Xylan is the major hemicellulosic polysaccharide in cereals and contributes to the recalcitrance of the plant cell wall toward degradation. Bacteroidetes, one of the main phyla in rumen and human gut microbiota, have been shown to encode polysaccharide utilization loci dedicated to the degradation of xylan. Here, we present the biochemical characterization of a xylanase encoded by a bacteroidetes strain isolated from the termite gut metagenome.
KEYWORDS: termite gut, lignocellulose, glycoside hydrolase, carbohydrate-binding module, xylanase, PUL, GH10, CBM4, protein domain insertion, functional genomics
ABSTRACT
The functional screening of a Pseudacanthotermes militaris termite gut metagenomic library revealed an array of xylan-degrading enzymes, including P. militaris 25 (Pm25), a multimodular glycoside hydrolase family 10 (GH10). Sequence analysis showed details of the unusual domain organization of this enzyme. It consists of one catalytic domain, which is intercalated by two carbohydrate binding modules (CBMs) from family 4. The genes upstream of the genes encoding Pm25 are susC-susD-unk, suggesting Pm25 is a Xyn10C-like enzyme belonging to a polysaccharide utilization locus. The majority of Xyn10C-like enzymes shared the same interrupted domain architecture and were vastly distributed in different xylan utilization loci found in gut Bacteroidetes, indicating the importance of this enzyme in glycan acquisition for gut microbiota. To understand its unusual multimodularity and the possible role of the CBMs, a detailed characterization of the full-length Pm25 and truncated variants was performed. Results revealed that the GH10 catalytic module is specific toward the hydrolysis of xylan. Ligand binding results indicate that the GH10 module and the CBMs act independently, whereas the tandem CBM4s act synergistically with each other and improve enzymatic activity when assayed on insoluble polysaccharides. In addition, we show that the UNK protein upstream of Pm25 is able to bind arabinoxylan. Altogether, these findings contribute to a better understanding of the potential role of Xyn10C-like proteins in xylan utilization systems of gut bacteria.
IMPORTANCE Xylan is the major hemicellulosic polysaccharide in cereals and contributes to the recalcitrance of the plant cell wall toward degradation. Members of the Bacteroidetes, one of the main phyla in rumen and human gut microbiota, have been shown to encode polysaccharide utilization loci dedicated to the degradation of xylan. Here, we present the biochemical characterization of a xylanase encoded by a Bacteroidetes strain isolated from the termite gut metagenome. This xylanase is a multimodular enzyme, the sequence of which is interrupted by the insertion of two CBMs from family 4. Our results show that this enzyme resembles homologues that were shown to be important for xylan degradation in rumen or human diet and show that the CBM insertion in the middle of the sequence seems to be a common feature in xylan utilization systems. This study shed light on our understanding of xylan degradation and plant cell wall deconstruction, which can be applied to several applications in food, feed, and bioeconomy.
INTRODUCTION
Xylan is the most abundant hemicellulose present in cell walls of higher plants, especially cereal grains and hardwoods (1). The xylan main chain is composed of β-1,4-linked d-xylopyranosyl (d-Xylp) residues that can bear substitutions at O-2 and/or O-3 positions. l-arabinofuranosyl (l-Araf), 4-O-methyl glucuronyl (d-MeGlcAp), and acetyl residues are frequent main-chain substituents, and l-Araf moieties can be esterified by ferulate at their O-5 position. The nature of xylan backbone decorations varies depending on the species, the tissue, and the stage of development of the plant (2). Generally, graminaceous plants are rich in glucuronoarabinoxylan (GAX), while glucuronoxylan (GX) is found in dicots, the difference between these two categories being the relative amounts of l-Araf and d-MeGlcAp present. Complete xylan degradation requires an extensive arsenal of enzymes that can act synergistically (3). The main chain is depolymerized by β-d-xylanases (EC 3.2.1.8) that hydrolyze internal β-1,4 bonds, while decorations are removed by a variety of accessory enzymes, including α-l-arabinofuranosidases (EC 3.2.1.55), α-d-glucuronidases (EC 3.2.1.139), feruloyl esterases (EC 3.1.1.73), and acetyl xylan esterases (EC 3.1.1.72). Finally, β-d-xylosidases (EC 3.2.1.37) break down xylooligosaccharides, removing d-Xylp from the nonreducing end (4).
Xylanases are mainly found in the glycoside hydrolase (GH) families 5, 8, 10, 11, 30, and 43 in the CAZy database (www.cazy.org) (5). The GH10 family constitutes a monospecific family that includes only endo-xylanases. Enzymes from this family perform catalysis via a retaining mechanism (6), and their canonical three-dimensional (3D) structure is a TIM barrel, (β/α)8, which is the most commonly known (2,077 occurrences) protein fold in the Protein Data Bank (PDB) and which forms an active cleft able to accommodate up to seven xylosyl backbone units (7). In addition, according to the Pfam database (http://pfam.xfam.org/), 20 to 30% of β-d-xylanases are multidomain proteins, comprising catalytic domains associated with accessory or helper domains, such as carbohydrate binding modules (CBMs). The latter have been attributed various roles, including the ability to target specific regions in substrates (8), disrupt polysaccharide structure (9), or anchor enzymes to bacterial surfaces (10). In multidomain proteins, individual domains are defined as the structural, functional, or evolutionary units of proteins (11) and can be regarded as biological equivalents of components in complex devices whose parts can be interchanged. Mostly, domains in proteins are sequentially organized, with one domain following another one. However, around 10% to 20% of domain combinations are discontinuous, with one domain being inserted into another one (12).
Termites are wood-feeding animals that are considered an abundant source of biomass-degrading enzymes (13). Termites produce very few endogenous lignocellulose-degrading enzymes, and their gut microbiome is mainly responsible for their ability to capture nutrients and energy from plant biomass (14, 15). Over the last decade, numerous metagenomics studies revealed enzyme arsenals of termite gut microbiomes and detected promising enzymes for industrial use (16–20). Notably, Gram-negative Bacteroidetes, the dominant phylum in many animal digestive systems (21–25), utilize finely tuned glycan utilization systems. The paradigm for this type of system was provided by the well-studied starch utilization system (Sus) (26). In Sus-like systems, several proteins are encoded by genes found in a cluster (known as polysaccharide utilization loci, or PUL) and act in a coordinated manner to bind and hydrolyze complex sugars and utilize them for their metabolism (27). A xylan utilization system (Xus) that is composed of two outer membrane polysaccharide-binding proteins (XusB and XusD), two transporter proteins (XusA and XusC), and two outer membrane proteins (XusE and Xyn10C) was previously described in rumen and human digestive systems (28). Each of these proteins is expressed from a cluster of tandem genes that are organized as xusA-xusB-xusC-xusD (or sometimes only xusC-xusD), followed by xusE and xyn10C, the latter encoding a CBM-containing GH10 β-d-xylanase (28).
According to previous data, the CBMs in Xyn10C are inserted into the polypeptide sequence of the GH10 catalytic domain between structural elements β3 and α3 of the TIM barrel (29). The expression of Xyn10C was shown to be induced by xylan (30) along with the other xylanases, XynA and XynB. The most effective inducer is demonstrated to be a xylooligosaccharide with a degree of polymerization (DP) around 35, similar to the hydrolysates of Xyn10C (31). Altogether, this is consistent with the hypothesis that Xyn10C serves as a functional homologue of the Bacteroides thetaiotaomicron VPI-5482 SusG protein, initiating xylan metabolism through extracellular hydrolysis of polymeric substrates (28). In this regard, it has been proposed that Xyn10C is used as a functional marker of xylan degradation in the human gut (28, 30, 32). The potential roles and distributions of Xyn10C have recently attracted considerable attention but have not yet been fully described (29, 30, 32, 33).
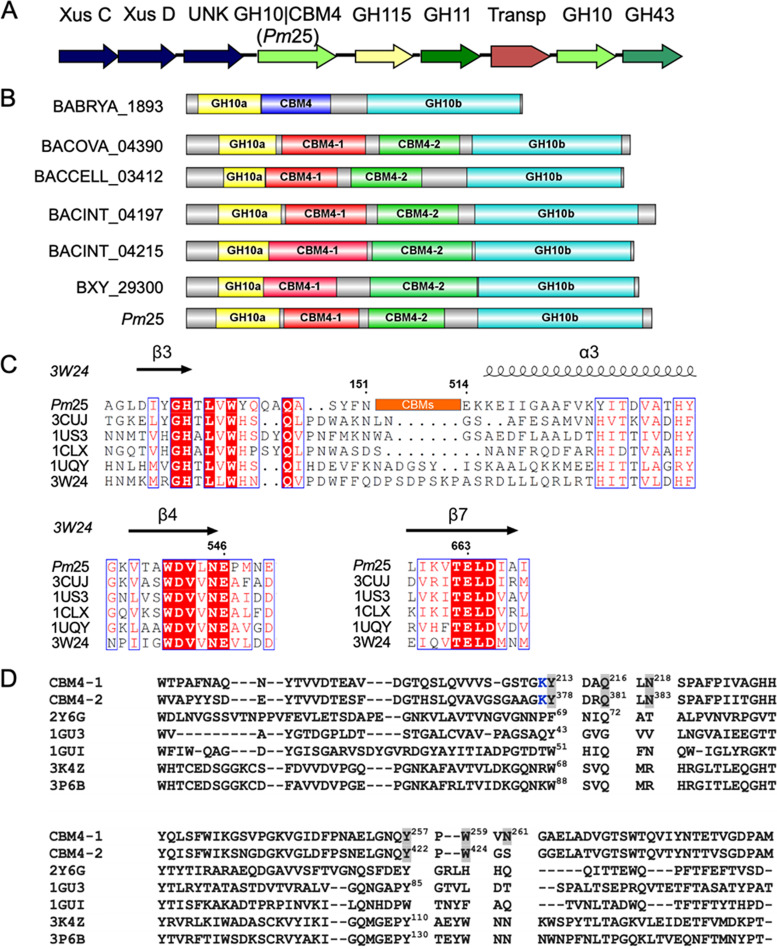
Previously, a putative xus locus assigned to the genus Bacteroides, Gram-negative anaerobic bacteria, was identified in a metagenomic library from the microbiome of a fungus-growing termite, Pseudacanthotermes militaris (17). This xus is composed of eight different open reading frames (ORFs) encoding putative XusC/D-like proteins, unknown protein (UNK), GH10 containing an insertion of two CBM4s (GH10|CBM4), GH115, GH11, a putative transporter protein, GH10, and GH43 (Fig. 1A). The GH10|CBM4 protein, designated P. militaris 25 (Pm25) here, presents an insertional modular structure homologous to Xyn10C protein (Fig. 1B). Here, we describe the characterization of Pm25 and discuss its activity with respect to its unusual multidomain organization. In addition, the potential function of the UNK was also investigated.
FIG 1.
Modular architecture of Xyn10C-like protein. (A) Schematic representation of the putative xylan-active gene cluster. (B) Insertional architecture of Xyn10C-like proteins; the length of the amino acid is proportional to the length of the colored box. Shown are BABRYA_1893 (29), BACOVA_04390 (71), BACCELL_03412 (72), BACINT_04197 and BACINT_04215 (33), BXY_29300 (30), and Pm25 (17). (C) Amino acid alignment of Pm25 with other native GH10 xylanases, designated by their PDB codes: 3CUJ, Cellulomonas fimi xylanase/cellulase Cex (CfXyn10A); 1US3, Cellvibrio japonicus xylanase 10C; 1CLX, Pseudomonas fluorescens xylanase A; 1UQY, Cellvibrio mixtus xylanase B; 3W24, Thermoanaerobacterium saccharolyticum xylanase A. The β strands and α helices are shown above the aligned sequences. Numbering corresponds to the sequences of Pm25. Residues in red background are conserved, and those within blue frames are conservatively substituted. (D) Amino acid alignment of CBM4s in Pm25 with well-characterized CBM4s in the literature. CBM4-1 is the first CBM4 in Pm25; CBM4-2 is the second CBM4 in Pm25. PDB 2Y6G, CBM4 from Rhodothermus marinus xylanase; 1GU3, CBM4 from Cellulomonas fimi endoglucanase C; 1GUI, CBM4 from Thermotoga maritima laminarinase 16A; 3K4Z, CBM4 from Clostridium thermocellum cellulase CbhA; 3P6B, CBM4 from Clostridium thermocellum cellulase CelK. Residues highlighted in gray were used for mutagenesis. Numbered residues in PDB sequences indicate that they are responsible for ligand binding according to the literature. The K in blue is potentially labeled with N-hydroxysuccinimide dye, which was described in Materials and Methods for the MST experiment.
