Alphabaculoviruses are used as biological insecticides and expression vectors in biotechnology and medical applications. We demonstrate that in caterpillars infected with particular mixtures of viruses, the genomes of different baculovirus species can be enveloped together within individual virions and occluded within proteinaceous occlusion bodies.
KEYWORDS: alphabaculovirus, Baculoviridae, multinucleocapsid virions, transmission of virus mixtures, host range, plaque purification, qPCR, AcMNPV, SfMNPV, SeMNPV, coenvelopment, transmission
ABSTRACT
Alphabaculoviruses (Baculoviridae) are pathogenic DNA viruses of Lepidoptera that have applications as the basis for biological insecticides and expression vectors in biotechnological processes. These viruses have a characteristic physical structure that facilitates the transmission of groups of genomes. We demonstrate that coinfection of a susceptible insect by two different alphabaculovirus species results in the production of mixed-virus occlusion bodies containing the parental viruses. This occurred between closely related and phylogenetically more distant alphabaculoviruses. Approximately half the virions present in proteinaceous viral occlusion bodies produced following coinfection of insects with a mixture of two alphabaculoviruses contained both viruses, indicating that the viruses coinfected and replicated in a single cell and were coenveloped within the same virion. This observation was confirmed by endpoint dilution assay. Moreover, both viruses persisted in the mixed-virus population by coinfection of insects during several rounds of insect-to-insect transmission. Coinfection by viruses that differed in genome size had unexpected results on the length of viral nucleocapsids, which differed from those of both parental viruses. These results have unique implications for the development of alphabaculoviruses as biological control agents of insect pests.
IMPORTANCE Alphabaculoviruses are used as biological insecticides and expression vectors in biotechnology and medical applications. We demonstrate that in caterpillars infected with particular mixtures of viruses, the genomes of different baculovirus species can be enveloped together within individual virions and occluded within proteinaceous occlusion bodies. This results in the transmission of mixed-virus populations to the caterpillar stages of moth species. Once established, mixed-virus populations persist by coinfection of insect cells during several rounds of insect-to-insect transmission. Mixed-virus production technology opens the way to the development of custom-designed insecticides for control of different combinations of caterpillar pest species.
INTRODUCTION
Interactions among viruses are pervasive and highly diverse and have the potential to modulate key aspects of virus phenotype, including virulence (1–3). Within-host interactions include complementation, superinfection exclusion, and the production of pseudotypes from different viruses replicating in a shared cell, whereas interactions that occur during virus dispersal and transmission to a new host involve the physical association of viruses in groups that can be considered multivirion infectious units (4). Examples of multivirion infectious units include aggregates of virions that are formed following release from the host cell (5, 6), whereas in other cases, particles are released as multivirion structures such as extracellular vesicles (7, 8) or proteinaceous occlusion bodies (9). The production of groups of virions for dispersal and transmission is costly but may be offset by fitness advantages such as increased infectivity (through reduced dilution and high multiplicity of infection), reduced exposure to host immune responses, increased stability, and improved persistence outside the host (10, 11).
The genetic diversity and relatedness among coinfecting variants can influence key aspects of virus fitness, including virulence, opportunities for recombination, competition among genotypes, and the evolution of cheater variants such as defective interfering particles that survive through complementation with complete genotypes (12, 13). Different viruses that coinfect and replicate within a shared host cell will have low relatedness and may be expected to compete for host resources, although cooperation may evolve if that results in higher fitness for each of the viruses involved (3, 14).
Alphabaculoviruses (Baculoviridae, genus Alphabaculovirus) that infect Lepidoptera are an unusual group of double-stranded DNA (dsDNA) viruses with a circular genome of 80 to 180 kbp (15). Alphabaculoviruses produce two types of collective infectious units. Individual nucleocapsids, each containing a single genome, are enveloped singly or in groups (typically clusters of 2 to 8 nucleocapsids), to form occlusion-derived virions (ODVs). Groups of ODVs are then occluded within a matrix of polyhedrin protein and wrapped by a polyhedron envelope protein to form resistant occlusion bodies (OBs) that can persist in the environment for extended periods (16).
Viral OBs are released from virus-killed insects for dispersal over plant surfaces (17). When OB-contaminated foliage is consumed by a caterpillar, the OB dissolves in the alkaline environment of the insect midgut, releasing ODVs that infect midgut epithelial cells and subsequently disperse to other tissues within the infected insect (18). Alphabaculovirus infections are genetically heterogeneous (19, 20), and each cell within a caterpillar is usually infected by several budded virions, each of which delivers a single genome to the host cell (21). As a result, OBs contain multiple variants that reflect the genetic composition of the host cell in which they were produced (22). OBs also ensure that multiple virions are delivered simultaneously to susceptible insects, thus increasing the likelihood of transmission, whereas multiple-nucleocapsid ODVs increase the likelihood that individual cells are infected by multiple variants during the initial establishment of infection in the caterpillar gut.
As natural agents of mortality in agricultural and forest insect pests, alphabaculoviruses are used as the basis for potent biological insecticides in many countries (23). Their favorable biosecurity profile and amenability to genetic manipulation have also favored their use as expression vectors (24), in the production of vaccines (25), and as potential gene therapy vectors (26). Our previous studies have demonstrated that the construction of specific mixtures of genotypic variants can markedly improve the insecticidal characteristics of these pathogens, in terms of their ability to cause lethal disease in insect pests (22, 27–29) and to increase the number of OBs produced in lethally infected insects (30, 31). The key to this technology is that the multivirion infectious units (OBs) favor transmission of the genotypic diversity present in the inoculum.
The host range of these viruses affects their usefulness in pest control and their ecological risk profile (32–34). Some alphabaculovirus species, such as Autographa californica multiple nucleopolyhedrovirus (AcMNPV), are able to infect over 30 different species of Lepidoptera (35), whereas the majority of these viruses are highly host specific (33, 36). Similarly, insect species differ in their susceptibilities to a given virus and are often classified as permissive, semipermissive, or nonpermissive depending on the quantity of inoculum required to initiate a productive infection. Multiple infections by different alphabaculoviruses have been studied in vitro (37, 38), although the implications of coinfection on virus structure and transmission are largely unknown.
In the present study, we examined the consequences of coinfection by distinct alphabaculovirus species on the genomic composition of virions, virion formation, and species persistence in a mixed-virus population. This issue was addressed from different perspectives. First, we investigated the effect of coinfection by two phylogenetically distant viruses on the composition of the virions and OBs produced in individual host insects. Second, we studied the size of nucleocapsids enveloped in virions produced after coinfection. Finally, we examined evidence for heterotypic cooperation between two species of alphabaculoviruses that are closely related phylogenetically. The results offer potential insights into the evolution and maintenance of diversity of these viruses. More importantly, these results open the way to technologies for the development of novel bioinsecticides based on multivirion mixtures of alphabaculoviruses.
RESULTS
Production of mixed-virus occlusion bodies.
The viruses used in the present study were Autographa californica multiple nucleopolyhedrovirus (AcMNPV), Mamestra brassicae multiple nucleopolyhedrovirus (MbMNPV), Spodoptera exigua multiple nucleopolyhedrovirus (SeMNPV), and Spodoptera frugiperda multiple nucleopolyhedrovirus (SfMNPV), all of which have the multiple-nucleocapsid phenotype of occlusion-derived virions (ODVs). Four different mixed-virus OBs were produced to achieve the different objectives of this study: (i) AcMNPV+SfMNPV, (ii) AcMNPV+MbMNPV, (iii) SfMNPV+MbMNPV, and (iv) SfMNPV+SeMNPV.
First, AcMNPV genotype C6 (AcC6) and a complete orally infectious genotype of SfMNPV (SfB) were chosen because they replicate well in the Sf9 cell line and share a permissive host, Spodoptera frugiperda. Peroral inoculation of S. frugiperda fourth-stage caterpillars with a 1:1 mixture of OBs of AcC6 and SfB resulted in 97.2% ± 2.0% (mean ± standard deviation [SD]) mortality of insects. SfB was the majority virus in the progeny OBs and comprised 99.95% ± 0.01% of the genomes present in the mixed-virus preparation, as determined by quantitative PCR (qPCR) (Table 1). This result was consistent across a series of preliminary studies in which we were able to modify the proportions of each virus in progeny OBs, either by altering their ratio in the inoculum or by altering the interval between inoculation of one virus and the other (data not shown), thereby precluding the possibility that the low-level detection of AcC6 was due to a contamination event.
TABLE 1.
Composition of mixed-virus preparations determined by qPCR quantification of virus genomes
| Virus mixture | Composition, % (mean ± SD) |
|
|---|---|---|
| Component 1 | Component 2 | |
| AcMNPV + SfMNPV | 0.05 ± 0.01 AcC6 | 99.95 ± 0.01 SfB |
| AcMNPV + MbMNPV | 34.9 ± 3.6 AcC6 | 65.1 ± 3.6 Mb |
| SfMNPV + MbMNPV | 53.4 ± 2.3 SfB | 46.7 ± 2.3 Mb |
| SeMNPV + SfMNPV | 95.1 ± 1.3 SeA | 4.9 ± 1.3 SfC |
Second, by inoculating fourth-stage S. frugiperda larvae we produced two different mixed-virus preparations comprising viruses that differed in the lengths of their genomes. The AcC6+MbMNPV (Mb) mixed-virus preparation comprised 34.9% ± 3.6% AcC6 genomes, whereas the SfB+Mb mixed-virus preparation comprised 53.4% ± 2.3% SfB genomes, as determined by qPCR (Table 1).
