactive CTL in vitro by selected Melan-A/MART-1 immunodominant peptide analogues. J. Immunol. 160, 1750–1758 (1998). [PubMed] [Google Scholar]
- 5.Parkhurst M. R., et al. , Improved induction of melanoma-reactive CTL with peptides from the melanoma antigen gp100 modified at HLA-A*0201-binding residues. J. Immunol. 157, 2539–2548 (1996). [PubMed] [Google Scholar]
- 6.Rosenberg S. A., Yang J. C., Restifo N. P., Cancer immunotherapy: Moving beyond current vaccines. Nat. Med. 10, 909–915 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cole D. K., et al. , Modification of MHC anchor residues generates heteroclitic peptides that alter TCR binding and T cell recognition. J. Immunol. 185, 2600–2610 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Speiser D. E., et al. , Unmodified self antigen triggers human CD8 T cells with stronger tumor reactivity than altered antigen. Proc. Natl. Acad. Sci. U.S.A. 105, 3849–3854 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clay T. M., et al. , Changes in the fine specificity of gp100(209-217)-reactive T cells in patients following vaccination with a peptide modified at an HLA-A2.1 anchor residue. J. Immunol. 162, 1749–1755 (1999). [PubMed] [Google Scholar]
- 10.Wieckowski S., et al. , Fine structural variations of alphabetaTCRs selected by vaccination with natural versus altered self-antigen in melanoma patients. J. Immunol. 183, 5397–5406 (2009). [DOI] [PubMed] [Google Scholar]
- 11.Buhrman J. D., Slansky J. E., Improving T cell responses to modified peptides in tumor vaccines. Immunol. Res. 55, 34–47 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Borbulevych O. Y., Baxter T. K., Yu Z., Restifo N. P., Baker B. M., Increased immunogenicity of an anchor-modified tumor-associated antigen is due to the enhanced stability of the peptide/MHC complex: Implications for vaccine design. J. Immunol. 174, 4812–4820 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Borbulevych O. Y., et al. , Structures of MART-1(26/27-35) peptide/HLA-A2 complexes reveal a remarkable disconnect between antigen structural homology and T cell recognition. J. Mol. Biol. 372, 1123–1136 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Webb A. I., et al. , Functional and structural characteristics of NY-ESO-1-related HLA A2-restricted epitopes and the design of a novel immunogenic analogue. J. Biol. Chem. 279, 23438–23446 (2004). [DOI] [PubMed] [Google Scholar]
- 15.Chen J. L., et al. , Ca2+ release from the endoplasmic reticulum of NY-ESO-1-specific T cells is modulated by the affinity of TCR and by the use of the CD8 coreceptor. J. Immunol. 184, 1829–1839 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Insaidoo F. K., et al. , Loss of T cell antigen recognition arising from changes in peptide and major histocompatibility complex protein flexibility: Implications for vaccine design. J. Biol. Chem. 286, 40163–40173 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Madura F., et al. , Structural basis for ineffective T-cell responses to MHC anchor residue-improved “heteroclitic” peptides. Eur. J. Immunol. 45, 584–591 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Adegoke A. O., Grant M. D., Enhancing human immunodeficiency virus-specific CD8(+) T cell responses with heteroclitic peptides. Front. Immunol. 6, 377 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Slansky J. E., Nakayama M., Peptide mimotopes alter T cell function in cancer and autoimmunity. Semin. Immunol. 47, 101395 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Duan F., et al. , Genomic and bioinformatic profiling of mutational neoepitopes reveals new rules to predict anticancer immunogenicity. J. Exp. Med. 211, 2231–2248 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Capietto A.-H., et al. , Mutation position is an important determinant for predicting cancer neoantigens. J. Exp. Med. 217, 1–18 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim S., et al. , Neopepsee: Accurate genome-level prediction of neoantigens by harnessing sequence and amino acid immunogenicity information. Ann. Oncol. 29, 1030–1036 (2018). [DOI] [PubMed] [Google Scholar]
