Seagrasses are important coastal species that are declining globally, and transplantation can be used to combat these declines. However, the bacterial communities associated with seagrass rhizospheres and roots (the microbiome) are often disturbed or removed completely prior to transplantation.
KEYWORDS: seagrass, eelgrass, restoration, rhizosphere, rhizoplane, microbial diversity, succession
ABSTRACT
Seagrasses can form mutualisms with their microbiomes that facilitate the exchange of energy sources, nutrients, and hormones and ultimately impact plant stress resistance. Little is known about community succession within the belowground seagrass microbiome after disturbance and its potential role in the plant’s recovery after transplantation. We transplanted Zostera marina shoots with and without an intact rhizosphere and cultivated plants for 4 weeks while characterizing microbiome recovery and effects on plant traits. Rhizosphere and root microbiomes were compositionally distinct, likely representing discrete microbial niches. Furthermore, microbiomes of washed transplants were initially different from those of sod transplants and recovered to resemble an undisturbed state within 14 days. Conspicuously, changes in the microbial communities of washed transplants corresponded with changes in the rhizosphere sediment mass and root biomass, highlighting the strength and responsive nature of the relationship between plants, their microbiome, and the environment. Potential mutualistic microbes that were enriched over time include those that function in the cycling and turnover of sulfur, nitrogen, and plant-derived carbon in the rhizosphere environment. These findings highlight the importance and resilience of the seagrass microbiome after disturbance. Consideration of the microbiome will have meaningful implications for habitat restoration practices.
IMPORTANCE Seagrasses are important coastal species that are declining globally, and transplantation can be used to combat these declines. However, the bacterial communities associated with seagrass rhizospheres and roots (the microbiome) are often disturbed or removed completely prior to transplantation. The seagrass microbiome benefits seagrasses through metabolite, nutrient, and phytohormone exchange and contributes to the ecosystem services of seagrass meadows by cycling sulfur, nitrogen, and carbon. This experiment aimed to characterize the importance and resilience of the seagrass belowground microbiome by transplanting Zostera marina with and without intact rhizospheres and tracking microbiome and plant morphological recovery over 4 weeks. We found the seagrass microbiome to be resilient to transplantation disturbance, recovering after 14 days. Additionally, microbiome recovery was linked with seagrass morphology, coinciding with increases in the rhizosphere sediment mass and root biomass. The results of this study can be used to include microbiome responses in informing future restoration work.
INTRODUCTION
The rhizosphere has long been recognized to have important impacts on plant growth and health (1). The microbes of the rhizosphere, which directly interact with and are influenced by the roots (2), can benefit their plant hosts through recycling and producing bioavailable nutrients (3–5), increasing disease resistance through competition with or inhibition of pathogens (6), and influencing plant growth and stress tolerance through the production of phytohormones (7, 8). The community composition within the rhizobiome is shaped by plant metabolism and physiology, which controls rhizodeposition, the exudation of organic carbon and nitrogen, and the release of defense compounds (7, 9, 10). The quantity and composition of exudates can impact microbial activity in the rhizosphere and vary as a result of many factors (11–14). While plant-rhizobiome interactions are relatively well defined for terrestrial plants, analogous interactions between aquatic plants and their microbiomes have only recently started to become known (15, 16).
Seagrasses are marine vascular plants that form key ecosystems in coastal areas worldwide, where they provide numerous ecosystem services (17). Recent evidence suggests that members of the seagrass microbiome may modulate host growth and responses to environmental stresses (15, 18, 19). In addition to fixing nitrogen and producing phytohormones (20, 21), the seagrass microbiome is proposed to mitigate the toxic effects of hydrogen sulfide in sediments, which have been linked to declines in seagrass health and localized die-back events (22–24). The seagrass rhizobiome is thought to be primarily influenced by the exudation of carbon compounds, which can provide up to 60% of the carbon assimilated by these microbes (25, 26), and by radial oxygen loss from roots, which may promote the colonization of the rhizosphere by distinct bacteria (24, 27).
The effect of rhizosphere disturbance on the composition of seagrass microbiomes and plant health has rarely been explored (28), but it may be important both for plant recovery after a disturbance and in the context of restoration outcomes, which are highly variable and dependent on methodology (29–32). Sod transplants, which transfer shoots with intact rhizospheres, have historically been one of the more successful methods, potentially because the intact rhizosphere sediment acts as a natural anchor and retains functional relationships between the plant and its rhizobiome (31). Conversely, bare-root transplants are generally less successful and could experience a decrease or lag in plant performance as the rhizobiome redevelops after transplantation. Importantly, microbial community succession after disturbance can strongly affect host health in several microbiome-host systems (e.g., algae, corals, and humans), whereby dysbiosis disrupts host functioning and increases susceptibility to disease (33–35). Thus, it is important to understand the recovery of seagrass microbiomes after disturbance, as this may impact seagrass health and resistance to environmental stresses.
In this study, we characterized the recovery of seagrass rhizobiomes after disturbance by transplanting Zostera marina, commonly referred to as eelgrass, with and without an intact rhizosphere and sampling for plant and microbiome characteristics over the course of 28 days. We expected to see the rhizobiome of eelgrass transplanted without an intact rhizosphere recover over time to resemble that of the control plants, with a corresponding delay in the response of plant growth traits.
RESULTS
Changes in Z. marina traits after transplantation.
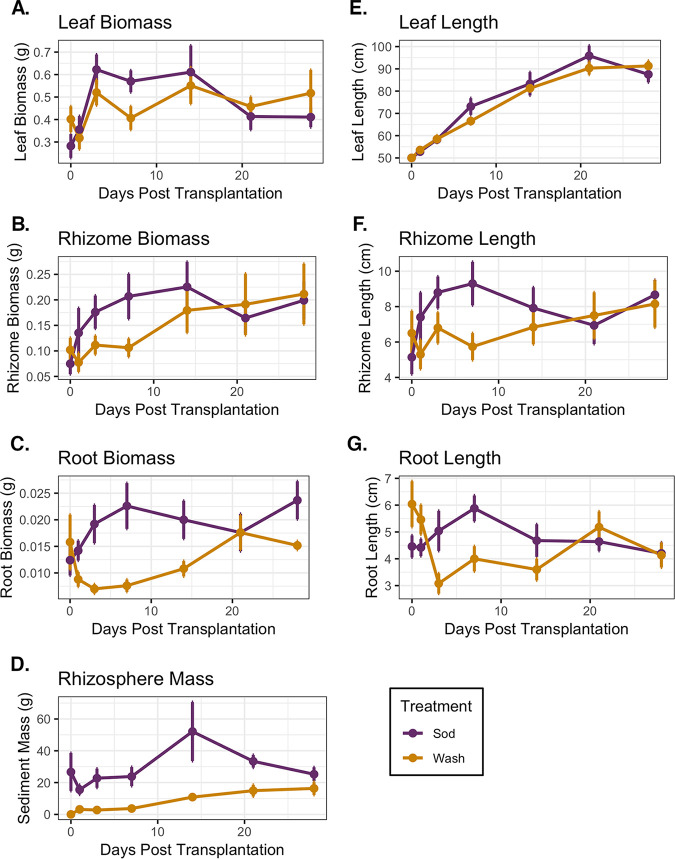
We quantified several traits to assess plant growth (i.e., biomass and lengths of leaves, rhizomes, and roots) and to measure the mass of rhizosphere sediment (i.e., the sediment firmly attached to roots after plant collection) (Fig. 1). Plant traits varied significantly due to an interaction between days posttransplantation (DPT) and treatment (DPT × treatment) (by permutational multivariate analysis of variance [PERMANOVA] for DPT × treatment, F1,61 = 2.85, P = 0.036, and R2 = 0.03). Plant traits began to exhibit overall differences within 7 days after transplantation, and those of the wash treatment began to more strongly resemble those of the sod treatment after 1 week (Fig. 2A). For sod transplants, the most variation in traits occurred within the first 7 days of the experiment, after which these measures stabilized and remained relatively constant (Fig. 1 and Fig. 2B). Conversely, changes in the traits of washed plants occurred more slowly, stabilizing only after 14 days. By the end of the experiment, no between-treatment variation in traits was evident (by PERMANOVA for day 28 treatment, F1,13 = 1.00, P = 0.422, and R2 = 0.07) (Fig. 1 and Fig. 2B).
FIG 1.
Z. marina morphometric data. (A) Leaf biomass; (B) rhizome biomass; (C) root biomass; (D) rhizosphere sediment biomass; (E) leaf length; (F) rhizome length; (G) root length. Colors designate treatment groups (mean values and standard errors are reported).
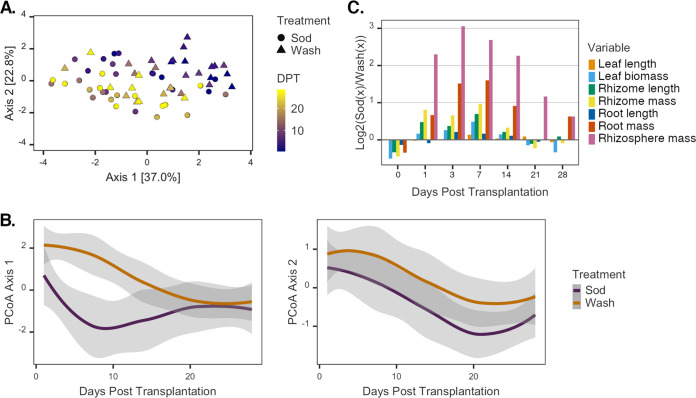
FIG 2.
