Significance
Wnt activation is critically important during both early development and throughout adulthood for the maintenance and regeneration of tissue-resident stem cells. However, the underlying mechanism regulating Wnt-dependent epithelial renewal and differentiation in the stomach remains poorly understood. Here we apply genome-scale CRISPR-Cas9 knockout screening to three-dimensional ex vivo stomach organoids to reveal important mechanistic insight into Wnt-dependent epithelial renewal in mouse gastric epithelia. Our organoid-based screening strategy can be readily applied to a wide variety of organoids derived from different sources, potentially providing important insights on multiple biological phenomena.
Keywords: CRISPR-Cas9, genome-wide screening, Wnt, tissue stem cells, Lgr5
Abstract
An ability to safely harness the powerful regenerative potential of adult stem cells for clinical applications is critically dependent on a comprehensive understanding of the underlying mechanisms regulating their activity. Epithelial organoid cultures accurately recapitulate many features of in vivo stem cell-driven epithelial renewal, providing an excellent ex vivo platform for interrogation of key regulatory mechanisms. Here, we employed a genome-scale clustered, regularly interspaced, short palindromic repeats (CRISPR) knockout (KO) screening assay using mouse gastric epithelial organoids to identify modulators of Wnt-driven stem cell-dependent epithelial renewal in the gastric mucosa. In addition to known Wnt pathway regulators, such as Apc, we found that KO of Alk, Bclaf3, or Prkra supports the Wnt independent self-renewal of gastric epithelial cells ex vivo. In adult mice, expression of these factors is predominantly restricted to non-Lgr5–expressing stem cell zones above the gland base, implicating a critical role for these factors in suppressing self-renewal or promoting differentiation of gastric epithelia. Notably, we found that Alk inhibits Wnt signaling by phosphorylating the tyrosine of Gsk3β, while Bclaf3 and Prkra suppress regenerating islet-derived (Reg) genes by regulating the expression of epithelial interleukins. Therefore, Alk, Bclaf3, and Prkra may suppress stemness/proliferation and function as novel regulators of gastric epithelial differentiation.
The mouse stomach comprises a proximal, nonglandular region not present in humans, and a more distal glandular region that can be anatomically and functionally subdivided into the pylorus and corpus regions (1). The epithelial lining of these glandular regions is composed of multiple tubular invaginations called gastric units, which each comprise a pit, isthmus, and base domain, and regularly self-renew throughout life. In the pylorus, active stem cell populations at the gland base marked by the Wnt target gene Lgr5 effect the homeostatic renewal of specialized epithelial cells responsible for secreting protective mucins and hormones, such as gastrin, that regulate acid and zymogen secretion in the corpus (2). Epithelial renewal in the corpus region is less well understood, but is thought to be largely driven by isthmus-resident stem cell populations during homeostasis (3). Quiescent Lgr5-expressing populations at the gland base are rapidly recruited to function as reserve stem cells following the injury-driven loss of the homeostatic stem cell pool (4). Lgr5 is instrumental in modulating R-spondin–mediated regulation of canonical Wnt signaling on stem cells in the glandular stomach as an essential prerequisite to balanced self-renewal and differentiation in vivo (5).
Development of three-dimensional epithelial culture systems capable of supporting the growth of highly stable, self-renewing primary tissues that more accurately recapitulate the composition and functionality of the tissue of origin has revolutionized the study of epithelial biology (6, 7). In the stomach, epithelial organoid formation is selectively driven by primary glandular Lgr5-expressing cells, further underscoring their endogenous stem cell identity (3, 4). Like their in vivo counterparts, the ex vivo stem cells display an absolute dependence on canonical Wnt signals to regulate their ability to orchestrate self-renewal and differentiation within the organoids. However, despite these observations, detailed mechanistic insight into the regulation of stem cell renewal versus differentiation by Wnt signaling in the stomach is currently lacking.
While the organoid culture system doesn’t fully mimic the homeostatic in vivo microenvironment, it is by far the most physiological ex vivo model of stem cell-driven epithelial renewal available (8). Together with the ready accessibility of its genome for performing phenotypic screening, the organoid provides a powerful model for dissecting the mechanism of Wnt-driven regulation of epithelial renewal in the stomach.
Genome-wide, targeted loss-of-function pooled screens using the clustered, regularly interspaced, short palindromic repeats (CRISPR)-associated nuclease Cas9 (CRISPR-Cas9) in human and mouse cells provide an alternative screening system to RNA interference (RNAi) (9). The genome-scale CRISPR knockout (GeCKO) pooled library was firstly reported by Sanjana et al. (10), and several libraries were subsequently developed (11–13). These genome-wide screening techniques have been applied to several cancer cell lines to clarify the mechanism driving malignancy (14, 15). Although CRISPR-Cas9-positive screens have been applied to colorectal organoids (16, 17), an application to normal gastric organoids to identify the mechanism that regulates stem cell-driven epithelial renewal has not been reported.
Here, we combined the GeCKO screening system and gastric organoid culture techniques to investigate the underlying mechanism by which Wnt signaling regulates self-renewal versus differentiation to maintain the integrity of the gastric epithelium. We established corpus and pylorus organoids from mouse normal stomach and confirmed that active Wnt signaling was necessary to maintain the self-renewal of those organoids. We subsequently applied the GeCKO screening method to those normal gastric organoids. Using next-generation sequencing, we found that anaplastic lymphoma kinase (Alk) Bclaf1 and Thrap3 family member 3 (Bclaf3), and Protein activator of interferon induced protein kinase Eif2ak2 (Prkra) are Wnt signal suppressors. In accordance with this, those factors present on non-stem cell compartments in the mouse stomach. We further reveal that these factors regulate the balance between proliferation and differentiation by suppressing Wnt pathway activity, and expression of Cd44 and regenerating islet-derived (Reg) family genes. Collectively, Alk, Bclaf3, and Prkra may directly influence cell renewal/cell death in vivo gastric epithelium.
Results
Establishment of a GeCKO Screening System Using Gastric Organoids.
To decipher the underlying mechanisms regulating Wnt-dependent epithelial renewal in the stomach, we established a GeCKO screen using normal mouse gastric organoids derived from both the corpus and pylorus regions (Fig. 1A).
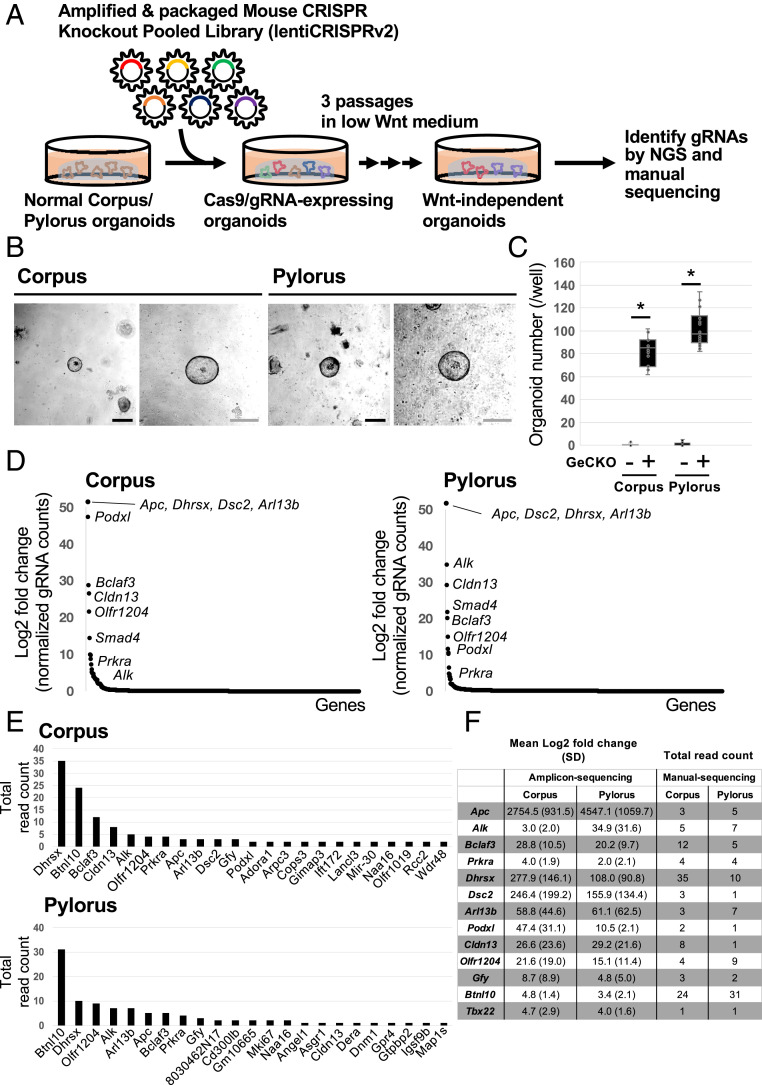
Fig. 1.
Establishment of GeCKO screening in normal gastric organoids. (A) Schematic of the GeCKO screening method to reveal novel regulators of Wnt-dependent epithelial renewal. (B) Images of representative gastric organoids grown in the low Wnt medium after infection with the GeCKO library. In total, six independent experiments were performed. (Scale bars, 50 µm.) (C) Wnt-independent organoid number per well (n = 6). *P < 0.05. (D) Enriched gRNA targeted genes from amplicon-sequencing analyses. (E) Enriched gRNA targeted genes from TA cloning/sanger sequencing analyses. Bar charts show total read count from three independent experiments. (F) Common genes identified from amplicon-sequencing and TA cloning-Sanger sequencing analyses.
