Abstract
Background
To inform researchers about the methodology and results of epidemic estimation studies performed for COVID-19 epidemic in Iran, we aimed to perform a rapid review.
Methods
We searched for and included published articles, preprint manuscripts and reports that estimated numbers of cumulative or daily deaths or cases of COVID-19 in Iran. We found 131 studies and included 29 of them.
Results
The included studies provided outputs for a total of 84 study-model/scenario combinations. Sixteen studies used 3–4 compartmental disease models. At the end of month two of the epidemic (2020-04-19), the lowest (and highest) values of predictions were 1,777 (388,951) for cumulative deaths, 20,588 (2,310,161) for cumulative cases, and at the end of month four (2020-06-20), were 3,590 (1,819,392) for cumulative deaths, and 144,305 (4,266,964) for cumulative cases. Highest estimates of cumulative deaths (and cases) for latest date available in 2020 were 418,834 on 2020-12-19 (and 41,475,792 on 2020-12-31). Model estimates predict an ominous course of epidemic progress in Iran. Increase in percent population using masks from the current situation to 95% might prevent 26,790 additional deaths (95% confidence interval 19,925–35,208) by the end of year 2020.
Conclusions
Meticulousness and degree of details reported for disease modeling and statistical methods used in the included studies varied widely. Greater heterogeneity was observed regarding the results of predicted outcomes. Consideration of minimum and preferred reporting items in epidemic estimation studies might better inform future revisions of the available models and new models to be developed. Not accounting for under-reporting drives the models’ results misleading.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12889-021-10183-3.
Keywords: COVID-19, Epidemic, Pandemic, Iran, Cases, Deaths, Model, Prediction, Estimation
Background
“On 31 December 2019, the World Health Organization (WHO) China Country Office was informed of cases of pneumonia unknown etiology (unknown cause) detected in Wuhan City, Hubei Province of China” [1]. The disease was officially designated as coronavirus disease 2019 or COVID-19 by WHO on 2020-02-11 [2]. Due to the rapid outbreak of the disease worldwide, WHO characterized the situation as a pandemic on 11 March [3]. The first two confirmed cases of COVID-19 in Iran were officially reported on 2020-02-19 in city of Qom, by the Ministry of Health and Medical Education (MOHME) (via [4, 5]). Since then, MOHME has officially reported number of cumulative and new confirmed cases, deaths, and recovered cases in a daily basis on press conferences. Those numbers are available by date on different web pages of the web site of the MOHME but are not compiled in one page. To our best knowledge, the most straightforward route to access cumulative and daily deaths and cases is the compilation of WHO situation reports or more comprehensive sources such as Johns Hopkins University dashboard for COVID-19 [4, 5]. Web site of Iranian MOHME was not accessible to us from Canada and United States most of the time, with an “Access Forbidden” massage from the “Security Department” of MOHME at several occasions.
Number of deaths and cases, in addition to other characteristics of epidemic are very important in decision-making and disease control. However, official reports suffer from undercounting in all countries. A relatively high percentage of patients with COVID-19 are asymptomatic or have a mild form of the disease which increases the chance of remaining undiagnosed. The emerging nature of COVID-19 has aggravated undercounting, as many countries are not prepared for conducting enough tests; as a result, many of suspected cases or deaths may not ever be confirmed by standard laboratory tests. Epidemiological studies of COVID-19 and model-based predictions and estimations are useful in assessing transmission rates, predicting epidemic trends and fatality rates with the inclusion of different intervention, environmental (seasonality), and virologic (mutations) scenarios, and thus can help policymakers for informed decision making in a timely way [6].
Despite the potential role of epidemic modeling and estimation studies in predicting outbreak size and trend, multiplicity of factors influencing viral disease transmission, relative uncertainty of data on model parameters, shifting disease dynamics in the setting of evolving epidemic, and suboptimality of model building methods and reporting, have been known to limit predictive models’ usefulness. A systematic review of individual-level prediction models for covid-19 concluded that many of the models suffer from poor reporting, high risk of bias, and optimistic reporting of performance [7].
Since the beginning of the outbreak in Iran, researchers inside and outside the country have used models to estimate or predict the size and trajectory of the epidemic of COVID-19 in Iran. All of them are introduced later in the methods and results section of this article. Some of these studies have been published in scientific journals, or available in their not yet peer-reviewed form or are presented as official or unofficial reports. We aimed to perform a rapid review mainly describing currently available COVID-19 estimates for Iran. We did not intend to scrutinize or criticize the studies or models at this point. Our objective was to review methods and results of COVID-19 epidemic estimation studies for Iran. The ultimate goal is to inform the audience, policy makers and researchers, for better decisions, as well as potential updates of their prediction or estimation studies or new studies being designed and conducted currently and in future.
Methods
Study design and outcomes of interest
This a rapid review, not a systematic review. The main outcomes of interest were the predicted values (and calendar dates) of (1) cumulative deaths, (2) cumulative cases, (3) daily deaths, and (4) daily cases of COVID-19 in Iran. Deaths are less dependent on testing than cases. Cumulative estimates do not show daily fluctuations. Daily estimates help demonstrate epidemic waves and peaks.
Place and time scope of target studies
We included all studies which their target population was or included Iran, and we found them from 2020-03-19 to 2020-04-12. We updated our search from 2020-10-02 to 2020-10-05.
Search strategy and selection criteria
There is not a customized protocol for the items that must be available in a report of epidemic estimation, prediction or epidemic model. Carrasco et al. proposed a ‘CCPV’ protocol (Characteristics, Construction, Parameterization and Validation aspects protocol) to standardize the reporting of Influenza pandemic models [8]. The items included in the “transparent reporting of a multivariable prediction model for individual prognosis or diagnosis” (TRIPOD) gives a general idea of the reporting items, however it is not specifically for the reason of epidemic prediction/ estimation models [9]. Wynants et al. review of prediction models for diagnosis and prognosis of covid-19 infection, even though focusing the individual-level modeling, can be considered in reporting epidemic estimation studies [7].
Based on our understanding of the TRIPOD statement, epidemic modelling literature, and the studies we reviewed, we think an epidemic estimation / prediction model is expected to report at least following items, that we call them ‘preferred reporting items’: (1) Epidemic start date and rationale, (2) Epidemic (disease) model type and description, (3) Statistical model type, description, and equation(s), (4) Model assumptions and their verification, (5) Model scenarios’ detailed description, (6) Validation process and findings, (7) List and sources of model parameters and input data, and (8) Model outputs preferably with uncertainty intervals for scenarios. Some of the reports that we found in our searching process were not the final versions; we included any study if met all the following ‘minimum reporting items’: (1) Provided estimates for at least one of the COVID-19 four outcomes of interests (cumulative deaths, cumulative cases, daily deaths or daily cases) in Iran in any period of time, (2) Provided a list of input data and their sources, (3) An explanation on methods of using input data and generation of model outputs was available.
Exclusion criteria were: (1) Absence of all four main outcomes of interest, or (2) Absence of the minimum reporting items, or (3) Elaboration on a previous modeling or estimation study without the aim (or content) of updating or improving the previous estimates.
By “report”, we mean studies results of which were not published as a journal article (or pre-print), but were released as short or long reports, available on the internet or shared with researchers. There are differences between epidemic modeling, prediction, and estimation; Modeling studies use explicit disease models and statistical models. Prediction studies do not use explicit disease models but predict (project) the number of cases and or deaths in future. Estimation studies provide estimates of cases for a recent point in time. The common feature of all these three study types is that they provide estimates of cases and or deaths in at least one point in calendar time. For pragmatic reasons, we call all of them as estimation studies.
Study revisions / updates: We actively searched and checked for revisions or updates of studies and their published formats (from pre-print, to journal pre-proof, to final published article). International studies update their estimations on a periodical basis: (1) DELPHI (Differential Equations Leads to Predictions of Hospitalizations and Infections) Epidemiological Case Predictions (DELPHI) [10], (2) Youyang Gu (YYG) [11], (3) Institute for Health Metrics and Evaluation (IHME) [12], (4) Imperial College (Imperial) [13], (5) Los Alamos National Laboratory (LANL) [14], and (6) Srivastava [15]. We stopped using new estimate updates on 2020-10-11, when we used the latest estimate by LANL (Los Alamos National Laboratories) [14]. As such, the latest estimation dates for international we finally included in our review are as follows (references are for studies’ data web sites): 2020-07-18 for DELPHI [16], 2020-10-05 for Gu (YYG) [17], 2020-10-09 for IHME [18], 2020-10-06 for Imperial College [19], 2020-10-11 for LANL [20], and 2020-09-12 for Srivastava [15]. We found a web site [21] and a published article [22] by Sabri and colleagues. We refer to them, as ‘Saberi (web site)’ and ‘Saberi (article)’ respectively. For the former, that included periodical updates, we used their 2020-03-30 version. Updates to their previous model [21] were later discontinued.
We searched PubMed and used Google Scholar and plain Google for articles (or reports) matching our study inclusion criteria. The used keywords were Iran, COVID, COVID-19, COVID 19, Corona, SARS-CoV-2, epidemic, outbreak, pandemic, case*, death*, fatal*, mortalit*, model*, estimat*, and predict*. The search syntax used in PubMed is shown in the Appendix. We performed the same search with keywords in Farsi in Google Scholar, Google, Scientific Information Database (of Iran) [23], and MAG-IRAN [24].
We also used studies or reports provided to us by our researcher colleagues. We report all the found, included, and excluded, studies using PRISMA 2009 flow diagram (Moher et al. 2009) [25] in Appendix Figure 1.
Data abstraction methods
We developed a spreadsheet for abstracting the items of methods and results from included studies – the items not restricted to the minimum required ones. Each study was reviewed independently by at least two authors, and discrepancies were resolved with involvement of a third reviewer. Two reviewers (MML and LJ) finalized the abstracted items for methodology of the target studies.
For abstraction of the results of the studies, we selected a set of six fixed calendar dates, and found and recorded the estimated / predicted values of main outcomes (cumulative deaths, cumulative cases, daily deaths, or daily cases) for each of those dates. To start with, we fixed the presumed epidemic start time on the date on which the first two cases were officially reported dead, on 2020-02-19 (1398-11-30 Hijri solar), although later official reports indicated the actual start date of the epidemic to be earlier. Rationale for this was that most of the studies used the official reports to start with, and most of the studies’ predictions also started from that date (2020-02-19).
We decided not to include the estimations for the end of the first, second and third weeks of the epidemic in our set of fixed dates, since we were already in month two of the epidemic, for sake of brevity, and considering the less robust nature of the predictions as early as the first month when the numbers were much smaller. The set of six fixed calendar dates were designated as the end of each Hijri solar month after the epidemic presumed start date, since the start date coincided with the last day of month 11 of the Hijri solar calendar, and as such, targeting the end of each solar month would enhance cross-study comparisons and further use of administrative data. We did not report predictions beyond the month six, for the sake of brevity, and given the more uncertainty regarding such longer-term intervention scenarios and outcomes. However, we demonstrated all the time span of the available predictions in our graphs to provide a visual overview.
Most of the reviewed studies had considered more than one scenario for the progression of the epidemic, based on different intervention options, and we treated them as study-scenarios. Some studies considered different statistical models for prediction of epidemic progress, and we treated them as study-models. Altogether, such arrangement provided multiple study-scenarios/models for which we abstracted the estimated / predicted values of the main outcomes. Occasional studies provided confidence limits for point estimates which were also recorded. Along with update of our search in October 2020, we included estimated values for latest dates available in 2020 (e.g. 2020-12-31) and in 2021 (e.g. 2021-01-31) where available.
Besides the six fixed dates, we also abstracted the data for the following items for each study-scenario/model: (a) predicted date of every peak in daily / new / active cases or deaths, and (b) predicted date of ‘epidemic control’ or equivalent (with the same study’s criteria or definition of control). Predicted values of main outcomes were recorded for these dates. We also recoded the methods used for as assessment of each statistical model’s validity (or fitness) and their findings.
Two reviewers (FP and MRH) abstracted the estimates from the target studies. For abstraction of the main outcomes’ values at the designated dates, we prioritized our sources and methods as, (1) mention in article text and tables, (2) digitization of article graphs. We used a web-based plot digitizer, “WebPlotDigitizer 4.2” [26]. Our error in digitizing data was less than 5%, as measured using the following formula: error in digitizing = ((digitized value – mentioned value) / (digitized value)), where the mentioned value means the value that was mentioned in study text or tables. We used the reported COVID-19 cases and deaths complied by the Johns Hopkins University [4, 5] for each calendar date, as equal to Iran’s official reported data compiled in WHO situation reports. For developing our graphs, we chose the median scenario/models for cumulative cases from each study in order to demonstrate the main level of the predictions in non-international studies. Only eight non-international studies estimated cumulative deaths, so that outcome could not be used for identification of median scenarios across all studies. In studies with even number of scenario/models, we chose the one (of the middle two) with the higher values of estimates. The same selected median scenario/models for cumulative cases were used to graph the outcomes where the predictions were available. We recorded the text, table number, or the graph number for each study where we extracted every single number or date used in our tables and graphs. “Additional file 2 - Target studies’ abstracted data” includes all the detailed data we abstracted from the studies, as well as detailed findings from the studies’ methods.
Results
General characteristics of reviewed studies: We found 114 articles, 10 non-peer reviewed reports, and seven web sites that described methods and present results of estimations; a total of 131 results.
We included 18 published articles, two medRxiv preprints, seven web sites of COVID-19 epidemic modeling studies, and two non-peer-reviewed reports; a total of 29 studies. The two non-peer-reviewed reports were in Farsi; Haghdoost [27], and Mashayekhi [28]. One published article was in Farsi; Rahimi Rise [29]. Among the seven included web sites, one estimated the outcomes in Iran (Saberi [21]) and the six others were international studies estimating the outcomes for multiple countries on a periodical basis; DELPHI [10], Gu (YYG) [17], IHME [12], Imperial College [13], LANL [14], and Srivastava [15]. Three studies, DELPHI, Gu (YYG), and Los Alamos did not have a publication, and whatever details about their study methods are available on their study web sites [10, 11, 14]. The three other international studies have at least one publication each (as of 2020-10-29): IHME, peer-review published [30], pre-print [31–33]; Imperial College, peer-review published [34], and Srivastava, pre-print [35]. For each of the six international studies, we mentioned the study web site, study data site, and their publications. For reporting the number of the reviewed component studies, we count each international study as one, rather than creating a hierarchical clustered structure with study publication / study web site at higher level and individual studies at lower level. Appendix Figure 1 shows the PRISMA studies flow diagram.
We report our findings following the ‘preferred reporting items’ mentioned above.
Place: 19 studies included only Iran; the other 13 studies included from 6 to 184 countries. Six studies included subnational level estimates: Haghdoost [27], Moghadami [36], Muniz-Rodriguez [37], Pourghasemi (PLoS ONE) [38], Pourghasemi (IJID) [39], and Zhan [40].
Epidemic start date and rationale
Twenty studies mentioned the epidemic start date, nine of which used presumed official start date of 2020-02-19. Epidemic start date ranged from 2020-01-02 (Ghaffarzadegan [41]) to 2020-02-20 (Moradi [42], Shen [43]).
Two studies (Moradi [42], Shen [43]) reported their estimates starting from 2020 to 02-20 without mention of the rationale. Ghaffarzadegan reported most of their estimates starting from 2020-01-02, based on unofficial reporting of suspected cases [41]. Haghdoost et al. designated their “Day-zero” as 2020-01-21 [Hijri solar date 1398-11-01], that is 20 days before the presumed official epidemic start date of 2020-02-19 [27]. Many of the predicted outcome values are zero or close to zero in the graphs prior to day 20 of the graphs. However, some of the graphs do seem to show non-zero values for cases or deaths before their day. They maintain that their start date of the epidemic in Iran (2020-01-21) was designated based on “available documentations and epidemiologic analyses”. Mashayekhi et al. did not mention their epidemic start date, and prediction graphs’ time axis showed day zero to 120 or 360 [28]. We made an assumption that their start date was 2020-02-19. Nine studies did not mention the epidemic start date.
Epidemic (disease) model type and description
Sixteen studies used compartmental models: SEIR or SEIR+ (nine studies), SIR or SIR+ (six studies), SLIR+ (one study). In model acronyms, ‘S’ stands for Susceptible, ‘E’ for Exposed, ‘I’ for Infected, ‘R’ for Removed or Recovered, and ‘L’ is for Latent. In any model with a + sign, there are other components for augmentation of model.
Statistical model type, description, and equation(s)
Some of the studies did not mentioned enough details about their statistical methods and did not clearly differentiate between the disease model and the statistical model. Statistical methods used included growth models (6 studies), dynamic models (4 studies), Auto-Regressive Integrated Moving Average (ARIMA) (3 studies), regression models (3 studies), and ‘curve fitting and functional analysis’, hyperparametric learning, machine learning, minimization, smoothing model, and time series (each in one study). Few studies provided formal representation (equation) of the model.
Model assumptions and their verification
None of the reviewed studies did explicitly mention all the assumptions, their verification methods, and results of the verification. Most studies did report some details about their assumptions.