Variance in Z. marina traits over time. (A) PCoA of Z. marina plants based on a Euclidean distance matrix relating plant traits. The color gradient represents the day of plant collection (DPT), and symbols represent the treatment assignment for each plant. (B) Locally estimated scatterplot smoothing (LOESS) of the first two principal-coordinate summary variables over time (cumulative variance = 59.8%) illustrates how variation in traits of plants from each treatment significantly diverges over time (left) and covaries over the course of the experiment (right). Shaded areas represent the 95% confidence intervals of the estimates. (C) Relative differences in log-transformed values of Z. marina morphometric data at sampling points over time. Positive values indicate higher values in sod transplants than in washed samples, and negative values indicate the opposite.
Principal-coordinate analysis (PCoA) showed that 22.8% of the overall variation of plant traits was synchronized between treatments (Fig. 2B). This variation likely relates to significant increases in the measurements of most traits over the course of the experiment in both treatment groups, indicating the overall growth of Z. marina shoots after transplantation regardless of rhizosphere presence (Fig. 1). For instance, upon the completion of the experiment, the total biomass of transplants had increased 1.5-fold on average, and the lengths of leaves and rhizomes had increased 1.5- and 1.8-fold, respectively (Fig. 1). Whereas differences in traits due to treatment were minimal at the beginning and end of the experiment, they were most pronounced from days 1 to 14 of the experiment when sod transplants consistently demonstrated greater increases than those of the washed transplants (Fig. 2C). For example, root biomass and root length were not significantly affected by rhizosphere removal at the beginning of the experiment [by Student’s t test for root biomass, wash mean (M) = 0.016 ± 0.012 g, sod M = 0.012 ± 0.006 g, t(8) = −0.57 [where 8 signifies degrees of freedom], and P = 0.58; by Student’s t test for root length, wash M = 6.04 ± 1.91 cm, sod M = 4.46 ± 0.92 cm, t(8) = −1.67, and P = 0.13]. Importantly, however, sod transplants increased 1.7-fold in root biomass on average, and wash transplants increased 1.1-fold by the end of the experiment (by analysis of covariance [ANCOVA] for treatment, F1,72 = 16.16 and P = 0.0001) (Fig. 1C). As expected, the rhizosphere sediment mass significantly varied with the interaction between the time covariate and the main treatment effect (by ANCOVA for DPT × treatment, F1,71 = 18.78 and P < 0.00005) (Fig. 1D). That is, the rhizosphere sediment mass attached to the roots of sod transplants did not change significantly during the experiment, whereas sediment accumulation on washed roots rapidly increased after 7 days posttransplantation and recovered to the levels observed on sod transplants by the end of the experiment [by Welch’s t test, wash M = 16.34 ± 10.03 g, sod M = 25.25 ± 13.26 g, t(13) = 1.48, and P = 0.16] (Fig. 1D).
Microbial community differences between the Z. marina rhizosphere and roots.
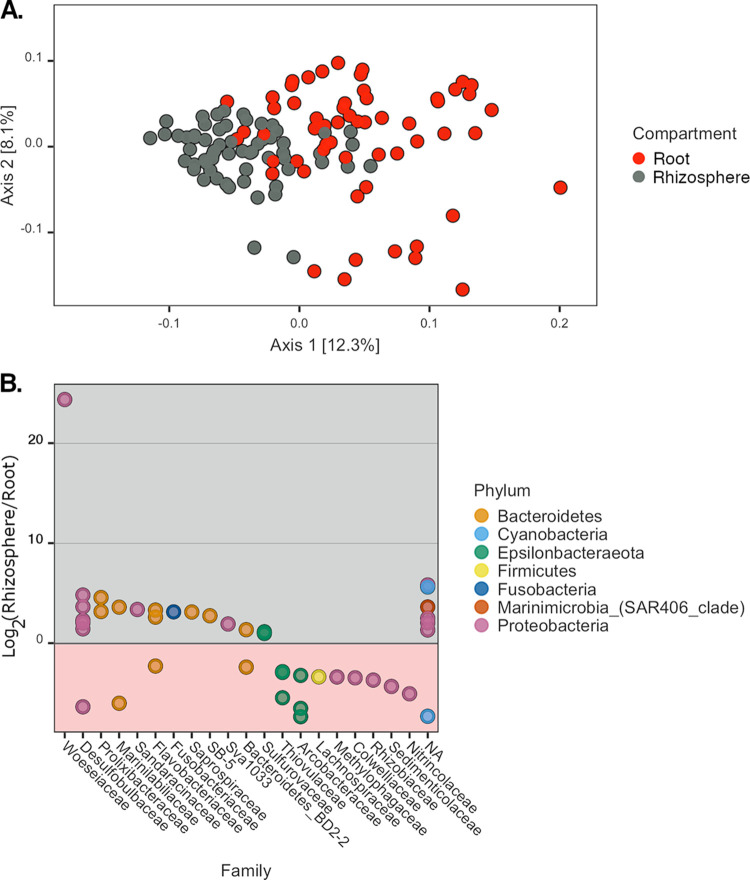
When considering all samples, microbial communities were most strongly clustered based on the belowground compartment (i.e., root versus rhizosphere compartment) (by PERMANOVA, F1,112 = 26.33, P = 0.001, and R2 = 0.16) (Fig. 3A). Forty-two prokaryotic amplicon sequence variants (ASVs) exhibited significantly different relative abundances in the rhizosphere versus roots (see Table S1 in the supplemental material). Twenty-five were enriched in the rhizosphere, while the remaining 17 were present at a higher relative abundance on roots (Fig. 3B). Significant ASVs were most commonly assigned to the Proteobacteria and Bacteroidetes phyla (n = 18 and 12, respectively), with 66% of the former taxon and 75% of the latter being detected at higher relative abundances in the rhizosphere than in root communities. Conversely, ASVs of the Epsilonbacteraeota phylum were typically present at higher relative abundances in root samples (five of seven ASVs). Due to the strong effect of compartment on microbial community structure, the remaining microbial diversity results are presented separately for rhizosphere and root samples.
FIG 3.
Microbial community differences between Z. marina compartments. (A) PCoA of all sampled Z. marina microbial communities based on a weighted UniFrac distance matrix. Colors indicate the compartment of each sample. (B) Taxa with significant relative abundance differences between compartments. Positive values indicate higher relative abundances of ASVs in rhizospheres than in roots, and negative values indicate the opposite. ASVs assigned to the same phylum have the same color. ASVs are grouped by column by taxonomic family. NA, unclassified at the family level.
Changes in rhizosphere microbiomes after transplantation.
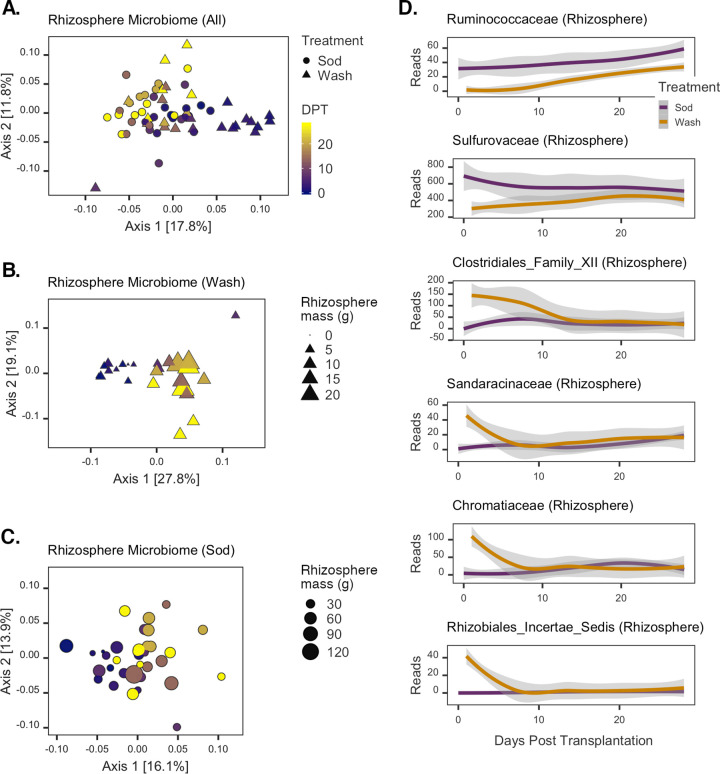
The temporal changes in the structure of rhizosphere microbial communities mirror the patterns observed for plant trait data. That is, initial differences were observed between rhizosphere communities from plants of different treatment groups, but communities became more similar in structure by the end of the experiment (Fig. 4A). The most variation was due to a shift of rhizosphere communities of washed transplants along the first principal coordinate to more strongly resemble those of sod samples after 7 days. As observed for plant traits, a significant interactive effect of treatment and time on the rhizosphere community structure was detected (by PERMANOVA for DPT × treatment, F1,58 = 2.53, P = 0.005, and R2 = 0.03).
FIG 4.
Changes in rhizosphere microbial communities posttransplantation. (A to C) PCoAs of all rhizosphere (A), washed rhizosphere (B), and sod transplant rhizosphere (C) communities. The color gradient represents the day of sample collection (DPT), and symbols represent the treatment assignment for each sample. Symbol sizes in panels B and C are scaled to the grams of rhizosphere sediment collected from each corresponding sampled plant. (D) Rhizosphere ASVs with significant time-treatment interaction effects. LOESS was applied to the sequence counts for each taxon; shaded areas represent the 95% confidence intervals of the estimates. Colors designate each treatment group.