Third, a mixture of two closely related viruses, SeMNPV and SfMNPV, was produced in a permissive host, Spodoptera exigua. For this, fourth-stage S. exigua larvae were injected with a suspension of ODVs comprising a 1:1 mixture of these viruses. The inoculum comprised a complete genotype of SeMNPV-US2A (SeA) and a defective genotype, SfMNPV-C (SfC), that was incapable of peroral infection because it lacked per os infection factors (PIFs). qPCR analysis indicated that 95.1% ± 1.3% (mean ± SD) of the genomes in the resulting mixed-virus OBs (named passage zero [P0] OBs) were of genotype SeA (Table 1).
Coenvelopment of AcC6 and SfB OBs the same virion.
The composition of OBs produced following coinoculation of S. frugiperda larvae with a 1:1 mixture of AcC6 OBs and SfB OBs was determined. A mean (±SD) of 87.3 ± 12.7 individual plaques were analyzed in each replicate by PCR using virus-specific primers (Table 2). Across the three replicates, the proportion of plaques that were positive for AcC6 varied from 1 to 8%, whereas 32.3 to 53.9% of plaques were positive for SfB and 43.9 to 59.7% of plaques were positive for both viruses (Fig. 1; see also Table S1 in the supplemental material). A procedure was performed to control for adhesion between different ODVs or cross-contamination during the plaque purification procedure. The control comprised ODVs released from a mixture of pure AcC6 OBs and pure SfB OBs in a ratio of ∼1:1,595, similar to that detected by qPCR on AcC6+SfB OBs. PCR analysis of control plaques indicated that just 10% of plaques (3 out of 30 plaques) were positive for both viruses, which was significantly lower than the prevalence of mixed-virus plaques (43.9 to 59.7%) in any of the three replicates (Fisher’s exact test, P < 0.001 in all cases) or the sum of the three replicates (Fisher’s exact test, P = 0.000098). As the proportion of plaques comprising both viruses in the mixed-virus OBs was much larger than in the control mixture of ODVs, it is likely that mixed-virus plaques originated from mixed-virus ODVs, in which nucleocapsids of both viruses were coenveloped in the same virion. This was further analyzed probabilistically by endpoint dilution assay.
TABLE 2.
Primers used for PCR and qPCR amplification in this study
| Primer | Sequencea | Amplification purpose | Nucleotide positions of the primer on the target genome | Expected product length (bp) |
|---|---|---|---|---|
| Seie0.F | 5′-CTATAGCTCGACGCTCGGTG-3′ | SeA detection (PCR) | 131937–131959 | 510 |
| Seie0.R | 5′-ATCGTCTTCGATACCGCGAG-3′ | SeA detection (PCR | 132447–132428 | |
| Sfie0.F | 5′-ATGAGTATTAATCATGATTC-3′ | SfC detection (PCR) | 129612–129630 | 535 |
| Sfie0.R | 5′-TCTTGGCAAATGTTACACT-3′ | SfC detection (PCR) | 130147–130128 | |
| qSe5.F | 5′-AGCAGCGAGCCAATGCAGTA-3′ | SeA DNA quantification (qPCR) | 6274–6293 | 99 |
| qSe5.R | 5′-CTTCTTGCAACCGCTCGTTC-3′ | SeA DNA quantification (qPCR) | 6354–6373 | |
| qSfCcath.F | 5′-ACGCCGCGTTTAGTAACAGC-3′ | SfC DNA quantification (qPCR) | 18717–18736b | 100 |
| qSfCsf36.R | 5′-TAAAACTATTTCTTGCAATC-3′ | SfC DNA quantification (qPCR) | 35185–35166b | |
| Ac.F | 5′-GATTTGTTGGCCGAATAACG-3′ | AcC6 DNA quantification (qPCR) | 84850–84869 | 108 |
| Ac.R | 5′-TGACTCTTTCACCCATTGCAG-3′ | AcC6 DNA quantification (qPCR) | 84958–84938 | |
| Sf.F | 5′-ACGCCGTTCAAAGACACGAG-3′ | SfB DNA quantification (qPCR) | 42832–42851 | 144 |
| Sf.R | 5′-CCGCTTTGCCTTCGACATAG-3′ | SfB DNA quantification (qPCR) | 42976–42957 | |
| AcDNApol.F | 5′-CAAATGTAGAATCTGTGTCG-3′ | AcC6 detection in plaques (PCR) | 53264–53283 | 704 |
| AcDNApol.R | 5′-CAGCCATCACAAACACGCGC-3′ | AcC6 detection in plaques (PCR) | 53968–53949 | |
| SfDNApol.F | 5′-CAACGACATCAATAGAGTGC-3′ | SfB detection in plaques (PCR) | 88358–88377 | 964 |
| SfDNApol.R | 5′-AAATATTGCTAAGCACATCG-3′ | SfB detection in plaques (PCR) | 89322–89303 |
Annealing temperatures were 50°C for all PCR and 60°C for all qPCRs.
Position of primers on the SfB genome that is the genotypic variant that has no deletions.
FIG 1.
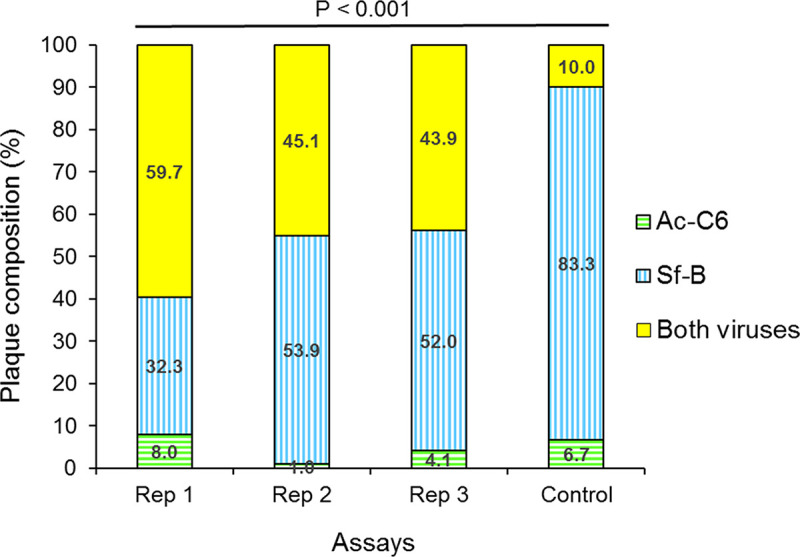
Composition of occlusion-derived virions produced in insects with mixed-virus infections. The composition of plaques derived from ODVs released from AcC6+SfB mixed-virus occlusion bodies (three replicates) was determined by PCR using virus-specific primers. The control consisted of ODVs released from a mixture of pure AcC6 OBs and pure SfB OBs in a ratio of ∼1:1,595 (similar to the relative prevalence of genomes detected by qPCR on AcC6+SfB mixed-virus OBs), to control for adhesion between different ODVs or cross-contamination during the plaque purification procedure. P value indicates the result of Fisher’s exact test.
Probabilistic support for coenvelopment of AcC6 and SfB in the same virion.
The presence of AcC6 and SfB genomes in mixed-virus ODVs was examined probabilistically in an endpoint dilution experiment. The probability with which wells were not inoculated with an infective dose of ODV in an endpoint dilution assay can be calculated according to the Poisson distribution (Table 3). A dilution (1/1,250) was selected that resulted in approximately 90% uninfected wells, which reflects a situation in which a single ODV will be responsible for initiating an infection in approximately 10% of the wells and two or more ODVs will be responsible for infection in approximately 1% of wells. All the wells that were infected at the selected dilution (10% infected wells and 90% uninfected wells) were analyzed by PCR. In all cases, the presence of both viruses was detected by PCR (Fig. 2), indicating the presence of both viruses within the single ODV that likely initiated the infection.
TABLE 3.
Poisson distribution-based probabilities of infection by 0, 1, 2 or 3 ODVs in wells by endpoint dilutiona
| Parameter | Value for: |
||
|---|---|---|---|
| Replicate 1 | Replicate 2 | Replicate 3 | |
| Positive wells/total wells | 12/80 | 11/90 | 4/86 |
| Probability 0 ODVs | 0.8500 | 0.8778 | 0.9535 |
| Probability 1 ODV | 0.1381 | 0.1144 | 0.0454 |
| Probability 2 ODVs | 0.0112 | 0.0075 | 0.0011 |
| Probability 3 ODVs | 0.0006 | 0.0003 | 0.0000 |
Positive wells were those that contained at least one cell with pathological signs of virus infection (OBs present in the nucleus). Probability 0 is the calculated probability of a well without any signs of infection. Probability 1 to probability 3 refer to the Poisson distribution probability of the cells in a particular well being infected by one, two, or three ODVs, respectively.
FIG 2.