We first established normal corpus and pylorus gastric organoids from Lgr5DTR-EGFP reporter mice, as shown in SI Appendix, Fig. S1A (4). Consistent with previous reports, these organoids could be cultured more than 3 mo without major morphological change, confirming their long-term stability (SI Appendix, Fig. S1B). Importantly, the organoids could be propagated from single cells, further underscoring the long-term maintenance of epithelial stem cells in this culture system (SI Appendix, Fig. S1C).
Next, to confirm which signal is essential for the maintenance of those organoids, we individually depleted the growth factors present in the culture medium. We confirmed that Wnt stimuli are essential for maintaining the growth of normal gastric organoids (SI Appendix, Fig. S1D). This is consistent with the in vivo situation, where it has been reported that the Wnt/R-spondin pathway is essential for the maintenance of stem cell-driven epithelial renewal (5). To better define the underlying mechanism by which Wnt/R-spondin regulates stem cell-driven epithelial renewal in the stomach, we decided to perform genome-wide GeCKO screening in gastric organoids under Wnt/R spondin-depleted conditions.
The GeCKO library comprises two libraries, A and B. Each library contains 3 independent gRNAs for 20,611 individual protein-coding genes and 1,000 control non-target guide RNAs (gRNAs). For this study, we used library A as it also contains gRNAs targeting 1,175 miRNAs and using library B doesn’t increase genome coverage. To perform the genome-wide KO screening, we determined multiplicity of infection (MOI) of lenti-GeCKO libraries for the screening (SI Appendix, Fig. S2). According to a previous report (14), we decided to use 50% lentivirus solution for the large-scale screen.
Identification of Candidates Regulating Wnt-Dependent Epithelial Renewal in Gastric Organoids.
To identify factors regulating Wnt-dependent renewal in gastric epithelia, we infected gastric corpus/pylorus organoids as single cells with GeCKO A lentivirus particles, and selected reconstituted organoids with puromycin for 2 d. We then transferred the puromycin-resistant organoids to a low WNT medium (5% L-WRN+Noggin) and cultured them for three passages according to standard procedures described in Materials and Methods and Fig. 1A. In contrast to parental corpus/pylorus organoids, which died within two passages under low Wnt conditions, 80 to 100 GeCKO-infected organoids continued to grow after more than three passages per well (Fig. 1 B and C). To identify the genes whose depletion facilitated the organoid growth under low Wnt conditions, we collected surviving organoids and examined gRNA sequences inserted in their genome by direct amplicon-sequencing and TA-cloning/manual sequencing methods. We accordingly identified a list of gRNA sequences that were commonly detected in both corpus and pylorus organoids (Fig. 1 D–F). Importantly, gRNA’s targeting KO of Apc, a critical negative regulator of Wnt signaling activity were identified in the screen, validating the utility of the GeCKO screening approach for revealing modulators of Wnt-driven epithelial renewal in the stomach. Other gRNA targets in the list identified have not previously been linked to the negative regulation of Wnt-dependent epithelial renewal, highlighting the novelty of our screening results (Fig. 1F).
KO of Alk, Bclaf3, and Prkra Induces Wnt-Independent Stem Cell Maintenance.
We considered genes listed in Fig. 1F as candidate repressors of Wnt-dependent epithelial renewal and activators of differentiation in gastric epithelia. Indeed, using immunohistochemistry (IHC) and qPCR, we found that Alk, Bclaf3, and Prkra are endogenously expressed within the upper regions of mouse corpus and pyloric gastric glands, outside the Lgr5-expressing stem cell zone (SI Appendix, Fig. S3). We accordingly chose these genes for further validation.
To validate the GeCKO screening results, we independently knocked out each gene using plasmid-based CRISPR/Cas9 (18). We cloned independent gRNAs into the pX330 plasmid, transfected them into both corpus and pylorus organoids by lipofection and subsequently selected for an ability to grow under Wnt limiting culture conditions (Fig. 2A). After three passages, we confirmed the gene KO efficiency by Western blotting (Fig. 2B) and evaluated whether the single gene KO is sufficient to confer Wnt independency (Fig. 2C). Apc KO abrogated exogenous Wnt dependency in both corpus and pylorus organoids. This result reconfirms that corpus and pylorus organoids critically depend on active Wnt signals for their proliferation (SI Appendix, Fig. S1D). By quantifying the number and size of surviving organoids, we confirmed that Alk, Bclaf3, and Prkra single KO was sufficient to confer Wnt independency in both corpus/pylorus organoids (Fig. 2 D and E).
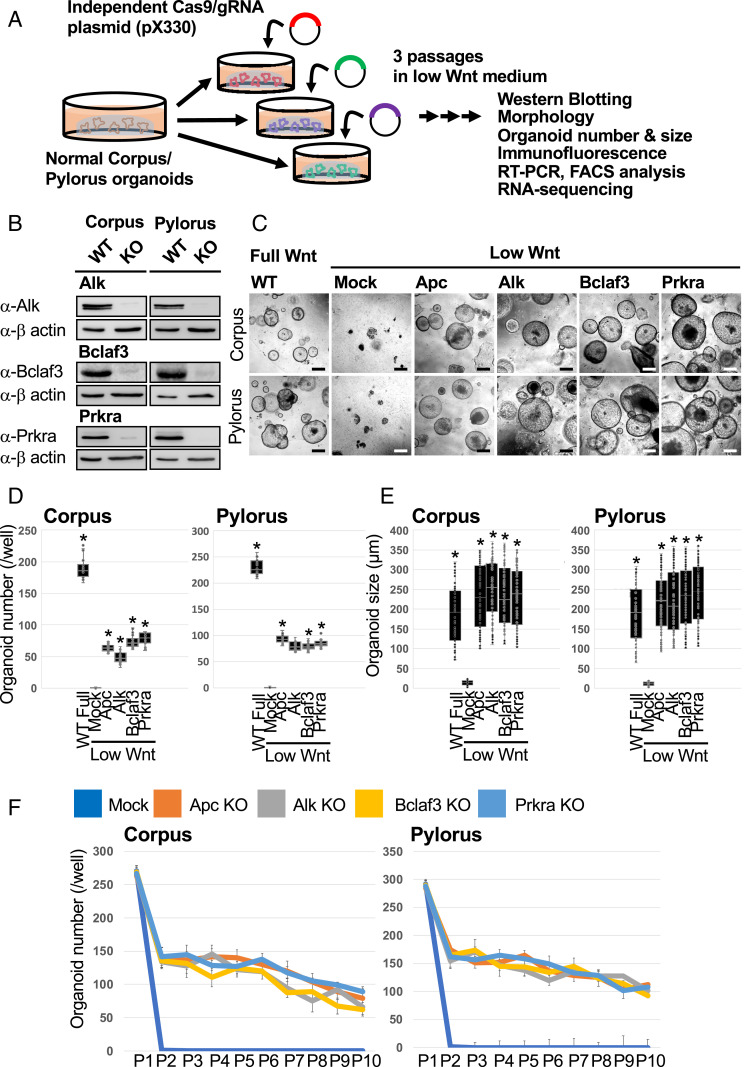
Fig. 2.
KO of Alk, Bclaf3, and Prkra supports organoid survival under low Wnt conditions. (A) Validation of GeCKO screening results. Individual gRNAs from GeCKO library were cloned into the CRISPR-Cas9 plasmid (pX330) and transduced into gastric organoids. Organoids were selected under low Wnt conditions for three passages. (B) Confirmation of the KO efficiency of each gene by Western blotting. (C) Representative images of single-gene KO organoids which proliferated under low Wnt conditions. (Scale bars, 200 µm.) (D) Organoid numbers under low Wnt conditions. n = 6. *P < 0.05 compared to Mock. (E) Organoid size under low Wnt conditions. n = 6. *P < 0.05 compared to Mock. (F) Organoid numbers during long-term passages. For each passage, one well was split into three wells.
Although the number of KO organoids generated in the Wnt-depleted media was lower in comparison to the Wnt-supplemented conditions, it was comparable to that obtained following Apc KO. Interestingly, once organoids were formed, the surviving KO organoids grew at the same rate as those cultured under Wnt-supplemented conditions, highlighting the ability of the individual gene KO to efficiently circumvent the strict dependence on exogenous Wnt ligands (Fig. 2E). This was further confirmed by the ability of these KO organoids to grow for more than 10 passages under low Wnt conditions (Fig. 2F).
To exclude any potential off-target CRISPR-Cas9 gRNA contributions to the observed phenotype, we repeated the gene KO experiments using independently designed gRNAs (SI Appendix, Fig. S4A and Table S1). The independent gRNA set also induced Wnt independency in the gastric organoids, confirming the role of Alk, Bclaf3, and Prkra in epithelial renewal ex vivo (SI Appendix, Fig. S4 B–D). Of note, KO of Axin1 or Axin2, known negative regulators of endogenous Wnt signaling, did not confer Wnt independency on gastric organoid cultures (SI Appendix, Fig. S4B). Axin1 and Axin2 are functionally redundant (19) and for the screen, we set the MOI at 0.4 to silence only one gene per cell. This explains why Axin1 or Axin2 were not identified as Wnt regulators in our KO-screen.