To further investigate the effect of rhizosphere disruption on the recovery of the rhizosphere communities, we analyzed the different treatment samples separately. Structural changes in the rhizosphere communities of the sod and wash treatment groups both demonstrated significant time effects, but a stronger temporal correlation was detected for the washed than for the sod transplant rhizosphere communities (by PERMANOVA for DPT [wash transplants], F1,25 = 6.47, P = 0.001, and R2 = 0.21; for DPT [sod transplants], F1,33 = 3.57, P = 0.001, and R2 = 0.10). A shift in the community structure of washed transplants occurred at 7 days and corresponded to the point of increasing sediment accumulation on washed roots (Fig. 1D). Additionally, overall community changes were significantly correlated with the rhizosphere sediment masses of all washed plants (Mantel test P = 0.004; Spearman’s ρ = 0.25) (Fig. 4B). For sod transplants, however, sediment mass was not correlated with rhizosphere community structure (Mantel test P = 0.43; Spearman’s ρ = 0.0001) (Fig. 4C) and was instead most strongly correlated with plant growth traits (leaf length, rhizome length, and leaf biomass, Mantel test P = 0.001; Spearman’s ρ = 0.27).
Using regression analyses with summarized sequence counts of family-level taxonomic units, we identified microbial taxonomic families that were specifically associated with Z. marina rhizosphere development during the experiment (generalized linear mixed model [GLMM]-adjusted P value of ≤0.05) (Table S2). Thirty-two families had significantly different modeled intercept coefficients between treatments, and 14 families exhibited significant differences in modeled slopes. Six families were found with significant differences in the coefficients of both slopes and intercepts (Fig. 4D; see also “Statistical analyses” in Materials and Methods for a description of the interpretation of these coefficients). Of these six families, the Ruminococcaceae and Sulfurovaceae had negative intercept and positive slope coefficients. For example, higher relative abundances of the Ruminococcaceae were detected in the sod samples on average, but the rate of increase of this taxon’s abundance was higher in washed samples over time. The Sulfurovaceae showed a similar temporal pattern of abundance in washed samples, but in sod transplants, this taxon generally demonstrated a decrease over time. The remaining four families (Clostridiales family XII, Sandaracinaceae, Chromatiaceae, and Rhizobiales [incertae sedis]) all showed similar patterns (Fig. 4D): in washed transplants, they rapidly decreased to low levels within the first 7 days of the experiment, whereas in sod transplants, there was little to no detection of them throughout the experiment.
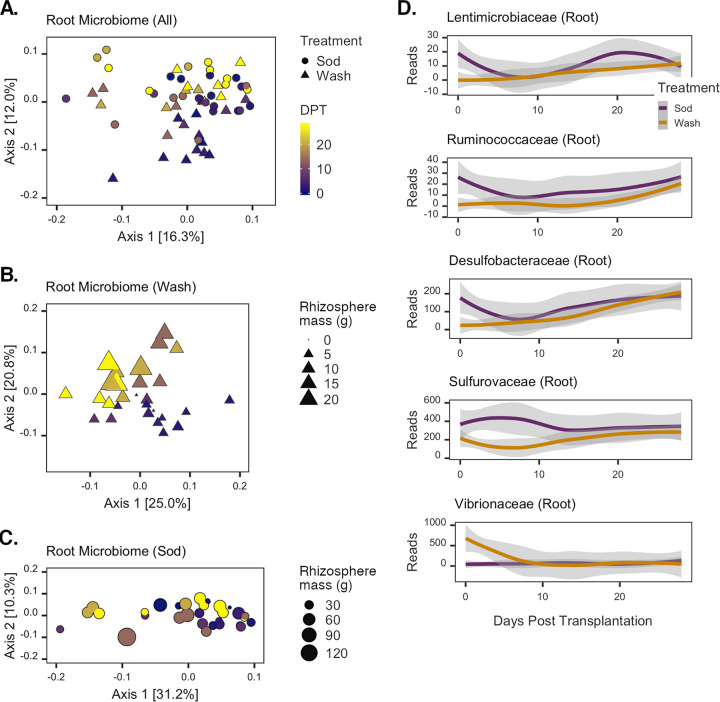
Changes in root microbiomes after transplantation.
The recovery dynamics of root microbiomes were largely similar to those observed for rhizosphere communities (Fig. 5A). A significant effect of the interaction between time and treatment on the structure of all root communities was detected (by PERMANOVA for DPT × treatment, F1,58 = 2.01, P = 0.043, and R2 = 0.03). A relatively strong effect of time was evident for communities from wash transplants (by PERMANOVA, F1,26 = 5.91, P = 0.001, and R2 = 0.19) (Fig. 5B) but not for sod transplants (by PERMANOVA, F1,28 = 1.78, P = 0.096, and R2 = 0.06) (Fig. 5C). Changes in the washed root microbiome community structure were not significantly correlated with sediment mass accumulation (Mantel test P = 0.085; Spearman’s ρ = 0.13) and were instead most strongly correlated with leaf length and rhizome mass (Mantel test P = 0.001; Spearman’s ρ = 0.32). In contrast, the root microbiomes of sod transplants were relatively stable over time (by PERMANOVA, F1,28 = 1.78, P = 0.096, and R2 = 0.06) (Fig. 5C) and not correlated with any single plant trait or combination thereof.
FIG 5.
Changes in root microbial communities posttransplantation. (A to C) PCoAs of all root (A), washed root (B), and sod transplant root (C) communities. The color gradient represents the day of sample collection (DPT), and symbols represent the treatment assignment for each sample. Symbol sizes in panels B and C are scaled to the grams of rhizosphere sediment collected from each corresponding sampled plant. (D) Root ASVs with significant time-treatment interaction effects. LOESS was applied to the sequence counts for each taxon; shaded areas represent the 95% confidence intervals of the estimates. Colors designate each treatment group.
Regression analyses identified 25 taxonomic families with significant differences in modeled intercept coefficients between treatments but no differences in modeled slopes (Table S3). Another four families were found to have no detectable differences in intercept coefficients but significant differences in slopes. Five families were found to have significant differences in both modeled intercept and slope coefficients (Fig. 5D). The Lentimicrobiaceae, Ruminococcaceae, and Desulfobacteraceae were all modeled to have largely similar dynamics, with negative intercept and positive slope coefficients. The abundances of these taxa on roots of sod transplants rapidly declined within 7 days of transplantation, followed by a more gradual increase in abundance over the last 2 weeks of the experiment (Fig. 5D). Conversely, on the roots of washed transplants, these taxonomic families were nearly undetectable initially, but their abundances recovered by the completion of the experiment. The Sulfurovaceae also exhibited gradual increases in relative abundances on washed roots, but in sod transplants, this taxon’s abundance increased after 7 days and subsequently decreased and leveled off (Fig. 5D). Vibrionaceae showed an altogether different pattern: high abundances were detected in the initial wash samples, but by day 7, these taxa were rarely detected. In sod transplants, the abundances of sequences assigned to the Vibrionaceae were generally absent throughout the experiment.
DISCUSSION
Our results indicate that Zostera marina rhizobiome communities are distinct, linked to seagrass performance, and resilient to disturbance. Indeed, eelgrass belowground root biomass suffered negatively from rhizosphere disruption but recovered after approximately 2 weeks. Concomitantly, the microbial communities in the rhizospheres of washed transplants resembled those of sod transplants by the end of the experiment, indicating that Z. marina and its belowground microbiome are resilient to stresses associated with transplantation.
The observation of consistently distinct microbial communities between compartments of Z. marina is in line with studies describing the structure of seagrass microbiomes from field-collected samples, where large differences are observed between plant microbial communities and those in the surrounding environment (15, 18, 19, 36–39). In a study by Cúcio et al. (15) where the rhizosphere compartment was specifically analyzed, significant differences were found between communities of bulk and rhizosphere sediments. Our work further distinguishes the root-attached microbiome as being different from the microbiota of the rhizosphere and suggests that these two compartments are separate microbial niches. These niches are likely shaped by prevailing redox and nutrient gradients formed across submillimeter ranges by plant metabolic processes (40). These results are supported by previous observations (19, 41) and a proposed model of microbiome assembly via the selection of bulk sediment microbes (19).
Although the mechanisms controlling the assembly of seagrass microbiomes are largely unknown, evidence from terrestrial plant studies suggests that they are based on metabolic interactions and nutrient exchange between plants and microbes. For instance, changes in abiotic factors and/or the presence of pathogens can induce or restrict the exudation of nutritional and allelopathic compounds, contributing to the selection of a root microbiome (9, 42). Root exudation is known to be metabolically costly for plants, however, and can result in significant losses of carbon and nitrogen (7). However, these costs are likely offset by the beneficial functions of the belowground microbiome (e.g., disease suppression, nutrient acquisition, stress tolerance, and growth enhancement) (7, 8, 43).
Similar to these terrestrial plant examples, we propose that exudation is an important factor modulating the belowground microbiomes of seagrasses. Seagrass exudation is known to change with environmental conditions (e.g., light restriction) (44) and can act as an important resource for sediment microbes (26, 45). Our concomitant observations of belowground root biomass loss in washed Z. marina plants and large-scale changes in the microbiome structure within the first week after transplantation may be related to changes in root exudation, which would imply a rapid and coordinated response by both the microbiome and plant to disturbance. An alternative explanation for our results is that root damage may have occurred during seawater rinses to remove the rhizosphere prior to transplantation. Our data suggest that no observable and significant root biomass loss occurred from initial washes, but these relatively coarse measures of biomass may not account for the loss of fine root biomass, which may have resulted in the observed treatment effects. Further experimentation specifically characterizing the exudation patterns of seagrasses after rhizosphere disturbance will be needed to definitively resolve these hypotheses.
When considering the timing of recovery between belowground compartments, it is notable that the change in the microbial community structure of washed roots was detected 3 days after transplantation, whereas a similar change in the rhizosphere was detected on day 7 (Fig. 4B and Fig. 5B). Interestingly, almost all of the root- and rhizosphere-associated taxonomic families that significantly changed in abundance over time demonstrated an inflection point in their abundance trajectories between 3 and 7 days after transplantation (Fig. 4D and Fig. 5D). When considered with the changes observed for plant traits, these data suggest that the first week after transplantation is a critical transition period for the plant and its associated microbiome.