Model scenarios’ detailed description
There were 77 study-scenarios (epidemic progression scenarios) and 38 study-models (statistical models), resulting in a total of 84 study-model/scenario combinations. Fourteen studies used only one scenario (that practically means no “scenario”). Fifteen other studies used two to 12 scenarios (median 3). Nine studies included policy intervention scenarios with different levels of details mentioned. Eight studies used three levels of interventions, that can be generally formulated as ‘current policies’, ‘more restrictions’, and ‘less restrictions’; Ahmadi [44], Ghaffarzadegan [41], Haghdoost [27], IHME [12], Imperial [13], Mashayekhi [28], Saberi (article) [22], Srivastava [15]. They varied substantially in the level of details provided about what was meant by more (or less) restrictions. Haghdoost also included a baseline scenario of no interventions [27]. Imperial had three additional ‘surged’ scenarios [13]. Mashayekhi and IHME provided detailed-enough description of their more (or less) restrictions [12, 28]. Policy interventions or restrictions included physical social distancing in Haghdoost [27], Mashayekhi [28], Rahimi Rise [29], and IHME [12]. Haghdoost also used patient detection and isolation [27]. Mashayekhi also included level of hygienic precautions practiced by the general population [28]. Rahimi Rise also included alterations in preparations for use of public transit [29]. IHME included use of masks by the general population [12]. Two studies motioned two levels of ‘no-policy’ and ‘actual policies’; Hsiang [45] and Rahimi Rise [29]. Saberi (web site) [21] and Srivastava [15] included under-reporting in official reports. Ghaffarzadegan, Haghdoost, and IHME included seasonality [12, 27, 41]. Tuite and Zhuang included air travel data scenarios early in the epidemic [46, 47]. Ahmadi started with statistical models and reasoned backwards about what intervention scenarios could match each statistical model [16, 44].
Ghaffarzadegan had two policy effect scenarios with different levels of efforts to decrease contact rate as well as three seasonality condition options, that amounted to six total scenarios [41]. Haghdoost had four final scenarios, each with levels of isolation for the infected and suspected patients, as they maintained that “to postpone the heavy wave of the disease, the most effective tool is isolation of patients, in a way that the infected and suspected patients would have the least contact with healthy people”. In the early stages of model building, they modeled “the effects of people’s behaviour change and seasonality on disease transmission”, to show the basic or worst model. Then three intervention scenarios with different levels of isolations were added. The people’s behaviour change and seasonality scenarios end only in the basic or worst scenario with no intervention [27]. Mashayekhi has three scenarios, each with different levels of social [physical] contacts and observation of sanitation cautions. As such, Mashayekhi and IHME were the only studies that considered two modalities of non-pharmacologic interventions [12, 28]. Details of studies’ scenarios are presented in the Appendix.
There were factors other than deaths and / or cases, one or more of which were considered in 11 studies. Eight studies included policy interventions e.g. distancing, quarantine, use of masks by general population; DELPHI [10], Ghaffarzadegan [41], Haghdoost [27], Hsiang [45], Imperial [13], Mashayekhi [28], Rahimi Rise [29], and Thu [48]. Seven studies included asymptomatic cases; DELPHI [10], Ghaffarzadegan [41], Gu (YYG) [17], Mashayekhi [28], Saberi (article) [22], Rahimi Rise [29], and Srivastava [15]. Six studies included under-reporting and / or delays in reporting; DELPHI [10], Ghaffarzadegan [41], Gu (YYG) [17], Saberi (web site) [21], Saberi (article) [22], and Srivastava [15]. Three studies included seasonality; Ghaffarzadegan [41], Haghdoost [27], and IHME [12]. Two studies included testing availability and / or number of tests performed; Ghaffarzadegan [41] and IHME [12]. Two studies included environmental and meteorological variables; Pourghasemi (PLoS ONE) [38] and Pourghasemi (IJID) [39]. One study included seroprevalence, as well as mobility in population samples; IHME [12]. One study included comorbidities, as well as age stratification for mortality; Imperial [13]. One study included changes to domestic COVID-19-testing regimes, such as case definitions or testing methodology; Hsiang [45]. The following items were not incorporated in any of the scenarios of the included studies: potential vaccine(s), potential pharmacological treatments, changes in cause of death definition, possibility of reinfection, and possibility of mutations or any change in virulence.
Validation process and findings
Eighteen studies provided one or more measures and results for model validation: Root Mean Squared Error (RMSE) or Mean Squared Error (MSE) (9 studies), Mean Absolute Percentage Error (MAPE) (4 studies), Mean Absolute Error (MAE) (3 studies), Ratio Error (RE) (2 studies), R square (2 studies), Area Under Curve (AUC) (2 studies), Akaike Information Criterion (AIC) (2 studies), and Root Mean Squared Relative Error (RMSRE) (1 study).
List and sources of model parameters and input data
Twenty-three studies used deaths and / or cases as input data, source of which included MOHME official reports (10 studies), Johns Hopkins University [4, 5] (6 studies), source not mentioned (6 studies), and Worldometers web site [49] (4 studies). Cases data were used in 23 studies and deaths data in 18 studies. Eleven studies used additional input variables other than deaths and or cases; eight studies used Non-Pharmaceutical Intervention (NPI) variables and three studies used testing data. List and sources of model parameters are available in the Supplementary electronic material (“Studies’ Methods” tab). In Haghdoost’s study, for number of deaths and cases to start with, assumptions were made that on day-zero, there had been 1080 persons exposed to the virus In Iran (including 75 in Tehran), from which 90 persons had become infected in Iran (including 5 in Tehran) [27]. Four studies did not report using number of confirmed cases or confirmed deaths as model input; Tuite [46], Zhuang [47], Haghdoost [27], and Mashayekhi [28]. Among other studies, Ghaffarzadegan used other sources of data, including unofficial reports for number of cases and death and number of performed tests [41].
Model outputs preferably with uncertainty intervals for scenarios
Primary outcomes: The most frequent type of main outcome was cumulative cases only (seven studies). Other studies reported a combination of cumulative or daily deaths or cases. Mashayekhi [28] reported estimates of symptomatic and symptomatic cases separately, and Saberi (article) [22] reported total number of confirmed and suspected cases together. Thirteen studies provided confidence intervals for the primary outcomes.
Forms of primary outcomes: The intended outcomes and the terminology used in the included studies for the same outcomes, varied across the studies. For daily cases, two distinct groups could be recognized: daily incident cases, and daily prevalent cases. Our designation of daily incident cases included “new cases” reported daily by MOHME (via [4, 5]), “new cases” predicted by Haghdoost [27], and “daily cases” by Zareie [50]. Our designation of daily prevalent cases included “current cases” by Ghaffarzadegan [41], “maximum number of cases per day” by Haghdoost [27], “daily cases” by Mashayekhi [28], and “daily active cases” by Saberi (web site) [21]. Active cases are the difference between total cumulative cases with cumulative number of deceased and recovered cases. Saberi (article) reported estimated sum of daily confirmed and suspected cases [22].
Other outcomes: Four studies reported outcomes other than deaths and / or cases. They reported different combinations of hospitalization demand estimates (all beds, intensive care unit beds, invasive ventilators); DELPHI [10], Haghdoost [27], IHME [12], and Imperial [13].
Date range of estimates for deaths and / or cases: Start dates of outputs ranged from 2019-12–31 (Ghaffarzadegan [41]) to 2020-09-19 (Srivastava [15]). End dates of outputs ranged from 2020-02-24 (Zhuang [47]) to 2021-02-02 (Saberi (web site) [21]). Outputs duration ranged from 11 days (Muniz-Rodriguez [37]) to 364 days (IHME [12]).
R0 estimation results: Eleven studies reported estimated Reproductive Number values, ranging from 0.69 (95% CI 0.68–0.70) on 15 April 2020 (after control measures that took place) by Saberi (article) [22], to 7.24 at the beginning of the epidemic by Haghdoost [27].
Twenty-one studies mentioned their study limitations, among which 12 studies really described the limitations, and the nine others touched the limitations very minimally.
Increase in percent population using masks from the current situation (i.e. current scenario) to 95% (i.e. best scenario, 95% mask usage in public in every location) might prevent 26,790 additional deaths (95% confidence interval 19,925–35,208) by the end of year 2020 (IHME [12]).
Table 1 summarizes the findings regarding the methodology used in the reviewed studies. The six fixed dates, the ends of months one to six, are used and shown in Table 2. Table 2 shows the estimates of cumulative deaths. Table 3 summarizes the outcomes at the end of month two (2020-04-19) and month four (2020-06-20) after the official epidemic start date, and the latest dates available in 2020 and 2021. Estimates of cumulative cases, daily deaths and daily cases are demonstrated in Appendix Tables 1, 2, and 3 respectively. Appendix Table 4 demonstrates predictions of peak dates and values of outcomes, and Appendix Table 5 shows predictions of epidemic control dates and values of outcomes.
Table 1.
Reported items of methodology of the reviewed studies
| Study first author | Ahmadi [44] | Al-Qaness [51] | Ayyoubzadeh [52] | DELPHI [10] | Ghaffarzadegan [41] | Gu (YYG) [17] | Haghdoost [27] | |
| Situation of study | Published paper | Published paper | Published paper | Web site | Published paper | Web site | Full report (Farsi) | |
| Epidemic start date | 20-02-19 | 20-01-22 | 20-02-11 | N/M a | 20-01-02 | 20-01-26 | 20-01-21 | |
| Inputs: population | N/M a | N/M a | N/M a | Yes | Yes | Yes | Yes | |
| Inputs: cases | Yes | Yes | Yes | Yes | Yes | No | No | |
| Inputs: cases (source) | MOHME b official reports | World Health Organization | Worldometers website c | Johns Hopkins University d | MOHME b official reports; unofficial reports | Johns Hopkins University d | N/A e | |
| Inputs: deaths | Yes | No | No | Yes | Yes | Yes | No | |
| Inputs: deaths (source) | MOHME b official reports | N/A e | N/A e | Johns Hopkins University d | MOHME b official reports; unofficial reports | Johns Hopkins University d | N/A e | |
| Other input data | Number of cured [recovered] cases | N/A e | N/A e | Nonpharmaceutical interventions | Number of tests, Detected infected travelers, Travel data | Case and hospitalization data f | Post-infection isolated persons, Hospitalized cases, Infected cases recovered without isolation or hospitalization | |
| Output date range (number of days) | 20-02-19 to 20-04-03 (45 days) | 20-01-22 to 20-04-07 (77 days) | 20-02-11 to 20-03-18 (37 days) | 20-06-01 to 20-07-15 (45 days) | 19-12-31 to 20-06-30 (183 days) | 20-01-26 to 20-11-01 (281 days) | 20-01-21 to 20-05-20 (121 days) | |
| Place | Iran | 4 countries | Iran | 148 countries | Iran | 70 countries | Iran and Tehran capital city | |
| Compartmental model g | SIR g | None | None | SEIR+ g, h | SEIR+ g | SEIR g | SEIR+ g | |
| Statistical method: name | 3 growth models i | 6 time-series models j | 2 Models k | Regression trees | Dynamic simulation model | Machine learning | Dynamic model | |
| R0 estimation results | 1.75 | None | None | None | 2.72 (before starting the interventions) | 4 estimates k | 3 estimates l | |
| Scenarios /models: number | 3 m | 1 | 1 | 1 | 6 n | 1 | 4 o | |
| Other factors | No | No | No | Yes. Asymptomatic cases, under-reporting | Yes p | Yes. Asymptomatic cases, under-reporting | Yes q | |
| Primary outcomes | Cumulative deaths, Cumulative cases | Cumulative cases | Normalized Daily cases | Cumulative and daily deaths and cases | Cumulative deaths, Cumulative cases, Current cases | Cumulative and daily deaths and cases, Daily prevalent cases | Cumulative and daily deaths and cases, Daily prevalent cases | |
| Primary outcomes interval estimates | No | No | No | No | No | Yes | No | |
| Other outcomes | None | None | None | Active, Active hospitalized, Cumulative hospitalized, Active ventilated | None | Reproduction Number | Needed hospital beds, ICU beds | |
| Other outcomes interval estimates | N/A e | N/A e | N/A e | No | N/A e | Yes | No | |
| Model validation | No | Yes r | Yes s | No t | Yes u | Yes v | No | |
| Study limitations mentioned | Yes | Yes | Yes | Yes | Yes | Yes | No | |
| Study limitations described | Yes | No | No | Yes | No | Yes | No | |
| Study first author | Hsiang [45] | IHME [12] | Imperial [13] | LANL [14] | Mashayekhi [28] | Moftakhar [53] | Moghadami [36] | |
| Situation of study | Published paper | Web site [12] and published paper [30] | Web site [13] and published paper [34] | Web site | Summary report (Farsi) | Published paper | medRxiv preprint | |
| Epidemic start date | N/M aa | N/M aa | 20-01-03 | N/M aa | 20-02-19 [?] | 20-02-19 | 20-02-19 | |
| Inputs: population | Yes | Yes | Yes | Yes | Yes | No | No | |
| Inputs: cases | Yes | Yes | Yes | Yes | No | Yes | Yes | |
| Inputs: cases (source) | Wikipedia bb | Johns Hopkins University cc | Johns Hopkins University cc | Johns Hopkins University cc | N/A dd | MOHME ee and Johns Hopkins cc | MOHME ee | |
| Inputs: deaths | Yes | Yes | Yes | Yes | No | No | Yes | |
| Inputs: deaths (source) | Wikipedia bb | Johns Hopkins University cc | Johns Hopkins University cc | Johns Hopkins University cc | N/A dd | N/A dd | MOHME ee | |
| Other input data | 3 variables ff | 4 variables gg | 5 variables hh | N/M aa | N/M aa | N/M aa | None | |
| Output date range (number of days) | ~ 20-02-28 to 20-04-06 (~ 39 days) | 20-02-04 to 21-02-01 (364 days) | 20-01-06 to 20-11-24 (324 days) | 20-03-14 to 20-11-07 (239 days) | N/M aa (360 days) | 20-03-21 to 20-04-20 (31 days) | 20-03-21 to 20-04-20 (31 days) | |
| Place | 6 countries | 165 countries | 164 countries | 157 countries | Iran | Iran | Iran and top 5 provinces | |
| Compartmental model ii | SIR+ ii | SEIR ii | SIR, SEIR, SEIR+ ii | SEIR+ ii | SLIR+ ii | None | None | |
| Statistical method: name | Multiple regression | Curve fitting (backcating) functional analysis (forecasting) | Regression trees | Dynamic growth parameter modeling | Dynamic model | Autoregressive Integrated Moving Average (ARIMA) | Exponential smoothing model | |
| R0 estimation results | Not used | N/M aa | N/M aa | N/M aa | Not used | Not used | Not used | |
| Scenarios /models: number | 2 jj | 3 kk | 6 ll | 1 | 3 mm | 1 | 1 | |
| Other factors | Yes. Under-reporting. | Yes nn | Yes. Under-reporting. | Yes. Under-reporting. | Yes oo | No | No | |
| Primary outcomes | Cumulative cases | Cumulative and daily deaths and cases | Cumulative and daily deaths and cases | Cumulative and daily deaths and cases | Cumulative and daily deaths, Daily symptomatic and asymptomatic cases | Daily cases | Cumulative deaths, cases, recovered cases | |
| Primary outcomes interval estimates | Yes | Yes | Yes | Yes | No | Yes | Yes | |
| Other outcomes | None | Yes pp | Yes qq | None | None | None | None | |
| Other outcomes interval estimates | N/A dd | Yes | Yes | N/A dd | N/A dd | N/A dd | N/A dd | |
| Model validation | (?) | Yes rr | Yes ss | Yes tt | No | Yes uu | Yes vv | |
| Study limitations mentioned | Yes | Yes | No | No | Yes | Yes | No | |
| Study limitations described | Yes | Yes | No | No | No | Yes | No | |
| Study first author | Moradi [42] | Muniz-Rodriguez [37] | Pourghasemi (PLoS ONE) [38] | Pourghasemi (IJID) [39] | Rafieenasab [54] | Rahimi Rise [29] | Saberi (web site) [21] | Saberi (paper) [22] |
| Situation of study | Published paper | Published paper | Published paper | Published paper | Published paper | Published paper (Farsi) | Web site [21] | Published paper |
| Epidemic start date | 20-02-20 | 20-02-19 | 20-02-25 [?] | 20-02-25 [?] | 20-02-19 | 20-02-01 | 20-02-19 | 20-02-19 |
| Inputs: population | No | N/M aaa | Yes | Yes | N/M aaa | Yes | N/M aaa | Yes |
| Inputs: cases | No | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| Inputs: cases (source) | N/A bbb | MOHME ccc official reports | MOHME ccc official reports | MOHME ccc official reports | MOHME ccc official reports | Worldometers website ddd | MOHME ccc official reports, WHO, Worldometers ddd | MOHME ccc official reports, WHO |
| Inputs: deaths | Yes | No | Yes | Yes | No | Yes | Yes | Yes |
| Inputs: deaths (source) | MOHME ccc official reports | N/A bbb | MOHME ccc official reports | MOHME ccc official reports | N/A bbb | Worldometers website ddd | MOHME ccc official reports, WHO, Worldometers ddd | MOHME ccc official reports, WHO, Worldometers ddd |
| Other input data | None | Travel data | Environmental and meteorological conditions | Environmental and meteorological conditions | None | Public transportation variables | None | None |
| Output date range (number of days) | 20-02-20 to 20-03-26 (36 days) | 20-02-19 to 20-02-29 (11 days) | ~ 20-02-25 to ~ 20-06-10 (~ 107 days) eee | ~ 20-02-25 to ~ 20-06-20 (~ 117 days) fff | 20-02-19 to 20-06-07 (110 days) | 20-02-01 to 20-08-01 (183 days) | 20-02-19 to 21-02-02 (350 days) | ~ 20-03-19 to 20-10-26 (~ 222 days) |
| Place | Iran | Iran and 2 multi-province regions | Iran and Fars Province | Iran, 31 Provinces of Iran, World | Iran | Iran | Iran | Iran |
| Compartmental model ggg | None | None | None | None | SIR+ ggg, hhh | SEIR ggg | SIR ggg | SEIR+ ggg, iii |
| Statistical method: name | Calculating number of cases based on different assumptions for case fatality rate (CFR) | Generalized growth mode; Based on the calculation of the epidemic doubling times | Autoregressive Integrated Moving Average (ARIMA) and polynomial regression | Fourth-degree polynomial regression | 3-steps model based on the SIR model | Dynamic model | Classical SIRggg mathematical model with homogenous mixing assumption | Ordinary least squares minimization |