Collectively, these observations strongly indicate that the single KO of Alk, Bclaf3, or Prkra is sufficient to circumvent the strict dependency of gastric organoids for exogenous Wnt ligands.
To further characterize these Wnt ligand-independent organoids, we examined the expression of cell lineage, proliferation, and apoptosis markers by qPCR and immunofluorescence (IF) staining. We found that KO of Alk, Bclaf3, and Prkra increased expression of Wnt target genes and stem cell markers Lgr5, Axin2, and Gif, while decreasing expression of a pit mucus cell marker, Muc5ac under low Wnt conditions in comparison to empty vector-transfected control organoids (Fig. 3A). Furthermore, KO of these genes enhanced cell proliferation, as visualized by increased Mki67 expression and elevated expression of the stem cell marker Cd44, while decreasing apoptotic cell death (Fig. 3 A and B and SI Appendix, Fig. S5).
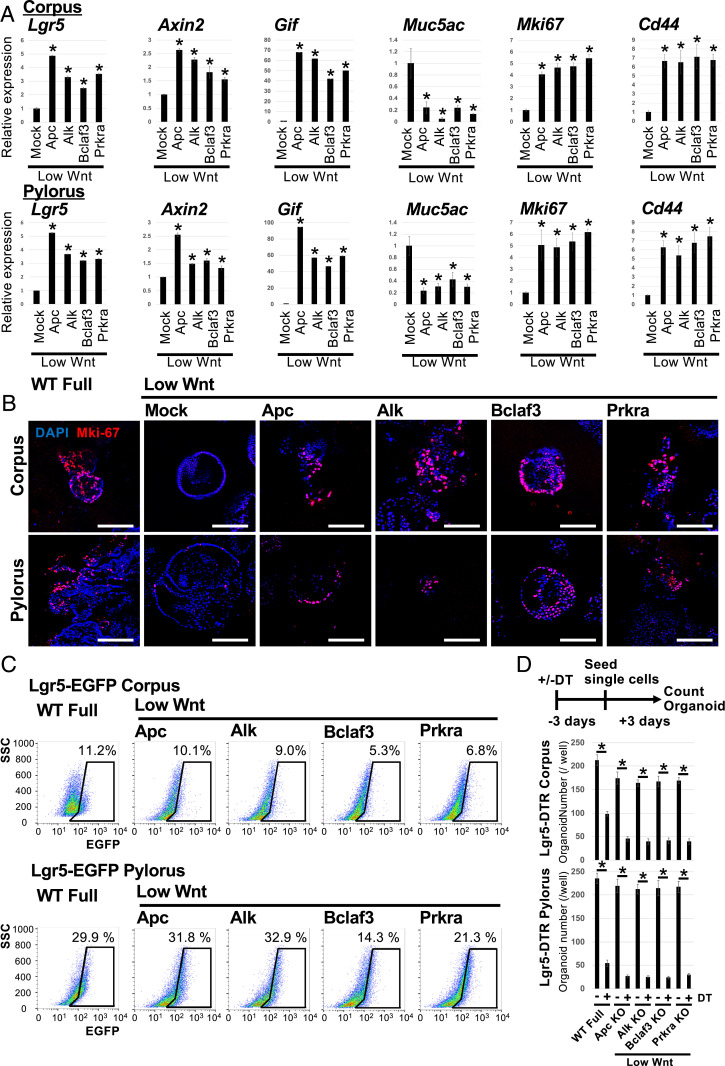
Fig. 3.
Characterization of Alk, Bclaf3, and Prkra KO organoids growing under low Wnt conditions. (A) The expression level of lineage markers and a proliferation marker, Mki-67. Mock-transfected organoids were cultured under low Wnt conditions for 3 d and Apc, Alk, Bclaf3, and Prkra KO organoids were maintained for three passages under low Wnt conditions. The expression level of lineage markers was confirmed by qPCR. *P < 0.05 compared to Mock. (B) Representative IF images of proliferation marker, Mki-67. (Scale bars, 200 µm.) (C) FACS analysis of Lgr5+ stem cells in organoids maintained under low Wnt conditions for three passages. (D) Results of organoid outgrowth assay combined with DT treatment. Candidate KO organoids were initially cultured under low Wnt conditions for more than three passages and subsequently treated with DT for 72 h before reseeding the organoids as single cells to evaluate their outgrowth efficiency relative to untreated controls. *P < 0.05.
Next, we directly evaluated the effect of Alk, Bclaf3, and Prkra KO on Lgr5+ stem cells via FACsorting and targeted ablation. FACs analysis of Lgr5-EGFP expression showed that KO organoids retained Lgr5+ gastric stem cells even under low Wnt conditions (Fig. 3C). To validate whether functional Lgr5+ stem cells are maintained in these Wnt-independent organoids, we performed organoid outgrowth assays following targeted ablation of Lgr5+ cells via diphtheria toxin (DT) treatment of Lgr5DTR organoids. The outgrowth efficacy from DT-treated KO organoids was dramatically reduced relative to untreated KO organoids, suggesting that Alk, Bclaf3, and Prkra KO maintains functional Lgr5+ stem cells to support organoid growth even under low Wnt conditions (Fig. 3D).
Collectively, these results indicate that KO of Alk, Bclaf3, or Prkra supports organoid growth under Wnt limiting conditions by maintaining the resident Lgr5+ stem cell population.
Deciphering Underlying Mechanisms of Wnt-Independent Cell Proliferation in Gastric Organoids.
Next, we examined the mechanism by which the novel factors regulate Wnt-driven, stem cell-dependent epithelial renewal in gastric organoids. First, to evaluate whether these factors simply function as negative regulators of Wnt signaling, we knocked out each gene and maintained organoids in low Wnt medium for three passages. The activation status of the Wnt pathway was subsequently determined by quantifying active β-catenin (nonphospho-β-catenin) levels via Western blotting. KO of Apc and Alk in both corpus and pylorus organoids increased active β-catenin levels even under low Wnt ligand conditions (Fig. 4A). Similar modulation of Wnt activity by APC and ALK was observed via TOPFlash reporter assays following KO in HEK293T cells (Fig. 4B and SI Appendix, Fig. S6A). These results indicate that Alk is a negative regulator of Wnt signaling in gastric epithelial cells, while Bclaf3 and Prkra are not. It has been reported that Alk phosphorylates and activates Gsk3β in mouse neural crest explants and neuroblastoma cell lines (20). Therefore, we tested whether Alk indeed phosphorylates Y279-GSK3β in stomach epithelial cells. As expected, KO of Alk decreased the level of phosphorylated Y279-GSK3β, suggesting that Alk negatively regulates Wnt signaling in the stomach by phosphorylating the tyrosine of GSK3β to facilitate its activation (Fig. 4C).
Fig. 4.
Alk, but not Bclaf3 or Prkra suppresses canonical Wnt signaling in gastric organoids. (A) Confirmation of active (nonphospho) β-catenin levels in each KO clone by Western blotting. (B) TOPFlash assay using HEK293T cells. Each gene was knocked out in HEK293T and Wnt pathway activity measured by TOPFlash assay. F, FOP signal; T, TOP signal. *P < 0.05. (C) Confirmation of phospho Y279 (active)-Gsk3β level in each KO clone by Western blotting. (D) β-Catenin (Ctnnb1) KD by siRNA. KD efficiency of Ctnnb1 in each KO clone was confirmed by qPCR. N; Nontarget siRNA, T; Ctnnb1-target siRNA. *P < 0.05. (E) Organoid number of candidate gene KO/Ctnnb1 KD lines under low Wnt conditions. *P < 0.05.
Although Bclaf3 and Prkra KO organoids could also adapt to growth in medium depleted for Wnt/R-spondin (SI Appendix, Fig. S6B), there was no associated increase in the level of active β-catenin in the gastric organoids, or concomitant activation of TOPFlash in HEK293T cells (Fig. 4 A and B). This suggests that Bclaf3 and Prkra regulate epithelial renewal independently of canonical Wnt signaling. To further substantiate this hypothesis, we additionally knocked down Ctnnb1 in Apc, Alk, Bclaf3, and Prkra KO organoids and maintained them under low Wnt conditions for three passages (Fig. 4D). We reasoned that if Bclaf3 and Prkra KO organoids still depend on the Wnt pathway, the knockdown (KD) of β-catenin should inhibit cell growth significantly. As expected, Ctnnb1 KD in Apc and Alk KO organoids robustly inhibited organoid growth, confirming their dependence on active Wnt signaling. In contrast, both Bclaf3 and Prkra KO organoids continued to proliferate and expand at rates comparable to nontarget small interfering RNA (siRNA) transfected control organoids, even under low Wnt/β-catenin conditions (Fig. 4E). We further evaluated whether the Wnt signal inhibitors, IWR-1-endo, XAV939, and IWP-2 can suppress the proliferation of Bclaf3 and Prkra KO organoids. We cultured Bclaf3 or Prkra KO organoids without Wnt/R-Spondin in the presence of the Wnt inhibitors for three passages. In contrast to the WT/full Wnt condition, Bclaf3 and Prkra KO organoids continued to proliferate and expand (SI Appendix, Fig. S6C). Collectively, these results imply that KO of Bclaf3 and Prkra stimulates renewal of normal gastric organoids independent of Wnt signaling.