Rapid and resilient responses of microbiomes to disturbance, such as those seen here, have also been observed for microbiomes of terrestrial plants (46, 47) and marine algae (34). Interestingly, the speed of recovery may be dependent on the physical route of microbial transmission, as microbiomes of the phyllosphere of Arabidopsis thaliana appear to be acquired from the air and converge to mature communities only after 60 days (46). In contrast, recovery of microbiomes colonizing biotic surfaces found in water-saturated environments (e.g., algal surfaces and rice roots) occurs within days to weeks. For example, community assembly on the surface of Delisea pulchra was found to be deterministic, recovering to a predisturbed state within 12 days. In this system, the production of antifouling chemicals (i.e., halogenated furanones), either by early-colonizing bacteria or by the algae, is an important factor controlling community succession. Seagrasses can also produce a diverse set of antifouling chemicals on their surfaces (48), although their precise role in modulating the epibiont community structure is currently unknown.
Our results also show that several ASVs assigned to taxonomic groups that have previously been proposed to benefit plants or enhance the turnover of nutrients in sediments are enriched in seagrass-associated compartments after transplantation. For instance, ASVs enriched in rhizosphere over root samples were assigned to taxonomic groups (e.g., Marinilabiliaceae, Bacteroidetes BD2-2 and SB-5, Flavobacteriaceae, and Sandaracinaceae) widely recognized to be important degraders of complex organic material such as rhizodeposits and algal cell wall polysaccharides (49–54). Notable taxa that were enriched on roots relative to the rhizosphere include ASVs with potentially important roles in the turnover of plant exudates. For example, all detected ASVs assigned to the methylotrophic lineages (i.e., Methylomonaceae, Methylophagaceae, and Methylophilaceae) were found at higher relative abundances on the root than in the rhizosphere, supporting a potential symbiotic role for these populations based on their described abilities to consume plant-derived methanol and produce plant phytohormones (55, 56). Additionally, ASVs of the Lachnospiraceae and Colwelliaceae families, which were enriched on roots relative to the rhizosphere, may have potential roles in the consumption of plant-derived polysaccharides and lignin (57, 58). In fact, the former group may have an additional symbiotic role with plants, as a novel species of Lachnospiraceae is proposed to be diazotrophic (59).
Other taxa found enriched in either the root or rhizosphere compartment appear to rely on respiratory metabolisms linked to sulfur and nitrogen cycles, a common feature of populations of the seagrass microbiome (20, 60). Seagrasses within the Yaquina estuary typically grow under eutrophic conditions, with NO2 and NO3 concentrations reaching ≥30 μM during summer months when upwelling is active along the Oregon coast (61). Additionally, sediments in the estuary are strongly anoxic, with high porewater concentrations of sulfide (10 to 80 μM) and total dissolved inorganic nitrogen (≥10 μM) (41, 62). Previous reports suggest that organic matter inputs from seagrass roots can stimulate microbial activities that control these cycles, ultimately leading to higher sulfate reduction, denitrification, and nitrogen fixation rates in seagrass bed sediments as well as stimulating the release of bioavailable phosphorus and iron (3, 41, 63–66). ASVs assigned to the Desulfobulbaceae, which can act as anaerobic sulfate reducers (e.g., Desulforhopalus sp.) (67) or as sulfide oxidizers that transfer electrons from reduced sulfur compounds to either oxygen or nitrate (e.g., “Candidatus Electrothrix sp.”) (68, 69), were commonly enriched in the rhizosphere relative to roots and have been frequently detected within rhizospheres of aquatic plants (24, 70). In addition, several ASVs found enriched on roots relative to the rhizosphere were designated known or putative sulfur-oxidizing bacteria (SOB), including Sedimenticolaceae (71), Thiovulaceae, and Arcobacteraceae (72), supporting the hypothesis that seagrasses facilitate the activities of SOB as a way to combat sulfide toxicity (73).
The Ruminococcaceae and the Sulfurovaceae were notable in our time course analyses, as both exhibited similar abundance differences initially and over time in both root and rhizosphere samples. ASVs of these taxa, along with those of the Lentimicrobiaceae and Desulfobacteraceae, were noticeably absent on washed roots at the start of the experiment, but all reached the relatively high levels found on roots of sod transplants by the end of the experiment. Many of these taxa are known to drive sulfur cycling in marine sediments (72, 74, 75), and their functional roles may be important in long-term associations with plants. In contrast, the Vibrionaceae were the only taxa that rapidly decreased from high relative abundances on washed roots to undetectable levels after 7 days. Given that many Vibrio species are fast-growing copiotrophs that rapidly form biofilms on marine surfaces (76, 77), it is possible that these ASVs may rapidly colonize the rhizosphere and root environment as a result of rhizosphere disturbance and that plants respond by changing their physiology as a way to discourage the growth of these of bacteria while encouraging the growth of beneficial microbes shortly after disturbance.
Seagrass health after transplantation is often unpredictable (30), and restoration success is thought to be dependent on many factors, with root growth and sediment anchoring being identified as keys to long-term success (31, 78, 79). Despite the importance of these belowground processes, few studies have explicitly examined the impact of microbiome community structure on transplantation success. A study by Milbrandt and colleagues is, perhaps, an instructive exception (28). Similar to our findings, washed and sod transplants of Thalassia testudinum showed few differences in plant traits several weeks after transplantation. Critically, however, transplants that were planted into autoclaved sediment demonstrated a strong and significant die-off starting at 7 weeks posttransplantation, leading the authors to suggest that an intact microbial community is essential for the plant’s ability to combat transplantation shock.
An important distinction of our work is that growth traits of washed transplants consistently lagged behind those of sod transplants during the first week of the experiment when microbiome recovery was most pronounced. Notably, root biomass showed rapid and significant decreases for plants assigned to the wash treatment versus those assigned to the sod treatment. The potential implications of root biomass loss after bare-root restoration attempts should be further considered and investigated due to the established roles of roots in physical anchoring (80), microbial recruitment (81), and resource acquisition (82). Although we do not see significant effects between transplantation methods at the end of our incubations, other factors not accounted for in our controlled tank experiments may also be influencing the ultimate success of restoration projects in the field. When considering our results in light of the highly variable nature of restoration outcomes, it is apparent that understanding the roles of the seagrass microbiome in optimizing plant physiology, combating transplantation shock, and contributing to anchoring effects at the bed scale will be essential for the development of best practices for future seagrass restoration programs.
MATERIALS AND METHODS
Experimental setup.
Sediment (top ∼15 cm) and 90 healthy Z. marina primary shoots were manually collected at low tide from intertidal eelgrass beds in Yaquina Bay, Oregon (lat 44.624518, long −124.044372), during July 2018. After collection, sediment was sieved through wire mesh with 0.25-cm2 openings and held in buckets filled with seawater for 24 h. Plants were manually extracted from the beds by excavating an ∼3-cm-radius sediment ball around the roots and collecting terminal shoots with attached rhizome fragments, a method that is similar to those previously used in studies on seagrass transplantation (83–85). Plants were placed in plastic bags and processed for transplantation within 3 h of collection.
Individual plants were randomly assigned to either the “wash” or the “sod” transplant treatment group. The rhizospheres of plants in the washed group were removed by a gentle seawater rinse, retaining the rhizoplane bacteria and replicating the potential rhizosphere loss in transplantation efforts. The rhizospheres of plants assigned to the sod treatment group were left undisturbed. The rhizomes of plants in the wash treatment group were trimmed to retain five internodes connected to the first five root bundles (86), and rhizomes of sod transplants were standardized by trimming to lengths matching those of the washed plants. Plant leaves were standardized across treatments by trimming to 50 cm (87).
PVC cylinders (18 by 7.6 cm) were filled with sediment, and the meristem of each plant was positioned near the top of each. Sediment was added to cover the rhizome, roots, and rhizosphere (if attached). Planters were randomly and evenly placed inside a 2,000-liter outdoor flowthrough tank filled with water from Yaquina Bay.
Plant sampling and morphometric analyses.
Whole-plant sampling was performed on the initial day of the experiment (t = 0) prior to transplantation and on days 1, 3, 7, 14, 21, and 28 posttransplantation. At least five plants from each treatment group were collected and destructively sampled at each time point. Methods outlined previously by Cúcio et al. (15) were followed to isolate the rhizosphere. For each sample, the plant and bulk sediment were extracted from the PVC planter as a single unit. A sterile metal spatula was used to excavate the rhizome from the unattached bulk sediment. Once all nonattached sediment had been removed, the plant was lifted from its base to minimize root breakage and then gently shaken 2 to 5 times until the majority of the residual loose sediment was dislodged. Sediment adhering to the roots was defined as the rhizosphere, which is in accordance with established operational definitions for both terrestrial plants (88, 89) and seagrasses (15, 90). The rhizosphere sediment was then washed from the plant roots in 25 ml of sterile seawater and collected in sterile tubes. One milliliter of the resulting slurry was transferred to a sterile microcentrifuge tube and stored at −80°C until DNA extraction. One pair of the youngest root cluster was then removed from the plant, transferred to a sterile microcentrifuge tube, and stored at −80°C for DNA extraction.
Roots not used for extractions were removed from the plants, counted, and measured to calculate average lengths. Rhizome lengths and longest leaf lengths were recorded for plants. Biomass measurements were recorded for the component parts of plants (i.e., leaves, rhizomes, and roots) after drying for 7 days at 40°C. The residual sediment slurries from plants (∼24 ml/plant) were vacuum filtered through preweighed glass fiber filter (GFF) membranes and dried as described above, and net weights were recorded as rhizosphere masses.
DNA extraction, PCR, and amplicon sequencing.