- 23.Eby J. M., et al. , Molecular properties of gp100-reactive T-cell receptors drive the cytokine profile and antitumor efficacy of transgenic host T cells. Pigment Cell Melanoma Res. 32, 68–78 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moore T. V., et al. , Relationship between CD8-dependent antigen recognition, T cell functional avidity, and tumor cell recognition. Cancer Immunol. Immunother. 58, 719–728 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ayres C. M., Corcelli S. A., Baker B. M., Peptide and peptide-dependent motions in MHC proteins: Immunological implications and biophysical underpinnings. Front. Immunol. 8, 935 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Batalia M. A., et al. , Class I MHC is stabilized against thermal denaturation by physiological concentrations of NaCl. Biochemistry 39, 9030–9038 (2000). [DOI] [PubMed] [Google Scholar]
- 27.Hellman L. M., et al. , Differential scanning fluorimetry based assessments of the thermal and kinetic stability of peptide-MHC complexes. J. Immunol. Methods 432, 95–101 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Blevins S. J., Baker B. M., Using global analysis to extend the accuracy and precision of binding measurements with T cell receptors and their peptide/MHC ligands. Front. Mol. Biosci. 4, 2 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Davis-Harrison R. L., Armstrong K. M., Baker B. M., Two different T cell receptors use different thermodynamic strategies to recognize the same peptide/MHC ligand. J. Mol. Biol. 346, 533–550 (2005). [DOI] [PubMed] [Google Scholar]
- 30.Armstrong K. M., Baker B. M., A comprehensive calorimetric investigation of an entropically driven T cell receptor-peptide/major histocompatibility complex interaction. Biophys. J. 93, 597–609 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Devlin J. R., et al. , Structural dissimilarity from self drives neoepitope escape from immune tolerance. Nat. Chem. Biol. 16, 1269–1276 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Armstrong K. M., Insaidoo F. K., Baker B. M., Thermodynamics of T-cell receptor-peptide/MHC interactions: Progress and opportunities. J. Mol. Recognit. 21, 275–287 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baker B. M., Murphy K. P., Prediction of binding energetics from structure using empirical parameterization. Methods Enzymol. 295, 294–315 (1998). [DOI] [PubMed] [Google Scholar]
- 34.Schmidt J., et al. , Prediction of neo-epitope immunogenicity reveals TCR recognition determinants and immunoediting in human cancer. Cell Reports Medicine (2021) In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Blevins S. J., et al. , How structural adaptability exists alongside HLA-A2 bias in the human αβ TCR repertoire. Proc. Natl. Acad. Sci. U.S.A. 113, E1276–E1285 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rossjohn J., et al. , T cell antigen receptor recognition of antigen-presenting molecules. Annu. Rev. Immunol. 33, 169–200 (2015). [DOI] [PubMed] [Google Scholar]
- 37.Hawse W. F., et al. , Peptide modulation of class I major histocompatibility complex protein molecular flexibility and the implications for immune recognition. J. Biol. Chem. 288, 24372–24381 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Borbulevych O. Y., et al. , T cell receptor cross-reactivity directed by antigen-dependent tuning of peptide-MHC molecular flexibility. Immunity 31, 885–896 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hopkins J. R., et al. , Peptide cargo tunes a network of correlated motions in human leucocyte antigens. FEBS J. 287, 3777–3793 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Boehr D. D., Nussinov R., Wright P. E., The role of dynamic conformational ensembles in biomolecular recognition. Nat. Chem. Biol. 5, 789–796 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ayres C. M., Riley T. P., Corcelli S. A., Baker B. M., Modeling sequence-dependent peptide fluctuations in immunologic recognition. J. Chem. Inf. Model. 57, 1990–1998 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ayres C. M., et al. , Dynamically driven allostery in MHC proteins: Peptide-dependent tuning of class I MHC global flexibility. Front. Immunol. 10, 966 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jiang J., et al. , Crystal structure of a TAPBPR-MHC I complex reveals the mechanism of peptide editing in antigen presentation. Science 358, 1064–1068 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McShan A. C., et al. , Peptide exchange on MHC-I by TAPBPR is driven by a negative allostery release cycle. Nat. Chem. Biol. 14, 811–820 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Borbulevych O. Y., Piepenbrink K. H., Baker B. M., Conformational melding permits a conserved binding geometry in TCR recognition of foreign and self molecular mimics. J. Immunol. 