To better define the mechanism of action of the factors, we performed RNA-sequencing (RNA-seq) of Apc, Alk, Bclaf3, and Prkra KO organoids. We generated Apc, Alk, Bclaf3, and Prkra KO corpus organoids via the plasmid-based CRISPR-Cas9 system and selected clones under low Wnt conditions for three passages before performing RNA-seq (Fig. 2A). Using RNA-seq data, we performed principal component analysis (PCA) and hierarchical cluster analyses (Fig. 5 A and B). We found that the profile of Alk KO is similar to Apc KO rather than Bclaf3 KO or Prkra KO. This further supports the observation that Alk KO regulates gastric stem cells mainly via activation of the Wnt pathway (Fig. 5 A and B). We also found that a set of genes and pathways (e.g., cadherin signaling, inflammation-mediated chemokine and cytokine signaling) were commonly up-regulated in those KO organoids (Fig. 5C and SI Appendix, Fig. S7A). Importantly, some of the genes identified in the Lgr5+ stem cell transcriptome were also up-regulated in Alk, Bclaf3, and Prkra KO organoids (Fig. 5D). Therefore, those genes and pathways may be important for the maintenance of epithelial stem cells, and endogenous Alk, Bclaf3, and Prkra could be instrumental in directing cell differentiation by suppressing those genes and pathways.
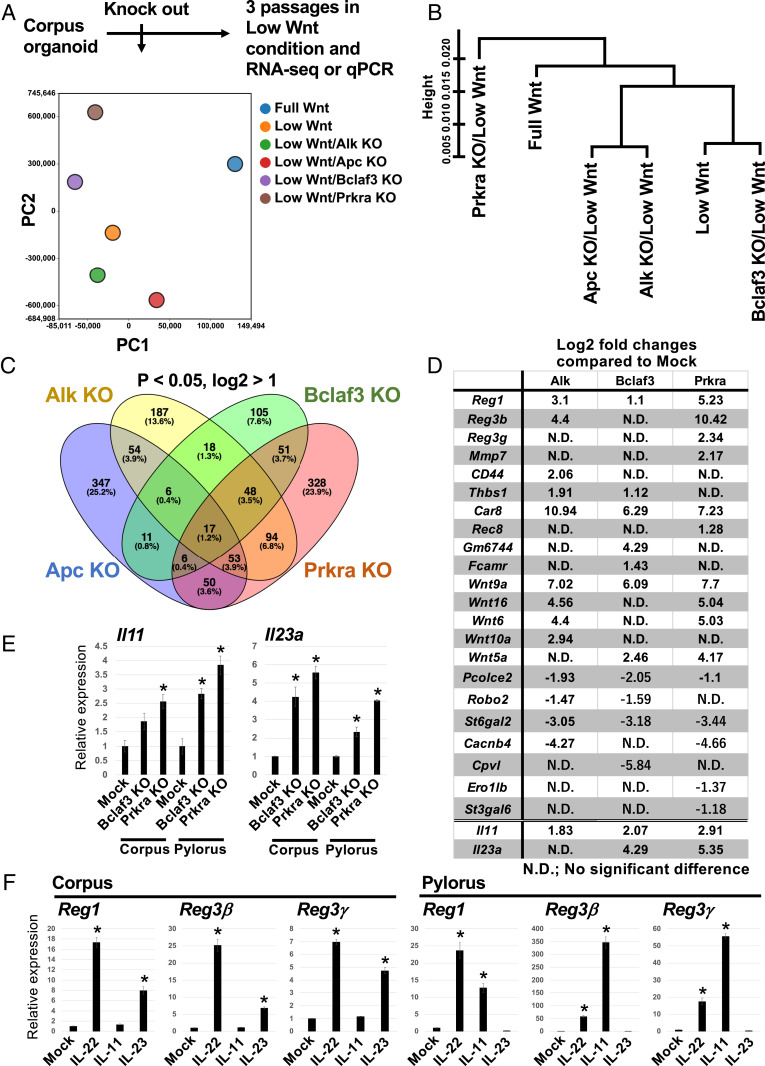
Fig. 5.
Defining the mechanism of action of Bclaf3 and Prkra KO on gastric organoid renewal/renewal. (A) Schematic of the RNA-seq experiment and results of PCA analysis and (B) hierarchical cluster analysis. (C) A Venn diagram summarizing the number of up-regulated genes in each KO clone. (D) An up-regulated gene list for each KO clone. Listed genes are selectively enriched on Lgr5+ stem cells (4). (E) Expression level of interleukins in each KO clone. *P < 0.05 compared to Mock. (F) Expression level of Reg genes induced by administration of interleukins. IL-22 is a positive control (42). *P < 0.05 compared to Mock.
Reg Family Genes Promote Cell Proliferation under Wnt-Limiting Conditions.
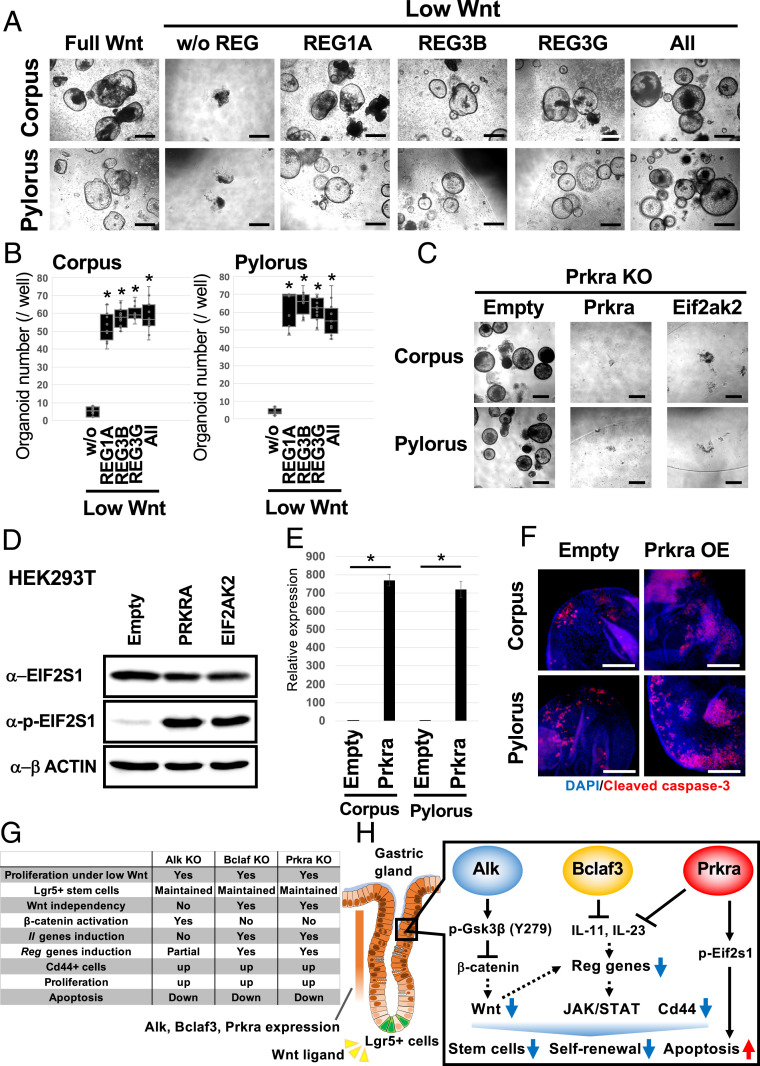
Among the commonly up-regulated genes, we focused on Reg family genes, which encode small secretory proteins. Several Reg family members have been shown to promote cell proliferation and prevent apoptosis in diverse cell types (21). Reg gene expression was significantly increased in the KO organoids compared to WT organoids when cultured for three passages under Wnt-limiting conditions (Fig. 5D and SI Appendix, Fig. S7B). Mechanistically, we found that epithelial interleukins (ILs), Il11 and Il23a, are robustly up-regulated by Bclaf3 and Prkra KO (Fig. 5 D and E). Although Wnt signaling partially regulates Reg gene expression (SI Appendix, Fig. S7C), IL-11 and Il-23 significantly induce Reg gene expression in stomach organoids (Fig. 5F). Additionally, endogenous Reg gene expression was strongest at the base of gastric glands in vivo, where adult stem cells reside (SI Appendix, Fig. S8). We therefore examined whether up-regulation of Reg gene family expression contributes to the sustenance of KO organoid growth under Wnt-limiting conditions. We cultured normal corpus and pylorus organoids for three passages in low Wnt/R-spondin medium supplemented with individual recombinant REG proteins (Fig. 6A). In contrast to the lack of organoid growth in low Wnt/R-Spondin media, both corpus and pylorus organoids continued to expand in REG protein-supplemented media (Fig. 6B). Notably, concomitant addition of the various recombinant REGs failed to further enhance organoid growth, suggesting that Reg1, Reg3β, and Reg3γ are functionally redundant for the growth of epithelial cells.
Fig. 6.
