Abstract
Background
Considering that the Lactobacillus casei group is strongly associated with caries progression, the use of lactobacilli as probiotics must be balanced due to their possible involvement in dental caries.
Objective
This study aimed to detect and quantify L. paracasei, L. rhamnosus, and L. casei group species in the active and arrested dentinal lesions of preschoolers. It also aimed to determine the expression profiles of lactobacilli genes related to adhesion, extracellular polymeric substance regulation, and pyruvate oxidation.
Methods
Total ribonucleic acid (RNA) was extracted from dentinal lesion samples (25 active, 13 arrested) of children between 2 and 5 years of age. The samples were converted to complementary deoxyribonucleic acid (cDNA), and quantitative polymerase chain reaction (qPCR) analyses were performed to quantify and determine the relative abundance (measured by percentage of total bacteria) of L. paracasei, L. rhamnosus, and L. casei group species. The expression profiles of L. paracasei/casei genes (spaC and spxB) and L. rhamnosus genes (spaE and wzb) were assessed. The Student t-test and the Mann-Whitney U test were used for comparisons.
Results
The L. casei group species were found to be part of the viable microbial community in dentinal caries. L. paracasei (p = 0.001), L. rhamnosus (p = 0.022), and L. casei (p = 0.004) group species were abundant in the active dentinal lesions compared to the arrested dentinal lesions. Only the wzb gene (p = 0.006) exhibited a statistically significant difference between the active and arrested lesions in terms of its expression profile; it was expressed to a higher extent in the active dentinal lesions.
Conclusions
The L. casei group species presented in large numbers in the active dentinal caries lesions, indicating that these microorganisms are related to caries activity, and the wzb gene may play an important role in caries progression.
Keywords: Dental caries, Child, RNA
1. Introduction
Dental caries is a polymicrobial biofilm-mediated disease (Fejerskov, 2004, Philip et al., 2018) resulting from a dysbiosis caused by frequent sugar consumption (Mira, 2018, Solbiati and Frias-Lopez, 2018, Zhan, 2018). It presents globally (Gao et al., 2016, Tanner et al., 2016) as one of the most common chronic childhood diseases (Jiang et al., 2014, Simón-Soro and Mira, 2015). Early childhood caries (ECC) affects children between 0 and 6 years of age and is a serious public health problem due to its early beginning, rapid clinical development, high treatment cost, and negative effects on preschool children ´s quality of life (Li et al., 2015, Colombo et al., 2017).
Low levels of Streptococcus mutans have been found in dentinal lesions, despite the recognition of its pathogenicity in dental caries (Aas et al., 2008, Simón-Soro et al., 2014). Similar prevalence and quantification of S. mutans have previously been reported in active and arrested dentinal lesions (Bezerra et al., 2016), suggesting that S. mutans may play an accessory role in dentinal caries progression. Lactobacillus spp., a late colonizer that is not necessary for caries initiation (Young and Featherstone, 2013, Obata et al., 2014, Takahashi and Nyvad, 2016), has been frequently detected in ECC, being more closely related to dentinal caries lesions than to enamel lesions (Badet and Thebaud, 2008, Li et al., 2015, Shimada et al., 2015, Mitrakul et al., 2017) and more numerous in children with ECC than in cavity-free children (Ledder et al., 2018). The L. casei group (a closely related taxonomic group) formed by L. rhamnosus, L. casei, and L. paracasei (Hill et al., 2019) is a predominant group in the biofilms and dentin lesions of children with ECC (Badet and Thebaud, 2008, Neves et al., 2017), and the L. rhamnosus and L. paracasei species seem to be present at all caries stages of deep carious lesions in deciduous molars (Kneist et al., 2010).
The Lactobacillus casei group is studied mainly due to its marketing, industrial, and health potential (Salvetti et al., 2012), and clear support was recently found for the short-term persistence of Lactobacillus in the saliva microbiome, showing that the ingestion of commercially accessible probiotics may affect the diversity and constitution of the saliva microbiome (Dassi et al., 2018). In addition, several L. paracasei and L. rhamnosus strains, which are used to ferment dairy products and have been widely studied as probiotics, are capable of surviving in the oral cavity (Smokvina et al., 2013, Toh et al., 2013, Surachat et al., 2017, Coqueiro et al., 2018, Pahumunto et al., 2019, Zaura and Twetman, 2019). L. rhamnosus is the most widely used and clinically researched L. casei group probiotic for caries prevention, followed by L. paracasei (Lebeer et al., 2007, Silva et al., 2008, Twetman and Keller, 2012, Cagetti et al., 2013, Seminario-Amez et al., 2017, Coqueiro et al., 2018, Pahumunto et al., 2019, Zaura and Twetman, 2019). However, although lactobacilli present a very restricted capacity to inhabit the healthy human oral cavity, it has been suggested that they might colonize cavity lesions and cause caries (Twetman and Keller, 2012), inducing mineral loss, especially in dentinal cavities, and contributing to in vitro caries processes (Schwendicke et al., 2014). This demonstrates that probiotic species can also be cariogenic under peculiar growth circumstances, such as low pH (Vuotto et al., 2014). Bacteria from the L. casei group can ferment glucose and produce lactic acid, and, depending on the pH, lactate can be metabolized into ethanol, acetic acid, and CO2 (Sharpe, 1979, Salvetti et al., 2012). Beyond acid production, the L. casei group species have a high tolerance for low pH (Obata et al., 2014) and H2O2 production when the spxB gene catalyzes pyruvate oxidation (Zotta et al., 2014, Savo Sardaro et al., 2016, Li et al., 2017). Furthermore, L. paracasei and L. rhamnosus play a role in microbial adhesion and biofilm formation, binding to mucin, collagen, and cultured epithelial cells (Lebeer et al., 2012, Smokvina et al., 2013, Toh et al., 2013, Miljkovic et al., 2015). These bacteria have genes related to transport and carbohydrate metabolism, biosynthesis of extracellular polysaccharides (EPS), production of bacteriocins, and pili (Toh et al., 2013, Ceapa et al., 2016). The protein pilin is encoded by genes (including spaC and spaE) related to adhesion, and the wzb gene is involved in the regulation of EPS synthesis (von Ossowski et al., 2010, Lebeer et al., 2012, Nadkarni et al., 2014, Rintahaka et al., 2014, Miljkovic et al., 2015). L. paracasei and L. rhamnosus have high capacity for adhesion, but few details are known about the adhesion mechanism of these bacteria in dental caries (Piwat et al., 2015, Ciandrini et al., 2017).