| R0 estimation results | Not used | Two methods: 3.6 and 3.58 | Not used | Not used | 2.8–3.3 (range) | Not used | 2.37 (for the last 7 days before 20-03-21) | 1.73 (20-03-01) and 0.69 (2004-15) jjj |
| Scenarios /models: number | 4 kkk | 2 lll | 1 | 1 | 1 | 2 | 12 mmm | 3 nnn |
| Other factors | No | No | No | No | No | Yes. Asymptomatic cases | Yes ooo | Yes ppp |
| Primary outcomes | Cumulative cases | Daily cases | Cumulative and daily deaths and cases qqq | Cumulative and daily deaths and cases rrr | Cumulative and daily deaths, Daily cases | Daily deaths and cases | Cumulative cases, Daily active cases | Fractions of national population estimated to be confirmed and suspected cases sss |
| Primary outcomes interval estimates | No | Yes | No | No | No | No | No | Yes |
| Other outcomes | Case Fatality Rate | None | None | None | None | None | None | Intensive Care Unit beds needed |
| Other outcomes interval estimates | No | N/A bbb | N/A bbb | N/A bbb | N/A bbb | N/A bbb | N/A bbb | Yes |
| Model validation | No | No | Yes ttt | Yes uuu | No | Yes vvv | No | Yes www |
| Study limitations mentioned | Yes | Yes | No | Yes | No | No | Yes | Yes |
| Study limitations described | No | Yes | No | No | No | No | No | Yes |
| Study first author | Shen [43] | Singh [55] | Srivastava [15] | Thu [48] | Tuite [46] | Zhan [40] | Zhuang [47] | |
| Situation of study | Published paper | Published paper | Web site [15] and preprint [35] | Published paper | Published paper | Published paper | Published paper | |
| Epidemic start date | 20-02-20 | N/M aaaa | N/M aaaa | N/M aaaa | N/M aaaa | 20-02-19 | N/M aaaa | |
| Inputs: population | No | No | N/M aaaa | No | N/M aaaa | N/M aaaa | Yes | |
| Inputs: cases | Yes | Yes | Yes | Yes | No | Yes | No | |
| Inputs: cases (source) | “WIND DATA” bbbb | Worldometers cccc | Johns Hopkins University dddd | WHO | N/A bbbb | MOHME eeee official reports | N/A ffff | |
| Inputs: Deaths | No | No | Yes | Yes | No | Yes | No | |
| Inputs: Deaths (source) | N/M aaaa | N/M aaaa | Johns Hopkins University dddd | WHO | N/A ffff | WHO | N/A ffff | |
| Other input data | None | None | None | Social distancing | Exported cases from Iran to other countries; Travel data | COVID-19 spreading profiles of 367 cities in China | Exported cases from Iran to other countries, Travel data | |
| Output date range (number of days) | 20-02-20 to 20-04-20 (61 days) | 20-04-24 to 20-07-07 (75 days) | 20-09-19 to 20-12-19 (every 7th day, 14 dates, 92 days duration) | 20-03-30 to 20-05-02 (34 days) | 20-01-01 to N/M aaaa | 20-02-22 to 20-06-24 (124 days) | 20-02-01 to 20-02-24 (24 days) | |
| Place | 9 countries and 11 provinces / municipalities in China | 15 countries | 184 countries | 10 countries | Iran | Iran and 12 provinces | Iran | |
| Compartmental model gggg | None | None | SIR+ gggg, hhhh | None | None | SEIR+ gggg | None | |
| Statistical method: name | Logistic growth | Autoregressive Integrated Moving Average (ARIMA) | Hyper-parametric learning | Linear growth rates iiii | N/M aaaa | Data-driven prediction algorithm kkkk | Binomial distributed likelihood framework | |
| R0 estimation results | Not used | Not used | 1.44 (20-03-21), 1.46 (20-03-28) | Not used | Not used | Not used | Not used | |
| Scenarios /models: number | 1 | 1 | 3 llll | 1 | 6 mmmm | 1 | 5 nnnn | |
| Other factors | No | No | Asymptomatic cases, under-reporting | No | No | No | No | |
| Primary outcomes | Cumulative cases | Cumulative cases | Cumulative deaths and cases | Daily cases | Cumulative cases | Cumulative and daily cases | Cumulative cases | |
| Primary outcomes interval estimates | No | Yes | No | No | Yes | Yes | Yes | |
| Other outcomes | None | None | None | None | None | None | None | |
| Other outcomes interval estimates | N/A ffff | N/A ffff | N/A ffff | N/A ffff | N/A ffff | N/A ffff | N/A ffff | |
| Model validation | Yes oooo | Yes pppp | Yes qqqq | No | No | Yes jjjj | No | |
| Study limitations mentioned | Yes | Yes | Yes | Yes | No | Yes | Yes | |
| Study limitations described | No | Yes | Yes | Yes | No | Yes | No | |
a N/M: Not Mentioned
b MOHME: Ministry of Health and Medical Education, Iran
c Worldometers Coronavirus [49]
d Johns Hopkins University, Coronavirus Resource Center ([4, 5])
e N/A: Not applicable
f “We do not use case-related data in our modeling. We do look at case and hospitalization data to help determine the bounds for our search grid, as changes in cases lead changes in deaths.” Gu (YYG) [17]
g Compartmental models: S: Susceptible, E: Exposed, I: Infected, R: Removed or Recovered, L: Latent. In any model with a + sign, there are other components for augmentation of model
h DELPHI model: The model underlying the predictions is DELPHI (Differential Equations Leads to Predictions of Hospitalizations and Infections), that is based on SEIR with augmentations for under-detection and governmental response. DELPHI [10]
i Three growth models: M1: Gompertz Differential Equation, M2: Von Bertalanffy differential growth equation, and M3: Cubic polynomial least squared errors
j Six time-series models: (1) Adaptive Neuro-Fuzzy Inference System (ANFIS) enhanced with Genetic Algorithm (GA), (2) Original Adaptive Neuro-Fuzzy Inference System (ANFIS), (3) Particle Swarm Optimizer (PSO), (4) Artificial Bee Colony (ABC), (5) hybridized of Flower Pollination Algorithm and SALP Swarm Algorithm (SSAFPA), (6) Sine-Cosine Algorithm (SCA)
h Two Models: Linear Regression, Long Short-Term Memory (LSTM)
k Four estimates: Initial R0 = 2.65. Reopen R = 1.17. Current R = 1.2. Post-mitigation R = 0.90
l Three estimates: 7.24 (at the beginning). 2.58 (after interventions). 1.82 (conditional to isolation of 50% within 3 days)
m Three scenarios based on 3 growth models: S1: Gompertz Differential Equation, S2: Von Bertalanffy differential growth equation, and S3: Cubic polynomial least squared errors
n Six scenarios based on combination of two factors: Seasonality (S), and Policy interventions (P). (1) S1P1: Seasonality conditions 1 (no effect or status quo) and Policy effect 1 (status quo contact rate). Estimates for 2020-03-19, the end of first month after the epidemic start date, are equal across the six scenarios. (2) S1P2: Seasonality conditions 1 (no effect or status quo) and Policy effect 2 (aggressive efforts to decrease contact rate by half of what it would be otherwise). (3) S2P1: Seasonality conditions 2 (moderate effect; infectivity of the virus decreases linearly from April 1st and halves by June 1st, then stays the same for the rest of the simulation) and Policy effect 1 (status quo contact rate). (4) S2P2: Seasonality conditions 2 (moderate effect; infectivity of the virus decreases linearly from April 1st and halves by June 1st, then stays the same for the rest of the simulation) and Policy effect 2 (aggressive efforts to decrease contact rate by half of what it would be otherwise). (5) S3P1: Seasonality conditions 3 (very strong mitigating effect; infectivity of the virus decreases from April 1st to a quarter of its base value by June 1st, then stays the same for the rest of the simulation) and Policy effect 1 (status quo contact rate). (6) S3P2: Seasonality conditions 3 (very strong mitigating effect; infectivity of the virus decreases from April 1st to a quarter of its base value by June 1st, then stays the same for the rest of the simulation) and Policy effect 2 (aggressive efforts to decrease contact rate by half of what it would be otherwise)
o Four scenarios: S0: Basic scenario (no intervention), only 10% isolation. S1: Worst scenario, minimum (25%) isolation. S2: Medium scenario, medium (32%) isolation. S3: Best scenario, maximum (40%) isolation
p Seven other factors included: Asymptomatic cases, Under-reporting / Completeness of reporting cases and deaths to MOHME, Delays in reporting cases and deaths to MOHME, Testing availability, Number of tests performed, Social distancing / Quarantine interventions, Seasonality
q Two other factors included: Seasonality, Social distancing / Quarantine interventions
r Root Mean Squared Error (RMSE), Mean Absolute Error (MAE), Mean Absolute Percentage Error (MAPE), Root Mean Squared Relative Error (RMSRE), and Coefficient of Determination (R square)
s Root Mean Squared Error (RMSE)
t Friedman [31] assessed predictive performance of international COVID-19 mortality forecasting models, using median absolute percent error (MAPE) and Median absolute errors (MAE)
u Root Mean Squared Error (RMSE)
v Mean Square Error (MSE), Mean Absolute Error (MAE), and Ratio Error (RE). Did not mention the results
aa N/M: Not Mentioned
bb Wikipedia. COVID-19 pandemic in Iran [56]
cc Johns Hopkins University, Coronavirus Resource Center ([4, 5])
dd N/A: Not Applicable
ee MOHME: Ministry of Health and Medical Education, Iran
ff Four variables: Cumulative recoveries, Active cases, Any changes to domestic COVID-19-testing regimes, such as case definitions or testing methodology, and Non-pharmaceutical interventions
gg Three variables: Mobility, Testing, and Seroprevalence (the latter for 41 locations)
hh Five variables: Interventions, Social contacts, Comorbidities, Hospital bed capacity, Intensive Care Unit bed capacity
ii Compartmental models: S: Susceptible, E: Exposed, I: Infected, R: Removed or Recovered, L: Latent. In any model with a + sign, there are other components for augmentation of model
jj Two scenarios: ‘No-policy scenario’ and ‘Actual policies’
kk Three scenarios: S1 Best (Masks): ‘Universal Masks’ scenario reflects 95% mask usage in public in every location. S2 Reference (Current): ‘Current projection’ scenario assumes social distancing mandates are re-imposed for 6 weeks whenever daily deaths reach 8 per million (0.8 per 100,000). S3 Worse (Easing): ‘Mandates easing’ scenario reflects continued easing of social distancing mandates, and mandates are not re-imposed
ll Six scenarios: S1: Additional 50% Reduction. S2: Maintain Status Quo. S3: Relax Interventions 50%. S4: Surged Additional 50% Reduction. S5: Surged Maintain Status Quo. S6: Surged Relax Interventions 50%
mm S1: Ideal scenario, serious distancing. People reduce their social [physical] contacts to 20% of regular level, voluntarily or on a forced basis, after number of cases and deaths have increased, plus close observation of sanitation cautions, so that transmission rate reduces by 65%. S2: Medium scenario, not serious distancing. People reduce their social [physical] contacts only to 20% of regular level, voluntarily, after number of cases and deaths have increased, and other settings are like scenario 1. S3: Worst scenario. People reduce their social [physical] contacts only to 50% of regular level, voluntarily, after number of cases and deaths have increased, plus inadequate observation of sanitation cautions, so that transmission rate reduces only by 40% (instead of 55%), and 60% of people do not observe the sanitation cautions
nn Five other factors included: Asymptomatic cases, Mobility, Testing, Seroprevalence, Seasonality
oo Two other factors included: Asymptomatic cases, Social distancing / Quarantine interventions
pp Six other outcomes: All beds needed, Intensive Care Unit beds needed, Invasive ventilators needed, Tests, Mobility, Seroprevalence
qq Five other outcomes: Hospital demand, Hospital incidence, Intensive Care Unit demand, Intensive Care Unit incidence, Rt (Effective Reproduction Number)
rr IHME web site [12] refers to Friedman [31], who assessed predictive performance of international COVID-19 mortality forecasting models, using median absolute percent error (MAPE) and Median absolute errors (MAE)
ss Mean Absolute Percentage Error (MAPE)
tt They validated the model “by looking at the coverage of the forecasts, i.e. the proportion of times that the number of confirmed cases/deaths fell within a specified lower and upper bound, X min and X max. Coverage plots can help visualize how well the model is doing”
uu Graphical residual assessment of the model
vv Mean Squared Error (MSE), Root Mean Squared Error (RMSE), Mean Absolute Error (MEA), Mean Absolute Percentage Error (MAPE), Akaike Information Criterion (AIC)
aaa N/M: Not Mentioned
bbb N/A: Not Applicable
ccc MOHME: Ministry of Health and Medical Education, Iran
ddd Worldometers Coronavirus [49]
eee Start and end dates mentioned in manuscript text, mentioned in title of their Fig. 14, and shown within their Fig. 14 do not seem to be congruent
fff Start and end dates mentioned in manuscript text, mentioned in title of their Fig. 15, and shown within their Fig. 15 do not seem to be congruent
ggg Compartmental models: S: Susceptible, E: Exposed, I: Infected, R: Removed or Recovered, L: Latent. In any model with a + sign, there are other components for augmentation of model
hhh SIR with exact and approximated solutions, extrapolation based on least squares model with three functions
iii SEIR+ Distinguishing between fatal and recovered cases combined with an estimate of the percentage of symptomatic cases using delay-adjusted Case Fatality Rate
jjj Estimated effective reproduction number that ranged from 0.66 to 1.73 between February and April 2020, with a median of 1.16. Estimated a reduction in the effective reproduction number during this period, from 1.73 (95% CI 1.60–1.87) on 1 March 2020 to 0.69 (95% CI 0.68–0.70) on 15 April 2020, due to various non-pharmaceutical interventions
kkk Four scenarios based on different values of Case Fatality Rate. S1: 0.3%, S2: 0.5%, S3: 1%, and S4: 2%
lll Based on two different methods to estimate R0
mmm (1) S1P10: Scenario 1 (Best scenario, based on official reports with correction factor of 1) with 10 million susceptible population. (2) 12 scenarios based on combination of three options for number of cases and deaths to start with, and four options for the susceptible population size. (1) S1P10: Scenario 1 (Best scenario, based on official reports with correction factor of 1) with 10 million susceptible population. (2) S1P30:Scenario 1 (Best scenario, based on official reports with correction factor of 1) with 30 million susceptible population. (3) S1P50: Scenario 1 (Best scenario, based on official reports with correction factor of 1) with 50 million susceptible population. (4) S1P80: Scenario 1 (Best scenario, based on official reports with correction factor of 1) with 80 million susceptible population. (5) S2P10: Scenario 2 (Medium scenario, based on official reports with correction factor of 5 (after Dr. Rick Brennan, Director of Emergency Operations, World Health Organization [57]) with 10 million susceptible population. (6) S2P30: Scenario 2 (Medium scenario, based on official reports with correction factor of 5 (after Dr. Rick Brennan, Director of Emergency Operations, World Health Organization [57]) with 30 million susceptible population. (7) S2P50: Scenario 2 (Medium scenario, based on official reports with correction factor of 5 (after Dr. Rick Brennan, Director of Emergency Operations, World Health Organization [57]) with 50 million susceptible population. (8) S2P80: Scenario 2 (Medium scenario, based on official reports with correction factor of 5 (after Dr. Rick Brennan, Director of Emergency Operations, World Health Organization [57]) with 80 million susceptible population. (9) S3P10: Scenario 3 (Worst scenario, based on official reports with correction factor of 10 (after Russell [58]) with 80 million susceptible population. (10) S3P30: Scenario 3 (Worst scenario, based on official reports with correction factor of 10 (after Russell [58]) with 30 million susceptible population. (11) S3P50: Scenario 3 (Worst scenario, based on official reports with correction factor of 10 (after Russell [58]) with 50 million susceptible population. (12) S3P80: Scenario 3 (Worst scenario, based on official reports with correction factor of 10 (after Russell [58]) with 10 million susceptible population
nnn Three scenarios: (1) maintaining the same level of control measures as of 12 April 2020, (2) reinforcing the control measures to increase physical distancing by a 20% increase in the reproduction number, and (3) partial lifting the restrictions to ease physical distancing by a 20% decrease in the reproduction number
ooo Completeness of reporting cases and deaths to MOHME
ppp Accounted for the under-reporting of the number of infected cases using delay-adjusted case fatality ratio (CFR) approach
qqq Cumulative deaths and cases (for Iran and Fars Province), Daily deaths and cases (for Fars Province)
rrr Cumulative deaths and cases (for Iran and World), Daily or cumulative cases in 30 days after the first day of infected cases in the 31 Iranian provinces)
sss We transformed their reported fractions of national population estimated to be confirmed and suspected cases to numbers of people estimated to be confirmed and suspected cases, using a total national population of 84,297,880 (used by IHME [12])
ttt Area Under Curve (AUC)
uuu Area Under Curve (AUC)
vvv Root Mean Squared Error (RMSE)
www Root Mean Squared Error (RMSE)
aaaa N/M: Not mentioned
bbbb Mentioned: “WIND DATA, a leading financial data services provider in China”
cccc Worldometers Coronavirus [49].
dddd Johns Hopkins University, Coronavirus Resource Center ([4, 5])
eeee MOHME: Ministry of Health and Medical Education, Iran
ffff N/A: Not Applicable
gggg SEIR+ Distinguishing between fatal and recovered cases combined with an estimate of the percentage of symptomatic cases using delay-adjusted Case Fatality Rate
hhhh SI-kJ alpha model: S: Susceptible. I: Infected. k: k sub-states of infection. J: J is a hyperparameter introduced for a smoothing effect to deal with noisy data. Alpha: an additional hyperparameter to minimizes the Root Mean Squared Error
iiii They have not named their method. It could be names as linear growth rates, according to their Eq. (1) and Eq. (2)
j Another study by Zhan and colleagues was cited for validity of their models
kkkk A data-driven prediction algorithm to find the most resembling growth curve from the historical profiles in China
llll Three scenarios: Current, Released, Restricted, each with 6 levels of putative under-ascertainment parameter
mmmm Six scenarios based on six sets of international travel destinations
nnnn Five scenarios based on selected combinations of (1) Effective catchment population, (2) Detection window 10 or 8 days, and (3) 90% or 70% load factors
oooo R Square
pppp Akaike Information Criterion (AIC)
qqqq Root Mean Squared Error (RMSE)
Table 2.