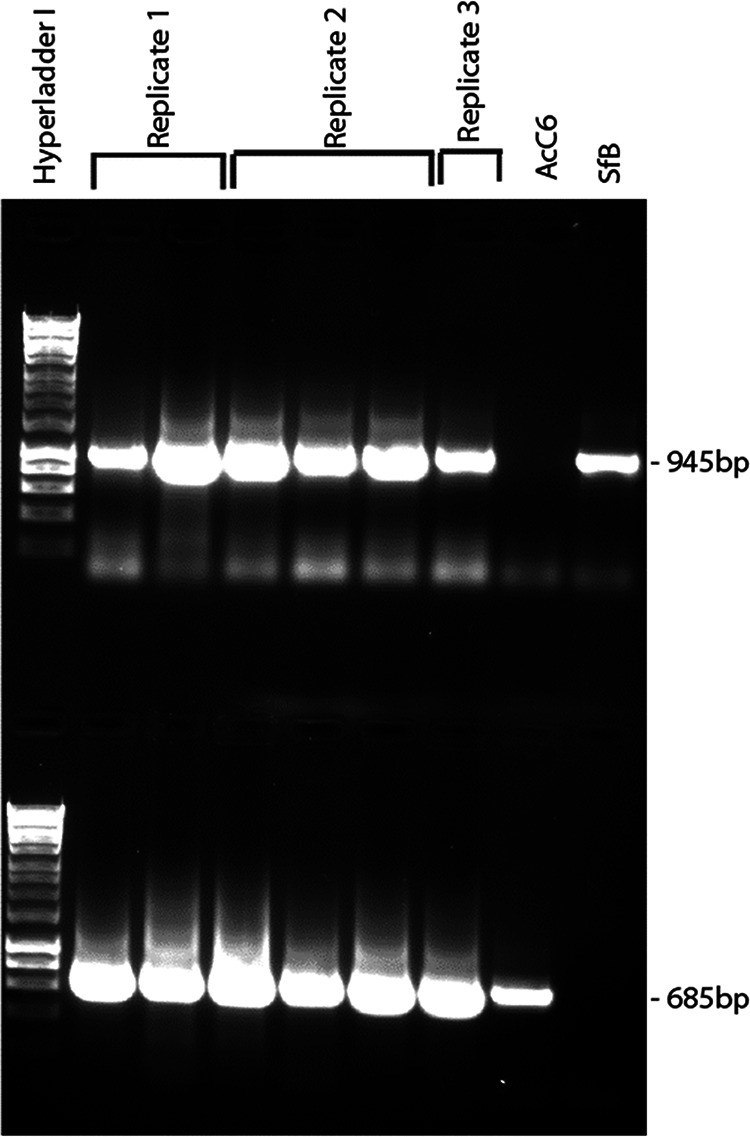
Frequency of single- and mixed-virus ODVs estimated by endpoint dilution. Shown are the results of PCR analysis of the presence of each or both viruses (AcC6 and SfB viruses) in Sf9 cells from virus-infected wells inoculated with a 1/1,250 dilution of ODV suspension. In all cases, infection of wells was likely to have originated from a single ODV (Table 2). Three independent replicate assays were performed. The final two lanes indicate positive controls for AcC6 and SfB genomic DNA. Upper lanes indicate amplifications using SfB-specific primers; lower lanes indicate amplifications using AcC6-specific primers (Table S1). Hyperladder I indicates molecular size marker (Bioline).
Electron microscope analysis of nucleocapsid length.
Nucleocapsids in multinucleocapsid ODVs, each comprising a minimum of two nucleocapsids (total = 68), were negatively stained and measured by transmission electron microscopy (TEM) using image analysis software (Fig. S1). Nucleocapsid lengths differed significantly among the five groups of nucleocapsids examined (F = 50.01; df = 4, 63; P < 0.001) (Fig. 3). The length of nucleocapsids reflected the size of the viral genome with the longest nucleocapsids (mean length ± SD: 387 ± 20 nm) in MbMNPV (153-kb genome), the shortest nucleocapsids (315 ± 9 nm) in SfMNPV variant B (∼131-kb genome), and intermediate length nucleocapsids (368 ± 17 nm) in the AcMNPV C6 variant (∼134-kb genome) (Fig. 3). However, nucleocapsids in multinucleocapsid ODVs obtained after mixed infection differed in length from nucleocapsids of the parental viruses. In mixed-virus OBs comprising AcC6 and Mb, nucleocapsids were significantly shorter than either of the parental viruses, whereas in OBs comprising SfB and Mb viruses, nucleocapsid length was intermediate between those of the parental viruses and statistically similar to the length of nucleocapsids produced in AcC6+Mb virus infections (Fig. 3).
FIG 3.
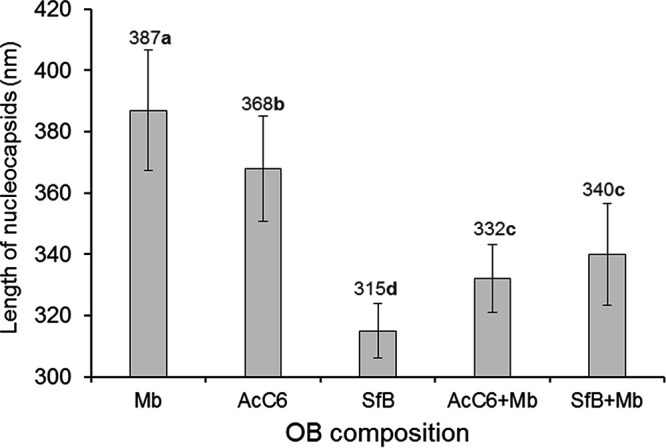
Mean length of nucleocapsids from multinucleocapsid ODVs released from individual and mixed-virus OBs. Nucleocapsids were visualized by negative staining. Vertical bars indicate SDs. Values above columns indicate mean values. Means followed by identical letters did not differ significantly (ANOVA, Tukey’s test, P > 0.05).
Peroral infectivity of SeA+SfC mixed-virus occlusion bodies.
To produce SeA+SfC mixed-virus OBs, S. exigua larvae were injected with an ODV suspension comprising SeA and SfC viruses (1:1). The resulting mixed-virus OBs (P0 inoculum, comprising 95.1% SeA genomes and 4.9% SfC genomes [Table 1]) were used to inoculate S. frugiperda second-instar larvae at two concentrations (107 and 109 OBs/ml). Of the inoculated insects, 28.0% ± 4.0% and 46.7% ± 3.2%, respectively, died of polyhedrosis disease (Fig. 4). In contrast, pure preparations of each of the parental virus OBs were not capable of infecting and killing S. frugiperda larvae, due to the absence of PIF-1 and PIF-2 in the ODV envelope in the case of SfC virus, or due to high host specificity in the case of SeA virus (Fig. 4). PCR analysis and restriction endonuclease (PstI) analysis of the OBs produced in SeA+SfC mixed-virus treated insects revealed that only SfC virus was present and SeA virus was not present (Fig. 5; Fig. S2). The P1 OBs were not infectious to S. frugiperda second instars, none of which died following peroral inoculation with 109 OBs/ml. None of the mock-infected control insects died from polyhedrosis disease.
FIG 4.

Virus-induced mortality in second-instar Spodoptera frugiperda larvae inoculated with 109 occlusion bodies (OBs)/ml of SeMNPV (SeA) or SfMNPV defective C genotype (SfC) virus and 107 or 109 OBs/ml of SeA+SfC mixed-virus inoculum. SeA does not replicate in S. frugiperda, and SfC cannot infect larvae per os due to an absence of the pif-1 and pif-2 genes (resulting in 0% mortality in both cases), but the coinfection of S. exigua larvae with both viruses allowed SfC to use PIFs produced by SeA in coinfected cells for successful peroral infection of S. frugiperda larvae, resulting in lethal polyhedrosis disease in a fraction of inoculated larvae. Vertical bars indicate SDs (n = 3). Values above columns indicate mean percentage of mortality.
FIG 5.
PCR amplification of SfC and SeA genomic DNA from initial inocula and mixed-virus OBs. Lanes 1 to 6 on the left indicate results of amplification using SfC-specific primers, whereas lanes 1 to 6 on the right indicate the results of amplification using SeA-specific primers (Table 2). The amplification products of SeA (lane 1) and SfC (lane 2) viruses are shown, as are the amplification products (lanes 3 and 5) of DNAs from OBs (P0) produced in S. exigua larvae coinoculated with a mixture of SeA and SfC ODVs. Also shown are amplification products (lanes 4 and 6) of DNAs extracted from OBs (P1) produced following inoculation of S. frugiperda larvae with P0 OBs that had been produced in S. exigua. Lanes 3 and 4 correspond to replicate 1, whereas lanes 5 and 6 correspond to replicate 2 of this experiment. Specific amplification of SfC DNA (535 bp) was observed in all samples except for SeA genomic DNA. Specific amplification of SeA DNA (510 bp) was observed only for SeA genomic DNA (control, lane 1) and OBs from coinfected S. exigua larvae (lanes 3 and 5), indicating that this virus was eliminated following infection in S. frugiperda larvae. Lane M indicates the molecular size marker (MWD1; Nippon Genetics).
These results demonstrate that the SfC genotype was capable of utilizing the PIF-1 and PIF-2 proteins produced by SeA virus to initiate primary infection in the midgut cells of perorally inoculated S. frugiperda larvae treated with P0 OBs. For this, both viruses must have replicated simultaneously in coinfected cells in the ODV-injected S. exigua larvae. In contrast, P1 OBs produced in S. frugiperda and comprising the SfC genotype alone lacked peroral infectivity, precluding a hypothetical recombination event in which a SfC genotype had acquired pif-1 and pif-2 genes from SeA virus.
Persistence of SfC virus in SeA+SfC mixed-virus population during serial passage.