Exogenous Reg protein reduces the Wnt dependency of gastric organoids, while Prkra overexpression promotes apoptosis. (A) Representative images of normal gastric organoids passaged three times in the presence of Reg recombinant proteins. (Scale bars, 200 µm.) (B) Organoid numbers in the presence of Reg proteins. n = 6. *P < 0.05 compared to without sample. (C) Induction of Prkra or a downstream gene, Eif2ak2 counteracts the effect of Prkra KO suggesting that Prkra KO inhibits the apoptosis induced by the Prkra-Eif2ak2-Eif2s1 axis. (Scale bars, 200 μm.) (D) Increased phospho-EIF2S1 levels following PRKRA and EIF2AK2 overexpression in HEK293T cells. Overexpression of Prkra and Eif2ak2 induced apoptosis in organoids; therefore, we tested the effect of overexpression in HEK293T cells. (E) Confirmation of Tet-on induction of Prkra expression in corpus and pylorus organoids. *P < 0.05. (F) Detection of an apoptosis marker, Cleaved caspase-3 after 24 h of Prkra induction. (Scale bars, 100 µm.) (G) Summary of ex vivo organoid studies and (H) a model of the in vivo situation. Alk, Bclaf3, and Prkra expressed on differentiated compartments suppress canonical Wnt signaling, expression of Reg genes and the stem cell marker Cd44. Additionally, Prkra regulates apoptosis. Concerted actions of these signals ensure the strict regulation of Wnt-driven, stem cell dependent epithelial renewal in the gastric gland.
Similar positive effects on epithelial proliferation were observed following overexpression of Reg family genes in gastric organoids (SI Appendix, Figs. S9 A and B and S10A). Those organoids could be maintained for at least five passages under low Wnt/R-Spondin conditions (SI Appendix, Fig. S9C). Of note, elevated Reg activity leads to increased expression of the epithelial stem cell marker gene Lgr5 and proliferation markers such as Mki67 (SI Appendix, Fig. S10B). Furthermore, we confirmed that KO of Reg genes significantly reduced organoid numbers even under full Wnt culture conditions, suggesting that Reg genes are indispensable for gastric organoid growth (SI Appendix, Fig. S9E).
Collectively, these observations identify Reg proteins as a family of IL-11/IL-23–responsive regulators of gastric epithelial renewal through their ability to partially substitute for canonical Wnt signaling.
Prkra KO Blocks Cell Death by Limiting Phosphorylation of Eif2s1 Proteins.
In gastric epithelia, apoptotic cell death is typically observed within the upper region of gastric glands under homeostatic conditions (22). It has been reported that Prkra induces the phosphorylation of Eif2s1, leading to cell death (23). We therefore reasoned that KO of Prkra might promote epithelial expansion ex vivo through suppression of cell death via inhibition of endogenous Eif2s1 phosphorylation. To examine this possibility, we overexpressed Prkra and Eif2ak2, a serine/threonine-protein kinase which transmits stress signals by phosphorylated Eif2s1 in Prkra KO gastric organoids, and cultured them for three passages under Wnt-limiting conditions. We showed that overexpression of Prkra and Eif2ak2 counteracted the organoid growth observed following Prkra KO (Fig. 6C). Since constitutive overexpression of Prkra and Eif2ak2 via PiggyBac vectors induced rapid cell death of normal gastric organoids, we overexpressed PRKRA and EIF2AK2 in HEK293T cells and confirmed associated phosphorylation of EIF2S1 (Fig. 6D). Furthermore, to test whether Prkra directly induces apoptosis, we temporally induced Prkra and stained those organoids with the apoptosis marker, cleaved caspase-3. As a result, we confirmed that induction of Prkra increased apoptosis in normal gastric organoids (Fig. 6 E and F). This observation is consistent with the KD data in SI Appendix, Fig. S5B. These results suggest that Prkra KO can support epithelial renewal by suppressing apoptotic cell death via limiting phospo-Eif2s1 levels.
Our data collectively reveal that Alk, Bclaf3, and Prkra function to suppress Wnt-related and Reg family genes, and enhance apoptosis to regulate epithelial renewal in the stomach. These observations readily explain why KO of these genes enhances epithelial expansion independently of exogenous Wnt signals in our organoid screen (Fig. 6G).
Discussion
Wnt signaling is of critical importance for maintaining the integrity of gastric epithelia via regulation of endogenous stem cell function (5). In mouse gastric epithelia, resident Lgr5-expressing populations at the gland base in the pylorus and corpus function as homeostatic and injury-activated stem cell populations respectively (3, 4). Lgr5, which is itself a Wnt target gene, functions as a facultative component of the Wnt receptor complex, where it is thought to facilitate binding of the Wnt agonist, R-spondin, to modulate Wnt signaling levels activated by Wnt ligands provided by surrounding mesenchyme to a level compatible with optimal stem cell function (24). However, detailed mechanistic insight into the regulation of Wnt signaling and the delicate balance between self-renewal and differentiation in the stomach is currently lacking. To try and address this knowledge gap, we established a GeCKO screening technique on normal gastric organoids and identified factors that potentially regulate either endogenous Wnt signaling or cell differentiation in mouse gastric epithelia. We found that KO of Alk, Bclaf3, and Prkra supports Wnt-independent renewal and reduces apoptosis in gastric epithelia, while concomitantly inducing expansion of the Lgr5+ stem cell population. Taken together with the observation that these factors are predominantly expressed outside the Lgr5+ stem cell zone in mouse glandular epithelia, this indicates they may function to suppress stemness and maintain/promote the differentiation status of gastric epithelial cells. Mechanistically, these factors suppress Wnt signaling, integrin signaling, and inflammation mediated by the chemokine and cytokine signaling pathways. Of note, we found that Alk can function as a kinase for Y279-Gsk3β to suppress canonical Wnt signaling in stomach epithelial cells. In contrast, Bclaf3 and Prkra may negatively regulate expression of Reg family genes via modulation of epithelial interleukins to suppress stemness/proliferation in gastric epithelia. Although further verification is needed, results here suggest that Alk, Bclaf3, and Prkra may function as regulators of gastric epithelial differentiation.
We show that Alk is predominantly expressed outside the Lgr5+ gastric stem cell zone and functions to suppress endogenous Wnt signaling. A similar result was observed in HEK293T cells, indicating that Alk may be a Wnt antagonist in different tissues. In cancer, Alk has been reported as a common fusion partner for other genes to promote tumor malignancy (25), although it is not known whether this involves modulation of the Wnt pathway. Our results indicate that Alk itself can suppress Wnt signaling in ex vivo gastric organoids, potentially by phosphorylating Y279-Gsk3β to promote the degradation of β-catenin as a mechanism to ensure terminal differentiation toward the various functional lineages that populate the epithelial glands.
Our data also implicate Bclaf3 and Prkra as regulators of gastric epithelial growth. Some Wnt ligands were up-regulated by Bclaf3 and Prkra KO, resulting in the Wnt signaling pathway being identified as one of the top up-regulated pathways. However, our IWP-2 treatment results indicate that these up-regulated Wnts may not be sufficient to maintain gastric renewal. It has been reported that Bclaf3 binds to the Oct-Sox binding motif and is necessary to maintain pluripotency of mouse embryonic stem cells (mESCs) (26). In contrast to this report, we found that Bclaf3 expression is present in non-stem cells and may induce differentiation in gastric glands by suppressing Wnt signaling and Reg family gene expression. Importantly, our results suggest that Bclaf3 and Prkra likely regulate epithelial differentiation by controlling expression of members of the Reg gene family, which are known to be potent modulators of epithelial renewal and stem cell maintenance as discussed below.
Several Reg family genes are expressed in the endogenous gastric stem cell compartment (4, 27). Indeed, Reg has a growth-promoting effect and may play an important role in the reconstitution of gastric mucosa (22). Reg KO mice present significantly decelerated growth and migration of intestinal epithelial cells toward the villus tip (28). In support of those observations, our Reg KO experiment revealed that Reg gene expression is indispensable for the growth of stomach organoids.
It has been reported that Reg expression is controlled by Wnt signaling in mESCs (29). Consistent with this, we confirmed that KD of β-catenin decreased Reg gene expression in stomach organoids. Furthermore, REG expression is regulated by Gastrin and also IL-6, IL-22 in human gastric cancer lines (30, 31). In this study, we found that Bclaf3/Prkra KO induced Il11 and Il23a expression, and the administration of those interleukins up-regulated the Reg gene expression in stomach organoids. Mechanistically, the gp130 transmembrane protein is thought to be a Reg receptor and the gp130-JAK2-STAT3 axis promotes cell growth and resistance to apoptosis, as well as inducing poorly differentiated cells in pancreatic cancer cells (32). Therefore, there is a possibility that the Wnt and interleukin-Regs-JAK/Stat3 signaling cascade is essential for the renewal of gastric epithelia.
While administration of REGs cannot fully compensate for Wnt withdrawal, Reg genes clearly supported organoid growth. Supplementation of gastric organoid media with all three recombinant REGs failed to show a synergistic effect on organoid growth. This result suggests that the Reg family gene displays functional redundancy during epithelial renewal.
Our data, together with the mutually exclusive expression pattern of Bclaf3/Prkra and Reg genes within the non-stem cell and stem cell compartments, respectively, of mouse gastric glands in vivo, supports a model in which the absence of Bclaf3/Prkra expression at the gland base facilitates robust Reg gene expression to support Lgr5+ stem cell function. Of note, we found that Bclaf3 and Prkra may inhibit the expression of Reg family genes by suppressing interleukins in stomach organoids. Therefore, expression of Bclaf3/Prkra outside the stem cell zone may help facilitate terminal differentiation toward the specialized lineages of the gastric epithelia. The role of Reg family genes in gastric epithelia warrants further investigation in future.