In this context, the consumption of lactobacilli probiotics based on the claim that they provide health benefits needs to be pondered, considering their possible contribution to dental caries when associated with sugar intake. It is hypothesized that species of Lactobacillus are associated with dentinal caries, especially in ECC, and have specific genetic elements that enable adhesion, biofilm formation, and production of biocide agents, thus modulating caries activity processes. Therefore, the objective of this study was to detect and quantify 1) the metabolically active cells of the L. casei group and the L. paracasei and L. rhamnosus species and 2) the expression of the spaC, spxB, spaE, and wzb genes of L. paracasei/casei and L. rhamnosus, respectively, in the active and arrested dentinal caries lesions of children with ECC.
2. Materials and methods
2.1. Study population and sample collection
The dentin samples used in this study were derived from a previous study (Bezerra et al., 2016), in which informed consent was obtained from the legally responsible parents, guardians, or custodians of the children after the research protocol was approved by the Ethics Committee of the Federal University of Ceará (protocol no. 548.405). The use of the stored samples was authorized by the Research Ethics Committee of the Federal University of Ceará (protocol no. 3.227.777).
In sum, a group of 32 children aged between 40 and 71 months from four public schools in Fortaleza (Ceará, Brazil) were diagnosed with ECC and selected according to the inclusion criteria: presence of a minimum of one cavitated dentinal carious lesion with visible dentin in teeth with pulpal health, assessed by clinical and radiographic exams. Patients who had any health problems, were not cooperative during clinical examination, or had used antibiotics 3 months before the study were excluded (Bezerra et al., 2016). The carious dentin samples were collected and rapidly transported to sterile RNAse free microtubes (Axygen, Union City, CA, USA), in which they were combined with ribonucleic acid (RNA) stabilization reagent (RNAlaterTM, Ambion Inc., Austin, TX, USA) for 18 h (at 4 °C), following the manufacturer's specifications. Prior to RNA extraction, the microtubes remained at −80 °C.
2.2. RNA extraction and purification
The dentin samples were processed according to the method described by Bezerra et al. (2016). Briefly, all the samples were thawed, centrifuged (11,000 × g/1 min/4 °C), and transferred to cryogenic tubes containing 0.16 g of zirconia beads (0.1 mm in diameter) (Biospec Products, Bartlesville, OK, USA) for mechanical disruption in a mini-beadbeater (BioSpec Products). Supernatants (350 μl) were submitted to RNA extraction using RNeasy Mini KitTM (Qiagen, Valencia, CA, USA) in accordance with the manufacturer’s guidance. Subsequently, genomic deoxyribonucleic acid (DNA) was removed using TurboTM DNAse (Applied Biosystems, Ambion, Austin, TX, USA), and the RNA solution was cleaned using the RNeasy MinieluteTM CleanUp Kit (Qiagen, Dus, Bundesland, Germany). Measurements of RNA concentration (A260/A280) and purity (A260/A230) were determined by absorbance ratios obtained in a spectrophotometer (Nanodrop 2000c, Thermo Scientific, Wilmington, DE, USA).
2.3. cDNA synthesis
The complementary DNA (cDNA) was synthesized from the RNA using the iScriptTM cDNA Synthesis Kit (Bio-Rad, Hercules, CA, USA). Reverse transcription reactions were prepared with 6 μl of 5x iScript reaction mix, 1 μl of iScript reverse transcriptase, 1 μg of purified RNA, and RNAse-free water in a sufficient amount to yield a final volume of 30 μl. The reaction cycle (5 min at 25 °C, 120 min at 42 °C, and 5 min at 85 °C) was carried out in a thermocycler (VeritiTM, Applied Biosystems, Foster City, CA, USA) (Bezerra et al., 2019). Afterward, the cDNA concentration for each sample was normalized to obtain a concentration of 10 ng/μl and then reserved at −20 °C.
2.4. Design of the primers
We designed specific primers for the expression of genes (spxB and spaC) from Lactobacillus casei/paracasei, using Primer3 (http://biotools.umassmed.edu/bioapps/primer3_www.cgi) and BLAST® (https://blast.ncbi.nlm.nih.gov/Blast.cgi). Gene sequences (Table 1) were deposited in the GenBank (https://www.ncbi.nlm.nih.gov/genbank/). The other genes used in this analysis (spaE and wzb) were previously described by Nadkarni et al. (2014).
Table 1.
Primers designed in this study.
| Gene | Locus tag | Description | Primer | Sequence 5́- 3́ | Product (bp) |
|---|---|---|---|---|---|
| spxB | LBPG_02063 | pyruvate oxidase | spxBLbp For spxBLbp Rev |
GTGCCGACGTTATTTCTTG ATCACAACAATCGCAGCTC |
200 |
| spaC | LBPG_02639 | pilus specific protein | spaCLbp For spaCLbp Rev |
GGTCAGGGAGAAGCGTACT CGGTGTGACGACTTACCAT |
202 |
All the primers used in this research (Table 2) were synthesized by Exxtend (Campinas, São Paulo, Brazil); they were analyzed in the BLAST® and Netprimer® (https://www.premierbiosoft.com/netprimer/) target, and the specificity was then confirmed experimentally with polymerase chain reaction (PCR) reactions. The individual standardization for the reactions of the primers was performed with genomic DNA obtained with strains of culture collections being positive controls (L. paracasei for the primers in Table 1 and L. rhamnosus to spaE and wzb). Amplifications were made in volumes of 25 µl with 200 µM of deoxyribonucleotide triphosphates (dNTPs), 2.5 mM of MgCl2, 0.3 µM of each primer, 1.25 U of Taq DNA polymerase (Invitrogen, Carlsbad, CA, USA), and approximately 10 ng of genomic DNA, using a thermocycler (VeritiTM, Applied Biosystems). All the primers were tested in accordance with the preliminary patterns for ideal thermal condition determinations (Table 3): DNA denaturation at 95 °C for 5 min, 35 cycles at 95 °C for 30 sec, primer hybridization at 55 °C (for the primers in Table 1), 58 °C (for spaE and wzb), and 60 °C (for the L. casei group, L. paracasei and L. rhamnosus primers) for 30 sec, extension at 72 °C for 30 sec, and conclusion of the process at 72 °C for 5 min. Tris-borate-EDTA buffer on 2% agarose gel was used for separating the PCR products by electrophoresis, and staining agent ethidium bromide (0.5 ug/ml) was employed for visualizing the bands under ultraviolet (UV) light (Gel Logic 100 Imaging System, Kodak, Tokyo, Japan). For a well-defined examination of all the bands, a 100 bp DNA ladder (Invitrogen, Carlsbad, CA, USA) was included on each gel.