A peroral serial passage experiment was performed in S. exigua larvae using SeA+SfC P0 OBs as the initial inoculum. The percentage of SfC genomic DNA extracted from OBs collected from insects that died at passages 1 to 6 (P1 to P6) was quantified by qPCR and declined progressively, with mean (±SD) values of 1.17% ± 0.45% at P1 to 0.010% ± 0.004% at P6 (Fig. 6). The persistence of SfC virus at low prevalence in OB progeny across passages would be possible only if nucleocapsids containing a genome of SfC were transmitted in ODVs that had PIF-1 and PIF-2 proteins produced by SeA virus in coinfected cells. The consistent decrease in the prevalence of SfC virus in the mixed-virus population likely reflects the higher efficiency with which SeA virus replicates in S. exigua than that of SfC virus.
FIG 6.
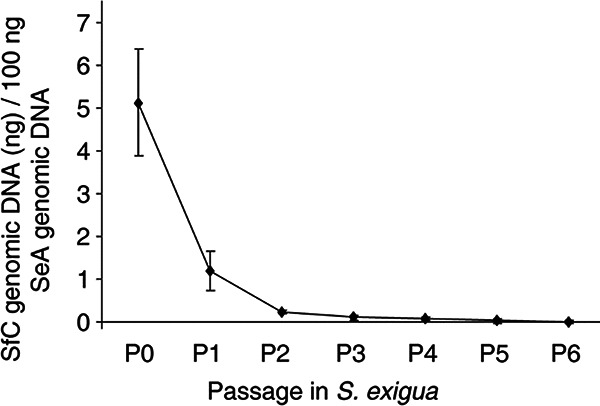
Persistence of defective virus (SfC) in mixtures with a homologous virus (SeA) during serial passage (P0 to P6) of mixed-virus OBs in Spodoptera exigua larvae. The presence of SfC was quantified by qPCR. The P0 initial inoculum was generated by injection of ODVs released from mixtures of SeA and SfC OBs (1:1 ratio) into S exigua larvae that subsequently died from virus disease. Error bars indicate SDs (n = 3).
DISCUSSION
We demonstrated that under specific experimental conditions, the genetic diversity in ODVs and OBs can extend beyond genotypic diversity to include interspecific diversity across closely related and more distantly related species of alphabaculoviruses. Although the evolutionary implications of coenvelopment and cotransmission of different alphabaculovirus species remain uncertain, this phenomenon provides a potential opportunity for the development of novel biological insecticides targeted at specific combinations of caterpillar pest species attacking particular crops.
To acquire evidence for coenvelopment of mixtures of viruses within ODVs, the plaque purification technique was applied, which has been universally adopted as a standard method for the isolation of individual infective units in a complex viral population (39, 40). A genotype of AcMNPV (AcC6) (41) and a complete orally infectious genotype of SfMNPV (SfB) (42) were selected for this experiment, as both viruses lethally infect S. frugiperda cells and larvae. Following coinfection of S. frugiperda larvae, the presence of both viruses within the same ODV in the progeny OBs was demonstrated by plaque purification. The presence of two phylogenetically distant alphabaculoviruses in the same ODV demonstrated that both viruses had replicated simultaneously in the same cell and their nucleocapsids had been coenveloped in the same ODV. Given their proximity in the cell nucleus, mixed-virus ODVs would inevitably be co-occluded in the same OB. Previous studies performed with these virus species in Sf9 cells indicated that both viruses were able to coinfect the same cell, but SfMNPV-infected cells were more prevalent when both viruses were coinoculated simultaneously in equal multiplicities of infection (MOIs) and in high concentrations (37). The results of cell culture-based studies largely depend on the susceptibility of a given cell line to each type of virus. We observed a high prevalence of plaques containing AcMNPV compared with the prevalence of this virus in the initial inoculum. This contrasts with our findings in a previous study (37) and was likely due to (i) the higher infectivity of BV inoculum used in the previous study compared to ODVs in the present study, as BVs are markedly more infectious than ODVs in cell culture (40), (ii) the high MOI used in the previous study, and (iii) the high infectivity and rapid replication of AcMNPV in S. frugiperda cells compared to those of SfMNPV, which tends to replicate more slowly and reach lower titers than AcMNPV in cell culture systems (43, 44). Nonetheless, although it was not possible to quantify the prevalence of coenveloped viruses in ODVs or the proportion of ODVs that comprised both viruses in the P0 population, the presence of both viruses in individual plaques leads us to conclude that both viruses were present in a significant fraction of the multinucleocapsid ODVs.
An alternative explanation would be that SfMNPV-only and AcMNPV-only ODVs may adhere to one another at a very high prevalence and act as a single infectious unit. We performed a control assay in which the proportion of plaques involving both viruses was 10.0%, much lower than the 43.9 to 59.7% mixed plaques obtained with mixed-virus OBs. Furthermore, we considered that a high prevalence of ODV-ODV adhesion was highly unlikely, as all plaque purification procedures performed to date using baculoviruses would be invalidated by frequent adhesion between ODVs.
Due to the high impact of the results, we corroborated the coenvelopment of viruses by adopting a statistical approach to the results of endpoint dilution assays in Sf9 cells (44). Both viruses were found to be present in wells in which the probability of being infected by a single ODV was approximately 10% and the probability of being infected by two or more ODVs was approximately 1%, a situation in which probabilistic theory (based on the Poisson distribution) requires that both viruses were present in each of the individual ODVs that initiated these infections.
It had been suggested that the length of capsids may be linked to genome size (45). When we produced mixed-virus OBs including MbMNPV, which has a larger genome than AcMNPV and SfMNPV, we expected to find nucleocapsids of different sizes within virions released from mixed-virus OBs. Surprisingly, the size of these nucleocapsids was quite uniform and different from that of the parental virus. The reason for this result remains unclear, but studies involving assembly of nucleocapsids in coinfected cells might provide novel insights into the complex baculovirus nucleocapsid assembly process (46).
Finally, we examined the consequences of coenvelopment and co-occlusion for interspecific complementation and transmission. The nucleopolyhedroviruses of Spodoptera exigua (SeMNPV) and S. frugiperda (SfMNPV) belong to the same phylogenetic group of nucleopolyhedroviruses (15) and are genetically similar (78% similarity) (36). However, these viruses differ in their host ranges. SeMNPV can infect only S. exigua, whereas SfMNPV is lethal to both species, although in S. exigua inoculation of SfMNPV OBs results in an atypical infection with almost no production of progeny OBs (36). In the present study, both viruses were transmitted to the viral progeny (SeA+SfC, P0) following intrahemocoelic coinjection of S. exigua larvae with a complete SeMNPV genotype (SeA) and a defective SfMNPV genotype (SfC), which lacked the pif-1 and pif-2 genes, which are essential for per os infection (47–49). The passage zero (SeA+SfC) OB inoculum, which had presence of both viruses, was orally infective to S. frugiperda larvae; the death of these insects was attributed exclusively to SfC virus, as it was the only virus present in the resulting viral progeny (P1). As described previously, SeMNPV infection in S. frugiperda larvae is blocked at the stage in which budded virions (that each comprise a single nucleocapsid and genome) leave the site of primary infection in midgut cells and disperse in the hemocoel and along the trachea. This block cannot be overcome by coinfection with SfMNPV (36). We obtained similar results in our experiment when the SeA+SfC P0 inoculum was administered to S. frugiperda larvae. As the defective SfC was not able to enter gut epithelial cells alone, due to the lack of the pif-1 and pif-2 genes (27), this result demonstrates that SfC ODVs had acquired PIF-1 and PIF-2 proteins produced by expression of the corresponding genes in the SeA genome. This occurred in cells simultaneously infected by both viruses in the parental host S. exigua used to produce the P0 OBs. This is a clear example of interspecific complementation.
Only in this manner would peroral transmission of the defective SfC variant have been possible, given its inability to produce PIF-1 and PIF-2 proteins, which are essential for peroral transmission (22, 48). An alternative hypothesis involving recombination of the pif-1–pif-2 region between the genomes of each virus was rejected because P1 OBs comprising the SfC genotype alone were devoid of peroral infectivity, indicating that the infective capacity of P0 OBs had not been produced by a recombinant SfC genotype expressing pif-1 and pif-2 from SeA virus. We can also preclude the possibility that the SfC genotype was able to acquire pif genes from a putative covert infection present in the insect colony because inoculation by intrahemocoelic injection of the SfC genotype alone in colony insects invariably resulted in accurate replication of the pure SfC genotype that remained noninfectious per os (data not shown).
In contrast to the results observed for S. frugiperda, serial passage of mixed-virus OBs in S. exigua demonstrated the persistence of SfC virus in OBs comprising SeA and SfC viruses. As recombination between viruses did not occur and SfC virus is only able to infect midgut epithelial cells if co-occluded with SeA virus (i.e., if SfC virus replicated in a cell coinfected by SeA virus that expressed pif-1 and pif-2 genes), the progressive decline in the frequency of SfC genomes in mixed-species OBs during serial passage was likely due to a combination of two factors. First, SfMNPV replicates poorly in S. exigua (36), so SeA virus had a marked replication advantage in the homologous host. Second, in each passage, host cells that were infected exclusively by SfC BVs would have produced OBs that were not perorally infectious and that will not be transmitted in the following passage step.