(This research was conducted by H. Wu in partial fulfillment of the requirements for a Ph.D. degree from Toulouse University [34].)
RESULTS
Bioinformatics analysis of the Pm25-encoding gene sequence.
Analysis of the primary amino acid sequence of Pm25 revealed a multimodular architecture composed of a signal peptide (residues 1 to 32), a GH10 catalytic domain (amino acid residues 66 to 151 and 514 to 753), and two putative tandem CBM4s (CBM4-1, residues 161 to 321, and CBM4-2, residues 324 to 486) that constitute insertion domains (Fig. 1B). Alignment of the amino acid sequence of Pm25 with those of other GH10 family members for which structural data are available revealed that CBM4-1 and CBM4-2 are inserted between strand β3 and helix α3 of the (β/α)8 structure (Fig. 1C). Moreover, this alignment allowed the identification of E546 and E663 as the putative catalytic acid/base and nucleophile, respectively.
Alignment of the amino acid sequences of CBM4-1/CBM4-2 with the well-characterized tandem CBMs, PDB entry 2Y6G from Rhodothermus marinus Xyn10A, 1GU3 from Cellulomonas fimi Cel9B, 1GUI from Thermotoga maritima Lam16, 3K4Z from Clostridium thermocellum Cbh9A, and 3P6B from Clostridium thermocellum Cel9K, revealed modest similarity (16/19,15/15, 16/19, 20/21, and 18/21% similarity, respectively). Moreover, using this similarity, we could assign possible ligand binding roles to residues Y213, Q216, and Y257 in CBM4-1 and Y378, Q381, and Y422 in CBM4-2 (35–40) (Fig. 1D). In addition, based on homolog modeling (unpublished data), W259 and W424 in CBM4-1 and CBM4-2, respectively, could be aligned to W102, located in the hydrophobic cleft of the Thermotoga maritima Lam16 CBM4, whose importance in binding was demonstrated by Boraston and colleagues (35).
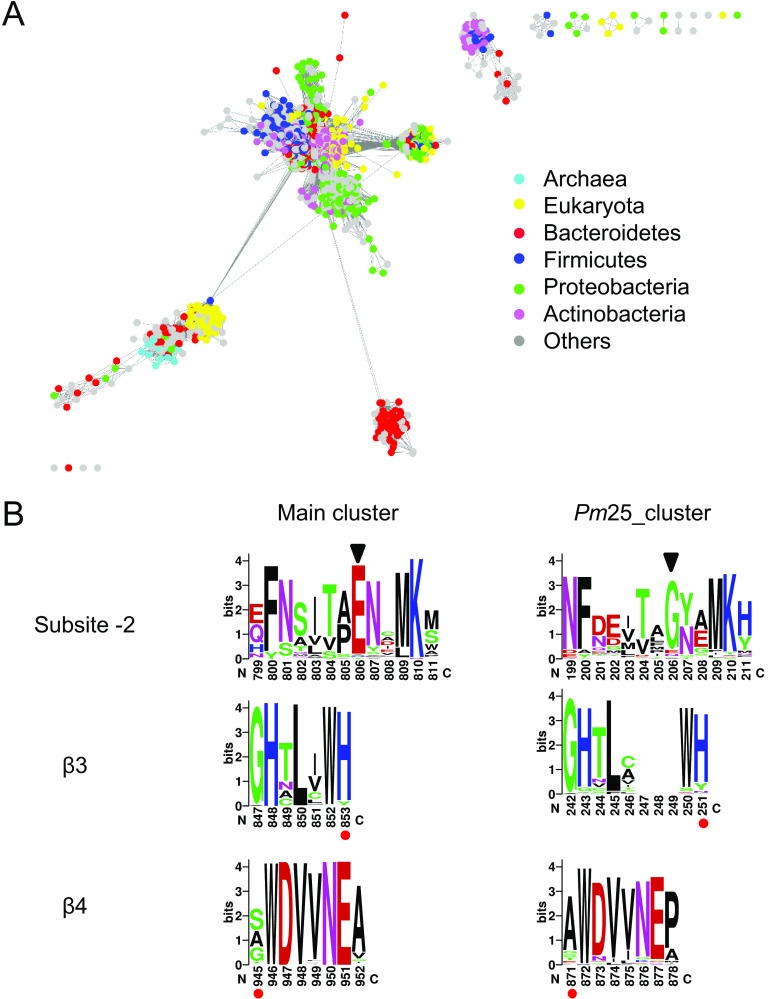
SSN analysis.
To identify Pm25 homologs and study their distribution, an SSN was created for all GH10 sequences (2,539 nonredundant sequences) present in the CAZy database (Fig. 2A). Archaea, Eukaryota, and Bacteria contribute 1, 16, and 81%, respectively, of GH10s, the remaining 3% being unassigned sequences. Bacterial GH10s belong mainly to four bacterial phyla, Actinobacteria (25%), Bacteroidetes (14%), Proteobacteria (14%), and Firmicutes (13%).
FIG 2.
Sequence analysis of GH10 xylanase. (A) SSN of GH10 xylanase in CAZy database. The nodes are colored by the taxonomic annotation listed on the right. (B) Sequence logos of different motifs located at subsite −2, β3, and β4 of GH10 sequences. The length of amino acid sequence between β3 and β4 can be deduced from the sequence number with red dots underneath.
The SSN is centered on the main cluster, with other minor clusters scattered around. Pm25 is located in the bottom right cluster (Pm25_cluster), and all classified sequences (61 nodes) thereof are from Bacteroidetes.
To investigate the sequence differences between the Pm25_cluster and main cluster, sequences in each cluster were aligned before the sequence logo was constructed (Fig. 2B). The −2 subsite motif in Pm25_cluster is glycine instead of glutamate in the main cluster, and both glycine and glutamate at the −2 subsite were found in other enzymes (41–43). Importantly, the distance between the β3 motif and β4 motif is wider in Pm25_cluster (620 amino acids) than in the main cluster (92 amino acids), which indicated interrupted catalytic domains in Pm25_cluster.
To verify the relationship of Pm25_cluster with PUL, each sequence was searched against PULDB (http://www.cazy.org/PULDB/) (44). The majority of sequences (45 out of 61) were found in PUL. Among these, 44 out of 45 are always located downstream of hypothetical susC-susD-unk (see Table S2 in the supplemental material), suggesting that sequences in Pm25_cluster are Xyn10C-like proteins from the xylan utilization system in gut Bacteroidetes. The remaining sequences (16 out of 61) were not found in PUL, perhaps because of poor assignment.
Enzyme optimal pH and temperature.
The optimal pH of Pm25 was determined using beechwood GX as the substrate. The purified enzyme retained greater than 80% of its activity in the pH range from 4.5 to 9.0 (Fig. 3A). The activity was also measured at temperatures from 50 to 75°C, with maximum activity being observed at 60°C (Fig. 3B). However, since Pm25 was rather unstable at 60°C, thermostability was measured at both pH 7.5 and pH 9 (Fig. 3C and D, respectively). Pm25 remained fully active for over 24 h at either 50°C (pH 7.5) or 45°C (pH 9). Overall, optimal conditions for routine Pm25 assays were defined as pH 7.5 and 50°C.
FIG 3.
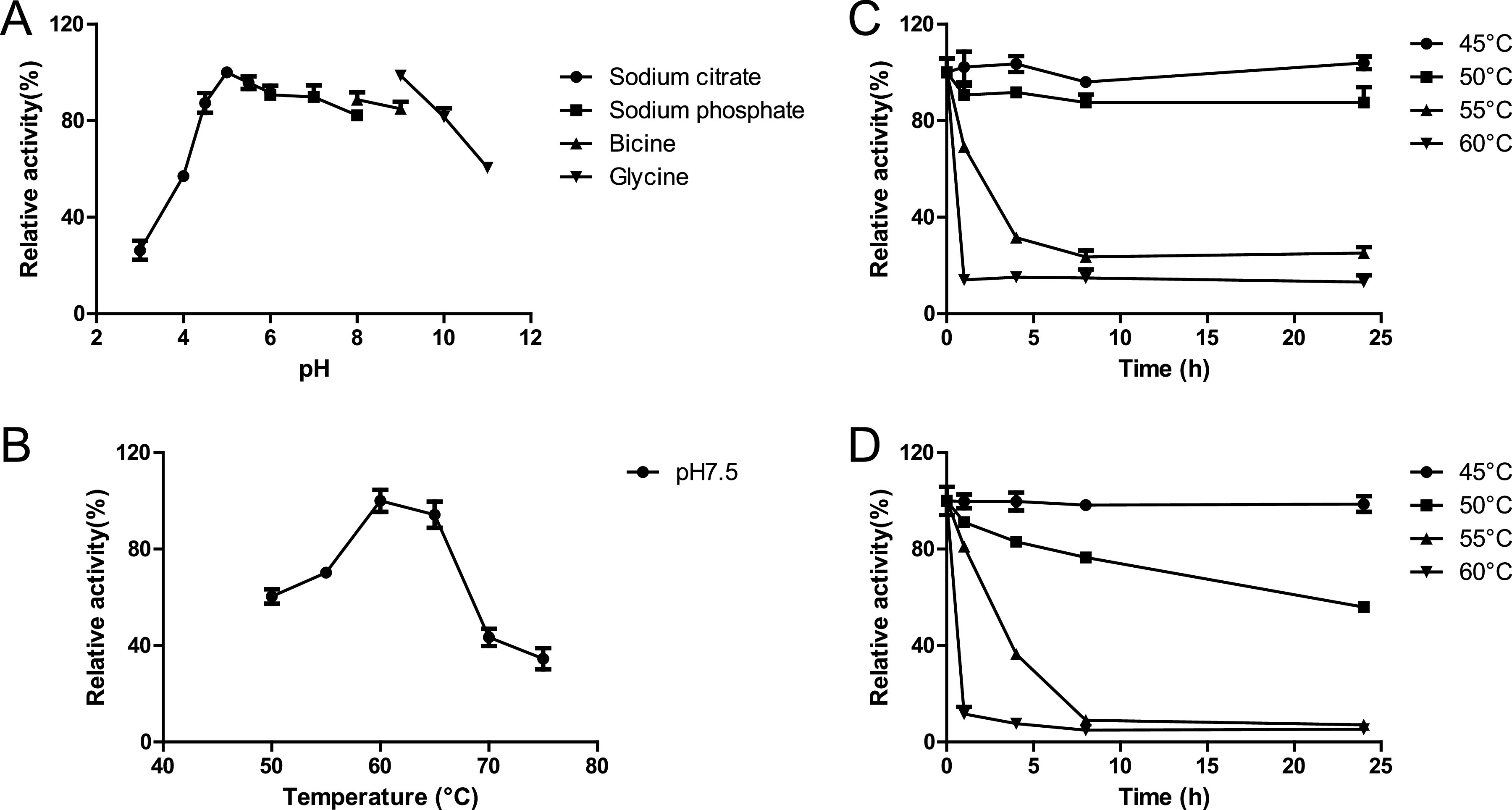
Optimal physiochemical parameters. (A) Optimum pH. (B) Thermoactivity at pH 7.5. (C) Thermostability at pH 7.5. (D) Thermostablity at pH 9.0.