Microbial community DNA was extracted from frozen roots and sediment slurries using a cetyltrimethylammonium bromide (CTAB) and phenol-chloroform extraction method (91) within 6 weeks of sample collection. DNA was extracted from the root phytoplane but may also include DNA from the root endosphere. Amplicon sequencing libraries were constructed from 25 to 100 ng of the template DNA using one-step PCR with barcoded 515F and 806R universal 16S rRNA (v3-v4) primers (92). PCRs were performed by using AccuStart II ToughMix polymerase according to the manufacturer’s instructions and performing a thermal cycle program of 94°C (3 min); 25 cycles of 94°C (45 s), 50°C (60 s), and 72°C (90 s); 72°C (10 min); and 4°C (hold).
Mixtures from successful amplification reactions (139 of 143 samples) were purified using Agencourt AMPure XP beads according to the manufacturer’s instructions, with the exception that a 1:1 ratio of the bead solution and PCR product was used. A Qubit 2.0 fluorometer (Thermo Fisher Scientific, Waltham, MA, USA) was used to quantify concentrations of purified amplicons, and these values were used to evenly pool libraries prior to sequencing with the Illumina MiSeq platform (Illumina Inc., San Diego, CA, USA).
The DADA2 package (v 1.10.1) (93) within the Bioconductor software environment (v 3.8) (94) of the R Project (v 3.5.2) (95) was used to process raw sequencing reads. All reads were initially quality filtered using the filterAndTrim command with default settings [“maxN=0, maxEE=c(2,2), truncQ=2”]. To avoid computational limitations resulting from the fact that multiple libraries contained >100,000 reads, the resulting high-quality reads of libraries were randomly down-sampled to 15,000 paired-end reads (BioProject accession number PRJNA591021). This resulted in 126 libraries with ≥8,891 high-quality paired-end reads used as inputs for the remaining DADA2 pipeline (i.e., error rate training, sample inference, paired-read merging, chimera removal, amplicon sequence variant [ASV] counting, and taxonomic assignment against the SILVA nonredundant reference database version 132 [Ref NR 132 database]) (96). An average of 7,819 ± 1,430 sequences were retained across all libraries (see Table S1 in the supplemental material), and sequence counts were rarefied to the library with the minimum count (n = 4,881) using the rrarefy function of vegan (v 2.5-5) (97). A final count table with individual samples containing 119 ± 27 ASVs and 2,296 ASVs detected across all samples was generated.
A filtered alignment of representative ASV sequences against the precomputed SILVA Ref NR 132 alignment was created using the align.seqs and filter.seqs commands of the mothur software package (v 1.40.5) (98). FastTreeMP (v 2.1.7) (99) calculated a phylogenetic tree from the filtered alignment applying a generalized time-reversible model of evolution (100). The resulting tree was midpoint rooted using reroot.pl (101).
Statistical analyses.
The phyloseq package (v 1.26.1) (102) was used to import the phylogenetic tree, count table, taxonomy table, sequence FASTA of ASVs, and a matrix containing plant trait data, sampling date, plant compartment information, and treatment assignments for each sequence library into R. Single pseudocounts were added to plant trait variables containing zeros, allowing for log2 transformation. All statistical testing was performed in R, and plots were created using ggplot2 (v 3.1.1) (103) and ggpubr (v 0.2.1) (104). Summary statistics are reported as means (M) ± standard deviations unless otherwise stated.
The vegdist function of vegan was used to create a Euclidean distance matrix of samples based on log2-transformed, centered, and scaled plant morphometric data. The UniFrac function of phyloseq created weighted UniFrac distance matrices (105) from count tables and the phylogenetic tree. To test for the significance of sample clustering, the adonis2 function of vegan was used with 1,000 permutations (106). Two- and three-way tests were performed multiple times with the order of the independent variables in the formula changed to ensure the consistency of test results, regardless of term precedence. To visualize sample distance relationships, principal-coordinates analyses (PCoAs) (107) were performed using the pcoa command of ape (v 5.3) (108). In the figures, percentages on axis labels of PCoA plots report the percent variation captured by each coordinate, and axis lengths are scaled to this number. Spearman’s rank correlations (ρ) between distance matrices of plant trait and ASV count data were determined using the bioenv and mantel functions of vegan. Significant effects of treatment and/or time on response variables were assessed with Student’s t tests and analyses of covariance (ANCOVAs) using the t.test and ancova functions of stats (v 3.5.2) and HH (v 3.1-37) (109). If no significant interactions between the treatment effect and the time covariate were detected, an analysis of variance (ANOVA) was performed on a reduced model without the interaction term using the Anova function of the car package and applying type II sum-of-squares calculations (110).
Significant differences in ASV abundances between plant compartments (α ≤ 0.01) were tested using the DESeq function of DESeq2 (v 1.22.2) (111). Generalized linear mixed models (GLMMs) (112) were used to determine significantly different temporal trends in abundance for microbial taxa. A Tweedie compound Poisson distribution was chosen for this model given that it best captures the nature of amplicon sequence data sets (e.g., overdispersion, zero-inflated data sets, and continuous values) (113). The cpglmm function of the cplm R package (114) was used for time series analyses according to a general procedure outlined previously (113). Summarized sequence count tables of family-level taxonomic units were created, and full GLMMs were fit relating counts to treatment, days posttransplantation, the interaction of main effects, and random effects of each taxon. Taxa detected in >25% of samples and with cumulative sequence counts of >100 reads were tested to focus on the most abundant, prevalent, statistically robust groups in our samples. P values of modeled slope and intercept coefficients were obtained via likelihood ratio tests between the full model and two reduced models where the interaction or the treatment variable was removed. Slope and intercept coefficient P values were adjusted using the Benjamini-Hochberg method (115), and adjusted values of ≤0.05 were considered significant. The resulting intercept coefficients with positive values indicated that a taxon’s initial abundance was higher in washed than in sod transplant samples, with negative intercept coefficients implying the opposite. Modeled slopes with positive coefficients indicated that the rate of increase for a given taxon’s abundance was higher over the course of the experiment in the wash treatment than in sod samples, and vice versa for negative slope coefficients.
Data availability.
The sequence reads from all samples collected from experiments were deposited in the NCBI data bank (BioProject accession number PRJNA591021).
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge Oregon State University’s Summer Undergraduate Research Experience program, HMSC’s Mamie Markham Research Award, and the Joan Countryman Suit Scholarship for financial support.
We acknowledge the CGRB core facility for sequencing support, C. Moffett and I. Cheung at HMSC for facility and administrative support, and E. Slick for help with sample collection.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Hiltner L. 1904. Über nevere Erfahrungen und Probleme auf dem Gebiet der Boden Bakteriologie und unter besonderer Beurchsichtigung der Gründüngung und Broche. Arb Dtsch Landwirtsch Ges Berl 98:59–78. [Google Scholar]
- 2.Bandyopadhyay P, Bhuyan SK, Yadava PK, Varma A, Tuteja N. 2017. Emergence of plant and rhizospheric microbiota as stable interactomes. Protoplasma 254:617–626. doi: 10.1007/s00709-016-1003-x. [DOI] [PubMed] [Google Scholar]
- 3.Brodersen KE, Koren K, Moßhammer M, Ralph PJ, Kühl M, Santner J. 2017. Seagrass-mediated phosphorus and iron solubilization in tropical sediments. Environ Sci Technol 51:14155–14163. doi: 10.1021/acs.est.7b03878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Welsh DT. 2000. Nitrogen fixation in seagrass meadows: regulation, plant-bacteria interactions and significance to primary productivity. Ecol Lett 3:58–71. doi: 10.1046/j.1461-0248.2000.00111.x. [DOI] [Google Scholar]
- 5.Welsh DT, Wellsbury P, Bourguès S, de Wit R, Herbert RA. 1996. Relationship between porewater organic carbon content, sulphate reduction and nitrogen fixation (acetylene reduction) in the rhizosphere of Zostera noltii. Hydrobiologia 329:175–183. doi: 10.1007/BF00034556. [DOI] [Google Scholar]
- 6.Bais HP, Fall R, Vivanco JM. 2004. Biocontrol of Bacillus subtilis against infection of Arabidopsis roots by Pseudomonas syringae is facilitated by biofilm formation and surfactin production. Plant Physiol 134:307–319. doi: 10.1104/pp.103.028712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Morgan JAW, Bending GD, White PJ. 2005. Biological costs and benefits to plant-microbe interactions in the rhizosphere. J Exp Bot 56:1729–1739. doi: 10.1093/jxb/eri205. [DOI] [PubMed] [Google Scholar]