Table 2.
List of primers used in this study.
| Target or genes | Sequence (5′ 3′) | References |
|---|---|---|
| Total Bacteria* | F: TCCTACGGGAGGCAGCAGT R: GGACTACCAGGGTATCTAATCCTGTT |
(Nadkarni et al., 2002) |
| L. casei group* | F: GCGGACGGGTGAGTAACACG R: GCTTACGCCATCTTTCAGCCAA |
(Furet et al., 2004) |
| L. paracasei* | F: GTGCTTGCACCGAGATTCAACATG R: TGCGGTTCTTGGATCTATGCG |
(Furet et al., 2004) |
| L. rhamnosus* | F: GTGCTTGCATCTTGATTTAATTTT R: TGCGGTTCTTGGATCTATGCG |
(Furet et al., 2004) |
| spxB | F: GTGCCGACGTTATTTCTTG R: ATCACAACAATCGCAGCTC |
(This study) |
| spaC | F: GGTCAGGGAGAAGCGTACT R: CGGTGTGACGACTTACCAT |
(This study) |
| wzb | F: CTTGAACGCTGCACTCATCTC R: CGGATTAACGGTCAGTTGTTAGA |
(Nadkarni et al., 2014) |
| spaE | F: TGGCCGTCAATTAACACAAA R: TATGACGCGTAAGCAAGCAC |
(Nadkarni et al., 2014) |
16S rDNA.
Table 3.
Ideal thermal conditions applied for qPCR analysis.
| Genes | Thermal conditions |
||||
|---|---|---|---|---|---|
| Pre-heating | Denaturation | Annealing | Elongation | Cycles | |
| L. rhamnosus* | 50 °C, 2 min/ 95 °C, 10 min | 95 °C, 15 sec | 60 °C, 30 sec | 60 °C, 30 sec | 40 |
| L. paracasei* | |||||
| L. casei group* | 95 °C, 10 min | 95 °C, 15 sec | 60 °C, 30 sec | 60 °C, 30 sec | 40 |
| spaC | 95 °C, 15 min | 95 °C, 15 sec | 55 °C, 30 sec | 60 °C, 60 sec | 40 |
| spxB | |||||
| spaE | 95 °C, 15 min | 95 °C, 15 sec | 58 °C, 30 sec | 60 °C, 60 sec | 40 |
| wzb | |||||
Not determined in this study.
2.5. Quantitative PCR
Quantitative PCR (qPCR) assays were performed both to quantify the 16S rRNA genes of L. rhamnosus, L. paracasei, and L. casei group species (Furet et al., 2004) and to assess the levels of expression of the spaC, spaE, spxB, and wzb genes. Assays of qPCR were also used to determine the ideal concentration and efficiency of all the primers used in this study. Standard curves used serial dilutions from 400 ng to 0.0004 ng (detection limit) (10-fold) of genomic DNA extracted from L. paracasei ATCC 335 and L. rhamnosus ATCC 53103. Standard amplification and melting-point curves were obtained for all the primer sets. The qPCR conditions are specified in Table 3.
The qPCR assays were made in duplicate in a MicroAmp® Fast Optical 48-Well Reaction Plate (Applied Biosystems, Ambion, Austin, TX, USA) covered with optical adhesive film (Applied Biosystems) in a StepOneTM Real-Time PCR System (Applied Biosystems). All the reaction mixtures (10 μl) contained 5 μl of Power SYBRTM Green PCR Master Mix (Applied Biosystems), nuclease-free water (3.4 μl), each forward/reverse primer (0.3 μl), and 1 μl of cDNA (10 ng/μl) or 1 μl of genomic DNA individually placed into the respective wells of a 48-well plate. The final analyses were obtained from the means of the two duplicates. The negative control consisted of reactions without the template. After the final qPCR cycle, analysis of the cycle threshold (CT) values, melting temperature (Tm) values, melting curve, and standard curve (correlation coefficient, R2, and efficiency %) was conducted for all the amplified samples. For all the bacteria, samples with CT and Tm values lower than the detection level in the standard DNA curves were counted as negative (Colombo et al., 2017).
2.6. Statistical analysis
Data analyses were accomplished using the BioEstat 5.3 program with a 95% confidence level. The D'Agostino-Pearson normality test was used to examine the sample distribution patterns, and the Student t-test or the Mann-Whitney U test was used to compare the groups in terms of parametric or nonparametric data, respectively. The response variables L. casei group abundance and wzb gene expression were compared using the Student t-test. Meanwhile, the Mann-Whitney U test was used to compare the abundance of L. paracasei and L. rhamnosus, the proportion of the L. casei group, L. paracasei, and L. rhamnosus in relation to the total bacteria, and the gene expression profiles of spaC, spxB, and spaE.
3. Results
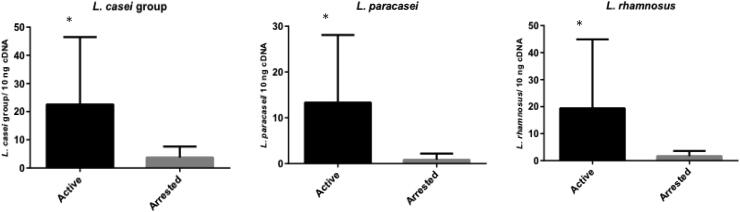
The results showed that L. paracasei and L. rhamnosus are part of the metabolically active community in dentinal caries lesions of children with ECC, since L. paracasei, L. rhamnosus, and L. casei group species were detected in all the samples, regardless of the lesion type. In addition, L. paracasei (p = 0.001), L. rhamnosus (p = 0.022), and L. casei (p = 0.004) group species were more abundant in active dentinal lesions than in inactive dentinal lesions (Fig. 1).
Fig. 1.
Quantification of L. casei group, L. paracasei and L. rhamnosus in active (n = 25) and arrested (n = 13) dentin caries lesions. Data are expressed in bars (means ± standard deviation). *Statistical difference (p < 0.05) between the groups according to Student t-test for L. casei group and Mann-Whitney test for L. paracasei and L. rhamnosus.