Enzyme assays and kinetic analysis.
Determining the hydrolytic activity of Pm25 on various substrates revealed that it is 3- to 4-fold more active on arabinoxylans (30 U·mg−1) than on beechwood GX (7.4 U·mg−1), indicating that the latter is the least suitable substrate among those tested (Table 1). When comparing the relative activities of Pm25 on different arabinoxylans, no significant differences were detected, although rye arabinoxylan (RAX), which possess more O-3 substitutions (45), qualifies as the best substrate tested. Testing the activity of the inactive mutants (M1 and M2) (Fig. 4) on different substrates revealed that they both displayed much lower (2 to 3 orders of magnitude) activity than Pm25 (Table S3).
TABLE 1.
Kinetic parameters of Pm25
| Substrate | Activity (U/mg) | Km appb (mg/ml) | Kcat (s−1) | Kcat/Km (min−1 mM−1) |
|---|---|---|---|---|
| Beechwood GX | 7.4 ± 0.1 | 3.1 ± 0.2 | 10.3 ± 0.2 | 3.4 ± 0.1a |
| LVWAX | 25.8 ± 0.8 | 2.0 ± 0.2 | 36.2 ± 1.1 | 18.0 ± 0.5a |
| ADWAX | 21.7 ± 0.5 | 1.0 ± 0.1 | 30.4 ± 0.7 | 29.4 ± 0.7a |
| EDWAX | 21.4 ± 0.4 | 1.7 ± 0.1 | 29.9 ± 0.5 | 18.1 ± 0.3a |
| RAX | 30.3 ± 0.6 | 1.4 ± 0.1 | 42.4 ± 0.8 | 31.3 ± 0.6a |
| X6 | —c | — | — | 292.8 ± 36.8 |
| X5 | — | — | — | 35.5 ± 2.4 |
| X4 | — | — | — | 3.75 ± 0.1 |
| pNPX4 | — | — | — | 2381.7 ± 381.7 |
| pNPX3 | — | — | — | 1041.4 ± 138.2 |
| pNPX2 | — | — | — | 14.09 ± 0.66 |
For polysaccharide, kcat/Km app is in s−1 mg·ml−1.
Km app, apparent Km.
—, Not determined due to inability to saturate the enzyme under experimental conditions.
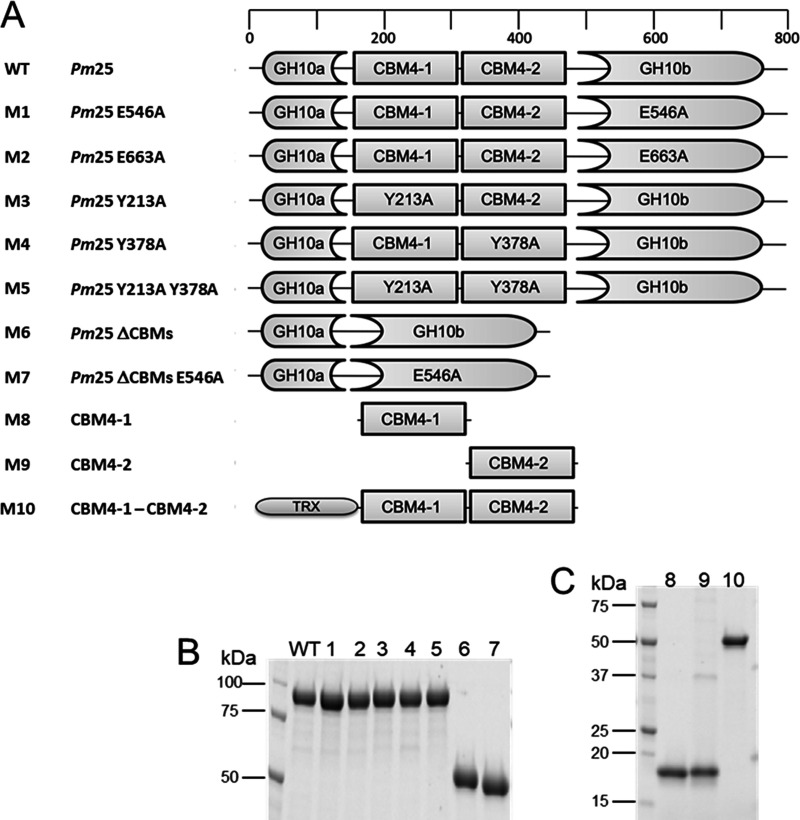
FIG 4.
Mutational and truncation constructs. (A) Domain organization of Pm25 (WT) and modified constructs. (B) SDS-PAGE of purified WT and M1 to M7 constructs. (C) SDS-PAGE of purified CBM constructs M8 to M10.
Hydrolysis product analysis with XOSs.
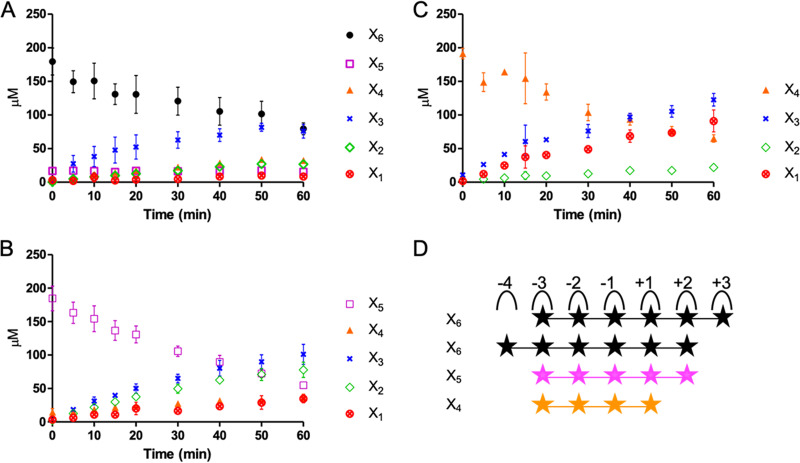
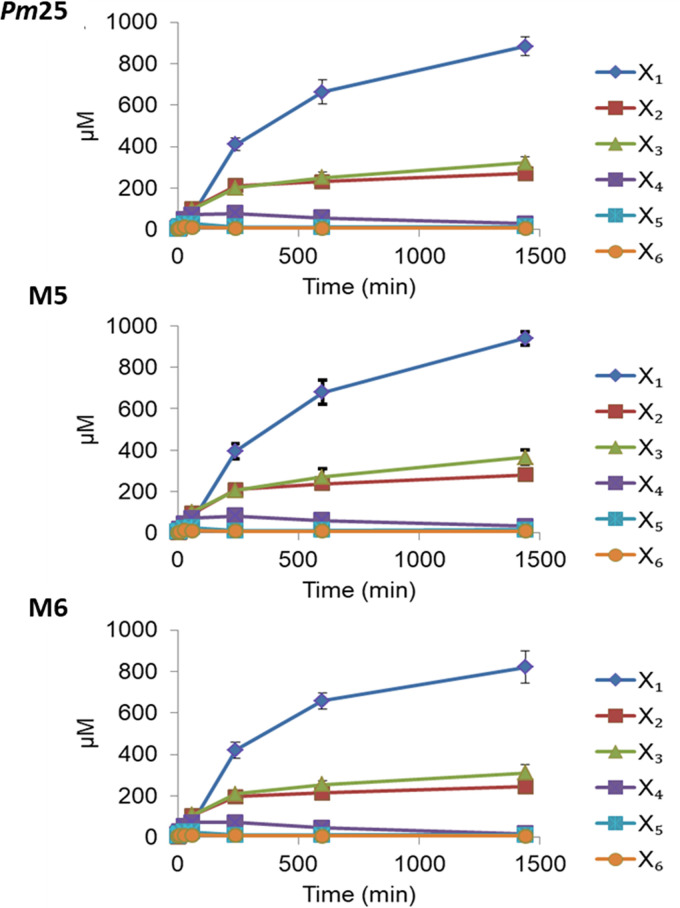
HPAEC-PAD analysis of reaction mixtures containing different XOSs and Pm25 revealed that the hydrolysis of X6 (after 60 min) produced a mixture of X2, X3, and X4 at a ratio of 1:2:1, reaching 74 μM (Fig. 5A). Likewise, the hydrolysis of X5 led to the production of X2 and X3 as major products at 75 and 96 μM, respectively (Fig. 5B), while the hydrolysis of X4 yielded X1 and X3 (Fig. 5C) at 81 and 112 μM, respectively. Moreover, the activity of Pm25 was directly correlated with the DP of the XOS used, with higher-DP XOS leading to higher activity (Table 1). This suggests that the Pm25 substrate binding cleft is quite large and can accommodate at least six xylosyl residues. To further study binding cleft subsite interactions, reactions were performed using Pm25 and different aryl-β-xylosides. Accordingly, the catalytic efficiency (kcat/Km) obtained when using p-nitrophenyl-β-d-xylotetraose (pNPX4) as the substrate was approximately 2.3-fold greater than that measured when using p-nitrophenyl-β-d-xylotriose (pNPX3) (Table 1). These data provided the basis to calculate the binding affinity at the −4 subsite, revealing a value of 0.53 kcal/mol. Likewise, the −3 subsite binding affinity is 2.76 kcal/mol, which correlates with the fact that the kcat/Km value for the hydrolysis of pNPX3 is over 70-fold higher than that measured for the hydrolysis of p-nitrophenyl-β-d-xylobiose (pNPX2) (Table 1). Furthermore, the use of construct M6 (i.e., Pm25 devoid of CBMs) (Fig. 4) to hydrolyze aryl-β-xylosides revealed similar affinities at subsites −4 and −3 (0.75 and 2.69 kcal/mol, respectively), inferring that the two CBM4 domains do not influence catalysis when using these substrates.
FIG 5.
Progressive degradation of 0.2 mM XOS. (A to C) X6 (A), X5 (B), and X4 (C) by Pm25. (D) The potential subsite mapping of Pm25 with X6, X5, and X4.
CBM and UNK ligand specificity.
To probe the function of CBM4-1 and -2, various constructions with CBM deletions or inactivations (M1 Pm25 E546A, M7 Pm25ΔCBMs E546A, M8 CBM4-1, M9 CBM4-2, and M10 CBM4-1-CBM4-2) were designed and expressed (Fig. 4), and affinity gel electrophoresis experiments were performed. Since the results of bioinformatics analysis putatively assign CBM4-1 and -2 to CBM family 4, initial tests were performed on xylan and glucan before performing complementary tests with other polysaccharides (Table 2). All proteins displayed strong affinity with the different xylans tested. However, very low or no affinity was detected for β-glucan and xyloglucan, and none of the proteins bound to arabinan, nanocellulose, and galactomannan (Table 2). Importantly, in the presence of different xylans, the dissociation constant (Kd) of M10 (CBM4-1 and -2 in tandem) was much lower than that of M8 (CBM4-1 alone) and M9 (CBM4-2 alone), ranging from 3.2 to 9, 10.9 to 31.9, and 8.0 to 17.5 10−2 mg·ml−1, respectively, suggesting that there is cooperativity between the two CBMs. Comparing the behavior of the inactive mutants M1 and M7 with that of M10 revealed that the affinity of the inactive Pm25 M1 for xylans was similar to that exhibited by M10 and the CBM4-1/-2 tandem, whereas the Kd values obtained for the inactive enzyme devoid of CBMs (i.e., M7) were significantly higher. This suggests that xylan binding is largely driven by the two CBM4 domains. Comparing M8 and M9 revealed that M9 systematically exhibited lower (1.4 to 1.8 times) Kd values for tests involving xylans, suggesting that its binding ability is stronger (Table 2). However, the results obtained with other polysaccharides indicate that CBM4-1 weakly binds to xyloglucan, while M9 (CBM4-2 alone) does not (Fig. S1). Overall, the results suggest that binding specificities of CBM4-1 and -2 are not identical.