- 8.Yang J, Kloepper JW, Ryu C-M. 2009. Rhizosphere bacteria help plants tolerate abiotic stress. Trends Plant Sci 14:1–4. doi: 10.1016/j.tplants.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 9.Lebeis SL, Paredes SH, Lundberg DS, Breakfield N, Gehring J, McDonald M, Malfatti S, del Rio TG, Jones CD, Tringe SG, Dangl JL. 2015. Salicylic acid modulates colonization of the root microbiome by specific bacterial taxa. Science 349:860–864. doi: 10.1126/science.aaa8764. [DOI] [PubMed] [Google Scholar]
- 10.Bais HP, Weir TL, Perry LG, Gilroy S, Vivanco JM. 2006. The role of root exudates in rhizosphere interactions with plants and other organisms. Annu Rev Plant Biol 57:233–266. doi: 10.1146/annurev.arplant.57.032905.105159. [DOI] [PubMed] [Google Scholar]
- 11.Tkacz A, Cheema J, Chandra G, Grant A, Poole PS. 2015. Stability and succession of the rhizosphere microbiota depends upon plant type and soil composition. ISME J 9:2349–2359. doi: 10.1038/ismej.2015.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chaparro JM, Badri DV, Vivanco JM. 2014. Rhizosphere microbiome assemblage is affected by plant development. ISME J 8:790–803. doi: 10.1038/ismej.2013.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carvalhais LC, Dennis PG, Fan B, Fedoseyenko D, Kierul K, Becker A, von Wiren N, Borriss R. 2013. Linking plant nutritional status to plant-microbe interactions. PLoS One 8:e68555. doi: 10.1371/journal.pone.0068555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lareen A, Burton F, Schäfer P. 2016. Plant root-microbe communication in shaping root microbiomes. Plant Mol Biol 90:575–587. doi: 10.1007/s11103-015-0417-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cúcio C, Engelen AH, Costa R, Muyzer G. 2016. Rhizosphere microbiomes of European seagrasses are selected by the plant, but are not species specific. Front Microbiol 7:440. doi: 10.3389/fmicb.2016.00440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garcias-Bonet N, Arrieta JM, Duarte CM, Marbà N. 2016. Nitrogen-fixing bacteria in Mediterranean seagrass (Posidonia oceanica) roots. Aquat Bot 131:57–60. doi: 10.1016/j.aquabot.2016.03.002. [DOI] [Google Scholar]
- 17.Orth RJ, Carruthers TJB, Dennison WC, Duarte CM, Fourqurean JW, Heck KL, Hughes AR, Kendrick GA, Kenworthy WJ, Olyarnik S, Short FT, Waycott M, Williams SL. 2006. A global crisis for seagrass ecosystems. Bioscience 56:987–996. doi: 10.1641/0006-3568(2006)56[987:AGCFSE]2.0.CO;2. [DOI] [Google Scholar]
- 18.Crump BC, Wojahn JM, Tomas F, Mueller RS. 2018. Metatranscriptomics and amplicon sequencing reveal mutualisms in seagrass microbiomes. Front Microbiol 9:388. doi: 10.3389/fmicb.2018.00388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fahimipour AK, Kardish MR, Lang JM, Green JL, Eisen JA, Stachowicz JJ. 2017. Global-scale structure of the eelgrass microbiome. Appl Environ Microbiol 83:e03391-16. doi: 10.1128/AEM.03391-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tarquinio F, Hyndes GA, Laverock B, Koenders A, Säwström C. 2019. The seagrass holobiont: understanding seagrass-bacteria interactions and their role in seagrass ecosystem functioning. FEMS Microbiol Lett 366:fnz057. doi: 10.1093/femsle/fnz057. [DOI] [PubMed] [Google Scholar]
- 21.Lehnen N, Marchant HK, Schwedt A, Milucka J, Lott C, Weber M, Dekaezemacker J, Seah BKB, Hach PF, Mohr W, Kuypers MMM. 2016. High rates of microbial dinitrogen fixation and sulfate reduction associated with the Mediterranean seagrass Posidonia oceanica. Syst Appl Microbiol 39:476–483. doi: 10.1016/j.syapm.2016.08.004. [DOI] [PubMed] [Google Scholar]
- 22.Borum J, Pedersen O, Greve TM, Frankovich TA, Zieman JC, Fourqurean JW, Madden CJ. 2005. The potential role of plant oxygen and sulphide dynamics in die-off events of the tropical seagrass, Thalassia testudinum. J Ecol 93:148–158. doi: 10.1111/j.1365-2745.2004.00943.x. [DOI] [Google Scholar]
- 23.Holmer M, Bondgaard E. 2001. Photosynthetic and growth response of eelgrass to low oxygen and high sulfide concentrations during hypoxic events. Aquat Bot 70:29–38. doi: 10.1016/S0304-3770(00)00142-X. [DOI] [Google Scholar]
- 24.Martin BC, Bougoure J, Ryan MH, Bennett WW, Colmer TD, Joyce NK, Olsen YS, Kendrick GA. 2019. Oxygen loss from seagrass roots coincides with colonisation of sulphide-oxidising cable bacteria and reduces sulphide stress. ISME J 13:707–719. doi: 10.1038/s41396-018-0308-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Donnelly A, Herbert R. 1998. Bacterial interactions in the rhizosphere of seagrass communities in shallow coastal lagoons. J Appl Microbiol 85:151S–160S. doi: 10.1111/j.1365-2672.1998.tb05294.x. [DOI] [PubMed] [Google Scholar]
- 26.Kaldy JE, Eldridge PM, Cifuentes LA, Jones WB. 2006. Utilization of DOC from seagrass rhizomes by sediment bacteria: C-13-tracer experiments and modeling. Mar Ecol Prog Ser 317:41–55. doi: 10.3354/meps317041. [DOI] [Google Scholar]
- 27.Brodersen KE, Nielsen DA, Ralph PJ, Kuhl M. 2015. Oxic microshield and local pH enhancement protects Zostera muelleri from sediment derived hydrogen sulphide. New Phytol 205:1264–1276. doi: 10.1111/nph.13124. [DOI] [PubMed] [Google Scholar]
- 28.Milbrandt EC, Greenawalt-Boswell J, Sokoloff PD. 2008. Short-term indicators of seagrass transplant stress in response to sediment bacterial community disruption. Bot Mar 51:103–111. doi: 10.1515/BOT.2008.020. [DOI] [Google Scholar]
- 29.Cunha AH, Marbá NN, van Katwijk MM, Pickerell C, Henriques M, Bernard G, Ferreira MA, Garcia S, Garmendia JM, Manent P. 2012. Changing paradigms in seagrass restoration. Restor Ecol 20:427–430. doi: 10.1111/j.1526-100X.2012.00878.x. [DOI] [Google Scholar]
- 30.van Katwijk MM, Thorhaug A, Marbà N, Orth RJ, Duarte CM, Kendrick GA, Althuizen IHJ, Balestri E, Bernard G, Cambridge ML, Cunha A, Durance C, Giesen W, Han Q, Hosokawa S, Kiswara W, Komatsu T, Lardicci C, Lee K‐S, Meinesz A, Nakaoka M, O’Brien KR, Paling EI, Pickerell C, Ransijn AMA, Verduin JJ, Österblom H. 2016. Global analysis of seagrass restoration: the importance of large‐scale planting. J Appl Ecol 53:567–578. doi: 10.1111/1365-2664.12562. [DOI] [Google Scholar]
- 31.van Katwijk MM, Bos AR, de Jonge VN, Hanssen LSAM, Hermus DCR, de Jong DJ. 2009. Guidelines for seagrass restoration: importance of habitat selection and donor population, spreading of risks, and ecosystem engineering effects. Mar Pollut Bull 58:179–188. doi: 10.1016/j.marpolbul.2008.09.028. [DOI] [PubMed] [Google Scholar]
- 32.Matheson FE, Reed J, Santos VMD, Mackay G, Cummings VJ. 2017. Seagrass rehabilitation: successful transplants and evaluation of methods at different spatial scales. N Z J Mar Freshw Res 51:96–109. doi: 10.1080/00288330.2016.1265993. [DOI] [Google Scholar]
- 33.Cochetière MFDL, Durand T, Lepage P, Bourreille A, Galmiche JP, Doré J. 2005. Resilience of the dominant human fecal microbiota upon short-course antibiotic challenge. J Clin Microbiol 43:5588–5592. doi: 10.1128/JCM.43.11.5588-5592.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Longford SR, Campbell AH, Nielsen S, Case RJ, Kjelleberg S, Steinberg PD. 2019. Interactions within the microbiome alter microbial interactions with host chemical defences and affect disease in a marine holobiont. Sci Rep 9:1363. doi: 10.1038/s41598-018-37062-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ritchie KB. 2006. Regulation of microbial populations by coral surface mucus and mucus-associated bacteria. Mar Ecol Prog Ser 322:1–14. doi: 10.3354/meps322001. [DOI] [Google Scholar]
- 36.Ettinger CL, Voerman SE, Lang JM, Stachowicz JJ, Eisen JA. 2017. Microbial communities in sediment from Zostera marina patches, but not the Z. marina leaf or root microbiomes, vary in relation to distance from patch edge. PeerJ 5:e3246. doi: 10.7717/peerj.3246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jensen H, McGlathery K, Marino R, Howarth R. 1998. Forms and availability of sediment phosphorus in carbonate sand of Bermuda seagrass beds. Limnol Oceanogr 43:799–810. doi: 10.4319/lo.1998.43.5.0799. [DOI] [Google Scholar]
- 38.Mejia AY, Rotini A, Lacasella F, Bookman R, Thaller MC, Shem-Tov R, Winters G, Migliore L. 2016. Assessing the ecological status of seagrasses using morphology, biochemical descriptors and microbial community analyses. A study in Halophila stipulacea (Forsk.) Aschers meadows in the northern Red Sea. Ecol Indic 60:1150–1163. doi: 10.1016/j.ecolind.2015.09.014. [DOI] [Google Scholar]
- 39.Ugarelli K, Laas P, Stingl U. 2019. The microbial communities of leaves and roots associated with turtle grass (Thalassia testudinum) and manatee grass (Syringodium filliforme) are distinct from seawater and sediment communities, but are similar between species and sampling sites. Microorganisms 7:4. doi: 10.3390/microorganisms7010004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brodersen KE, Siboni N, Nielsen DA, Pernice M, Ralph PJ, Seymour J, Kühl M. 2018. Seagrass rhizosphere microenvironment alters plant-associated microbial community composition. Environ Microbiol 20:2854–2864. doi: 10.1111/1462-2920.14245. [DOI] [PubMed] [Google Scholar]