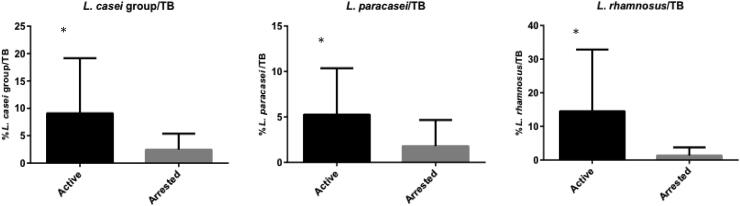
The proportion of L. paracasei (p = 0.022), L. rhamnosus (p = 0.009), and L. casei group species (p = 0.018) in relation to the total bacteria (TB) load was also significantly higher in active dentinal lesions, as illustrated in Fig. 2.
Fig. 2.
Proportion of L. casei group, L. paracasei and L. rhamnosus in relation to the total bacteria (TB). Data are expressed in bars (means ± standard deviation). *Statistical difference (p < 0.05) between the groups according to Mann-Whitney test.
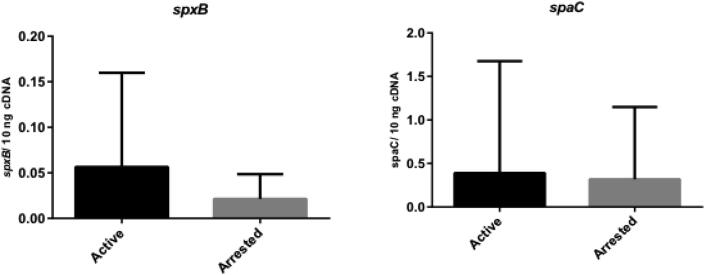
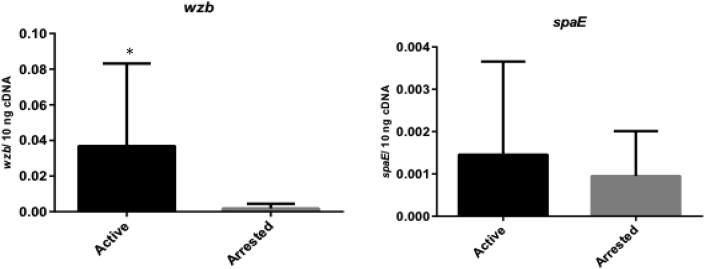
The spaC and spxB genes from the L. paracasei designed in this research were equally expressed in active and arrested lesions (p > 0.05) (Fig. 3). The spaE gene from the L. rhamnosus was also equally expressed in both types of lesions (p > 0.05), but the wzb gene exhibited greater expression in active lesions than in arrested lesions (p = 0.006) (Fig. 4).
Fig. 3.
The spaC and spxB from L. paracasei genes expression in active (n = 25) and arrested (n = 13) dentin caries lesions. Data are expressed in bars (means ± standard deviation). *Statistical difference (p < 0.05) between the groups according to Mann-Whitney test.
Fig. 4.
L. rhamnosus genes (wzb and spaE) expression in active (n = 25) and arrested (n = 13) dentin caries lesions. Data are expressed in bars (means ± standard deviation). *Statistical difference (p < 0.05) between the groups according to Student t-test for wzb and Mann-Whitney test for spaE.
4. Discussion
The current results corroborate those of previous studies, which have revealed that L. rhamnosus, L. paracasei, and L. casei group species are frequently isolated from dentine sites in ECC (Švec et al., 2009, Caufield et al., 2015, Takahashi, 2015), as they are part of the metabolically active bacteria in dentinal caries and probably related to caries progression (Kneist et al., 2010). Caries progression is associated with the increase in disease severity and the frequency of biofilm acidification due to pH reduction (Obata et al., 2014, Takahashi and Nyvad, 2016). Increased dental biofilm acidification results in the proliferation of acidogenic and acidogenic/aciduric strains (such as the L. casei group) in an adaptive manner and the suppression of acid-sensitive species (Jiang et al., 2014, Caufield et al., 2015).
In this research, a greater abundance of L. rhamnosus, L. paracasei, and L. casei group species was observed in active dentinal lesions than in arrested dentinal lesions; this was previously demonstrated in the case of the L. casei group (Neves et al., 2017) when DNA analyses were performed.
These microorganisms ferment glucose, producing lactic acid (Sharpe, 1979, Salvetti et al., 2012), which is the dominant acid in active dentinal lesions, and causing the low pH levels found (Hojo et al., 1994, Takahashi and Nyvad, 2016). By contrast, arrested dentinal lesions exhibit a weakly acidic pH, which can be related to the lower quantities of L. paracasei, L. rhamnosus, and L. casei group species in these lesions (Shimada et al., 2015, Takahashi and Nyvad, 2016).
The use of probiotics strains, especially L. rhamnosus and L. paracasei, has been identified as beneficial in maintaining oral health, playing an important role as an antagonistic agent for S. mutans growth (Twetman and Keller, 2012, Cagetti et al., 2013, Seminario-Amez et al., 2017, Coqueiro et al., 2018, Pahumunto et al., 2019). However, with the current concept of caries being the result of a dysbiosis, the eradication of a particular bacterial group, even one consisting of bacteria that are considered pathogens, would not be meaningful (Zaura and Twetman, 2019). In addition, clinical data are considered insufficient to demonstrate that such bacterial interference can be effective in controlling dental caries (Twetman and Keller, 2012, Cagetti et al., 2013, Seminario-Amez et al., 2017, Coqueiro et al., 2018). The inefficacy of bacteriotherapy may be due to the S. mutans associated with the caries rise; meanwhile, lactobacilli are opportunistic and are favored by the acidic environment created at the beginning of the lesion because they are more aciduric than S. mutans (Takahashi and Nyvad, 2011). Thus, in vitro studies where these microorganisms are grown together would seem to be an inappropriate way of testing the probiotic effects of Lactobacillus in controlling dental caries (Twetman and Keller, 2012), particularly in dentinal caries.
It is worth highlighting that infections such as abscesses related to L. paracasei and L. rhamnosus have been reported (Salminen et al., 2004, Burns et al., 2007, Chan et al., 2010, Sherid et al., 2016, Pararajasingam and Uwagwu, 2017, Harding-Theobald and Maraj, 2018), albeit rarely, demonstrating Lactobacillus as opportunistic pathogens.