Predictions of cumulative deaths for the end of months one to six after the official epidemic start date (2020-02-19) and the latest date available in 202
| Date1a | 20-03-19 | 20-04-19 | 20-05-20 | 20-06-20 | 20-07-21 | 20-08-21 | Latest date | |
|---|---|---|---|---|---|---|---|---|
| Date 2 b | 98-12-29 | 99-01-31 | 99-02-31 | 99-03-31 | 99-04-31 | 99-05-31 | in 2020 c | |
| - First Author, Outcome | S/M d | Value | Value | Value | Value | Value | Value | Value |
| - MOHME official via ([4, 5]) | ||||||||
| Cumulative deaths | N/A e | 1,284 | 5,118 | 7,183 | 9,507 | 14,634 | 20,376 | 30,712 |
| - Ahmadi [44] | ||||||||
| Cumulative deaths | M1 f | 1,264 | ·· | ·· | ·· | ·· | ·· | ·· |
| Cumulative deaths | M2 g | 1,322 | ·· | ·· | ·· | ·· | ·· | ·· |
| Cumulative deaths | M3 h | 1,263 | ·· | ·· | ·· | ·· | ·· | ·· |
| - DELPHI [10] | ||||||||
| Total detected deaths | S1i | ·· | ·· | ·· | 8,426 | ·· | ·· | ·· |
| - Ghaffarzadegan [41] | ||||||||
| Cumulative deaths | S1P1 j | 15,317 | 44,078 | 70,462 | 95,658 | ·· | ·· | ·· |
| Cumulative deaths | S1P2 k | 15,317 | 41,702 | 52,937 | 66,549 | ·· | ·· | ·· |
| Cumulative deaths | S2P1 l | 15,317 | 44,078 | 68,383 | 85,262 | ·· | ·· | ·· |
| Cumulative deaths | S2P2 m | 15,317 | 41,702 | 52,937 | 60,015 | ·· | ·· | ·· |
| Cumulative deaths | S3P1 n | 15,317 | 44,078 | 68,383 | 80,213 | ·· | ·· | ·· |
| Cumulative deaths | S3P2 o | 15,317 | 41,702 | 52,937 | 57,341 | ·· | ·· | ·· |
| - Gu (YYG) [17] | ||||||||
| Cumulative deaths, mean | S1i | ·· | ·· | ·· | ·· | ·· | 31,955 | ·· |
| Cumulative deaths, lower | S1i | ·· | ·· | ·· | ·· | ·· | 29,231 | ·· |
| Cumulative deaths, upper | S1i | ·· | ·· | ·· | ·· | ·· | 36,014 | ·· |
| - Haghdoost [27] | ||||||||
| Cumulative deaths | S0 p | ·· | ·· | 30,700 | ·· | ·· | ·· | ·· |
| Cumulative deaths | S1 q | 3,824 | 9,107 | 13,450 | ·· | ·· | ·· | ·· |
| Cumulative deaths | S2 r | 2,796 | 6,231 | 8,632 | ·· | ·· | ·· | ·· |
| Cumulative deaths | S3 s | ·· | ·· | 6,030 | ·· | ·· | ·· | ·· |
| - IHME [12] | ||||||||
| Cumulative deaths, mean t | S1 u | 1,215 | 5,150 | 7,183 | 9,495 | 14,642 | 20,369 | 44,087 |
| Cumulative deaths, lower t | S1 u | 1,215 | 5,150 | 7,183 | 9,495 | 14,642 | 20,369 | 38,031 |
| Cumulative deaths, upper t | S1 u | 1,215 | 5,150 | 7,183 | 9,495 | 14,642 | 20,369 | 51,027 |
| Cumulative deaths, mean t | S2 v | 1,215 | 5,150 | 7,183 | 9,495 | 14,642 | 20,369 | 67,186 |
| Cumulative deaths, lower t | S2 v | 1,215 | 5,150 | 7,183 | 9,495 | 14,642 | 20,369 | 57,913 |
| Cumulative deaths, upper t | S2 v | 1,215 | 5,150 | 7,183 | 9,495 | 14,642 | 20,369 | 72,170 |
| Cumulative deaths, mean t | S3 w | 1,215 | 5,150 | 7,183 | 9,495 | 14,642 | 20,369 | 70,877 |
| Cumulative deaths, lower t | S3 w | 1,215 | 5,150 | 7,183 | 9,495 | 14,642 | 20,369 | 57,956 |
| Cumulative deaths, upper t | S3 w | 1,215 | 5,150 | 7,183 | 9,495 | 14,642 | 20,369 | 86,235 |
| - Imperial [13] | ||||||||
| Cumulative deaths, mean | S1 x | 763 | 3,743 | 5,276 | 7,303 | 11,537 | 16,538 | 27,195 |
| Cumulative deaths, lower | S1 x | 434 | 2,095 | 3,067 | 4,203 | 6,537 | 9,895 | 17,638 |
| Cumulative deaths, upper | S1 x | 1,254 | 6,096 | 8,462 | 11,620 | 17,058 | 23,543 | 36,103 |
| Cumulative deaths, mean | S2 y | 763 | 3,743 | 5,276 | 7,303 | 11,537 | 16,538 | 32,372 |
| Cumulative deaths, lower | S2 y | 434 | 2,095 | 3,067 | 4,203 | 6,537 | 9,895 | 19,989 |
| Cumulative deaths, upper | S2 y | 1,254 | 6,096 | 8,462 | 11,620 | 17,058 | 23,543 | 45,124 |
| Cumulative deaths, mean | S3 z | 763 | 3,743 | 5,276 | 7,303 | 11,537 | 16,538 | 121,960 |
| Cumulative deaths, lower | S3 z | 434 | 2,095 | 3,067 | 4,203 | 6,537 | 9,895 | 42,697 |
| Cumulative deaths, upper | S3 z | 1,254 | 6,096 | 8,462 | 11,620 | 17,058 | 23,543 | 252,429 |
| Cumulative deaths, mean | S4 aa | 744 | 3,616 | 5,089 | 7,053 | 11,102 | 15,908 | 26,738 |
| Cumulative deaths, lower | S4 aa | 388 | 1,777 | 2,572 | 3,590 | 5,960 | 9,019 | 16,176 |
| Cumulative deaths, upper | S4 aa | 1,127 | 5,712 | 8,112 | 10,778 | 16,171 | 22,470 | 35,621 |
| Cumulative deaths, mean | S5 bb | 744 | 3,616 | 5,089 | 7,053 | 11,102 | 15,908 | 31,916 |
| Cumulative deaths, lower | S5 bb | 388 | 1,777 | 2,572 | 3,590 | 5,960 | 9,019 | 18,504 |
| Cumulative deaths, upper | S5 bb | 1,127 | 5,712 | 8,112 | 10,778 | 16,171 | 22,470 | 48,300 |
| Cumulative deaths, mean | S6 cc | 744 | 3,616 | 5,089 | 7,053 | 11,102 | 15,908 | 85,087 |
| Cumulative deaths, lower | S6 cc | 388 | 1,777 | 2,572 | 3,590 | 5,960 | 9,019 | 40,819 |
| Cumulative deaths, upper | S6 cc | 1,127 | 5,712 | 8,112 | 10,778 | 16,171 | 22,470 | 158,299 |
| - LANL [14] | ||||||||
| Cumulative deaths, median | S1i | 1,284 | 5,118 | 7,183 | 9,507 | 14,634 | 20,376 | 34,263 |
| Cumulative deaths, lower | S1i | 1,284 | 5,118 | 7,183 | 9,507 | 14,634 | 20,376 | 30,762 |
| Cumulative deaths, upper | S1i | 1,284 | 5,118 | 7,183 | 9,507 | 14,634 | 20,376 | 43,022 |
| - Mashayekhi [28] | ||||||||
| Cumulative deaths | S1 dd | 759 | 10,316 | 11,751 | 11,857 | ·· | ·· | ·· |
| Cumulative deaths | S2 ee | 1,285 | 33,349 | 61,322 | 77,302 | 86,931 | 92,620 | ·· |
| Cumulative deaths | S3 ff | 11,752 | 97,445 | 612,953 | 1,819,392 | 3,002,721 | 3,562,136 | ·· |
| - Moghadami [36] | ||||||||
| Cumulative deaths, meangg | S1i | 1,144 | 5,378 | ·· | ·· | ·· | ·· | ·· |
| Cumulative deaths, lowergg | S1i | 1,104 | 3,929 | ·· | ·· | ·· | ·· | ·· |
| Cumulative deaths, uppergg | S1i | 1,166 | 7,003 | ·· | ·· | ·· | ·· | ·· |
| - Rafieenasab [54] | ||||||||
| Cumulative deaths | S2 hh | 32,101 | 39,026 | ·· | ·· | ·· | ·· | ·· |
| Cumulative deaths | S3 ii | 69,583 | 388,951 | 402,569 | ·· | ·· | ·· | ·· |
| - Srivastava [15] | ||||||||
| Cumulative deaths | S1P1 jj | ·· | ·· | ·· | ·· | ·· | ·· | 43,631 |
| Cumulative deaths | S1P2 kk | ·· | ·· | ·· | ·· | ·· | ·· | 43,282 |
| Cumulative deaths | S1P5 ll | ·· | ·· | ·· | ·· | ·· | ·· | 42,289 |
| Cumulative deaths | S1P10 mm | ·· | ·· | ·· | ·· | ·· | ·· | 40,802 |
| Cumulative deaths | S1P20 nn | ·· | ·· | ·· | ·· | ·· | ·· | 38,324 |
| Cumulative deaths | S1P40 oo | ·· | ·· | ·· | ·· | ·· | ·· | 34,721 |
| Cumulative deaths | S2P1 pp | ·· | ·· | ·· | ·· | ·· | ·· | 418,834 |
| Cumulative deaths | S2P2 qq | ·· | ·· | ·· | ·· | ·· | ·· | 354,756 |
| Cumulative deaths | S2P5 rr | ·· | ·· | ·· | ·· | ·· | ·· | 241,214 |
| Cumulative deaths | S2P10 ss | ·· | ·· | ·· | ·· | ·· | ·· | 154,826 |
| Cumulative deaths | S2P20 tt | ·· | ·· | ·· | ·· | ·· | ·· | 87,664 |
| Cumulative deaths | S2P40 uu | ·· | ·· | ·· | ·· | ·· | ·· | 45,995 |
| Cumulative deaths | S3P1 vv | ·· | ·· | ·· | ·· | ·· | ·· | 27,959 |
| Cumulative deaths | S3P2 ww | ·· | ·· | ·· | ·· | ·· | ·· | 27,786 |
| Cumulative deaths | S3P5 xx | ·· | ·· | ·· | ·· | ·· | ·· | 27,327 |
| Cumulative deaths | S3P10 yy | ·· | ·· | ·· | ·· | ·· | ·· | 26,724 |
| Cumulative deaths | S3P20 zz | ·· | ·· | ·· | ·· | ·· | ·· | 25,909 |
| Cumulative deaths | S3P40 aaa | ·· | ·· | ·· | ·· | ·· | ·· | 25,043 |
a Date 1: Gregorian
b Date 2: Hijri
c Latest date in 2020: As of 2020-10-19 for MOHME official via ([4, 5]), 2020-11-01 for Gu (YYG) [17], 2020–12–31 for IHME [12] and Imperial [13] [2020-11-28 for LANL [14], and 2020-12-19 for Srivastava [15]
d S/M: Scenario / Model
e N/A: Not Applicable
f M1: Gompertz
g M2: Von Bertalanffy growth
h M3: Cubic Polynomial
i S1: Single scenario
j S1P1: Seasonality conditions 1 (no effect or status quo) and Policy effect 1 (status quo contact rate). Estimates for 2020-03-19, the end of first month after the epidemic start date, are equal across the six scenarios
k S1P2: Seasonality conditions 1 (no effect or status quo) and Policy effect 2 (aggressive efforts to decrease contact rate by half of what it would be otherwise)
l S2P1: Seasonality conditions 2 (moderate effect; infectivity of the virus decreases linearly from April 1st and halves by June 1st, then stays the same for the rest of the simulation) and Policy effect 1 (status quo contact rate)
m S2P2: Seasonality conditions 2 (moderate effect; infectivity of the virus decreases linearly from April 1st and halves by June 1st, then stays the same for the rest of the simulation) and Policy effect 2 (aggressive efforts to decrease contact rate by half of what it would be otherwise)
n S3P1: Seasonality conditions 3 (very strong mitigating effect; infectivity of the virus decreases from April 1st to a quarter of its base value by June 1st, then stays the same for the rest of the simulation) and Policy effect 1 (status quo contact rate)
o S3P2: Seasonality conditions 3 (very strong mitigating effect; infectivity of the virus decreases from April 1st to a quarter of its base value by June 1st, then stays the same for the rest of the simulation) and Policy effect 2 (aggressive efforts to decrease contact rate by half of what it would be otherwise)
p S0: Basic scenario (no intervention), only 10% isolation
q S1: Worst scenario, minimum (25%) isolation
r S2: Medium scenario, medium (32%) isolation
s S3: Best scenario, maximum (40%) isolation
t Smoothed estimates
u S1 Best (Masks): ‘Universal Masks’ scenario reflects 95% mask usage in public in every location
v S2 Reference (Current): ‘Current projection’ scenario assumes social distancing mandates are re-imposed for 6 weeks whenever daily deaths reach 8 per million (0.8 per 100,000)
w S3 Worse (Easing): ‘Mandates easing’ scenario reflects continued easing of social distancing mandates, and mandates are not re-imposed
x S1: Additional 50% Reduction
y S2: Maintain Status Quo
z S3: Relax Interventions 50%
aa S4: Surged Additional 50% Reduction
bb S5: Surged Maintain Status Quo
cc S6: Surged Relax Interventions 50%
dd S1: Serious distancing
ee S2: Not serious distancing
ff S3: Worse than Scenario 2
gg Dates for Moghadami [36] are 2020-03-21 and 2020-04-18, instead of 2020-03-19 and 2020-04-19 respectively
hh S2: Based on SIR model
ii S3: Approximation calculation
jj S1P1: Scenario Current, Parameter 1
kk S1P2: Scenario Current, Parameter 2
ll S1P5: Scenario Current, Parameter 5
mm S1P10: Scenario Current, Parameter 10
nn S1P20: Scenario Current, Parameter 20
oo S1P40: Scenario Current, Parameter 40
pp S2P1: Scenario Released, Parameter 1
qq S2P2: Scenario Released, Parameter 2
rr S2P5: Scenario Released, Parameter 5
ss S2P10: Scenario Released, Parameter 10
tt S2P20: Scenario Released, Parameter 20
uu S2P40: Scenario Released, Parameter 40
vv S3P1: Scenario Restricted, Parameter 1
ww S3P2: Scenario Restricted, Parameter 2
xx S3P5: Scenario Restricted, Parameter 5
yy S3P10: Scenario Restricted, Parameter 10
zz S3P20: Scenario Restricted, Parameter 20
aaa S3P40: Scenario Restricted, Parameter 40
Table 3.