TABLE 2.
Binding of the recombinant proteins to soluble substrates as determined by affinity gel electrophoresis
| Ligand |
Kd fora: |
||||
|---|---|---|---|---|---|
| M1 | M7 | M8 | M9 | M10 | |
| RAX | 2.8 ± 0.1 | 18.8 ± 3.5 | 31.9 ± 0.8 | 18.1 ± 2.2 | 5.1 ± 0.4 |
| LVWAX | 3.7 ± 0.3 | 27.1 ± 1.9 | 22.7 ± 1.1 | 15.9 ± 1.9 | 3.7 ± 0.2 |
| ADWAX | 2.3 ± 0.2 | 15.6 ± 0.2 | 10.9 ± 0.7 | 8.0 ± 0.1 | 6.4 ± 0 |
| EDWAX | 2.0 ± 0.1 | 16.7 ± 0.4 | 12.6 ± 0.8 | 9.2 ± 0.4 | 3.2 ± 0.0 |
| Beechwood GX | 4.9 ± 0.3 | 62.0 ± 1.9 | 30.3 ± 1.2 | 17.5 ± 0.1 | 9.0 ± 0.5 |
| Nanocellulose | − | − | − | − | − |
| Galactomannan | − | − | − | − | − |
| Xyloglucan | + | + | + | − | + |
| Arabinan | − | − | − | − | − |
| β-Glucan | + | − | + | + | + |
The unit for Kd is mg·ml−1; +, retardation observable; −, no retardation.
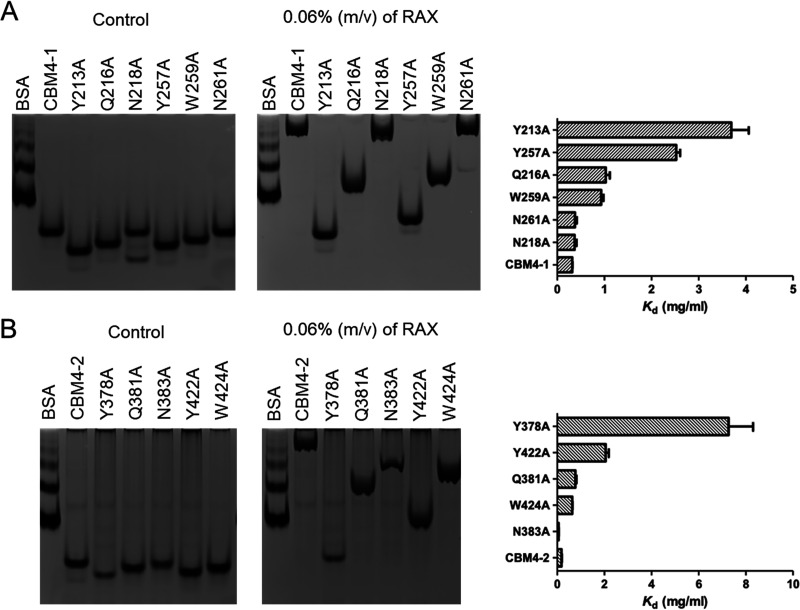
To further probe the ligand binding ability of the two Pm25-associated CBM4 domains, different residues putatively involved in ligand binding were mutated (Fig. 6) based on sequence alignment (Fig. 1D) and structural comparison of CBM4 (unpublished data). Accordingly, the binding abilities of both CBM4-1|Y213A and CBM4-2|Y378A were lost, confirming that these residues play essential roles in the CBM-ligand interaction (Fig. 6). The binding abilities of CBM4-1|Y257A and CBM4-2|Y422A were also diminished, but ligand interactions were still observable, indicating that these tyrosines play less critical roles than Y213 and Y378, respectively (Fig. 6A and B). Likewise, other mutants, such as CBM4-1|Q216A, CBM4-1|W259A, CBM4-2|Q381A, and CBM4-2|W424A, also diminished, to some extent, ligand binding, but the mutation of asparagines (N218 and N261 in CBM4-1 and N383 in CBM4-2) had no apparent effect on binding, although these residues are close to the essential ones.
FIG 6.
Affinity gel electrophoresis of different CBM mutations in Pm25. The control gel on the left lacks ligand, while the right one is with 0.06% (mol/vol) RAX. (A) Differential retardation of CBM4-1 (M8) and its mutants, CBM4-1 Y213A, Q216A, N218A, Y257A, W259A, and N261A. The Kd values for each construct are on the right. (B) Differential retardation of CBM4-2 (M9) and its mutants, CBM4-2 Y378A, Q381A, N383A, Y422A, and W424A. The Kd values for each construct are on the right.
The determination of the Kd values of M7, M8, and M9 for X6 was achieved using microscale thermophoresis (MST). The sigmoidal titration curves were used to calculate Kd values (Fig. S2). The Kd values obtained when using M7, M8, and M9 were 1.5 ± 0.2 mM, 4.7 ± 0.4 mM, and 1.8 ± 0.1 mM, respectively. Recalling that M7 is devoid of CBM domains, it is noteworthy that this construction displayed the highest binding ability, with M9 (i.e., CBM4-2) exhibiting a similar ligand binding ability.
Substrate depletion experiments performed using M1, M7, M8, and M9 on wheat bran revealed that the latter two constructions (CBM4-1 and CBM4-2, respectively) exhibited binding ability. Similarly, M1 (i.e., Pm25|E546A) was also able to bind to wheat bran despite its catalytic impotency (Fig. S3A). However, this was not the case for M7 (i.e., the inactivated catalytic domain alone), clearly demonstrating the role of the CBM in binding.
The affinity of UNK toward low-viscosity wheat arabinoxylan (LVWAX) was investigated. The retardation of the UNK band suggested the UNK can bind to xylan (Fig. S3B).
Analysis of polysaccharide and wheat bran hydrolysis.
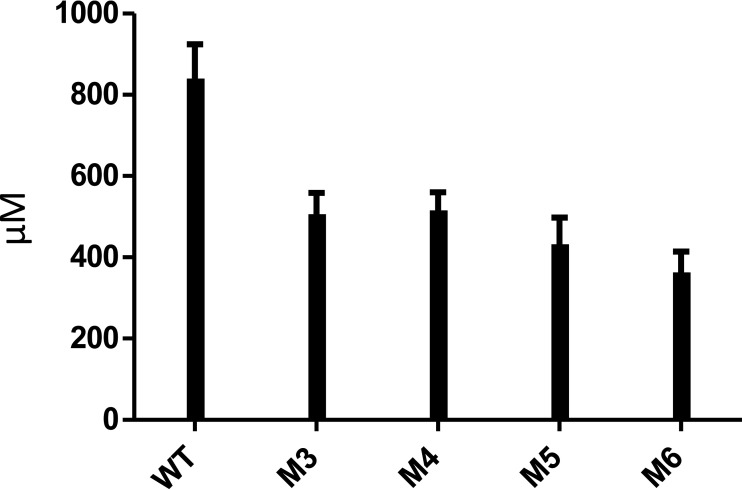
HPAEC-PAD of soluble polysaccharide hydrolysis mediated by Pm25 and its variants M5 and M6 failed to reveal significant differences in d-Xyl and XOS release (Fig. 7). This suggests that the CBM4 domains do not enhance degradation of soluble polysaccharides, since M6 is devoid of CBM4 domains. Performing a similar analysis using wheat bran as the substrate revealed that (after 14 h of incubation) d-Xyl and XOS release by Pm25 was approximately twice that of the variants M3, M4, M5, and M6 (Fig. 8). Significantly, in this experiment the consequences of the point mutations CBM4-1|Y213A and CBM4-1|Y378A were approximately equivalent to those produced by the ablation of the two CBM4 domains.
FIG 7.
Hydrolysis of LVWAX by Pm25 and its mutants. The time-dependent degradation of LVWAX by Pm25 (WT), M5 (Pm25 Y213A Y378A), and M6 (Pm25ΔCBMs).
FIG 8.
Hydrolytic patterns of Pm25 (WT) and mutants toward wheat bran. The release of xylooligosaccharides from wheat bran after overnight incubation with wild-type Pm25 compared to that of the mutants.
DISCUSSION
Unlike the vast majority of multimodular enzymes that display a sequential arrangement of their modules, the enzyme described here is characterized by a discontinuous organization that involves the insertion of two CBM domains into one GH10 xylanase domain. In this regard, it is significant that the SSN analysis performed using the amino acid sequence of M6 replacing Pm25 located the sequence within the same cluster, even though the CBMs were omitted (data not shown). This suggests that the Pm25 GH10 domain forms part of a distinct group and implies that the intercalated GH10 arrangement is robust from an evolutionary standpoint. Moreover, the biochemical data described here demonstrate that, despite its discontinuous organization, Pm25 is a fully functional xylanase.
The first Pm25 analog was identified in a rumen-based member of the Bacteroidetes phylum (29). More have since been found in human gut bacteria (30, 32, 46), with Pm25 being the first described in termite gut. Several studies have revealed the importance of Pm25-like GH10 in xylan utilization systems (29, 30, 32, 33). Using SSN analysis, we have shown that Pm25-like xylanases are exclusively linked to Bacteroidetes and are mostly (44 out of 61 based on SSN analysis) adjacent to an susC-susD-(unk) cluster. This evidence of strong conservation is consistent with the fact that in their native host, the genes encoding Pm25 homologs are highly induced/expressed during growth on xylan (30, 46). In addition, our data show that the UNK protein upstream of Pm25 is a xylan-binding protein that strengthens the xylan utilization function of this core cluster (46), suggesting it is an analogue of SusE, which is also supported by the fact that like SusE, UNK is predicted to have a lipoprotein peptide signal by SignalP (47). Taken together, one can conclude that each component in the core cluster is essential for xylan utilization by members of the Bacteroidetes phylum in the gut ecosystem.
The in vivo function of Pm25 homologs in gut Bacteroidetes has not yet been fully established, although it has been suggested that it is a functional homolog of SusG (28). SusG is a cell surface-bound GH13 α-amylase that catalyzes the initial cleavage of polysaccharides (48). In our study, we also predict that Pm25 bears an N-terminal signal peptide that directs it to the cell surface, consistent with a proposal that was previously made for a Pm25 homolog (32). Moreover, SusG displays negligible activity compared to periplasmic α-amylases (48), an observation that is consistent with our findings. Indeed, compared to other xylanases (41, 49), both Pm25 and similar elements display quite poor catalytic efficiency toward polysaccharides (33, 50) and oligosaccharides (32). This trend is also observed in other polysaccharide-degrading systems, such as mannan utilization loci from members of the Bacteroidetes phylum (51) and the xylan-degrading system in the Proteobacteria (43) phylum. The underlying reason for such low activity most likely reflects its function. SusG-like proteins probably have a carbohydrate surveillance function, while highly active intracellular enzymes are charged with complete oligosaccharide breakdown prior to sugar catabolism. This clever and “selfish” strategy ensures that readily metabolizable sugars are not released into the environment, where they could be used by other bacteria that lack a specific glycan utilization machinery (52).
Remarkably, we found that Pm25 remains active over a broad pH range, maintaining more than 80% of its maximum activity at pH 9.0. This observation correlates well with results obtained for the Pm25 homologs Bacteroides intestinalis Xyn10C (BiXyn10C) and BiXyn10A, which were identified in the human gut microbiome (50). Accounting for the fact that alkaline-stable xylanases are sought after for use in applications such as paper pulp biobleaching, Pm25 might constitute a useful starting point for enzyme engineering aimed at improving its hydrolytic properties.