186, 2950–2958 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Abualrous E. T., et al. , The carboxy terminus of the ligand peptide determines the stability of the MHC class I molecule H-2Kb: A combined molecular dynamics and experimental study. PLoS One 10, e0135421 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Romero P., et al. , CD8+ T-cell response to NY-ESO-1: Relative antigenicity and in vitro immunogenicity of natural and analogue sequences. Clin. Cancer Res. 7(suppl. 3)766s–772s (2001). [PubMed] [Google Scholar]
- 48.Chen J.-L., et al. , Structural and kinetic basis for heightened immunogenicity of T cell vaccines. J. Exp. Med. 201, 1243–1255 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cooper A., Dryden D. T. F., Allostery without conformational change. A plausible model. Eur. Biophys. J. 11, 103–109 (1984). [DOI] [PubMed] [Google Scholar]
- 50.Smock R. G., Gierasch L. M., Sending signals dynamically. Science 324, 198–203 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Motlagh H. N., Wrabl J. O., Li J., Hilser V. J., The ensemble nature of allostery. Nature 508, 331–339 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hawse W. F., et al. , Cutting edge: Evidence for a dynamically driven T cell signaling mechanism. J. Immunol. 188, 5819–5823 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rangarajan S., et al. , Peptide-MHC (pMHC) binding to a human antiviral T cell receptor induces long-range allosteric communication between pMHC- and CD3-binding sites. J. Biol. Chem. 293, 15991–16005 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Natarajan K., et al. , An allosteric site in the T-cell receptor Cβ domain plays a critical signalling role. Nat. Commun. 8, 15260 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Garstka M. A., et al. , Tapasin dependence of major histocompatibility complex class I molecules correlates with their conformational flexibility. FASEB J. 25, 3989–3998 (2011). [DOI] [PubMed] [Google Scholar]
- 56.van Hateren A., Bailey A., Elliott T., Recent advances in major histocompatibility complex (MHC) class I antigen presentation: Plastic MHC molecules and TAPBPR-mediated quality control. F1000 Res. 6, 158 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kim J., et al. , A dynamic hydrophobic core orchestrates allostery in protein kinases. Sci. Adv. 3, e1600663 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ahuja L. G., Aoto P. C., Kornev A. P., Veglia G., Taylor S. S., Dynamic allostery-based molecular workings of kinase:peptide complexes. Proc. Natl. Acad. Sci. U.S.A. 116, 15052–15061 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Borbulevych O. Y., Santhanagopolan S. M., Hossain M., Baker B. M., TCRs used in cancer gene therapy cross-react with MART-1/Melan-A tumor antigens via distinct mechanisms. J. Immunol. 187, 2453–2463 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rödström K. E., Elbing K., Lindkvist-Petersson K., Structure of the superantigen staphylococcal enterotoxin B in complex with TCR and peptide-MHC demonstrates absence of TCR-peptide contacts. J. Immunol. 193, 1998–2004 (2014). [DOI] [PubMed] [Google Scholar]
- 61.Wiseman T., Williston S., Brandts J. F., Lin L.-N., Rapid measurement of binding constants and heats of binding using a new titration calorimeter. Anal. Biochem. 179, 131–137 (1989). [DOI] [PubMed] [Google Scholar]
- 62.Beechem J. M., Global analysis of biochemical and biophysical data. Methods Enzymol. 210, 37–54 (1992). [DOI] [PubMed] [Google Scholar]
- 63.Turnbull W. B., Daranas A. H., On the value of c: Can low affinity systems be studied by isothermal titration calorimetry? J. Am. Chem. Soc. 125, 14859–14866 (2003). [DOI] [PubMed] [Google Scholar]
- 64.Heemskerk M. H. M., et al. , Redirection of antileukemic reactivity of peripheral T lymphocytes using gene transfer of minor histocompatibility antigen HA-2-specific T-cell receptor complexes expressing a conserved alpha joining region. Blood 102, 3530–3540 (2003). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Crystallographic structure data have been deposited in Protein Data Bank (6VM7, 6VMA, 6VM9, and 6VMC). All study data are included in the article and supporting information.