Although the presence of diverse genotypes of an alphabaculovirus in an individual ODV has been demonstrated (22), interspecific co-occlusion and coenvelopment of alphabaculoviruses provide a new tool for the study of virus-cell interactions and host specificity during replication and virion assembly. Our findings also suggest that the envelopment process is nonselective during ODV assembly. A physical association between different viruses has also been suggested after observing structures resembling iflavirus particles within SeMNPV OBs produced in iflavirus-infected insects (50), resulting in the transmission and persistence of both viruses (51). Exploitation of common host resources by two virus species and, more importantly, the envelopment of different species’ genomes within transmissible virions appear paradoxical given that no clear advantages were detected in replication or transmission metrics for either of the interacting viruses, except in the special case of transmission achieved by the noninfective SfC virus via complementation with SeA virus. Despite the phylogenetic distances among alphabaculoviruses, the majority of proteins produced by these viruses in a coinfected cell are expected to be compatible and likely interchangeable for some of the highly conserved proteins (52), thus resulting in the production of viable and transmissible virions. The ability of such mixed-species virions to infect hosts in situations where one or other of the viruses is not easily transmitted will likely determine the transient persistence of mixed species virus populations in situations where two or more susceptible insect species are present on a particular crop or host plant.
Simultaneous infections by different virus species within the same host have been documented but are usually considered infrequent in natural insect populations (53, 54), although coinfection of baculoviruses and small RNA viruses may be common in natural populations of some species of Lepidoptera (55). Indeed, Roy et al. (56) have pointed out that inapparent infections in low-density nonpest insects may provide a largely unrecognized reservoir of insect-virus and potentially virus-virus interactions, of which almost nothing is known. There is, however, clear phylogenetic evidence for horizontal gene transfer between different alphabaculovirus species (57). If confirmed to occur in natural insect populations, the replication of distinct baculoviruses provides a mechanism for genomic interactions between virus species that have implications for baculovirus evolution. The exchange of genetic material by recombination, or through the action of transposable elements, between different baculoviruses is likely to generate novel genotypes and potentially novel traits in each of the viruses involved (58, 59). The transmission of SfC virus by complementation with SeA virus observed in the present study represents a clear example of heterotypic complementation (3), although the ability of SfC virus to survive in mixtures with other SfMNPV genotypes (homotypic complementation), mediated by the production of mixed-genotype ODVs and OBs (22), was well established in our previous studies (60, 61).
One of the most important applications of these viruses is their use as biological insecticides (62). Therefore, co-occlusion and coenvelopment of different virus species constitute a novel technology by which virus-based insecticides could be designed to control two or more species of caterpillar pests present on a particular crop following a single application of formulated mixed-species OBs (63). The narrow host range of these viruses has previously been recognized as a factor limiting their attractiveness to farmers that need to control complexes of lepidopterous pests (64), such as Pseudoplusia includens and Helicoverpa armigera on soybean and cotton in Brazil (65) and armyworms such as Mythimna unipuncta and S. frugiperda on maize in North America (66, 67). Consequently, novel virus insecticides with an expanded host range are likely to be more commercially viable if they can control multiple pests (34, 64). There are many other potential combinations of alphabaculoviruses that could be coenveloped to produce mixed-virus insecticides, provided that virulent viruses can be identified for each pest and both viruses can replicate in a shared host species for production purposes. The need for such innovative pest control tools is becoming increasingly urgent given the recent range expansion of many lepidopteran pests (68).
Viral species present in co-occluded virions are expected to share the battery of proteins responsible for the host range, so co-occlusion may facilitate the entry or replication of some viruses in some hosts that otherwise would be nonpermissive, such as the case of SfMNPV and SpliNPV, which facilitated the replication of SeMNPV in S. frugiperda and Spodoptera littoralis larvae, respectively (36), or the observations in which replication of SeMNPV in S. frugiperda cells was improved in the presence of coinfection by AcMNPV, possibly through complementation of transcripts produced by AcMNPV (69). This interspecific association should assist in mitigating the marked host specificity of many baculovirus-based insecticides, and it may improve the commercial prospects for the uptake of these products by growers concerned with the levels of synthetic pesticide residues in their produce.
We conclude that the production of mixed-alphabaculovirus virions containing the genomes of each of the parental viruses offers a new window to study the evolution of these pathogens, in addition to providing a unique mechanism for the production of custom-designed biological insecticides, with low environmental impact, to control specific complexes of pests on agricultural crops or in forests.
MATERIALS AND METHODS
Insects, cells, and viruses.
Larvae of Spodoptera frugiperda and S. exigua were obtained from laboratory colonies maintained on a semisynthetic diet (70) at 25°C. Sf9 cells (Thermo Fisher Scientific, Waltham, MA) were maintained in TC100 medium (Lonza Bioscience, Cologne, Germany) containing 10% fetal calf serum (Lonza Bioscience) at 28°C (44). The five viruses used in the experiments were as follows: (i) the C6 clone (AcC6) of AcMNPV (41); a (ii) a plaque-purified variant of SfMNPV named SfMNPV-B (SfB), (iii) a plaque-purified variant, SfMNPV-C (SfC), that lacked the pif-1 and pif-2 genes (47, 48); (iv) an in vivo-isolated variant of SeMNPV-US2A, named SeA (71); and (v) a strain of MbMNPV (Mb) isolated from the microbial insecticide Mamestrin (NPP, Nogueres, France) (72). SfB has the largest genome of all the variants present in a Nicaraguan isolate of SfMNPV, and the OBs are perorally infectious (42). SeMNPV is infectious only to S. exigua larvae. AcMNPV and MbMNPV are able to cause productive lethal infections in both S. exigua and S. frugiperda larvae. SfMNPV is lethal to S. exigua and S. frugiperda but produces an atypical infection in S. exigua, in which larvae produce almost no OBs and do not undergo postmortem liquefaction (36). Occlusion bodies (OBs) of SfC are not perorally infectious, but the virus is capable of replication and production of OBs in cell culture or following intrahemocoelic injection of occlusion-derived virions (ODVs) or budded virions (BVs) into S. frugiperda larvae.
Production of mixed-virus occlusion bodies.
Four experimental mixed-virus preparations were generated. (i) AcC6+SfB OBs were obtained following peroral inoculation (73) of fourth-instar S. frugiperda larvae with a 1:1 mixture of AcC6 and SfB OBs at a total concentration of 108 OBs/ml. (ii) AcC6+Mb OBs were obtained following peroral infection of fourth-instar S. frugiperda larvae with a 1:10 mixture of AcC6 and Mb OBs at a total concentration of 108 OBs/ml. (iii) SfB+Mb OBs were obtained by perorally inoculating fourth-instar S. frugiperda larvae with MbMNPV OBs at a concentration of 109 OBs/ml. At 24 h post-first inoculation, larvae were inoculated with SfB OBs at 108 OBs/ml. This two-step procedure was necessary because preliminary experiments indicated that these viruses differed in their efficiencies of replication in S. frugiperda (74). (iv) The passage zero (P0) inoculum of SeA+SfC OBs was produced by intrahemocoelic injection of fourth-instar S. exigua larvae with 10 μl of a mixed-virus ODV suspension released from a 1:1 mixture of SeA and SfC OBs (5 × 107 OBs of each virus in a volume of 50 μl). In all cases, inoculated larvae were reared individually on diet at 25 ± 1°C until death.
The ODVs obtained from mixed-virus OBs were studied in three ways. First, the coenvelopment of two different alphabaculovirus species within the same ODV was examined by plaque assay or the endpoint dilution of ODVs released from AcC6+SfB mixed-virus OBs. Second, mixed-virus OBs in which MbMNPV was present were designed to examine the effects of the mixed infection on the length of nucleocapsids enveloped within mixed-virus ODVs. MbMNPV was selected because it has a much larger genome (∼153 kb) than AcMNPV (∼134 kb) or SfMNPV (∼131 kb) and was expected to assemble in longer nucleocapsids. Third, SeA+SfC mixed OBs were used to study the coenvelopment of these viral species in vivo, as SfC is not perorally infective and coenvelopment with a helper virus (SeA) that provides PIF-1 and PIF-2 through complementation is required for per os transmission.
Genome proportions in mixed-virus occlusion bodies.
Genomic DNA was extracted from samples of 106 OBs of each mixed-virus OB preparation as described previously (74, 75). DNA was subjected to restriction endonuclease analysis with PstI (New England BioLabs [NEB], Hitchin, UK). The relative proportion of the genomes of each virus in the different progeny OBs was estimated by using qPCR. Four sets of primers were designed for specific amplification of characteristic sequences from each virus genome (Table 2). PCR products were cloned into the pGEM-T Easy vector (Promega, Madison, WI). Plasmid DNA was quantified by measuring optical absorbance (A260) and by gel electrophoresis and used as an internal standard for each qPCR. The qPCR assays were performed in a volume of 10 μl, containing 5 μl of iQ SYBR green supermix (Bio-Rad Laboratories, Hercules, CA), 0.2 μl of each forward and reverse primer (10 μM), and 1 μl of DNA template. The amplification reaction comprised a polymerase activation and DNA denaturation step at 95°C for 150 s, followed by 45 amplification cycles at 95°C for 15 s and 60°C for 30 s. Finally, a melting-curve analysis, involving a dissociation stage of 60°C for 15 s and 95°C for 5 s, was performed to confirm the presence of a single peak of the target product. Each virus was quantified in independent qPCRs. Data acquisition and analysis were handled by Sequence Detector version 2.2.2 software (Applied Biosystems, Foster City, CA). All standards and samples were measured in triplicate. The proportions of the viruses in each sample were calculated independently, and the relative proportion of each virus in each replicate (mean ± SD) was then calculated from these results.