Lowest and highest predictions at the end of month 2 (2020-04-19), month 4 (2020-06-20) after the official epidemic start date (2020-02-19), and the latest dates available in 2020 and 2021
| Outcomes: | Lowest | Study | MOHME | Highest | Study |
|---|---|---|---|---|---|
| - End of month 2 (20–04-19) | |||||
| Cumulative deaths | 1,777 | Imperial a | 5,118 | 388,951 | Rafieenasab b |
| Daily deaths | 30 | Imperial a | 87 | 11,289 | Rahimi Rise c |
| Cumulative cases | 20,588 | Al-Qaness d | 82,211 | 2,310,161 | IHME e |
| Incident daily cases | 93 | Thu f | 1,343 | 216,262 | Rahimi Rise c |
| Incident daily total cases g k | 72,950 | Saberi (paper) h | .. | 1,616,385 | Saberi (paper) i |
| - End of month 4 (20–06-20) | |||||
| Cumulative deaths | 3,590 | Imperial a | 9,507 | 1,819,392 | Mashayekhi j |
| Daily deaths | 5 | Mashayekhi k | 115 | 44,934 | Mashayekhi j |
| Cumulative cases | 144,305 | DELPHI l | 202,584 | 4,266,964 | IHME e |
| Incident daily cases | 211 | DELPHI l | 2,322 | 138,892 | Gu (YYG) m |
| Incident daily total cases g | 9,625 | Saberi (paper) h | .. | 1,255,012 | Saberi (paper) i |
| - Latest date available in 2020 | |||||
| Cumulative deaths | 16,176 | Imperial n | 30,712 o | 418,834 | Srivastava p |
| Daily deaths | 0 | Imperial a | 373 o | 3,984 | Imperial q |
| Cumulative cases | 3,588,293 | Imperial n | 534,631o | 41,475,792 | Imperial q |
| Incident daily cases | 0 | Imperial a | 4,251o | 486,745 | Imperial e |
| Incident daily total cases g | 9,625 | Saberi (paper) h | .. | 169,110 | Saberi (paper) i |
| - Latest date available in 2021 | |||||
| Cumulative deaths | 40,151 | IHME r | .. | 125,690 | IHME s |
| Daily deaths | 55 | IHME r | .. | 1,093 | IHME s |
| Cumulative cases | 19,799,934 | IHME r | .. | 34,417,912 | IHME s |
| Incident daily cases | 14,818 | IHME r | .. | 236,781 | IHME s |
a Imperial, S4: Surged Additional 50% Reduction. Lower 95% uncertainty limit [13]
b Rafieenasab, S3: Approximation calculation. Mean estimate [54]
c Rahimi Rise, S2: No interventions. Mean estimate [29]
d Al-Qaness, M1: Adaptive Neuro-Fuzzy Inference System (ANFIS) enhanced with Genetic Algorithm (GA). Mean estimate [51]
e IHME, S2 Reference (Current): ‘Current projection’ scenario assumes social distancing mandates are re-imposed for 6 weeks whenever daily deaths reach 8 per million (0.8 per 100,000). Upper 95% uncertainty limit [12]
f Thu, M1: Linear growth rate, eq. 1. Mean estimate [48]
g Saberi (paper), Incident daily total cases (confirmed and suspected) [22]
h Saberi (paper), S1: 20% more distancing. Mean estimate [22]
i Saberi (paper), S3: 20% less distancing. Upper 95% uncertainty limit [22]
j Mashayekhi, S3: Worse than Scenario 2 (S2: Not serious distancing). Mean estimate [28]
k Mashayekhi, S1: S1: Serious distancing. Mean estimate [28]
l DELPHI, S1: Single scenario. Mean estimate [10]
m Gu (YYG) S1, Single scenario. Upper 95% uncertainty limit [17]
n Imperial, S4: Surged Additional 50% Reduction. Lower 95% uncertainty limit. For 2020-12-31 [13]
o MOHME official via ([4, 5]), as of 2020-10-19
p Srivastava, S2P1: Scenario Released, Parameter 1. mean estimate For 2020-12-19 [15]
q Imperial, S3: Relax Interventions 50%. Upper 95% uncertainty limit. For 2020-12-31 [13]
r IHME, S1Best (Masks): ‘Universal Masks’ scenario reflects 95% mask usage in public in every location. Lower 95% uncertainty limit. For 2021-01-31 [12]
s IHME, S3 Worse (Easing): ‘Mandates easing’ scenario reflects continued easing of social distancing mandates, and mandates are not re-imposed. Upper 95% uncertainty limit. For 2021-01-31 [12]
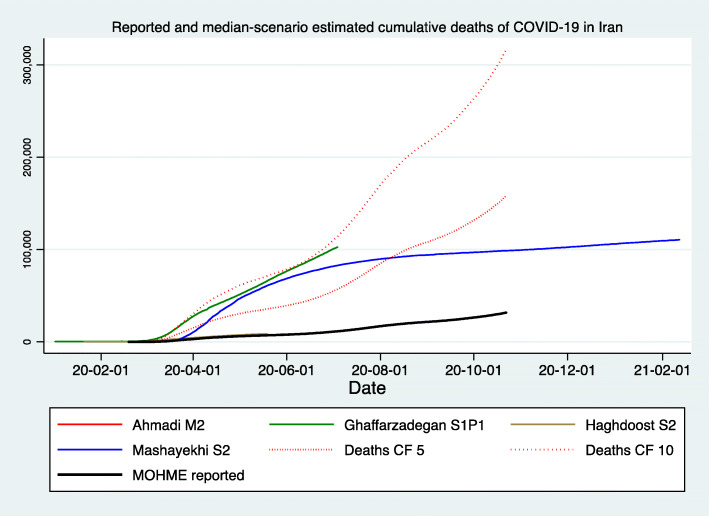
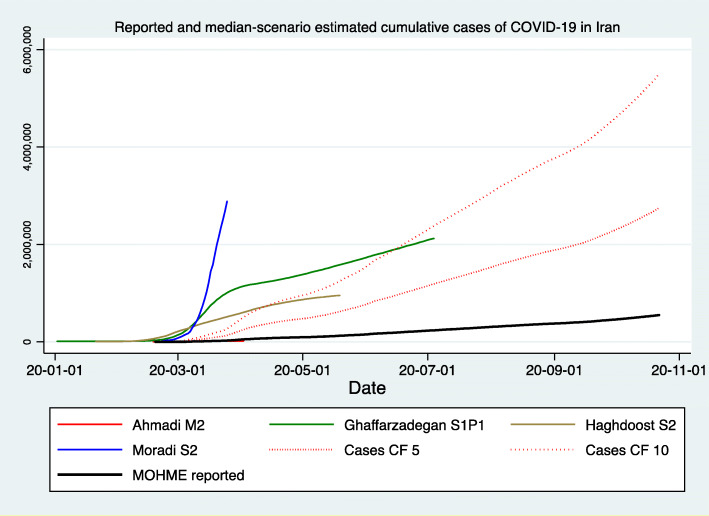
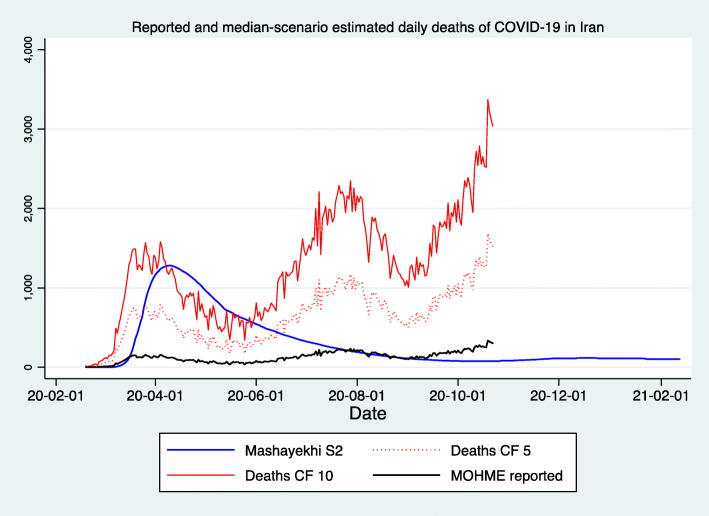
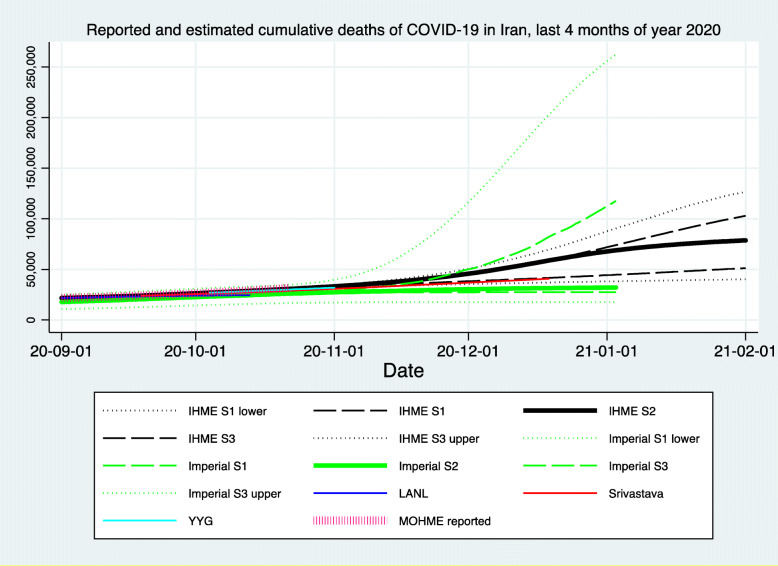
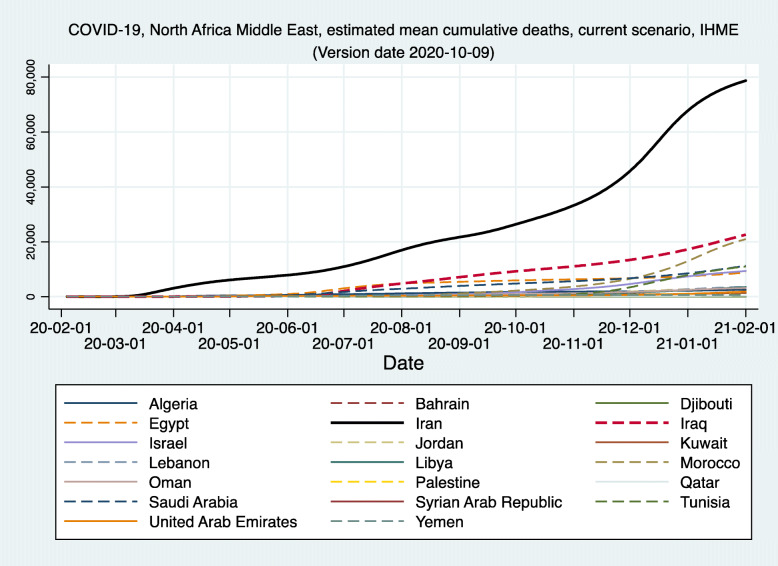
Figures 1, 2, and 3 demonstrate the reported and estimated outcomes in median scenarios. Figures 1 and 2 show the cumulative deaths and cumulative cases respectively. Figure 3 shows the daily deaths. In general, international studies tend to replicate or imitate the official national reports of deaths and cases up to the date that they are available and estimate the future trajectories for when the official reports seize to be available. Therefore, adding such estimates from the international studies does not provide added value. Hence, international studies’ estimates are not added to Figs. 1, 2 and 3. We created two graphs for the international studies’ estimates for Iran. The first one (Appendix Figure 8) shows officially reported and “current scenario” estimates of cumulative deaths for the whole time period available, starting on 2020-01-03 (Imperial [13]) and ending with 2021-02-01 (IHME [12]). The second graph for the international studies’ estimates for Iran is Fig. 4 and shows officially reported and “current scenario” estimates of cumulative deaths for the last 4 months of 2020 and January 2021, as well as highest and lowest estimates from other scenarios. Figure 5 shows the “current scenario” estimates of cumulative deaths by IHME for Iran and 20 other countries in North Africa Middle East region.
Fig. 1.
Reported and median-scenario estimated cumulative deaths of COVID-19 in Iran . (1) Ahmadi M2: Model 2, Von Bertalanffy (Curve lies behind MOHME reported) [44]. (2) Ghaffarzadegan S1P1: Seasonality conditions 1 (no effect or status quo) and Policy effect 2 (aggressive efforts to decrease contact rate by half of what it would be otherwise) [41]. (3) Haghdoost S2: Medium scenario, medium (32%) isolation (Curve lies behind MOHME reported) [27]. (4) Mashayekhi S2: Medium scenario, not serious distancing; People reduce their social [physical] contacts only to 20% of regular level, voluntarily, after number of cases and deaths have increased, and other settings are like scenario one [28]. (5) Deaths CF 5: Reported deaths with a Correction Factor of 5, after Dr. Rick Brennan, Director of Emergency Operations, World Health Organization [57]. (6) Deaths CF 10: Reported deaths with a Correction Factor of 10, after Russell [58]. (7) MOHME reported: Official reported deaths via [4, 5]
Fig. 2.
Reported and median-scenario estimated cumulative case of COVID-19 in Iran. (1) Ahmadi M2: Model 2, Von Bertalanffy (Curve lies behind MOHME reported) [44]. (2) Ghaffarzadegan S1P1: Seasonality conditions 1 (no effect or status quo) and Policy effect 2 (aggressive efforts to decrease contact rate by half of what it would be otherwise) [41]. (3) Haghdoost S2: Medium scenario, medium (32%) isolation [27]. (4) Moradi S2: Scenario 2, Case Fatality Rate, 0.5% [42]. (5) Cases CF 5: Reported cases with a Correction Factor of 5, after Dr. Rick Brennan, Director of Emergency Operations, World Health Organization [57]. (6) Cases CF 10: Reported cases with a Correction Factor of 10, after Russell [58]. (7) MOHME reported: Official reported cases via [4, 5]
Fig. 3.
Reported and median-scenario estimated daily deaths of COVID-19 in Iran. (1) Mashayekhi S2: Medium scenario, not serious distancing; People reduce their social [physical] contacts only to 20% of regular level, voluntarily, after number of cases and deaths have increased, and other settings are like scenario 1 [28]. (2) Deaths CF 5: Reported deaths with a Correction Factor of 5, after Dr. Rick Brennan, Director of Emergency Operations, World Health Organization [57]. (3) Deaths CF 10: Reported deaths with a Correction Factor of 10, after Russell [58]. (4) MOHME reported: Official reported deaths via [4, 5]
Fig. 4.
Reported and current (median) scenario cumulative deaths of COVID-19 in Iran, last 4 months of year 2020 and January 2021, International studies. (1) DELPHI: DELPHI (Differential Equations Leads to Predictions of Hospitalizations and Infections) Epidemiological Case Predictions. Mean estimate [10]. (2) IHME: Institute for Health Metrics and Evaluation (IHME) Mean estimate [12]. (3) Imperial: Imperial College COVID-19 LMIC Reports. Mean estimate [13]. (4) LANL: Los Alamos National Laboratory (LANL) COVID-19 Cases and Deaths Forecasts. Mean estimate [14]. (5) Srivastava: ReCOVER- Accurate Predictions and Resource Management for COVID-19 Epidemic Response. Mean estimate [15]. (6) YYG (Youyang Gu): COVID-19 Projections Using Machine Learning. Mean estimate [17]. (7) MOHME reported: Ministry of Health and Medical Education, Iran via [4, 5]
Fig. 5.
Current (median) scenario estimated cumulative deaths of COVID-19 in Iran and 20 other countries of North Africa Middle East, IHME. IHME: Institute for Health Metrics and Evaluation (IHME) Mean estimate. The Reference or ‘Current projection’ scenario assumes social distancing mandates are re-imposed for 6 weeks whenever daily deaths reach 8 per million (0.8 per 100,000) [12]
Appendix Figure 1 shows the PRISMA study flow diagram. Appendix Figure 2 demonstrates the officially reported cumulative confirmed cases, deaths, and recovered cases, and Appendix Figure 3 shows the daily equivalents. Appendix Figures 4 and 5 show the estimated daily prevalent cases, with and without the estimate form Saberi (web site) [21]. That estimate by Saberi, even in the median scenario, had high values compared to other studies. To visualize the quantitative diversity of the studies’ results, we also graphed the reported and worst-scenario estimated cumulative deaths in Appendix Figures 6 and 7, with and without the estimate form Mashayekhi [28]. That estimate by Mashayekhi, was the most extreme prediction among all the studies. Appendix Figure 8 shows officially reported and “current scenario” estimates of cumulative deaths by international studies for Iran.
MOHME
Official reports of MOHME for cumulative deaths and cases at 2020-10-19 were 30,712 and 534,631 respectively (via [4, 5]). Peaks in daily deaths (and dates) were 158 (2020-04-04), 235 (2020-07-28), and 337 (2020-10-19). Peaks in daily cases (and dates) were 3,186 (2020-03-30), 3,574 (2020-06-04), and 6,191 (2020-10-25).
Cumulative deaths
Lowest and highest predicted cumulative deaths for the end of the second month (2020-04-19) were 1777 (Imperial [13]) and 388,951 (Rafieenasab [54]) respectively, when the official number was 5,118. At the end of month four (2020-06-20), they were 3,590 (Imperial [13]) and 1,819,392 (Mashayekhi [28]), and the official number was 9,507. Those estimates for latest date available in 2020 were 16,176 (Imperial [13] for 2020-12-31) and 418,834 (Srivastava [15], for 2020-12-19). For the latest date available in 2021, those estimates were 40,151 (IHME [12], for 2021-01-31) and 125,690 (IHME [12], for 2021-01-31).
Cumulative cases
Lowest and highest predicted cumulative cases for the end of the second month (2020-04-19) were 20,588 (Al-Qaness [51]) and 2,310,161 (IHME [12]) respectively. Where the official number was 82,211. At the end of month four (2020-06-20), those estimates were 144,305 (DELPHI [10]) and 4,266,964 (IHME [12]), and the official number was 202,584. Those estimates for latest date available in 2020 were 3,588,293 (Imperial [13], for 2020-12-31) and 41,475,792 (Imperial [13], for 2020-12-31). For the latest date available in 2021, those estimates were 19,799,934 (IHME [12], for 2021-01-31) and 34,417,912 (IHME [12], for 2021-01-31).