- 41.Wang L, Tomas F, Mueller RS. 2020. Nutrient enrichment increases size of Zostera marina shoots and enriches for sulfur and nitrogen cycling bacteria in root-associated microbiomes. FEMS Microbiol Ecol 96:fiaa129. doi: 10.1093/femsec/fiaa129. [DOI] [PubMed] [Google Scholar]
- 42.Lakshmanan V, Castaneda R, Rudrappa T, Bais HP. 2013. Root transcriptome analysis of Arabidopsis thaliana exposed to beneficial Bacillus subtilis FB17 rhizobacteria revealed genes for bacterial recruitment and plant defense independent of malate efflux. Planta 238:657–668. doi: 10.1007/s00425-013-1920-2. [DOI] [PubMed] [Google Scholar]
- 43.Wei Z, Yang T, Friman V-P, Xu Y, Shen Q, Jousset A. 2015. Trophic network architecture of root-associated bacterial communities determines pathogen invasion and plant health. Nat Commun 6:8413. doi: 10.1038/ncomms9413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Martin BC, Gleeson D, Statton J, Siebers AR, Grierson P, Ryan MH, Kendrick GA. 2018. Low light availability alters root exudation and reduces putative beneficial microorganisms in seagrass roots. Front Microbiol 8:2667. doi: 10.3389/fmicb.2017.02667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Säwström C, Serrano O, Rozaimi M, Lavery PS. 2016. Utilization of carbon substrates by heterotrophic bacteria through vertical sediment profiles in coastal and estuarine seagrass meadows. Environ Microbiol Rep 8:582–589. doi: 10.1111/1758-2229.12406. [DOI] [PubMed] [Google Scholar]
- 46.Maignien L, DeForce EA, Chafee ME, Eren AM, Simmons SL. 2014. Ecological succession and stochastic variation in the assembly of Arabidopsis thaliana phyllosphere communities. mBio 5:e00682-13. doi: 10.1128/mBio.00682-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Edwards J, Johnson C, Santos-Medellín C, Lurie E, Podishetty NK, Bhatnagar S, Eisen JA, Sundaresan V. 2015. Structure, variation, and assembly of the root-associated microbiomes of rice. Proc Natl Acad Sci U S A 112:E911–E920. doi: 10.1073/pnas.1414592112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Papazian S, Parrot D, Buryskova B, Weinberger F, Tasdemir D. 2019. Surface chemical defence of the eelgrass Zostera marina against microbial foulers. Sci Rep 9:3323. doi: 10.1038/s41598-019-39212-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Garcia R, La Clair JJ, Müller R. 2018. Future directions of marine myxobacterial natural product discovery inferred from metagenomics. Mar Drugs 16:303. doi: 10.3390/md16090303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mohr KI, Garcia RO, Gerth K, Irschik H, Müller R. 2012. Sandaracinus amylolyticus gen. nov., sp. nov., a starch-degrading soil myxobacterium, and description of Sandaracinaceae fam. nov. Int J Syst Evol Microbiol 62:1191–1198. doi: 10.1099/ijs.0.033696-0. [DOI] [PubMed] [Google Scholar]
- 51.Coskun ÖK, Pichler M, Vargas S, Gilder S, Orsi WD. 2018. Linking uncultivated microbial populations and benthic carbon turnover by using quantitative stable isotope probing. Appl Environ Microbiol 84:e01083-18. doi: 10.1128/AEM.01083-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang F-Q, Chen Z-J, Yang J-M, Wang W-J, Feng Y-W, Li Z, Sun G-H. 2020. Labilibacter sediminis sp. nov., isolated from marine sediment. Int J Syst Evol Microbiol 70:321–326. doi: 10.1099/ijsem.0.003758. [DOI] [PubMed] [Google Scholar]
- 53.McIlroy S, Nielsen P. 2014. The family Saprospiraceae, p 863–889. In Rosenberg E, DeLong EF, Lory S, Stackebrandt E, Thompson F (ed), The prokaryotes: other major lineages of bacteria and the archaea. Springer-Verlag, Berlin, Germany. [Google Scholar]
- 54.McBride MJ. 2014. The family Flavobacteriaceae, p 643–676. In Rosenberg E, DeLong EF, Lory S, Stackebrandt E, Thompson F (ed), The prokaryotes: other major lineages of bacteria and the archaea. Springer-Verlag, Berlin, Germany. [Google Scholar]
- 55.Kumar M, Kour D, Yadav AN, Saxena R, Rai PK, Jyoti A, Tomar RS. 2019. Biodiversity of methylotrophic microbial communities and their potential role in mitigation of abiotic stresses in plants. Biologia (Bratisl) 74:287–308. doi: 10.2478/s11756-019-00190-6. [DOI] [Google Scholar]
- 56.Trotsenko YA, Ivanova EG, Doronina NV. 2001. Aerobic methylotrophic bacteria as phytosymbionts. Microbiology 70:623–632. doi: 10.1023/A:1013167612105. [DOI] [Google Scholar]
- 57.Boutard M, Cerisy T, Nogue P-Y, Alberti A, Weissenbach J, Salanoubat M, Tolonen AC. 2014. Functional diversity of carbohydrate-active enzymes enabling a bacterium to ferment plant biomass. PLoS Genet 10:e1004773. doi: 10.1371/journal.pgen.1004773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Woo HL, Hazen TC. 2018. Enrichment of bacteria from eastern Mediterranean Sea involved in lignin degradation via the phenylacetyl-CoA pathway. Front Microbiol 9:922. doi: 10.3389/fmicb.2018.00922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Igai K, Itakura M, Nishijima S, Tsurumaru H, Suda W, Tsutaya T, Tomitsuka E, Tadokoro K, Baba J, Odani S, Natsuhara K, Morita A, Yoneda M, Greenhill AR, Horwood PF, Inoue J-I, Ohkuma M, Hongoh Y, Yamamoto T, Siba PM, Hattori M, Minamisawa K, Umezaki M. 2016. Nitrogen fixation and nifH diversity in human gut microbiota. Sci Rep 6:31942. doi: 10.1038/srep31942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cúcio C, Overmars L, Engelen AH, Muyzer G. 2018. Metagenomic analysis shows the presence of bacteria related to free-living forms of sulfur-oxidizing chemolithoautotrophic symbionts in the rhizosphere of the seagrass Zostera marina. Front Mar Sci 5:171. doi: 10.3389/fmars.2018.00171. [DOI] [Google Scholar]
- 61.Brown CA, Ozretich RJ. 2009. Coupling between the coastal ocean and Yaquina Bay, Oregon: importance of oceanic inputs relative to other nitrogen sources. Estuaries Coasts 32:219–237. doi: 10.1007/s12237-008-9128-6. [DOI] [Google Scholar]
- 62.Morse JW, Dimarco SF, Sell KS, Hebert AB. 2003. Determination of the optimum sampling intervals in sediment pore waters using the autocovariance function. Aquat Geochem 9:41–57. doi: 10.1023/B:AQUA.0000005657.18821.46. [DOI] [Google Scholar]
- 63.Hansen J, Udy J, Perry C, Dennison W, Lomstein B. 2000. Effect of the seagrass Zostera capricorni on sediment microbial processes. Mar Ecol Prog Ser 199:83–96. doi: 10.3354/meps199083. [DOI] [Google Scholar]
- 64.Deborde J, Abril G, Mouret A, Jézéquel D, Thouzeau G, Clavier J, Bachelet G, Anschutz P. 2008. Effects of seasonal dynamics in a Zostera noltii meadow on phosphorus and iron cycles in a tidal mudflat (Arcachon Bay, France). Mar Ecol Prog Ser 355:59–71. doi: 10.3354/meps07254. [DOI] [Google Scholar]
- 65.Chisholm J, Moulin P. 2003. Stimulation of nitrogen fixation in refractory organic sediments by Caulerpa taxifolia (Chlorophyta). Limnol Oceanogr 48:787–794. doi: 10.4319/lo.2003.48.2.0787. [DOI] [Google Scholar]
- 66.Cole LW, McGlathery KJ. 2012. Nitrogen fixation in restored eelgrass meadows. Mar Ecol Prog Ser 448:235–246. doi: 10.3354/meps09512. [DOI] [Google Scholar]
- 67.Isaksen MF, Teske A. 1996. Desulforhopalus vacuolatus gen. nov., sp. nov., a new moderately psychrophilic sulfate-reducing bacterium with gas vacuoles isolated from a temperate estuary. Arch Microbiol 166:160–168. doi: 10.1007/s002030050371. [DOI] [Google Scholar]
- 68.Kessler AJ, Wawryk M, Marzocchi U, Roberts KL, Wong WW, Risgaard‐Petersen N, Meysman FJR, Glud RN, Cook PLM. 2019. Cable bacteria promote DNRA through iron sulfide dissolution. Limnol Oceanogr 64:1228–1238. doi: 10.1002/lno.11110. [DOI] [Google Scholar]
- 69.Pfeffer C, Larsen S, Song J, Dong M, Besenbacher F, Meyer RL, Kjeldsen KU, Schreiber L, Gorby YA, El-Naggar MY, Leung KM, Schramm A, Risgaard-Petersen N, Nielsen LP. 2012. Filamentous bacteria transport electrons over centimetre distances. Nature 491:218–221. doi: 10.1038/nature11586. [DOI] [PubMed] [Google Scholar]
- 70.Scholz VV, Müller H, Koren K, Nielsen LP, Meckenstock RU. 2019. The rhizosphere of aquatic plants is a habitat for cable bacteria. FEMS Microbiol Ecol 95:fiz062. doi: 10.1093/femsec/fiz062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bourque AS, Vega-Thurber R, Fourqurean JW. 2015. Microbial community structure and dynamics in restored subtropical seagrass sediments. Aquat Microb Ecol 74:43–57. doi: 10.3354/ame01725. [DOI] [Google Scholar]
- 72.Waite DW, Vanwonterghem I, Rinke C, Parks DH, Zhang Y, Takai K, Sievert SM, Simon J, Campbell BJ, Hanson TE, Woyke T, Klotz MG, Hugenholtz P. 2017. Comparative genomic analysis of the class Epsilonproteobacteria and proposed reclassification to Epsilonbacteraeota (phyl. nov.). Front Microbiol 8:682. doi: 10.3389/fmicb.2017.00682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jensen SI, Kühl M, Priemé A. 2007. Different bacterial communities associated with the roots and bulk sediment of the seagrass Zostera marina. FEMS Microbiol Ecol 62:108–117. doi: 10.1111/j.1574-6941.2007.00373.x. [DOI] [PubMed] [Google Scholar]