Although conditional KO of Alk, Bclaf3, and Prkra in the stomach has not been reported, constitutive KO of each gene does not generate obvious gastric phenotypes. Individual KO of those genes in gastric organoids up-regulates common pathways, suggesting that Alk, Bclaf3, and Prkra likely exhibit some functional redundancy in the stomach. One of those pathways may be up-regulation of Cd44. Cd44 is a nonkinase transmembrane glycoprotein thought to be induced in several types of stem cells including cancer-initiating cells (33, 34). Cd44 activates cell proliferation, survival and modulates cytoskeletal changes to enhance cellular motility (35). Therefore, Cd44 may also have a role in regulating stemness in the normal gastric gland.
KO of the factors may also influence gastric epithelial growth via suppression of apoptosis. It has been reported that Prkra induces apoptosis via phosphorylation of Eif2s1 in both human and mouse cell lines (24, 36). Furthermore, Prkra is expressed in colonic epithelial cells, where it activates apoptosis (37). Here, we confirm that Prkra induces apoptosis in ex vivo gastric organoids (Fig. 5 E and F). Although massive induction of Prkra caused apoptosis, endogenous expression of Prkra in the gastric epithelium was modest. Given that Alk and Bclaf3 may also influence apoptosis, these factors may work cooperatively to balance epithelial renewal/cell death in vivo.
Our genome-wide organoid screen has revealed factors involved in regulating Wnt signaling, Reg gene expression and proliferation/apoptosis in the mouse gastric epithelium (Fig. 6G). Future studies will be directed at further refining the role of these factors in regulating the balance between self-renewal and differentiation in healthy gastric epithelium, and the potential contributions of these factors to gastric cancer. Given that healthy/cancer organoids can now be readily generated from multiple tissues of mouse and human origin, the screening procedure detailed should prove useful in deriving clinically relevant biological insights across a range of healthy and disease settings.
Materials and Methods
Mice.
Lgr5DTR-EGFP mice have been described previously (38). The line is bred as heterozygotes. All animal experimental protocols were approved by the Committee on Animal Experimentation of Kanazawa University.
Preparation of L-WRN–Conditioned Medium.
L-WRN cell line was purchased from ATCC (ATCC CRL-3276) and maintained in DMEM (Wako) with 10% FBS and 1× penicillin/streptomycin (Nacalai Tesque) (39). L-WRN–conditioned medium was prepared according to the protocol provided by ATCC.
Organoid Culture.
Organoid culture was performed as described previously with minor modifications (4). Briefly, 2- to 3-mm tissue fragments from murine corpus and pylorus were incubated in 1× PBS supplemented with 5 mM and 10 mM EDTA (Corning) at 4 °C for 2 and 3 h, respectively. Glands were isolated by repeated pipetting of finely chopped tissues in ice-cold 1× PBS supplemented with EDTA. Next, 1× PBS containing isolated glands was filtered through 100-µm filter mesh and centrifuged at 720 × g at 4 °C for 3 min. Whole gastric glands were embedded in GFR Matrigel (Corning) and cultured in basal media (Advanced DMEM/F-12 media with 10 mM Hepes, 2 mM Glutamax, 1× N2, 1× B27 (Invitrogen), N-acetyl-cysteine (Sigma-Aldrich) supplemented with growth factors, EGF (Peprotech), Gastrin (Wako), FGF10 (Peprotech), L-WRN–conditioned medium and passaged every 3 d. For passage, organoids were collected with Recovery Solution (Corning), dissociated by gentle pipetting and transferred to Matrigel. For low Wnt medium, the L-WRN–conditioned medium was reduced to 5% and supplemented with Noggin (Peprotech) and FBS.
For organoid out-growth assays, organoids were cultured for three passages in 5% L-WRN supplemented with Noggin. Parental organoids were cultured in 50% L-WRN conditions. Lgr5+ cells were ablated by treatment with 50 ng/mL DT for 3 d, before being dissociated. Next, 5,000 cells were seeded in a well (three technical triplicates × two biological replicates). DT treatment was continued throughout the assay.
Canonical Wnt signal inhibitors, IWR-1-endo (Wako) and XAV939 (Sigma-Aldrich) were added to the medium at 5 µM and 1 µM, respectively. The porcupine inhibitor, IWP-2 (Wako) was added to the medium at 2 µM. Recombinant Human REG1A, REG3b, and REG3G (R&D Systems) were added at 500 µg/mL Recombinant Human IL-11, IL-22, and IL-23 (R&D Systems) were added at 10 ng/mL
HEK293T Cell Culture.
HEK293T cell line was maintained in DMEM (Wako) with 10% FBS (Sigma-Aldrich) and 1× penicillin/streptomycin (Nacalai Tesque). Passaging was performed using TrypLE (ThermoFisher Scientific) according to standard protocols.
Preparation of Lenti-GeCKO Library.
Mouse GeCKOv2 CRISPR knockout pooled library was a gift from Feng Zhang (Addgene #1000000052). To prepare lentivirus particles, the Mouse GeCKOv2 CRISPR KO pooled library plasmids were cotransfected with packaging plasmids pCMV-VSV-G and pCMV-dR8.2 dvpr (gifts from Bob Weinberg, Addgene plasmid #8485 and #8455) into HEK293T cells. Briefly, 80% confluent HEK293T cells in 100 mm dishes were transfected in OptiMEM (Life Technologies) using 6 µg of mouse GeCKOv2 CRISPR knockout pooled library plasmids, 1.5 µg pCMV-VSV-G, 4.5 µg pCMV-dR8.2 dvpr, 60 µL of 1 µg/mL PEI MAX (Polysciences). After 16 h, media was changed to Advanced DMEM/F-12 with 1× B-27 and 1× GlutaMax. After 72 h, viral supernatants were harvested and centrifuged at 1,000 rpm at 4 °C for 5 min to pellet cell debris. The supernatant was filtered through a 0.45-µm membrane filter (Sartorius) and aliquots were stored at −80 °C until use.
The GeCKO library is divided into two sublibraries, A and B. Both libraries comprise three gRNAs targeting each gene, as well as 1,000 non-targeting control gRNAs. Library A also contains gRNAs targeting noncoding micro RNAs. Normal corpus and pylorus organoids were infected at a low MOI (<0.4) to ensure that most cells receive only one viral construct with high probability (40). Briefly, organoids were dissociated by TrypLE (ThermoFisher Scientific) and cells were mixed with the lentivirus solution and 8 µg/mL polybrene (Sigma Aldrich). The sample was centrifuged at 600 × g at 32 °C for 60 min and incubated at 37 °C for 6 h. After incubation, cells were collected and embedded in matrigel with medium containing Y-27632 (Wako) and CHIR99021 (Cayman Chemical).
To determine MOI, cells were counted and each well was split into duplicate wells. One replicate received 1 µg/mL puromycin (Invivogen). After 3 d, cells were counted to calculate percent transduction. Percent transduction is calculated as cell count from the replicate with puromycin divided by cell count from the replicate without puromycin multiplied by 100. The virus volume yielding a MOI closest to 0.4 was chosen for large-scale screening.
For the large-scale screening, 24 h after transduction, organoids were selected with 1 µg/mL puromycin and 5% L-WRN supplemented media for three passages. After three passages, only organoids transduced with lenti-CRISPR constructs and can proliferate in the absence of Wnt signal were preserved. These organoids were collected for use in subsequent analyses. We independently repeated this screening steps three times.
Identification of Integrated gRNA.
Amplicon sequencing.
The primer sets HT-gLibrary-Miseq_150bp-PE-F: TCGTC GGCAGCGTCAGATGTGTATAAGAGACAGcttgaaagtatttcgatttcttgg and HT-gLibrary-Miseq_150bp-PE-R: GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGactcggtgccactttttcaa were used to amplify the gRNA locus. The PCR condition was 95 °C (20 s), 55 °C (20 s), and 72 °C (30 s) for 26 cycles. Q5 Taq polymerase (New England Biolabs) was used. Secondary PCR was performed using Nextra XT index primers (Illumina). The secondary PCR condition was 95 °C (20 s), 55 °C (20 s), and 72 °C (30 s) for 12 cycles. PCR products were purified using a PCR purification kit (Hokkaido System Science) and sequenced on a Next-Seq. 500 (Illumina) system (75-bp single-end reads). Sequence reads were trimmed and quality-filtered using Trim Galore! (v0.4.4), Trimmomatic (v0.36), and Cutadapt (v1.16). Frequency of occurrence of each contig sequence was calculated using the table function of R (v3.4.1).
Manual sequencing.
Inserted gRNA sequences were amplified with the following primers: LentiCRISPR v2 gRNA scaffold F; 5′- TGCATGCTCTTCAACCTCAA -3′, LentiCRISPR v2 gRNA scaffold R; 5′-CCAATTCCCACTCCTTTCAA-3′. PCR fragments were gel extracted and cloned with the pGEM-T easy Vector system (Promega) and transfected into XL10Gold competent cells (Agilent). After blue/white selection, only white colonies which contain an amplified sequence were picked and plasmids subsequently extracted. Cloned fragments were sequenced by the standard sanger sequencing method using T7 and SP6 universal primers. We sequenced at least 100 plasmids from each screening.
KO of individual genes using CRISPR-Cas9.