Daily deaths
Lowest and highest predicted daily deaths for the end of the second month (2020-04-19) were 30 (Imperial [13]) and 11,289 (Rahimi Rise [29]) respectively, where the official number was 87. At the end of month four (2020-06-20), they were 5 (Mashayekhi [28]) and 44,934 (Mashayekhi [28]), and the official number was 115. Those estimates for latest date available in 2020 were zero (Imperial [13], for 2020-12-31) and 3,984 (Imperial [13], for 2020-12-31). For the latest date available in 2021, those estimates were 55 (IHME [12], for 2021-01-31) and 1,093 (IHME [12], for 2021-01-31).
Daily cases
Lowest and highest predicted daily incident cases for the end of the second month (2020-04-19) were 93 (Thu [48]) and 216,262 (Rahimi Rise [29]) respectively, where the official number was 1,343. At the end of month four (2020-06-20), they were 211 (DELPHI [10]) and 138,892 (Gu (YYG) [11]), and the official number was 2,322. Those estimates for latest date available in 2020 were zero (Imperial [13], for 2020-12-31) and 486,745 (Imperial [13], for 2020-12-31). For the latest date available in 2021, those estimates were 14,818 (IHME [12], for 2021-01-31) and 236,781 (IHME [12], for 2021-01-31). Lowest and highest predicted daily incident confirmed and suspected cases for the end of the second month (2020-04-19) were 72,950 (Saberi (article) [22]) and 1,616,385 (Saberi (article) [22]) respectively; there are no correspondent official numbers for this measure. At the end of month four (2020-06-20), they were 9,625 (Saberi (article) [22]) and 1,255,012 (Saberi (article) [22]).
Peak dates and control dates
In general, studies with shorter (or longer) durations of outputs estimated lower (or higher) numbers of peaks. For daily deaths, official reports showed the first peak of 158 (2020-04-04), second peak of 235 (2020-07-28), and third peak of 337 (2020-10-19). For estimates of daily deaths, size (and date) of the lowest first peak was 81 (2020-06-01) (LANL [14]), and the highest first peak was 44,934 (2020-06-20) (Mashayekhi [28]). For the third peak, lowest estimate was 134 (2020-09-29) (Imperial [13]) and highest estimate was 4,968 (2020-12-11) (Imperial [13]). For daily cases, official reports showed the first peak of 3,186 (2020-03-30), second peak of 3,574 (2020-06-04), and third peak of 4,830 (2020-10-14). For estimates of daily cases, size (and date) of the lowest first peak was 1,050 (2020-03-12) (Shen [43]), and the highest first peak was 470,229 (2020-03-31) (Rahimi Rise [29]). For the second peak, lowest estimate was 3,825 (2020-10-010) (LANL [14]) and highest estimate was 14,272 (2020-03-11) (Haghdoost [27]). For the third peak, lowest estimate was 6,526 (2020-10-25) (LANL [14]) and highest estimate was 717,356 (2020-12-04) (Imperial [13]).
Three studies predicted the epidemic control (or end) dates and outcome’s values. Two studies predicted the potential date for epidemic to be controlled in April; Ahmadi et al. predicted the “end of the epidemic” on 2020-05-13 with 87,000 cumulative cases or on 2020-06-01 with 4,900 cumulative deaths (using Von Bertalanffy model) or 11,000 cumulative deaths (using Gompertz model) [44]. Haghdoost predicted that with their either medium or best scenarios, the epidemic would be well controlled in month 2 of Hijri solar year 1,399 (2020-04-20 to 2020-05-20). Their ‘maximum number of infected people in day’ would be 92,100 in middle scenario and 9150 in best scenario [27]. Zhan et al. predicted that if the “authorities continue to impose strict control measures, the epidemic will come under control by the end of April and is expected to end before June 2020, and as the quality of treatment improves, more rapid recovery will be expected” [40]. Beyond the correspondent values of the predicted outcomes, no further criteria or definition of epidemic end or control was provided.
Discussion
There were lots of heterogeneity in methods and findings of the COVID-19 prediction models and estimation studies for Iran. After the presumed official start date of the COVID-19 epidemic in Iran, i.e. 2020-02-19, and at the end of month two (2020-04-19), the lowest (and highest) values of predictions were 1777 (388,951) for cumulative deaths, 20,588 (2,310,161) for cumulative cases, and at the end of month four (2020-06-20), they were 3,590 (1,819,392) for cumulative deaths, and 144,305 (4,266,964) for cumulative cases. Lowest and highest predicted cumulative deaths for latest date available in 2020 were 16,176 (Imperial [13], for 2020-12-31) and 418,834 (Srivastava [15], for 2020-12-19). For the latest date available in 2021, those estimates were 40,151 (IHME [12], for 2021-01-31) and 125,690 (IHME [12], for 2021-01-31). Lowest and highest predicted cumulative cases for latest date available in 2020 were 3,588,293 (Imperial [13], for 2020-12-31) and 41,475,792 (Imperial [13], for 2020-12-31). For the latest date available in 2021, those estimates were 19,799,934 (IHME [12], for 2021-01-31) and 34,417,912 (IHME [12], for 2021-01-31).
Part of the heterogeneity observed in component studies’ methods originates from the actual lack of universal consensus-based standards for epidemic modeling study methodology and reporting. Another part originates from studies’ adherence to principles of reporting their methods and findings. For instance, eight of the reviewed studies did not mention their study limitations at all, and nine studies did mention limitations but just touched the limitations very minimally.
Part of the heterogeneity observed in studies’ findings originates from choices of methods and assumptions, including the untrue assumptions of homogeneous population mixing, no role of asymptomatic cases in disease spread, and no under-reporting of deaths and cases. Another part originates from quality, availability, and completeness of data, including, epidemic start date, and numbers of cases and deaths.
The epidemic start date and the reported number of deaths and cases are the most important starting points for epidemic estimation studies. There are uncertainties about the epidemic start date and real numbers of deaths and cases. The presumed official start date of the COVID-19 epidemic in Iran was 2020-02-19, when the first two tandem cases were reported as dead. Report of the first case or cases as dead on the same date they were diagnosed is not the most frequent type of reporting in this pandemic. Haghdoost et al. study, dated 2020-03-15 [1,398-12-25 Hijri solar], maintains that their start date of the epidemic in Iran for their modeling purpose was designated as 2020-01-21 [1,398-11-01 Hijri solar] based on “available documentations and epidemiologic analyses” [27]. No description or references were provided for their “available documentations and epidemiologic analyses”. Two days later, MOHME announced in 2020-03-27 that the epidemic had probably started in month 11 of Hijri solar year 1,398 (2020-01-21 to 2020-02-19) [59]. As such, the models that use the official start date of 2020-02-19 start with an inaccurate start date of the epidemic to begin with.
WHO Country Support Mission to Iran (2–11 March 2020) reported the following: “On 20 February, the Islamic Republic of Iran IHR [International Health Regulations] National Focal Point (IHR-NFP) notified WHO of five cases, including two deaths, of laboratory-confirmed COVID-19 cases. Three of the cases were from Qom City, and the fourth had a travel history to Qom. In the following days, the investigation concluded that the virus was probably circulating in Qom for several weeks, based on the following observations: Among 186 patients with severe acute severe acute respiratory infection (SARI) hospitalized during February, 8 deaths were observed (0 deaths for the same month last year). Samples taken in February in patients with influenza-like illness (ILi) symptoms that tested negative for Influenza were also tested for COVID-19. Among workers of the Salafchegan free zone located 50 km from Qom city centre, 5 tests were positive for COVID-19; their onset of symptoms was 10 February. In late February, of 17 Chinese workers who had not traveled back to China for the Chinese New Year, 5 tested positive.” [57]. As such, most of the models start with an inaccurate start date of the epidemic to begin with, and most of the studies rely on the officially reported numbers of cases and deaths. Iran’s MOHME stopped reporting provincial cases and deaths on 2020-03-23 [45].
In addition to Ghaffarzadegan [41] that used both official and unofficial data as input, one study (Saberi [21]), also used correction factors of 5 and 10 taken from other sources [57] or studies [58] applied to the officially reported numbers of cases and deaths. A correction factor of 20 has been proposed for the epidemic in Iran [60]. We do not know when or where were the results of the ‘investigation’ referred to in the above quoted “In the following days, the investigation concluded that …” , were announced or published. The first COVID-19 patient “with a definite diagnosis” was reported in an article by Ghadir and colleagues on 2020-04-06 [61]. On 2020-07-30, clinical and virologic characteristics of the first seven cases of COVID-19 in Iran were reported by Yavarian and colleagues, academics from Tehran University of Medical Sciences and officials from Iran MOHME [62]. Numbers and dates of cases and deaths do not match with correspondent numbers and dates of cases and deaths officially announced by MOHME. Date of symptoms onset and date of encounter for the first patient reported by Ghadir [61] and by Yavarian [62] do not match.
A study on all-cause excess mortality and COVID-19-related deaths in Iran found a correction factor of two for reported COVID-19 deaths in Iran [63]. Deputy Minister of Health in Iran announced on 2020-10-14 that the real numbers of COVID-19 deaths In Iran are on average about two times higher than the official reports (ranging 1.7 from 2.2 depending on the province), because they follow the WHO protocols that requires positive PCR test result [64]. World Health Organization maintains that’ “Countries have varying approaches to COVID-19 case definitions. Consequently, the numerator and the denominator of any formula used to calculate fatality rate will vary according to how they are defined. WHO recommends using the surveillance case definitions which are available in the WHO interim guidance on Global surveillance for COVID-19. A COVID-19 death is defined for surveillance purposes as a death resulting from a clinically compatible illness in a probable or confirmed COVID-19 case, unless there is a clear alternative cause of death that cannot be related to COVID-19 disease (e.g. trauma). There should be no period of complete recovery between the illness and death.” [65]. Moreover, WHO’s “International Guidelines for Certification and Classification (Coding) of Covid-19 as Cause of Death” maintains the immediately-above-mentioned for a “COVID-19 death”, and also provide the following codes. “U07.1 COVID-19, virus identified” for when the virus was identified, and “U07.2 COVID-19, virus not identified. Clinically-epidemiologically diagnosed COVID-19. Probable COVID-19. Suspected COVID-19” for when the virus was NOT identified [66]. However, WHO maintains in its “COVID-19 Weekly Epidemiological Update” of 25 October 2020, that “Data presented are based on official laboratory-confirmed COVID-19 case and deaths reported to WHO by country/territories/areas, largely based upon WHO case definitions and surveillance guidance.” and “A small number of countries/territories/areas report combined probable and laboratory-confirmed cases; efforts are underway to identify these for notation in the data table. Differences are to be expected between information products published by WHO, national public health authorities, and other sources.” [67].
Another deputy Minister of Health in Iran announced on 2020-10-18 that the real numbers of COVID-19 deaths and cases of COVID-19 In Iran are on average about 2.5 times more than the official reports because the diagnostic test kits have only 30 to 50% ability to diagnose the disease [68]. A member of High Council of Iranian Medial Council mentioned on 2020-10-25 that the real number of COVID-19 deaths is 3–4 times more than the official reports due to low number of PCR testing [69]. Euro-news quoted Dr. Michael Ryan of WHO on 2020-10-05, “Around 10% of the world’s population may have had COVID-19” on about 2020-10-05 [70]. There were 37,738,990 reported global cumulative cases on 2020-10-05 [49], and 10% of 7.8 billion global population is 780 million, and hence the correction factor for going from reported cases to the total cases is 20.67 at the global level. If the global average Infection Fatality Rate (IFR) is about 0.5% (0.2–1.0%) [71–74], then with 780 million expected global cumulative deaths on 2020-10-05, the expected deaths could be 3,900,000 (1,560,000-7,800,000). With 1,054,089 reported global cumulative deaths on 2020-10-05, the correction factor for deaths would be about 3.70 (1.48–7.40) at the global level. Therefore, at the global average level, the correction factor for cases is about 20.67, and correction factor for deaths is about 3.70 (1.48–7.40). Deputy Minister of Health in Iran announced on 2020-10-25 [75] that based on a national seroprevalence study, 30 million people were infected at about the end of Hijri month 2 (about 2020-05-20). If the global average Infection Fatality Rate (IFR) is about 0.5% (0.2–1.0%) [71–74], and applies to Iran, then 30 million infections translates to 150,000 (60,000-300,000) deaths. Official report of cumulative deaths on 2020-05-20 was 7183, and all of these end up in three possibilities: (1) correction factor of 21 (8–42) for COVID-19 deaths in Iran, or (2) the IFR in Iran was 0.024% at that time, or (3) that seroprevalence finding was wrong. While some politicians and researchers may wish, advocate, or publish for herd immunity – knowingly or unknowingly [76–79], research evidence yet describe why “COVID-19 herd immunity is unethical and unachievable” [80, 81]. On a rather daily basis, more and more announcements are being made by authorities and officials about higher correction factors for COVID-19 deaths and cases in Iran, or unpreparedness of testing system or hospital infrastructure for combating the epidemic; for which continued quotation and referencing here would not further help anyone.
Undercounting, under-ascertainment, or under-reporting is a known issue with the number of official confirmed cases and deaths, almost in all countries. Factors such as health system capacity for performing tests, access of people to testing services, on-time availability of test results, precision of diagnostic or screening tests, performance of surveillance systems, and transparency of health systems affect number of cases and deaths in official reports. In addition to such factors, SARS-CoV-2 itself has characteristics that might aggravate undercounting. A study on Santa Clara county in the United States revealed that prevalence, based on testing antibodies to SARS-CoV-2, is 50–85-fold more than the confirmed cases [82]. This pattern is different from other viruses of the Coronaviridae family, such as Middle East Respiratory Syndrome (MERS-CoV), with an estimated 25–50% asymptomatic to mild cases [83]. Some of the reviewed studies have estimated number of infected cases without excluding or mentioning asymptomatic cases. An implicit conclusion is that their numbers mainly refer to symptomatic cases, similar to the case mix of their input data. Different approaches have been used in studies to the issue of undercounting, under-ascertainment, or under-reporting. (1) Some studies have accounted for undercounting in their models: DELPHI [10], Ghaffarzadegan [41], Gu (YYG) [17], Saberi (web site) [21], Saberi (article) [22], and Srivastava [15]. (2) Some studies have not used number of confirmed deaths and / or cases as input data; Haghdoost [27], Mashayekhi [28], Rahimi Rise [29], Tuite [46], and Zhuang [47]. (3) In order to conclude that “In emerging epidemics, CFR indicator must not be used as a basis to judge the performance of a health system unless that epidemic condition has been clarified”, Moradi estimated the “actual number of COVID-19 cases in Iran based on different proposed scenarios for Case Fatality Rate [42]. (4) Other studies did nothing about the issue of undercounting, with or without mentioning it. To our best understanding, the most important issue that can drive the prediction models’ results misleading and misinforming is ignoring the issue of undercounting, under-ascertainment, and under-reporting. Role of the symptomatic cases in spread of the disease was accounted for only in some studies; DELPHI [10], Ghaffarzadegan [41], Gu (YYG) [17], Mashayekhi [28], Saberi (article) [22], Rahimi Rise [29], Srivastava [15].
Some studies provided some sort of subnational estimates as well as national; Haghdoost [27], Moghadami [36], Muniz-Rodriguez [37], Pourghasemi (PLoS ONE) [38], Pourghasemi (IJID) [39], and Zhan [40]. Access to COVID-19 data at provincial and subnational level has obviously been an important limitation for most researchers. This threatens the usability of models. Naturally, all the provinces are not at the same stage of epidemic growth, they have different conditions that affect disease transmission and their capacities to respond to the epidemic are different. This means that centralized strategies for estimation of the epidemic extent and intervention options might not fit all the needs of the subnational levels. Spatial heterogeneity in propagation of epidemics should be taken into account [84]. MOHME has a subnational level defined between the national and the provincial levels; some of the studies or operational plans have used these conglomerates of provinces in Iran, labeled as “climes”, which share relatively homogenous epidemiologic profiles within the climes before the COVID-19 era. Such conglomerates of provinces in Iran, or newly designed conglomerates, might be considered usable in estimation of the epidemic propagation in Iran with a smaller number of subnational geographic units (i.e. climes), compared to studying all the provinces, which is more resource-intensive. Alternatively, there are potential modeling approaches for simultaneous modeling of different subnational levels accounting for different stages of epidemic progression, both with and without access to detailed province data; for instance, Rojas [85] and Xiong [86].
The international studies, i.e. DELPHI [10], Gu (YYG) [17], IHME [12], Imperial [13], LANL [14], and Srivastava [15], generally provide updated estimates on a periodical basis, mostly weekly. This means that all estimations (from the start date of the epidemic) change in each version of running model. The usual explanation for this approach is to feed model with more input data that can improve prediction and provide an opportunity for improving methods as well.
Among the models that have used number of confirmed cases or deaths as input, only two studies, Ghaffarzadegan [41] and Saberi (article) [22], have considered delayed diagnosis in their calculations; this might be quite important. The Wuhan municipal headquarters for COVID-19 epidemic prevention and control released a notification and revised the total number of fatalities up by around 50% to 3869 after reviewing all available sources of data [87]. There not an unusual practice in many of death registries that physicians use more general terms as the final diagnosis or cause of death when they cannot or do not have time to match patients’ characteristics with exact definitions. It is more common in situations like epidemics that all health care workers are overwhelmed with number of patients and preoccupied with treating patients. Also, field hospitals and COVID-19 specific hospices might not be linked properly to health information systems to share data. Some cases that were initially classified under more general terms (such as pneumonia or acute respiratory syndrome) or even more specified but incorrect diagnoses (such as seasonal flu) might be re-classified to COVID-19 after reviewing all clinical data, test results, or autopsies. Five studies (Ayyoubzadeh [52], Ghaffarzadegan [41], Haghdoost [27], Mashayekhi [28], Rahimi Rise [29]) forecasted more than one peak for the epidemic. International studies generally ‘back-cast’ (replicate or imitate) the officially reported epidemic curves up to the time where reported data is available, and ‘forecast’ the epidemic curve under different scenarios for the future. Prediction of time and magnitude of future waves of the epidemic is an important aspects of modeling studies. Also, there is an implicit assumption in all studies or models, that the socio-economic response capacity will remain constant over the timespan of the epidemic and the calendar time period for which estimations are performed; this is not necessarily correct.