So far, we have been unable to obtain structural data pertaining to Pm25, and none is available for its closest homologs. Therefore, at this stage it is tricky to speculate on the exact topology and molecular determinants of its active site. Nevertheless, to gain some understanding, we have examined similarities with the family GH10 xylanase Cellvibrio japonicus Xyn10C (CjXyn10C), which displays approximately 30% identity to Pm25 and whose structure is known (PDB entry 1US3). Like Pm25, CjXyn10C exhibits rather poor activity on XOS, ascribed to weak substrate binding in subsite −2 (43). Unlike most other GH10 enzymes, CjXyn10C subsite −2 contains G295 in the place of E, whose side chain can hydrogen bond to the substrate. According to sequence alignment, Pm25 also lacks the vital E residue in subsite −2, an observation that might explain its poor ability to hydrolyze X4 (43, 53). Therefore, the −3 subsite with rather strong affinity value (2.76 kcal/mol) compared to others (53) is probably involved in the glycine subsite in the degradation of X4 to compensate for the poor −2 subsite. Taken together, a hypothetical subsite mapping of the active site of Pm25 with XOS is proposed for Pm25 (Fig. 5D).
The two CBM4s that are inserted into Pm25 clearly contribute to the binding and degradation of complex biomass. Our results reveal that this is especially true when both CBMs are functional and suggest that binding of large ligands involves a cooperativity phenomenon (Fig. 8). However, based on the PULDB database, the number of CBM domains in Pm25 homologs varies from one to three, and the CBMs are from different families, CBM4, CBM22, or unclassified. This suggests that the SusG-assimilated functions can be fulfilled by enzymes that are not configured in an identical way. Moreover, it also confirms that the TIM-barrel fold in the GH10 family is quite accommodating in terms of insertions at the β3/α3 loop.
Apparently, unlike many highly active periplasmic endoglucanases, such as SusA (48) and CjXyn10D (43), extracellular enzymes such as SusG, CjXyn10A, and CjXyn10C are generally appended to CBMs (43). Therefore, it is of interest to discuss the reason for this. CBM58 in SusG (54) and the CBM4s in Pm25 appear to improve the ability of the enzymes to hydrolyze insoluble substrates (Fig. 8), while CBM15 in CjXyn10C does not play an important role in catalysis, irrespective of whether the substrate is soluble or not (43). However, our data suggest that the affinity of Pm25 for soluble substrates was mostly derived from the binding ability of the CBMs (Table 2). In light of this observation, we propose that CBMs in membrane-associated enzymes temporarily withhold soluble oligosaccharides before their importation into the cell. This implies that the function of the CBM4 domains would be relatively independent of that of the GH10 domain. In this regard, it is noteworthy that the first structure of a SusG protein (54), which reveals that a CBM58 domain is inserted into the B domain of the GH13 α-amylase domain, reports that CBM58 does not form hydrogen bonds with the catalytic domain, an observation that argues in favor of an independent function. Regarding Pm25, evidence for an independent function of the CBM4 domains is provided by the fact that the xylan-degrading profile of the Pm25 wild type was almost identical to that of the CBM-deleted version, M6 (Fig. 7), and the fact that the xylan binding affinity of CBMs was relatively unaltered when the CBM domains were separated from the GH10 domain (Table 2). Finally, it is also useful to recall that the affinity values determined for subsites −4 and −3 of Pm25 and M6 were nearly identical. Therefore, we believe that the catalytic center of Pm25 and the binding surfaces of the CBM4 domains are disconnected, an organization that corresponds to independent functions and contributes to low enzyme reaction rates (55).
In conclusion, focusing on a termite gut-derived enzyme, we have provided further insight into the properties and function of Xyn10C-like enzymes that form part of core xylan utilization systems. This system seems to be rather efficient in terms of evolution, since it is conserved in termite gut, rumen, and human gut. Therefore, the role of the CBM insertion is an interesting question. In this respect, we have thoroughly succeeded in characterizing the enzyme and shown that the CBM4 domains can be successfully excised without loss of catalytic function. Regarding the enzyme’s substrate specificity, although it is difficult to speculate on the group of polysaccharides that might be preferential substrates in the termite gut environment, we have shown that it is better adapted for the hydrolysis of arabinoxylans than glucuronoxylans, which is consistent with the fact that the host termite feeds on crops such as sugarcane rather than wood.
MATERIALS AND METHODS
Materials.
Beechwood GX, d-Xyl, and most other reagents were purchased from Sigma-Aldrich (Darmstadt, Germany). Low-viscosity wheat arabinoxylan (Ara:Xyl = 38:62; LVWAX), acid-debranched wheat arabinoxylan (Ara:Xyl = 22:78; ADWAX), enzyme-debranched wheat arabinoxylan (Ara:Xyl = 30:70; EDWAX), rye arabinoxylan (Ara:Xyl = 38:62; RAX), galactomannan (carob; low viscosity), xyloglucan (tamarind), arabinan (sugar beet), β-glucan (barley; medium viscosity), xylobiose (X2), xylotriose (X3), xylotetraose (X4), xylopentaose (X5), xylohexaose (X6), p-nitrophenyl-β-d-xylopyranoside (pNPX), p-nitrophenyl-β-d-xylobiose (pNPX2), p-nitrophenyl-β-d-xylotriose (pNPX3), and p-nitrophenyl-β-d-xylotetraose (pNPX4) were all purchased from Megazyme (Bray, Ireland). Cellulose nanocrystals (nanocellulose) from cotton linters were prepared as previously described (56). Oligonucleotide primers were purchased from Eurogentec (Liège, Belgium) (Table 3).
TABLE 3.
Primers used in this study
| Target | Orientationa | Sequence (5′→3′) |
|---|---|---|
| Primers for site-directed mutagenesis | ||
| Pm25|E546A | F | GTGGGATGTGCTGAACGCGCCCATGAACGAGAAC |
| R | GTTCTCGTTCATGGGCGCGTTCAGCACATCCCAC | |
| Pm25|E663tga | F | GGCAAACTCATCAAGGTGACGTGACTCGATATTGCCATATCCAC |
| R | GTGGATATGGCAATATCGAGTCACGTCACCTTGATGAGTTTGCC | |
| Pm25|tga663A | F | GCAAACTCATCAAGGTGACGGCGCTCGATATTGCCATATCC |
| R | GGATATGGCAATATCGAGCGCCGTCACCTTGATGAGTTTGC | |
| Pm25|H634A | F | GCATCGGCACGCAAATGGCGCTCAACCTCAACTGG |
| R | CCAGTTGAGGTTGAGCGCCATTTGCGTGCCGATGC | |
| CBM4-1|Y213A | F | GGCTCGACCGGAAAAGCGGACGCACAACTCAACTCTCC |
| R | GGAGAGTTGAGTTGTGCGTCCGCTTTTCCGGTCGAGCC | |
| CBM4-1|Q216A | F | CGGAAAATACGACGCAGCGCTCAACTCTCCGGCG |
| R | CGCCGGAGAGTTGAGCGCTGCGTCGTATTTTCCG | |
| CBM4-1|N218A | F | TACGACGCACAACTCGCGTCTCCGGCGTTCCCG |
| R | CGGGAACGCCGGAGACGCGAGTTGTGCGTCGTA | |
| CBM4-1|Y257A | F | CGGAACTGGGTAATCAAGCGCCATGGGTGAACGGC |
| R | GCCGTTCACCCATGGCGCTTGATTACCCAGTTCCG | |
| CBM4-1|W259A | F | GGTAATCAATATCCAGCCGTGAACGGCGCAGAACTGGC |
| R | GCCAGTTCTGCGCCGTTCACGGCTGGATATTGATTACC | |
| CBM4-1|N261A | F | CAATATCCATGGGTGGCGGGCGCAGAACTGGCCG |
| R | CGGCCAGTTCTGCGCCCGCCACCCATGGATATTG | |
| CBM4-2|Y378A | F | GGTGCGGCTGGCAAAGCGGACCGCCAACTCAAC |
| R | GTTGAGTTGGCGGTCCGCTTTGCCAGCCGCACC | |
| CBM4-2|Q381A | F | GGCAAATACGACCGCGCGCTCAACAGCCCGGCATTTCC |
| R | GGAAATGCCGGGCTGTTGAGCGCGCGGTCGTATTTGCC | |
| CBM4-2|N383A | F | GCAAATACGACCGCCAACTCGCGAGCCCGGCATTTCCG |
| R | CGGAAATGCCGGGCTCGCGAGTTGGCGGTCGTATTTGC | |
| CBM4-2|Y422A | F | GAACTCGGCAACCAGGCGCCGTGGGGAAGCGGC |
| R | GCCGCTTCCCCACGGCGCCTGGTTGCCGAGTTC | |
| CBM4-2|W424A | F | GGCAACCAGTATCCGGCCGGAAGCGGCGGCGAG |
| R | CTCGCCGCCGCTTCCGGCCGGATACTGGTTGCC | |
| Primers for in-fusion cloning | ||
| Pm25_CBM4-1_L167-L320 | F | CAGCCATATGGCTAGCTTGTGGACGGAAGTTTCGAAG |
| R | GGTGGTGGTGCTCGATTAGAGGTCTGTGATCTCGACG | |
| Pm25_CBM4-2_D332-D484 | F | CAGCCATATGGCTAGCCTTGTGGACGGCAATTTCGAAGGCGGAG |
| R | GGTGGTGGTGCTCGAGTTATTAGAGGTCGGTGACCTCAACGGCGTC | |
| Pm25_CBM4-1 + 2_L167-D484 | F | GACGACGACGACAAGCTTGTGGACGGAAGTTTCGAAGAGGGGATG |
| R | GTGGTGGTGCTCGAGGTCGGTGACCTCAACGGCGTCGATC | |
| pET32a_linearization | F | CTCGAGCACCACCACCACCACCAC |
| R | CTTGTCGTCGTCGTCGCCAGAACCAGAACCGGCCAGGTTAG |
F, forward; R, reverse.
The GenBank accession number for the clone containing Pm25 is HF548280.1, and the protein ID for Pm25 is CCO21036.1.
Bioinformatics analysis.
Putative signal peptide sequence analysis was performed using the SignalP 4.1 server (47). The domain annotation of Pm25 was done using InterPro protein sequence analysis (https://www.ebi.ac.uk/interpro/) with accession number S0DFK9. Multiple-protein sequence alignment of CBM4s was done using Clustal Omega (https://www.ebi.ac.uk/Tools/msa/clustalo/), and the alignment of the secondary structure elements of Pm25 with other structurally characterized GH10 family members was achieved using both Clustal Omega and ESPript 3 at http://espript.ibcp.fr/ESPript/ESPript/ (57).
SSNs.
Amino acid sequences of GH10 family members were extracted from the CAZy database (http://www.cazy.org/GH10_all.html), updated on 29 May 2020. To remove redundant sequences, the 4,936 sequences were winnowed down to 2,539 by a sequence identity cutoff of 0.9 (58), length cutoff of 250, and fragment exclusion (59). Sequence similarity networks (SSNs) were constructed using the Enzyme Function Initiative Enzyme Similarity Tool (EFI-EST) (59) and visualized using Cytoscape 3.6 (60). The alignment score threshold was set to 35% sequence similarity, since nodes are linked with the edge when they share over 35% identity and each node represents one protein sequence. Multiple-sequence alignment of GH10s in different clusters was done by MAFFT (https://www.ebi.ac.uk/Tools/msa/mafft/), and sequence logos were constructed via WebLoGo (https://weblogo.berkeley.edu/logo.cgi).
Cloning and site-directed mutagenesis.
Cloning of the plasmid pDEST17 containing Pm25 was achieved as described previously (61). All mutants were constructed using the QuikChange site-directed mutagenesis kit (Strategene, La Jolla, CA, USA) with oligonucleotide primers (Table 3). The M6 construct, which corresponds to Pm25 deprived of its CBMs, was obtained by gene synthesis (NZYTech, Lda, Portugal). The mutation of E546 to A in M6 yielded M7, while M8 and M9 were constructed by cloning the sequences encoding CBM4-1 and CBM4-2, respectively, into pET28a(+) expression vector. Likewise, the construct M10 is the pET32a(+) expression vector containing the sequence encoding both CBM4-1 and CBM4-2 cloned in frame with the thioredoxin tag using the In-Fusion cloning kit (Clontech, TaKaRa, Shiga, Japan). The DNA sequence of UNK (UniProt ID S0DDM9) deprived of its signal peptide sequence, as identified by the SignalP 4.1 server (47), was synthesized and subcloned into pET28a between the NheI and XhoI restriction sites.