The in vitro inhibitory activity presented by strains of L. rhamnosus and L. paracasei in decreasing streptococci (S. mutans and S. oralis) biofilm formation (Ciandrini et al., 2017) may be related to the production and delivery of hydrogen peroxide (H2O2), which is an interspecific competition mechanism in oral biofilms (Reis et al., 2012, Zhu et al., 2014). In this study, the spxB gene was expressed in dentinal lesions, but although it presented a numerical trend toward greater expression in active lesions, no statistically significant difference was observed between arrested and active dentinal lesions. This might be as a result of the conditions for both types of lesion, since this gene catalyzes the transformation of pyruvate to acetyl phosphate, CO2, and H2O2 in aerobic environments (Zotta et al., 2014, Zhu et al., 2014). Moreover, a recent study concluded that dental caries progression seems to be related to Lactobacillus spp. strains that are not capable of producing H2O2 and that the H2O2 released by Lactobacillus strains acts antagonistically toward mutans streptococci (Szkaradkiewicz-Karpinska et al., 2018). Expression of spxB has been suggested to play an active role in the biofilm physiology (Zotta et al., 2014), preventing the growth of other bacterial species by inhibiting DNA synthesis via H2O2.
The adhesion mechanism mediated by the spaC pilus in L. paracasei and L. rhamnosus is the major binding factor to mucus and collagen, stimulating biofilm formation for these bacteria (Lebeer et al., 2012, Toh et al., 2013, Tripathi et al., 2013, von Ossowski et al., 2013). Thus, the function of the pilus is essentially to facilitate cell adhesion to the “first contact” (von Ossowski et al., 2010, Rintahaka et al., 2014). The spaC gene was equally expressed in active and arrested lesions, suggesting that this surface component employs the same mechanism of adhesion for both types of dentinal lesions (arrested and active). No relationship can be suggested between the spaC and spaE genes, which encode the protein pilin, in Lactobacillus and the caries activity in dentinal lesions.
Although low values of wzb gene expression were presented, a higher wzb expression in active dentinal lesions than in arrested dentinal lesions was recorded. It can be hypothesized that the regulation of EPS biosynthesis (Lebeer et al., 2007, Nadkarni et al., 2014), which is the function of this gene, is more crucial in active lesions. This result was expected because the presence of simpler biofilms is more associated with caries arrestment processes (Cury and Tenuta, 2009, Maltz et al., 2010). Furthermore, it is important to note that inverse processes are related to active lesions, since caries progression is strongly associated with dental biofilm presence (Wolff and Larson, 2009). Research using clinical isolates of L. rhamnosus from dental pulp infection has shown that the silencing of wzb expression resulted in the largest reduction in biofilm formation and that the L. rhamnosus biofilm is strongly modulated by environmental factors, such as low pH (Lebeer et al., 2007, Lebeer et al., 2012, Nadkarni et al., 2014), commonly found in active dentinal lesions (Hojo et al., 1994).
An important limitation of this study was to distinguish the L. paracasei from the L. casei species using the spaC and spxB genes, since in silico comparisons revealed the alignment of both species with these genes. However, although no dietary data was collected, this study demonstrated that genes described in probiotic strains (Lebeer et al., 2007, von Ossowski et al., 2010, Rintahaka et al., 2014, Savo Sardaro et al., 2016) are expressed in isolated clinical strains of L. paracasei and L. rhamnosus. Thus, it is possible that microorganisms from dairy/probiotic products can survive in the mouth (Wolff and Larson, 2009) and eventually become associated with caries progression.
5. Conclusion
The L. casei group and the L. paracasei and L. rhamnosus species are part of the metabolically active community of the dentinal caries of children with ECC and are related to caries activity. The higher wzb gene expression in active lesions may be associated with the higher activity of microorganisms present in lesions with progressive caries processes.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgements
The authors thank the Professors Dr. José Roberto V. Silva and Rodrigo Maranguape S. da Cunha for technical assistance.
Funding
This research was supported by the Brazilian National Council for Scientific and Technological Development (CNPq) [Process #475346/2011-4MCT/CNPq 14/2011].
Footnotes
Peer review under responsibility of King Saud University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.sdentj.2020.01.006.
Contributor Information
Ana Catarina Martins Reis, Email: acmr_bio@hotmail.com.
Daniela da Silva Bezerra, Email: danysbezerra@yahoo.com.br.
Erika Nikitza Shiauha Hart-Chú, Email: erikanikitza@yahoo.com.
Rafael Nóbrega Stipp, Email: rafaelns@fop.unicamp.br.
Sarah Florindo de Figueiredo Guedes, Email: sarahffguedes@yahoo.com.br.
Beatriz Gonçalves Neves, Email: beatrizneves@ufc.br.