As some of the studies have not mentioned enough details on their epidemic model and statistical methods, the largest gap was related to not mentioning the methods used to assess model validity, accuracy, or fitness, and the findings of these methods. Eleven studies did not report any method used to verify model validity. Absence of reporting uncertainty intervals for model output was another downside in statistical methods. Poor reporting has been one of the common issues in many of the COVID-19 prediction and estimation studies [7]. Although guidelines such as TRIPOD are not specifically designed for this type of studies but can be used as a base to remind researchers about the standards of reporting [9]. There have been a few systematic reviews on COVID-19 epidemiological studies. None of the studies included in this research were among the included studies in Park’s systematic review [88].
Eight studies (DELPHI [10], Ghaffarzadegan [41], Haghdoost [27], Hsiang [45], Imperial [13], Mashayekhi [28], Rahimi Rise [29], Thu [48]) have considered scenarios to assess the effects of social distancing policies; this increases the usability of these models, however social distancing policies have a wide range of methods and effectiveness. They need to be clarified with more details in scenarios to increase practical usability of models for decision making. We expect to see postponement in the first peak date or reduction of its height (i.e. flattening the curve) in scenarios based on appropriate interventions [35]. Most of the studies have ignored availability and numbers of performed tests to detect COVID-19 cases; such data were not publicly available in the first few weeks after start of the epidemic in Iran. Only two studies (Ghaffarzadegan [41], IHME [12]) included test coverage in their model. Evidence on potential seasonality effects is not conclusive yet, but it has been proposed by some researchers [36, 37]. This factor has been considered in three of the models; Ghaffarzadegan [41], Haghdoost [27], and IHME [12]. Among the 84 study-model/scenarios, the median scenario of Haghdoost [27] was the best model that perfectly forecasted the cumulative deaths officially reported by MOHME (Fig. 1).
Non-COVID causes of mortality and morbidity are also important in epidemic modeling and intervention planning. Increase in non-COVID all-cause mortality and morbidity is a tandem phenomenon running alongside the COVID epidemic, that goes on with less drawn attention compared to the epidemic. European mortality monitoring (EuroMOMO) Network has assessed excess all-cause mortality overall for the participating European countries and estimated a marked increase [38]. As the cold season will come, taking into account the influenza season in estimations, and in particular regarding the caseload burden to be imposed on the health care system. Some COVID cases or deaths might be misclassified as non-COVID Acute Respiratory Distress Syndrome (ARDS), influenza, or pneumonia, and analyses of expected levels of such cases deaths could illuminate and improve COVID estimates. Some countries have expedited release of reports of provisional counts of death and excess deaths in January to March 2019 and January to March 2020, e.g. Iran [39]. Excess deaths in provinces of Iran have been assessed in non-peer-reviewed report [40].
Models differ in their mathematical configuration, designated start date of the epidemic for a given population, use of parameter values as input (e.g. Basic reproduction number, R0; or Case Fatality Ratio, CFR), use of data with varying time lengths to calibrate the model, and interventions formulated in scenarios. As Panovska-Griffiths explains, the ultimate questions of “can mathematical modelling solve the current Covid-19 crisis” or “which model is correct” evolve to realization that “no one model can give all the answers” and that “we need more models that answer complementary subquestions that can piece together the jigsaw and halt COVID- 19 spread” [41]. Estimation and use of a single correction factor for both the cases and deaths all across the time for any given country assumes invariance of diagnosis, detection and reporting completeness for infections and mortality during the epidemic; which is not necessarily true.
We believe that increasing public access to data on number of confirmed or suspected cases (and their outcomes of death and recovery), healthcare utilization by patients with COVID-19 (such as hospital admission, intensive care utilization and performed tests) both at national and province levels may improve accuracy and usability of models, and eventually lead to prevention of more cases and deaths. Estimates of cumulative deaths are less dependent on testing compared to cumulative cases. Estimates of daily deaths and daily cases look at possible waves and peak heights. Estimation of all these four outcomes can depict a better trajectory and extent of the epidemic. We suggest researcher to consider subnational estimations, as well as factors such as non-pharmacological interventions, test availability, age-stratification, and delayed diagnosis for updating their models. Some of the reports do not have enough details on methods and results that reduces their usage in disease control. We recommend researchers to consider standard items for reporting their models to increase practical use of the findings. It would also be vitally desirable if the veteran and current researchers of epidemic modeling could develop a consensus-based declaration on Preferred Reporting Items of Epidemic Modeling Studies, preferably before the next pandemic.
The results of epidemic modeling studies are certainly uncertain. The way in which the problem of uncertainty is handled by some of the national-level and international models of COVID-19 epidemic has been criticized in Davey Smith’s ‘Covid-19’s known unknowns’ [89], albeit without suggestion of any solutions beyond “respecting uncertainty”. Beyond the considerations of usability and certainty of epidemic models, the immense problem for people on the ground who suffer from the direct and indirect mortality, morbidity, and economic hardship from COVID-19, is the actually materialized accountability and the public health and economic interventions the governments have undertaken, are doing, and will perform – with or without the best or worst estimation models’ results. As demonstrated in Fig. 5, Iran would have the highest estimated death toll among the 21 countries in in North Africa Middle East region, with 78,693 estimated cumulative deaths under the current scenario on 2021-02-01, that is about 3.5 times more than the second highest death toll of 22,614 in Iraq [12]. Model estimates predict an ominous course of epidemic progress in Iran. Wisdom, political and economic commitment, and effective policy making, and management are needed indeed.
Study limitations
We did not search international databases other than PubMed, did not describe subnational results, and did not assess other outcomes such as utilization of intensive care unit (ICU) beds. We did not have access to the full report of one of the studies (Mashayekhi [28]). In the abridged study report we found, we noticed some discrepancies in outcome prediction values in different graphs [26]. None of our digitized outcome predictions reported here are 100% accurate. All of them are wrong in terms of having non-prefect accuracy. However, the error range is mostly around 1 or 2% points and below 5% for almost all instances.
Conclusions
We believe that COVID-19 models which consider scenarios for policy options, include key influencing factors such as role of asymptomatic cases in spread of the disease, under-reporting of deaths and cases, testing availability, and delayed diagnosis, and provide estimates for subnational regions are more useful for epidemic control. Not accounting for under-reporting drives the models’ results misleading. Increasing public access to COVID-19 related data is very important for improving quality of models and enhancing evidence-informed decisions to prevent more deaths. To increase the usability of reports, researchers should consider requirements of reporting a prediction or estimation model.
Supplementary Information
Additional file 1: Appendix Text 1. Search syntax used in PubMed. Appendix Text 2. Details of studies’ scenario. Appendix Table 1. Predictions of cumulative cases for the end of months one to six after the official epidemic start date (2020-02-19) and the latest date available in 2020. Appendix Table 2. Predictions of daily deaths at end of months one to six after the official epidemic start date (2020-02-19) and the latest date available in 2020. Appendix Table 3. Predictions of daily cases for the end of months one to six after the official epidemic start date (2020-02-19) and the latest date available in 2020. Appendix Table 4. Predictions of epidemic peak dates and values of outcomes. Appendix Table 5. Predictions of epidemic control dates and values of outcomes. Appendix Figure 1. PRISMA 2009 study flow diagram. Appendix Figure 2. Officially reported cumulative confirmed cases, deaths, and recovered cases of COVID-19 in Iran. Appendix Figure 3. Reported daily confirmed cases, deaths, and recovered cases of COVID-19 in Iran. Appendix Figure 4. Reported and median-scenario estimated daily prevalent cases of COVID-19 in Iran, including predictions by Saberi. Appendix Figure 5. Reported and median-scenario estimated daily prevalent case of COVID-19 in Iran, without predictions by Saberi. Appendix Figure 6. Reported and worst-scenario estimated cumulative deaths of COVID-19 in Iran, including predictions by Mashayekhi. Appendix Figure 7. Reported and worst-scenario estimated cumulative deaths of COVID-19 in Iran, without predictions by Mashayekhi. Appendix Figure 8. Reported and current (median) scenario estimated cumulative deaths of COVID-19 in Iran, International studies.
Additional file 2. Target studies’ abstracted data.
Acknowledgements
Not applicable.
Abbreviations
- CCPV
Characteristics, Construction, Parameterization and Validation
- CFR
Case Fatality Ratio
- COVID-19
Coronavirus Disease 2019
- ICU
Intensive Care Unit
- DELPHI
-
Differential Equations Leads to Predictions of Hospitalizations and Infections (DELPHI)
Epidemiological Case Predictions
- Gu (YYG)
(First author, Youyang Gu) COVID-19 Projections Using Machine Learning
- IHME
Institute for Health Metrics and Evaluation. COVID-19 projections
- IHR-NFP
International Health Regulations - National Focal Point
- Ili
Influenza-like illness
- Imperial
Imperial College COVID-19 LMIC Reports
- LANL
Los Alamos National Laboratory (LANL) COVID-19 Cases and Deaths Forecasts
- MERS-CoV
Middle East Respiratory Syndrome – Corona Virus
- MOHME
Ministry of Health and Medical Education (of Iran)
- PCR
Polymerase Chain Reaction (test)
- PRISMA
Preferred Reporting Items for Systematic Reviews and Meta-Analyses
- R0
Basic reproduction number
- SARI
Acute Severe acute Respiratory Infection
- SARS-CoV-2
Severe acute respiratory syndrome coronavirus 2
- TRIPOD
Transparent Reporting of a multivariable prediction model for Individual Prognosis Or Diagnosis
- WHO
World Health Organization
Authors’ contributions
MML and FP conceived the study. MML and LJ produced Table 1. FP developed the data abstraction spreadsheet, reviewed and digitized the studies’ graphs and produced Table 2 and 3 and Appendix Tables. FP and MML drafted the manuscript. MRH reviewed and digitized the studies’ graphs. All authors contributed to finding the studies, reviewed three to all of the articles, and contributed substantial input to finalization of results and the manuscript. All authors read and approved the manuscript.
Funding
No funding.
Availability of data and materials
All data generated or analysed during this study are included in this published article and its supplementary information files.
Ethics approval and consent to participate
All data are publicly available on an aggregate basis. Consequently, no ethics approval was necessary.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Farshad Pourmalek, Email: pourmalek_farshad@yahoo.com.
Mohsen Rezaei Hemami, Email: Mohsen.rezaeihemami@abdn.ac.uk.
Leila Janani, Email: leila_janani@yahoo.com.
Maziar Moradi-Lakeh, Email: moradilakeh.m@iums.ac.ir.
References
- 1.World Health Organization. Novel coronavirus (2019-nCoV) situation report – 1. https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200121-sitrep-1-2019-ncov.pdf?sfvrsn=20a99c10. Accessed 4 May 2020.
- 2.World Health Organization. WHO director-general’s remarks at the media briefing on 2019-nCoV on 11 February 2020. https://www.who.int/dg/speeches/detail/who-director-general-s-remarks-at-the-media-briefing-on-2019-ncov-on-11-february-2020. Accessed 4 May 2020.
- 3.World Health Organization. WHO director-general’s opening remarks at the media briefing on COVID-19 - 11 March 2020. https://www.who.int/dg/speeches/detail/who-director-general-s-opening-remarks-at-the-media-briefing-on-covid-19%2D%2D-11-march-2020. Accessed 4 May 2020.
- 4.Johns Hopkins University. Coronavirus resource center. https://coronavirus.jhu.edu/map.html. Accessed 4 May 2020.
- 5.Johns Hopkins University. COVID-19 data repository by the Center for Systems Science and Engineering (CSSE) at Johns Hopkins University. https://github.com/CSSEGISandData/COVID-19. Accessed 19 Oct 2020.
- 6.Anderson RM, Heesterbeek H, Klinkenberg D, Hollingsworth TD. How will country-based mitigation measures influence the course of the COVID-19 epidemic? Lancet. 2020;395:931–934. doi: 10.1016/S0140-6736(20)30567-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wynants L, Calster BV, Bonten MMJ, Collins GS, Debray TPA, Vos MD, et al. Prediction models for diagnosis and prognosis of covid-19 infection: systematic review and critical appraisal. BMJ. 2020;369:m1328. doi: 10.1136/bmj.m1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carrasco LR, Jit M, Chen MI, Lee VJ, Milne GJ, Cook AR. Trends in parameterization, economics and host behaviour in influenza pandemic modelling: a review and reporting protocol. Emerg Themes Epidemiol. 2013;10:3. doi: 10.1186/1742-7622-10-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moons KGM, Altman DG, Reitsma JB, Ioannidis JPA, Macaskill P, Steyerberg EW, et al. Transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD): explanation and elaboration. Ann Intern Med. 2015;162:W1–73. doi: 10.7326/M14-0698. [DOI] [PubMed] [Google Scholar]
- 10.COVID Analytics. DELPHI epidemiological case predictions. Cambridge: Operations Research Center, Massach Institut Technol https://www.covidanalytics.io/projections. Accessed 18 July 2020.
- 11.Youyang Gu. COVID-19 projections using machine learning. https://covid19-projections.com. Accessed 5 Oct 2020.
- 12.Institute for Health Metrics and Evaluation (IHME). COVID-19 mortality, infection, testing, hospital resource use, and social distancing projections. Seattle: Institute for Health Metrics and Evaluation (IHME), University of Washington. http://www.healthdata.org/covid/. Accessed 9 Oct 2020.
- 13.MRC Centre for Global Infectious Disease Analysis (MRC GIDA). Future scenarios of the healthcare burden of COVID-19 in low- or middle-income countries. London: MRC Centre for Global Infectious Disease Analysis, Imperial College London. https://mrc-ide.github.io/global-lmic-reports/. Accessed 6 Oct 2020.
- 14.Los Alamos National Laboratory (LANL). COVID-19 cases and deaths forecasts. Los Alamos: Los Alamos National Laboratory (LANL). https://covid-19.bsvgateway.org. Accessed 11 Oct 2020.
- 15.University of Southern California (USC). COVID-19 forecast. Los Angeles: University of Southern California. https://scc-usc.github.io/ReCOVER-COVID-19. Accessed 12 Sept 2020.
- 16.DELPHI. The epidemiological model underlying COVID analytics. [Data site]. https://github.com/COVIDAnalytics/DELPHI. Accessed 18 July 2020.
- 17.Youyang Gu. COVID-19 projections using machine learning. [Data site]. https://github.com/youyanggu/covid19_projections. Accessed 5 Oct 2020.
- 18.Institute for Health Metrics and Evaluation (IHME). COVID-19 estimate downloads. [Data site]. http://www.healthdata.org/covid/data-downloads. Accessed 9 Oct 2020.
- 19.MRC Centre for Global Infectious Disease Analysis (MRC GIDA). Future scenarios of the healthcare burden of COVID-19 in low- or middle-income countries. [Data site]. https://github.com/mrc-ide/global-lmic-reports/raw/master/data. Accessed 6 Oct 2020.
- 20.Los Alamos National Laboratory (LANL). COVID-19 cases and deaths forecasts. [Data site]. https://covid-19.bsvgateway.org. Accessed 11 Oct 2020.
- 21.Saberi M, Hamedmoghadam H. IRAN COVID-19. Scenario analysis of COVID-19 epidemic in Iran. Sydney: Research Centre for Integrated Transport Innovation (rCITI), University of New South Wales (UNSW) https://www.irancovid-19.com/. Accessed 4 May 2020.
- 22.Saberi M, Hamedmoghadam H, Madani K, Dolk HD, Morgan A, Morris JK, et al. Accounting for underreporting in mathematical modelling of transmission and control of COVID-19 in Iran. Front Physics. 2020;8(Article 289). 10.3389/fphy.2020.00289.
- 23.Scientific Information Database of Iran (SDI). https://www.sid.ir. Accessed 5 Oct 2020.
- 24.MAG-IRAN National Database of Periodicals (Farsi). https://www.magiran.com. Accessed 5 Oct 2020.
- 25.Moher D, Liberati A, Tetzlaff J, Altman DG. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. Ann Intern Med. 2009;151:264–269. doi: 10.7326/0003-4819-151-4-200908180-00135. [DOI] [PubMed] [Google Scholar]
- 26.Ahmadi A, Fadaei Y, Shirani M, Rahmani F. Modeling and forecasting trend of COVID-19 epidemic in Iran until May 13, 2020. Med J Islam Repub Iran. 2020;34:27. [DOI] [PMC free article] [PubMed]
- 27.Ghaffarzadegan N, Rahmandad H. Simulation-based estimation of the early spread of COVID-19 in Iran: actual versus confirmed cases. Syst Dyn Rev. 2020;36(1):101–29. [DOI] [PMC free article] [PubMed]
- 28.Haghdoost AA. First report of modeling COVID-19 epidemic in Iran. Ministry of Health and Medical Education (MOHME), Tehran, Iran, COVID-19 epidemic analysis workgroup. 1398-12-25 [2020-03-15]. 20 pages. Found via colleagues; could not be retrieved on the internet; not an article or preprint. Accessed 17 Mar2020.