Protein expression and purification.
Wild-type Pm25 and the mutants M1, M2, M3, M4, and M5 (Fig. 4A) were expressed in Escherichia coli Rosetta(DE3) pLysS grown in ZYP autoinduction medium (62) at 25°C overnight. Constructs M6 to M10 and UNK were transformed into E. coli Tuner(DE3) and cultured for 2 h at 37°C until the optical density at 600 nm (OD600) reached 0.6. At this point, isopropyl-β-d-thiogalactopyranoside (IPTG; 200 μM final concentration) was added and growth was pursued at 16°C overnight. Cell pellets were collected by centrifugation, washed, and lysed using sonication (Fisherbrand Q700; tip diameter, 13 mm; output, 40 W), and the clarified cell lysates were applied to TALON metal affinity resin (Clontech, Mountain View, CA, USA). After elution, protein purity was estimated by SDS-PAGE to be 95%. Protein concentrations were determined by measuring absorbance at 280 nm and applying the Beer-Lambert equation. Theoretical molar extinction coefficients were calculated using ProtParam online software (63).
Determination of pH and temperature optima.
The apparent optimal pH of Pm25 was determined in the pH range of 3.0 to 11.0, measuring the enzyme activity (0.4 μM final enzyme concentration) on 1% (mol/vol) beechwood GX at 37°C. The buffers used were 50 mM citrate buffer for pH 3.0 to 6.0, 50 mM phosphate buffer for pH 6.0 to 8.0, 20 mM bicine buffer for pH 8.0 to 9.0, and 20 mM glycine-NaOH for pH 9.0 to 11.0. Xylanase activity was determined by measuring the release of reducing sugars using the 3,5-dinitrosalicylic acid (DNS) assay (64, 65). Reactions were performed in triplicate at 37°C in the different buffers from pH 3.0 to 11.0, containing bovine serum albumin (BSA; 1 mg/ml). At regular intervals (0, 3, 6, 9, 12, 15, 18, 21, and 24 min), 100 μl of the reaction mixture was removed and added to 100 μl of DNS and kept on ice until all samples were ready. All samples then were heated at 95°C for 10 min and cooled on ice before adding 1 ml of deionized water and recording the absorbance at 540 nm using a spectrophotometer. A d-xylose series (0 to 1 mg/ml) was used to prepare a standard curve. The apparent optimal temperature was determined over the range of 21 to 90°C in 50 mM phosphate buffer (pH 7.5). Thermostability was monitored by preincubating the enzyme in the absence of substrate in 50 mM phosphate buffer (pH 7.5) at 45, 50, 55, and 60°C from 0 to 24 h. Residual enzyme activity in each case was then assayed as described above.
Enzyme specificity and kinetics.
Enzyme kinetics were measured using a Pm25 concentration of 0.4 μM and 0.08 μM to degrade beechwood GX and other soluble polysaccharides, respectively. Initial rates (the concentration of d-Xyl equivalent released, in milligrams per milliliter per minute) were determined using a range of substrate concentrations (from 0.5 to 40 g/liter beechwood GX and 0.25 to 10 g/liter for other soluble substrates) under optimal conditions. The DNS assay was used to monitor reducing sugar release as described earlier. The kinetic parameters (kcat and apparent Km) were calculated using nonlinear regression in SigmaPlot 11.0 (Systat Software, San Jose, CA, USA). One unit of xylanase activity was defined as the amount of enzyme that catalyzes the release of 1 μmol of d-Xyl equivalents per min.
To study the hydrolysis of xylooligosaccharides (XOS) with a degree of polymerization of 4 to 6 (X4 to X6), reactions were performed using various concentrations (0.05 to 0.8 mM) and the optimal reaction conditions. Assays began upon the addition of enzyme, its final concentration being fixed to account for the nature of the substrate. Accordingly, 2.60, 0.26, and 0.026 μM enzyme were used for X4, X5, and X6, respectively. At regular intervals (0, 5, 10, 15, 20, 30, 40, 50, and 60 min), aliquots were removed and immediately heated at 95°C for 10 min to stop the reaction. The hydrolyzed products were then analyzed by high-performance anion-exchange chromatography with pulsed amperometric detection (HPAEC-PAD) using an ICS 3000 dual device (Dionex, France) equipped with Carbo-Pac PA-100 guard and analytical columns (2 by 50 mm and 2 by 250 mm, respectively) as described before (65). Ten microliters of sample was injected, and separation was achieved by applying a gradient of 0 to 85 mM sodium acetate, 150 mM NaOH from 0 to 30 min, isocratic elution with 500 mM sodium acetate, 150 mM NaOH from 30 to 33 min, and reequilibration of the column with 50 mM sodium acetate, 150 mM NaOH for another 10 min at a flow rate of 0.25 ml/min. Calibration was achieved using d-Xyl and XOS (X2, X3, X4, X5, and X6) at concentrations from 5 to 50 μM. Plotting the hydrolysis rate (micromolars per minute) versus oligosaccharide substrate concentration (micromolars) yielded a linear relationship, meaning that the catalytic constant kcat/Km could be calculated from the slope k using equation 1, where [E] is the final concentration of enzyme.
| (1) |
All experiments were performed in triplicate, and reported values are the means from three experiments.
Determination of subsite affinities.
The binding affinities of glycone subsites were calculated using equation 2 (66, 67).
| (2) |
where A−i is the subsite affinity at −i subsite, kcat/Km of pNPXi is the performance constant for pNP-labeled XOS with a DP of i (where i is a whole number), and R is the universal gas constant (8.314 J mol−1 K−1).
To determine the catalytic parameters of reaction mixtures containing pNP-XOS, the final concentration of Pm25 used was 6, 27, and 136 nM for pNPX4, pNPX3, and pNPX2, respectively. Similarly, the final concentration of M6 was 10 nM for pNPX4 and pNPX3 and 54 nM for pNPX2. The concentration range of substrate was 0.025, 0.05, and 0.1 mM for pNPX3 and pNPX4 and 0.5, 2, and 5 mM for pNPX2. All experiments were performed in duplicate. The plot of hydrolysis rate against pNP substrate concentration was linear, which indicated that substrate concentration was far below the Km. Therefore, the kcat/Km of reaction mixtures containing aryl β-xylosides was determined under optimum conditions using equation 3 (53, 66). Briefly, the substrate concentrations at the beginning of the reaction ([S0]) and at specific times ([St]) were fitted to equation 3, where k = (kcat/Km) [Enzyme] and [Enzyme] is the final concentration of enzyme.
| (3) |
The molar extinction coefficient of pNP (15,570 M−1·cm−1) was determined experimentally by measuring the absorbance at 404 nm for a standard curve ranging from 0 to 0.12 mM pNP at pH 7.5 and 50°C.
AGE.
The binding of CBM4-1 and CBM4-2 to soluble polysaccharides was evaluated by affinity gel electrophoresis (AGE), using 7.5% (mol/vol) acrylamide gels containing various amounts of polysaccharide (for the concentration range of RAX, refer to Table S1 in the supplemental material). ADWAX, EDWAX, and LVWAX samples and beechwood GX were used at 0.006 to 0.06% (mol/vol), while other polysaccharides were used at 0.5% (mol/vol). Pure protein (6 μg) was migrated (10 mA/gel for about 1 h at room temperature) on gels in 25 mM Tris, 250 mM glycine buffer, pH 8.3. BSA (15 μg) was also included in the experiment as a negative, noninteracting control. Proteins were visualized by Coomassie blue staining. The dissociation constant Kd was calculated as previously described (68). In equation 4,
| (4) |
R0 is the relative protein migration distance compared to that of BSA in the control gel (without ligand). The variable r is the relative protein migration distance compared to that of BSA in ligand-containing gels. Rc is the relative protein migration distance of complex between protein and ligand. c is the concentration of ligand. When equation 4 was plotted, taking 1/(R0 − r) as the ordinate and 1/c as the abscissa, a straight line was obtained. The intercept of the line on the abscissa provided a negative reciprocal value of the dissociation constant (−1/K). All experiments were performed in triplicate, and reported values are the means from three experiments.
MST.
Microscale thermophoresis (MST) (69) was carried out on a Monolith NT115 (NanoTemper Technologies GmbH, Munich, Germany) at 25°C, 20% light-emitting diode (LED) power, and 40% MST power. Protein samples were labeled at a final concentration of 10 μM, as previously described (68). An aliquot of 0.6 μM labeled protein was mixed with decreasing concentrations of cellulose nanocrystals (from 312.5 mg·liter−1 to 0.3 mg·liter−1) in either buffer 1 (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 10 mM MgCl2, 0.05% Tween 20) or buffer 2 (50 mM sodium phosphate buffer, pH 7, and 0.05% pluronic acid). Data analysis was performed with MO Affinity software (NanoTemper). The Hill equation was chosen to determine a value for the 50% effective concentration (EC50).
To fluorescently label the amine groups of exposed lysines (Fig. 1D) lying in the vicinity of the ligand binding clefts in M7, M8, and M9, 100 μl of pure proteins (20 μM) was treated with the reagents in the protein labeling kit (RED-NHS) by following the manufacturer’s instructions. The labeled proteins were recovered and purified using TALON metal affinity resin, and the concentration of the labeled proteins was estimated using SDS-PAGE and serial dilutions of a protein solution of known concentration. A total of 16 dilutions (350 to 11 mM) of a solution of X6 containing either 0.075 μM M7, 0.13 μM M8, or 0.07 μM M9 in 50 mM phosphate buffer, pH 7, 0.05% Pluronic F-127 were loaded onto 16 standard capillaries. The initial fluorescence of all 16 samples was obtained by performing a capillary scan with LED power of 25% for M8 and 20% for M7 and M9. The dissociation constant Kd was calculated by selecting the tab “Initial Fluorescence Analysis Set” in the Affinity Analysis software (70). To perform the SDS denaturation (SD) test, 10 μl of samples 1 to 3 and 14 to 16 were mixed with 10 μl of 4% SDS, 40 mM dithiothreitol (DTT) after 10 min centrifugation at 15,000 × g, followed by a 5-min incubation of the mixture at 95°C to denature the protein. The samples then were loaded into the capillaries to measure their fluorescence intensities.
Solid depletion assay.
The ability of inactivated M1, M7, M8, and M9 to bind wheat bran was investigated by incubating 100 μg of protein with 4 mg of wheat bran in 200 μl of reaction buffer (50 mM sodium phosphate, pH 7). Reactions were performed in 0.2-ml PCR tubes and incubated at 10°C for 2 h with agitation in an Eppendorf Thermomixer R at 1,400 rpm. For each reaction, the supernatant containing the unbound enzyme fraction was recovered after centrifugation using a benchtop microcentrifuge. The pelleted substrate was washed 3 times with reaction buffer. Finally, 20 μl of Laemmli sample buffer was added to the pellet and heated at 95°C for 10 min to denature the protein (bound fraction). All the fractions were verified by SDS-PAGE. BSA was used as a negative control.
Hydrolysis of wheat arabinoxylan and wheat bran.