- 74.Probandt D, Knittel K, Tegetmeyer HE, Ahmerkamp S, Holtappels M, Amann R. 2017. Permeability shapes bacterial communities in sublittoral surface sediments. Environ Microbiol 19:1584–1599. doi: 10.1111/1462-2920.13676. [DOI] [PubMed] [Google Scholar]
- 75.Rodriguez-Mora MJ, Edgcomb VP, Taylor C, Scranton MI, Taylor GT, Chistoserdov AY. 2016. The diversity of sulfide oxidation and sulfate reduction genes expressed by the bacterial communities of the Cariaco Basin, Venezuela. Open Microbiol J 10:140–149. doi: 10.2174/1874285801610010140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lilburn TG, Gu J, Cai H, Wang Y. 2010. Comparative genomics of the family Vibrionaceae reveals the wide distribution of genes encoding virulence-associated proteins. BMC Genomics 11:369. doi: 10.1186/1471-2164-11-369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mueller RS, McDougald D, Cusumano D, Sodhi N, Kjelleberg S, Azam F, Bartlett DH. 2007. Vibrio cholerae strains possess multiple strategies for abiotic and biotic surface colonization. J Bacteriol 189:5348–5360. doi: 10.1128/JB.01867-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Suykerbuyk W, Govers LL, Bouma TJ, Giesen WBJT, de Jong DJ, van de Voort R, Giesen K, Giesen PT, van Katwijk MM. 2016. Unpredictability in seagrass restoration: analysing the role of positive feedback and environmental stress on Zostera noltii transplants. J Appl Ecol 53:774–784. doi: 10.1111/1365-2664.12614. [DOI] [Google Scholar]
- 79.Thom R, Gaeckle J, Buenau K, Borde A, Vavrinec J, Aston L, Woodruff D, Khangaonkar T, Kaldy J. 2018. Eelgrass (Zostera marina L.) restoration in Puget Sound: development of a site suitability assessment process. Restor Ecol 26:1066–1074. doi: 10.1111/rec.12702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zenone A, Alagna A, D’Anna G, Kovalev A, Kreitschitz A, Badalamenti F, Gorb SN. 2020. Biological adhesion in seagrasses: the role of substrate roughness in Posidonia oceanica (L.) Delile seedling anchorage via adhesive root hairs. Mar Environ Res 160:105012. doi: 10.1016/j.marenvres.2020.105012. [DOI] [PubMed] [Google Scholar]
- 81.Garcia-Martinez M, Kuo J, Kilminster K, Walker D, Rossello-Mora R, Duarte C. 2005. Microbial colonization in the seagrass Posidonia spp. roots. Mar Biol Res 1:388–395. doi: 10.1080/17451000500443419. [DOI] [Google Scholar]
- 82.Hemminga M. 1998. The root/rhizome system of seagrasses: an asset and a burden. J Sea Res 39:183–196. doi: 10.1016/S1385-1101(98)00004-5. [DOI] [Google Scholar]
- 83.Novak AB, Plaisted HK, Hays CG, Hughes RA. 2017. Limited effects of source population identity and number on seagrass transplant performance. PeerJ 5:e2972. doi: 10.7717/peerj.2972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Davis RC, Short FT. 1997. Restoring eelgrass, Zostera marina L., habitat using a new transplanting technique: the horizontal rhizome method. Aquat Bot 59:1–15. doi: 10.1016/S0304-3770(97)00034-X. [DOI] [Google Scholar]
- 85.Zhou Y, Liu P, Liu B, Liu X, Zhang X, Wang F, Yang H. 2014. Restoring eelgrass (Zostera marina L.) habitats using a simple and effective transplanting technique. PLoS One 9:e92982. doi: 10.1371/journal.pone.0092982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lundberg DS, Lebeis SL, Paredes SH, Yourstone S, Gehring J, Malfatti S, Tremblay J, Engelbrektson A, Kunin V, del Rio TG, Edgar RC, Eickhorst T, Ley RE, Hugenholtz P, Tringe SG, Dangl JL. 2012. Defining the core Arabidopsis thaliana root microbiome. Nature 488:86–90. doi: 10.1038/nature11237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Costa R, Götz M, Mrotzek N, Lottman J, Berg G, Smalla K. 2006. Effects of site and plant species on rhizosphere community structure as revealed by molecular analysis of microbial guilds. FEMS Microbiol Ecol 56:236–249. doi: 10.1111/j.1574-6941.2005.00026.x. [DOI] [PubMed] [Google Scholar]
- 88.Shieh WY, Yang JT. 1997. Denitrification in the rhizosphere of the two seagrasses Thalassia hemprichii (Herenb.) Aschers and Halodule uninervis (Forsk.) Aschers. J Exp Mar Biol Ecol 218:229–241. doi: 10.1016/S0022-0981(97)00076-2. [DOI] [Google Scholar]
- 89.Kaldy JE. 2012. Influence of light, temperature and salinity on dissolved organic carbon exudation rates in Zostera marina L. Aquat Biosyst 8:19. doi: 10.1186/2046-9063-8-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tomas F, Abbott JM, Steinberg C, Balk M, Williams SL, Stachowicz JJ. 2011. Plant genotype and nitrogen loading influence seagrass productivity, biochemistry, and plant-herbivore interactions. Ecology 92:1807–1817. doi: 10.1890/10-2095.1. [DOI] [PubMed] [Google Scholar]
- 91.Crump BC, Kling GW, Bahr M, Hobbie JE. 2003. Bacterioplankton community shifts in an Arctic lake correlate with seasonal changes in organic matter source. Appl Environ Microbiol 69:2253–2268. doi: 10.1128/aem.69.4.2253-2268.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kieft B, Li Z, Bryson S, Crump BC, Hettich R, Pan C, Mayali X, Mueller RS. 2018. Microbial community structure-function relationships in Yaquina Bay estuary reveal spatially distinct carbon and nitrogen cycling capacities. Front Microbiol 9:1282. doi: 10.3389/fmicb.2018.01282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP. 2016. DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods 13:581–583. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Morgan M. 2018. BiocManager: access the Bioconductor Project Package Repository. https://rdrr.io/cran/BiocManager/.
- 95.R Core Team. 2018. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- 96.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glöckner FO. 2013. The SILVA ribosomal RNA gene database project: improved data processing and Web-based tools. Nucleic Acids Res 41:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Oksanen J, Blanchet FG, Friendly M, Kindt R, Legendre P, McGlinn D, Minchin PR, O’Hara RB, Simpson GL, Solymos P, Stevens MHH, Szoecs E, Wagner H. 2019. vegan: community ecology package. https://cran.r-project.org/web/packages/vegan/index.html.
- 98.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Horn DJV, Weber CF. 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Price MN, Dehal PS, Arkin AP. 2010. FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS One 5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Tavaré S. 1986. Some probabilistic and statistical problems in the analysis of DNA sequences. Lect Math Life Sci 17:57–86. [Google Scholar]
- 101.Junier T, Zdobnov EM. 2010. The Newick utilities: high-throughput phylogenetic tree processing in the Unix shell. Bioinformatics 26:1669–1670. doi: 10.1093/bioinformatics/btq243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.McMurdie PJ, Holmes S. 2013. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 8:e61217. doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wickham H. 2016. ggplot2: elegant graphics for data analysis. Springer-Verlag, New York, NY. [Google Scholar]
- 104.Kassambara A. 2019. ggpubr: “ggplot2” based publication ready plots. https://rdrr.io/cran/ggpubr/.
- 105.Lozupone CA, Stombaugh JI, Gordon JI, Jansson JK, Knight R. 2012. Diversity, stability and resilience of the human gut microbiota. Nature 489:220–230. doi: 10.1038/nature11550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Anderson MJ. 2001. A new method for non-parametric multivariate analysis of variance. Austral Ecol 26:32–46. doi: 10.1046/j.1442-9993.2001.01070.x. [DOI] [Google Scholar]
- 107.Borg I, Groenen PJF. 2005. Modern multidimensional scaling: theory and applications, 2nd ed Springer-Verlag, New York, NY. [Google Scholar]
- 108.Paradis E, Schliep K. 2019. ape 5.0: an environment for modern phylogenetics and evolutionary analyses in R. Bioinformatics 35:526–528. doi: 10.1093/bioinformatics/bty633. [DOI] [PubMed] [Google Scholar]
- 109.Heiberger RM. 2019. HH: statistical analysis and data display: Heiberger and Holland. https://rdrr.io/cran/HH/.
- 110.Fox J, Weisberg S. 2019. An R companion to applied regression, 3rd ed Sage, Thousand Oaks, CA. [Google Scholar]
- 111.Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Bolker BM, Brooks ME, Clark CJ, Geange SW, Poulsen JR, Stevens MHH, White J-SS. 2009. Generalized linear mixed models: a practical guide for ecology and evolution. Trends Ecol Evol 24:127–135. doi: 10.1016/j.tree.2008.10.008. [DOI] [PubMed] [Google Scholar]
- 113.Sharpton T, Lyalina S, Luong J, Pham J, Deal EM, Armour C, Gaulke C, Sanjabi S, Pollard KS. 2017. Development of inflammatory bowel disease is linked to a longitudinal restructuring of the gut metagenome in mice. mSystems 2:e00036-17. doi: 10.1128/mSystems.00036-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Zhang Y. 2013. Likelihood-based and Bayesian methods for Tweedie compound Poisson linear mixed models. Stat Comput 23:743–757. doi: 10.1007/s11222-012-9343-7. [DOI] [Google Scholar]
- 115.Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol 57:289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequence reads from all samples collected from experiments were deposited in the NCBI data bank (BioProject accession number PRJNA591021).