Identified gRNAs in organoids which survived under low Wnt conditions were cloned into pX330-U6-Chimeric_BB-CBh-hSpCas9 (a gift from Feng Zhang, Broad Institute of MIT and Harvard, Cambridge, MA, Addgene #42230) and transfected into organoids and HEK293T cells with Lipofectamine LTX or Lipofectamine 2000 (ThermoFisher Scientific), respectively, according to the manufacturer’s instructions. Plasmids were cotransfected with a CAG-EGFP-IRES-Puror vector and cells were selected by 1 µg/mL puromycin for 3 d. Additional gRNAs which were not listed in the GeCKO library were designed by the CHOPCHOP software. KO efficiency was confirmed by Western blotting. For Reg1, Reg3β, and Reg3γ triple KO, three pX330 plasmids harboring Reg-targeting gRNAs were transfected to organoids together with the CAG-EGFP-IRES-Puror vector. gRNAs used this study are listed in SI Appendix, Table S1.
Overexpression of Prkra, Eif2ak2, Reg1, Reg3β, and Reg3γ.
Prkra and Eif2ak2 genes were amplified from a cDNA pool prepared from mouse normal gastric organoids. Reg1, Reg3β, and Reg3γ cDNA plasmids were provided by the RIKEN BRC through the National BioResource Project of the MEXT/AMED, Japan. cDNAs were cloned into pPBCAG-IRES-Puror, a PiggyBac-based puromycin resistant constitutive expression vector or pPBhCMV*1-pA, a PiggyBac-based inducible expression vector (gifts from H. Niwa, Kumamoto University, Kumamoto, Japan). These vectors were transfected into organoids derived from normal stomach of Lgr5DTR-EGFP mice, together with pPyCAG-PBase vector and pPBCAG-rtTA3G-IN vector as described previously (4). After 1 wk of Puromycin (1 μg/mL; InVivogen) or G418 (500 μg/mL; Wako) selection, pooled clones were used for subsequent experiments.
Histology.
IHC and IF.
IHC was performed according to standard protocols. Briefly, tissues were fixed in 4% paraformaldehyde/PBS (wt/vol) overnight and processed into paraffin blocks. Four-micrometer sections from the paraffin blocks were deparaffinated and rehydrated, followed by antigen retrieval via heating in a microwave in standard 10 mM citric acid pH6 buffer. Primary antibodies used were anti-Reg1α (1:100; Abcam), anti-Reg3γ (1:100; LSBio). To detect signals, ImmPRESS Reagent (Vector Laboratories) was used. IHC sections were dehydrated, cleared, and mounted with EUKITT mounting medium (ORSAtec). Immunostaining was performed on a minimum of three biological replicates and representative images of the replicates were included in the manuscript. Sections were imaged on a microscope (Leica).
For organoid staining, IF was performed in Matrigel. Briefly, organoids were fixed in 4% paraformaldehyde/PBS (wt/vol) for 20 min and permeabilized in 0.5% Triton X-100/PBS at 4 °C for 1 h. After blocking with 10% normal donkey serum, primary antibodies used were anticleaved caspase-3 (1:200; Cell Signaling), anti-Ki67 (1:100; BioScience), anti-CD44 (1:200; Millipore). To detect signals, an Alexa 594-conjugated second antibody (1:500; Invitrogen) was used. Stained organoids were mounted with Vectashield antifade containing DAPI (Dako) for imaging. Immunostainings were performed on a minimum of three biological replicates and representative images of the replicates were included in the manuscript. Sections were imaged on a confocal microscope (Leica).
H&E.
H&E staining was performed on FFPE sections according to standard protocols.
RNA In situ hybridization.
RNA in situ hybridization was performed using RNAscope 2.5 High Definition Brown Assay (Advanced Cell Diagnostics) according to the manufacturer’s instructions. DapB was used as a negative control for all the RNAscope experiments. In situ hybridization and imaging was performed on a minimum of three biological replicates and representative images of the replicates were included in the report. Probes for each gene were obtained from Advanced Cell Diagnostics.
RNA-seq.
RNA-seq library preparation, sequencing, mapping, gene expression and gene ontology (GO) enrichment analysis were performed by DNAFORM. Quality of total RNA was assessed by Bioanalyzer (Agilent) to ensure that RNA integrity number was above 7.0. Double-stranded cDNA libraries (RNA-seq libraries) were prepared using SMARTer Stranded Total RNA-seq Kit v2 – Pico Input Mammalian (Clontech) according to the manufacturer’s instruction. RNA-seq libraries were sequenced using a HiSeq (Illumina), as 150-bp PE. Obtained reads were mapped to the mouse mm9 genome using STAR (v2.7.2b). Reads on annotated genes were counted using featureCounts (v1.6.1). Fragments per kilobase of transcript per million mapped reads (FPKM) values were calculated from mapped reads by normalizing to total counts and transcript. Differentially expressed genes were detected using the DESeq2 package (v1.20.0). The list of differentially expressed genes detected by DESeq2 (basemean > 5 and fold-change < 0.25, or basemean > 5 and fold-change > 4) were used for GO enrichment analysis by clusterProfiler package (41). Pathway analysis was performed using PANTHER software (http://pantherdb.org). PCA analysis was performed by BioVinci software (https://vinci.bioturing.com).
β-Catenin KD.
ON-TARGET plus SMARtpool siRNAs for Ctnnb1 and ON-TARGET plus SMARTpool nontargeting siRNAs pool (Horizon Discovery) were transfected into organoids at 1 µM. Briefly, organoids were dissociated by TrypLE (ThermoFisher Scientific) and cells were mixed with siRNA mixtures and Lipofectamine RNAi Max (ThermoFisher Scientific). The cells were centrifuged at 600 × g at 32 °C for 60 min and incubated at 37 °C for 6 h. After incubation, cells were collected and embedded in Matrigel with medium containing Y-27632 (Wako) and CHIR99021 (Cayman Chemical). After 24 h, KD efficiency was confirmed by qPCR. To evaluate cell growth, organoids were maintained for 3 passages without Y-27632 and CHIR99021.
TOPFlash assay.
M50 Super 8x TOPFlash and M51 Super 8x FOPFlash plasmids (gifts from Randall Moon, Addgene #12456 and 12457) were transfected into HEK293T cells with Lipofectamine 2000 (ThermoFisher Scientific) according to the manufacturer’s instructions. The next day, luciferase activity was measured using the ONE-Glo luciferase assay system (Promega). Protein quantification was also performed using Pierce 660 nm protein assay reagents (ThermoFisher Scientific). Luciferase activity was normalized against total protein.
qPCR.
Total RNA was extracted from organoids by RNeasy Mini Kit or Micro Kit (Qiagen) and cDNA generated with PrimeScript RT reagent Kit (Perfect Real Time) (Takara) according to the manufacturer’s instructions. qPCR was performed with a minimum of three biological replicates per gene using THUNDERBIRD SYBR qPCR Mix (Toyobo) according to the manufacturer’s instructions and ran on Mx3000P Real-Time QPCR System (Agilent). Analysis was carried out using double CT method. Primers used this study are listed in SI Appendix, Table S1.
Western blotting.
Western blotting was performed according to standard protocols. Cells were lysed in lysis buffer supplemented with protease inhibitors (Sigma-Aldrich) and a phosphatase inhibitor (ThermoFisher Scientific). Lysates were centrifuged at 12,000 rpm for 5 min at 4 °C. Supernatants were quantified using Pierce 660 nm Protein Assay Reagent (ThermoFisher Scientific). Lysates were denatured at 95 °C for 5 min before gel electrophoresis on 8 to 12% acrylamide gels. Proteins were then transferred to PVDF membranes at 20 V, 400 mA for 150 min at 4 °C. Membranes were blocked by incubation with 5% skimmed milk/1× TBS-T. Primary antibodies used: anti–β-Actin (1:2,000; Sigma Aldrich), anti-Alk (1:100; SantaCruz), anti-Bclaf3 (1:300; Biorbyt), anti-Prkra (1:1,000; GeneTex), antinon-Phospho (active) β-catenin (1:1,000; Cell Signaling), anti–β-Catenin (1:1,000; Cell Signaling), anti-elF2α (1:1,000; Cell Signaling), antiphospho-elF2α (1:1,000; Cell Signaling), anti-Gsk3β (1:5000; R&D Systems), anti-phopho Y279-Gsk3β (1:500; Abcam). Membranes were developed by Amersham ECL select (GE Lifesciences) and imaged using ImageQuant LAS 4000 (GE Lifesciences).
FACS analysis.
Organoids were dissociated in TrypLE (ThermoFisher Scientific) for 5 min at 37 °C. Digestion was quenched by DMEM 10% FCS medium. The suspension was centrifuged at 300 × g for 5 min. The pellet was resuspended in 1× PBS supplemented with 2% FBS (HyClone) and 1 µg/mL 7-AAD (Life Technologies), filtered through a 40-µm strainer, and analyzed by BD FACS CantoII or BD FACS AriaIII (BD Biosciences).
Statistical Methods.
Data were quantified and depicted as the mean ± SD. Statistical analysis was performed using an unpaired t test. Significant difference is denoted as *P < 0.05.