Product profiles were generated with Pm25, M3, M4, M5, and M6 on either LVWAX (0.5% [mol/vol]) or wheat bran (20 mg/ml of wheat bran prehydrated for 12 h at 37°C, 1,400 rpm using the Eppendorf Thermomixer R). The enzymes (final concentration, 0.5 μM) were incubated with the respective substrate in 50 mM phosphate buffer (pH 7.5) and 1 mg/ml BSA. Enzymatic reaction mixtures were incubated at 37°C for either 24 h for LVWAX or 14 h for wheat bran, and aliquots were removed at regular time intervals and heated at 95°C for 10 min to terminate the reaction. Each sample was centrifuged at 20,000 × g for 5 min and quantified by HPAEC-PAD on a Dionex PA1 column equipped with a Carbo-Pac PA-1 guard and analytical columns (4 by 50 mm and 4 by 250 mm, respectively). Separation of oligosaccharides was achieved by isocratic elution with 100 mM NaOH at a flow rate of 1 ml/min from 0 to 5 min, a gradient of 0 to 120 mM sodium acetate in 100 mM NaOH from 5 min to 25 min, and isocratic elution with 500 mM sodium acetate in 100 mM NaOH from 25 min to 35 min. The column then was reequilibrated with 100 mM NaOH for another 10 min. Calibration was achieved using d-Xyl and XOS (X2, X3, X4, X5, and X6) at concentrations from 5 to 100 μM.
Supplementary Material
ACKNOWLEDGMENTS
The research was supported by CSC (China Scholarship Council) (H.W.) and the Climate-KIC ADMIT BIOSUCCINOVATE project (E.I.). The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
We thank the ICEO facility dedicated to enzyme screening and discovery, part of the Integrated Screening Platform of Toulouse (PICT, IBiSA), for providing access to high-performance liquid chromatography and protein purification systems. G. Arnal is gratefully acknowledged for insightful reading of the manuscript.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Stephen AM. 1983. Other plant polysaccharides, p 97–193. In Aspinall GO. (ed), The polysaccharides, vol 2 Elsevier Inc, New York, NY. [Google Scholar]
- 2.Scheller HV, Ulvskov P. 2010. Hemicelluloses. Annu Rev Plant Biol 61:263–289. doi: 10.1146/annurev-arplant-042809-112315. [DOI] [PubMed] [Google Scholar]
- 3.Malgas S, Thoresen M, van Dyk JS, Pletschke BI. 2017. Time dependence of enzyme synergism during the degradation of model and natural lignocellulosic substrates. Enzyme Microb Technol 103:1–11. doi: 10.1016/j.enzmictec.2017.04.007. [DOI] [PubMed] [Google Scholar]
- 4.Merino ST, Cherry J. 2007. Progress and challenges in enzyme development for biomass utilization. Adv Biochem Eng Biotechnol 108:95–120. doi: 10.1007/10_2007_066. [DOI] [PubMed] [Google Scholar]
- 5.Lombard V, Ramulu HG, Drula E, Coutinho PM, Henrissat B. 2014. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res 42:490–495. doi: 10.1093/nar/gkt1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Collins T, Gerday C, Feller G. 2005. Xylanases, xylanase families and extremophilic xylanases. FEMS Microbiol Rev 29:3–23. doi: 10.1016/j.femsre.2004.06.005. [DOI] [PubMed] [Google Scholar]
- 7.Chu Y, Tu T, Penttinen L, Xue X, Wang X, Yi Z, Gong L, Rouvinen J, Luo H, Hakulinen N, Yao B, Su X. 2017. Insights into the roles of non-catalytic residues in the active site of a GH10 xylanase with activity on cellulose. J Biol Chem 292:19315–19327. doi: 10.1074/jbc.M117.807768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bolam DN, Ciruela A, McQueen-Mason S, Simpson P, Williamson MP, Rixon JE, Boraston A, Hazlewood GP, Gilbert HJ. 1998. Pseudomonas cellulose-binding domains mediate their effects by increasing enzyme substrate proximity. Biochem J 331:775–781. doi: 10.1042/bj3310775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Din N, Damude HG, Gilkes NR, Miller RC, Warren RA, Kilburn DG. 1994. C1-Cx revisited: intramolecular synergism in a cellulase. Proc Natl Acad Sci U S A 91:11383–11387. doi: 10.1073/pnas.91.24.11383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Montanier C, van Bueren AL, Dumon C, Flint JE, Correia MA, Prates JA, Firbank SJ, Lewis RJ, Grondin GG, Ghinet MG, Gloster TM, Herve C, Knox JP, Talbot BG, Turkenburg JP, Kerovuo J, Brzezinski R, Fontes CA, Davies GJ, Boraston AB, Gilbert HJ. 2009. Evidence that family 35 carbohydrate binding modules display conserved specificity but divergent function. Proc Natl Acad Sci U S A 106:3065–3070. doi: 10.1073/pnas.0808972106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aroul-Selvam R, Hubbard T, Sasidharan R, Aroul-Selvam R, Hubbard T, Sasidharan R. 2004. Domain insertions in protein structures. J Mol Biol 338:633–641. doi: 10.1016/j.jmb.2004.03.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xue Z, Jang R, Govindarajoo B, Huang Y, Wang Y. 2015. Extending protein domain boundary predictors to detect discontinuous domains. PLoS One 10:e0141541. doi: 10.1371/journal.pone.0141541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ohkuma M. 2003. Termite symbiotic systems: efficient bio-recycling of lignocellulose. Appl Microbiol Biotechnol 61:1–9. doi: 10.1007/s00253-002-1189-z. [DOI] [PubMed] [Google Scholar]
- 14.Brune A. 2007. Woodworker’s digest. Nature 450:487–488. doi: 10.1038/450487a. [DOI] [PubMed] [Google Scholar]
- 15.Scharf ME, Tartar A. 2012. Termite digestomes as sources for novel lignocellulases. Biofuels, Bioprod Biorefining 6:246–256. doi: 10.1002/bbb.107. [DOI] [Google Scholar]
- 16.Warnecke F, Luginbühl P, Ivanova N, Ghassemian M, Richardson TH, Stege JT, Cayouette M, McHardy AC, Djordjevic G, Aboushadi N, Sorek R, Tringe SG, Podar M, Martin HG, Kunin V, Dalevi D, Madejska J, Kirton E, Platt D, Szeto E, Salamov A, Barry K, Mikhailova N, Kyrpides NC, Matson EG, Ottesen EA, Zhang X, Hernández M, Murillo C, Acosta LG, Rigoutsos I, Tamayo G, Green BD, Chang C, Rubin EM, Mathur EJ, Robertson DE, Hugenholtz P, Leadbetter JR. 2007. Metagenomic and functional analysis of hindgut microbiota of a wood-feeding higher termite. Nature 450:560–565. doi: 10.1038/nature06269. [DOI] [PubMed] [Google Scholar]
- 17.Bastien G, Arnal G, Bozonnet S, Laguerre S, Ferreira F, Fauré R, Henrissat B, Lefèvre F, Robe P, Bouchez O, Noirot C, Dumon C, O'Donohue M. 2013. Mining for hemicellulases in the fungus-growing termite Pseudacanthotermes militaris using functional metagenomics. Biotechnol Biofuels 6:78. doi: 10.1186/1754-6834-6-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Otani S, Mikaelyan A, Nobre T, Hansen LH, Koné NA, Sørensen SJ, Aanen DK, Boomsma JJ, Brune A, Poulsen M. 2014. Identifying the core microbial community in the gut of fungus-growing termites. Mol Ecol 23:4631–4644. doi: 10.1111/mec.12874. [DOI] [PubMed] [Google Scholar]
- 19.Su L, Yang L, Huang S, Su X, Li Y, Wang F, Wang E, Kang N, Xu J, Song A. 2016. Comparative gut microbiomes of four species representing the higher and the lower termites. J Insect Sci 16:97. doi: 10.1093/jisesa/iew081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu N, Li H, Chevrette MG, Zhang L, Cao L, Zhou H, Zhou X, Zhou Z, Pope PB, Currie CR, Huang Y, Wang Q. 2019. Functional metagenomics reveals abundant polysaccharide-degrading gene clusters and cellobiose utilization pathways within gut microbiota of a wood-feeding higher termite. ISME J 13:104–117. doi: 10.1038/s41396-018-0255-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kudo T. 2009. Termite-microbe symbiotic system and its efficient degradation of lignocellulose. Biosci Biotechnol Biochem 73:2561–2567. doi: 10.1271/bbb.90304. [DOI] [PubMed] [Google Scholar]
- 22.Bignell DE. 2010. Morphology, physiology, biochemistry and functional design of the termite gut: an evolutionary wonderland, p 375–412. In Biology of termites: a modern synthesis. Springer Netherlands, Dordrecht, the Netherlands. [Google Scholar]
- 23.Bryant MP, Small N, Bouma C, Chu H. 1958. Bacteroides ruminicola n. sp. and Succinimonas amylolytica; the new genus and species; species of succinic acid-producing anaerobic bacteria of the bovine rumen. J Bacteriol 76:15–23. doi: 10.1128/JB.76.1.15-23.1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dehority BA. 1966. Characterization of several bovine rumen bacteria isolated with a xylan medium. J Bacteriol 91:1724–1729. doi: 10.1128/JB.91.5.1724-1729.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chassard C, Goumy V, Leclerc M, Del'homme C, Bernalier-Donadille A. 2007. Characterization of the xylan-degrading microbial community from human faeces. FEMS Microbiol Ecol 61:121–131. doi: 10.1111/j.1574-6941.2007.00314.x. [DOI] [PubMed] [Google Scholar]
- 26.Bolam DN, Koropatkin NM. 2012. Glycan recognition by the Bacteroidetes Sus-like systems. Curr Opin Struct Biol 22:563–569. doi: 10.1016/j.sbi.2012.06.006. [DOI] [PubMed] [Google Scholar]
- 27.Martens EC, Koropatkin NM, Smith TJ, Gordon JI. 2009. Complex glycan catabolism by the human gut microbiota: the bacteroidetes sus-like paradigm. J Biol Chem 284:24673–24677. doi: 10.1074/jbc.R109.022848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dodd D, Mackie RI, Cann IKO. 2011. Xylan degradation, a metabolic property shared by rumen and human colonic Bacteroidetes. Mol Microbiol 79:292–304. doi: 10.1111/j.1365-2958.2010.07473.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Flint HJ, Whitehead TR, Martin JC, Gasparic A. 1997. Interrupted catalytic domain structures in xylanases from two distantly elated strains of Prevotella ruminicola. Biochim Biophys Acta 1337:161–165. doi: 10.1016/S0167-4838(96)00213-0. [DOI] [PubMed] [Google Scholar]
- 30.Despres J, Forano E, Lepercq P, Comtet-Marre S, Jubelin G, Chambon C, Yeoman CJ, Berg Miller ME, Fields CJ, Martens E, Terrapon N, Henrissat B, White BA, Mosoni P. 2016. Xylan degradation by the human gut Bacteroides xylanisolvens XB1AT involves two distinct gene clusters that are linked at the transcriptional level. BMC Genomics 17:326. doi: 10.1186/s12864-016-2758-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miyazaki K, Hirase T, Kojima Y, Flint HJ. 2005. Medium- to large-sized xylo-oligosaccharides are responsible for xylanase induction in Prevotella bryantii B14. Microbiology 151:4121–4125. doi: 10.1099/mic.0.28270-0. [DOI] [PubMed] [Google Scholar]
- 32.Rogowski A, Briggs JA, Mortimer JC, Tryfona T, Terrapon N, Lowe EC, Baslé A, Morland C, Day AM, Zheng H, Rogers TE, Thompson P, Hawkins AR, Yadav MP, Henrissat B, Martens EC, Dupree P, Gilbert HJ, Bolam DN. 2015. Glycan complexity dictates microbial resource allocation in the large intestine. Nat Commun 6:1–15. doi: 10.1038/ncomms8481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang M, Chekan JR, Dodd D, Hong P-Y, Radlinski L, Revindran V, Nair SK, Mackie RI, Cann I. 2014. Xylan utilization in human gut commensal bacteria is orchestrated by unique modular organization of polysaccharide-degrading enzymes. Proc Natl Acad Sci U S A 111:E3708–E3717. doi: 10.1073/pnas.1406156111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu H. 2018. Characterizing xylan-degrading enzymes from a putative xylan utilization system derived from termite gut metagenome. PhD dissertation. INSA de Toulouse, Toulouse, France. [Google Scholar]