Plaque purification of virions from mixed-virus occlusion bodies.
Coenvelopment of viruses in the same ODV was studied in Sf9 cells, which are permissive to both AcMNPV and SfMNPV. Plaque purification was performed after release of ODVs from samples of 5 × 107 AcC6+SfB OBs following a 30-min treatment with 0.1 M sodium carbonate. This suspension was filtered through a 0.45-μm filter and serially diluted (1/5) in TC100 medium. Six-well tissue culture plates were seeded with 106 cells/well and were inoculated with 200 μl of each dilution. The plates were incubated for 1 h at room temperature on an orbital shaker to allow ODVs to adsorb to the cells. Virus inoculum was then removed and 2 ml of 2% SeaPlaque agarose (Lonza, Rockland, ME) was added to the wells. Inoculated cell culture plates were incubated at 28°C for 5 to 6 days. Only wells containing 30 or fewer plaques were subjected to analysis. Individual plaques were picked and suspended in 500 μl of TC100. The identity of each plaque was determined by PCR amplification of virus-specific products using virus-specific primers for each of the target viruses (Table 2). The presence of both viruses in the same plaque indicated that nucleocapsids of both viruses had been enveloped in a shared ODV that produced the plaque. SfMNPV-only and AcMNPV-only BVs were used as controls in order to monitor nonspecific amplification or false positives in PCRs. The experiment was performed in triplicate. A control experiment was performed in which ODVs were released from a mixture of 3.13 × 104 AcC6 OBs and 4.99 × 107 SfB OBs, i.e., the ratio (∼1:1,595) in which genomes were present in AcC6+SfB mixed-virus OBs as determined by qPCR (Table 1). The resulting ODVs were mixed and subjected to plaque assay to control for potential adhesion or clumping of AcC6 and SfB ODVs, which could have generated an erroneous result in the plaque assay of mixed-virus OBs.
Confirmation of mixed-virus virion composition by endpoint dilution.
Endpoint dilution (44) was applied to ODV suspension from AcC6+SfB mixed-virus OBs in order to confirm the results of the previous plaque purification experiment. For this, ODVs were released from samples of 5 × 108 mixed-virus OBs and were subjected to 5-fold serial dilution (1/50, 1/250, 1/1,250, and 1/6,250). Sf9 cells (1 × 104 cells/well) in 96-well plates were inoculated with one dilution of the ODV suspension. Plates were incubated at 28 ± 1°C for 7 days and then examined for signs of infection. The assay was performed in triplicate. Plates inoculated with the ODV dilution of 1/1,250 presented ∼10% of wells with virus infection. Infected wells in these plates were counted and individually subjected to PCR using specific primers (Table 2) to determine the identities of viruses in each infected well.
Electron microscope analysis of nucleocapsid length.
The length of individual nucleocapsids enveloped within multinucleocapsid ODVs was determined by transmission electron microscopy (TEM) and image analysis. ODVs were released from OBs by treatment with an alkaline solution (0.1 M Na2CO3, 166 mM NaCl, 10 mM EDTA [pH 10.5]) for 10 min at 4°C. The ODV membrane was then removed by mild treatment with 0.5% Triton X-100. Particles were negatively stained with uranyl acetate. Nucleocapsids in groups (i.e., those derived from multinucleocapsid ODVs), in which both ends of the nucleocapsids were clearly defined, were measured from between 22 and 51 microphotographs per sample using ImageJ software (76). A total of 12 to 14 nucleocapsids per treatment were measured. Results were subjected to analysis of variance (ANOVA) and Tukey’s test using the R-based package Jamovi (77).
Peroral inoculation of S. frugiperda with mixed-virus occlusion bodies.
To determine whether mixed-virus OBs comprising SeA virus and the nonperorally infectious variant SfC virus were perorally infective to S. frugiperda, the following experiment was performed. SeA virus is not infectious to S. frugiperda but expresses the pif-1 and pif-2 genes, which might facilitate peroral infection by SfC virus if these proteins were present in the ODV envelope of ODVs produced in cells coinfected by both viruses. To test this, groups of 24 second-instar S. frugiperda larvae were inoculated with one of two concentrations—1 × 107 and 1 × 109 OBs/ml—of SeA+SfC mixed-virus OBs, designated passage zero OBs (P0), using the droplet feeding method (73). Other groups of larvae were inoculated with each of the parental viruses, SeA and SfC viruses, at a concentration of 109 OBs/ml. Larvae that drank OB suspension within 10 min were reared individually, and virus mortality was recorded daily until larvae had died or pupated. Control larvae consumed sucrose and food dye solution without OBs. OBs collected from virus-killed insects (designated passage one [P1] OBs) were pooled and purified, and DNA was extracted as described previously (74, 75). Restriction endonuclease analysis using PstI and qPCR (Table 2) were performed to determine the presence and estimate the relative abundance of virus genomes in OB samples. The P1 OBs were then used for the oral inoculation of S. frugiperda second instars at a concentration of 1 × 109 OBs/ml. Inoculated larvae were monitored daily until death or pupation to determine whether P1 OBs retained peroral infectivity and preclude a possible recombination event by which SfC had acquired the pif-1 and pif-2 genes from SeA virus. The experiment was performed in triplicate.
Persistence of SfC virus in SeA+SfC mixed-virus population during serial passage.
Following positive results in the previous experiment, six serial passages were performed in fourth-instar S. exigua larvae inoculated with P0 SeA+SfC OBs as the starting inoculum. At each passage, groups of 24 larvae were orally inoculated with 1 × 106 OBs/ml by the droplet feeding method and individually reared at 25 ± 1°C until death. Control larvae were treated identically but did not consume OBs. OBs were recovered from virus-killed insects, pooled, purified by centrifugation, and used to orally inoculate groups of larvae for the following passage. Genomic DNA was extracted from OBs taken at each passage and subjected to restriction profile (PstI) examination and PCR analysis using virus-specific primers (Table 2). The relative abundance of viral genomes in OBs in each passage was quantified by qPCR as described in the section on quantification of genome proportions in mixed-virus OBs. The experiment was performed in triplicate.
Supplementary Material
ACKNOWLEDGMENTS
We thank I. Ibáñez and N. Gorría (Universidad Pública de Navarra) for insect rearing. T.W. acknowledges logistical support from L. Hernández (INECOL).
This research was supported by the projects AGL2014-57752-C2-1-R and AGL2017-83498-C2-1-R (Spanish Ministry for Science and Technology). I.B. received a Torres Quevedo grant CSIC doctoral studentship.