Lidiany Karla Azevedo Rodrigues, Email: lidianykarla@ufc.br.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
References
- Aas J.A., Griffen A.L., Dardis S.R., Lee A.M., Olsen I., Dewhirst F.E., Leys E.J., Paster B.J. Bacteria of dental caries in primary and permanent teeth in children and young adults. J. Clin. Microbiol. 2008;46:1407–1417. doi: 10.1128/JCM.01410-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badet C., Thebaud N.B. Ecology of lactobacilli in the oral cavity: a review of literature. Open Microbiol. J. 2008;2:38–48. doi: 10.2174/1874285800802010038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bezerra D.S., Neves B.G., Guedes S.F.F., Regis W.F.M., Stipp R.N., Rodrigues L.K.A. Extraction and purification of RNA from human carious dentine: an approach to enable bacterial gene expression studies. J. Health Biol. Sci. 2019;7:145–151. [Google Scholar]
- Bezerra, D.S., Stipp, R.N., Neves, B.G., Guedes, S.F.F., Nascimento, M.M., Rodrigues, L,K,A., 2016. Insights into the virulence traits of Streptococcus mutans in dentine carious lesions of children with early childhood caries. Caries Res. 50, 279-87. [DOI] [PubMed]
- Burns D., Hurst J.R., Hopkins S., Patch D., Burroughs A.K., Agarwal B. Purpura fulminans associated with Lactobacillus paracasei liver abscess. Anaesth. Intensive Care. 2007;35:121–123. doi: 10.1177/0310057X0703500120. [DOI] [PubMed] [Google Scholar]
- Cagetti M.G., Mastroberardino S., Milia E., Cocco F., Peter Lingström P., Campus G. The use of probiotic strains in caries prevention: a systematic review. Nutrients. 2013;5:2530–2550. doi: 10.3390/nu5072530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caufield P.W., Schön C.N., Saraithong P., Li Y., Argimón S. Oral lactobacilli and dental caries: a model for niche adaptation in humans. J. Dent. Res. 2015;94:110–118. doi: 10.1177/0022034515576052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceapa C., Davids M., Ritari J., Lambert J., Wels M., Douillard F.P., Smokvina T., de Vos W.M., Knol J., Kleerebezem M. The variable regions of Lactobacillus rhamnosus genomes reveal the dynamic evolution of metabolic and host-adaptation repertoires. Genome Biol. Evol. 2016;8:1889–1905. doi: 10.1093/gbe/evw123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J.F., Lau S.K., Woo P.C., Fan R.Y., Ip J.J., Chan C.F., Luk J.K., Yuen K.Y. Lactobacillus rhamnosus hepatic abscess associated with Mirizzi syndrome: a case report and review of the literature. Diagn. Microbiol. Infect. Dis. 2010;66:94–97. doi: 10.1016/j.diagmicrobio.2009.08.009. [DOI] [PubMed] [Google Scholar]
- Ciandrini E., Campana R., Baffone W. Live and heat-killed Lactobacillus spp. interfere with Streptococcus mutans and Streptococcus oralis during biofilm development on titanium surface. Arch. Oral Bio. 2017;78:48–57. doi: 10.1016/j.archoralbio.2017.02.004. [DOI] [PubMed] [Google Scholar]
- Colombo N.H., Kreling P.F., Ribas L.F.F., Pereira J.A., Kressirer C.A., Klein M.I., Tanner A.C.R., Duque C. Quantitative assessment of salivary oral bacteria according to the severity of dental caries in childhood. Arch. Oral Bio. 2017;83:82–288. doi: 10.1016/j.archoralbio.2017.08.006. [DOI] [PubMed] [Google Scholar]
- Coqueiro A.Y., Bonvini A., Raizel R., Tirapegui J., Rogero M.M. Probiotic supplementation in dental caries: is it possible to replace conventional treatment? Nutrire. 2018;43:6. [Google Scholar]
- Cury J.A., Tenuta L.M.A. Enamel remineralization: controlling the caries disease or treating early caries lesions. Braz. Oral Res. 2009;23:23–30. doi: 10.1590/s1806-83242009000500005. [DOI] [PubMed] [Google Scholar]
- da Silva, A.C., Cruz, J.dos.S., Sampaio, F.C., de Araújo, D.A., 2008. Detection of oral streptococci in dental biofilm from caries-active and caries-free children. Braz. J. Microbiol. 39, 648-651. [DOI] [PMC free article] [PubMed]
- Dassi, E., Ferretti, P., Covello G., HTM-CMB-2015, Bertorelli, R., Denti, M.A., De Sanctis V., Tett, A., Segata, N., 2018. The short-term impact of probiotic consumption on the oral cavity microbiome. Sci. Rep. 11, 110s-118s. [DOI] [PMC free article] [PubMed]
- Fejerskov O. Changing paradigms in concepts on dental caries: consequences for oral health care. Caries Res. 2004;38:182–191. doi: 10.1159/000077753. [DOI] [PubMed] [Google Scholar]
- Furet J., Quéneé P., Tailliez P. Molecular quantification of lactic acid bacteria in fermented milk products using real-time quantitative PCR. Int. J. Food Microbiol. 2004;97:197–207. doi: 10.1016/j.ijfoodmicro.2004.04.020. [DOI] [PubMed] [Google Scholar]
- Gao, X., Jiang, S., Koh, D., Hsu, C.S., 2016. Salivary biomarkers for dental caries. Periodontol. 2000. 70, 128–141. [DOI] [PubMed]
- Harding-Theobald E., Maraj B. Case Rep. Gastrointest. Med.; 2018. Spontaneous Bacterial Peritonitis due to Lactobacillus paracasei in Cirrhosis; p. 5714053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill D., Sugrue I., Tobin C., Hill C., Stanton C., Ross R.P. The Lactobacillus casei group: history and health related applications. Front. Microbiol. 2018;9:2107. doi: 10.3389/fmicb.2018.02107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hojo S., Komatsu M., Okuda R., Takahashi N., Yamada T. Acid profiles and pH of carious dentin in active and arrested lesions. J. Dent. Res. 1994;73:1853–1857. doi: 10.1177/00220345940730121001. [DOI] [PubMed] [Google Scholar]
- Jiang W., Ling Z., Lin X., Chen Y., Zhang J., Yu J., Xiang C., Chen H. Pyrosequencing analysis of oral microbiota shifting in various caries states in childhood. Microb. Ecol. 2014;67:962–969. doi: 10.1007/s00248-014-0372-y. [DOI] [PubMed] [Google Scholar]
- Kneist S., Schmidt F., Callaway A., Willershausen B., Rupf S., Wicht M., Thiede B. Diversity of Lactobacillus species in deep carious lesions of primary molars. Eur. Arch. Paediatr. Dent. 2010;11:181–186. doi: 10.1007/BF03262741. [DOI] [PubMed] [Google Scholar]