- 35.Boraston AB, Nurizzo D, Notenboom V, Ducros V, Rose DR, Kilburn DG, Davies GJ. 2002. Differential oligosaccharide recognition by evolutionarily-related β-1,4 and β-1,3 glucan-binding modules. J Mol Biol 319:1143–1156. doi: 10.1016/S0022-2836(02)00374-1. [DOI] [PubMed] [Google Scholar]
- 36.von Schantz L, Håkansson M, Logan DT, Walse B, Osterlin J, Nordberg-Karlsson E, Ohlin M. 2012. Structural basis for carbohydrate-binding specificity-A comparative assessment of two engineered carbohydrate-binding modules. Glycobiology 22:948–961. doi: 10.1093/glycob/cws063. [DOI] [PubMed] [Google Scholar]
- 37.Alahuhta M, Xu Q, Bomble YJ, Brunecky R, Adney WS, Ding SY, Himmel ME, Lunin VV. 2010. The unique binding mode of cellulosomal CBM4 from Clostridium thermocellum cellobiohydrolase A. J Mol Biol 402:374–387. doi: 10.1016/j.jmb.2010.07.028. [DOI] [PubMed] [Google Scholar]
- 38.Alahuhta M, Luo Y, Ding SY, Himmel ME, Lunin VV. 2011. Structure of CBM4 from Clostridium thermocellum cellulase K. Acta Crystallogr Sect F Struct Biol Cryst Commun 67:527–530. doi: 10.1107/S1744309111003307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kormos J, Johnson PE, Brun E, Tomme P, McIntosh LP, Haynes CA, Kilburn DG. 2000. Binding site analysis of cellulose binding domain CBD N1 from endoglucanse C of Cellulomonas fimi by site-directed mutagenesis. Biochemistry 39:8844–8852. doi: 10.1021/bi000607s. [DOI] [PubMed] [Google Scholar]
- 40.Fisher SZ, Von Schantz L, Hakansson M, Logan DT, Ohlin M. 2015. Neutron crystallographic studies reveal hydrogen bond and water-mediated interactions between a carbohydrate-binding module and its bound carbohydrate ligand. Biochemistry 54:6435–6438. doi: 10.1021/acs.biochem.5b01058. [DOI] [PubMed] [Google Scholar]
- 41.Pell G, Taylor EJ, MGloster MT, Turkenburg JP, Fontes MGAC, Ferreira MAL, Nagy T, Clark SJ, Davies JG, Gilbert JH, Gloster TM, Turkenburg JP, Fontes CA, Ferreira LA, Nagy T, Clark SJ, Davies GJ, Gilbert HJ. 2004. The mechanisms by which family 10 glycoside hydrolases bind decorated substrates. J Biol Chem 279:9597–9605. doi: 10.1074/jbc.M312278200. [DOI] [PubMed] [Google Scholar]
- 42.Han X, Gao J, Shang N, Huang CH, Ko TP, Chen CC, Chan HC, Cheng YS, Zhu Z, Wiegel J, Luo W, Guo RT, Ma Y, Sl-Ys JW, Han X, Gao J, Shang N, Huang CH, Ko TP, Chen CC, Chan HC, Cheng YS, Zhu Z, Wiegel J, Luo W, Guo RT, Ma Y. 2013. Structural and functional analyses of catalytic domain of GH10 xylanase from Thermoanaerobacterium saccharolyticum JW/SL-YS485. Proteins Struct Funct Bioinforma 81:1256–1265. doi: 10.1002/prot.24286. [DOI] [PubMed] [Google Scholar]
- 43.Pell G, Szabo L, Charnock SJ, Xie H, Gloster TM, Davies GJ, Gilbert HJ. 2004. Structural and biochemical analysis of Cellvibrio japonicus xylanase 10C: how variation in substrate-binding cleft influences the catalytic profile of family GH-10 xylanases. J Biol Chem 279:11777–11788. doi: 10.1074/jbc.M311947200. [DOI] [PubMed] [Google Scholar]
- 44.Terrapon N, Lombard V, Drula É, Lapébie P, Al-Masaudi S, Gilbert HJ, Henrissat B. 2018. PULDB: the expanded database of polysaccharide utilization loci. Nucleic Acids Res 46:D677–D683. doi: 10.1093/nar/gkx1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Comino P, Collins H, Lahnstein J, Beahan C, Gidley MJ. 2014. Characterisation of soluble and insoluble cell wall fractions from rye, wheat and hull-less barley endosperm flours. Food Hydrocoll 41:219–226. doi: 10.1016/j.foodhyd.2014.04.005. [DOI] [Google Scholar]
- 46.Dodd D, Moon Y-HH, Swaminathan K, Mackie RI, Cann IKO. 2010. Transcriptomic analyses of xylan degradation by Prevotella bryantii and insights into energy acquisition by xylanolytic bacteroidetes. J Biol Chem 285:30261–30273. doi: 10.1074/jbc.M110.141788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Petersen TN, Brunak S, von Heijne G, Nielsen H. 2011. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 48.Shipman JA, Cho KH, Siegel HA, Salyers AA. 1999. Physiological characterization of SusG, an outer membrane protein essential for starch utilization by Bacteroides thetaiotaomicron. J Bacteriol 181:7206–7211. doi: 10.1128/JB.181.23.7206-7211.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chakdar H, Kumar M, Pandiyan K, Singh A, Nanjappan K, Kashyap PL, Srivastava AK. 2016. Bacterial xylanases: biology to biotechnology. 3 Biotech 6:1–15. doi: 10.1007/s13205-016-0457-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang K, Pereira GV, Cavalcante JJV, Zhang M, Mackie R, Cann I. 2016. Bacteroides intestinalis DSM 17393, a member of the human colonic microbiome, upregulates multiple endoxylanases during growth on xylan. Sci Rep 6:34360–34311. doi: 10.1038/srep34360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bågenholm V, Reddy SK, Bouraoui H, Morrill J, Kulcinskaja E, Bahr CM, Aurelius O, Rogers T, Xiao Y, Logan DT, Martens EC, Koropatkin NM, Stålbrand H. 2017. Galactomannan catabolism conferred by a polysaccharide utilisation locus of Bacteroides ovatus: enzyme synergy and crystal structure of a β-mannanase. J Biol Chem 292:229–243. doi: 10.1074/jbc.M116.746438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Martens EC, Kelly AG, Tauzin AS, Brumer H. 2014. The devil lies in the details: how variations in polysaccharide fine-structure impact the physiology and evolution of gut microbes. J Mol Biol 426:3851–3865. doi: 10.1016/j.jmb.2014.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Charnock SJ, Spurway TD, Xie H, Beylot MH, Virden R, Warren RA, Hazlewood GP, Gilbert HJ. 1998. The topology of the substrate binding clefts of glycosyl hydrolase family 10 xylanases are not conserved. J Biol Chem 273:32187–32199. doi: 10.1074/jbc.273.48.32187. [DOI] [PubMed] [Google Scholar]
- 54.Koropatkin NM, Smith TJ. 2010. SusG: a unique cell-membrane-associated alpha-amylase from a prominent human gut symbiont targets complex starch molecules. Structure 18:200–215. doi: 10.1016/j.str.2009.12.010. [DOI] [PubMed] [Google Scholar]
- 55.Khosla C, Harbury PB. 2001. Modular enzymes. Nature 409:247–252. doi: 10.1038/35051723. [DOI] [PubMed] [Google Scholar]
- 56.Martinez T, Texier H, Nahoum V, Lafitte C, Cioci G, Heux L, Dumas B, O’Donohue M, Gaulin E, Dumon C. 2015. Probing the functions of carbohydrate binding modules in the cbel protein from the oomycete phytophthora parasitica. PLoS One 10:e0137481. doi: 10.1371/journal.pone.0137481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Robert X, Gouet P. 2014. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res 42:320–324. doi: 10.1093/nar/gku316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Huang Y, Niu B, Gao Y, Fu L, Li W. 2010. CD-HIT Suite: a web server for clustering and comparing biological sequences. Bioinformatics 26:680–682. doi: 10.1093/bioinformatics/btq003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zallot R, Oberg N, Gerlt JA. 2019. The EFI web resource for genomic enzymology tools: leveraging protein, genome, and metagenome databases to discover novel enzymes and metabolic pathways. Biochemistry 58:4169–4182. doi: 10.1021/acs.biochem.9b00735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. 2003. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vincentelli R, Romier C. 2013. Expression in Escherichia coli: becoming faster and more complex. Curr Opin Struct Biol 23:326–334. doi: 10.1016/j.sbi.2013.01.006. [DOI] [PubMed] [Google Scholar]
- 62.Studier FW. 2005. Protein production by auto-induction in high-density shaking cultures. Protein Expr Purif 41:207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 63.Gasteiger E, Hoogland C, Gattiker A, Duvaud S, Wilkins MR, Appel RD, Bairoch A. 2005. Protein identification and analysis tools on the ExPASy server, p 571–607. In The proteomics protocols handbook. Humana Press, Totowa, NJ. [Google Scholar]
- 64.Miller GL. 1959. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal Chem 31:426–428. doi: 10.1021/ac60147a030. [DOI] [Google Scholar]
- 65.Arnal G, Bastien G, Monties N, Abot A, Anton Leberre V, Bozonnet S, O'Donohue M, Dumon C. 2015. Investigating the function of an arabinan utilization locus isolated from a termite gut community. Appl Environ Microbiol 81:31–39. doi: 10.1128/AEM.02257-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Matsui I, Ishikawa K, Matsui E, Miyairi S, Fukui S, Honda K. 1991. Subsite structure of Saccharomycopsis alpha-amylase secreted from Saccharomyces cerevisiae. J Biochem 109:566–569. doi: 10.1093/oxfordjournals.jbchem.a123420. [DOI] [PubMed] [Google Scholar]
- 67.Song L, Dumon C, Siguier B, André I, Eneyskaya E, Kulminskaya A, Bozonnet S, O'Donohue MJ. 2014. Impact of an N-terminal extension on the stability and activity of the GH11 xylanase from Thermobacillus xylanilyticus. J Biotechnol 174:64–72. doi: 10.1016/j.jbiotec.2014.01.004. [DOI] [PubMed] [Google Scholar]
- 68.Takeo K. 1984. Affinity electrophoresis: principles and applications. Electrophoresis 5:187–195. doi: 10.1002/elps.1150050402. [DOI] [Google Scholar]
- 69.Wu H, Montanier CY, Dumon C. 2017. Quantifying CBM carbohydrate interactions using microscale thermophoresis, p 129–141. In Methods in molecular biology. Humana Press, New York, NY. [DOI] [PubMed] [Google Scholar]
- 70.Jerabek-Willemsen M, André T, Wanner R, Roth HM, Duhr S, Baaske P, Breitsprecher D. 2014. MicroScale thermophoresis: interaction analysis and beyond. J Mol Struct 1077:101–113. doi: 10.1016/j.molstruc.2014.03.009. [DOI] [Google Scholar]
- 71.Martens EC, Lowe EC, Chiang H, Pudlo NA, Wu M, McNulty NP, Abbott DW, Henrissat B, Gilbert HJ, Bolam DN, Gordon JI. 2011. Recognition and degradation of plant cell wall polysaccharides by two human gut symbionts. PLoS Biol 9:e1001221. doi: 10.1371/journal.pbio.1001221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.McNulty NP, Wu M, Erickson AR, Pan C, Erickson BK, Martens EC, Pudlo NA, Muegge BD, Henrissat B, Hettich RL, Gordon JI. 2013. Effects of diet on resource utilization by a model human gut microbiota containing Bacteroides cellulosilyticus WH2, a symbiont with an extensive glycobiome. PLoS Biol 11:e1001637. doi: 10.1371/journal.pbio.1001637. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.