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
I.B., O.S., T.W., M.L.-F., and P.C. are coinventors of a patent describing coenvelopment and co-occlusion of different alphabaculovirus species for pest control.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Alizon S, de Roode JC, Michalakis Y. 2013. Multiple infections and the evolution of virulence. Ecol Lett 16:556–567. doi: 10.1111/ele.12076. [DOI] [PubMed] [Google Scholar]
- 2.Díaz-Muñoz SL. 2017. Viral coinfection is shaped by host ecology and virus-virus interactions across diverse microbial taxa and environments. Virus Evol 3:vex011. doi: 10.1093/ve/vex011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Díaz-Muñoz SL, Sanjuán R, West S. 2017. Sociovirology: conflict, cooperation, and communication among viruses. Cell Host Microbe 22:437–441. doi: 10.1016/j.chom.2017.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sanjuán R, Thoulouze M-I. 2019. Why viruses sometimes disperse in groups. Virus Evol 5:vez014. doi: 10.1093/ve/vez014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cuevas JM, Durán-Moreno M, Sanjuán R. 2017. Multi-virion infectious units arise from free viral particles in an enveloped virus. Nat Microbiol 2:17078. doi: 10.1038/nmicrobiol.2017.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Anschau V, Sanjuán R. 2020. Fibrinogen gamma chain promotes aggregation of vesicular stomatitis virus in saliva. Viruses 12:282. doi: 10.3390/v12030282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feng Z, Hensley L, McKnight KL, Hu F, Madden V, Ping L, Jeong SH, Walker C, Lanford RE, Lemon SM. 2013. A pathogenic picornavirus acquires an envelope by hijacking cellular membranes. Nature 496:367–371. doi: 10.1038/nature12029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen Y-H, Du WLi, Hagemeijer MC, Takvorian PM, Pau C, Cali A, Brantner CA, Stempinski ES, Connelly PS, Ma H-C, Jiang P, Wimmer E, Altan-Bonnet G, Altan-Bonnet N. 2015. Phosphatidylserine vesicles enable efficient en bloc transmission of enteroviruses. Cell 160:619–630. doi: 10.1016/j.cell.2015.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chiu E, Coulibaly F, Metcalf P. 2012. Insect virus polyhedra, infectious protein crystals that contain virus particles. Curr Opin Struct Biol 22:234–240. doi: 10.1016/j.sbi.2012.02.003. [DOI] [PubMed] [Google Scholar]
- 10.Sanjuán R. 2017. Collective infectious units in viruses. Trends Microbiol 25:402–412. doi: 10.1016/j.tim.2017.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sanjuán R. 2018. Collective properties of viral infectivity. Curr Opin Virol 33:1–6. doi: 10.1016/j.coviro.2018.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Segredo-Otero E, Sanjuán R. 2019. The effect of genetic complementation on the fitness and diversity of viruses spreading as collective infectious units. Virus Res 267:41–48. doi: 10.1016/j.virusres.2019.05.005. [DOI] [PubMed] [Google Scholar]
- 13.Rezelj VV, Levi LI, Vignuzzi M. 2018. The defective component of viral populations. Curr Opin Virol 33:74–80. doi: 10.1016/j.coviro.2018.07.014. [DOI] [PubMed] [Google Scholar]
- 14.Syller J. 2012. Facilitative and antagonistic interactions between plant viruses in mixed infections. Mol Plant Pathol 13:204–216. doi: 10.1111/j.1364-3703.2011.00734.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harrison RL, Herniou EA, Jehle JA, Theilmann DA, Burand JP, Becnel JJ, Krell PJ, van Oers MM, Mowery JD, Bauchan GR. 2018. ICTV virus taxonomy profile: Baculoviridae. J Gen Virol 99:1185–1186. doi: 10.1099/jgv.0.001107. [DOI] [PubMed] [Google Scholar]
- 16.Sajjan DB, Hinchigeri SB. 2016. Structural organization of baculovirus occlusion bodies and protective role of multilayered polyhedron envelope protein. Food Environ Virol 8:86–100. doi: 10.1007/s12560-016-9227-7. [DOI] [PubMed] [Google Scholar]
- 17.D'Amico V, Elkinton JS. 1995. Rainfall effects on transmission of gypsy moth (Lepidoptera: Lymantriidae) nuclear polyhedrosis virus. Env Entomol 24:1144–1149. doi: 10.1093/ee/24.5.1144. [DOI] [Google Scholar]
- 18.Clem RJ, Passarelli AL. 2013. Baculoviruses: sophisticated pathogens of insects. PLoS Pathog 9:e1003729. doi: 10.1371/journal.ppat.1003729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Erlandson MA. 2009. Genetic variation in field populations of baculoviruses: mechanisms for generating variation and its potential role in baculovirus epizootiology. Virol Sin 24:458–469. doi: 10.1007/s12250-009-3052-1. [DOI] [Google Scholar]
- 20.Chateigner A, Bézier A, Labrousse C, Jiolle D, Barbe V, Herniou EA. 2015. Ultra deep sequencing of a baculovirus population reveals widespread genomic variations. Viruses 7:3625–3646. doi: 10.3390/v7072788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bull JC, Godfray HCJ, O’Reilly DR. 2001. Persistence of an occlusion-negative recombinant nucleopolyhedrovirus in Trichoplusia ni indicates high multiplicity of cellular infection. Appl Environ Microbiol 67:5204–5209. doi: 10.1128/AEM.67.11.5204-5209.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Clavijo G, Williams T, Muñoz D, Caballero P, López-Ferber M. 2010. Mixed genotype transmission bodies and virions contribute to the maintenance of diversity in an insect virus. Proc Biol Sci 277:943–951. doi: 10.1098/rspb.2009.1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grzywacz D, Moore S. 2017. Production, formulation, and bioassay of baculoviruses for pest control, p 109–124. In Lacey LA. (ed), Microbial control of insect and mite pests: from theory to practice. Academic Press, San Diego, CA. [Google Scholar]
- 24.van Oers MM. 2011. Opportunities and challenges for the baculovirus expression system. J Invertebr Pathol 107:S3–S15. doi: 10.1016/j.jip.2011.05.001. [DOI] [PubMed] [Google Scholar]
- 25.Premanand B, Wee PZ, Prabakaran M. 2018. Baculovirus surface display of immunogenic proteins for vaccine development. Viruses 10:298. doi: 10.3390/v10060298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kwang TW, Zeng X, Wang S. 2016. Manufacturing of AcMNPV baculovirus vectors to enable gene therapy trials. Mol Ther Methods Clin Dev 3:15050. doi: 10.1038/mtm.2015.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.López-Ferber M, Simón O, Williams T, Caballero P. 2003. Defective or effective? Mutualistic interactions between virus genotypes. Proc Biol Sci 270:2249–2255. doi: 10.1098/rspb.2003.2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Arrizubieta M, Simón O, Torres-Vila LM, Figueiredo E, Mendiola J, Mexia A, Caballero P, Williams T. 2016. Insecticidal efficacy and persistence of a co-occluded binary mixture of Helicoverpa armigera nucleopolyhedrovirus (HearNPV) variants in protected and field-grown tomato crops on the Iberian peninsula. Pest Manag Sci 72:660–670. doi: 10.1002/ps.4035. [DOI] [PubMed] [Google Scholar]
- 29.Del-Angel C, Lasa R, Rodríguez-del-Bosque LA, Mercado G, Beperet I, Caballero P, Williams T. 2018. Anticarsia gemmatalis nucleopolyhedrovirus from soybean crops in Tamaulipas, Mexico: diversity and insecticidal characteristics of individual variants and their co-occluded mixtures. Fla Entomol 101:404–410. doi: 10.1653/024.101.0319. [DOI] [Google Scholar]
- 30.Simón O, Williams T, López-Ferber M, Taulemesse JM, Caballero P. 2008. Population genetic structure determines speed of kill and occlusion body production in Spodoptera frugiperda multiple nucleopolyhedrovirus. Biol Control 44:321–330. doi: 10.1016/j.biocontrol.2007.12.005. [DOI] [Google Scholar]
- 31.Barrera G, Williams T, Villamizar L, Caballero P, Simón O. 2013. Deletion genotypes reduce occlusion body potency but increase occlusion body production in a Colombian Spodoptera frugiperda nucleopolyhedrovirus population. PLoS One 8:e77271. doi: 10.1371/journal.pone.0077271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cory JS. 2003. Ecological impacts of virus insecticides: host range and non-target organisms, p 73–91. In Hokkanen HMT, Hajek A (ed), Environmental impacts of microbial insecticides. Springer, Dordrecht, Netherlands. [Google Scholar]
- 33.Thiem SM, Cheng XW. 2009. Baculovirus host-range. Virol Sin 24:436–457. doi: 10.1007/s12250-009-3058-8. [DOI] [Google Scholar]
- 34.Wu C, Deng Z, Long Z, Cai Y, Ying Z, Yin H, Yuan M, Clem RJ, Yang K, Pang Y. 2016. Generating a host range-expanded recombinant baculovirus. Sci Rep 6:28072. doi: 10.1038/srep28072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vail P, Hostetter D, Hoffmann D. 1999. Development of the multi-nucleocapsid nucleopolyhedroviruses (MNPVs) infectious to loopers (Lepidoptera: Noctuidae: Plusiinae) as microbial control agents. Integr Pest Manag Rev 4:231–257. doi: 10.1023/A:1009601212375. [DOI] [Google Scholar]
- 36.Simón O, Williams T, López-Ferber M, Caballero P. 2004. Virus entry or the primary infection cycle are not the principal determinants of host specificity of Spodoptera spp. nucleopolyhedroviruses. J Gen Virol 85:2845–2855. doi: 10.1099/vir.0.80179-0. [DOI] [PubMed] [Google Scholar]
- 37.Beperet I, Irons SL, Simón O, King LA, Williams T, Possee RD, López-Ferber M, Caballero P. 2014. Superinfection exclusion in alphabaculovirus infections is concomitant with actin reorganization. J Virol 88:3548–3556. doi: 10.1128/JVI.02974-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Salem T, Cheng X-H, Cheng X-W. 2012. AcMNPV enhances infection by ThorNPV in Sf21 cells and SeMNPV in Hi5 cells. Arch Virol 157:1875–1885. doi: 10.1007/s00705-012-1347-2. [DOI] [PubMed] [Google Scholar]
- 39.Brown M, Faulkner P. 1978. Plaque assay of nuclear polyhedrosis viruses in cell culture. Appl Environ Microbiol 36:31–35. doi: 10.1128/AEM.36.1.31-35.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Volkman LE, Knudson DL. 1986. In vitro replication of baculoviruses, p 109–127. In Granados RR, Federici BA (ed), The biology of baculoviruses, vol I CRC Press, Boca Raton, FL. [Google Scholar]