- Lebeer S., Claes I., Tytgat H.L., Verhoeven T.L., Marien E., von Ossowski I., Reunanen J., Palva A., Vos W.M., Keersmaecker S.C., Vanderleyden J. Functional analysis of Lactobacillus rhamnosus GG pili in relation to adhesion and immunomodulatory interactions with intestinal epithelial cells. Appl. Environ. Microbiol. 2012;78:185–193. doi: 10.1128/AEM.06192-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebeer S., Verhoeven T.L.A., Vélez M.P., Vanderleyden J., Keersmaecker S.C.J. Impact of environmental and genetic factors on biofilm formation by the probiotic strain Lactobacillus rhamnosus GG. Appl. Environ. Microbiol. 2007;73:6768–6775. doi: 10.1128/AEM.01393-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledder R.G., Kampoo K., Teanpaisan R., McBain A.J. Oral microbiota in severe early childhood caries in thai children and their families: a pilot study. Front. Microbiol. 2018;9:2420. doi: 10.3389/fmicb.2018.02420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Chen X., Tu Y., Wang S., Chen H. Effect of probiotic lactobacilli on the growth of Streptococcus mutans and multispecies biofilms isolated from children with active caries. Med. Sci. Monit. 2017;23:4175-¬4181. doi: 10.12659/MSM.902237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Argimón S., Schön C.S., Saraithong P., Caufield P.W. Characterizing diversity of Lactobacilli associated with severe early childhood caries: a study protocol. Adv. Microbiol. 2015;5:9–20. doi: 10.4236/aim.2015.51002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maltz M., Jardim J.J., Alves L.S. Health promotion and dental caries. Braz. Oral Res. 2010;24:18–25. doi: 10.1590/s1806-83242010000500004. [DOI] [PubMed] [Google Scholar]
- Miljkovic, M., Strahinic, I., Tolinacki, M., Zivkovic, M., Kojic, S., Golic, N., Kojic, M., 2015. AggLb Is the Largest Cell-Aggregation Factor from Lactobacillus paracasei Subsp. paracasei BGNJ1-64, Functions in Collagen Adhesion, and Pathogen Exclusion In Vitro. PLoS One. 10, e0126387. [DOI] [PMC free article] [PubMed]
- Mira A. Oral microbiome studies: potential diagnostic and therapeutic implications. Adv. Dent. Res. 2018;29:71–77. doi: 10.1177/0022034517737024. [DOI] [PubMed] [Google Scholar]
- Mitrakul K., Chanvitan S., Jeamset A., Vongsawan K. Quantitative analysis of S. mutans, Lactobacillus and Bifidobacterium found in initial and mature plaques in Thai children with early childhood caries. Eur. Arch. Paediatr. Dent. 2017;18:251–261. doi: 10.1007/s40368-017-0295-7. [DOI] [PubMed] [Google Scholar]
- Nadkarni M.A., Chen Z., Wilkins M.R., Hunter N. Comparative genome analysis of Lactobacillus rhamnosus clinical isolates from initial stages of dental pulp infection: identification of a new exopolysaccharide cluster. PLoS ONE. 2014;9 doi: 10.1371/journal.pone.0090643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadkarni M.A., Martin F.E., Jacques N.A., Hunter N. Determination of bacterial load by real-time PCR using a broad-range (universal) probe and primers set. Microbiology. 2002;148:257–266. doi: 10.1099/00221287-148-1-257. [DOI] [PubMed] [Google Scholar]
- Neves B.G., Stipp R.N., Bezerra D.S., Guedes S.F.F., Rodrigues L.K.A. Molecular detection of bacteria associated to caries activity in dentinal lesions. Clin. Oral Invest. 2017;21:2053–2061. doi: 10.1007/s00784-016-1995-9. [DOI] [PubMed] [Google Scholar]
- Obata J., Takeshita T., Shibata Y., Yamanaka W., Unemori M., Akifumi Akamine A., Yamashita Y. Identification of the microbiota in carious dentin lesions using 16S rRNA gene sequencing. PLoS ONE. 2014;9 doi: 10.1371/journal.pone.0103712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pahumunto N., Sophatha B., Piwat S., Teanpaisan R. Increasing salivary IgA and reducing Streptococcus mutans by probiotic Lactobacillus paracasei SD1: A double-blind, randomized, controlled study. J. Dent. Sci. 2019 doi: 10.1016/j.jds.2019.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pararajasingam A., Uwagwu J. Lactobacillus: the not so friendly bacteria. Case Rep. 2017 doi: 10.1136/bcr-2016-218423. bcr-2016-218423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philip N., Suneja B., Walsh L.J. Ecological Approaches to dental caries prevention: paradigm shift or shibboleth? Caries Res. 2018;52:153–165. doi: 10.1159/000484985. [DOI] [PubMed] [Google Scholar]
- Piwat S., Sophatha B., Teanpaisan R. An assessment of adhesion, aggregation and surface charges of Lactobacillus strains derived from the human oral cavity. Lett. Appl. Microbiol. 2015;61:98–105. doi: 10.1111/lam.12434. [DOI] [PubMed] [Google Scholar]
- Reis J.A., Paula A.T., Casarotti S.N., Penna A.L.B. Lactic acid bacteria antimicrobial compounds: characteristics and applications. Food Eng. Rev. 2012;4:124–140. [Google Scholar]
- Rintahaka J., Yu X., Kant R., Palva A., von Ossowski I. Phenotypical analysis of the Lactobacillus rhamnosus GG fimbrial spaFED operon: surface expression and functional characterization of recombinant SpaFED pili in Lactococcus lactis. PLoS ONE. 2014;9 doi: 10.1371/journal.pone.0113922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salminen M.K., Rautelin H., Tynkkynen S., Poussa T., Saxelin M., Valtonen V., Järvinen A. Lactobacillus bacteremia, clinical significance, and patient outcome, with special focus on probiotic L. rhamnosus GG. Clin. Infect. Dis. 2004;38:62–69. doi: 10.1086/380455. [DOI] [PubMed] [Google Scholar]
- Salvetti E., Torriani S., Felis G.E. The genus Lactobacillus: a taxonomic update. Probiotics Antimicrob. Proteins. 2012;4:217–226. doi: 10.1007/s12602-012-9117-8. [DOI] [PubMed] [Google Scholar]
- Savo Sardaro M.L., Levante A., Bernini V., Gatti M., Neviani E., Lazzi C. The spxB gene as a target to identify Lactobacillus casei group species in cheese. Food Microbiol. 2016;59:57–65. doi: 10.1016/j.fm.2016.05.004. [DOI] [PubMed] [Google Scholar]
- Schwendicke F., Dörfer C., Kneist S., Meyer-Lueckel H., Paris S. Cariogenic effects of probiotic Lactobacillus rhamnosus GG in a dental biofilm model. Caries Res. 2014;48:186–192. doi: 10.1159/000355907. [DOI] [PubMed] [Google Scholar]
- Seminario-Amez M., López-López J., Estrugo-Devesa A., Ayuso-Montero R., Enric Jané-Salas E. Probiotics and oral health: a systematic review. Med. Oral Patol. Oral Cir. Bucal. 2017;22:e282–e288. doi: 10.4317/medoral.21494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharpe M.E. Lactic acid bacteria in the dairy industry. Int. J. Dairy Technol. 1979;32:9–18. [Google Scholar]