Supplementary Material
Acknowledgments
We thank Dr. Fred de Sauvage (Genentech) for the Lgr5DTR-EGFP mice; Dr. Hitoshi Niwa (Kumamoto University) for PiggyBac vectors; Dr. Feng Zhang (Broad Institute of MIT and Harvard) for the Mouse GeCKOv2 CRISPR knockout pooled library and the pX330-U6-Chimeric_BB-CBh-hSpCas9 plasmid; Dr. Bob Weinberg (Whitehead Institute for Biomedical Research) for pCMV-VSV-G and pCMV-dR8.2 dvpr plasmids; Dr. Randall Moon (University of Washington School of Medicine) for M50 Super 8x TOPFlash and M51 Super 8x FOPFlash plasmids; Dr. Dominic Voon (Kanazawa University) for recombinant human IL-23; and Ms. Ayako Tsuda and Ms. Manami Watanabe for technical assistance. K.M. is supported by Japan Society for the Promotion of Science Grant 17K07161. Y.T. is supported by Japan Society for the Promotion of Science Grant 17H06710. H.T. is supported by Japan Society for the Promotion of Science Grant 17H03586. N.B. is supported by the Agency for Science, Technology and Research (A*STAR); the National Research Foundation, Prime Minister’s Office, 510 Singapore under its Investigatorship Program (Award NRF-NRF12017-03); and Japan Society for the Promotion of Science Grant 17H01399. Funding for the open access charge is from Japan Society for the Promotion of Science Grant 17H01399.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2016806118/-/DCSupplemental.
Data Availability.
Amplicon sequencing and RNA-seq data have been deposited in the National Center for Biotechnology Information BioProject database (accession nos. PRJNA645063 and PRJNA646064). All study data are included in the article and supporting information.
References
- 1.Kim T.-H., Shivdasani R. A., Stomach development, stem cells and disease. Development 143, 554–565 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tan S., Barker N., Epithelial stem cells and intestinal cancer. Semin. Cancer Biol. 32, 40–53 (2015). [DOI] [PubMed] [Google Scholar]
- 3.Barker N., et al. , Lgr5(+ve) stem cells drive self-renewal in the stomach and build long-lived gastric units in vitro. Cell Stem Cell 6, 25–36 (2010). [DOI] [PubMed] [Google Scholar]
- 4.Leushacke M., et al. , Lgr5-expressing chief cells drive epithelial regeneration and cancer in the oxyntic stomach. Nat. Cell Biol. 19, 774–786 (2017). [DOI] [PubMed] [Google Scholar]
- 5.Tan S. H., Barker N., Wnt signaling in adult epithelial stem cells and cancer. Prog. Mol. Biol. Transl. Sci. 153, 21–79 (2018). [DOI] [PubMed] [Google Scholar]
- 6.Clevers H., Modeling development and disease with organoids. Cell 165, 1586–1597 (2016). [DOI] [PubMed] [Google Scholar]
- 7.Fatehullah A., Tan S. H., Barker N., Organoids as an in vitro model of human development and disease. Nat. Cell Biol. 18, 246–254 (2016). [DOI] [PubMed] [Google Scholar]
- 8.Huch M., Koo B.-K., Modeling mouse and human development using organoid cultures. Development 142, 3113–3125 (2015). [DOI] [PubMed] [Google Scholar]
- 9.Shalem O., Sanjana N. E., Zhang F., High-throughput functional genomics using CRISPR-Cas9. Nat. Rev. Genet. 16, 299–311 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sanjana N. E., Shalem O., Zhang F., Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods 11, 783–784 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tzelepis K., et al. , A CRISPR dropout screen identifies genetic vulnerabilities and therapeutic targets in acute myeloid leukemia. Cell Rep. 17, 1193–1205 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Doench J. G., et al. , Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat. Biotechnol. 34, 184–191 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Henriksson J., et al. , Genome-wide CRISPR screens in T helper cells reveal pervasive crosstalk between activation and differentiation. Cell 176, 882–896.e18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shalem O., et al. , Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 343, 84–87 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Takeda H., et al. , CRISPR-Cas9-mediated gene knockout in intestinal tumor organoids provides functional validation for colorectal cancer driver genes. Proc. Natl. Acad. Sci. U.S.A. 116, 15635–15644 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ringel T., et al. , Genome-scale CRISPR screening in human intestinal organoids identifies drivers of TGF-β resistance. Cell Stem Cell 26, 431–440.e8 (2020). [DOI] [PubMed] [Google Scholar]
- 17.Michels B. E., et al. , Pooled in vitro and in vivo CRISPR-Cas9 screening identifies tumor suppressors in human colon organoids. Cell Stem Cell 26, 782–792.e7 (2020). [DOI] [PubMed] [Google Scholar]
- 18.Cong L., et al. , Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chia I. V., Costantini F., Mouse axin and axin2/conductin proteins are functionally equivalent in vivo. Mol. Cell. Biol. 25, 4371–4376 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gonzalez Malagon S. G., et al. , Glycogen synthase kinase 3 controls migration of the neural crest lineage in mouse and Xenopus. Nat. Commun. 9, 1126 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen Z., Downing S., Tzanakakis E. S., Four decades after the discovery of regenerating islet-derived (Reg) proteins: Current understanding and challenges. Front. Cell Dev. Biol. 7, 235 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miyaoka Y., et al. , Transgenic overexpression of Reg protein caused gastric cell proliferation and differentiation along parietal cell and chief cell lineages. Oncogene 23, 3572–3579 (2004). [DOI] [PubMed] [Google Scholar]
- 23.Patel C. V., Handy I., Goldsmith T., Patel R. C., PACT, a stress-modulated cellular activator of interferon-induced double-stranded RNA-activated protein kinase, PKR. J. Biol. Chem. 275, 37993–37998 (2000). [DOI] [PubMed] [Google Scholar]
- 24.de Lau W., Peng W. C., Gros P., Clevers H., The R-spondin/Lgr5/Rnf43 module: Regulator of Wnt signal strength. Genes Dev. 28, 305–316 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Soda M., et al. , Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature 448, 561–566 (2007). [DOI] [PubMed] [Google Scholar]
- 26.Chakraborty D., et al. , lncRNA Panct1 maintains mouse embryonic stem cell identity by regulating TOBF1 recruitment to Oct-Sox sequences in early G1. Cell Rep. 21, 3012–3021 (2017). [DOI] [PubMed] [Google Scholar]
- 27.Tan S. H., et al. , AQP5 enriches for stem cells and cancer origins in the distal stomach. Nature 578, 437–443 (2020). [DOI] [PubMed] [Google Scholar]
- 28.Ose T., et al. , Reg I-knockout mice reveal its role in regulation of cell growth that is required in generation and maintenance of the villous structure of small intestine. Oncogene 26, 349–359 (2007). [DOI] [PubMed] [Google Scholar]
- 29.Jing D., Kehoe D. E., Tzanakakis E. S., Expression of Reg family proteins in embryonic stem cells and its modulation by Wnt/β-catenin signaling. Stem Cells Dev. 19, 1307–1319 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ashcroft F. J., Varro A., Dimaline R., Dockray G. J., Control of expression of the lectin-like protein Reg-1 by gastrin: Role of the Rho family GTPase RhoA and a C-rich promoter element. Biochem. J. 381, 397–403 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tsuchida C., et al. , Expression of REG family genes in human inflammatory bowel diseases and its regulation. Biochem. Biophys. Rep. 12, 198–205 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu X., et al. , REG3A accelerates pancreatic cancer cell growth under IL-6-associated inflammatory condition: Involvement of a REG3A-JAK2/STAT3 positive feedback loop. Cancer Lett. 362, 45–60 (2015). [DOI] [PubMed] [Google Scholar]
- 33.Al-Hajj M., Wicha M. S., Benito-Hernandez A., Morrison S. J., Clarke M. F., Prospective identification of tumorigenic breast cancer cells. Proc. Natl. Acad. Sci. U.S.A. 100, 3983–3988 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dalerba P., et al. , Phenotypic characterization of human colorectal cancer stem cells. Proc. Natl. Acad. Sci. U.S.A. 104, 10158–10163 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen C., Zhao S., Karnad A., Freeman J. W., The biology and role of CD44 in cancer progression: Therapeutic implications. J. Hematol. Oncol. 11, 64 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee E.-S., Yoon C.-H., Kim Y.-S., Bae Y.-S., The double-strand RNA-dependent protein kinase PKR plays a significant role in a sustained ER stress-induced apoptosis. FEBS Lett. 581, 4325–4332 (2007). [DOI] [PubMed] [Google Scholar]
- 37.Gupta V., Patel R. C., Proapoptotic protein PACT is expressed at high levels in colonic epithelial cells in mice. Am. J. Physiol. Gastrointest. Liver Physiol. 283, G801–G808 (2002). [DOI] [PubMed] [Google Scholar]
- 38.Tian H., et al. , A reserve stem cell population in small intestine renders Lgr5-positive cells dispensable. Nature 478, 255–259 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miyoshi H., Stappenbeck T. S., In vitro expansion and genetic modification of gastrointestinal stem cells in spheroid culture. Nat. Protoc. 8, 2471–2482 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Konermann S., et al. , Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature 517, 583–588 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yu G., Wang L.-G., Han Y., He Q.-Y., clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS 16, 284–287 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zheng Y., et al. , Interleukin-22 mediates early host defense against attaching and effacing bacterial pathogens. Nat. Med. 14, 282–289 (2008). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Amplicon sequencing and RNA-seq data have been deposited in the National Center for Biotechnology Information BioProject database (accession nos. PRJNA645063 and PRJNA646064). All study data are included in the article and supporting information.