- 41.Ayres MD, Howard SC, Kuzio J, López-Ferber M, Possee RD. 1994. The complete DNA sequence of Autographa californica nuclear polyhedrosis virus. Virology 202:586–605. doi: 10.1006/viro.1994.1380. [DOI] [PubMed] [Google Scholar]
- 42.Simón O, Palma L, Beperet I, Muñoz D, López-Ferber M, Caballero P, Williams T. 2011. Sequence comparison between three geographically distinct Spodoptera frugiperda multiple nucleopolyhedrovirus isolates: detecting positively selected genes. J Invertebr Pathol 107:33–42. doi: 10.1016/j.jip.2011.01.002. [DOI] [PubMed] [Google Scholar]
- 43.Danyluk GM, Maruniak JE. 1987. In vivo and in vitro host range of Autographa californica nuclear polyhedrosis virus and Spodoptera frugiperda nuclear polyhedrosis virus. J Invertebr Pathol 50:207–212. doi: 10.1016/0022-2011(87)90084-x. [DOI] [PubMed] [Google Scholar]
- 44.King LA, Possee RD. 1992. The baculovirus expression system: a laboratory guide. Chapman & Hall, London, United Kingdom. [Google Scholar]
- 45.Kool M, Voncken JW, van Lier FL, Tramper J, Vlak JM. 1991. Detection and analysis of Autographa californica nuclear polyhedrosis virus mutants with defective interfering properties. Virology 183:739–746. doi: 10.1016/0042-6822(91)91003-y. [DOI] [PubMed] [Google Scholar]
- 46.Zhao S, He G, Yang Y, Liang C. 2019. Nucleocapsid assembly of baculoviruses. Viruses 11:595. doi: 10.3390/v11070595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Simón O, Williams T, López-Ferber M, Caballero P. 2004. Genetic structure of a Spodoptera frugiperda nucleopolyhedrovirus population: high prevalence of deletion genotypes. Appl Environ Microbiol 70:5579–5588. doi: 10.1128/AEM.70.9.5579-5588.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Clavijo G, Williams T, Simón O, Muñoz D, Cerutti M, López-Ferber M, Caballero P. 2009. Mixtures of complete and pif-1- and pif-2-deficient genotypes are required for increased potency of an insect nucleopolyhedrovirus. J Virol 83:5127–5136. doi: 10.1128/JVI.02020-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Boogaard B, Van Oers MM, Van Lent JW. 2018. An advanced view on baculovirus per os infectivity factors. Insects 9:84. doi: 10.3390/insects9030084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jakubowska AK, Murillo R, Carballo A, Williams T, van Lent JW, Caballero P, Herrero P. 2016. Iflavirus increases its infectivity and physical stability in association with baculovirus. PeerJ 4:e1687. doi: 10.7717/peerj.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Carballo A, Williams T, Murillo R, Caballero P. 2020. Iflavirus covert infection increases susceptibility to nucleopolyhedrovirus disease in Spodoptera exigua. Viruses 12:509. doi: 10.3390/v12050509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.van Oers MM, Vlak JM. 2007. Baculovirus genomics. Curr Drug Targets 8:1051–1068. doi: 10.2174/138945007782151333. [DOI] [PubMed] [Google Scholar]
- 53.Kemp EM, Woodward DT, Cory JS. 2011. Detection of single and mixed covert baculovirus infections in eastern spruce budworm, Choristoneura fumiferana populations. J Invertebr Pathol 107:202–205. doi: 10.1016/j.jip.2011.05.015. [DOI] [PubMed] [Google Scholar]
- 54.Nakai M, Takeda M, Kunimi Y. 1997. Seasonal changes in prevalence of viral disease and parasitism by parasitic insects in a larval population of the smaller tea tortrix, Adoxophyes sp. (Lepidoptera: Tortricidae) in a tea field. Appl Entomol Zool 32:609–615. doi: 10.1303/aez.32.609. [DOI] [Google Scholar]
- 55.Virto C, Navarro D, Tellez MM, Herrero S, Williams T, Murillo R, Caballero P. 2014. Natural populations of Spodoptera exigua are infected by multiple viruses that are transmitted to their offspring. J Invertebr Pathol 122:22–27. doi: 10.1016/j.jip.2014.07.007. [DOI] [PubMed] [Google Scholar]
- 56.Roy HE, Hails RS, Hesketh H, Roy DB, Pell JK. 2009. Beyond biological control: non-pest insects and their pathogens in a changing world. Insect Conserv Divers 2:65–72. doi: 10.1111/j.1752-4598.2009.00046.x. [DOI] [Google Scholar]
- 57.Barrera GP, Belaich MN, Patarroyo MA, Villamizar LF, Ghiringhelli PD. 2015. Evidence of recent interspecies horizontal gene transfer regarding nucleopolyhedrovirus infection of Spodoptera frugiperda. BMC Genomics 16:1008. doi: 10.1186/s12864-015-2218-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Loiseau V, Herniou EA, Moreau Y, Lévêque N, Meignin C, Daeffler L, Federici B, Cordaux R, Gilbert C. 2020. Wide spectrum and high frequency of genomic structural variation, including transposable elements, in large double-stranded DNA viruses. Virus Evol 6:vez060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Thézé J, Lopez-Vaamonde C, Cory JS, Herniou EA. 2018. Biodiversity, evolution and ecological specialization of baculoviruses: a treasure trove for future applied research. Viruses 10:366. doi: 10.3390/v10070366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Simón O, Williams T, Caballero P, López-Ferber M. 2006. Dynamics of deletion genotypes in an experimental insect virus population. Proc Biol Sci 273:783–790. doi: 10.1098/rspb.2005.3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Simón O, Williams T, Cerutti M, Caballero P, López-Ferber M. 2013. Expression of a peroral infection factor determines pathogenicity and population structure in an insect virus. PLoS One 8:e78834. doi: 10.1371/journal.pone.0078834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lacey LA, Grzywacz D, Shapiro-Ilan DI, Frutos R, Brownbridge M, Goettel MS. 2015. Insect pathogens as biological control agents: back to the future. J Invertebr Pathol 132:1–41. doi: 10.1016/j.jip.2015.07.009. [DOI] [PubMed] [Google Scholar]
- 63.Caballero-Murillo P, Beperet-Arive I, Simón-de-Goñi O, Williams T, López-Ferber M. July 2018. Production of virus occlusion bodies that occlude virions comprising genomes of different species of baculoviruses that can be used to combat insect pests. European patent EP 3 049 516 B1.
- 64.Grzywacz D. 2017. Basic and applied research: baculovirus, p 27–46. In Lacey LA. (ed), Microbial control of insect and mite pests: from theory to practice. Academic Press, San Diego, CA. [Google Scholar]
- 65.Muraro DS, Giacomelli T, Stacke RF, Godoy DN, Marçon P, Popham HJ, Bernardi O. 2019. Baseline susceptibility of Brazilian populations of Chrysodeixis includens (Lepidoptera: Noctuidae) to C. includens nucleopolyhedrovirus and diagnostic concentration for resistance monitoring. J Econ Entomol 112:349–354. doi: 10.1093/jee/toy361. [DOI] [PubMed] [Google Scholar]
- 66.Escribano A, Williams T, Goulson D, Cave RD, Chapman JW, Caballero P. 1999. Selection of a nucleopolyhedrovirus for control of Spodoptera frugiperda (Lepidoptera: Noctuidae): structural, genetic, and biological comparison of four isolates from the Americas. J Econ Entomol 92:1079–1085. doi: 10.1093/jee/92.5.1079. [DOI] [PubMed] [Google Scholar]
- 67.Keathley CP, Harrison RL, Potter DA. 2012. Baculovirus infection of the armyworm (Lepidoptera: Noctuidae) feeding on spiny-or smooth-edged grass (Festuca spp.) leaf blades. Biol Contr 61:147–154. doi: 10.1016/j.biocontrol.2012.01.013. [DOI] [Google Scholar]
- 68.Suckling DM, Conlong DE, Carpenter JE, Bloem KA, Rendon P, Vreysen MJB. 2017. Global range expansion of pest Lepidoptera requires socially acceptable solutions. Biol Invasions 19:1107–1119. doi: 10.1007/s10530-016-1325-9. [DOI] [Google Scholar]
- 69.Yanase T, Yasunaga C, Hara T, Kawarabata T. 1998. Coinfection of Spodoptera exigua and Spodoptera frugiperda cell lines with the nuclear polyhedrosis viruses of Autographa californica and Spodoptera exigua. Intervirology 41:244–252. doi: 10.1159/000024946. [DOI] [PubMed] [Google Scholar]
- 70.Greene GL, Leppla NC, Dickerson WA. 1976. Velvetbean caterpillar: a rearing procedure and artificial medium. J Econ Entomol 69:487–488. doi: 10.1093/jee/69.4.487. [DOI] [Google Scholar]
- 71.Munoz D, Castillejo JI, Caballero P. 1998. Naturally occurring deletion mutants are parasitic genotypes in a wild-type nucleopolyhedrovirus population of Spodoptera exigua. Appl Environ Microbiol 64:4372–4377. doi: 10.1128/AEM.64.11.4372-4377.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Murillo R, Muñoz D, Lipa JJ, Caballero P. 2001. Biochemical characterization of three nucleopolyhedrovirus isolates of Spodoptera exigua and Mamestra brassicae. J Appl Entomol 125:267–270. doi: 10.1046/j.1439-0418.2001.00533.x. [DOI] [Google Scholar]
- 73.Hughes PR, Wood HA. 1986. In vivo and in vitro bioassay methods for baculoviruses, p 1–30. In Granados RR, Federici BA (ed), The biology of baculoviruses, vol II CRC Press, Boca Raton, FL. [Google Scholar]
- 74.Beperet-Arive I. 2014. Regulation of multiple infection in alphabaculoviruses: critical factors that determine success. Doctoral thesis Universidad Pública de Navarra, Navarra, Spain. [Google Scholar]
- 75.Beperet I, Barrera G, Simón O, Williams T, López-Ferber M, Gasmi L, Herrero S, Caballero P. 2013. The sf32 unique gene of Spodoptera frugiperda multiple nucleopolyhedrovirus (SfMNPV) is a non-essential gene that could be involved in nucleocapsid organization in occlusion-derived virions. PLoS One 8:e77683. doi: 10.1371/journal.pone.0077683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ferreira T, Rasband W. 2012. ImageJ user guide. ImageJ/Fiji 1.46r, p 155–161. https://imagej.nih.gov/ij/docs/guide/user-guide.pdf. Accessed 24 July 2020.
- 77.Jamovi. 2020. Jamovi project (version 1.2). https://www.jamovi.org. Accessed 24 July 2020.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.