- 29.Mashayekhi A, Aghaei M. Prevalence of Corona disease in Iran. Sharif University of Technology. 1398-12-23 [2020-03-13]. https://static1.tinn.ir/servev2/tgRHjZeywi26/hhF3zQUm1m4,/647098_288.pdf .Accessed 4 May 2020.
- 30.Moghadami M, Moghadami M, Hassanzadeh M, Wa K, Hedayati A, Malekolkalami M. Modeling the Corona virus outbreak in Iran. medRxiv. Posted 31 March 31 2020. 10.1101/2020.03.24.20041095. [DOI]
- 31.Rafieenasab S, Zahiri A-P, Roohi E. Prediction of peak and termination of novel coronavirus COVID-19 epidemic in Iran. Int J Mod Phys C. 2020;0(0):2050152.
- 32.WebPlotDigitizer 4.2. Web-based plot digitizer, WebPlotDigitizer - extract data from plots, images, and maps. https://automeris.io/WebPlotDigitizer/. Accessed 20 Oct 2020.
- 33.Rahimi Rise Z, Ershadi MM, Shahabi Haghighgi SH. Scenario-based analysis about COVID-19 outbreak in Iran using systematic dynamics modeling - with a focus on the transportation system. Transport Res J. 2020;17(2):33–47.
- 34.Reiner RC, Barber RM, Collins JK, et al. Modeling COVID-19 scenarios for the United States. Nat Med. 2020. 10.1038/s41591-020-1132-9. [DOI] [PMC free article] [PubMed]
- 35.Friedman J, Liu P, Gakidou E, IHME COVID19 Model Comparison Team. Predictive performance of international COVID-19 mortality forecasting models.medRxiv. Posted 26 August 2020. 10.1101/2020.07.13.20151233. [DOI]
- 36.IHME COVID-19 Forecasting Team, Hay SI. COVID-19 scenarios for the United States. medRxiv. Posted 14 July 2020. 10.1101/2020.07.12.20151191. [DOI]
- 37.IHME COVID-19 health service utilization forecasting team, Murray CJ. Forecasting the impact of the first wave of the COVID-19 pandemic on hospital demand and deaths for the USA and European Economic Area countries. medRxiv. Posted 26 April 2020. 10.1101/2020.04.21.20074732. [DOI]
- 38.Walker PGT, Whittaker C, Watson OJ, Baguelin M, Winskill P, Hamlet A, Djafaara BA, Cucunuba Z, Olivera Mesa D, Green W, et al. The impact of COVID-9 and strategies for mitigation and suppression in low- and middle-income countries. Science. 2020;369(6502):413–22. [DOI] [PMC free article] [PubMed]
- 39.Srivastava A, Xu T. Fast and accurate forecasting of COVID-19 deaths using the SIkJα model. arXiv:200705180. Submitted on 10 Jul 2020 (v1), last revised 13 Jul 2020 (this version, v2).
- 40.Muniz-Rodriguez K, Fung IC, Ferdosi SR, Ofori SK, Lee Y, Tariq A, Chowell G. Severe acute respiratory syndrome coronavirus 2 transmission potential, Iran, 2020. Emerg Infect Dis. 2020;26(8):1915–7. [DOI] [PMC free article] [PubMed]
- 41.Pourghasemi HR, Pouyan S, Farajzadeh Z, Sadhasivam N, Heidari B, Babaei S, Tiefenbacher JP. Assessment of the outbreak risk, mapping and infection behavior of COVID-19: application of the autoregressive integrated-moving average (ARIMA) and polynomial models. PLoS One. 2020;15(7):e0236238. [DOI] [PMC free article] [PubMed]
- 42.Pourghasemi HR, Pouyan S, Heidari B, Farajzadeh Z, Fallah Shamsi SR, Babaei S, Khosravi R, Etemadi M, Ghanbarian G, Farhadi A, et al. Spatial modeling, risk mapping, change detection, and outbreak trend analysis of coronavirus (COVID-19) in Iran (days between February 19 and June 14, 2020). Int J Infect Dis. 2020;98:90–108. [DOI] [PMC free article] [PubMed]
- 43.Zhan C, Tse CK, Lai Z, Hao T, Su J. Prediction of COVID-19 spreading profiles in South Korea, Italy and Iran by data-driven coding. PLoS One. 2020;15(7):e0234763. [DOI] [PMC free article] [PubMed]
- 43.Moradi G, Piroozi B, Mohamadi-Bolbanabad A, Safari H, Shokri A, Rahimi R. Can judgments according to case fatality rate be correct all the time during epidemics? Estimated cases based on CFR in different scenarios and some lessons from early case fatality rate of coronavirus disease 2019 in Iran. Med J Islam Repub Iran. 2020;34:26.. [DOI] [PMC free article] [PubMed]
- 45.Shen CY. Logistic growth modelling of COVID-19 proliferation in China and its international implications. Int J Infect Dis. 2020;96:582–9. [DOI] [PMC free article] [PubMed]
- 46.Hsiang S, Allen D, Annan-Phan S, Bell K, Bolliger I, Chong T, Druckenmiller H, Huang LY, Hultgren A, Krasovich E, et al. The effect of large-scale anticontagion policies on the COVID-19 pandemic. Nature. 2020;584(7820):262–7. [DOI] [PubMed]
- 47.Tuite AR, Bogoch II, Sherbo R, Watts A, Fisman D, Khan K. Estimation of coronavirus disease 2019 (COVID-19) burden and potential for international dissemination of infection from Iran. Ann Intern Med. 2020;172(10):699–701. [DOI] [PMC free article] [PubMed]
- 48.Zhuang Z, Zhao S, Lin Q, Cao P, Lou Y, Yang L, He D. Preliminary estimation of the novel coronavirus disease (COVID-19) cases in Iran: a modelling analysis based on overseas cases and air travel data. Int J Infect Dis. 2020;94:29–31. [DOI] [PMC free article] [PubMed]
- 49.Thu TPB, Ngoc PNH, Hai NM, Tuan LA. Effect of the social distancing measures on the spread of COVID-19 in 10 highly infected countries. Sci Total Environ. 2020;742:140430. [DOI] [PMC free article] [PubMed]
- 50.Worldometers. COVID-19 coronavirus pandemic. https://www.worldometers.info/coronavirus/. Accessed 19 Oct 2020.
- 51.Zareie B, Roshani A, Mansournia MA, Rasouli MA, Moradi G. A model for COVID-19 prediction in Iran based on China parameters. Archives of Iranian Medicine. 2020;23(4):244–8. [DOI] [PubMed]
- 52.Al-Qaness MAA, Ewees AA, Fan H, Abualigah L, Abd Elaziz M. Marine predators algorithm for forecasting confirmed cases of COVID-19 in Italy, USA, Iran and Korea. Int J Environ Res Public Health. 2020;17(10). [DOI] [PMC free article] [PubMed]
- 53.Ayyoubzadeh SM, Ayyoubzadeh SM, Zahedi H, Ahmadi M, Niakan Kalhori RS. Predicting COVID-19 incidence thro [DOI] [PMC free article] [PubMed]
- 54.Moftakhar L, Seif M. The exponentially increasing rate of patients infected with COVID-19 in Iran. Arch Iran Med. 2020;23(4):235–8. [DOI] [PubMed]
- 55.Singh RK, Rani M, Bhagavathula AS, Sah R, Rodriguez-Morales AJ, Kalita H, Nanda C, Sharma S, Sharma YD, Rabaan AA, et al. Prediction of the COVID-19 pandemic for the top 15 affected countries: advanced autoregressive integrated moving average (ARIMA) model. JMIR Public Health Surveill. 2020;6(2):e19115. doi: 10.2196/19115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wikipedia. COVID-19 pandemic in Iran. https://en.wikipedia.org/wiki/COVID-19_pandemic_in_Iran.
- 57.World Health Organization (WHO), Eastern Mediterranean Regional Office (EMRO). Coronavirus disease – 2019 (COVID-19) country support missions, Iran mission report, 2–11 March 2020. https://issuu.com/iranintl/docs/6028353855. Accessed 4 May 2020.
- 58.Russell TW, Hellewell J, Abbott S, Golding N, Gibbs H, Jarvis CI, et al. Using a delay-adjusted case fatality ratio to estimate under-reporting. Available at the Centre for Mathematical Modelling of Infectious Diseases Repository https://cmmid.github.io/topics/covid19/global_cfr_estimates.html. Accessed 19 Oct 2020.
- 59.Faradeed News. From which cities in Iran the corona virus has become prevalent in Iran? News code 78522. 1399-01-08 [2020-03-27]. http://faradeed.ir/000KQU. Accessed 28 Mar 2020.
- 60.Setareh Sobh Newspaper. Corona statistics in Iran, real or unreal. Interview with Dr. Masoud Younesian. News code 58862. 2020-04-25. http://www.setaresobh.ir/fa/news/main/58862. Accessed 27 Apr 2020.
- 61.Ghadir MR, Ebrazeh A, Khodadadi J, Zamanlu M, Shams S, Nasiri M, Koohpaei A, Abbasinia M, Sharifipour E, Golzari SE. The COVID-19 outbreak in Iran; the first patient with a definite diagnosis. Arch Iran Med. 2020;23(7):503–504. doi: 10.34172/aim.2020.48. [DOI] [PubMed] [Google Scholar]
- 62.Yavarian J, Shafiei-Jandaghi N-Z, Sadeghi K, Shatizadeh Malekshahi S, Salimi V, Nejati A, et al. First cases of SARS-CoV-2 in Iran, 2020: case series report. Iran J Public Health. 2020;49(8):1564–8. [DOI] [PMC free article] [PubMed]
- 63.Tadbiri H, Moradi-Lakeh M, Naghavi M. All-cause excess mortality and COVID-19-related deaths in Iran. Med J Islam Repub Iran. 2020;34(1):561–566. doi: 10.34171/mjiri.34.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mehr News Agency. Real numbers of deaths are two times higher than the official reports. News code 5047851. 2020-10-14. http://mehrnews.com/xSVqp. Accessed 14 Oct 2020.
- 65.World Health Organization. Estimating mortality from COVID-19 - scientific brief. 4 Aug 2020. https://apps.who.int/iris/rest/bitstreams/1289937/retrieve. Accessed 15 Oct 2020.
- 66.World Health Organization. International guidelines for certification and classification (coding) of Covid-19 as cause of death. WHO/HQ/DDI/DNA/CAT. 2020. https://www.who.int/classifications/icd/Guidelines_Cause_of_Death_COVID-19-20200420-EN.pdf?ua=1. Accessed 15 Oct 2020.
- 67.World Health Organization. COVID-19 weekly epidemiological update, 25 October 2020. https://www.who.int/docs/default-source/coronaviruse/situation-reports/weekly-epi-update-11.pdf?sfvrsn=f5e837f3_2&download=true. Accessed 15 Oct 2020.
- 68.Tabnak News Agency. Real numbers of deaths and vases are 2.5 times higher than the official reports. News code 1009679. 2020-10-18. http://tabnak.ir/004Ef9. Accessed 19 Oct 2020.
- 69.Asriran News. Real numbers of deaths are 3–4 times higher than the official reports. News code 752937. 2020-10-25. http://asriran.com/0039s9. Accessed 26 Oct 2020.
- 70.Euro News. Around 10% of the world’s population may have had COVID-19, according to WHO. 2020-10-05. https://www.euronews.com/2020/10/05/around-10-of-the-world-s-population-may-have-had-covid-19-according-to-who. Accessed 7 Oct 2020.
- 71.Neil M, Fenton N, Osman M, McLachlan S. Bayesian network analysis of Covid-19 data reveals higher infection prevalence rates and lower fatality rates than widely reported. J Risk Res. 2020;23(7–8):866–79.
- 72.Grewelle R, De Leo G. Estimating the global infection fatality rate of COVID-19. medRxiv. Posted 18 May 2020. doi: 10.1101/2020.05.11.20098780.
- 73.Ioannidis J. The infection fatality rate of COVID-19 inferred from seroprevalence data. medRxiv. Posted 14 July 2020. doi: 10.1101/2020.05.13.20101253. [DOI] [PMC free article] [PubMed]
- 74.Levin AT, Hanage WP, Owusu-Boaitey N, Cochran KB, Walsh SP, Meyerowitz-Katz G. Assessing the age specificity of infection fatality rates for COVID-19: systematic review, meta-analysis, and public policy implications. medRxiv. Posted 31 Oct 2020. doi: 10.1101/2020.07.23.20160895. [DOI] [PMC free article] [PubMed]
- 75.SALAMATTV. 30 million Iranians have acquired Corona. 2020-10-25. https://salamattv.ir/news/208900. Accessed 12 Oct 2020.
- 76.Great Barrington Declaration. 4 Oct 2020. https://gbdeclaration.org. Accessed 12 Oct 2020.
- 77.Gollier C. If the objective is herd immunity, on whom should it be built? Environ Resour Econ. 2020;76(4):671–83. [DOI] [PMC free article] [PubMed]
- 78.James JJ. COVID-19: yin and yang and herd immunity. Disaster Med Public Health Prep. 2020;7:1–5. [DOI] [PMC free article] [PubMed]
- 79.Neipel J, Bauermann J, Bo S, Harmon T, Jülicher F. Power-law population heterogeneity governs epidemic waves. PLoS One. 2020;15(10):e0239678. doi: 10.1371/journal.pone.0239678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Griffin S. Covid-19: herd immunity is “unethical and unachievable,” say experts after report of 5% seroprevalence in Spain. BMJ. 2020;370:m2728. doi: 10.1136/bmj.m2728. [DOI] [PubMed] [Google Scholar]
- 81.Abbasi K. Covid-19: the fatal attraction of herd immunity. BMJ. 2020;370:m3714. doi: 10.1136/bmj.m3714. [DOI] [Google Scholar]
- 82.Bendavid E, Mulaney B, Sood N, Shah S, Ling E, Bromley-Dulfano R, Lai C, Weissberg Z, Saavedra-Walker R, Tedrow J, et al. COVID-19 antibody seroprevalence in Santa Clara county, California. medRxiv. Posted 30 April 2020. doi: 10.1101/2020.04.14.20062463. [DOI] [PMC free article] [PubMed]
- 83.Memish ZA, Perlman S, Van Kerkhove MD, Zumla A. Middle East respiratory syndrome. Lancet. 2020;395:1063–1077. doi: 10.1016/S0140-6736(19)33221-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Capasso V. Mathematical structures of epidemic systems. Berlin Heidelberg: Springer-Verlag; 1993. [Google Scholar]
- 85.Rojas I, Rojas F, Valenzuela O. Estimation of COVID-19 dynamics in the different states of the United States using Time-Series Clustering. medRxiv. Posted 29 June 2020. doi: 10.1101/2020.06.29.20142364.
- 86.Xiong D, Zhang L, Watson GL, Sundin P, Bufford T, Zoller JA, Shamshoian J, Suchard MA, Ramirez CM. Pseudo-likelihood based logistic regression for estimating COVID-19 infection and case fatality rates by gender, race, and age in California. medRxiv. Posted 1 July 2020. doi: 10.1101/2020.06.29.20141978. [DOI] [PMC free article] [PubMed]
- 87.English.news.cn, Xinhua. Full text of Wuhan’s notification on revising numbers of confirmed COVID-19 cases, deaths. http://www.xinhuanet.com/english/2020-04/17/c_138984653.htm. Accessed 4 May 2020.
- 88.Park M, Cook AR, Lim JT, Sun Y, Dickens BL. A systematic review of COVID-19 epidemiology based on current evidence. J Clin Med. 2020;9(4):967. [DOI] [PMC free article] [PubMed]
- 89.Davey Smith G, Blastland M, Munafò M. Covid-19’s known unknowns. BMJ. 2020;371:m3979. doi: 10.1136/bmj.m3979. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Appendix Text 1. Search syntax used in PubMed. Appendix Text 2. Details of studies’ scenario. Appendix Table 1. Predictions of cumulative cases for the end of months one to six after the official epidemic start date (2020-02-19) and the latest date available in 2020. Appendix Table 2. Predictions of daily deaths at end of months one to six after the official epidemic start date (2020-02-19) and the latest date available in 2020. Appendix Table 3. Predictions of daily cases for the end of months one to six after the official epidemic start date (2020-02-19) and the latest date available in 2020. Appendix Table 4. Predictions of epidemic peak dates and values of outcomes. Appendix Table 5. Predictions of epidemic control dates and values of outcomes. Appendix Figure 1. PRISMA 2009 study flow diagram. Appendix Figure 2. Officially reported cumulative confirmed cases, deaths, and recovered cases of COVID-19 in Iran. Appendix Figure 3. Reported daily confirmed cases, deaths, and recovered cases of COVID-19 in Iran. Appendix Figure 4. Reported and median-scenario estimated daily prevalent cases of COVID-19 in Iran, including predictions by Saberi. Appendix Figure 5. Reported and median-scenario estimated daily prevalent case of COVID-19 in Iran, without predictions by Saberi. Appendix Figure 6. Reported and worst-scenario estimated cumulative deaths of COVID-19 in Iran, including predictions by Mashayekhi. Appendix Figure 7. Reported and worst-scenario estimated cumulative deaths of COVID-19 in Iran, without predictions by Mashayekhi. Appendix Figure 8. Reported and current (median) scenario estimated cumulative deaths of COVID-19 in Iran, International studies.
Additional file 2. Target studies’ abstracted data.
Data Availability Statement
All data generated or analysed during this study are included in this published article and its supplementary information files.