- Sherid M., Samo S., Sulaiman S., Husein H., Sifuentes H., Sridhar S. Liver abscess and bacteremia caused by lactobacillus: role of probiotics? BMC Gastroenterol. 2016;16:138. doi: 10.1186/s12876-016-0552-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada A., Noda M., Matoba Y., Kumagai T., Katsuyuki K., Sugiyama M. Oral lactic acid bacteria related to the occurrence and/or progression of dental caries in Japanese preschool children. Biosci. Microbiota Food Health. 2015;34:29–36. doi: 10.12938/bmfh.2014-015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simón-Soro A., Guillen-Navarro M., Mira A. Metatranscriptomics reveals overall active bacterial composition in caries lesions. J. Oral Microbiol. 2014;6:25443. doi: 10.3402/jom.v6.25443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simón-Soro A., Mira A. Solving the etiology of dental caries. Trends Microbiol. 2015;23:76–82. doi: 10.1016/j.tim.2014.10.010. [DOI] [PubMed] [Google Scholar]
- Smokvina T., Wels M., Polka J., Chervaux C., Brisse S., Boekhorst J., van Hylckama Vlieg J.E., Siezen R.J. Lactobacillus paracasei comparative genomics: towards species pan-genome definition and exploitation of diversity. PLoS ONE. 2013;8 doi: 10.1371/journal.pone.0068731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solbiati J., Frias-Lopez J. Metatranscriptome of the oral microbiome in health and disease. J. Dent. Res. 2018;00:1–9. doi: 10.1177/0022034518761644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surachat K., Sangket U., Deachamag P., Chotigeat W. In silico analysis of protein toxin and bacteriocins from Lactobacillus paracasei SD1 genome and available online databases. PLoS ONE. 2017;12 doi: 10.1371/journal.pone.0183548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Švec P., Sedlácek I., Žácková L., Nováková D., Kukletová M. Lactobacillus spp. associated with early childhood caries. Folia Microbiol. 2009;54:53–58. doi: 10.1007/s12223-009-0008-7. [DOI] [PubMed] [Google Scholar]
- Szkaradkiewicz-Karpinska A.K., Zeidler A., Goslinska-Kuzniarek O., Uram K., Szkaradkiewicz A. Oral Lactobacilli and salivary acidic proline-rich proteins (APRP-1/2) in dental caries. J. Physiol. Pharmacol. 2018;1:139–144. doi: 10.26402/jpp.2018.1.15. [DOI] [PubMed] [Google Scholar]
- Takahashi N. Oral microbiome metabolism: from “who are they?” to “what are they doing?”. J. Dent. Res. 2015;94:1628–1637. doi: 10.1177/0022034515606045. [DOI] [PubMed] [Google Scholar]
- Takahashi N., Nyvad B. The role of bacteria in the caries process: ecological perspectives. J. Dent. Res. 2011;90:294–303. doi: 10.1177/0022034510379602. [DOI] [PubMed] [Google Scholar]
- Takahashi N., Nyvad B. Ecological hypothesis of dentin and root caries. Caries Res. 2016;50:422–431. doi: 10.1159/000447309. [DOI] [PubMed] [Google Scholar]
- Tanner A.C., Kressirer C.A., Faller L.L. Understanding caries from the oral microbiome perspective. J. Calif. Dent. Assoc. 2016;44:437–446. [PubMed] [Google Scholar]
- Toh H., Oshima K., Nakano A., Takahata M., Murakami M., Takaki T., Nishiyama H., Igimi S., Hattori M., Morita H. Genomic adaptation of the Lactobacillus casei group. PLoS ONE. 2013;8 doi: 10.1371/journal.pone.0075073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi P., Beaussart A., Alsteens D., Dupres V., Claes I., von Ossowski I., de Vos W.M., Palva A., Lebeer S., Vanderleyden J., Dufrêne Y.F. Adhesion and nanomechanics of pili from the probiotic Lactobacillus rhamnosus GG. ACS Nano. 2013;7:3685–3697. doi: 10.1021/nn400705u. [DOI] [PubMed] [Google Scholar]
- Twetman S., Keller M.K. Probiotics for caries prevention and control. Adv. Dent. Res. 2012;24:98–102. doi: 10.1177/0022034512449465. [DOI] [PubMed] [Google Scholar]
- von Ossowski I., Pietila T.E., Rintahaka J., Nummenmaa E., Makinen V.M., Reunanen J., Satokari R., Vos W.M., Palva I., Palva A. Using recombinant Lactococci as an approach to dissect the immunomodulating capacity of surface piliation in probiotic Lactobacillus rhamnosus GG. PLoS ONE. 2013;8 doi: 10.1371/journal.pone.0064416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuotto C., Longo F., Donelli G. Probiotics to counteract biofilm-associated infections: promising and conflicting data. Int. J. Oral Sci. 2014;6:189–194. doi: 10.1038/ijos.2014.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Ossowski I., Reunanen J., Satokari R., Vesterlund S., Kankainen M., Huhtinen H., Tynkkynen S., Salminen S., de Vos W.M., Palva A. Mucosal adhesion properties of the probiotic Lactobacillus rhamnosus GG SpaCBA and SpaFED pilin subunits. Appl. Environ. Microbiol. 2010;76:2049–2057. doi: 10.1128/AEM.01958-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff M.S., Larson C. The cariogenic dental biofilm: good, bad or just something to control? Braz. Oral. Res. 2009;23:31–38. doi: 10.1590/s1806-83242009000500006. [DOI] [PubMed] [Google Scholar]
- Young D.A., Featherstone J.D. Caries management by risk assessment. Commun. Dent. Oral Epidemiol. 2013;41:53–63. doi: 10.1111/cdoe.12031. [DOI] [PubMed] [Google Scholar]
- Zaura E., Twetman S. Critical appraisal of oral pre- and probiotics for caries prevention and care. Caries Res. 2019;4:1–13. doi: 10.1159/000499037. [DOI] [PubMed] [Google Scholar]
- Zhan L. Rebalancing the caries microbiome dysbiosis: targeted treatment and sugar alcohols. Adv. Dent. Res. 2018;29:110–116. doi: 10.1177/0022034517736498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L., Xu Y., Ferretti J.J., Kreth J. Probing oral microbial functionality-expression of spxB in plaque samples. PLoS ONE. 2014;9 doi: 10.1371/journal.pone.0086685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zotta T., Ricciardi A., Ianniello R.G., Parente E., Reale A., Rossi F., Iacumin L., Comi G., Coppola R. Assessment of aerobic and respiratory growth in the Lactobacillus casei group. PLoS ONE. 2014;9 doi: 10.1371/journal.pone.0099189. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.